Abstract
Local translation of mRNAs in the synapse has a major role in synaptic structure and function. Chronic alcohol use causes persistent changes in synaptic mRNA expression, possibly mediated by microRNAs localized in the synapse. We profiled the transcriptome of synaptoneurosomes (SN) obtained from the amygdala of mice that consumed 20% ethanol (alcohol) in a 30-day continuous two-bottle choice test to identify the microRNAs that target alcohol-induced mRNAs. SN are membrane vesicles containing pre- and post-synaptic compartments of neurons and astroglia and are a unique model for studying the synaptic transcriptome. We previously showed that chronic alcohol regulates mRNA expression in a coordinated manner. Here, we examine microRNAs and mRNAs from the same samples to define alcohol-responsive synaptic microRNAs and their predicted interactions with targeted mRNAs. The aim of the study was to identify the microRNA–mRNA synaptic interactions that are altered by alcohol. This was accomplished by comparing the effect of alcohol in SN and total homogenate preparations from the same samples. We used a combination of unbiased bioinformatic methods (differential expression, correlation, co-expression, microRNA-mRNA target prediction, co-targeting, and cell type-specific analyses) to identify key alcohol-sensitive microRNAs. Prediction analysis showed that a subset of alcohol-responsive microRNAs was predicted to target many alcohol-responsive mRNAs, providing a bidirectional analysis for identifying microRNA–mRNA interactions. We found microRNAs and mRNAs with overlapping patterns of expression that correlated with alcohol consumption. Cell type-specific analysis revealed that a significant number of alcohol-responsive mRNAs and microRNAs were unique to glutamate neurons and were predicted to target each other. Chronic alcohol consumption appears to perturb the coordinated microRNA regulation of mRNAs in SN, a mechanism that may explain the aberrations in synaptic plasticity affecting the alcoholic brain.
Introduction
Local translation of synaptic mRNAs is essential for the functional properties of brain cells (Raab-Graham et al, 2006; Wang et al, 2009). The extensive neuroadaptations associated with alcohol dependence are likely caused by persistent changes in the expression of hundreds of mRNAs (Mayfield et al, 2002; Ponomarev et al, 2012; Nunez et al, 2013). Many of the alcohol-responsive adaptations are related to synaptic structure and function and may be caused by coordinated changes in local mRNA translation (Wang et al, 2010; Mayfield and Nunez, 2012). MicroRNAs are short, non-coding RNAs that can regulate the translation of many target mRNAs, and this process is known to occur in the synaptic compartments of the cell (Lugli et al, 2005, 2008; Smalheiser and Lugli, 2009; Sosanya et al, 2013). The ability of microRNAs to regulate mRNAs provides a localized regulatory system that may be important in the treatment of alcoholism.
Exposure to alcohol and other drugs of abuse modulates microRNA expression in the brain (Pietrzykowski et al, 2008; He et al, 2010; Eipper-Mains et al, 2011; Lewohl et al, 2011; Tapocik et al, 2013). MicroRNAs have important roles in learning and memory (Konopka et al, 2010), and are also altered in addiction-related behaviors, such as cocaine-conditioned place preference (Chandrasekar and Dreyer, 2011), cocaine-seeking behavior (Novak et al, 2010), and self-administration of alcohol (Tapocik et al, 2014). Little is known about the microRNAs involved in the regulation of synaptic mRNA translation during alcohol dependence. Studies investigating the effects of chronic alcohol consumption in standard tissue (total homogenate, TH) preparations found a persistent change in the expression of microRNAs and their target mRNAs in humans, mice, and rats (Lewohl et al, 2011; Gorini et al, 2013; Nunez et al, 2013; Tapocik et al, 2013). However, the standard TH preparation likely underestimates the number and magnitude of alcohol-responsive transcripts localized in the synapse (Lewohl et al, 2011; Most et al, 2014).
Synaptoneurosomes (SN) (Hollingsworth et al, 1985; Quinlan et al, 1999; Raab-Graham et al, 2006; Sosanya et al, 2013) contain membrane vesicles of pre- and post-synaptic compartments of neurons as well as peri-synaptic compartments of astrocytes and microglia and offer an improved model for studying the synaptic transcriptome. We recently showed that alcohol-induced mRNA changes are greater in SN compared with TH (Most et al, 2014). Here, we profiled the microRNA transcriptomes from paired SN and TH samples of mouse amygdala after a voluntary alcohol consumption paradigm. We identified alcohol-induced microRNAs that were correlated with alcohol consumption and also identified their predicted mRNA synaptic targets.
Materials and Methods
Animal Housing and Alcohol Consumption
Adult (two-month-old) C57BL/6J female mice were maintained at the University of Texas at Austin Animal Resources Center. Mice were group-housed and given a 1-week acclimation period in combined housing and another week to acclimate to the bottle positions in individual housing. Food and water were provided ad libitum and monitored daily, as were the temperature and reverse light/dark cycle. Mice underwent a 30-day two-bottle choice paradigm with continuous (24-h) access to one bottle of 20% ethanol (referred to as alcohol) and one bottle of water (Blednov et al, 2012). A control group of mice received two bottles of water. Bottle weights were recorded daily, mice were weighed every four days, and the amount of alcohol consumed was calculated as g/kg/24 h (see Supplementary Figure 1 for amounts of alcohol consumed). Alcohol bottle positions were alternated daily to control for position preferences. The stage of the menstrual cycle was not determined. All procedures were approved by the Institutional Animal Care and Use Committee at the University of Texas at Austin and adhere to NIH Guidelines for the ethical care and use of animals in research.
RNA Extraction
As described previously (Most et al, 2014), after 30 days of the two-bottle choice drinking, 8 alcohol-consuming and 12 control mice were killed by cervical dislocation and decapitated at the beginning of the light phase of the light/dark cycle. Brains were removed and washed for 1 min with 1 ml of ice-cold homogenizing buffer (HB) containing 20 mM HEPES, 1 mM EDTA (pH 7.4), 40 U/ml RNAseOut (Invitrogen, CA), phosphatase inhibitor cocktail 3 (Sigma, MO) and protease inhibitors ‘Complete' (Roche, IN). Brains were then placed in a coronal Zivic mouse brain slicer with a 0.5 mm resolution (Zivic Instruments, PA) and sliced in the following coordinates in order to isolate extended amygdala: coronal level 56-66 (Bregma (−0.18)-(−1.155)) and 66–80 (Bregma (−1.155)-(−2.55)). The extended amygdala was dissected, placed in ice-cold HB (250 μl), and homogenized for 1 min using a VWR homogenizer and pestle (VWR, PA). To minimize homogenate loss, pestles were washed with 50 μl HB after use and the wash was collected and added to the sample. Ten percent of the homogenate (30 μl) were snap frozen in liquid nitrogen and stored at −80 °C for subsequent RNA TH analysis. Paired SN samples were isolated from the remaining 270 μl of the homogenate. Homogenates were filtered through a 100-μm-pore filter (Millipore, MA) and subsequently through a 5-μm-pore filter (Millipore); filters were washed with HB before use for protection from RNAse. To maximize yield, the filters were washed with 50 μl HB after use and the wash was collected and added to the homogenate. The homogenate was then centrifuged at 14 000 g for 20 min at 4 °C in order to pellet the cell fraction containing SN. The supernatant was removed and the pellet was snap frozen and stored at −80 °C for SN RNA analysis. Microscopy was used to further characterize the SN preparation (Most et al, 2014).
RNA was extracted from 20 paired SN and TH samples using the Direct-Zol RNA extraction kit (Zymo, Japan) with small IC extraction columns according to the manufacturer's instructions. The RNA was quantified using a NanoDrop1000 (Thermo Fisher Scientific Inc., IL) and assayed for quality using an Agilent 2100 Tape Station (Agilent Technologies, CA; Supplementary Figure 2A). The criteria for RNA quantity and quality were as follows: total amount of RNA >500 ng, 280/260>1.7, and RIN>6.5. We measured RIN of five of the control samples using the Bioanalyzer Nano kit (Agilent). To ensure that samples with high RINs also included high quality and quantity of microRNAs, the same five control samples were subjected to a small RNA analysis using the Bioanalyzer Small kit (Agilent). MicroRNAs comprised >10% of the small RNA population in the samples (Supplementary Figure 2B).
Microarray Hybridization, Data Quality Assessment, and Analysis
Total RNA was extracted from the TH and SN samples. The homogenates were divided into two parts (90% was used for SN and 10% for TH). The 20 samples from the SN and 20 from the TH (40 total) were hybridized to 40 microRNA microarrays. Previously, we used 40 mRNA microarrays from these same mice (Most et al, 2014). Four samples per mouse were hybridized (SN microRNA, SN mRNA, TH microRNA, and TH mRNA). The RNA samples were divided for mRNA (Most et al, 2014) and microRNA (this study) analyses. The RNA targeted for microRNA analysis was labeled with flash-tag biotin HSR (Affymetrix, CA) and hybridized to GeneChip microRNA 3.0 Arrays (Affymetrix) at the University of Texas Southwestern Medical Center microarray facility in Dallas. Affymetrix microarrays show a high correlation with results obtained from other platforms such as qPCR (Kolbert et al, 2013; Mestdagh et al, 2014). This microarray platform uses annotations from miRBase version 17 and contains 19 724 probes for mature microRNAs from 153 organisms. For this study, we focused on the 1111 mature mouse microRNAs detected on the array. The mRNA and microRNA data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al, 2002) and are accessible through GEO Series accession number GSE51730.
We analyzed the array data using R programming language and Bioconductor packages (http://www.bioconductor.org). Preprocessing (RMA, background correction, and quantile normalization) of the microRNA data was performed with the Bioconductor Oligo package. Quality measures were taken before and after preprocessing using the Array Quality Metrics package (Kauffmann et al, 2009; Kauffmann and Huber, 2010) to generate the principal component analysis (PCA). For all further analyses, only the mouse mature microRNA data were used. The microarray expression data were then analyzed using the Bioconductor Limma package according to the author instructions (Smyth, 2004). This analysis is the main approach currently used for studying thousands of genes from microarrays. We used differential expression analysis between paired SN and TH samples (paired/dependent t-test; n=20 per group). For comparison of SN and TH expression levels, fold changes were calculated as the ratio of SN to TH. Fold changes greater than 1 are referred to as ‘SN-enriched' and fold changes less than 1 are referred to as ‘SN-depleted'.
Independent t-tests were used to compare the alcohol (n=8) and control (n=12) groups to determine the effect of alcohol within SN and TH groups. These tests were performed in two separate analyses (one for each preparation). Equal sample sizes are not required for this analysis. For comparison of alcohol and control expression levels, fold changes were calculated as the ratio of alcohol to control expression levels. Fold changes greater than 1 are referred to as ‘upregulated' and fold changes less than 1 are referred to as ‘downregulated'. P-values <0.05 were considered significant for all analyses in the study.
For microRNA co-expression analyses, we used weighted gene correlation (co-expression) network analysis (WGCNA; Langfelder and Horvath, 2008). Individual mice can consume slightly different amounts of ethanol, and we were interested in the time period that represented a consistent amount of drinking and chose the average consumption per mouse from day 10 to day 30. The alcohol consumption was correlated with the expression data using Pearson correlation (n=8). However, a WGCNA network was calculated using the 20 SN samples, including both alcohol and control samples. We then determined which of the microRNA modules correlated with alcohol consumption. The WGCNA parameters were as follows for the SN: power 12, signed network, cutHeight 0.995, minModsize 60. For cell type enrichment analysis, we used lists from CamkIIa+ glutamate neurons and Gad+, somatostatin+ and parvalbumin+ GABA neurons (He et al, 2012).
We combined the microRNA data (see Table 1 for experimental design) with our previously published mRNA results (Most et al, 2014), which were obtained from the same samples. To determine alcohol-sensitive SN and TH modules, we used the ‘alcohol-responsive mRNA' lists from Most et al, 2014. All P-values from the bioinformatic analyses were adjusted using the Benjamini–Hochberg method. For cell types and immune response enrichment, we used the following lists of genes: neurons, astrocytes and oligodendrocytes (Cahoy et al, 2008), microglia (cured from Oldham et al, 2008), and glutamate/GABA (Sugino et al, 2006). For cross species comparison, we used lists from alcohol vapor-treated rats (Tapocik et al, 2013) and human alcoholics (Lewohl et al, 2011) to identify conserved alcohol-responsive microRNAs. Drugs that potentially target the alcohol-responsive mRNAs and the ‘mRNA targets of the alcohol-responsive microRNAs' were found using Ingenuity Pathway Analysis (IPA, Ingenuity Systems, Qiagen, CA). See Supplementary Table 1 for a detailed flowchart and the extensive battery of bioinformatic analyses used in the study.
Table 1. Experimental Design of the Study.
| A | B | C |
|---|---|---|
| Treatment | Preparation | 40 microRNA microarray samples |
| Alcohol-consuming mice, N=8 | Synaptoneurosome | N=8 × 2=16 |
| Total homogenate | ||
| Control mice, N=12 | Synaptoneurosome | N=12 × 2=24 |
| Total homogenate |
Each mouse was allocated to either the alcohol or control group (A) and each contributed an amygdala sample that was split into two parts for SN and TH (B). Total RNA was extracted from each of these preparations and was hybridized to microRNA microarrays (C). The total number of samples was 40 (8 SN and 8 TH for a total of 16 alcohol samples) and 24 control samples (12 SN and 12 TH), and these were individually hybridized to 40 microarrays.
Classification of microRNA–mRNA Interactions
Mouse target predictions for microRNAs and mRNAs were extracted from microRNA.org (version 8-2010). The microRNA.org resource comprises predictions computed by the miRanda algorithm (John et al, 2004; Betel et al, 2010). The algorithm predicts microRNAs according to the number of putative target sites and the sum of alignment scores determined by both seed match type and seed match context. The predictions that were considered were those annotated as ‘conserved microRNA' and ‘good mirSVR score'. The individual binding locations can be found in http://www.microRNA.org. We then overlapped the list of mRNAs that were predicted to be targeted by alcohol-responsive microRNAs with the list of microRNAs that were predicted to target the alcohol-responsive mRNAs. Prediction analysis showed that a subset of alcohol-responsive microRNAs were predicted to target certain alcohol-responsive mRNAs and vice versa, providing a bidirectional analysis for identifying microRNA–mRNA interactions using a novel approach.
Quantitative Real-Time PCR (qPCR)
qPCR was used to validate the array data of microRNA expression levels in the same SN samples. First, RNA was DNase treated using the DNA-free Kit (Ambion, TX) and reverse transcribed into cDNA using the iScript Select cDNA Synthesis Kit (Bio-Rad, CA). Samples were then evaluated for the presence of genomic DNA by comparing GAPDH Cq values from RT+ and RT− reactions, using single threshold Cq determination. Then, RNA (4.5 ng) from four of the SN alcohol samples and five of the SN control samples was used to synthesize cDNA, using the TaqMan MicroRNA Reverse Transcription Kit and the TaqMan PreAmp Master Mix, according to the manufacturer's protocol for creating custom RT and preamplification pools (Applied Biosystems, NY). qPCR was performed in triplicates using TaqMan Universal PCR Master Mix (no AmpErase UNG). FAM-labeled TaqMan MicroRNA Assays (Applied Biosystems) were used to amplify mmu-miR-137 (Assay ID 001129) and hsa-miR-9* (Assay ID 002231). Normalized relative expression was determined with respect to the most stably expressed small nucleolar RNAs, 234 (Assay ID 001234) and 202 (Assay ID 001232), as determined by geNorm analysis (Vandesompele et al, 2002). qPCR results were imported into qBase+ software, version 2.5 (Biogazelle, BE), where the ΔΔCt method was used (Hellemans et al, 2007). Statistical analysis was completed in GraphPad Prism software, version 6.
Results
SN and TH microRNA Transcriptomes are Different in Control and Alcohol Samples
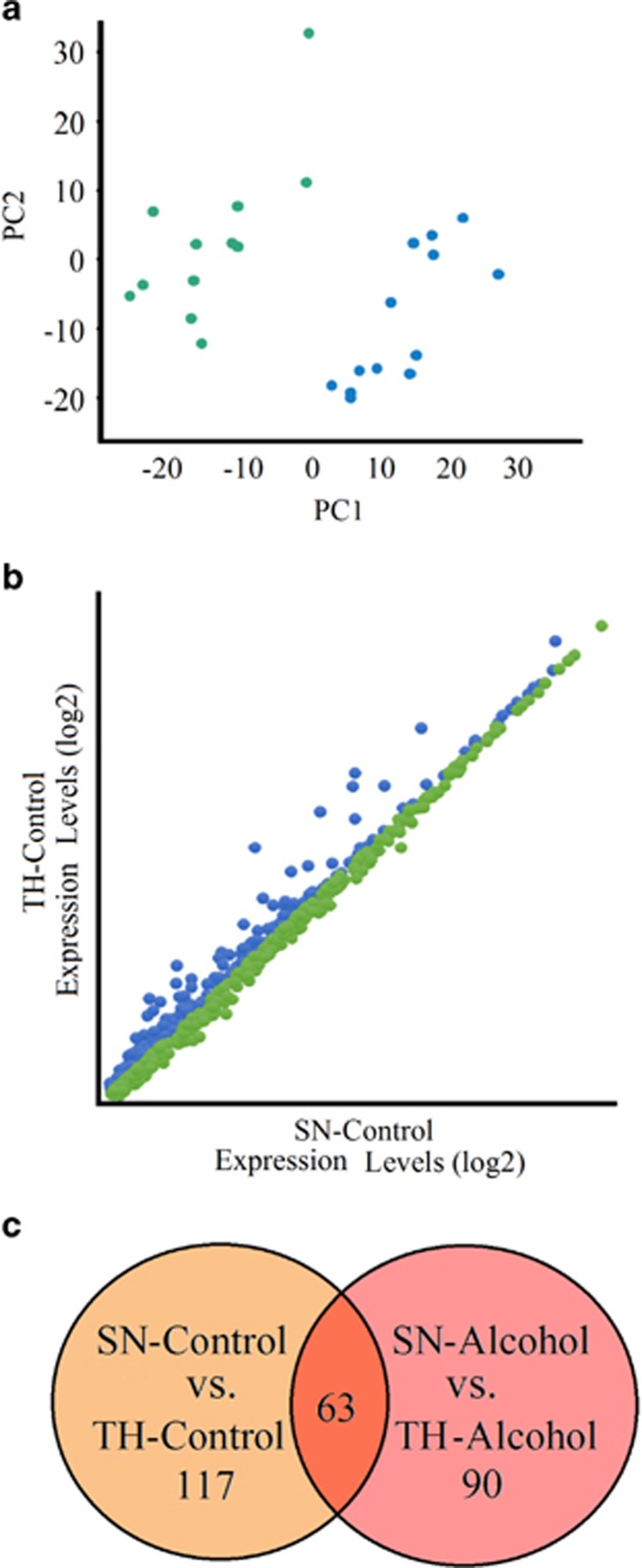
We studied the microRNA transcriptomes in paired SN and TH preparations to compare synaptic vs total cell microRNA expression. PCA revealed a distinct clustering of microRNA expression in the two preparations, while showing a homogenous sample population (no outliers detected) within each preparation (Figure 1a). The clustering was evident along the first principal component, indicating the largest variation stems from the distinct expression profiles of the preparations. A comparison of the individual microRNA expression levels on the arrays showed similar values in the SN and TH preparations as well as microRNAs enriched in SN (‘SN-enriched') and microRNAs depleted in SN (‘SN-depleted' Figure 1b). We compared the mature mouse microRNA expression levels in SN and TH (non-treated control) and found 180 differentially expressed microRNAs. Eighty-one microRNAs were ‘SN-enriched' showing up to an eightfold change in expression, and ninety-nine were ‘SN-depleted' (Supplementary Table 2, columns I and J).
Figure 1.
Difference in expression profiles in SN and TH preparations. (a) Principal component analysis of paired SN (green) and TH samples (blue). All microRNAs on the array (including those from different species) were used for this analysis (25 119 probes). The primary purpose for this analysis is quality control to show the overall detection of transcripts on the array and to facilitate comparison of the preparations. Only the mouse mature microRNAs will be discussed and presented in subsequent tables and figures. (b) Expression levels of microRNAs in SN and TH preparations (control group only). For comparison of SN and TH expression levels, fold changes were calculated as the ratio of SN to TH. MicroRNAs below the diagonal are enriched in SN relative to TH and have a fold change greater than 1 (referred to as ‘SN-enriched' shown in green). MicroRNAs above the diagonal are depleted in the SN relative to the TH and have a fold-change less than 1 (referred to as ‘SN-depleted' shown in blue). (c) Venn diagram showing the number of differentially expressed microRNAs from the SN-control/TH-control analysis and the SN-alcohol/TH-alcohol analysis, and the overlap between them. P-values <0.05 were considered significant.
We then compared SN and TH samples (referred to as SN-alcohol and TH-alcohol) from mice that chronically consumed 20% ethanol in a two-bottle choice paradigm and found 153 differentially expressed microRNAs, 96 of which were enriched in SN (Supplementary Table 2, columns K and L). It is possible that alcohol exposure changes the expression or trafficking of microRNAs to the synapse, resulting in different SN-enriched microRNAs in alcohol samples compared with control. We assessed the overlap between the differentially expressed microRNAs in SN and TH under alcohol and control conditions and found 63 overlapping microRNAs present in both conditions (Figure 1c).
Alcohol Consumption Alters microRNA Expression in SN and TH
We investigated alcohol's effects on SN microRNA expression and identified 65 mature mouse microRNAs that were differentially expressed between the alcohol and control samples (Table 2; Supplementary Table 2, columns E and F). Seventy-seven alcohol-sensitive microRNAs were differentially expressed in TH samples (Supplementary Table 2, columns G and H). Twenty microRNAs with the greatest fold changes are shown for SN and TH in Table 3.
Table 2. Alcohol-Sensitive microRNAs in SN and TH.
| A | B | C | D | E |
|---|---|---|---|---|
| MicroRNA type | Number of microRNAs investigated | Number of alcohol-responsive microRNAs in SN | Number of alcohol-responsive microRNAs in TH | Overlap between SN and TH |
| All species | 25 119 | 1377 | 1623 | 77 |
| Mouse mature | 1111 | 65 | 77 | 1 |
The number of microRNAs from all species (mature and premature/precursor mRNA) and the mouse (mature) microRNAs (A), the number of microRNAs in each group (B), the number of significant (P-values<0.05) alcohol-responsive microRNAs in SN (C) and TH (D), and the total number of microRNAs that were alcohol-sensitive in both preparations (E) are shown.
Table 3. Alcohol-sensitive microRNAs with the Largest Fold Change in SN (left) and TH (right).
| A | B | C | D | E | F | G | H | I | J |
|---|---|---|---|---|---|---|---|---|---|
| Alcohol-responsive microRNAs in SN | SN fold change | SN P-value | TH fold change | TH P-value | Alcohol-responsive microRNAs in TH | SN fold change | SN P-value | TH fold change | TH P-value |
| mmu-miR-1893 | 1.62 | 2.71E-02 | 0.91 | 6.36E-01 | mmu-miR-1965 | 1.06 | 7.66E-01 | 1.63 | 1.44E-02 |
| mmu-miR-875-3p | 1.61 | 4.45E-03 | 0.78 | 9.24E-02 | mmu-miR-207 | 0.99 | 9.48E-01 | 1.61 | 1.98E-04 |
| mmu-miR-187* | 1.56 | 2.87E-02 | 1.44 | 1.67E-01 | mmu-miR-467d* | 0.68 | 1.04E-01 | 1.57 | 1.38E-02 |
| mmu-miR-187 | 1.51 | 1.68E-02 | 1.33 | 1.76E-01 | mmu-miR-193* | 1.27 | 2.42E-01 | 1.53 | 1.44E-02 |
| mmu-miR-92a-2* | 1.47 | 4.53E-02 | 1.18 | 4.59E-01 | mmu-miR-3113* | 0.84 | 2.07E-01 | 1.45 | 1.98E-04 |
| mmu-miR-466n-3p | 1.43 | 3.83E-04 | 0.87 | 2.84E-01 | mmu-miR-574-5p | 0.63 | 8.91E-02 | 1.39 | 1.61E-02 |
| mmu-miR-669d-2* | 1.38 | 1.67E-02 | 1.06 | 4.32E-01 | mmu-miR-200c | 1.28 | 8.78E-02 | 1.35 | 2.95E-02 |
| mmu-miR-216b | 1.38 | 8.53E-03 | 0.98 | 8.67E-01 | mmu-let-7b* | 0.78 | 9.23E-02 | 1.32 | 2.11E-02 |
| mmu-miR-501-5p | 1.37 | 4.41E-02 | 0.80 | 1.09E-01 | mmu-miR-322 | 0.99 | 9.32E-01 | 1.30 | 3.78E-02 |
| mmu-miR-5115 | 1.34 | 3.51E-02 | 0.79 | 2.15E-01 | mmu-miR-30c-2* | 1.03 | 8.43E-01 | 1.28 | 2.79E-02 |
| mmu-miR-18a | 0.65 | 1.41E-02 | 1.00 | 9.98E-01 | mmu-miR-9 | 1.18 | 3.64E-01 | 0.63 | 1.93E-02 |
| mmu-miR-377* | 0.65 | 1.73E-02 | 1.13 | 4.23E-01 | mmu-miR-539-5p | 0.90 | 5.57E-01 | 0.62 | 1.40E-03 |
| mmu-miR-466g | 0.65 | 4.11E-02 | 1.33 | 1.97E-01 | mmu-miR-411* | 0.83 | 1.73E-01 | 0.62 | 7.47E-03 |
| mmu-miR-135a-2* | 0.64 | 4.82E-03 | 0.74 | 2.09E-01 | mmu-miR-3068 | 0.95 | 8.25E-01 | 0.61 | 2.32E-02 |
| mmu-miR-466f | 0.64 | 3.33E-02 | 0.85 | 3.12E-01 | mmu-miR-3473 | 1.03 | 8.39E-01 | 0.60 | 1.67E-02 |
| mmu-miR-5099 | 0.63 | 4.18E-02 | 0.90 | 3.74E-01 | mmu-miR-5100 | 1.28 | 9.73E-02 | 0.60 | 5.64E-04 |
| mmu-miR-344c | 0.59 | 8.78E-04 | 0.83 | 2.63E-01 | mmu-miR-5097 | 1.23 | 2.25E-01 | 0.58 | 1.07E-02 |
| mmu-miR-34c* | 0.58 | 3.78E-02 | 0.77 | 2.98E-01 | mmu-miR-132* | 0.96 | 6.99E-01 | 0.54 | 9.32E-03 |
| mmu-miR-1187 | 0.58 | 2.09E-03 | 1.48 | 1.75E-01 | mmu-miR-720 | 1.20 | 3.46E-01 | 0.52 | 3.52E-03 |
| mmu-miR-466f-3p | 0.43 | 3.83E-04 | 1.59 | 1.37E-01 | mmu-miR-1298 | 0.56 | 8.19E-02 | 0.37 | 1.78E-02 |
The top 10 most upregulated microRNAs (top rows) and the top 10 most downregulated microRNAs (bottom rows) in SN (columns A–E) and TH (columns F–J) are shown. Columns B and G show the fold changes and columns C and H show the P-values in SN. Columns D and I show the fold changes and columns E and J show the P-values in TH. For comparison of alcohol and control expression levels, fold changes were calculated as the ratio of alcohol to control expression levels. Fold changes greater than 1 are referred to as ‘upregulated' and fold changes less than 1 are referred to as ‘downregulated'. P-values<0.05 were considered significant and are in bold. The microRNAs are organized from top to bottom by the top 10 up- and downregulated microRNAs. For example, miR-1893 is upregulated in SN, but is downregulated in TH following alcohol treatment.
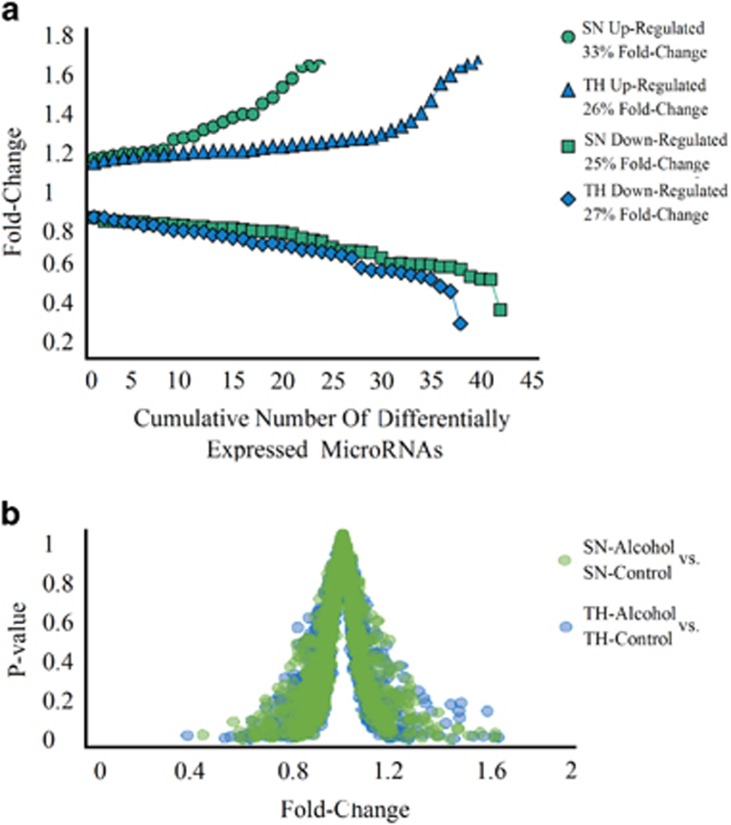
There were 23 and 39 upregulated microRNAs with average fold-change magnitudes of 33% and 26% in SN and TH, respectively (Figure 2a). There were 42 and 38 downregulated microRNAS with average fold-change magnitudes of 25% and 27% in SN and TH, respectively. The average fold changes between the preparations were not statistically significant (Supplementary Table 3). The fold changes in SN and TH were not influenced by inherent bias associated with either preparation, as shown in a volcano plot of the fold changes and P-values for SN and TH (Figure 2b).
Figure 2.
Alcohol-induced microRNAs are different in SN and TH. (a) The number of alcohol-responsive microRNAs in SN and TH. Alcohol induced fold changes are shown on the y-axis for microRNAs. For comparison of alcohol and control expression levels in each of the preparations, fold changes were calculated as the ratio of alcohol to control expression levels (SN-alcohol/SN-control and TH-alcohol/TH-control). Fold changes greater than 1 are referred to as ‘upregulated' and fold-changes less than 1 are referred to as ‘downregulated'. (b) Volcano plot (scatter plot) of fold changes and P-values of the effects of alcohol on microRNAs in SN and TH.
Two microRNAs were chosen for qPCR validation: miR-137, which was found to be alcohol-responsive in SN, and miR-9*, which was not alcohol responsive in SN. Pearson correlation for expression levels between the qPCR and arrays for miR-137 showed r=0.69. A one-tailed Students' t-test was used to test the significance of the correlation between the array and qPCR data. The correlation for miR-137 was significant (p=0.021). Pearson correlation for expression levels between qPCR and microarrays for miR-9* showed r=0.70 and this was also significant (p=0.018).
Synaptic microRNAs Coordinately Regulate Synaptic mRNAs Following Alcohol Consumption
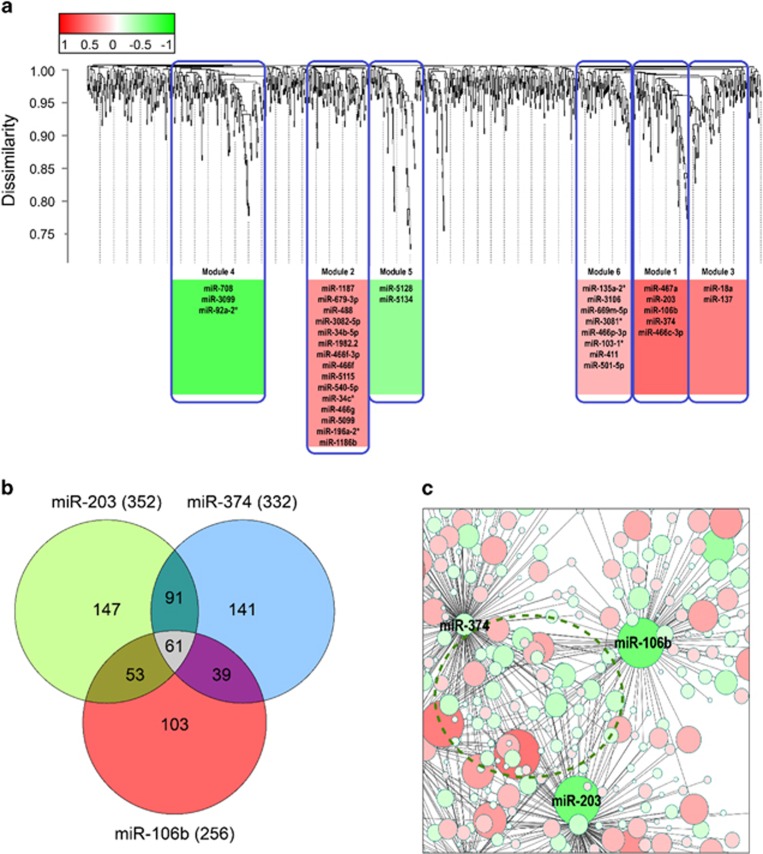
Co-expressed microRNAs may regulate their mRNA targets in response to alcohol treatment. We used WGCNA to create a co-expression network to group microRNAs with similar patterns of expression into modules. We then identified the modules that were significantly correlated with alcohol consumption and found 610 microRNAs that were co-expressed within six different modules (Supplementary Table 2, column B). Sixty-five microRNAs were differentially expressed in SN (Table 2, columns E and F), and 35 were co-expressed within these modules (Figure 3a). We identified microRNAs that correlated with alcohol consumption and found 74 in SN (Supplementary Table 2, columns C and D; average correlation: r>0.78), 48 of which were co-expressed within the six microRNA modules.
Figure 3.
Alcohol induces coordinated expression of microRNAs that are correlated with alcohol consumption. (a) Hierarchical clustering of microRNAs from SN, including both alcohol and control data. The microRNAs are arranged by covariance similarity; thus, microRNAs under the same branch have greater expression similarity than those outside the branch. The dissimilarity among microRNAs is represented in the y-axis. The six different modules are shown in boxes. The microRNAs represent the co-expressed microRNAs overlapping with alcohol-responsive microRNAs. The width of the box represents the number of microRNAs co-expressed in that module. The correlation of each module with alcohol consumption is shown as a heat map (red represents positive correlation with consumption and green represents negative). The microRNAs in the gap between the modules are ones that did not pass the co-expression threshold as defined by the WGCNA and were not included in any module. (b) Examples of three co-expressed microRNAs and the number of overlapping predicted alcohol-responsive mRNAs. (c) Examples of microRNA predicted interactions. The greater the color intensity, the greater the fold-change magnitude (red is upregulated and green is downregulated). The unmarked circles represent mRNAs. The dotted circle emphasizes mRNAs that are co-targeted by the illustrated microRNAs.
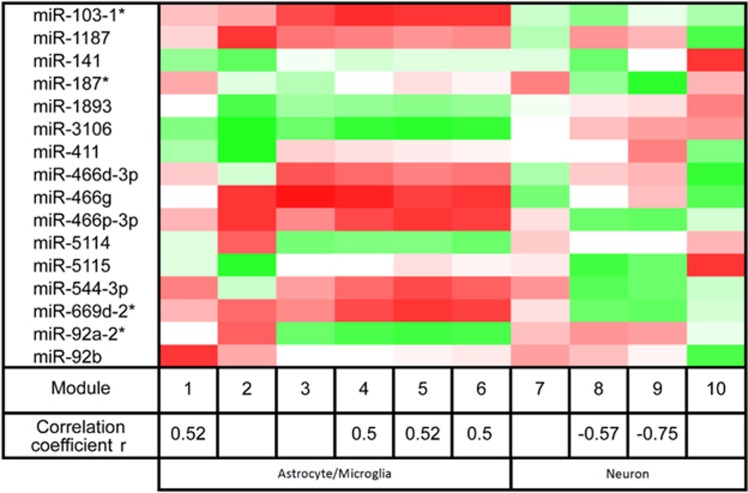
We next utilized mRNA expression data (alcohol-responsive modules of co-expressed mRNAs) from our previous study (Most et al, 2014) to identify the alcohol-responsive microRNAs co-expressed with the mRNA modules. The modules were defined as astrocytic, microglial, or neuronal if they contained a significant number of the cell type-associated mRNAs based on a hypergeometric distribution. The astrocyte modules overlapped with the microglia ones and were therefore combined. Cell type-specific analysis revealed 16 microRNAs that were significantly correlated with the alcohol-responsive mRNA modules (Figure 4).
Figure 4.
MicroRNA–mRNA interactions are coordinately expressed in response to alcohol and are associated with specific cell types. Shown are the alcohol-responsive mRNA modules and their correlation to individual microRNAs found in SN. The 10 alcohol-responsive mRNA modules are shown. The modules' correlation with consumption for the six alcohol-responsive modules is shown as r-values (Pearson's correlation coefficient). Alcohol-responsive mRNA module correlation to individual microRNAs is represented as a heatmap, with red representing a positive correlation and green representing a negative. Cell type mRNA enrichment is also shown. Modules 1–6 were enriched with astrocytic/microglial mRNAs, whereas modules 7–10 were enriched with neuronal mRNAs. The 16 microRNAs that were significantly correlated with at least one mRNA module are shown (mRNA data are from Most et al, 2014).
Synaptic microRNA–mRNA Interactions are Regulated by Alcohol
MicroRNA–mRNA interactions were constructed using target predictions from the 2010 miRanda database for ‘good mirSVR scores' and ‘conserved microRNAs' (http://www.microrna.org). From these predicted interactions, the alcohol-responsive microRNAs were predicted to target 1039 target mRNAs of the 1531 that were identified as alcohol-sensitive in Most et al, 2014. The alcohol-responsive mRNAs were predicted to be targeted by 15 of the 65 alcohol-responsive microRNAs (Table 4). The 15 microRNAs showed 15–51% change. We found that 250 of the mRNAs were predicted to be targeted by more than five of the alcohol-responsive microRNAs (Supplementary Table 4).
Table 4. Number of Alcohol-Sensitive mRNAs, microRNAs and their Predicted Interactions in SN.
| A | B | C |
|---|---|---|
| Number of alcohol-responsive RNAs | Number of predicted targets | Overlap between alcohol-responsive and predicted tragets |
| 1531 mRNA | 13 857 mRNAs | 1039 mRNAs |
| 65 microRNAs | 238 microRNAs | 15 microRNAs |
(A) The number of alcohol-responsive RNAs. (B) The number of predicted participants as defined by the miRanda mouse database for interactions. (C) The number of predicted participants that overlap with the alcohol-responsive mRNAs/microRNAs. The overlapping participants between the alcohol-responsive list and the predicted interaction list are termed ‘bidirectional'. The data for the alcohol-responsive mRNAs are from Most et al, 2014. P-values<0.05 were considered significant.
We identified mRNAs cooperatively targeted by co-expressed microRNAs by combining the mRNA predicted target analysis with the microRNA co-expression data (Table 5). MiR-106b, miR-203, and miR-374 are examples of co-expressed microRNAs that were predicted to target 61 overlapping targets (Figure 3b). MicroRNAs can be negatively or positively correlated with their mRNA targets; eg, miR-106b, miR-203, and miR-374 were all downregulated and were predicted to target both up- and downregulated mRNAs (Figure 3c).
Table 5. Co-expressed Alcohol-Responsive microRNAs and their Overlapping mRNA Targets.
| A | B | C | D | E | F | G |
|---|---|---|---|---|---|---|
| MicroRNA | MicroRNA fold change | MicroRNA P-value | Number of mRNA targets | Sum of scores | Co-expression | Overlapping targets for the co-expressed microRNAs |
| miR-203 | 0.66 | 1.63E-02 | 393 | −157.76 | Module 1 | 6330408A02Rik, Aebp2, Ank2, Ankfy1, Apc Asph, Camta1, Cdh8, Cobll1, Crbn, Dclk2, Dph3, Dpp3, Eif4a2, Etl4, Fam135a, Foxj3, Gria2, Hace1, Hnrnpa2b1, Hsd17b11, Huwe1, Ivns1abp, Maf, Mbd3l2, Mbd5, Mef2c, Mll3, Mtdh, Myt1l, Napepld, Narg2, Ndrg3, Necap1, Neto2, Oat, Ocrl, Osbp2, Pbrm1, Pbx1, Pja2, R3hdm1, Rgs7bp, Runx1t1, Scn1a, Sgk3 Ski, Slc30a1, Slc39a10, Srpk2, Syap1, Syncrip, Tmem209, Tmem87b, Trim37, Tshz3, Ube3a, Unc80, Zc3h6, Zfp644, Zxda |
| miR-374 | 0.85 | 2.90E-02 | 366 | −181.00 | ||
| miR-106b | 0.83 | 3.43E-02 | 282 | −129.25 | ||
| miR-488 | 0.83 | 1.20E-02 | 290 | −123.16 | Module 2 | 1600014C10Rik, 4833424O15Rik, AI314180, Acot11, Ahcyl1, Ankfy1, Ap2b1, Atl2, Camta1, Capn6, Car10, Cdc37, Cdc37l1, Cinp, Crbn, Cyld, Dgcr6, Etl4, Fam126b, Fbxo3, Gabrb2, Glce, Grm7, Gtf3c2, Hps5, Kif1b, Med18, Mllt3, Msi2, Mtdh, Myt1l, Ndst1, Pcdh10, Pja2, Pkp4, Prkd1, R3hdm1, Rab21, Rapgef4, Rarb, Rbbp9, Rftn2, Rnf4, Sec16a, Sesn1, Slc22a17, Slc35e3, Smc6, Snx12, Spnb2, Stx8, Tbxas1, Tcf25, Tle1, Tmem209, Tprkb, Tshz1, Tshz3, Tspan7, Unc80, Usp13, Utp6, Vps41, Vwa5b2, Zc3h14, Zfp644 |
| miR-34b-5p | 0.66 | 1.48E-02 | 246 | −84.36 | ||
| miR-137 | 0.65 | 2.03E-02 | 243 | −133.62 | Module 3 | 2700060E02Rik, 4732418C07Rik, Aebp2, Ahcyl1, Ank2, Ank3, Asph, Atl2, Cdk13, Crtc3, Ctdspl, Dgcr6, Dut, E2f6, Fry, Gatad1, Gigyf1, Hnrpdl, Kdm5b, Khdrbs3, Lingo2, Lrrc16a, Maf, Mbnl2, Mfsd6, Mll1, Msi2, Nme7, Nrg1, Osbp2, Ppp2r5c, Rab8a, Rin2, Sae1, Scamp2, Scn1a, Seh1l, Slc13a3, Smc6, Sntg2, Snurf, Spag9, Stx8, Syncrip, Tmem87b, Ybx1, Zfp804a |
| miR-18a | 0.65 | 1.35E-02 | 198 | −75.92 | ||
| miR-708 | 0.84 | 1.51E-02 | 192 | −70.24 | Module 4 | Asph, Azi2, E2f6, Foxj3, Gpt2, Gucy1a3, Hspa9, Lipa, Mia3, Mtpap, Ocrl, Palld, Phf17, Pja2, Prkar1a, Pros1, Rapgef6, Rfx1, Robo2, Seh1l, Sgms1, Socs5, Txnip, Unc5c, Ybx1, Zfp804a |
| miR-92b | 1.19 | 4.40E-02 | 190 | −90.99 | ||
| miR-365 | 1.18 | 4.45E-02 | 156 | −58.25 | Module 5 | |
| miR-411 | 0.84 | 3.89E-02 | 153 | −66.06 | Module 6 | |
| miR-141 | 0.87 | 4.10E-02 | 338 | −144.24 | NA | |
| miR-216b | 1.35 | 1.49E-02 | 230 | −101.41 | ||
| miR-92a | 1.19 | 3.62E-02 | 201 | −94.12 | ||
| miR-187 | 1.50 | 1.85E-02 | 64 | −22.28 |
Fifteen microRNAs that are regulated by alcohol (A), their fold change (B) and P-values (C) are shown. For comparison of alcohol and control expression levels, fold changes were calculated as the ratio of alcohol to control expression levels. Fold changes greater than 1 are referred to as ‘upregulated' and fold changes less than 1 are referred to as ‘downregulated'. P-values<0.05 were considered significant. The number of predicted target mRNAs (alcohol-responsive mRNAs identified in Most et al, 2014) is shown for mRNAs that were also regulated by alcohol (column D) and was calculated using the miRanda database. The sum of each of the mirSVR scores per microRNA is shown for all predicted interactions per microRNA (column E), with the scores representing the relative probability of occurrence of the microRNA–mRNA interactions. The WGCNA co-expression module for each of the microRNAs (column F) and the number of overlapping predicted mRNA targets for the microRNAs in the same module (column G) are shown.
The relationship among the alcohol-responsive microRNAs and the cell type specific glutamate and GABA microRNAs was examined using microRNA records (He et al, 2012). We identified 10 differentially expressed microRNAs that were unique to CamkIIa+ glutamate neurons, five unique to Gad+ GABA neurons, 9 unique to somatostatin+ GABA neurons but none unique to parvalbumin+ GABA neurons. In addition, 9 of the 15 alcohol-sensitive microRNAs were previously shown to be specific to glutamate neurons, and 8 of the 9 were highly predicted to target the alcohol-responsive glutamate mRNAs (Gria2, Grina, Grm7, and Grip1; Table 6).
Table 6. Alcohol-Sensitive microRNAs and their mRNA Targets that are Specific to Glutamate Neurons.
| MicroRNA | Grm7 | Gria2 | Grina | Grip1 |
|---|---|---|---|---|
| miR-106b | −0.58 | −1.03 | NA | NA |
| miR-137 | −0.69 | NA | NA | NA |
| miR-18a | NA | −0.23 | −0.34 | −0.25 |
| miR-203 | NA | −0.52 | NA | −0.39 |
| miR-374 | −0.77 | −0.27 | NA | NA |
| miR-411 | NA | −0.68 | NA | NA |
| miR-708 | NA | NA | NA | NA |
| miR-92a | −0.70 | NA | NA | NA |
| miR-92b | −0.70 | NA | NA | NA |
Nine glutamate-specific microRNAs of the 15 predicted microRNAs that are alcohol-responsive are shown. The numbers in the table represent the mirSVR scores for the predicted interactions between a microRNA and its target mRNA (as defined by the miRanda mouse database for interactions). NA means no interactions were predicted. mRNA gene symbols are shown. The data for the alcohol-responsive mRNAs are from Most et al, 2014. Grm7 (glutamate receptor, metabotropic 7), Gria2 (glutamate receptor, ionotropic, AMPA 2), Grina (glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1, glutamate binding), and Grip1 (glutamate receptor-interacting protein 1).
Discussion
The compartmentalization of RNA in cells allows for rapid responses to stimuli and may be important for the neuroadaptations in response to chronic alcohol consumption. The aim of this study was to identify the synaptic microRNAs that are altered by alcohol consumption and to propose microRNA–mRNA synaptic interactions that may be changed by chronic alcohol. We compared the effect of alcohol in paired SN vs TH samples and identified those changes that were specific to the SN. Our data indicate that the microRNAs in the SN and TH respond differently to alcohol exposure: there was only one common alcohol-responsive microRNA, miR-411, between the preparations (Supplementary Table 3). Such a small overlap was unexpected and underscores the advantage of the SN in examining discrete, localized responses to alcohol. We identified microRNAs that were SN enriched in alcohol but not control samples and vice versa. If only the magnitude of treatment fold changes for the same microRNA was larger in SN compared with TH, this might suggest that synaptic enrichment was responsible for the differences. However, changes in magnitude do not explain all of the differences that we observed which appear to be both qualitative (different microRNAs) and quantitative (different amounts of the same microRNA). The differences between SN and TH are likely due to localized effects of alcohol and it is possible that alcohol changes the trafficking of microRNAs to the synapse, resulting in unique SN-enriched microRNAs.
This regimen of alcohol consumption causes extensive and coordinated changes in gene expression in the brain, suggesting a network regulator such as a microRNA may be involved (Lewohl et al, 2011). The question remains regarding how, or if, alcohol affects synaptic pathways through synaptic microRNA regulation.
This is the first study to use SN profiling of microRNA and mRNA obtained from the same samples, enabling detection of alcohol-responsive synaptic microRNAs and mRNAs and the predicted interactions between them. We used a combination of unbiased methods to reveal key microRNAs and their targets. Chronic alcohol consumption caused robust changes in synaptic microRNA expression levels consistent with those seen in human alcoholics (Lewohl et al, 2011) and in other animal models of dependence (Gorini et al, 2013; Nunez et al, 2013; Tapocik et al, 2013). We further identified microRNAs with overlapping patterns of expression that correlated with alcohol consumption.
Previous studies used different experimental conditions, such as alcohol paradigms, species, gender, and brain regions, and we were interested in identifying potential conserved microRNAs that extend across all these different conditions. We suggest that a conserved microRNA could potentially be important in human disease. We employed many different bioinformatic approaches, such as co-expression and co-targeting, to identify the overlapping microRNAs and found some evidence that these are conserved among different species and genders. Further studies will be needed to validate the individual interactions. Nevertheless, the combined approaches provide a list of potential alcohol-sensitive interactions in the synapse that are candidates for further study.
We found the following conserved microRNAs in our study and that of an alcohol vapor exposure study in rats: miR-137, miR-187, miR-18a, miR-34c*, miR-369*, miR-374, miR-382*, miR-423, miR-488, and miR-92b (Tapocik et al, 2013). Differentially expressed SN microRNAs in the current study also overlapped with differentially expressed microRNAs from human alcoholics (Lewohl et al, 2011). These were miR-18a, miR-203, miR-369, miR-374, miR-92a, and miR-423.
We analyzed the differentially expressed mRNAs as potential targets for known drugs using IPA software and found three of the few drugs currently used to treat alcohol dependence (baclofen, disulfiram, and acamprosate) in the list. Using the differentially expressed microRNAs for the IPA analysis, we identified 26 drugs, 10 of which were also in the list of drugs identified using differentially expressed mRNAs. Seven of the ten drugs are FDA approved (Table 7), with several of these having links with alcohol actions. For example, aminophylline blocks the behavioral effects of alcohol in mice (Soares et al, 2009) and has antidepressant effects after alcohol exposure (Escudeiro et al, 2013). Theophylline blocks alcohol withdrawal-induced hyperalgesia (Gatch and Selvig, 2002). Other drugs discovered from IPA were rasagiline (treatment for Parkinson's disease) and vorinostat, regorafenib, gemcitabine (paclitaxel), and romidepsin (treatments for cancer). The mechanisms of these drugs include histone deacetylase inhibition, adenosine receptor antagonism, phosphodiesterase inhibition, anti-inflammatory actions, and inhibition of monoamine oxidase and tyrosine kinase.
Table 7. Drugs Identified by IPA that Potentially Target Alcohol-Responsive mRNAs.
| A | B | C |
|---|---|---|
| Drugs for alcohol targets | Molecular mechanisms | Potential uses |
| Belinostat | Histone deacetylase inhibitor | Cancer |
| Pyroxamide | ||
| Romidepsin | ||
| Vorinostat | ||
| Gemcitabine/paclitaxel | DNA polymerase inhibitor | |
| Tributyrin | Adenosine receptor antagonist | |
| Regorafenib | Tyrosine kinase receptor inhibitor | |
| Rasagiline | Monoamine oxidase inhibitor | Parkinson's disease |
| Aminophylline | Phosphodiesterase inhibitor and adenosine receptor antagonist | Respiratory diseases |
| Theophylline |
The alcohol-responsive mRNAs and the ‘mRNA targets of the alcohol-responsive microRNAs' were overlapped to identify drugs known to affect these targets. Drug names (A), molecular mechanisms (B), and their current potential uses (C) are shown. The data for the alcohol-responsive mRNAs are from Most et al, 2014. The seven drugs in bold are FDA approved.
We used a bidirectional approach to predict synaptic microRNA–mRNA interactions that were sensitive to alcohol (1039 mRNAs and 15 microRNAs). Of these RNAs, 99 mRNAs and 9 microRNAs were unique to glutamate neurons. Notably, eight of these nine microRNAs were predicted to target alcohol-responsive mRNAs, such as CamkII, Gria2, Grina, Grm7, and Grip 1. Moreover, miR-203, miR-18a, and miR-374 were among the glutamate microRNAs that overlapped with the human data set (Lewohl et al, 2011). These results suggest that alcohol regulates synaptic microRNAs, which in turn affect the expression of mRNAs in glutamate synapses and may partially explain the glutamate system dysregulation seen in alcoholics (Tsai and Coyle, 1998). We used the miRanda database to examine the most probable interactions that are regulated by alcohol consumption. This provides further support for the predicted interactions, but it is important to note that these require direct validation to define their role in alcohol consumption.
A single microRNA has the potential to target many alcohol-responsive mRNAs (Mayfield and Nunez, 2012; Tapocik et al, 2013). This mechanism may be of particular importance in the synaptic proteome where slight adaptions can greatly impact synaptic plasticity. Alcohol-responsive microRNAs were significantly correlated with astrocytic, microglial, and neuronal modules. The co-expression of a microRNA with a network of alcohol-responsive mRNAs supports the role of microRNAs as master regulators in the synapse. The biological pathways associated with the mRNA modules include long-term potentiation and depression, glutamate and neuroimmune signaling, RNA processing, etc., suggesting the regulation of microRNAs in multiple processes. As for many other diseases (Maciotta et al, 2013), the neuroadaptations associated with alcohol dependence likely rely on many mRNAs. A subset of the mRNA changes may be driven by only a small number of microRNAs, each with the ability to target multiple mRNAs, thereby impacting alcohol-mediated responses and therapeutic strategies.
Funding and Disclosure
The work published in this paper represents original research, not previously submitted or published elsewhere. This work was supported by the National Institutes of Health grants AA022557, AA013520, AA012404, RC2AA019382, and AA020683. The authors declare no conflict of interest.
Acknowledgments
We thank Jody Mayfield for many valuable comments and scientific editing.
Author contributions
Dana Most designed, performed experiments, and wrote the paper; Courtney Leiter performed the PCR experiments; Yuri A. Blednov designed the behavioral experiments; R. Adron Harris designed the experiments and edited the paper; R Dayne Mayfield designed experiments and edited the paper.
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
Supplementary Material
References
- Betel D, Koppal A, Agius P, Sander C, Leslie C (2010). Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites'. Genome Biol 11: R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Mayfield RD, Belknap J, Harris RA (2012). Behavioral actions of alcohol: phenotypic relations from multivariate analysis of mutant mouse data. Genes Brain Behav 11: 424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS et al (2008). A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci 28: 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekar V, Dreyer JL (2011). Regulation of MiR-124, Let-7d, and MiR-181a in the accumbens affects the expression, extinction, and reinstatement of cocaine-induced conditioned place preference. Neuropsychopharmacology 36: 1149–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eipper-Mains JE, Kiraly DD, Palakodeti D, Mains RE, Eipper BA, Graveley BR (2011). MicroRNA-Seq reveals cocaine-regulated expression of striatal microRNAs. RNA 17: 1529–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudeiro SS, Soares PM, Almeida AB, de Freitas Guimaraes Lobato R, de Araujo DP, Macedo DS et al (2013). Antidepressant effect of aminophylline after ethanol exposure. Sci Pharm 81: 211–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatch MB, Selvig M (2002). Theophylline blocks ethanol withdrawal-induced hyperalgesia. Alcohol Alcohol 37: 313–317. [DOI] [PubMed] [Google Scholar]
- Gorini G, Nunez YO, Mayfield RD (2013). Integration of miRNA and protein profiling reveals coordinated neuroadaptations in the alcohol-dependent mouse brain. PLoS One 8: e82565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Liu Y, Wang X, Zhang MQ, Hannon GJ, Huang ZJ (2012). Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron 73: 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Yang C, Kirkmire CM, Wang ZJ (2010). Regulation of opioid tolerance by let-7 family microRNA targeting the mu opioid receptor. J Neurosci 30: 10251–10258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth EB, McNeal ET, Burton JL, Williams RJ, Daly JW, Creveling CR (1985). Biochemical characterization of a filtered synaptoneurosome preparation from guinea pig cerebral cortex: cyclic adenosine 3':5'-monophosphate-generating systems, receptors, and enzymes. J Neurosci 5: 2240–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS (2004). Human MicroRNA targets. PLoS Biol 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffmann A, Gentleman R, Huber W (2009). arrayQualityMetrics—a bioconductor package for quality assessment of microarray data. Bioinformatics 25: 415–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffmann A, Huber W (2010). Microarray data quality control improves the detection of differentially expressed genes. Genomics 95: 138–142. [DOI] [PubMed] [Google Scholar]
- Kolbert CP, Feddersen RM, Rakhshan F, Grill DE, Simon G, Middha S et al (2013). Multi-platform analysis of microRNA expression measurements in RNA from fresh frozen and FFPE tissues. PLoS ONE 8: e52517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka W, Kiryk A, Novak M, Herwerth M, Parkitna JR, Wawrzyniak M et al (2010). MicroRNA loss enhances learning and memory in mice'. J Neurosci 30: 14835–14842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewohl JM, Nunez YO, Dodd PR, Tiwari GR, Harris RA, Mayfield RD (2011). Up-regulation of microRNAs in brain of human alcoholics. Alcohol Clin Exp Res 35: 1928–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli G, Larson J, Martone ME, Jones Y, Smalheiser NR (2005). Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J Neurochem 94: 896–905. [DOI] [PubMed] [Google Scholar]
- Lugli G, Torvik VI, Larson J, Smalheiser NR (2008). Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J Neurochem 106: 650–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciotta S, Meregalli M, Torrente Y (2013). The involvement of microRNAs in neurodegenerative diseases. Front Cell Neurosci 7: 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield RD, Lewohl JM, Dodd PR, Herlihy A, Liu J, Harris RA (2002). Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. J Neurochem 81: 802–813. [DOI] [PubMed] [Google Scholar]
- Mayfield RD, Nunez YO (2012). Understanding alcoholism through microRNA signatures in brains of human alcoholics. Front Genetics 3: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C et al (2014). Evaluation of quantitative miRNA expression platforms in the microRNA quality control(miRQC) study. Nat Methods 11: 809–815. [DOI] [PubMed] [Google Scholar]
- Most D, Ferguson L, Blednov Y, Mayfield RD, Harris RA (2014). The synaptoneurosome transcriptome: a model for profiling the synaptic molecular effects of alcohol. J Pharmacogenomics 15: 177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak M, Halbout B, O'Connor EC, Rodriguez Parkitna J, Su T, Chai M et al (2010). Incentive learning underlying cocaine-seeking requires mGluR5 receptors located on dopamine D1 receptor-expressing neurons. J Neurosci 30: 11973–11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez YO, Truitt JM, Gorini G, Ponomareva ON, Blednov YA, Harris RA et al (2013). Positively correlated miRNA-mRNA regulatory networks in mouse frontal cortex during early stages of alcohol dependence. BMC Genomics 14: 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S et al (2008). ‘Functional organization of the transcriptome in human brain.'. Nat Neurosci 11: 1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrzykowski AZ, Friesen RM, Martin GE, Puig SI, Nowak CL, Wynne PM et al (2008). Posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron 59: 274–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD (2012). Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci 32: 1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan EM, Philpot BD, Huganir RL, Bear MF (1999). Rapid, experience-dependent expression of synaptic NMDA receptors in visual cortex in vivo. Nat Neurosci 2: 352–357. [DOI] [PubMed] [Google Scholar]
- Raab-Graham KF, Haddick PC, Jan YN, Jan LY (2006). Activity- and mTOR-dependent suppression of Kv1.1 channel mRNA translation in dendrites. Science 314: 144–148. [DOI] [PubMed] [Google Scholar]
- Smalheiser NR, Lugli G (2009). microRNA regulation of synaptic plasticity. Neuromol Med 11: 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3 Article3. [DOI] [PubMed] [Google Scholar]
- Soares PM, Patrocinio MC, Assreuy AM, Siqueira RC, Lima NM, Arruda Mde O et al (2009). Aminophylline (a theophylline-ethylenediamine complex) blocks ethanol behavioral effects in mice. Behav Pharmacol 20: 297–302. [DOI] [PubMed] [Google Scholar]
- Sosanya NM, Huang PP, Cacheaux LP, Chen CJ, Nguyen K, Perrone-Bizzozero NI et al (2013). Degradation of high affinity HuD targets releases Kv1.1 mRNA from miR-129 repression by mTORC1. J Cell Biol 202: 53–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C et al (2006). Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci 9: 99–107. [DOI] [PubMed] [Google Scholar]
- Tapocik JD, Barbier E, Flanigan M, Solomon M, Pincus A, Pilling A et al (2014). microRNA-206 in rat medial prefrontal cortex regulates BDNF expression and alcohol drinking. J Neurosci 34: 4581–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapocik JD, Solomon M, Flanigan M, Meinhardt M, Barbier E, Schank JR et al (2013). Coordinated dysregulation of mRNAs and microRNAs in the rat medial prefrontal cortex following a history of alcohol dependence. Pharmacogenomics J 13: 286–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai G, Coyle JT (1998). The role of glutamatergic neurotransmission in the pathophysiology of alcoholism. Annu Rev Med 49: 173–184. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A et al (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DO, Kim SM, Zhao Y, Hwang H, Miura SK, Sossin WS, Martin KC (2009). Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science 324: 1536–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DO, Martin KC, Zukin RS (2010). Spatially restricting gene expression by local translation at synapses. Trends Neurosci 33: 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.