Summary
The last steps in mRNA export and remodeling are performed by the Nup82 complex, a large conserved assembly at the cytoplasmic face of the nuclear pore complex (NPC). By integrating diverse structural data, we have determined the molecular architecture of the native Nup82 complex at subnanometer precision. The complex consists of two compositionally identical multiprotein subunits that adopt different configurations. The Nup82 complex fits into the NPC through the outer ring Nup84 complex. Our map shows that this entire 14 MDa Nup82-Nup84 complex assembly positions the cytoplasmic mRNA export factor docking sites and mRNP remodeling machinery right over the NPC's central channel, rather than on distal cytoplasmic filaments as previously supposed. We suggest that this configuration efficiently captures and remodels exporting mRNP particles immediately upon reaching the cytoplasmic side of the NPC.
Graphical abstract
Introduction
The nuclear pore complex (NPC) is a large cylindrical structure with eight symmetrically arranged spokes embedded in the nuclear envelope (NE), being composed of multiple copies of ~30 different nucleoporins (Nups). Discrete Nup subcomplexes associate to form the different substructures of the NPC, consisting of coaxial outer, inner, and membrane rings surrounding a central channel, and linked to peripheral components such as the nuclear basket. Approximately one-third of all Nups, termed FG-Nups, contain intrinsically disordered domains comprising multiple Phe-Gly (FG) repeats between hydrophilic spacers. These FG repeat regions populate the NPC central channel and, through their specific interaction with cargo-carrying transport factors, mediate transport (Knockenhauer and Schwartz, 2016).
Although much of transport across the NPC is mediated by the karyopherin family of transport factors, the export of mRNAs follows a different mechanism that requires a special platform located at the cytoplasmic face of the NPC, called the Nup82 complex in budding yeast (Oeffinger and Zenklusen, 2012), which in turn associates with Dyn2, Nup116, Gle2 and Gle1 (Folkmann et al., 2011). The central role of this complex is underscored by the fact that its mammalian homolog, the Nup88 complex, is a nexus for disease-associated mutations (Kaneb et al., 2015; Nousiainen et al., 2008). The Nup82 complex and its associated proteins have proven challenging for structural analyses due to their flexibility and the presence of intrinsically disordered domains. The core of the Nup82 complex is composed of the proteins Nup82, Nup159, and Nsp1. Fragments of each have been solved crystallographically (Chug et al., 2015; Stuwe et al., 2015a; Yoshida et al., 2011), and negative stain EM revealed this complex to have an overall “P-shaped” morphology (Gaik et al., 2015), but no structures exist for either the whole complex or how it interacts with its associated proteins and the NPC.
mRNA export is achieved in several stages. First, mRNAs, packaged into export-competent mRNP particles, are docked into the nuclear basket; the mRNP particle then travels across the NPC through interaction of the non-karyopherin transport factors Mex67-Mtr2 with FG repeats that fill the NPC's central channel (Oeffinger and Zenklusen, 2012). Once the mRNP particle reaches the cytoplasmic face of the NPC, the coordinated action of the DEAD-box RNA helicase Dbp5, the nucleoporin Gle1 as well as the N-terminal β-propeller of Nup159 lead to active remodeling of the mRNP (Folkmann et al., 2011; Montpetit et al., 2011). Mex67-Mtr2 and other transport factors are removed during remodeling (Lund and Guthrie, 2005), preventing the mRNA from travelling back to the nucleus. In the final stage, the remodelled mRNA is released into the cytoplasm for translation.
Unfortunately, the precise coordination of these processes at the molecular scale has not been elucidated, in large part due to the lack of sufficiently detailed information on the spatial arrangement of transport and remodeling components relative to each other and the NPC. Localization studies have led to the proposal that the Nup82 complex forms filaments that project orthogonally from the cytoplasmic face of the NPC; such a location would imply that exporting mRNPs must first transit the central channel of the NPC before being transferred out to these peripheral cytoplasmic filaments, where the final stages of mRNP remodeling and export would occur distally from the central channel of the NPC (reviewed in (Folkmann et al., 2011; Knockenhauer and Schwartz, 2016; Oeffinger and Zenklusen, 2012)). However, exactly how this transfer would be accomplished, and how central channel transit and mRNP processing could be coordinated, remained unclear.
To understand these processes, we solved the structure of the endogenous Nup82 complex by using an integrative approach that relies on multiple structural and proteomic data (Alber et al., 2007b; Shi et al., 2014). We also determined how the Nup82 complex is anchored to the cytoplasmic face of the NPC via the Nup84 complex - a seven member assembly forming the outer rings. In addition, we used a combined structural and functional mapping analysis to elucidate the major mechanism responsible for mRNA export defects affecting Nup84 complex components. Finally, we integrate our data into a detailed map of the whole cytoplasmic mRNA export and remodelling machinery. We show that, surprisingly, the Nup82 complex positions the cytoplasmic FG repeats and mRNP remodeling machinery right over the NPC's central channel, rather than on distal cytoplasmic filaments as previously supposed.
Results
Solving the structure of the endogenous Nup82 holo-complex
We solved the structure of the endogenous native Nup82 holo-complex (Figure 1) by an integrative modeling approach that has previously allowed us and others to successfully determine the molecular architecture of numerous other large native assemblies (Sali et al., 2015). Such integrative strategies have proven to be suited for the structural analysis of large endogenous complexes such as the Nup82 holo-complex that are by nature flexible, contain unstructured regions, and are conformationally heterogeneous (Shi et al., 2015).
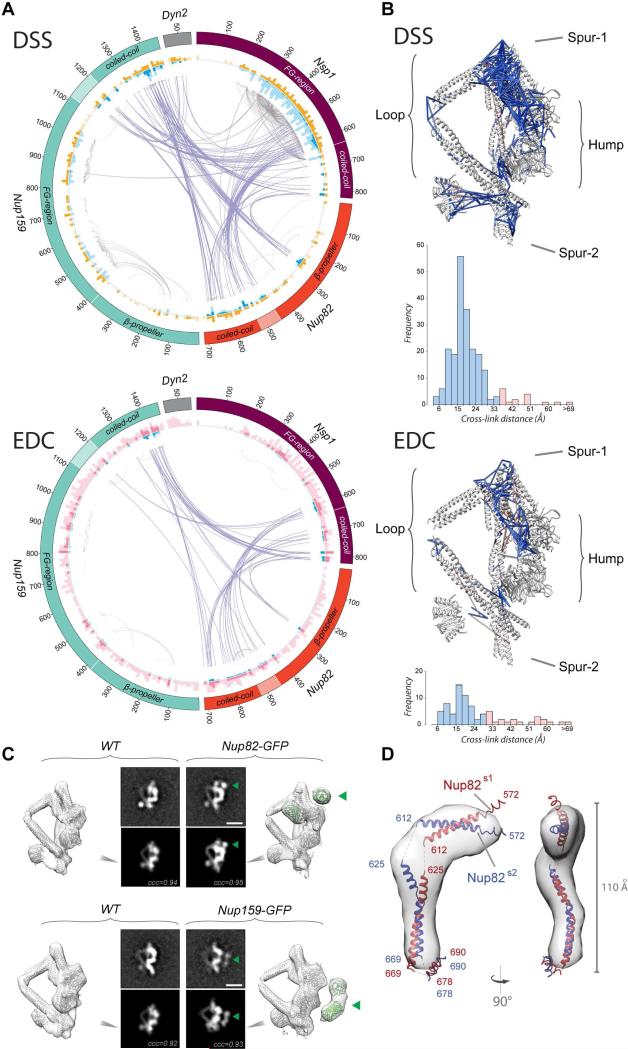
Figure 1. Structure of the core Nup82 holo-complex.
(A) Three views of the localization probability density map corresponding to the Nup82 holo-complex ensemble are shown (light gray), with a single representative ribbon structure embedded; the proteins, subunits, and different structural features of the complex are indicated. Subunit assignment is indicated with a superscript “s1” (subunit 1) or “s2” (subunit 2). In all views the components of each subunit are colored in tones of red (subunit 1) or blue (subunit 2) (see also panel B).
(B) Exploded view of the Nup82 holo-complex subunits and protein components, with the whole complex shown in the center and the two subunits and the different components shown on the right (subunit 1, colored in red tones) or the left side (subunit 2, colored in blue tones). CCS indicates “Coiled Coil Segment” as described in the main text. See also Figures S1, S2, S3 and Tables S1 and S2.
We measured the native stoichiometry of the purified Nup82 holo-complex by a combination of QConCAT-MS (Pratt et al., 2006) and classical Siegel and Monte biophysical measurements (Figure S1 and STAR Methods). The consensus of our analyses results in a stoichiometry of 2:2:2:2 (Nup159:Nup82:Nsp1:Dyn2), consistent with that previously measured (Gaik et al., 2015) for a truncated over-expressed version of the complex, with the exception of the Dyn2 dimer, a labile component that, unless over-expressed (Figure S1D), we found present as a single dimer in the average native complex. The morphology and dimensions of the complex were determined by negative stain electron microscopy (EM), where 4,266 particles were classified into 23 class averages (Figure S2C); a majority of these (21) showed what appears to be a single dimer of Dyn2, in agreement with a previous study (Gaik et al., 2015) and with our stoichiometry (see above), and were thus included in the calculation. Interestingly, two of the class averages seemingly presented two consecutive dimers of Dyn2 (Figure S2C, arrowheads), underscoring the previously observed heterogeneity of the complex in vivo (Gaik et al., 2015). Instead of using a highly uncertain 3D map computed via single particle reconstruction based on a heterogenous set of images, we relied on much more robustly computed 2D class averages, following a previously demonstrated procedure (Shi et al., 2014). Only the structured portions of the complex were constrained by the EM data, because we showed that the unstructured FG repeats are not revealed by negative stain EM (Figure S2D).
All components of the complex were used in the final calculation, including FG repeats to account for their excluded volume and emanating points. Protein representations were derived either from the atomic structures in the Protein Data Bank where available, or comparative models were built with MODELLER 9.13 (Sali and Blundell, 1993) based on the closest homolog with a known structure detected by HHPred (Soding, 2005) (Figure S3 and Table S1); disordered FG repeat containing regions were modeled as flexible strings of beads, guided by our recent NMR data (Hough et al., 2015). Finally, the residue-specific spatial proximity and orientation of the different subunits were determined by a comprehensive chemical cross-linking with mass spectrometry readout (CX-MS) method, using two complementary cross-linkers (Figure 2A and S2A) (Shi et al., 2014). To reduce the intrinsic ambiguity of cross-link data arising from the presence of two copies of each protein, we also analyzed a strain expressing an exogenous homolog of Nup82 (skNup82) from the yeast Saccharomyces kudriavzevii (Borneman et al., 2012) (Figure S2A and STAR Methods), whose distinct protein sequence allows crosslinks to it to be distinguished from the endogenous Nup82. We identified a total of 1,131 cross-links (Table S2) that include 662 unique DSS and 126 unique EDC cross-links from the wild type yeast strain and 343 unique DSS cross-links from the skNup82-containing complex (Figure S2A). The majority of the identified inter-molecular cross-links mapped to the coiled-coil, C-terminal regions of Nup159 and Nsp1, and the whole Nup82 and Dyn2 proteins. Few inter-molecular cross-links were found to connect to the FG regions of Nup159 or Nsp1, and none to the β-propeller domain of Nup159, strongly indicating that those domains are dynamic, peripheral and not located in proximity to the core of the complex (Gaik et al., 2015).
Figure 2. Nup82 holo-complex structure validation.
(A) Circos-XL plots showing the distribution of all DSS (upper plot) or EDC (lower plot) cross-links mapping within the core of the Nup82 holo-complex. Each protein is represented as a colored segment, with the amino acid residue indicated on the outside of the plot and relevant domains indicated inside each segment; regions without reliable fold assignment are identified by lighter shading. Inter-molecular cross-links are depicted as purple lines and intra-molecular cross-links as gray lines. The internal circles include bars representing the density of cross-links per 10 residues in DSS and EDC (blue and light blue color for inter-molecular cross-links and intra-molecular cross-links, respectively) and the density of lysines in DSS (orange and light orange bars for cross-linked and uncross-linked residues, respectively) or the density of lysine/carboxylic acid in EDC (pink and light pink bars for cross-linked and uncross-linked residues, respectively).
(B) Structure of the Nup82 holo-complex showing the cross-links falling within the expected Cα–Cα maximum distance threshold (blue) or outside of that threshold (orange). Below the structure, a bar graph shows the Cα–Cα distance distribution of all DSS or EDC cross-links in the structure. DSS threshold = 35 Å and EDC threshold = 30 Å.
(C) GFP mass-tagging analysis of the Nup82 holo-complex: Analyses of a Nup82-GFP tagged version (upper diagram) or a Nup159-GFP tagged version (lower diagram) of the holo-complex are shown. For each diagram, a view of the native Nup82 holocomplex structure is shown (WT), and the tagged version of the structure shown on the right side. The upper panels show a representative negative stain 2D class average of the native complex (left panel) and the tagged version (right panel; green arrowhead, GFP). The lower panels show 2D projections of the native structure (left panel) and the calculated GFP-tagged version (right panel; green arrowhead, GFP). ccc, cross correlation coefficient; Scale bar, 10 nm.
(D) SAXS analysis of the Nup82(572-690) fragment, showing two views of the computed ab initio shape (gray envelope), with ribbon representations of the equivalent Nup82 fragments in the conformation they adopt within the Nup82 holo-complex; subunits 1 (red) and 2 (blue) are indicated. See also Figure S5D-F and Table S4.
We computed the structure of the Nup82 complex (Figure 1) through our integrative modeling approach as implemented in the IMP program (Russel et al., 2012), using the data described above. A detailed assessment of the input data and the resulting model are shown in Table 1 and the STAR Methods. In summary, the 463 best-scoring solutions satisfy within stringent tolerances the data used to compute them. The clustering analysis of the best-scoring solutions identified a single dominant cluster of 370 similar structures. The corresponding localization probability density map represents the probability of any volume element being occupied by a given protein (Figure 1A). The 9.0 Å precision of the core structured region is sufficiently high to pinpoint the locations and orientations of the constituent proteins and domains, demonstrating the quality of the input data, including the cross-links and EM 2D class averages (Figure S4 and Table 1).
Table 1.
Summary of Integrative structure determination of the Nup82 complex
| Modeling programs | - Python Modeling Interface (PMI), version
c7411c3 - Integrative Modeling Platform (IMP), version 2.5 - MODELLER 9.13 |
| Homology detection and structure prediction | HHPred, PSIPRED, DISOPRED, DomPred, COILS/PCOILS, and Multicoil2. See Figure S3 and Table S1. |
| Spatial restraints | Chemical cross-links, electron microscopy 2D, excluded volume, sequence connectivity, and 5 homo-dimer cross-links restraints. See STAR Methods. |
| Sampling method | Replica Exchange Gibbs sampling, based on the Metropolis Monte Carlo algorithm. 8 to 16 replicas were used through 270 (initial step) and 80 (refinement step) independent runs, at the temperature range of 1.0 - 2.5. |
| Monte Carlo moves | - random translation and rotation of rigid
bodies (up to 2 Å and 0.04 radians, respectively). - random translation of individual beads in the flexible segments (up to 3 Å). |
| Number of structures generated | - 1,350,000 (initial step) and 10,000
(refinement step) structures. - 463 top-scoring structures were subjected to the clustering analysis. |
| Clustering analysis | 2 clusters of 370 (80%) and 93 (20%) structures. See Figs S4/S5. |
| Sampling exhaustiveness | p-value = 0.972 |
| Precision of the clusters | 9.0 Å (cluster 1: 370 structures) / 16.3 Å (cluster 2: 93 structures) |
| Stoichiometry | 2 : 2 : 2 : 2 (= Nup82 : Nup159 : Nsp1 : Dyn2). See Figure S1. |
| Chemical cross-links satisfied in the cluster | 88.5% combined (93.3% DSS and 74.1% EDC within 35 and 30 Å distances, respectively). See Figures 2B and S4D. |
| EM 2D class averages | Average ccc for 21 class averages is 0.931. See Figures 2C and S2C. |
| GFP mass-tagging EM 2D class averages | ccc = 0.932 (GFP mass-tagging at the Nup159
C-termini). ccc = 0.953 (GFP mass-tagging at the Nup82 C-termini). See Figure 2C. |
| Small angle X-ray scattering (SAXS) | Χ = 1.66 (Nup824-220), 2.55
(Nup824-452), and 6.47
(Nup82572-690). See Figures 2D and S5D-F, and Table S4. |
| human NPC cryo-EM map | ccc = 0.72 (wild-type) and 0.81 (mutant). See Figures 5 and S7. |
| Visualization and plotting | UCSF Chimera 1.10, CX-Circos, matplotLib, GNUPLOT |
Our structure is validated by seven considerations as follows. First, the EDC and DSS cross-links are highly consistent with each other, despite different chemistries, and there is significant highly non-random clustering of both EDC and DSS cross-links into equivalent “cliques” (Figure 2A). These represent immediately adjacent regions in the complex, as validated by those cliques that coincide with known crystallographic interface regions, such as Nup159:Dyn2 (PDB: 4DS1) (Romes et al., 2012) and Nup159:Nup82 (PDB: 3PBP) (Yoshida et al., 2011) (Figure 2B); indeed, in our final calculated structure these cliques represent immediately adjacent regions in the complex. Second, those few cross-links in violation of strict distance limits in our structure are nevertheless right next to one of the cliques; they are thus consistent with the structure when locally limited flexibility is taken into account (Figures 2A and S4D). Third, mass tagging of our structure is consistent with the localization of GFP tags on both the Nup82 and Nup159 C-termini (Figure 2C). Fourth, our structure is consistent with the previously published data, including an independent negative stain 3D density map (Figure S5A) (Gaik et al., 2015). Fifth, the trimeric coiled-coil structure is recapitulated even when computed using the chemical cross-linking data alone (Figure S5C). Sixth, our structure is in agreement with SAXS profiles and ab initio shapes of Nup82 constructs spanning residues 4-220, 4-452, and 572-690 (Figures 2D and S5D-F and Table S4). Notably, the Nup82 coiled-coil (572-690) forms a kinked structure and the corresponding SAXS profile shows a monotonous increase in the Kratky plot (Figures 2D and S5F), indicating a high degree of flexibility between coiled-coil segments in solution, as would be expected for coiled-coils that form two different conformers seen in the final structure. Finally, our structure is also validated by the non-random and clustered distribution of cross-links connecting the Nup82 holo-complex to other parts of the NPC, revealing interaction sites, as described below.
Features of the Nup82 holo-complex
The C-termini of Nup82, Nup159 and Nsp1 share a common domain arrangement, formed by consecutive helical coiled-coil regions of different length, connected by flexible linkers. They assemble (together with Dyn2) to form the Nup82 holo-complex, a roughly “D”-shaped particle, which is formed by the asymmetric assembly of two compositionally identical subunits (termed subunit 1(s1) and subunit 2 (s2) in Figure 1). Each subunit consists mainly of parallel, three-stranded, hetero-trimeric coiled-coils connected by flexible linkers, consisting of a single copy of the C-termini of Nup82, Nup159 and Nsp1. However, the two subunits adopt different configurations, mainly due to the different degree of flexion of the hinges between hetero-trimeric coiled-coil segments (termed CCSs) and the relative position of the Nup82 β-propellers. Subunit 1 mainly forms the “rod” while subunit 2 forms the “loop” of the holo-complex, with both subunits contributing to the spurs (Figure 1). The CCS1s2 and CCS2s2 trimers constitutes the extended loop that can be observed in certain orientations of the particle (Figure 1A, left and center). The denser region of the complex is formed by trimeric parallel CCS domains that form the slightly bent, elongated central rod. Both Nup82 β-propellers are located side by side on top of the rod formed by subunit s1, with Nup82 β-propellers2 located in trans in a distal position from the CCS1-2s2 loop. The two ends of the central rod are each formed by the C-terminal (spur-1) and the N-terminal (spur-2) bundles of the CCS domains. Two copies of Dyn2 form a dimer that is perpendicular with spur-2 and seems to help lock the two subunits into their asymmetric arrangement. Dyn2 also helps to orient the two Nup159 copies, so that their FG regions emanate in parallel from that end of the complex. Interestingly, the FG regions of Nsp1 also project from spur-2, forming, together with the Nup159 FGs, an intrinsically disordered plume. In agreement with prior work, the hump formed by the Nup82 β-propellers helps to lock down the C-termini of Nup159, as well as forming the attachment site for two Nup116 copies (Yoshida et al., 2011) (see below).
Structure of the Nup82-Nup84 complex assembly and the cytoplasmic mRNA export platform
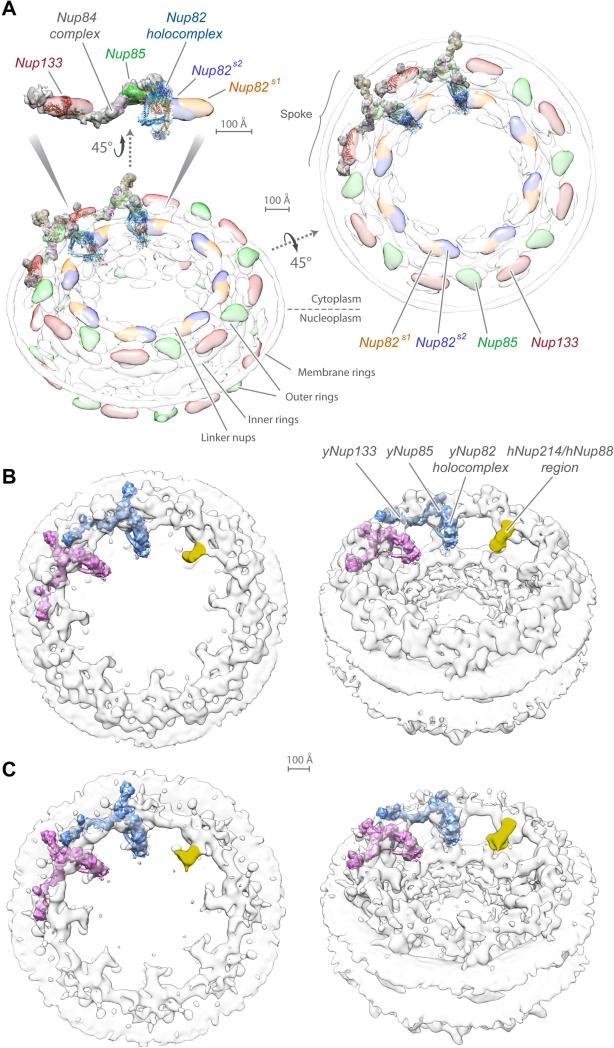
To understand how the Nup82 holo-complex is associated with the whole NPC, we isolated it under conditions that preserved its interaction with other Nups (Fernandez-Martinez et al., 2012). CX-MS was used to analyze those proteins proximally associated with each of the Nup82 holo-complex's components (Table S3). Notably, most of the identified cross-links connected the spur-1 region of the Nup82 holo-complex to components of the Nup84 complex hub (Figure 3 and Table S3) (Shi et al., 2014); indeed, a direct physical connection between the Nup82 and Nup84 complexes was recently demonstrated in Chaetomium thermophilum (Kellner et al., 2016). Our data, together with our prior map of the Nup84 complex (Shi et al., 2014), crystallographic data on the Nup84 complex (Kelley et al., 2015; Stuwe et al., 2015b), and the previous map of the entire NPC (Alber et al., 2007b), were sufficient to allow us to dock the two complexes together to generate a map of the entire ~1.3 MDa, 15 protein, Nup82-Nup84 complex assembly (Figure 3A). All our solutions were similar, differing only in the degree of rotation along the Nup82 complex long axis relative to the Nup84 complex (Figure S7). The Nup82 holo-complex body associates through its spur-1 region with the Nup85/Seh1 arm on the Y-complex hub and the N-terminal side of Nup145C (Figure 3A), with the two complexes oriented orthogonally with respect to their long axis (Figure 3A). Our arrangement is supported by the tight clustering of cross-links between the Nup82 and Nup84 complexes mainly to two discrete locations, one on spur-1 and the other on a single region of the Nup85-Seh1 arm, respectively.
Figure 3. Molecular architecture of the cytoplasmic mRNA export and remodeling platform.
(A) Structure of the Nup82-Nup84 complex assembly. Three views of the structural arrangement formed by the Y-shaped Nup84 complex (light gray density) and the Nup82 holo-complex (light blue density) calculated using CX-MS data. Each component and structural feature of the different complexes are labeled and shown as a density with fitted ribbon representations of their component nups. A circos plot shows the distribution of cross-links (dashed, light blue lines) identified between components of the Nup82 complex, Nup84 complex, and mRNP export/remodeling machinery, used for the calculation of the assembly and the map described in panel B.
(B) Molecular architecture of the cytoplasmic mRNA export and remodeling platform. An exploded view of the the different platform components is presented (solid blue lines, covalent attachment; dashed blue lines, CX-MS identified associations). When available, components are represented as crystal structures (Dbp5, Gle1 and Nup159 N-termini, pdb: 3RRM (Montpetit et al., 2011); Gle2/RAE1, pdb: 3MMY (Ren et al., 2010); Nup116 C-termini, pdb: 3PBP (Yoshida et al., 2011) and 3NF5 (Sampathkumar et al., 2012)). The Gle1 N-terminus is represented with a homology model of its predicted coiled-coil region inside a light red density of the approximate expected size for the domain. See also Table S3.
It has been previously shown that the Nup84 complex long axis orientation is approximately parallel to the plane of the NE in the NPC's outer ring (Alber et al.; Bui et al., 2013). Consequently, our structure reveals that the Nup82 holo-complex long axis is orthogonal to that of the Nup84 complex, forming a potential linker between the outer and inner ring. The coiled-coil bundles of the Nup82 holo-complex body form a scaffold, and their downward orientation makes it so that the FG plume in spur-2 projects from the bottom of the complex. The FG regions of Nsp1 and Nup159 would thus face the central transport channel and be adjacent to the Nsp1 FG regions emanating from the inner ring Nic96 complex (Figure 3).
Our CX-MS analysis of the higher order assembly also identified cross-links connecting other known components of the mRNA export machinery - Gle1, Nup42 and Nup116 - to the Nup82 holo-complex (Figure 3 and Table S3). The identified cross-links are fully consistent with previous work showing physical connections between some of these components, such as the C-termini of Gle1 and Nup42 (Strahm et al., 1999) and the C-terminus of Nup116 to Nup82 (Yoshida et al., 2011), indicating that our CX-MS analysis is targeting bona fide physical connections within the mRNA export machinery. In combination with published crystal structures of labile components of this machinery (Montpetit et al., 2011; Ren et al., 2010), our data allowed us to assemble a physical map of the whole cytoplasmic mRNA export platform, made of 16 different proteins (some in multiple copies, so comprising 24 subunits) and summing to ~1.8 MDa of mass (Figure 3B). The organization of the assembly reveals that the components actively involved in the mRNP remodeling process (Dbp5, Gle1, Nup159 N-terminus) and associated FG regions (Nup42, Nup116, Nup159, Nsp1) are localized around the Nup82 holo-complex and the short arms of the Nup84 complex. Remarkably, we identified 10 cross-links connecting Gle1 to the Nup82 holo-complex, delineating for the first time the position and orientation of Gle1 in the NPC, adjacent to the Nup82 holo-complex and oriented with its N-terminus towards the holo-complex hump, its middle region running parallel to spur 2, and its C-terminal and the Dbp5-interacting domain facing downwards towards the NPC central channel (Figure 3). Through its interaction with Gle1, Nup42 is also seemingly localized towards the central channel, in agreement with a recent report that showed how the FG region of Nup42 is fully functional if fused to the Gle1 C-terminus (Adams et al., 2014). Thus, in our map, both the core of the Nup82 holo-complex and the Nup84 complex form a flexible scaffold, which organizes and properly orients the two functional ends (FG regions and enzymatic activities) of the cytoplasmic mRNA export machinery.
Functional relationship between the Nup82 holo-complex and the Nup84 complex
To functionally annotate our Nup82-Nup84 complex assembly structure, we sought to investigate its relationship to mRNA export. Mutations affecting both Nup84 and Nup82 complex components have previously been shown to display characteristic mRNA export defects (Fabre and Hurt, 1997). Although the direct involvement of components of the Nup82 holo-complex in mRNA export has been long established (Fabre and Hurt, 1997), until now the association of mRNA defects with the Nup84 complex has remained unclear. Thus, to identify regions of the Nup84 complex that are most relevant for mRNA export, we analyzed a collection of truncation mutants (Fernandez-Martinez et al., 2012). The mRNA export defect of each mutant was quantified and heat-mapped into the Nup84 complex structure (Figure 4). We detected a clear hot spot mapping to the Nup85/Seh1 arm (Figure 4), different from those determined for other Nup84 complex phenotypes (Fernandez-Martinez et al., 2012). Notably, this hot spot maps to where the Nup85-Seh1 arm connects to the Nup82 holo-complex (Figure 3). This significant structure/function correlation supports the idea that the mRNA export phenotype, focused to this part of the Nup84 complex, is largely associated with a defective incorporation of the Nup82 complex into the NPC. To test this idea, we analyzed the in vivo localization of Nup82-GFP in several Nup84 complex truncation mutants affecting different parts of the Y-shaped complex. As shown in Figure 4, the Nup82-GFP construct is indeed significantly mislocalized to the cytoplasm only in mutations affecting the Nup85/Seh1 arm, while a control Nup49-GFP reporter did not show a similar behavior (Fernandez-Martinez et al., 2012). Thus, we conclude that the mRNA export phenotype found in Nup84 complex mutants is mainly the consequence of a defective or weakened incorporation of the Nup82 holo-complex into the NPC.
Figure 4. mRNA export phenotype in Nup84 complex mutants is associated to defective incorporation of the Nup82 holo-complex into the NPC.
(A) The mRNA export defect phenotype was quantified and plotted (mean value; n = 4) for each Nup84 complex component mutant in order of increasing level of nuclear polyA mRNA accumulation as observed by FISH (see STAR Methods and Figure S6 for details), and assigned to five divisions of increasing level of accumulation (white to dark purple)(Fernandez-Martinez et al., 2012). Representative examples of strains included in each division are shown on the top. AU, arbitrary unit; Errors = SEM; Scale bar, 5 microns.
(B) Mapping of the color code described in (A) into the Nup84 complex components. Horizontal lines represent the amino acid residue length of each protein and truncated version; amino acid residue positions are shown on top of the lines.
(C) The severity of nuclear mRNA accumulation phenotype (detailed in panels A and B) for specific truncations of the Nup84 complex components are shown mapped into the Nup82-Nup84 complex assembly. The color code is the same as the one described in panel A. The Nup82 holo-complex density is shown in light blue.
(D) Subcellular localization of Nup82-GFP in Nup84 complex truncation mutants. Top, diagrams representing the Nup84 complex, with the corresponding truncated region of the complex shown. Middle panels show the localization of the genomically tagged Nup82-GFP reporter as determined by fluorescence microscopy. Lower panels show the differential interference contrast (DIC) image of the same cells. Scale bar, 5 microns. See also Figure S6.
Conservation of the Cytoplasmic mRNA Export Platform in Opisthokonts
We tested whether or not our current structure was consistent with previous maps of the whole NPC. When the Nup82-Nup84 complex assembly is docked into our yeast NPC map (Alber et al., 2007b), the arrangement of their common components is fully consistent, as shown in Figure 5A. The Nup82 holo-complex overlaps with the localization density of Nup82, facing down into the central channel, and is in close proximity to the Nup85 arm of the Nup84 complex.
Figure 5. Position of the Nup82-Nup84 complex assembly within the NPC.
(A) Fitting to the yeast NPC map. Two views of the optimized alignment of two S. cerevisiae Nup82-Nup84 complex assemblies into the S. cerevisiae NPC localization probability density map (transparent gray), together with a side view of the detailed alignment (Alber et al., 2007b); Nup85 (green), Nup133 (red), and two Nup82 units (blue and orange) are indicated. Scale bars, 100 Å.
(B) Comparison with the human NPC tomographic cryo-EM map (EMDB 2444) (Bui et al., 2013). Two views of the optimized alignment of two S. cerevisiae Nup82-Nup84 complex assemblies (pink and blue) into the human NPC map (CCC = 0.72). One suggested localization for the human Nup214/Nup88 complex is colored in yellow.
(C) Comparison with the mutant human NPC tomographic cryo-EM map (EMDB 3104) (von Appen et al., 2015), lacking an outer cytoplasmic Y-complex ring (CCC = 0.81). See also Figure S7.
Previous attempts to align a single EM envelope for the yeast Nup82 complex to a human cryo-EM NPC map (Bui et al., 2013) led to divergent and ambiguous results (Gaik et al., 2015). However, we were able to unambiguously dock the yeast Nup82-Nup84 complex assembly into the available human cryo-EM maps (Bui et al., 2013; von Appen et al., 2015). When the Nup84 complex was aligned to the corresponding inner copy of its homolog (the Nup107-160 complex), the Nup82 holo-complex aligned with a density projecting only from the cytoplasmic ring, pointing towards the central channel (Figures 5B and 5C). It has been suggested that this protrusion might indeed represent some aspect of the Nup88-Nup214 complex, the vertebrate counterpart to the Nup82 holo-complex (Bui et al., 2013). The yeast and human alignments both support an overall conservation for certain major features of NPC architecture between fungi and metazoa, and provides further independent validation of our Nup82-Nup84 complex assembly structure. Importantly, the position of the Nup82 holo-complex FG repeat regions with respect to the whole NPC is suggestive of an organized arrangement of transport factor docking sites (see Discussion).
Discussion
Structure and evolution of the Nup82 holo-complex
We present the structure of the Nup82 holo-complex and show how it assembles with the Nup84 complex and other proteins to form the 24 subunit, ~1.8 MDa cytoplasmic mRNA export platform in the NPC. Our structural analysis therefore covers close to one-third of the yeast NPC mass (Alber et al., 2007b), which is now mapped in molecular detail. Unexpectedly, the Nup82 holo-complex and its associated machinery do not form any kind of cytoplasmic filament, in contrast to how it has been pictured in the literature. On the contrary, it forms a strut that faces the central channel. The Nup82 holo-complex exhibits an unusual architecture, with two compositionally identical trimers forming an asymmetric structure. Hinges in coiled coils allow flexibility to convert two otherwise identically arranged subunits into two similar, but morphologically distinct subunits. This structural arrangement, with flexibility in the subunits permitting alternate assemblies, is reminiscent of how vesicle coating proteins form variable architectures within the same coat complex, such as the hexagonal versus pentagonal architectures observed in clathrin coated vesicles (Cheng et al., 2007). Perhaps this variability is another echo of the evolutionary origin of the NPC in an ancient coating complex (Devos et al., 2004), and may also contribute to the observed flexibility of the NPC as a whole.
Another feature shared by the NPC and its related coating complexes is the presence of compositionally distinct but structurally and evolutionarily related modules within the entire assembly that arose from ancient duplication events (Alber et al., 2007b; Devos et al., 2004; Fernandez-Martinez et al., 2012). Indeed, there is another NPC subcomplex that also uses a trimeric bundle, and appears to be homologous and evolutionarily related to the Nup82 holo-complex. We discovered this relationship through a homolog detection search using HHPred (Soding, 2005), aiming to find structures comparable to the coiled-coil regions of the three core Nup82 complex components. Remarkably, the top and highly significant hit (HHpred P-value of 4.5E-60, 3.3E-9, and 0.0053 for Nup82, Nsp1, and Nup159, respectively) was another complex from the NPC, also containing a heterotrimer of coiled-coils: the Xenopus laevis Nup93:Nup62:Nup58:Nup54 complex (Chug et al., 2015) and its Chaetomium thermophilum Nic96:Nsp1:Nup57:Nup49 complex homolog (Stuwe et al., 2015a) (Figure S3). This similarity aided in generating high confidence comparative models for our calculations (STAR Methods). The C-termini of both complexes share a common domain arrangement, formed by three consecutive helical coiled-coil regions of different lengths, connected by flexible linkers (Figure 1), and both complexes even share a common component, Nsp1. Collectively, these observations further support the idea that both complexes evolved from a single common precursor structure, providing yet another example of an ancient duplication generating now diverse modules within the NPC, as postulated by our original protocoatomer hypothesis (Devos et al., 2004).
Spatial organization of the FG repeats
A common architecture and evolutionary origin might also imply a degree of shared functionality. In the case of the Nup82 holo-complex, the coiled-coil region serves as a strut to position various transport factor docking sites out from the core scaffold and towards the central channel of the NPC, where nucleocytoplasmic exchange is mediated (Figure 6). We therefore suggest that the coiled-coil trimeric region of the homologous Nic96:Nsp1:Nup57:Nup49 complex and that of the Nup82 holo-complex perform analogous functions, namely to serve as struts for the correct positioning of transport factor docking sites along the nucleocytoplasmic axis of the central transport channel (Figure 6). Being intrinsically disordered, the FG repeat regions themselves cannot form ordered structures to span the central channel. But, by providing a semi-rigid support, the coiled-coil regions of the two complexes may act as flexible struts, placing the FG nup docking sites so that they efficiently occupy the central channel to form an effective selective barrier, perhaps such that the struts plus FG repeats together comprise the observed “central transporter” (Yang et al., 1998). Indeed, space-filling models based on size data for FG repeats (Yamada et al., 2010) (Figure 6) show that the FG regions would project from the Nup82 holo-complex in such a manner as to essentially span the NPC's central channel and essentially form the top, cytoplasmic part of the central transporter.
Figure 6. The Nup82-Nup84 complex assembly acts as a scaffold to organize the FG region conveyor belts and mRNP remodeling sites in the NPC.
Upper: model for the arrangement of the FG regions associated to the Nup82 holo-complex. FG regions were modeled using molecular dynamics. The position of the Nup116 FG regions is based on the position of their C-termini (PDB: 3PBP (Yoshida et al., 2011), but could vary significantly, depending on the orientation of the unstructured region connecting the FG domains (dotted blue line). N-termini of Nup159 can interact during mRNP remodeling with Dbp5 as indicated by dashed blue line. Sequential mRNP export and remodeling steps associated with each region of the complex are shown on the left. Lower left: mapping of disease-associated Gle1 mutations into our model for the mRNA export platform. The yeast Gle1 region equivalent to where disease-related mutations have been found in human Gle1 were colored in purple (lethal congenital contracture syndrome 1 (LCCS-1)), gold (lethal arthrogryposis with anterior horn cell disease (LAAHD)) and cyan (amyotrophic lateral sclerosis (ALS)), based on data described in (Folkmann et al., 2014; Kaneb et al., 2015; Kendirgi et al., 2003). Proteins are represented as described in Figure 3B. Dashed blue lines indicate identified protein-protein associations. Lower right: schematic representation comparing the previous view (left) of the Nup82 complex as components of cytoplasmically oriented filaments, with the new view (right) of how it instead forms struts projecting towards the NPC central channel, positions the FG regions to fill the channel and forms the top part of the central transporter region. See also Table S3.
The Nup82 complex projects into the NPC's central channel to coordinate efficient export and remodeling of mRNPs
The FG repeats associated with the Nup82 holo-complex project from the end of the complex adjacent to the Nic96 complex, towards the midplane of the central channel (Figure 6); there, they would neighbor the Nsp1, Nup57, and Nup49 FG repeats at the equator of the NPC (Kosinski et al., 2016; Lin et al., 2016; Stuwe et al., 2015a). It is known that the relative position of FG repeats in the Nup82 holo-complex are crucial (Adams et al., 2014), and that the Mex67/Mtr2 dimer mediating mRNA export directly engages the FG repeats associated with the Nup82 holo-complex (Strasser et al., 2000; Trahan and Oeffinger, 2016) (Figure 3). Collectively, these results suggest that the type and position of FG repeats in the Nup82 holo-complex is key for an efficient mRNA export mechanism.
Surprisingly, we show that the Nup82 holo-complex does not project outward from the cytoplasmic face of the NPC, as previously assumed. Instead, it projects inwards, both radially and vertically. This arrangement has several important functional consequences. First, based on the organization of the Nup82 holo-complex, this places the associated cytoplasmically-disposed FG repeat regions in intimate contact with the symmetrically positioned FG repeat regions in the central channel, forming a continuous conduit of transport factor docking sites from the nuclear to cytoplasmic sides of the NPC. Second, this arrangement also places the mRNP remodeling machineries at the immediate cytoplasmic end of this channel (Figures 5 and 6). We suggest that the Nup82 holo-complex and Nup84 complex position these cytoplasmic docking and remodeling sites right over the central channel to efficiently capture exporting mRNP particles immediately upon reaching the cytoplasmic end of the central channel; once captured, they can be directly processed by the proximally tethered Gle1/Dbp5/Nup159N remodeling machinery, rather than requiring a transfer mechanism to previously supposed distal processing sites on cytoplasmic filaments. Third, the transport factors released during remodeling are also potentially well positioned to be recycled back into the nucleus, while the now translationally-primed mRNP exits to the cytoplasm (Figure 6). Our molecular architecture is fully consistent with proposed mRNP remodeling models (Folkmann et al., 2011; Montpetit et al., 2011), as well as with the observation that cytoplasmic release - but not translocation - is a rate limiting step during mRNA export (Oeffinger and Zenklusen, 2012). When translated into the overall NPC architecture, the presence of eight remodeling hubs surrounding the central channel ensures a highly efficient system consistent with the fast mRNA export rates observed in vivo (Grunwald and Singer, 2010; Mor et al., 2010; Smith et al., 2015). Other types of ribonucleoproteins are also actively exported through the NPC, using pathways and components that largely overlap with those of mRNA export (Nerurkar et al., 2015). It is thus reasonable to expect that our structural analysis would also serve as a framework for revealing the mechanisms governing their transit and maturation through the NPC.
While the Nup82 holo-complex is a major nexus for RNA export and remodeling processes, its human homolog when altered is also a major nexus for numerous diseases, as underscored by the fact that the mammalian orthologues of Nup82 (Nup88), Nup159 (Nup214) and Nup116 (Nup98) represent the Nups most prevalent in cancer and developmental diseases (Simon and Rout, 2014). Hence, our structure may also help rationalize the modifications in this machinery that lead to severe human diseases. For example, mutations in the human homolog of Gle1 are associated with lethal congenital contracture syndrome-1, lethal arthrogryposis with anterior horn cell disease (Nousiainen et al., 2008) and amyotrophic lateral sclerosis (Kaneb et al., 2015). We have been able to localize and orient Gle1 at the hump and spur 2 region of the Nup82 holo-complex, facing the NPC central channel (Figure 3). Our data (Table S3) also indicates that the C-terminus of Nup42 is associated with the C-terminus of Gle1 (Strahm et al., 1999), where the Nup159 N-terminal β-propellers are dynamically associated, with the Nup159 FG regions oriented towards the arms of the Nup84 complex (Figure 6), and Dbp5 physically associated with its ATPase cycle modulators Gle1 and the Nup159 β-propeller (Montpetit et al., 2011; Noble et al., 2011). Strikingly, the residues equivalent to those causing disease states in human Gle1 (Folkmann et al., 2014) all map to sites that anchor the yeast protein to either the Nup82 holo-complex or to Nup42 and Dbp5-Nup159N (Figure 6). These results, taken together with our structural and functional analyses, underscore the importance of the Nup82 complex as a hub for anchoring both the mRNA transport and processing machineries into the heart of the NPC itself, and help explain why this complex is a focus for so many developmental, oncogenic and viral diseases.
STAR METHODS
Methods and Resources
Detailed methods are provided in the online version of this paper and include the following:
KEY RESOURCES TABLE
CONTACT FOR REAGENT AND RESOURCE SHARING
- EXPERIMENTAL MODEL AND SUBJECT DETAILS
- ○ Yeast strains
- METHOD DETAILS
- ○ Affinity purification of protein complexes
- ○ Stoichiometry of the Nup82 holo-complex
- ○ Chemical cross-linking and mass spectrometry
- ○ CX-MS analysis of S. cerevisiae-S. kudriavzevii Nup82 holo-complex
- ○ Negative stain electron microscopy
- ○ Fluorescence in situ hybridization
- ○ Fluorescence Microscopy
- ○ Integrative Structure Determination
- ○ GFP mass-tagging electron microscopy
- ○ Docking of the Nup82 holo-complex and the Y-shape Nup84 complex
QUANTIFICATION AND STATISTICAL ANALYSIS
- DATA AND SOFTWARE AVAILABILITY
- ○ Software
- ○ Data Resources
METHODS AND RESOURCES
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to and will be fulfilled by the Lead Contact author Michael P. Rout (rout@rockefeller.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Yeast strains
All Saccharomyces cerevisiae strains used in this study are listed in the Key Resources Table, with the exception of the Nup84 complex truncation mutants that were described in detail in (Fernandez-Martinez et al., 2012). The Nup82 complex tagged strains were constructed in a W303 (Mata/alpha ade2-1 ura3-1 his3-11,15 trp 1-1 leu2- 3,112 can1-100) background. Otherwise stated, strains were grown at 30°C in YPD media (1% yeast extract, 2% bactopeptone, and 2% glucose). The Saccharomyces kudriavzevii strain was obtained from the American Type Culture Collection (ATCC 2601) and grown in the same conditions as referred above for S. cerevisiae.
METHODS DETAILS
Affinity purification of protein complexes
To purify the native Nup82 complex, that we will call from now on Nup82 holo-complex (as it includes all its intact, full-length endogenous components), we constructed strains in which the NUP encoding gene was genomically tagged with a variant of the Staphylococcus aureus Protein-A, preceded by the human rhinovirus 3C protease (ppx) target sequence (GLEVLFQGPS). The sequence was introduced by PCR amplification of the transformation cassette from the plasmid pProtA/HIS5. Harvested yeast cells, grown in YPD at 30°C to mid-log phase were frozen i n liquid nitrogen and cryogenically lysed in a Retsch PM100 planetary ball mill (http://lab.rockefeller.edu/rout/protocols). A total of 10-20 g of frozen cell powder were resuspended in 9 volumes of IP buffer (20mM Hepes pH 7.4, 300mM NaCl, 2mM MgCl2, 0.1% Tween 20, 1mM DTT). Cell lysate was clarified by centrifugation at 20,000 g for 10 minutes. IgG Ab conjugated magnetic beads (Invitrogen) at a concentration of 50 μl slurry/g of frozen powder were added to the clarified cell lysate and incubated for 30 minutes at 4°C. Beads were washed three times with 1 ml of IP buffer without protease inhibitors. The native complex was released from the affinity matrix by PreScission protease digestion in the same buffer. The recovered sample was then centrifuged at 20.000 g for 10 min. The supernatant (50-100 μl) was loaded on top of a 5-20% sucrose gradient made in IP buffer without Tween 20 plus 1/1000 of protease inhibitors. Gradients were ultracentrifuged on a SW55 Ti rotor (Beckman) at 42.000 rpm and 5°C for 17 hours. Gradients were manually unloaded from the top in 12 fractions of 410 μl. Fractions were analyzed by SDS-PAGE and R250 Coomassie or Sypro Ruby staining.
A higher order complex, containing the Nup84 complex plus several other nups, including the Nup82 holo-complex components, was identified previously (Fernandez-Martinez et al., 2012). The complex was affinity purified from a Nup84-ppx-PrA strain (Key Resources Table) as described above using as IP buffer 20mM Hepes pH 7.4, 20mM NaCl, 150mM potassium acetate, 2mM MgCl2, 0.5% Triton-X100, 0.1% Tween 20, 1mM DTT, and processed for cross-linking and mass spectrometry analysis.
Stoichiometry of the Nup82 holo-complex
Diploid strains, carrying one wild-type and one Protein-A-tagged version of each of the major Nup82 holo-complex components were analyzed by affinity purification as described above and the identity of the bands verified by mass spectrometry (Figure S1). To determine the Stokes radius (Rs) of the Nup82 holo-complex, the natively eluted complex was run through a calibrated Superose 6 GL 30/100 column in 20mM Hepes pH 7.4, 150mM NaCl, 0.1% Tween 20 buffer, and the results plotted against reference protein standards (Ovalbumin, Rs: 3.05; Aldolase, Rs: 4.81; Ferritin, Rs: 6.1; Thyroglobulin, Rs: 8.5). The sedimentation coefficient (S20,w) of the Nup82 holo-complex was estimated from the peak of the complex banded in sucrose gradients, run as described above, using the formula S20,w = ΔI/(ω2*t), where ΔI is the time integral, ω the angular velocity (seconds−1) and t is time (seconds) (see also (Griffith, 1994)). The mass of the holo-complex was then calculated using the Siegel-Monte equation (Figure S1A-B) (Erickson, 2009). Quantification of the relative amounts of each protein in the purified complex was performed using a synthetic concatamer of tryptic peptides or QconCAT (Pratt et al., 2006) based on the Nup82 complex components (Figure S1D). Quantotypic peptides for each of the four nucleoporins of the Nup82 complex were selected based on their mass spectrometric behavior (Nup82: 7-LSALPIFQASLSASQSPR-24, 636-NQILQFNSFVHSQK-649; Nup159: 301-TNAFDFGSSSFGSGFSK-717, 948-TSESAFDTTANEEIPK-963; Nsp1: 779-TTNIDINNEDENIQLIK-795, 806-SLDDNSTSLEK-816; Dyn2: 64-NFGSYVTHEK-73, 53-YGNTWHVIVGK-63). A synthetic gene (called Nup82 QconCAT) was designed by concatenation of the sequences encoding the referred peptides and addition of a 6xHis c-terminal tag: (MKEIRNQILQFNSFVHSQKTNAFDFGSSSFGSGFSKNFGSYVTHEKTTNIDINNEDENIQLIKLSALPIFQASLSASQSPRTSESAFDTTANEEIPKYGNTWHVIVGKSLDDNSTSLEKQINSIKHHHHHH).
The E. coli codon optimized sequence was cloned into plasmid pGEX6p-1, resulting in the expression of a protein with an n-terminal GST tag that was used both as a purification tag and sacrificial peptide (Ding et al., 2011). The Nup82-QconCAT protein was expressed by growing 300ml of BL21 E. coli cells at 37°C to OD 600 = 0.6 in minimal M9 media (Pratt et al., 2006) supplemented with heavy arginine and lysine (L-arginine:HCl 13C6; L-lysine:2HCl 13C6, Cambridge Isotope Laboratories Inc.). IPTG (1mM) was used to induce expression of the construct for 3 hours at 37°C. Harvested cells were processed using BugBuster Extraction Reagent (Novagen) as indicated by the manufacturer. The full-length Nup82 QconCAT was then purified using a two-step method that ensures a final full-length product by consecutive purification from the n and c-terminal tags: i) Clarified soluble material was incubated with 500 μl of gluthatione sepharose 4b (GE Healthcare) at room temperature for 1 hour at 4°C, and the retained proteins eluted using 2x 1ml of elution buffer (20mM Hepes pH 7.4, 150mM NaCl, 45mM imidazole, 6M guanidinium hydrochloride, 1mM TCEP, 1/500 protease inhibitor cocktail (PIC) (Sigma)). ii) The elution volume was then passed through an equilibrated His-Trap HP (GE Healthcare) at room temperature. The retained Nup82 QconCAT was then eluted in 20mM Hepes pH 7.4, 500mM imidazole, 150mM NaCl, 6M guanidinium hychloride, 1mM TCEP, 1/500 PIC. The resulting elution was analyzed by SDS-PAGE to ensure the presence of a full-length, pure protein.
For the MS analysis, the Nup82 holo-complex was purified as described above. The gradient fractions containing the complex were collected and concentrated by centrifugation at 355,000 g for 6 hours in a TLA 120.1 rotor at 4°C. The concentrated complex was then resuspended in a final 1x Nupage LDS Sample buffer (Thermo Fisher Scientific), 10mM TCEP (Thermo Fisher Scientific). The Nup82-QconCAT was ethanol precipitated and washed to eliminate the guanidinium chloride and resuspended in 1x Nupage LDS Sample buffer, 10mM TCEP. Approximately equimolar amounts of complex and Nup82-Qconcat were combined to give a final protein amount of 1 μg. The combined sample was heated at 72°C for 10 minutes a nd then alkylated using a final concentration of 30mM iodoacetamide (Sigma). The sample was then loaded into a 4% (37.5:1) in-house prepared stacking acrylamide SDS-PAGE gel. The resulting band, containing a mixture of Nup82 complex and stable-isotopically labeled Nup82 QconCAT proteins, was excised and sequentially digested by endoproteinase LysC (Roche) and trypsin (Roche) inside gel matrix, followed by LC-MS analyses to determine L/H ratio of standard peptides. LC-MS analyses were performed on an Orbitrap Fusion Mass Spectrometer (Thermo Scientific), with an Easy-nLC 1000 HPLC (Thermo Scientific) and an Easy-Spray electrospray source (Thermo Scientific). L/H ratios of standard peptides were determined using the MaxQuant software (version 1.2.2.5) (Cox and Mann, 2008).
Overexpression of Dyn2 was performed mimicking the conditions described in (Gaik et al., 2015): the S. cerevisiae Dyn2 coding sequence was cloned into the 2-micron plasmid p424-Gal1, under the control of the Gal-1 promoter. Overexpression was achieved by growing the transformed yeast cells in yeast synthetic minimal media supplemented with 2% glucose, 1% raffinose, harvesting the cells in mid-log phase, washing them with with ddH2O and then transferring them to yeast synthetic minimal media supplemented with 2% galactose, 1% raffinose for 3 hours at 30°C. Cells were then harvested and cryo-milled and the endogenous Nup82 holo-complex was purified as described above using Nup82-PrA as the handle. Purified complexes were run in SDS-PAGE gels, stained with Sypro-Ruby (Thermo Fisher Scientific) and the relative intensity of the different bands were quantified using ImageJ (www.imagej.net).
Chemical cross-linking and mass spectrometry
The natively eluted complex (250 μl, in buffer 1- 20mM Hepes pH 7.4, 300mM NaCl, 0.1% Tween, 2mM MgCl2, 1mM DTT) was cross-linked via the addition of DSS-H12/D12 (DiSuccinimidylSuberate) cross-linker (Creative Molecules) to yield a final concentration of 0.25 mM and incubated for 45 min at 25°C with gentle agitation in a shaker (900 rpm). The reaction was then quenched by 50 mM ammonium bicarbonate. In the case of cross-linking using EDC reagent (Pierce), the sample was equilibrated and natively eluted in EDC cross-linking buffer (10mM BisTris pH 6.5, 100mM Nacl, 2mM MgCl2, 0.1% Tween, 1mM DTT). EDC (20 mM) and N-hydroxysulfosuccinimide (0.4 mM) (i.e. 2% molar ratio with respect to EDC) were then added to cross-link the sample. The sample was incubated for 45 min at 25°C with gentle agitation. After cross-linking, Tris-HCl pH 8.0 (50 mM) and β-mercaptoethanol (20 mM) were added to the cross-linked sample to quench the reaction. After Cysteine reduction and alkylation, cross-linked samples were separated in a 4-12% NuPage SDS-PAGE (Invitrogen). Gels were briefly stained by GelCode Blue Stain Reagent (Thermo Fisher Scientific) to enable the visualization of the cross-linked protein complexes. The cross-linked complexes were then digested in-gel with trypsin or chymotrypsin to generate cross-linked peptides as previously described (Shi et al., 2014). After in-gel digestion, the cross-linked peptide mixtures were fractionated by peptide SEC (Superdex Peptide PC 3.2/30, GE Healthcare) by an off-line HPLC (Agilent Technologies). Two or three SEC fractions covering the molecular mass range of ~2.5 kD to ~10 kD were subsequently collected and analyzed by LC/MS. For cross-link identifications, the purified peptides were dissolved in the sample loading buffer (5% MeOH, 0.2% FA) and analyzed by a LTQ Velos Orbitrap Pro mass spectrometer or an Orbitrap Q Exactive (QE) Plus mass spectrometer (Thermo Fisher). For the analysis by the Velos Orbitrap mass spectrometer, briefly, the dissolved peptides were pressure loaded onto a self-packed PicoFrit® column with integrated electrospray ionization emitter tip (360 O.D, 75 I.D with 15 μm tip, New Objective). The column was packed with 10 cm reverse-phase C18 material (3 μm porous silica, 200 Å pore size, Dr. Maisch GmbH). Mobile phase A consisted of 0.5% acetic acid and mobile phase B of 70% ACN with 0.5% acetic acid. The peptides were eluted in a 120 or a 140 minute LC gradient (8% B to 50% B, 0-93 minutes, followed by 50% B to 100% B, 93-110 minutes and equilibrated with 100% A until 120 or 140 minutes) using a HPLC system (Agilent), and analyzed with a LTQ Velos Orbitrap Pro mass spectrometer. The flow rate was ~200-250 nL/min. The spray voltage was set at 1.9-2.3 kV. The instrument was operated in the data-dependent mode, where the top eight-most abundant ions were fragmented by higher energy collisional dissociation (HCD) (normalized collisional energy 27-29) and analyzed in the Orbitrap mass analyzer. The target resolution for MS1 was 60,000 and 7,500 for MS2. The QE instrument was directly coupled to an EASY-nLC™ 1200 System (Thermo Fisher) and experimental parameters were similar to those of the Velos Orbitrap. The cross-linked peptides were loaded onto an Easy-Spray column heated at 35°C (C18, 3mm particle size, 200 Å pore size, and 50 micrometer X 15cm, Thermo fisher). The top 8 or 10 most abundant ions (with charge stage of 3-7) were selected for fragmentation by HCD. The raw data were searched by pLink (Yang et al., 2012a) using a FASTA database containing protein sequences of the complexes. An initial MS1 search window of 5 Da was allowed to cover all isotopic peaks of the cross-linked peptides. The data were automatically filtered using a mass accuracy of MS1 ≤ 10 ppm (parts per million) and MS2 ≤ 20 ppm of the theoretical monoisotopic (A0) and other isotopic masses (A+1, A+2, A+3, and A+4) as specified in the software. Other search parameters include cysteine carbamidomethyl as a fixed modification, and methionine oxidation as a variable modification. A maximum of two trypsin missed-cleavage sites was allowed. The initial search results were obtained using a default 5% false discovery rate (FDR) – expected by target-decoy search strategy. All spectra were manually verified. ~ 94% of the cross-link identifications have a MS1 mass accuracy within 6 ppm. The cross-link data was visualized and analyzed by the CX-Circos software (manuscript in preparation).
Chemical cross-linking and mass spectrometry analysis of S. cerevisiae-S. kudriavzevii Nup82 holo-complex
To define the relative orientation of the two copies of Nup82 present in the Nup82 holo-complex we expressed an exogenous copy of Nup82 from the yeast Saccharomyces kudriavzevii (called from now on skNup82). We selected S. kudriavzevii because it is a closely related species that forms natural hybrids with S. cerevisiae, some of them used for wine fabrication (Borneman et al., 2012), and the level of conservation at the amino acid level between both species is particularly high, ensuring functionality of the skNup82 version and enough sequence variation to identify the specific peptides from each species protein version. S. kudriavzevii strain was obtained from ATCC (ATCC 2601) and genomic DNA was prepared using standard methods. The 3’ UTR and open reading frame for skNup82 was amplified and sequenced to account for potential mutations detected in the sequence available in the public database (GenBank: EHN01740.1). The wild type verified skNup82 sequence was found to encode a 716 amino acid protein with 75% identity to the scNup82 primary sequence (alignment available upon request). The upstream 190 nucleotides (promoter) region and the gene sequence were amplified using primers skN82Prom-F(5’-CACCGAAAGTTTATAGATTCAT-3’) and skN82GTW_R2 (5’-GCTGGGCCCCTGGAACAGAACTTCCAGGCCGTTTTTTGGCTGAGTATTAGTG-3’) that introduces an in-frame prescission protease cleavage site at the end of the skNup82 coding sequence. The PCR product was cloned using the pENTR™/D-TOPO® Cloning Kit (ThermoFisher Scientific) and then transferred to a modified pAG305GPD-ccdb-EGFP plasmid (Addgene), where the GPD promoter had been eliminated through a SacI-XbaI (New England Biolabs) cleavage and refill. The resulting integrative plasmid, pAG305-skNup82ppx-EGFP, was linearized using ClaI (New England Biolabs) and transformed into a diploid w303 S. cerevisiae strain. Successful integrations were assessed by PCR; correct expression and localization of the skNup82-EGFP construct were confirmed by western-blot and fluorescence microscopy, that showed the characteristic nuclear rim staining of a properly localized nucleoporin. Affinity purification of the Nup82 complex using skNup82-EGFP as a handle showed all the components of the native Nup82 complex, including a substoichiometric amount of scNup82, showing correct incorporation of the construct into the native Nup82 complex. The isolated, purified complex (see above for details on purification) was analyzed by CX-MS (see above).
Negative stain electron microscopy
Purified endogenous Nup82 holo-complex samples were applied to glow-discharged carbon-coated copper grids and stained with 1% uranyl formate. Images were collected on a Tecnai F20 (FEI Inc., USA) transmission electron microscope operating at an acceleration voltage of 80 kV at 50,000x magnification and underfocus ~1.5 μm. Images were recorded on a Tietz F224 4096×4096 CCD camera (15 μm pixels) at 2x binning. The pixel size at the specimen level was 3.23 Å. Particles were selected using Boxer from EMAN (Ludtke et al., 1999). The contrast transfer function (CTF) of the normalized images was determined using ctfit from EMAN and the phases were flipped accordingly. After that, the particles were subjected to Iterative Stable Alignment and Clustering (ISAC; (Yang et al., 2012b)) technique. A pixel error of 2√3 was used for the stability threshold. For comparison, the Nup82 holo-complex class averages were aligned and paired with Nsp1-FGΔ class averages or with GFP-tagged Nup82 complex class averages using the modified Spider ‘AP SH’ operation. Then the Nsp1-FGΔ class averages were subtracted from the Nup82 holo-complex class averages and the Nup82 holo-complex class averages were subtracted from the GFP-tagged Nup82 complex class averages and difference maps generated.
Fluorescence in situ hybridization
FISH on wild type and Nup84 complex truncation mutant strains was performed in 96-well plates. A 35 nucleotide long oligo dT probe (synthetized by Exiqon) and labeled post-synthesis with cy5 was used to detect poly A+ RNA [TT+TTT+TTTT+TTT+TTT+TT TT+TTT+TTT+TTT+TTT+TTTT, T+ represents locked nucleic acids (LNA). Cells were grown in SD complete at 25°C to OD 600=0.5-0.6 and fixed by the addition of para-formaldehyde at a final concentration of 4% for 45min at room temperature. Cells were washed 3x with buffer B (1.2M Sorbitol, 100mM KHPO4 pH7.5), suspended in spheroplast buffer [1.2M Sorbitol, 100mM KHPO4 pH7.5, 20mM Ribonucleoside-vanadyl complex (NEB #S1402S), 20mM β-mercaptoethanol, 25U lyticase / 1OD600 of cells (Sigma cat # L2524)] and incubated at 37°C un til cell walls were digested. Digested cells were washed 2x with cold buffer B, attached to polyA lysine (0.01%) treated 96 glass bottom MicroWell plate (MGB096-1-2-LG-L #0325289L2L) and stored in 70% ethanol at −20°C. For hybridization cells we re washed twice with 2× saline sodium citrate (SSC) and 1x 35% formamide/2× SSC. 20ng of labeled dT LNA probe was resuspended in 35% (v/v) formamide, 2× SSC, 1 mg ml−1 BSA, 10 mM ribonucleoside vanadyl complex (NEB #S1402S), 5 mM NaHPO4, pH 7.5, 0.5 mg ml−1 Escherichia coli tRNA and 0.5 mg ml−1 single-stranded DNA and denatured at 95°C for 3 min and cells hybridized overnight in the dark at 37°C. Cells were then washed in 35% formamide/2× SSC at 37°C 2x 30 min, followed by a 1 min wash in 1× PBS at room temperature followed by the addition of DAPI containing mounting medium to each well (Prolong Gold - Invitrogen #P36935). Images were acquired using a Zeiss Z1 inverted microscope, a 100x 1.43 NA oil objective and a AxioCam mRm CCD camera and the following filter sets: Zeiss 488050-9901-000 (Cy5), Zeiss 488049-9901-000 (DAPI). Three-dimensional datasets were generated by acquiring multiple 200 nm z-stacks spanning the entire volume of cells, 3D datasets reduced to 2D datasets by applying a maximum projection function in FiJi. The polyA accumulation phenotype was quantified by determining the fraction of cells showing strong nuclear polyA accumulation. For each strain, at least 200 cells from at least 3 different fields were quantified.
Fluorescence Microscopy
Nup82 was genomically tagged with GFP on selected Nup84 complex truncation yeast mutant strains using standard techniques. Cells were grown in YPD media at 30°C and visualized with a 63x 1.4 numerical aperture plan-apochromat objective using a Carl Zeiss Axioplan 2 microscope equipped with a Hamamatsu Orca ER-cooled CCD camera. The system was controlled with Openlab imaging software (Perkin Elmer). Images were treated with ImageJ (www.imagej.net/Welcome) and Adobe Photoshop (Adobe) softwares.
Integrative Structure Determination
Our integrative structure determination of the Nup82 holo-complex proceeded through four stages (Figure S3D) (Alber et al., 2007a; Alber et al., 2007b): (1) gathering of data, (2) representation of subunits and translation of the data into spatial restraints, (3) configurational sampling to produce an ensemble of structures that satisfies the restraints, and (4) analysis and validation of the ensemble structures. The modeling protocol (i.e., stages 2, 3, and 4) was scripted using the Python Modeling Interface (PMI), version c7411c3, a library for modeling macromolecular complexes based on our open-source Integrative Modeling Platform (IMP) package, version 2.5 (http://integrativemodeling.org) (Russel et al., 2012). Further details of the integrative modeling procedures are provided in Table 1, as well as previous publications (Shi et al., 2014). Files containing the input data, scripts, and output structures are available online (http://salilab.org/nup82; https://github.com/salilab/nup82).
Stage 1: Gathering of Data
The stoichiometry was determined via biochemical quantitation of the density-gradient purified Nup82 complex (Figure S1). 1,131 cross-links were identified via mass spectrometry (Fig 2A and Table S2). The atomic structures for some of the yeast Nup82 complex components had been previously determined via X-ray crystallography (Table S1) (Romes et al., 2012; Sampathkumar et al., 2012; Weirich et al., 2004; Yoshida et al., 2011). Their close homologs were identified by HHPred (Table S1) (Soding, 2005). Secondary structure and disordered regions were predicted by PSIPRED (Jones, 1999) and DISOPRED (Ward et al., 2004), respectively (Table S1). Coiled-coil regions of Nup82, Nsp1, and Nup159 were predicted by COILS/PCOILS (Lupas et al., 1991) and Multicoil2 (Trigg et al., 2011) (Table S1). 21 EM class averages and 3 SAXS profiles were obtained as described in Experimental Procedures.
Stage 2: Representation of Subunits and Translation of the Data into Spatial Restraints
The domains of the Nup82 complex subunits were coarse-grained using beads of varying sizes representing either a rigid body or a flexible string, based on the available crystallographic structures and comparative models (Table S1). In a rigid body, the beads have their relative distances constrained during configurational sampling, whereas in a flexible string the beads are restrained by the sequence connectivity (Shi et al., 2014). The residues in the rigid bodies and flexible strings corresponded to 37.3% and 62.7% of the Nup82 complex, respectively. To maximize computational efficiency while avoiding using too coarse a representation, we represented the Nup82 complex in a multi-scale fashion, as follows.
First, the crystallographic structures of each Nup82 complex domain were coarse-grained using two categories of resolution, where beads represented either individual residues or segments of up to 10 residues. For the one-residue bead representation, the coordinates of a bead were those of the corresponding Cα atoms. For the 10-residue bead representation, the coordinates of a bead were the center of mass of all atoms in the corresponding consecutive residues (each residue was in one bead only). The crystallographic structures covered 25.6% of the residues in the Nup82 complex.
Second, for predicted non-disordered domains of the remaining sequences, comparative models were built with MODELLER 9.13 (Sali and Blundell, 1993) based on the closest known structure detected by HHPred (Soding, 2005) and the literature (Table S1) (Chug et al., 2015; Stuwe et al., 2015a). Notably, structurally defined remote homologs (PDB codes 5C3L and 5CWS) (Chug et al., 2015; Stuwe et al., 2015a) were detected for the C-terminal coiled-coil regions of Nup82, Nup159, and Nsp1 (Figure S3 and Table S1). Similarly to the X-ray structures, the modeled regions were also coarse-grained using two categories of resolution, resulting in the 1-residue and 10-residue bead representations. The comparative models covered 11.7% of the residues in the Nup82 complex.
Finally, the remaining regions without a crystallographic structure or a comparative model (i.e., regions predicted to be disordered without a known homolog) were represented by a flexible string of beads corresponding to up to 100 residues each. We used the low-resolution representation (100 residues per bead) only for the unstructured FG repeats, whose structure is “decoupled” from the configurations of the core of the Nup82 holo-complex (Alber et al., 2007a). The residues in these beads corresponded to 62.7% of the Nup82 complex.
To improve the accuracy and precision of the structure ensemble obtained through the satisfaction of spatial restraints (below), we also imposed constraints based on crystallographically defined interfaces: Dyn27-92-Nup1591117-1126 (PDB: 4DS1) (Romes et al., 2012) and Nup827-452-Nup1591429-1456-Nup116966-1111 (PDB: 3PBP) (Yoshida et al., 2011). The latter interface of ScNup116966-1111 was compared with the structure of CgNup116882–1034 (PDB: 3NF5) (Sampathkumar et al., 2012), leading to the conclusion that the Nup116 interfaces are consistent among different species. Subcomplexes including these interfaces were simply represented as rigid bodies.
With this representation in hand, we next encoded the spatial restraints into a Bayesian scoring function (Shi et al., 2014) based on the information gathered in Stage 1, as follows.
First, the collected DSS and EDC cross-links were used to construct the Bayesian scoring function that restrained the distances spanned by the cross-linked residues (Shi et al., 2014), taking into account the ambiguity due to multiple copies of identical subunits; the ambiguous cross-link restraint considers all possible pairwise assignments in multiple copies of identical subunits, weighting more the least violated distance(s). Second, the excluded volume restraints were applied to each bead in 10-residue (or the closest) bead representations, using the statistical relationship between the volume and the number of residues that it covered (Alber et al., 2007a).
Third, we applied the sequence connectivity restraint, using a harmonic upper bound on the distance between consecutive beads in a subunit, with a threshold distance equal to four times the sum of the radii of the two connected beads. The bead radius was calculated from the excluded volume of the corresponding bead, assuming standard protein density (Alber et al., 2007a; Shi et al., 2014).
Fourth, 5 homo-dimer DSS cross-links between Nup159 residues of 1384-1384, 1387-1387, 1414-1414, 1417-1417, and 1432-1432 as well as one homo-dimer DSS cross-link between Nup82 residues of 517-517 were transformed to upper-harmonic distance restraints (up to 30 Å), enforcing the homo-dimer formation of the helices.
Finally, the EM 2D restraint (Shi et al., 2014) was imposed on the highest resolution representation of each subunit, using a negative logarithm of the cross-correlation coefficient between the EM class average density and the best-matching density projection of the structure as the em2D score (Stage 3). For sufficient precision, 100 projections were generated by uniform sampling of the unit sphere (Shi et al., 2014). The pixel size of the resulting projection image was equal to the pixel size of the class average (3.23Å). The relative weight of the final EM 2D restraint in the total score of a structure was set to 104, so that the scale of the em2D score matched those of the other restraint types.
Most of the remaining information (stoichiometry, crystallographic structures of the subunits, their homologs, and the two crystallographic interfaces) is included in the representation, whereas the SAXS profiles, immuno-EM class averages, and the density map from single-particle EM reconstruction (Gaik et al., 2015) were used only for validating our final structures. See the IMP scripts for details (http://salilab.org/nup82; https://github.com/salilab/nup82).
Stage 3: Conformational Sampling
Structural models of the Nup82 complex were computed using Replica Exchange Gibbs sampling, based on the Metropolis Monte Carlo algorithm (Shi et al., 2014). The Monte Carlo moves included random translation and rotation of rigid bodies (up to 2 Å and 0.04 radians, respectively) and random translation of individual beads in the flexible segments (up to 3 Å). 8 to 16 replicas were used for each run, with temperatures ranging between 1.0 and 2.5 (Table 1). A structure model was saved every 10 Gibbs sampling steps, each consisting of a cycle of Monte Carlo steps that moved every rigid body and flexible bead once. The entire sampling procedure (Steps 1 to 3) took ~4 weeks on a cluster of ~5,000 cores.
(Step 1) Initial modeling against each corresponding EM 2D class
21 subsets of independent sampling runs were performed, each sampling run starting with a random initial configuration and sampled against the EM 2D restraint of the corresponding class. The calculations were repeated 10 to 20 times per subset, producing a total of ~1,350,000 structures through the 270 independent runs.
(Step 2) Application of the EM 2D filter
From the ~1,350,000 structures from Step 1, we selected 650 structures whose em2D cross-correlation coefficient was at least 0.89 for at least 10 of the 21 class averages (Figure S4B).
(Step 3) Refinement against all 21 EM 2D class averages
80 independent refinement runs were performed, each one starting with one of the 650 structures from Step 2. The scoring function included em2D scores for all 21 class averages as well as other restraints listed above. The sampling produced a total of ~10,000 structures. 463 top-scoring structures from Step 3 were subjected to the subsequent analysis in Stage 4.
Stage 4: Analysis and Validation of the Ensemble Structures
Input information and output structures need to be analyzed to estimate structure precision and accuracy, detect inconsistent and missing information, and to suggest more informative future experiments. Assessment begins with structural clustering of the modeled structures produced by sampling, followed by assessment of the thoroughness of structural sampling, estimating structure precision based on variability in the ensemble of good-scoring structures, quantification of the structure fit to the input information, structure assessment by cross-validation, and structure assessment by data not used to compute it. These validations are based on the nascent wwPDB effort on archival, validation, and dissemination of integrative structure models, which we lead (Sali et al., 2015). We now discuss each one of these points in turn.
Clustering
A prerequisite for structure analysis is the clustering of the structures generated by satisfying the input data (Alber et al., 2007b; Shi et al., 2014). We used Cα root-mean-square deviation (RMSD) quality-threshold clustering (Shi et al., 2014). In general, there are three possible modeling outcomes, based on the number of clusters of models and consistency between the models and information (Shi et al., 2014). First, if only a single model (or a cluster of similar models) satisfies all restraints and all input information, there is likely sufficient information for determining the structure (with the precision corresponding to the variability within the cluster). Second, if two or more different models are consistent with the input restraints, the information is insufficient to define the single state or there are multiple significantly populated states. If the number of distinct models is small, structural differences between models may suggest additional experiments to narrow down the number of possible solutions. Third, if no model satisfies all input information, the information or its interpretation in terms of the inferred spatial restraints is incorrect, in which case the representation needs to be modified to include additional degrees of freedom, and/or sampling needs to be improved.
In the case of the Nup82 complex, the clustering analysis identified a single dominant cluster of 370 similar structures (Figures S4A and S5B), corresponding to the most favorable outcome of the three possibilities described above. The average RMSD between the major (370 structures) and minor clusters (93 structures) is relatively low at approximately 20Å, considering the resolution of the data, the resolution of the coarse-grained molecular representation, and the variation within each cluster (Shi et al., 2014) (Figure S4A). As a result, localization of all components is effectively identical between the major and minor clusters, differing only in the orientation of the Nup82 β-propeller (Figure S5B). Most importantly, our functional interpretation of the structure is completely robust with regard to the differences between the means of the two clusters.
Convergence of sampling
Any structure determination or computational modeling exercise can be described as a structural sampling process, guided by a scoring function (Alber et al., 2007a). Generally, good-scoring structures need to be found by a sampling, optimization, or enumeration scheme. Unless structures are enumerated, the very first test needs to estimate the thoroughness of structural sampling or optimization (Shi et al., 2014), which is often stochastic (e.g., Monte Carlo and Molecular Dynamics simulations). For stochastic methods, thoroughness of sampling can be assessed by showing that two independent runs (e.g., using random starting configurations or different random number generator seeds) do not result in significantly different solutions (Alber et al., 2007a; Fernandez-Martinez et al., 2012; Shi et al., 2014). Given two or more sets of structures from independent runs, we first cluster structures from all sets together, followed by assessing whether or not the runs contribute evenly to the population of each cluster, using the p-value from the [g2767]-square contingency test for homogeneity of proportions (McDonald, 2014).
For the Nup82 complex, the highly significant p-value of 0.972 (Table 1) indicated that our Monte Carlo algorithm sampled all top-scoring solutions at the resolution better than the precision of the dominant cluster. The caveat is that passing this sampling test is not absolute evidence of thorough sampling; a positive outcome of the test may be misleading if, for example, the landscape contains only a narrow, and thus difficult to find, pathway to the pronounced minimum corresponding to the correct structure.
Estimating structure precision based on variability in the ensemble of good-scoring structures
The ensemble of the top-scoring structures is analyzed in terms of the precision of its structural features (Alber et al., 2007a; Alber et al., 2007b). In general, commonly-used features include particle positions, distances, and contacts. Precision is defined by the feature variability in the ensemble with a measure similar to the crystallographic isotropic temperature factor (Biso) (Figure S4C), and likely provides the lower bound on its accuracy. Of particular interest are features present in most configurations in the ensemble that have a single maximum in their probability distribution. The spread around the maximum describes how precisely the feature is determined from the input information. The precision of component position is quantified as the average root-mean-square fluctuation (RMSF) across all pairs of structures in the cluster, after least-squares superposition onto the centroid structure (Shi et al., 2014).
For the Nup82 complex, the 9.0 Å precision of the core structured region in the dominant cluster was sufficiently high to pinpoint the locations and orientations of the constituent proteins and domains (Figures 1 and S4C, and Table 1), demonstrating the quality of the data including the cross-links and EM 2D class averages. The localization probability density maps of every Nup82 subunit as well as the whole complex were computed from the dominant cluster of the 370 solutions (Figures 1 and S4A).
Fit to input information
An accurate structure needs to satisfy the input information used to compute it. The ensemble of solutions was assessed in terms of how well they satisfied information from which they were computed, including the cross-links, the excluded volume, sequence connectivity, and the EM two-dimensional restraints.
First, the dominant cluster satisfied 88.5% of all combined cross-links (93.3% and 74.1% of the DSS and EDC cross-links, respectively) (Figures 2B and S4D, and Table 1); a cross-link restraint was satisfied by the cluster ensemble if the median Cα-Cα distance of the corresponding residue pairs (considering restraint ambiguity) was < 35 Å and 30 Å for the DSS and the EDC cross-links, respectively. Our cross-link data (10 DSS and 1 EDC cross-links) is in complete agreement with the crystal structure of Nup827-452-Nup1591429-1456-Nup116966-1111 (PDB: 3PBP) (Figure 3).
Second, the EDC and DSS cross-links are highly consistent with each other, despite different chemistries, and there is significant highly non-random clustering of both EDC and DSS cross-links into equivalent “cliques” (Figure 2A). These represent adjacencies, as validated by those cliques that coincide with known crystallographic interface regions, such as Nup159:Dyn2 (PDB: 4DS1) (Romes et al., 2012) and Nup159:Nup82 (PDB: 3PBP) (Yoshida et al., 2011); indeed, in our final calculated structure these cliques represent immediately adjacent regions in the complex (Figure 2B).
Third, considering the more abundant DSS cross-links, as can be seen from Fig. S4D (left), relatively few cross-links (<7%) remain unsatisfied by our structures. Of those that are not satisfied, most involve relatively modest distance violations that can clearly be rationalized by locally limited flexibility of the proteins, as shown in Figure 2B (cross-link distance distributions). Moreover, those few cross-links in violation of strict distance limits in our structure are nevertheless right next to one of the cliques; they are thus consistent with the structure when locally limited flexibility is taken into account (Figures 2A and S4D) (Shi et al., 2014).
Fourth, the solutions also fit the EM class averages, with an average cross-correlation coefficient of 0.931 (Figure 2C and Table 1).
Finally, 99% of the top 463 solutions satisfied the excluded volume and sequence connectivity restraints under the combined score threshold of 500.
Satisfaction of data that were not used to compute structures
In principle, our Bayesian modeling already effectively includes cross-validation via its Bayesian scoring function and sampling (Shi et al., 2014). However, the most direct test of a modeled structure is by comparing it to the data that were not used to compute it (a generalization of cross-validation). A structure can be validated directly against experimental data deliberately omitted from the structural model calculation (Degiacomi et al., 2013). This goal is achieved by excluding a subset of the experimental data from structure calculation, followed by evaluation of the resulting structures against the omitted subset of data. This procedure is analogous to the one used for calculating the crystallographic Rfree parameter and can be used to assess both the structure and the input data.
First, mass tagging of our structure is consistent with the localization of GFP tags on both the Nup82 and Nup159 C-termini (See “GFP mass-tagging analysis of the Nup82 holo-complex by immuno-EM” below and Figure 2C).
Second, our structure is consistent with the previously published data, including an independent negative stain 3D density map (Figure S5A) (Gaik et al., 2015). Our asymmetric ~19 nm long structure bears a general resemblance to the Nup82 complex class averages by Gaik et al., except for having mostly one Dyn2 dimer at its end instead of five dimers (Gaik et al., 2015).
Third, the trimeric coiled-coil structure between the helical Nup82-Nup159-Nsp1 regions is recapitulated even when computed using the chemical cross-linking data alone, without using the EM class averages (Figure S5C). We modeled the trimer using the available crystallographic structures, the helical regions predicted by PSIPRED (Jones, 1999), and the cross-links. All crystallographic structures and predicted helical regions were kept rigid. We used an ideal helix template to construct the coordinates of the predicted helical regions. We adopted the same multi-scale approach used to represent the entire Nup82 complex described above. The 500 best-scoring solutions satisfied all cross-links. The structural clustering of the 500 best-scoring solutions revealed that regions Nup82522-612 - Nup1591211-1321 - Nsp1637-727 were consistently arranged into a trimeric helical bundle.
Fourth, our structure is in agreement with SAXS profiles and ab initio shapes of Nup82 constructs spanning residues 4-220, 4-452, and 572-690 (Figures 2D and S5D-F, and Table S4). Notably, the Nup82 coiled-coil (572-690) forms a kinked structure and the corresponding SAXS profile shows a tendency of monotonous increase in the Kratky plot (Figure S5F), indicating a high degree of flexibility between coiled-coil segments in solution, as would be expected for coiled-coils that form two different conformers as seen in the final structure.
Finally, our structure is also validated by the non-random and clustered distribution of cross-links connecting the Nup82 holo-complex to other parts of the NPC, revealing interaction sites, as described in “Docking of the Nup82 holo-complex and the Y-shape Nup84 complex” below.
GFP mass-tagging electron microscopy
Two different types of GFP-tagged structures of the Nup82 holo-complex were generated by attaching a rigid-body GFP structure (PDB 1GFL) to either the Nup82 or Nup159 C-termini via the 14 linker residues of DPLALPVATPGIPM. For the Nup82 complex, the best-scoring structure was used. The configuration of the GFP tags was optimized using the replica exchange Gibbs sampling as described above using IMP. In summary, 10 independent sampling runs were performed, each run starting with a random initial configuration of the GFP tags. 4 replicas were used for each run, with temperatures ranging between 1.0 and 2.5. We produced a total of 50,000 structures each for the Nup82 and Nup159 GFP tags, using the EM 2D restraint of the corresponding immuno-EM class average. As a result, the best-scoring model structures are consistent with the localization of the GFP tags on both the Nup82 (ccc=0.953) and Nup159 (ccc=0.932) C-termini (Figure 2C).
Docking of the Nup82 holo-complex and the Y-shape Nup84 complex
A structure of the Nup82 holo-complex interacting with the Y-shape Nup84 complex was obtained by rigid-body docking restrained by 9 chemical cross-links identified at the interface (Table S3), using the replica exchange Gibbs sampling using IMP, as described above. For the Nup82 complex, the best-scoring structure was used. For the Nup84 complex, our previous structure (Shi et al., 2014) was refined by using new crystallographic structures of the complex subunits (PDB 4XMM and 4YCZ) (Kelley et al., 2015; Stuwe et al., 2015b). Next, 20 independent sampling runs were performed, each run starting with a random initial configuration. 6 replicas were used for each run, with temperatures ranging between 1.0 and 2.5. We produced a total of 100,000 structures using the cross-link restraints spanning the interface between the Nup82 holo-complex and the Nup84 complex. Subsequently, 200 top-scoring structures were subjected to the clustering analysis, identifying 3 clusters (clusters A, B, and C; 86, 70, and 44 structures, respectively) of solution structures (Figure S7A). At least 7 out of the 9 chemical cross-links were satisfied by the 200 top-scoring structures, within the distance threshold of 35 Å. All our solutions were similar, differing only in the degree of the Nup82 complex rotation along its long axis, relative to the Nup84 complex (Figure S7B). Precisions of the Nup82 holo-complex in the 3 clusters were 30.2, 11.0, and 39.0 Å, respectively.
Among the three clusters, only cluster C satisfied the cross-links used to compute them (Table S3) and the S. cerevisiae NPC localization probability density map (fit score by overlapping volume = 0.46, Figures 5A and S7C) (Alber et al., 2007b). Notably, this cluster of solutions is also the only one that aligns with the wild-type human NPC tomographic cryo-EM map (Figures 5B and S7D, EMDB 2444) (Bui et al., 2013) and the mutant one lacking an outer cytoplasmic Y-complex ring (Figure 5C, EMDB 3104) (von Appen et al., 2015). The cross-correlation coefficients between the Nup82 holo-complex structure and the human NPC tomographic cryo-EM maps are 0.72 (wild-type, Figures 5B and S7D) and 0.81 (mutant, Figure 5C) in cluster C (Table 1). The cross-correlation coefficients were calculated using the measure correlation command in the UCSF Chimera software (https://www.cgl.ucsf.edu/chimera/).
QUANTIFICATION AND STATISTICAL ANALYSIS
See METHODS DETAILS for details on the statistical analyses.
DATA AND SOFTWARE AVAILABILITY
Software
The modeling protocol (i.e., stages 2, 3, and 4) was scripted using the Python Modeling Interface (PMI), version c7411c3, a library for modeling macromolecular complexes based on our open-source Integrative Modeling Platform (IMP) package, version 2.5 (http://integrativemodeling.org) (Russel et al., 2012).
To display the CX-MS data we used the software CX-Circos (http://cx-circos.net).
Data Resources
The chemical cross-linking with mass spectrometric readout data used in this study was deposited in the Chorus database (https://chorusproject.org/pages/index.html).
Files containing the input data, modeling scripts, and output structures are available online (http://salilab.org/nup82; https://github.com/salilab/nup82).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit IgG Protein A Purified | Innovative Research | Cat.# IR-RB-GF |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Uranyl formate | Electron Microscopy Sciences | Cat. #22451 Cas. #16984-59-1 |
| PreScission protease | GE Healthcare Life Sciences | Cat.# 27-0843-01 |
| Coomassie R250 | MP Biomedicals | Cat.# 190682 |
| GelCode Blue Stain Reagent | Thermo Fisher Scientific | Cat.# 24592 |
| Trypsin Sequencing Grade, modified | Roche | Cat.# 11418033001 |
| Endoproteinase Lys-C Sequencing Grade | Roche | Cat.# 11047825001 |
| DSS(DiSuccinimidylSuberate)-H12/D12 | Creative molecules | Cat.# 001S |
| EDC (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride) | Thermo Fisher Scientific | Cat.# PI22980 |
| Sulfo-NHS (N-hydroxysulfosuccinimide) | Thermo Fisher Scientific | Cat.# P124510 |
| Trypsin Sequencing Grade, modified | Roche | Cat.# 11418033001 |
| Endoproteinase Lys-C Sequencing Grade | Roche | Cat.# 11047825001 |
| Iodoacetamide | Sigma | Cat.# I6125-10g |
| L-arginine:HCl 13C6 | Cambridge Isotope Laboratories Inc. | Cat.# CNLM-539-H- |
| L-lysine:2HCl 13C6 | Cambridge Isotope Laboratories Inc. | Cat.# CNLM-291-H |
| TCEP | Thermo Fisher Scientific | Cat.# PI20491 |
| Nupage LDS Sample buffer | Life Technologies | Cat.# NP0007 |
| Poly-L-lysine Solution | Sigma-Aldrich | Cat.# P8920 |
| Formamide | Sigma-Aldrich | Cat.# F9037 |
| Lyticase from Arthrobacter luteus | Sigma-Aldrich | Cat.# L2524 |
| tRNA from E. coli MRE 600 | Roche | Cat.# 10109541001 |
| 32% Paraformaldehyde (formaldehyde) aqueous solution | Electron Microscope Sciences | Cat.# 15714 |
| Ribonucleoside-vanadyl complex | New England Biolabs | Cat.# S1402S |
| Ultrapure Salmon Sperm DNA Solution | Invitrogen | Cat.# 15632011 |
| Prolong Gold | Invitrogen | Cat.# P36935 |
| Critical Commercial Assays | ||
| Dynabeads M270 Epoxy | Thermo Fisher Scientific | Cat # 143.02D |
| SYPRO® Ruby Protein Gel Stain | Thermo Fisher Scientific | Cat.# S12000 |
| Gel Filtration HMW Calibration Kit | GE Healthcare Life Sciences | Cat.# 28-4038-42 |
| BugBuster Extraction Reagent | EMD Millipore | Cat# 70921-4 |
| Gluthatione Sepharose 4b | GE Healthcare Bioscience | Cat# 17-0756-05 |
| His-Trap HP | GE Healthcare Biosciences | Cat# 17-5247-01 |
| Deposited Data | ||
| Chemical Cross-linking with Mass Spectrometry readout datasets | Chorus | https://chorusproject.org/pages/index.html |
| Saccharomyces kudriavzevii Nup82 cds genomic DNA | Genbank | Submission # 1896113 (pending accession number) |
| Files containing the input data, scripts, and output structures | N/A |
http://salilab.org/nup82
https://github.com/salilab/nup82 |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| MATa/MATα ade2-1/ade2-1 ura3-1/ura3-1 his3-11,15/his3-11,15 trp1-1/trp1-1 leu2-3,112/leu2-3,112 can1-100/can1-100 | wt | W303 |
| MATα ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 | wt | W303α |
| MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 | wt | W303a |
| MATa/MATα ura3-52/ura3-52 his3-Δ200/his3-Δ200 trp1-1/trp1-1 leu2-3,112/leu2-3,112 lys2-801/lys2-801 | wt | DF5 |
| MATα ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 NUP82-PPX-ProteinA::HIS5 | This study | Nup82-PPX-PrA |
| MATα ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 NUP159-PPX-ProteinA::HIS5 | This study | Nup159-PPX-PrA |
| MATa/MATα ura3-52/ura3-52 his3-Δ200/his3-Δ200 trp1-1/trp1-1 leu2-3,112/leu2-3,112 lys2-801/lys2-801 NUP82/NUP82-ProteinA::HIS5 | This study | Nup82-PrA 2n |
| MATa/MATα ura3-52/ura3-52 his3-Δ200/his3-Δ200 trp1-1/trp1-1 leu2-3,112/leu2-3,112 lys2-801/lys2-801 NUP159/NUP159-ProteinA::HIS5 | Rout et al. 2000 | Nup159-PrA 2n |
| MATa/MATα ura3-52/ura3-52 his3-Δ200/his3-Δ200 trp1-1/trp1-1 leu2-3,112/leu2-3,112 lys2-801/lys2-801 NSP1/NSP1-ProteinA::HIS5 | Rout et al. 2000 | Nsp1-PrA 2n |
| MATα ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 NUP82-PPX-ProteinA::HIS5 NUP159-GFP-3xFlag-6xHis::klURA3 | This study | Nup82-PPX-PrA/Nup159-GFP3xF6xH |
| MATα ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 NUP159-PPX-ProteinA::HIS5 NUP82-GFP-3xFlag-6xHis::klURA3 | This study | Nup159-PPX-PrA/Nup82-GFP3xF6xH |
| MATα ade2-1 ura3-1 his3-11,15 trp1-1 lys2 leu2-3,112 can1-100 Flag-LoxP-nsp1ΔFXFG-ΔFG NUP82-PPX-ProteinA::HIS5 | This study and Strawn et al. 2004 | Nup82-PPX-PrA/Nsp1ΔFXFG-ΔFG |
| MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 LoxP-NUP120Δ (aa1-396)-ProteinA::HIS5 | This study | Nup120(397-1037)/Nup82-GFP3xF6xH |
| MATa ura3-52 his3-Δ200 trp1-1 leu2-3,112 lys2-801 LoxP-NUP85Δ(aa 1-232)-ProteinA::HIS5 NUP82-GFP-3xFlag-6xHis::klURA3 | This study | Nup85(233-744)/Nup82-GFP3xF6xH |
| MATa ura3-52 his3-Δ200 trp1-1 leu2-3,112 lys2-801 NUP85-ProteinA::HIS5 NUP82-GFP-3xFlag-6xHis::klURA3 | This study | Nup85wt-PrA/Nup82-GFP3xF6xH |
| MATa ura3-52 his3-Δ200 trp1-1 leu2-3,112 lys2-801 NUP84Δ(aa574-726)-ProteinA::HIS5 NUP82-GFP-3xFlag-6xHis::klURA3 | This study | Nup84(1-573)/Nup82-GFP3xF6xH |
| MATa ura3-52 his3-Δ200 trp1-1 leu2-3,112 lys2-801 NUP145cΔ(aa317-326)-ProteinA::HIS5 NUP82-GFP-3xFlag-6xHis::klURA3 | This study | Nup145c(1-316)-(327-712)/ Nup82-GFP3xF6xH |
| MATα ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 NUP84-PPX-ProteinA::HIS5 | Fernandez-Martinez et al. 2012 | Nup84wt-PPX-PrA |
| Saccharomyces kudriavzevii | American Type Culture Collection (ATCC) | ATCC2601 |
| Recombinant DNA | ||
| Nup82 QconCat (synthetic concatamer of tryptic peptides from Nup82 complex components used as an internal standard for quantitative mass spectrometry) | Integrated DNA Technologies | N/A |
| pGEX6p-1 | (Ding et al., 2011) | N/A |
| p424-Gal1-Dyn2 | This study | S. cerevisiae Dyn2 uniprot Q02647 |
| pProtA/HIS5 | (Fernandez-Martinez et al., 2012) | N/A |
| pAG305GPD-ccdb-EGFP | Addgene | #14186 |
| pAG305-skNup82ppx-EGFP | This study | skNup82 GenBank: EHN01740.1 |
| Sequence-Based Reagents | ||
| Oligo skN82Prom-F (see STAR Methods for sequence) | This study | N/A |
| Oligo skN82GTW_R2 (see STAR Methods for sequence) | This study | N/A |
| oligo dT probe (see STAR Methods for sequence) | Exigon | N/A |
| Software and Algorithms | ||
| EMAN | Ludtke et al., 1999 | http://blake.bcm.edu/emanwiki/EMAN1 |
| Iterative Stable Alignment and Clustering (ISAC) | Yang et al., 2012b | http://sparx-em.org/sparxwiki/sxisac |
| Spider | Frank et al., 1996 | http://spider.wadsworth.org/spider_doc/spider/docs/spider.html |
| CX-Circos | N/A | http://cx-circos.net |
| MaxQuant (version 1.2.2.5) | Cox et al., 2008 | http://www.coxdocs.org/doku.php?id=maxquant:start |
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| pLink | Yang et al., 2012 | http://pfind.ict.ac.cn/software/pLink/ |
| Openlab | Perkin Elmer | http://cellularimaging.perkinelmer.com/support/openlab_resources/ |
| Integrative Modeling Platform (IMP), version 2.5 and Python Modeling Interface (PMI), version c7411c3 | Russel et al., 2012 | http://integrativemodeling.org |
| MODELLER 9.13 | Sali and Blundell, 1993 | http://salilab.org/modeller/ |
| HHPred | Soding et al., 2005 | https://toolkit.tuebingen.mpg.de/hhpred |
| PSIPRED | Jones, 1999 | http://bioinf.cs.ucl.ac.uk/psipred/ |
| DISOPRED | Ward et al., 2004 | http://bioinf.cs.ucl.ac.uk/psipred/?disopred=1 |
| DomPred | Marsden et al., 2002 | http://bioinf.cs.ucl.ac.uk/psipred/?dompred=1 |
| COILS/PCOILS | Lupas et al., 1991 | https://toolkit.tuebingen.mpg.de/pcoils |
| Multicoil2 | Trigg et al., 2011 | http://groups.csail.mit.edu/cb/multicoil2/cgi-bin/multicoil2.cgi |
| SeaView, version 4.6 | Gouy et al., 2010 | http://doua.prabi.fr/software/seaview |
| UCSF Chimera, version 1.10 | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| MatplotLib, version 1.5 | Hunter, 2007 | http://matplotlib.org/ |
| GNUPLOT, version 4.8 | Open software maintained by the developer community | http://www.gnuplot.info/ |
| FoXS | Schneidman-Duhovny et al., 2010 | http://salilab.org/foxs/ |
| ATSAS package (DAMMIF/ DAMMIN/DAMAVER/PRIMUS), version 2.6 | Petoukhov et al., 2012 | https://www.embl-hamburg.de/biosaxs/software.html |
| SASTOOL, version 0.9.5.3 | SSRL beamline 4-2 at SLAC | http://ssrl.slac.stanford.edu/~saxs/analysis/sastool.htm |
| Scatter | SIBYLS beamline 12.3.1 at LBNL | https://bl1231.als.lbl.gov/scatter/ |
| SAXS MOW, version 1.0 | Fischer et al., 2010 and SAXS beam line at the Brazilian Synchrotron Light National Laboratory | http://www.if.sc.usp.br/~saxs/obsolete/saxsmow.html |
| Other | ||
| Superose 6 GL 30/100 | GE Healthcare Life Sciences | Cat.# 17-5172-01 |
| Orbitrap Fusion Mass Spectrometer | Thermo Fisher Scientific | N/A |
| Easy-nLC 1000 HPLC | Thermo Fisher Scientific | N/A |
| Easy-Spray electrospray source | Thermo Fisher Scientific | N/A |
| NuPage 4-12% Bis-Tris Gel 1.0mm × 10 well | Thermo Fisher Scientific | Cat.# NP0321 Box |
Supplementary Material
Acknowledgements
We would like to thank S. R. Wente, R. Sadeh and A. Krutchinsky for sharing yeast strains and plasmids; K. Uryu and the EMRC Resource Center at The Rockefeller University for assistance with negative stain EM; NYSGRC for providing samples for SAXS; T. Matsui and T.M. Weiss at SSRL, SLAC National Accelerator Laboratory for assistance with collecting SAXS data; B. Raveh at UCSF for computing FG nup models. Support was provided by the NSERC, the Canadian Institute for Health Research (MOP-232642) and the Canadian Foundation for Innovation (D.Z.); an NSF GRF (#1650113) (I.E.C.); and the NIH from grants U54 RR022220 to B.C., A.S. and M.R.; R01 GM112108 to M.R.; P41 GM109824 to M.R., A.S. and B.C.; P41 GM103314 to B.C.; and R01 GM083960 to A.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, J.F-M., S.J.K., Y.S., R.P., B.T.C., A.S., and M.P.R.; Investigation, J.F-M., S.J.K., P.U., Y.S., R.W., I.N., W. Z., W.J.R., M.G. and D.Z.; Formal Analysis, S.J.K., R.P., J.W., I.E.C. and A.S.; Writing, J.F-M., S.J.K., A.S., B.T.C., and M.P.R.; Funding Acquisition, D.L.S., D.Z., A.S., B.T.C., and M.P.R.; Supervision, D.L.S., D.Z., A.S., B.T.C., and M.P.R.
References
- Adams RL, Terry LJ, Wente SR. Nucleoporin FG domains facilitate mRNP remodeling at the cytoplasmic face of the nuclear pore complex. Genetics. 2014;197:1213–1224. doi: 10.1534/genetics.114.164012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. Determining the architectures of macromolecular assemblies. Nature. 2007a;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007b;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Borneman AR, Desany BA, Riches D, Affourtit JP, Forgan AH, Pretorius IS, Egholm M, Chambers PJ. The genome sequence of the wine yeast VIN7 reveals an allotriploid hybrid genome with Saccharomyces cerevisiae and Saccharomyces kudriavzevii origins. FEMS yeast research. 2012;12:88–96. doi: 10.1111/j.1567-1364.2011.00773.x. [DOI] [PubMed] [Google Scholar]
- Bui KH, von Appen A, DiGuilio AL, Ori A, Sparks L, Mackmull MT, Bock T, Hagen W, Andres-Pons A, Glavy JS, et al. Integrated structural analysis of the human nuclear pore complex scaffold. Cell. 2013;155:1233–1243. doi: 10.1016/j.cell.2013.10.055. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Boll W, Kirchhausen T, Harrison SC, Walz T. Cryo-electron tomography of clathrin-coated vesicles: structural implications for coat assembly. Journal of molecular biology. 2007;365:892–899. doi: 10.1016/j.jmb.2006.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chug H, Trakhanov S, Hulsmann BB, Pleiner T, Gorlich D. Crystal structure of the metazoan Nup62*Nup58*Nup54 nucleoporin complex. Science. 2015;350:106–110. doi: 10.1126/science.aac7420. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Degiacomi MT, Iacovache I, Pernot L, Chami M, Kudryashev M, Stahlberg H, van der Goot FG, Dal Peraro M. Molecular assembly of the aerolysin pore reveals a swirling membrane-insertion mechanism. Nature chemical biology. 2013;9:623–629. doi: 10.1038/nchembio.1312. [DOI] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS biology. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C, Li Y, Kim BJ, Malovannaya A, Jung SY, Wang Y, Qin J. Quantitative analysis of cohesin complex stoichiometry and SMC3 modification-dependent protein interactions. Journal of proteome research. 2011;10:3652–3659. doi: 10.1021/pr2002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP. Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biological procedures online. 2009;11:32–51. doi: 10.1007/s12575-009-9008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre E, Hurt E. Yeast genetics to dissect the nuclear pore complex and nucleocytoplasmic trafficking. Annu Rev Genet. 1997;31:277–313. doi: 10.1146/annurev.genet.31.1.277. [DOI] [PubMed] [Google Scholar]
- Fernandez-Martinez J, Phillips J, Sekedat MD, Diaz-Avalos R, Velazquez-Muriel J, Franke JD, Williams R, Stokes DL, Chait BT, Sali A, et al. Structure-function mapping of a heptameric module in the nuclear pore complex. J Cell Biol. 2012;196:419–434. doi: 10.1083/jcb.201109008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkmann AW, Dawson TR, Wente SR. Insights into mRNA export-linked molecular mechanisms of human disease through a Gle1 structure-function analysis. Advances in biological regulation. 2014;54:74–91. doi: 10.1016/j.jbior.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkmann AW, Noble KN, Cole CN, Wente SR. Dbp5, Gle1-IP6 and Nup159: a working model for mRNP export. Nucleus. 2011;2:540–548. doi: 10.4161/nucl.2.6.17881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaik M, Flemming D, von Appen A, Kastritis P, Mucke N, Fischer J, Stelter P, Ori A, Bui KH, Bassler J, et al. Structural basis for assembly and function of the Nup82 complex in the nuclear pore scaffold. J Cell Biol. 2015;208:283–297. doi: 10.1083/jcb.201411003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular biology and evolution. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Griffith OM. In: Techniques of Preparative, Zonal, and Continuous Flow Ultracentrifugation. S.D. Applications Research Department, Beckman Instruments, Inc., editor. Beckman Instruments, Inc.; Fullerton, California, USA: 1994. p. 50. [Google Scholar]
- Grunwald D, Singer RH. In vivo imaging of labelled endogenous beta-actin mRNA during nucleocytoplasmic transport. Nature. 2010;467:604–607. doi: 10.1038/nature09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough LE, Dutta K, Sparks S, Temel DB, Kamal A, Tetenbaum-Novatt J, Rout MP, Cowburn D. The molecular mechanism of nuclear transport revealed by atomic-scale measurements. eLife. 2015;4 doi: 10.7554/eLife.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. Journal of molecular biology. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Kaneb HM, Folkmann AW, Belzil VV, Jao LE, Leblond CS, Girard SL, Daoud H, Noreau A, Rochefort D, Hince P, et al. Deleterious mutations in the essential mRNA metabolism factor, hGle1, in amyotrophic lateral sclerosis. Human molecular genetics. 2015;24:1363–1373. doi: 10.1093/hmg/ddu545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley K, Knockenhauer KE, Kabachinski G, Schwartz TU. Atomic structure of the Y complex of the nuclear pore. Nature structural & molecular biology. 2015;22:425–431. doi: 10.1038/nsmb.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner N, Schwarz J, Sturm M, Fernandez-Martinez J, Griesel S, Zhang W, Chait BT, Rout MP, Kuck U, Hurt E. Developing genetic tools to exploit Chaetomium thermophilum for biochemical analyses of eukaryotic macromolecular assemblies. Scientific reports. 2016;6:20937. doi: 10.1038/srep20937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendirgi F, Barry DM, Griffis ER, Powers MA, Wente SR. An essential role for hGle1 nucleocytoplasmic shuttling in mRNA export. J Cell Biol. 2003;160:1029–1040. doi: 10.1083/jcb.200211081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knockenhauer KE, Schwartz TU. The Nuclear Pore Complex as a Flexible and Dynamic Gate. Cell. 2016;164:1162–1171. doi: 10.1016/j.cell.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosinski J, Mosalaganti S, von Appen A, Teimer R, DiGuilio AL, Wan W, Bui KH, Hagen WJ, Briggs JA, Glavy JS, et al. Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science. 2016;352:363–365. doi: 10.1126/science.aaf0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DH, Stuwe T, Schilbach S, Rundlet EJ, Perriches T, Mobbs G, Fan Y, Thierbach K, Huber FM, Collins LN, et al. Architecture of the symmetric core of the nuclear pore. Science. 2016;352:aaf1015. doi: 10.1126/science.aaf1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Lund MK, Guthrie C. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Molecular cell. 2005;20:645–651. doi: 10.1016/j.molcel.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- McDonald JH. Handbook of Biological Statistics 3rd ed. Sparky House Publishing; Baltimore: 2014. [Google Scholar]
- Montpetit B, Thomsen ND, Helmke KJ, Seeliger MA, Berger JM, Weis K. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature. 2011;472:238–242. doi: 10.1038/nature09862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor A, Suliman S, Ben-Yishay R, Yunger S, Brody Y, Shav-Tal Y. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nature cell biology. 2010;12:543–552. doi: 10.1038/ncb2056. [DOI] [PubMed] [Google Scholar]
- Nerurkar P, Altvater M, Gerhardy S, Schutz S, Fischer U, Weirich C, Panse VG. Eukaryotic Ribosome Assembly and Nuclear Export. International review of cell and molecular biology. 2015;319:107–140. doi: 10.1016/bs.ircmb.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Noble KN, Tran EJ, Alcazar-Roman AR, Hodge CA, Cole CN, Wente SR. The Dbp5 cycle at the nuclear pore complex during mRNA export II: nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1. Genes & development. 2011;25:1065–1077. doi: 10.1101/gad.2040611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousiainen HO, Kestila M, Pakkasjarvi N, Honkala H, Kuure S, Tallila J, Vuopala K, Ignatius J, Herva R, Peltonen L. Mutations in mRNA export mediator GLE1 result in a fetal motoneuron disease. Nature genetics. 2008;40:155–157. doi: 10.1038/ng.2007.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Zenklusen D. To the pore and through the pore: a story of mRNA export kinetics. Biochimica et biophysica acta. 2012;1819:494–506. doi: 10.1016/j.bbagrm.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt JM, Simpson DM, Doherty MK, Rivers J, Gaskell SJ, Beynon RJ. Multiplexed absolute quantification for proteomics using concatenated signature peptides encoded by QconCAT genes. Nature protocols. 2006;1:1029–1043. doi: 10.1038/nprot.2006.129. [DOI] [PubMed] [Google Scholar]
- Ren Y, Seo HS, Blobel G, Hoelz A. Structural and functional analysis of the interaction between the nucleoporin Nup98 and the mRNA export factor Rae1. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10406–10411. doi: 10.1073/pnas.1005389107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romes EM, Tripathy A, Slep KC. Structure of a yeast Dyn2-Nup159 complex and molecular basis for dynein light chain-nuclear pore interaction. The Journal of biological chemistry. 2012;287:15862–15873. doi: 10.1074/jbc.M111.336172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel D, Lasker K, Webb B, Velazquez-Muriel J, Tjioe E, Schneidman-Duhovny D, Peterson B, Sali A. Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS biology. 2012;10:e1001244. doi: 10.1371/journal.pbio.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Berman HM, Schwede T, Trewhella J, Kleywegt G, Burley SK, Markley J, Nakamura H, Adams P, Bonvin AM, et al. Outcome of the First wwPDB Hybrid/Integrative Methods Task Force Workshop. Structure. 2015;23:1156–1167. doi: 10.1016/j.str.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. Journal of molecular biology. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Sampathkumar P, Kim SJ, Manglicmot D, Bain KT, Gilmore J, Gheyi T, Phillips J, Pieper U, Fernandez-Martinez J, Franke JD, et al. Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata. Proteins. 2012;80:2110–2116. doi: 10.1002/prot.24102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic acids research. 2010;38:W540–544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Pellarin R, Sali A. Uncertainty in integrative structural modeling. Current opinion in structural biology. 2014;28:96–104. doi: 10.1016/j.sbi.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Fernandez-Martinez J, Tjioe E, Pellarin R, Kim SJ, Williams R, Schneidman-Duhovny D, Sali A, Rout MP, Chait BT. Structural characterization by cross-linking reveals the detailed architecture of a coatomer-related heptameric module from the nuclear pore complex. Molecular & cellular proteomics : MCP. 2014;13:2927–2943. doi: 10.1074/mcp.M114.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Pellarin R, Fridy PC, Fernandez-Martinez J, Thompson MK, Li Y, Wang QJ, Sali A, Rout MP, Chait BT. A strategy for dissecting the architectures of native macromolecular assemblies. Nature methods. 2015;12:1135–1138. doi: 10.1038/nmeth.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DN, Rout MP. Cancer and the nuclear pore complex. Advances in experimental medicine and biology. 2014;773:285–307. doi: 10.1007/978-1-4899-8032-8_13. [DOI] [PubMed] [Google Scholar]
- Smith C, Lari A, Derrer CP, Ouwehand A, Rossouw A, Huisman M, Dange T, Hopman M, Joseph A, Zenklusen D, et al. In vivo single-particle imaging of nuclear mRNA export in budding yeast demonstrates an essential role for Mex67p. J Cell Biol. 2015;211:1121–1130. doi: 10.1083/jcb.201503135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics (Oxford, England) 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- Strahm Y, Fahrenkrog B, Zenklusen D, Rychner E, Kantor J, Rosbach M, Stutz F. The RNA export factor Gle1p is located on the cytoplasmic fibrils of the NPC and physically interacts with the FG-nucleoporin Rip1p, the DEAD-box protein Rat8p/Dbp5p and a new protein Ymr 255p. The EMBO journal. 1999;18:5761–5777. doi: 10.1093/emboj/18.20.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Bassler J, Hurt E. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J Cell Biol. 2000;150:695–706. doi: 10.1083/jcb.150.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn LA, Shen T, Shulga N, Goldfarb DS, Wente SR. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nature cell biology. 2004;6:197–206. doi: 10.1038/ncb1097. [DOI] [PubMed] [Google Scholar]
- Stuwe T, Bley CJ, Thierbach K, Petrovic S, Schilbach S, Mayo DJ, Perriches T, Rundlet EJ, Jeon YE, Collins LN, et al. Architecture of the fungal nuclear pore inner ring complex. Science. 2015a;350:56–64. doi: 10.1126/science.aac9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuwe T, Correia AR, Lin DH, Paduch M, Lu VT, Kossiakoff AA, Hoelz A. Nuclear pores. Architecture of the nuclear pore complex coat. Science. 2015b;347:1148–1152. doi: 10.1126/science.aaa4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trahan C, Oeffinger M. Targeted cross-linking-mass spectrometry determines vicinal interactomes within heterogeneous RNP complexes. Nucleic acids research. 2016;44:1354–1369. doi: 10.1093/nar/gkv1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigg J, Gutwin K, Keating AE, Berger B. Multicoil2: predicting coiled coils and their oligomerization states from sequence in the twilight zone. PloS one. 2011;6:e23519. doi: 10.1371/journal.pone.0023519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Appen A, Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull MT, Banterle N, Parca L, Kastritis P, et al. In situ structural analysis of the human nuclear pore complex. Nature. 2015;526:140–143. doi: 10.1038/nature15381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JJ, McGuffin LJ, Bryson K, Buxton BF, Jones DT. The DISOPRED server for the prediction of protein disorder. Bioinformatics (Oxford, England) 2004;20:2138–2139. doi: 10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- Weirich CS, Erzberger JP, Berger JM, Weis K. The N-terminal domain of Nup159 forms a beta-propeller that functions in mRNA export by tethering the helicase Dbp5 to the nuclear pore. Molecular cell. 2004;16:749–760. doi: 10.1016/j.molcel.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Yamada J, Phillips JL, Patel S, Goldfien G, Calestagne-Morelli A, Huang H, Reza R, Acheson J, Krishnan VV, Newsam S, et al. A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Molecular & cellular proteomics : MCP. 2010;9:2205–2224. doi: 10.1074/mcp.M000035-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Wu YJ, Zhu M, Fan SB, Lin J, Zhang K, Li S, Chi H, Li YX, Chen HF, et al. Identification of cross-linked peptides from complex samples. Nature methods. 2012a;9:904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- Yang Q, Rout MP, Akey CW. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Molecular cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- Yang Z, Fang J, Chittuluru J, Asturias FJ, Penczek PA. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure. 2012b;20:237–247. doi: 10.1016/j.str.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Seo HS, Debler EW, Blobel G, Hoelz A. Structural and functional analysis of an essential nucleoporin heterotrimer on the cytoplasmic face of the nuclear pore complex. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16571–16576. doi: 10.1073/pnas.1112846108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.