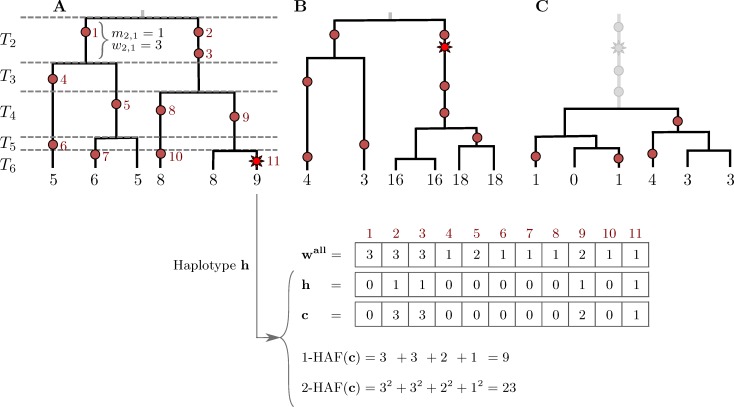
In panel B of Fig 1, the two haplotypes on the right have incorrect scores of 17. The correct scores of these haplotypes are 18. Please view the correct Fig 1 here.
Fig 1. The HAF score.
Genealogies of three samples (n = 6) progressing through a selective sweep, from left to right. Neutral mutations are shown as red circles, and are numbered in red; the favored allele is shown as a red star. The 1-HAF score of each haplotype is shown below its corresponding leaf, in black. For the rightmost haplotype in (A), the binary haplotype vector h is shown along with its HAF-vector c, and 1-HAF and 2-HAF scores. Vector wall lists the frequencies of all mutations. (A) The favored allele appears on a single haplotype. At this point in time, both the genealogy and the HAF score distribution are largely neutral. Coalescence times (T2, …, T6) are shown on the left, where Tk spans the epoch with exactly k lineages. (B) Carriers of the favored allele are distinguished by high HAF scores (in large part due to the long branch of high-frequency hitchhiking variation); non-carriers have low HAF scores. (C) After fixation, there is a sharp loss of diversity causing low HAF scores across the sample.
Reference
- 1.Ronen R, Tesler G, Akbari A, Zakov S, Rosenberg NA, Bafna V (2015) Predicting Carriers of Ongoing Selective Sweeps without Knowledge of the Favored Allele. PLoS Genet 11(9): e1005527 doi:10.1371/journal.pgen.1005527 [DOI] [PMC free article] [PubMed] [Google Scholar]