Abstract
Ginseng is one of the most widely used natural medicines in the world. Recent studies have suggested Panax ginseng has a wide range of beneficial effects on aging, central nervous system disorders, and neurodegenerative diseases. However, knowledge about the specific bioactive components of ginseng is still limited. This work aimed to screen for the bioactive components in Panax ginseng that act against neurodegenerative diseases, using the target cell-based bioactivity screening method. Firstly, component analysis of Panax ginseng extracts was performed by UPLC-QTOF-MS, and a total of 54 compounds in white ginseng were characterized and identified according to the retention behaviors, accurate MW, MS characteristics, parent nucleus, aglycones, side chains, and literature data. Then target cell-based bioactivity screening method was developed to predict the candidate compounds in ginseng with SH-SY5Y cells. Four ginsenosides, Rg2, Rh1, Ro, and Rd, were observed to be active. The target cell-based bioactivity screening method coupled with UPLC-QTOF-MS technique has suitable sensitivity and it can be used as a screening tool for low content bioactive constituents in natural products.
Keywords: Panax ginseng, component analysis, drug screening, target cell extraction, UPLC-QTOF-MS, SH-SY5Y cell
Graphical abstract
1. Introduction
Panax ginseng, the root of Panax ginseng C.A. Meyer (family Araliaceae), has been used as a traditional medicine for thousands of years in East Asian countries, and now is one of the most widely used natural medicines in the world. The chemical constituents in Panax ginseng include triterpene saponins, polysaccharides, peptides, polyacetylenic alcohols, phenolic compounds and fatty acids [1,2]. Ginsenosides, also called panaxosides or ginseng saponins, a type of triterpene glycosides, are the major active components in Panax ginseng. Recent studies have suggested Panax ginseng has a wide range of beneficial effects on aging, central nervous system disorders, and neurodegenerative diseases [2–5]. Panax ginseng extract [6], ginsenosides Rg1 [7–9], Rb1 [9,10], and Rd [11] have been found to protect SH-SY5Y cells against 1-methyl-4-phenylpyridinium-induced injury. In our preliminary experiments, ginseng extracts and the ginsenosides fractions were found to be capable of facilitating the proliferation of SH-SY5Y cells and protecting the cells against H2O2-induced injury. But what are the active constituents in the extracts and fractions? And are there any ginsenosides that can exert protective benefits other than Rg1, Rb1 and Rd? It is necessary to screen the active compounds in ginseng further.
The classic screening procedures for bioactive components in natural products include isolation, purification, and then pharmacological evaluation. This conventional method is time-consuming, arduous, and has a low efficiency. Modern pharmacological investigation has shown that most drugs should enter the target cells or bind with some receptors, enzymes or channels on cell membranes to elicit activity [12,13]. A target cell-based screening method coupled with modern chromatographic technique has been developed using the principles of this theory [12,13]. In the screening procedures, the extracts are added into the target cells, the potential bioactive components may selectively bind to the cells, the unbound compounds can be washed away, and finally the active components released by digestion are analyzed by the hyphenated chromatography technique. Recently, the target cell-based screening method has been successfully applied to find the bioactive compounds in natural products [14–20]. Meanwhile, the analytical tools used in the screening method have undergone evolution from common liquid chromatography (LC) [16], LC-MS [14,15,20], to LC-MS/MS with low energy collision dissociation (CID) [18], especially UPLC-QTOF-MS (Ultra-high-performance liquid chromatography coupled with Quadrupole-time-of-flight mass spectrometry) [17]. The UPLC-QTOF-MS technique possesses high resolution, high sensitivity, high mass accuracy, and abundant fragment information, and has become a powerful analytical tool for complicated samples.
This work aims to screen for the bioactive components in Panax ginseng that act against neurodegenerative disease with the target cell-based bioactivity screening method. SH-SY5Y human neuroblastoma cells have been widely used as in vitro models of neuronal function, differentiation, and neurodegenerative diseases. Thus, SH-SY5Y cells were selected as the target cells. Firstly, a component analysis of Panax ginseng extracts was performed by UPLC-QTOF-MS, and 54 compounds were identified. Secondly, target cell extraction was carried out, and the extracted compounds were analyzed by UPLC-QTOF-MS. Four potential neural active components in ginseng were identified in this work.
2. Experiment
2.1. Herbal materials and chemicals
Panax ginseng was collected from Tonghua County, Jilin Province, China, and was authenticated by associate professor Yuye Zhu from Jiangxi University of Traditional Chinese Medicine (JXUTCM). A voucher specimen was preserved in the Key Lab of Modern Preparation of TCM, JXUTCM, Nanchang, China. HPLC grade acetonitrile (ACN) was from Fisher (USA). Purified water was obtained by a Milli-Q system (Millipore, Bedford, MA, USA). Other reagents and chemicals were of analytical grade. SH-SY5Y cells were obtained from the China Center for Type Culture Collection in Beijing University. Dulbecco’s Modified Eagle Medium (DMEM, LOT: 1471272), Ham’s F12 Nutrient Mixture (F12, LOT: 21127022), fetal bovine serum (FBS, LOT: 1414426), trypsin (LOT: 25200056) were purchased from Invitrogen Gibco (Carlsbad, CA, USA).
2.2. Sample preparation
Ginseng samples were pulverized into powder. 60 grams of the powder were extracted twice by heat-reflux with 70% ethanol (600 mL, 2h; 480 mL, 1h). The combined extract was evaporated under vacuum and lyophilized. Two lyophilized samples were accurately weighed. One was re-diluted in 70% ethanol, and then passed through a 0.22 µm filter prior to analysis by UPLC-QTOF-MS. The other was dissolved in DMEM under a sterile environment. The final concentration of ginseng extract was 5 mg/mL, which was used for cell culture.
2.3. Cell culture
SH-SY5Y cells were seeded into cell culture flasks and were incubated in DMEM/F12 (1:1, v/v) medium supplemented with 10% FBS, 100 U/mL penicillin, and 100 µg/mL streptomycin. They were maintained in a humidified 5% CO2 incubator (SANYO MCO-175, Osaka, Japan) at 37 °C. The medium was replaced at 24 hour intervals. The cells were subcultured (1:3) until 80% confluence. The cells in the logarithmic growth phase were used for the following experiments. A phase-contrast microscope (Nikon XDS-1B) and Tyrpan blue exclusion test were used to determine cell viability.
2.4 Cell-based bioactivity screening
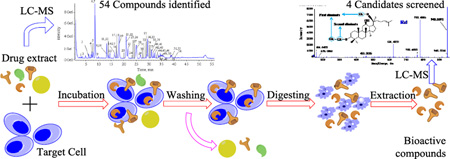
The screening scheme is shown in Fig. 1. The process includes incubation (drug-cell interaction), washing, digestion and LC-MS analysis.
Figure 1.
Schematic drawing of target cell-based bioactivity screening process.
The cells in the logarithmic growth phase were incubated in a cell culture bottle until a density of 5.0 × 105 cells/mL, and then were cultured in DMEM medium free of serum in a humidified 5% CO2 incubator at 37 °C for 24 h. The culture medium was discarded, and the cells were resuspended in DMEM within the presence of ginseng extract (final concentration of 5 mg/mL), and incubated at 37 °C for 4 h. The suspension was centrifuged at 1,000 ×g for 5 min. The precipitate was then washed five times with PBS to remove unbounded components. The eluates were discarded except for the last one, which was collected as a control for LC-MS analysis. Finally, the cells were denatured with 2 mL of 40% acetic acid, and were extracted with 2 mL of methanol by ultrasonic extraction (80 Hz, 15 min). After centrifugation at 15,800 × g for 15 min, the obtained supernatant was evaporated to dryness under a stream of nitrogen gas at 40 °C. The residue was reconstituted in 100 µL of methanol by vortexing for 2 min and centrifuging at 15,800 × g for 10 min at 4 °C, and the suspension was used for UPLC-QTOF-MS analysis. The control samples free-of-drugs were prepared using the same procedures as the above.
2.5 UPLC-QTOF-MS analysis
Ultra-performance liquid chromatography analysis was performed on a Nexera X2 series LC system (Nexera Technologies, Japan) equipped with a binary pump, micro-degasser, an auto-sampler, and a thermostatically controlled column compartment. The chromatographic separation was performed on a ZORBAX Eclipse plus C18 column (2.1×100 mm, 1.8-Micron) at 35 °C. The mobile phase was composed of water (A) and acetonitrile (B) both containing 0.1% formic acid, using the following gradient procedures: 15–30% (B) at 0–10 min, 30–30% (B) at 10–25 min, 30–50% (B) at 25–35 min, 50–70% (B) at 35–45 min, 70–90% (B) at 45–50 min, 90–5% (B) at 50–55 min. The sample volume injected was set at 5 µL.
Mass detection was performed by on a Triple TOF 5600 plus Mass spectrometer (AB SCIEX, Framingham, USA) equipped with an ESI source. QTOF-MS analysis was performed in negative ion modes using full scan mode with a mass range of 100–2000 Da for both the TOF-MS and TOF-MS/MS scans. The following parameter settings were used: ion spray voltage, +/−4500 V; ion source heater, 500 °C; curtain gas (CUR, N2), 35 psi; nebulizing gas (GS1, Air), 60 psi; Tis gas (GS2, Air), 60 psi. The declustering potential (DP) was +/−100 V; collision energy (CE) was +/−45 V with a collision energy spread (CES) of 15 V.
2.6. Data analysis
The accurate mass and composition for the precursor ions and fragment product ions were analyzed using the Peakview Software (AB SCIEX, version 1.2.0.3) integrated with the instrument. The exact mass calibration was performed automatically before each analysis employing the Automated Calibration Delivery System. Molecular formulas were generated by the molecular formula generator algorithm whose parameters were set as the following: C [0–80], H [0–150], O [0–60]. Other elements such as N, P, S, Br and Cl were not considered because of their rare presence in the ginseng. The empirical molecular formula was deduced from Peakview by comparing the theoretical mass of molecular ions and/or adduct ions with the determined values based on the limitation errors: mass accuracy <5 ppm, retention time <5.0%, and isotope abundance <10%. Components reported in the literature [1,2,21–31] in ginseng were summarized to establish a small-scale library for the rapid identification of non-target compounds. In a summary, retention time, accurate molecular weight, isotope abundance, fragment product ions, and literature data were reviewed to identify the compounds.
3. Results and discussions
3.1 UPLC-QTOF-MS conditions optimization
In the studied ESI-MS the analytical conditions were optimized in order to enhance and achieve better resolution, higher sensitivity, and formation of abundant fragment ions. The addition of 0.1% formic acid in mobile phase appeared to significantly improve the detection sensitivity. In the negative ion mode, clearer ESI-MS were obtained with lower background noise, clearer mass spectrum, and higher detect sensitivity, so the ESI in the negative ion mode was chosen for the target-cell extract experiments. Other parameters, such as eluating gradient, ion source parameters, and mass analyzer settings, were also investigated. The optimum analytical conditions were set as described in the section “UPLC-QTOF-MS analysis”.
3.2. Identification of ginsenosides and component analysis of ginseng
With respect to the structural characteristics of aglycone, ginseng saponins can be divided into several groups. The two major groups are the protopanaxadiol (PPD) group with sugar moieties attached to the -3 and/or C-20 and the protopanaxatriol (PPT) group with sugar moieties at C-6 and/or at C-20 [26,31–33]. Another family, the malonyl ginseng saponins, also called the acidic ginseng saponins, has a malonyl group attached at the 6-position of the glucosyl moiety. The minor group is oleanane (OLE) group with a nonsteroidal. The structure of other types of ginsenosides has some changes, but they still essentially belong to the original type of ginsenosides, only the side chain of the parent nucleus part is slightly different. The structures of ginseng saponins are shown in Fig. 2 and summarized in Table S1.
Figure 2.
Structures of skeleton (A) and monosaccharide & substituent group (B) of gensenosides.
The identification process of the detected compounds in ginseng was as follows. Firstly, the accurate molecular mass was obtained with the high-resolution QTOF-MS technique. When formic acid was added into the mobile phase, ginsenosides easily formed the deprotonated ion [M−H]− and adduct ion [M+HCOO]−, providing information about the molecular mass. Secondly, the formula was obtained by Peakview software according to the accurate molecular mass, element constituent, and isotope abundance. Thirdly, the differentiation and characterization of ginsenosides were completed according to the literature [20,26,32–34], and the types of ginsenosides were confirmed. The aglycone type can be distinguished according to the literatures in ESI+ mode [32–34]. The types could also be determined by characteristic fragmentations in ESI− mode. The relative characteristic aglycone ions at m/z 459, 477, 475, and/or 473 correspond to the PPD-type aglycone; m/z 475, 457, 493, and/or 473 to PPT-type; and m/z at 455.37 to OLE-type. In the low-energy CID-MS/MS (negetaive ion mode), the precursor deprotonated molecules formed characteristic sugar fragments product ions by the successive or simultaneous losses of the various sugar moieties: 162 Da (-Glc), and/or 146 Da (-Rha), and/or 132 Da (-Ara or-Xyl). Finally, the extracted compounds were identified according to their retention time behaviors, accurate MW, MS/MS fragmentation pathways, formation of aglycones product ions, and product ions produced by losses of side chains, and literature data. In this work, 54 compounds were identified in Panax ginseng extract (Fig. 3A).
Figure 3.
Typical TIC chromatogram of Panax ginseng extract by UPLC-QTOF-MS (A), MS/MS spectrum of the deprotonated ion and the proposed fragmentation pathway of Ra1 (B); MS/MS spectrum of Ra1 at m/z 70–400 and corresponding relationship between sugar moieties and its residue ions (C).
Ginsenoside Ra1 (PPD) was selected as an example to elucidate the analysis process. In the full scan ESI-MS of the UPLC peak #14 (at 21.708 min in Fig. 3A) gave high abundance ions at m/z 1255.6248 and m/z 1209.6272 (Fig. 3B); they were tentatively attributed as the [M+HCOO]− and [M−H]− according to the mass differences between the ions. So, the ion at m/z 1209.6272 is its quasi-molecular ion. Fig. 3B is the MS/MS spectrum of the peak 14. The fragment product ions at m/z 1077, 945, 783, 621, and 459 indicate the successive losses of 2 pentoses (−132 Da) and 3 hexoses (−162 Da) from the quasi-molecular ion at m/z 1209.6272. The product ion at m/z 459 is the characteristic ion of PPD type aglycone. Thus, the compound is a PPD ginsenoside that has a MW of 1210.6248 Da, and contains 2 pentoses (132 Da) and 3 hexoses (162 Da). Furthermore, relative high collision-induced dissociation (CID) energy was applied to determine sugar chain compositions in the range of m/z 70–400. As shown in Fig. 3C, m/z 161.04, 131.03, 119.03, and 113.02 indicate the presence of Glc; m/z 203.05, 323.09 indicate Glc–Glc composition; m/z 191.05 and 293.08 refer to Glc–Arap/Xly composition; and m/z 131.03, 149.04 indicate the presence of Arap/Xly. Therefore, the molecular mass, aglycone type, sugar numbers, and sugar composition were deduced. The results are consistent with the literature 28, so it is identified as Ra1.
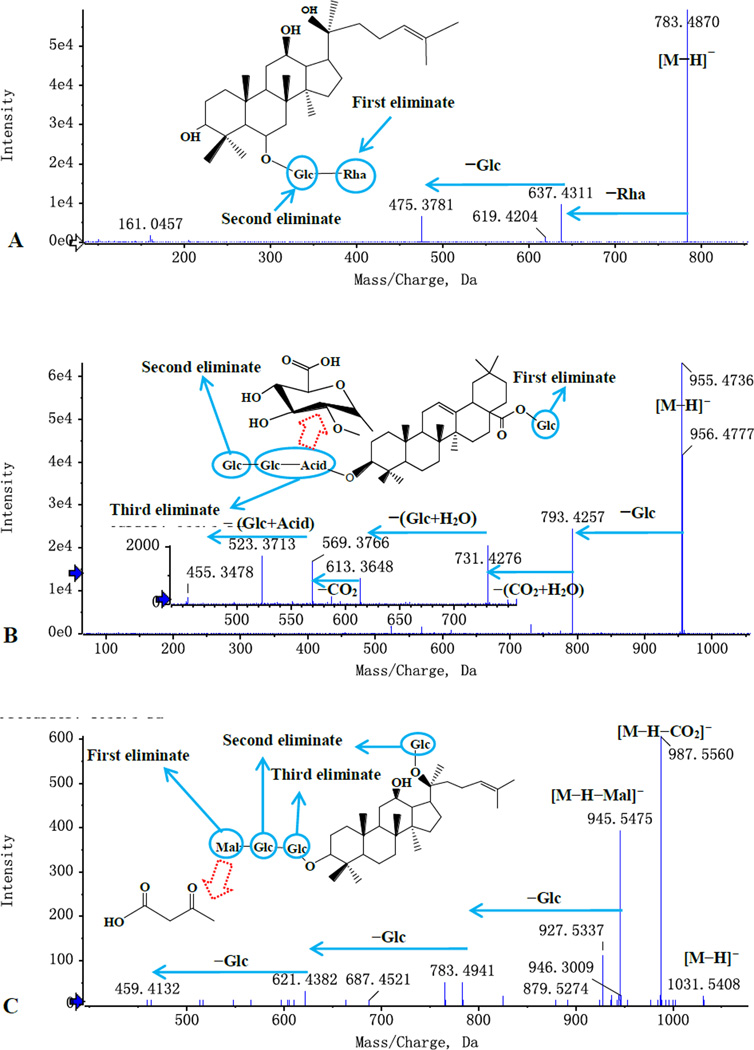
The typical MS/MS spectra of other types of ginsenosides are shown in Fig. 4. In Fig. 4A, the [M−H]− ion produced fragmentation at m/z 637, representing glycosidic cleavage by loss of one rhamnose residue (146 Da). The fragment ion at m/z 475, indicating the possibility of PPT-type aglycone, was produced by loss of glucose–rhamnose residue (162 Da +146 Da). The aglycone type was further verified with the characteristic ions at m/z 441.37, 423.36 and 405.35 in ESI+ mode. The results are consistent with the literature 23, so it is identified as as Rg2.
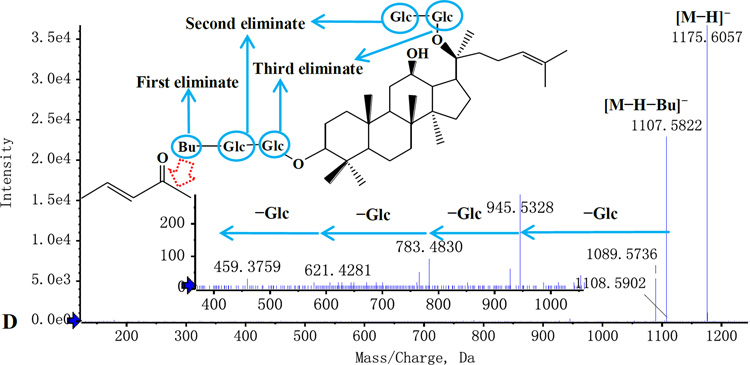
Figure 4.
Typical MS/MS spectra and the proposed fragmentation pathways. A: Rg2; B: Ro; C: Malonyl-Rd; D: Ra6.
In Fig. 4B, the [M−H]− ion at m/z 955 produced fragmentations at m/z 793, representing glycosidic cleavage by loss of one glucose residue (162 Da) in sugar chain at C-20 of Ro. The 6-position of the glucosyl moiety was attached to formic acid, which is easy to lose as CO2 and H2O (44 Da+18 Da). The ion at m/z 613 was generated by the loss of two glucose residue (162 Da) and one H2O residue (162 Da+162 Da+18 Da); The ions at m/z 731 and 569 were generated by the loss of CO2 and H2O; The ion at m/z 455, indicating OLE-type aglycone, was generated by loss of two glucose residues and glucose acid (162 Da+162 Da+176 Da).
Malonyl ginsenosides are a special type of compounds, with a malonyl group attached at the 6-position of the glucosyl moiety. Most malonyl ginsenosides are derived from PPD-type ginsenosides. As shown in Fig. 4C, malonyl-ginsenosides Rd exhibited characteristic ions [M-H-malonyl]− by the loss of 86 Da and ions [M−H-CO2]− by the loss of 44 Da from [M-H]−. The other observed fragments are consistent with their corresponding ginsenosides.
In addition, there is a special kind of ginsenosides, the 6-position of the glucosyl moiety was attached by butenoyl. These ginsenosides were derived from PPD-type ginsenosides or PPT-type ginsenosides. As shown in Fig. 4D, butenoyl-ginsenosides Rb2 exhibit characteristic ions [M-H-butenoyl]− by loss of 68 Da from [M-H]−. The other observed fragments are consistent with their corresponding ginsenosides.
With the UPLC-QTOF-MS method, 54 compounds in white ginseng were identified and their MS characteristics are listed in Table 1. Many ginsenoside isomers are found in ginseng, and 8 isomers cannot be differentiated in this work (Table 1). Recent IonKey/MS Ion Mobility technique developed by Waters Corporation provide the feasibility of differentiating the isomers [35]. Compounds Rk1, Rg5, Rh2, Gypenoside XVII, Compound Mc-1, Compound O, Gypenoside IX, Majonoside R1, notoginsenoside N1 and notoginsenoside A were detected in white ginseng for the first time, and they were found in red ginseng [26] or the leaves of ginseng [27].
Table 1.
Compounds identified from ginseng extract
| No . |
Name | RT (min ) |
Formul a |
MW (Da) |
Measu red (Da)a |
Err or (pp m) |
Main MS/MS fragment ions |
Ref . |
|---|---|---|---|---|---|---|---|---|
| 1 | Majonoside R1/isomer |
3.21 1 |
C42H72 O15 |
816.48 72 |
815.48 15 |
2.5 | 653.4313 [M−H−Glc]−; 635.4169 [M−H−Glc−H2O]−; 491.3742 [M−H−2Glc]− |
27 |
| 2 | Notoginsenoside N1/isomer |
3.64 3 |
C48H82 O19 |
962.54 5 |
961.54 05 |
3.4 | 799.4956 [M−H−Glc]−;781.4812 [M−H−Glc−H2O]− |
27 |
| 3 | Majonoside R1/isomer |
6.31 1 |
C42H72 O15 |
816.48 72 |
815.48 00 |
0.7 | 653.4299 [M−H−Glc]−; 635.4299 [M−H−Glc−H2O]−; 491.3761 [M−H−2Glc]− |
27 |
| 4 | 20-glc-Rf | 7.43 2 |
C48H82 O19 |
962.54 50 |
961.53 85 |
1.4 | 799.4893 [M−H−Glc]−; 637.4316 [M−H−2Glc]−; 475.3767 [M−H−3Glc]− |
23, 25 |
| 5 | Notoginsenoside R1 |
7.85 1 |
C47H80 O18 |
932.53 45 |
931.52 75 |
0.8 | 799.4871 [M−H−Glc]−; 637.4315 [M−H−Araf−Glc]−; 475.3794 [M−H−Araf−2Glc]− |
23, 28 |
| 6 | Ginsenoside Rg1 | 8.58 5 |
C42H72 O14 |
800.49 22 |
799.48 68 |
2.4 | 637.4321 [M−H−Glc]−; 475.3798 [M−H−2Glc]− |
23, 28 |
| 7 | Ginsenoside Re | 8.64 5 |
C48H82 O18 |
946.55 01 |
945.54 58 |
3.2 | 799.4918 [M−H−Rha]−; 783.4956 [M−H−Glc]−; 637.4348 [M−H−Rha−Glc]−; 475.3806 [M−H−Rha−Glc]− |
23, 28 |
| 8 | Notoginsenoside A/isomer |
9.92 3 |
C54H92 O24 |
1124.5 979 |
1123.5 901 |
0 | 961.5431 [M−H−Glc]−; 799.4781 [M−H−2Glc]−; 781.4802 [M−H−2Glc−H2O]− |
27 |
| 9 | Ginsenoside Rf | 13.2 36 |
C42H72 O14 |
800.49 22 |
799.48 19 |
−3.1 | 637.432 [M−H−Glc]−; 475.3768 [M−H−2Glc]− |
23, 28 |
| 10 | Bu-Rg1 | 13.3 57 |
C46H76 O15 |
868.51 84 |
867.51 34 |
3.2 | 799.4899 [M−H−Bu]−; 637.4337 [M−H−Bu−Glc]−; 475.3741 [M−H−Bu−2Glc]− |
22 |
| 11 | Ginsenoside Ra3 | 13.8 98 |
C59H100 O27 |
1240.6 452 |
1239.6 330 |
−3.5 | 1107.6207 [M−H−Xyl]−; 945.5357 [M−H−Xyl−Glc]−; 783.5173 [M−H−Xyl−2Glc]−; 621.4370 [M−H−Xyl−3Glc]−; 459.3882 [M−H−Xyl−4Glc]− |
28 |
| 12 | Notoginsenoside Fa |
13.9 64 |
C59H100 O27 |
1240.6 452 |
1239.6 319 |
−4.4 | 1107.5786 [M−H−Xyl]−; 945.5450 [M−H−Xyl−Glc]−; 783.4880 [M−H−Xyl−2Glc]−; 621.4380 [M−H−Xyl−3Glc]−; 459.3845 [M−H−Xyl−4Glc]− |
24 |
| 13 | Notoginsenoside R2 |
14.5 32 |
C41H70 O13 |
770.48 16 |
769.47 20 |
− 2.3 |
637.4273 [M−H−Xyl]−; 475.3757 [M−H−Xyl−Glc]− |
23, 28 |
| 14 | Ginsenoside Rg2 | 16.3 16 |
C42H72 O13 |
784.49 73 |
783.48 70 |
−3.2 | 637.4311 [M−H−Rha]−; 475.3781 [M−H−Rha−Glc]− |
23, 28 |
| 15 | Ginsenoside Rh1 | 16.5 47 |
C36H62 O9 |
638.43 94 |
637.43 30 |
1.4 | 475.3765 [M−H−Glc]− |
23, 28 |
| 16 | Bu-Re | 16.8 54 |
C52H86 O19 |
1014.5 763 |
1013.5 708 |
2.3 | 945.5451 [M−H−Bu]−; 799.4853 [M−H-Bu−Rha]−; 637.4331 [M−H-Bu−Rha−Glc]−; 475.3772 [M−H-Bu−Rha−2Glc]− |
22 |
| 17 | Ginsenoside Ra1 | 17.3 41 |
C58H98 O26 |
1210.6 346 |
1209.6 272 |
0.3 | 1077.6158 [M−H−Xyl]−; 945.5423 [M−H−Xyl−Arap]−; 783.4917 [M−H−Xyl−Arap−Glc]−; 765.4602 [M−H−Xyl−Arap−Glc−H2O] −; 621.4410 [M−H−Xyl−Arap−2Glc]−; 459.3818 [M−H−Xyl−Arap−3Glc]− |
28 |
| 18 | Bu-Rf | 17.6 88 |
C46H76 O15 |
868.51 84 |
867.51 13 |
0.8 | 799.5486[M−H−Bu]−; 637.4282 [M−H−Bu−Glc]−; 475.3801 [M−H−Bu−2Glc]− |
22 |
| 19 | Ginsenoside Rb1 | 18.1 90 |
C54H92 O23 |
1108.6 029 |
1107.5 911 |
−3.6 | 945.5507 [M−H−Glc]−; 783.4896 [M−H−2Glc]−; 621.4381 [M−H−3Glc]−; 459.3838 [M−H−4Glc]− |
23, 28 |
| 20 | Malonyl-Rb1 | 20.3 06 |
C57H94 O26 |
1194.6 033 |
1193.5 993 |
2.8 | 1107.6072 [M−H−mal]−; 945.5593 [M−H−mal−Glc]−; 783.4674 [M−H−mal−2Glc]−; 765.4901 [M−H−mal-2Glc−H2O]−; 621.4435 [M−H−mal−3Glc]− |
23, 28 |
| 21 | Ginsenoside Rc | 20.7 34 |
C53H90 O22 |
1078.5 924 |
1077.5 846 |
0 | 945.0000 [M−H−Araf]−; 783.0000 [M−H−Araf−Glc]−; 621.0000 [M−H−Araf−2Glc]−; 459.3812 [M−H−Araf−3Glc]− |
23, 28 |
| 22 | Ginsenoside Ro | 20.9 29 |
C48H76 O19 |
956.49 81 |
955.47 36 |
−1.7 | 955.4828 [M−H]−; 793.4456 [M−H−Glc]−; 713.427[M−H−Glc−CO2−H2 O]−; 613.3784 [M−H-2Glc−H2O]−; 569.3766 [M−H−2Glc−CO2−H2O]−; 455.3549 [M−H-3Glc−Acid]− |
23, 28 |
| 23 | Ginsenoside Ra2 | 21.7 08 |
C58H98 O26 |
1210.6 346 |
1209.6 202 |
1.7 | 1077.6158 [M−H−Xyl]−; 945.5381 [M−H−Xyl−Araf]−; 783.4853 [M−H−Xyl−Araf−Glc]−; 765.4739 [M−H−Xyl−Araf−Glc−H2O] −; 621.4294 [M−H−Xyl−Araf−Glc−H2O] −; 459.3787 [M−H−Xyl−Araf−3Glc]− |
28 |
| 24 | Ginsenoside F1 | 23.5 59 |
C36H62 O9 |
638.43 94 |
637.43 39 |
3.6 | 475.3816 [M−H−Glc]− | 28 |
| 25 | Malonyl-Rc | 23.8 07 |
C56H92 O25 |
1164.5 928 |
1163.5 887 |
2.7 | 1119.5898 [M−H−CO2]−; 1077.577 [M−H−mal]−; 945.5308 [M−H−mal-Araf]−; 783.4829 [M−H−mal-Araf−Glc]−; 621.4227 [M−H−mal-Araf−2Glc]−; 459.3813 [M−H−mal-Araf−3Glc]− |
23, 28 |
| 26 | Malonyl-Rb2 | 23.9 27 |
C56H92 O25 |
1164.5 928 |
1163.5 867 |
1.0 | 1119.5893 [M−H−CO2]−; 1077.5760 [M−H−mal-Arap]−; 945.5363 [M−H−mal-Arap]−; 783.4904 [M−H−mal-Arap−Glc]−; 621.4354 [M−H−mal-Arap−2Glc]− |
23, 28 |
| 27 | Malonyl-Ra1/ Malonyl-Ra2 |
24.5 01 |
C61H100 O29 |
1296.6 350 |
1295.6 299 |
1.7 | 1209.6272 [M−H−mal]−; 1077.6158 [M−H−mal−Xyl]−; 945.5423 [M−H−mal−Xyl−Ara]−; 783.4917[M−H−mal−Xyl−A ra−Glc]−; 765.4602 [M−H−mal]−Xyl−Ara−Glc− H2O]−; 621.4410[M−H−mal]−Xyl− Ara−2Glc]−; 459.3818[M−H−mal−Xyl−A ra−3Glc]− |
28 |
| 28 | Ginsenoside Rb2 | 25.0 78 |
C53H90 O22 |
1078.5 824 |
1077.5 761 |
0.9 | 945.5345 [M−H−Arap]−; 783.4844 [M−H−Arap−Glc]−; 621.4313 [M−H−Arap−2Glc]−; 459.3813 [M−H−Arap−3Glc]− |
23, 28 |
| 29 | Ginsenoside Rb3 | 26.7 16 |
C53H90 O22 |
1078.5 924 |
1077.5 846 |
0 | 945.0000 [M−H−Xyl]−; 783.0000 [M−H−Xyl−Glc]−; 621.4186 [M−H−Xyl−2Glc]−; 459.3711 [M−H−Xyl−3Glc]− |
23, 28 |
| 30 | Malonyl-Rb3 | 27.8 90 |
C56H92 O25 |
1164.5 928 |
1163.5 896 |
3.5 | 1119.5832 [M−H−CO2]−; 1077.5738 [M−H−mal]−; 945.5557 [M−H−mal−Xyl]−; 783.4785 [M−H−mal−Xyl−Glc]−; 621.4281 [M−H−mal−Xyl−2Glc]− |
23, 28 |
| 31 | Notoginsenoside Fc |
28.2 78 |
C58H98 O26 |
1210.6 346 |
1209.6 222 |
−3.8 | 1077.6158 [M−H−Xyl]−; 945.5410 [M−H−Xyl−Arap]−; 783.4793 [M−H−Xyl−Arap−Glc]−; 621.4312 [M−H−Xyl−Arap−2Glc]−; 459.3830 [M−H−Xyl−Arap−3Glc]− |
24 |
| 32 | Quinquenoside R1 | 28.7 63 |
C56H94 O24 |
1150.6 135 |
1149.6 118 |
−2.8 | 1107.5836 [M−H−AC]−; 1089.5745 [M−H−AC−H2O]−; 945.5285 [M−H−AC−Glc]−; 783.4795 [M−H−AC−2Glc]−; 621.4282 [M−H−AC−3Glc]−; 459.3848 [M−H−AC−4Glc]− |
24 |
| 33 | Chikusetsusaponi n IVa |
29.2 61 |
C42H66 O14 |
794.44 53 |
793.44 06 |
3.9 | 631.3770 [M−H−Glc]−; 613.7845 [M−H−Glc−H2O]−; 569.3789 [M−H−2Glc−CO2−H2O]−; 455.3485 [M−H−2Glc−Acid]− |
21, 29 |
| 34 | Ginsenoside Rd | 29.5 32 |
C48H82 O18 |
946.55 01 |
945.54 57 |
3.0 | 783.4816 [M−H−Glc]−; 621.4299 [M−H−2Glc]−; 459.3781 [M−H−3Glc]− |
23, 28 |
| 35 | Ginsenoside Rs1 | 29.8 93 |
C55H92 O23 |
1120.6 029 |
1119.5 993 |
3.3 | 1077.5668 [M−H−Ac]−; 945.5472 [M−H−Ac−Arap]−; 783.4679 [M−H−Ac−Arap−Glc]−; 621.4302 [M−H−Ac−Arap−2Glc]−; 459.3709 [M−H−Ac−Arap−3Glc]− |
28 |
| 36 | Malonyl-Rd | 30.2 5 |
C51H84 O21 |
1032.5 505 |
1031.5 408 |
−1.8 | 987.5372 [M−H−CO2]−; 945.5301 [M−H−mal]−; 783.4829 [M−H−mal−Glc]−; 621.4292 [M−H−mal−2Glc]−; 459.3823 [M−H−mal−3Glc]− |
23, 28 |
| 37 | Ginsenoside Rs2 | 30.6 49 |
C55H92 O23 |
1120.6 029 |
1119.6 003 |
4.6 | 1077.5742 [M−H−Ac]−; 945.5422 [M−H−Ac−Araf]−; 915.5121 [M−H−Ac−Glc]−; 783.4852 [M−H−Ac−Araf−Glc]−; 621.4312 [M−H−Ac−Araf−2Glc]−; 459.3818 [M−H−Ac−Araf−3Glc]− |
28 |
| 38 | Gypenoside XVII | 31.0 41 |
C48H82 O18 |
946.55 01 |
945.54 24 |
−0.4 | 783.4831 [M−H−Glc]−; 621.4288 [M−H−2Glc]−; 459.3804 [M−H−3Glc]− |
26 |
| 39 | ginsenoside Ra6 | 31.1 36 |
C58H96 O24 |
1176.6 075 |
1175.6 025 |
2.0 | 1107.5847 [M−H−Bu]−; 945.5311 [M−H-Bu−Glc]−; 783.4826 [M−H-Bu−2Glc]−; 621.4338 [M−H-Bu−3Glc]−; 459.3887 [M−H-Bu−4Glc]− |
22 |
| 40 | ginsenoside Ra7 | 31.7 48 |
C57H94 O23 |
1146.6 186 |
1145.6 150 |
3.7 | 1077.5731 [M−H−Bu]−; 945.5274 [M−H−Bu−Araf]−; 783.4809 [M−H−Bu−Araf−Glc]−; 621.4315 [M−H−Bu−Araf−2Glc]−; 459.3795 [M−H−Bu−Araf−3Glc]− |
22 |
| 41 | Compound Mc-1 | 31.8 68 |
C47H80 O17 |
916.53 90 |
915.52 99 |
−2.0 | 783.4833 [M−H−Araf]−; 621.4284 [M−H−Araf−Glc]−; 459.3816 [M−H−Araf−2Glc]− |
26 |
| 42 | Compound O | 32.0 33 |
C47H80 O17 |
916.53 90 |
915.53 00 |
−1.9 | 783.4795 [M−H−Arap]−; 621.4306 [M−H−Arap−Glc]−; 459.3778 [M−H−Arap−2Glc]− |
26 |
| 43 | Pseudoginsenosid e Rc1 |
32.1 24 |
C50H84 O19 |
988.56 07 |
987.55 61 |
3.2 | 945.5316 [M−H−AC]−; 783.4837 [M−H−AC−Glc]−; 621.4302 [M−H−AC−2Glc]−; 459.3802 [M−H−AC−3Glc]− |
26 |
| 44 | ginsenoside Ra8/Ra9 |
32.3 05 |
C57H94 O23 |
1146.6 18 |
1145.6 155 |
4.2 | 1077.5731 [M−H−Bu]−; 945.5274 [M−H−Bu−Araf]−; 783.4809 [M−H−Bu−Araf−Glc]−; 621.4315 [M−H−Bu−Araf−2Glc]−; 459.3795 [M−H−Bu−Araf−3Glc]− |
22 |
| 45 | Gypenoside IX | 32.4 46 |
C47H80 O17 |
916.53 90 |
915.53 52 |
3.8 | 961.5394 [M+HCOO]−; 783.4812 [M−H−Xyl]−; 621.4284 [M−H−Xy−Glc]−; 459.3864 [M−H−Xy−2Glc]− |
26 |
| 46 | Bu-Gypenoside XVII |
33.7 09 |
C52H86 O19 |
1014.5 757 |
1013.5 650 |
−3.4 | 945.5287 [M−H−Bu]−; 783.4796 [M−H-Bu−Glc]−; 621.4307 [M−H-Bu−2Glc]−; 459.3760 [M−H-Bu−3Glc]− |
22 |
| 47 | 20(S)-Ginsenosid e Rg3/isomer |
34.0 69 |
C42H72 O13 |
784.49 73 |
783.48 86 |
−1.8 | 621.4291 [M−H−Glc]−; 459.3784 [M−H−2Glc]− |
23, 28 |
| 48 | Zingibroside R1 | 34.6 13 |
C42H66 O14 |
794.44 53 |
793.44 10 |
4.4 | 631.3742 [M−H−Glc]−; 613.3672 [M−H−Glc−H2O]−; 569.3779 [M−H−2Glc−CO2−H2O]−; 455.3474 [M−H−2Glc−Acid]− |
26, 29 |
| 49 | Bu-Rd | 34.7 09 |
C52H86 O19 |
1014.5 763 |
1013.5 672 |
−1.3 | 945.5314 [M−H−Bu]−; 783.4778 [M−H-Bu−Glc]−; 621.4255 [M−H-Bu−2Glc]−; 459.3775[M−H-Bu−3Glc]− |
22 |
| 50 | 20(R)-Ginsenosid e Rg3/isomer |
35.5 36 |
C42H72 O13 |
784.49 73 |
783.49 27 |
3.4 | 621.4245 [M−H−Glc]−; 459.3759 [M−H−2Glc]− |
23, 28 |
| 51 | 24(R)-pseudogins enoside RT5 |
38.6 59 |
C33H58 O14 |
678.38 27 |
677.37 60 |
1.6 | 677.3823 [M−H]−; | 24 |
| 52 | Ginsenoside Rk1 | 39.3 51 |
C42H70 O12 |
766.48 67 |
765.47 69 |
−2.6 | 03.4125 [M−H−Glc]− |
23, 28 |
| 53 | Ginsenoside Rg5 | 39.7 82 |
C42H70 O12 |
766.48 67 |
765.47 59 |
−3.9 | 603.4125 [M−H−Glc]− |
23, 28 |
| 54 | Ginsenoside Rh2 | 40.1 17 |
C36H62 O8 |
622.44 45 |
621.43 86 |
3.1 | 459.3792 [M−H−Glc]− |
23, 28 |
[M−H]−;
Glc=β-D-glucopyranosyl (162 Da); Rha=α-L-rhamnopyranosyl (146 Da);
Ara=β-D-arabopyranosyl (132 Da); Xyl=β-D-Xylopyranosyl (132 Da);
Glc-Acid=β-D-glucopyranosylacetyl acid (176 Da); Ac=acetyl (42 Da); Bu=trans-but-2-enoyl (68 Da); Mal=malonyl (86 Da).
3.3 Optimization of the conditions of cell extraction
It is well known that the drug absorption of the cells plays a very important role in drug–cell interaction [15,20]. Influencing factors such as drug concentration, incubation time and digestion method were investigated carefully. The optimized conditions were: concentration of ginseng, 5 mg/mL; incubation time, 4 h; five washes with PBS buffer; digestion with methanol ultrasonic.
3.4. Find of bioactive candidates
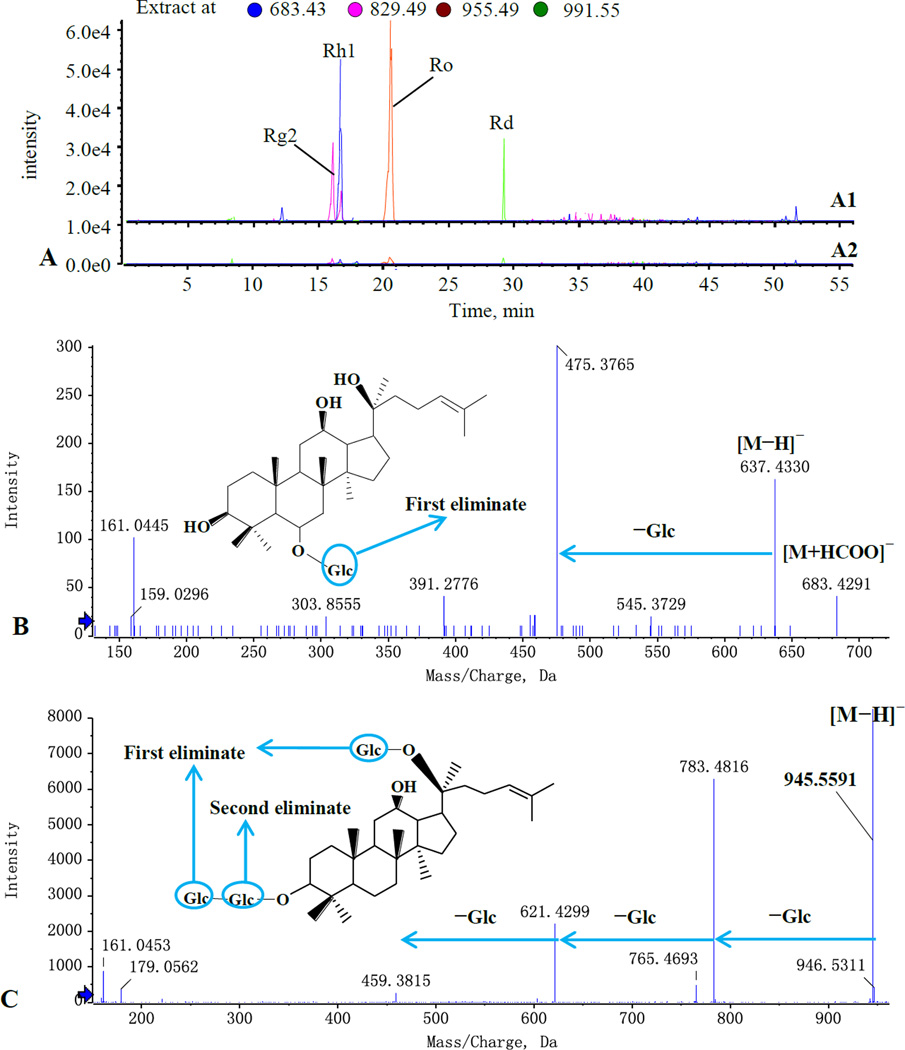
The typical total ion chromatograms (TIC) are shown in Fig. 5. Four compounds (peak No. 14, 15, 22, and 34) were found to be potential bioactive candidates by comparing the chromatograms between target cell-extract group (Fig. 5D) with the control samples (Fig. 5A, 5B and 5C). Furthermore, the detected four peaks were further confirmed by using a narrow mass window of 0.01 Da to restructure the extracted ion chromatograms (EICs) (Fig. 6A) with the main fragmentations of Rg2 (peak 14), Rh1 (peak 15), Ro (peak 22) and Rd (peak 34). The four candidates were characterized in Fig. 4A (Rg2), Fig. 4B (Ro), Fig. 6B (Rh1) and Fig. 6C (Rd), and their LC–MS data are summarized in Table 1.
Figure 5.
Typical TIC chromatograms from various samples. A, ginseng extract treated with DMEM; B, the fifth eluate; C, the extract of cells incubated without ginseng extract; D, the extract of cells incubated with ginseng extract.
Figure 6.
The extracted ion chromatogram (EIC) of the cells incubated with ginseng extract, A (A1, the experimental group; A2, the control group). The negative MS/MS spectra and the proposed fragmentation pathways. B, Rh1; C, Rd.
In general, when cells are incubated with drugs, the bioactive molecules may selectively bind with the cell or be transported into the cell. As can be seen from Fig. 4, some relatively high abundance compounds (peak no. 3 (Majonoside R1), 6 (Rg1), 7 (Re), 19 (Rb1) and 28 (Rb2), Fig. 5A) were not detected in the target cell-extract samples (Fig. 5D), which may indicate these compounds have no selective affinity for the target cells. Ginsenosides Rg1 [7–9], Rb1 [9,10], and Rd [11] have been found to protect SH-SY5Y cells, and Ro was shown to have a neuroprotective effect in animal experiments, but only Rd and Ro were verified in this work. Our experiments do not defy the pharmacological effects of the reported ginsenosides in the literature, and they may exert effects through other pathways. Except for Ro and Rd, the relatively low abundance Rg2 and Rh1 were found to be bioactive candidates, which indicates that the proposed method has suitable sensitivity and can be used as a screening tool for low content constituents in natural products. This case is interesting because most traditional pharmacological screening methods require relatively large amounts of purified compound.
4. Conclusion
In this study, a total of 54 compounds in white ginseng were characterized and identified with the proposed UPLC-QTOF-MS. A target cell-based bioactivity screening method was developed and successfully applied to the predication of potential candidates in panax ginseng with SH-SY5Y cells as target cells, and four ginsenosedes, Rg2, Rh1, Ro, and Rd, were found to be the bioactive components. The target cell-based bioactivity screening method coupled with UPLC-QTOF-MS technique has suitable sensitivity and can be used as a screening tool for low content constituents in natural products.
Supplementary Material
Highlights.
-
✧
A total of 54 compounds in white ginseng were characterized and identified with UPLC-QTOF-MS.
-
✧
Four candidates (Rg2, Rh1, Ro, and Rd) in ginseng were found with the target cell-based bioactivity screening method.
-
✧
The target cell-based bioactivity screening method coupled with UPLC-QTOF-MS technique has suitable sensitivity and can be used as a screening tool for low content constituents in natural products.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China [grant numbers 81260605, 81560648] and the National Institutes of Health [grant numbers AT004418, AT005362].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ru WW, Wang DL, Xu YP, He XX, Sun YE, Qian LY, Zhou XS, Qin YF. Drug Discov. Ther. 2015;9:23–32. doi: 10.5582/ddt.2015.01004. [DOI] [PubMed] [Google Scholar]
- 2.Liu XF, Hao JY, Tang Y, Li JK, Yan ZH, Chen HF, Yuan JB. Mod. Chin. Med. 2016;18:76–81. [Google Scholar]
- 3.Vaibhav R, Juan SM, Sylvain D. Front. Cell. Neurosci. 2015;8:1–13. [Google Scholar]
- 4.Kim HJ, Kim P, Shin CY. J. Ginseng Res. 2013;37:8–29. doi: 10.5142/jgr.2013.37.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho IH. J. Ginseng Res. 2012;36:342–353. doi: 10.5142/jgr.2012.36.4.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu S, Han R, Mak S, Han Y. J. Ethnopharmacol. 2011;135:34–42. doi: 10.1016/j.jep.2011.02.017. [DOI] [PubMed] [Google Scholar]
- 7.Chen XC, Zhou YC, Chen Y, Zhu YG, Fang F, Chen LM. Acta Pharmacol. Sin. 2005;26:56–62. doi: 10.1111/j.1745-7254.2005.00019.x. [DOI] [PubMed] [Google Scholar]
- 8.Shi C, Zheng DD, Fang L, Wu F, Kwong WH, Xu J. Biochim. Biophys. Acta. 2012;1820:453–460. doi: 10.1016/j.bbagen.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Li NJ, Zhou L, Li W, Liu Y, Wang JH, He P. J. Chromatogr. B. 2015;985:54–61. doi: 10.1016/j.jchromb.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 10.Ni N, Liu Q, Ren H, Wu D, Luo C, Li P, Wan JB, Su H. Molecules. 2014;19:3012–3024. doi: 10.3390/molecules19033012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Zhang RY, Zhao J, Dong Z, Feng DY, Wu R, Shi M, Zhao G. Int. J. Mol. Sci. 2015;16:14395–14408. doi: 10.3390/ijms160714395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li SL, Li P, Sheng LH, Li RY, Qi LW, Zhang LY. J. Pharm. Biomed. Anal. 2006;41:576–581. doi: 10.1016/j.jpba.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 13.Zhang HY, Hu CX, Liu CP, Li HF, Wang JS, Yuan KL, Tang JW, Xu GW. J. Pharm. Biomed. Anal. 2007;43:151–157. doi: 10.1016/j.jpba.2006.06.033. [DOI] [PubMed] [Google Scholar]
- 14.Su SL, Yu L, Hua YQ, Duan JA, Deng HS, Tang YP, Lu Y, Ding AW. Biomed. Chromatogr. 2008;22:1385–1392. doi: 10.1002/bmc.1070. [DOI] [PubMed] [Google Scholar]
- 15.Qu FN, Qi LW, Wei YJ, Wen XD, Yi L, Luo HW, Li P. Biol. Pharm. Bull. 2008;31:501–506. doi: 10.1248/bpb.31.501. [DOI] [PubMed] [Google Scholar]
- 16.Hong M, Wang XZ, Wang L, Hua YQ, Wen HM, Duan JA. J. Pharm. Biomed. Anal. 2011;54:87–93. doi: 10.1016/j.jpba.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 17.Li YH, Wang PX, Xiao W, Zhao L, Wang ZH, Yu L. Am. J. Chin. Med. 2013;41:221–229. doi: 10.1142/S0192415X1350016X. [DOI] [PubMed] [Google Scholar]
- 18.Qiu JY, Chen X, Zheng XX, Jiang XL, Yang DZ, Yu YY, Du Q, Tang DQ, Yin XY. Biomed. Chromatogr. 2015;29:226–232. doi: 10.1002/bmc.3264. [DOI] [PubMed] [Google Scholar]
- 19.Sun M, Huang LM, Zhu JL, Bu WJ, Sun J, Fang ZH. Arch. Phar. Res. 2015;38:1044–1053. doi: 10.1007/s12272-014-0469-3. [DOI] [PubMed] [Google Scholar]
- 20.Otvos RA, Nierop PV, Niessen WMA, Kini RM, Somsen GW, Smit AB, Kool J. Anal. Chem. 2016;88:4825–4832. doi: 10.1021/acs.analchem.6b00455. [DOI] [PubMed] [Google Scholar]
- 21.Li SL, Lai SF, Song JZ, Qiao CF, Liu X, Zhou Y, Cai H, Cai BC, Xu HX. J. Pharm. Biomed. Anal. 2010;53:946–957. doi: 10.1016/j.jpba.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 22.Zhu GY, Li YW, Hau DK, Jiang ZH, Yu ZL, Fong WF. Chem. Biodivers. 2011;8:1853–1863. doi: 10.1002/cbdv.201000196. [DOI] [PubMed] [Google Scholar]
- 23.Zhang HM, Li SL, Zhang H, Wang Y, Zhao ZL, Chen SL, Xu HX. J. Pharm. Biomed. Anal. 2012;62:258–273. doi: 10.1016/j.jpba.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Chu C, Xu SJ, Li XN, Yan J, Liu L. J. Food Sci. 2013;78:653–659. doi: 10.1111/1750-3841.12102. [DOI] [PubMed] [Google Scholar]
- 25.Yang XB, Yang XW, Liu JX. Mod. Chin. Med. 2013;15:349–358. [Google Scholar]
- 26.Wang HY, Hua HY, Liu XY, Liu JH, Yu BY. J. Pharm. Biomed. Anal. 2014;98:296–306. doi: 10.1016/j.jpba.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 27.Mao Q, Bai M, Xu JD, Kong M, Zhu LY, Zhu H, Wang Q, Li SL. J. Pharm. Biomed. Anal. 2014;97:129–140. doi: 10.1016/j.jpba.2014.04.032. [DOI] [PubMed] [Google Scholar]
- 28.Wu W, Sun L, Zhang Z, Guo Y, Liu S. J. Pharm. Biomed. Anal. 2015;107:141–150. doi: 10.1016/j.jpba.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 29.Yang H, Lee DY, Kang KB, Kim SO, Yoo YH, Sung SH. J. Pharm. Biomed. Anal. 2015;109:91–104. doi: 10.1016/j.jpba.2015.02.034. [DOI] [PubMed] [Google Scholar]
- 30.Qiu S, Yang WZ, Shi XJ, Yao CL, Yang M, Liu X, Jiang BH, Wu WY, Guo DA. Anal. Chim. Acta. 2015;893:65–76. doi: 10.1016/j.aca.2015.08.048. [DOI] [PubMed] [Google Scholar]
- 31.Bai HR, Wang SJ, Liu JJ, Gao D, Jiang YY, Liu HX, Cai ZW. J. Chromagrogr. B. 2016;1026:263–271. doi: 10.1016/j.jchromb.2015.09.024. [DOI] [PubMed] [Google Scholar]
- 32.Qi LW, Wang HY, Zhang H, Wang CZ, Li P, Yuan CS. J. Chromatogr. A. 2012;1230:93–99. doi: 10.1016/j.chroma.2012.01.079. [DOI] [PubMed] [Google Scholar]
- 33.Wan JY, Liu P, Wang HY, Qi LW, Wang CZ, Li P, Yuan CS. J. Chromatogr. A. 2013;1286:83–92. doi: 10.1016/j.chroma.2013.02.053. [DOI] [PubMed] [Google Scholar]
- 34.Wang LL, Han LF, Yu HS, Sang MM, Liu EW, Zhang Y, Fang SM, Wang T, Gao XM. Molecules. 2015;20:20518–20537. doi: 10.3390/molecules201119712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCullagh M, Rao R, Chipperfield J, Douce D. Waters Corporation, Application Note 720005423EN. http://www.waters.com/webassets/cms/library/docs/720005423en.pdf. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.