Abstract
Neutrophil extracellular traps (NETs) released from dead neutrophils at the site of inflammation represent webs of neutrophilic DNA stretches dotted with granule-derived antimicrobial proteins, including lactoferrin, and play important roles in innate immunity against microbial infection. We have shown the coexistence of NETs and fibrin meshwork in varied fibrinopurulent inflammatory lesions at both light and electron microscopic levels. In the present study, correlative light and electron microscopy (CLEM) employing confocal laser scanning microscopy and scanning electron microscopy was performed to bridge light and electron microscopic images of NETs and fibrin fibrils in formalin-fixed, paraffin-embedded, autopsied lung sections of legionnaire’s pneumonia. Lactoferrin immunoreactivity and 4'-6-diamidino-2-phenylindole (DAPI) reactivity were used as markers of NETs, and fibrin was probed by fibrinogen gamma chain. Of note is that NETs light microscopically represented as lactoferrin and DAPI-colocalized dots, 2.5 μm in diameter. CLEM gave super-resolution images of NETs and fibrin fibrils: “Dotted” NETs were ultrastructurally composed of fine filaments and masses of 58 nm-sized globular materials. A fibrin fibril consisted of clusters of smooth-surfaced filaments. NETs filaments (26 nm in diameter) were significantly thinner than fibrin filaments (295 nm in diameter). Of note is that CLEM was applicable to formalin-fixed, paraffin-embedded sections of autopsy material.
Keywords: neutrophil extracellular traps, fibrin, correlative light and electron microscopy, scanning electron microscopy, confocal laser scanning microscopy
I. Introduction
Neutrophils are one of the effector cells in the innate immune system involved in host defense against microbial infections [14, 17, 21]. In 2004, Brinkmann et al. reported a new protective function of neutrophils, called neutrophil extracellular traps (NETs) [3]. NETs consist of extracellular spider’s web-like structures, resulting from cell death of activated neutrophils [3, 30]. The framework is formed by neutrophilic DNA stretches, 15 to 17 nm in diameter [3, 30]. The DNA-based filaments are dotted with globular materials, around 50 nm in diameter, composed of varied neutrophilic granule-derived antimicrobial proteins such as neutrophil elastase, myeloperoxidase, gelatinase and lactoferrin (LF) [3, 17, 30]. NETs capture and kill bacteria and fungi [3, 26, 30], to prevent them from spreading and colonizing host cell surfaces [28]. NETs are commonly formed in fibrinopurulent inflammatory lesions such as pneumonia and abscess, and often coexist with fibrin meshwork [20, 23].
Fluorescence microscopy and confocal laser scanning microscopy (CLSM) visualize the localization of target proteins in cells and tissues at the light microscopic level [8, 18], but the background tissue structure is often unclear and the optical resolution is not high enough [7, 10, 18]. Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) map the cellular fine structure with a nanometer scale resolution [18], but the cellular ultrastructure is displayed as a black and white image [7]. Correlative light and electron microscopy (CLEM) is a unique microscopic method that can combine the advantages of light and electron microscopy by examining the same area of the specimen simultaneously using both methods [2, 18]. CLEM has received much attention among micromorphologists in recent years [6, 11, 18, 22, 24, 32]. The cellular ultrastructure-function association analysis by CLEM is investigated firstly by observing with fluorescence microscopy and CLSM, followed by imaging with SEM or TEM. A super-resolution fluorescent image can be obtained finally by overlaying the light and electron microscopic pictures [11, 32].
In our previous reports [20, 23], we successfully observed NETs and fibrin meshwork in formalin-fixed, paraffin-embedded sections at both the light and electron microscopic levels. LF served as a reliable marker of NETs, and LF positivity often coexisted with fibrin fibrils, demonstrated as fibrinogen gamma chain (FGG) immunoreactivity [20, 23]. In the present study, an autopsied lung tissue of legionnaire’s pneumonia, the same sample used in our previous study [20], was analyzed by CLEM to co-visualize light and electron microscopic features of NETs and fibrin fibrils. This is the first report describing the fine structural features of NETs and fibrin fibrils with CLEM.
II. Materials and Methods
Sample
A lung tissue of legionnaire’s pneumonia was obtained at autopsy in Fujita Health University Hospital, Toyoake, Japan. The fresh lung tissue was cut to confirm the complication of lobar pneumonia, and then routinely fixed in 10% formalin and embedded in paraffin wax. We have focused to analyze the area of fibrinopurulent inflammation accompanying deposition of fibrillar meshwork structures, which were confirmed with hematoxylin-eosin (HE) staining, as described in our previous studies [20, 23].
Immunoperoxidase staining
A paraffin section at 3 μm thickness was mounted on a coated glass slide New Silane II (Muto Pure Chemicals, Tokyo, Japan), deparaffinized with xylene, and rehydrated through graded ethanol. For amino acid polymer immunohistochemical staining, endogenous peroxidase activity was quenched with 0.3% hydrogen peroxide in methanol for 30 min at room temperature. After a brief dip in tap water, the antigenicity was retrieved by heating with a pressure pan cooker (Delicio 6L, T-FAL, Rumily, France) in 1 mM ethylenediamine tetraacetic acid solution, pH 8.0, for 10 min, and the section was left for 30 min at room temperature for cooling. A phosphate-buffered saline (PBS, pH 7.2) rinse was interposed between every step. Anti-Legionella pneumophila serogroup 1 rabbit polyclonal antibody (diluted at 1:500, Denka Seiken, Tokyo, Japan) was incubated overnight at room temperature. As the second layer reagent, Simple Stain MAX-PO (Nichirei Bioscience, Tokyo, Japan) was incubated for 30 min at room temperature. The site of antigen localization was visualized in 50 mM Tris-HCl buffer, pH 7.6, containing 20 mg/dl diaminobenzidine tetrahydrochloride and 0.006% hydrogen peroxide. Finally, the nuclei were lightly counterstained with Mayer’s hematoxylin.
Correlative light and electron microscopy (CLEM) using confocal laser scanning microscopy (CLSM) and scanning electron microscopy (SEM)
A paraffin section at 3 μm thickness was mounted on a coated glass slide New Silane II, deparaffinized with xylene, rehydrated through graded ethanol, heat-treated with a pressure pan cooker in 10 mM citrate buffer, pH 6.0, for 10 min, and left for 30 min at room temperature for cooling. A PBS rinse was interposed between every step. The section was incubated with a mixture of anti-LF rabbit antiserum (diluted at 1:300, GenWay Biotech, San Diego, CA, USA) and anti-FGG mouse monoclonal antibody (clone: 1F2, diluted at 1:300, Abnova, Taipei, Taiwan) overnight at room temperature, followed by incubation with a mixture of Alexa Fluor 488 (green)-labeled goat anti-rabbit IgG antibody (diluted 1:300, Molecular Probes, Tokyo, Japan), Alexa Fluor 568 (red)-labeled goat anti-mouse IgG antibody (diluted 1:300; Molecular Probes) and 4'-6-diamidino-2-phenylindole (DAPI) solution (diluted at 1:1,000, Thermo Fisher Scientific, Yokohama, Japan) for 1 hr at room temperature. The section was dehydrated in graded ethanol and tertial-butyl alcohol, dried in a freeze-drying apparatus (JFD-310, JEOL, Tokyo, Japan), and observed without using a cover slip on a confocal laser scanning microscope (LSM710, Carl Zeiss, Oberkochen, Germany). The section was then sputter-coated with gold palladium by JFC-1500 (JEOL), and observed on a scanning electron microscope (S-4000, Hitachi, Tokyo, Japan). Confocal laser scanning microscopic and scanning electron microscopic images were merged by using an imaging software (Adobe Photoshop CS5.1, Adobe Systems, San Jose, USA). The fluorescent signal strength of Alexa Fluor 488 (green), Alexa Fluor 568 (red) and DAPI (blue) was adjusted using the same imaging software (Adobe Photoshop CS5.1).
Measurement of the size of fibrils and globular structures
On light microscopic CLSM images, the diameter of randomly selected 30 LF and DAPI-colocalized dots (representing NETs-related structures) and 30 FGG-positive fibrils (representing fibrin fibrils) was measured. On ultrastructural CLEM images, the diameter of randomly selected 30 DAPI-positive filaments (representing NETs filaments), 30 FGG-positive filaments (representing fibrin filaments) and 30 LF-positive NETs-related globular materials was evaluated. Values were presented as the mean±standard error of the mean. The diameters of DAPI-positive filaments and FGG-positive filaments on the CLEM images were statistically compared with two-tailed Student’s t-test. Values of p<0.05 were considered to indicate statistical significance.
Ethical issue
The use of human material was approved by the ethical review board for clinical and epidemiological investigations at Fujita Health University, Toyoake (approval number: HM15-583).
III. Results
Light microscopic observation
HE and immunohistochemical stains were performed for evaluating the localization of the microbe, Legionella pneumophila serotype 1, LF and FGG in consecutive paraffin sections sampled from legionnaire’s pneumonia (Fig. 1). DNA was detected as DAPI fluorescence. Inflammatory cells (neutrophils) phagocytizing Legionella pneumophila were dispersed among eosinophilic fibrin deposits (Fig. 1a, b). The cytoplasm and nuclei of neutrophils were densely labeled for LF (green in color) and DAPI (blue in color), respectively. In addition, LF and DAPI co-existed extracellularly as a dotted structure (Fig. 1c–f). It is known that DNA is a major structural component of NETs [3, 29], and LF forms a complex with DNA on NETs [13, 29]. At the site we analyzed, LF and DAPI reactivities did not form a fibrillar network at light microscopic level, while FGG (red in color) formed a meshwork structure of thick fibrils (Fig. 1d, e, f).
Fig. 1.
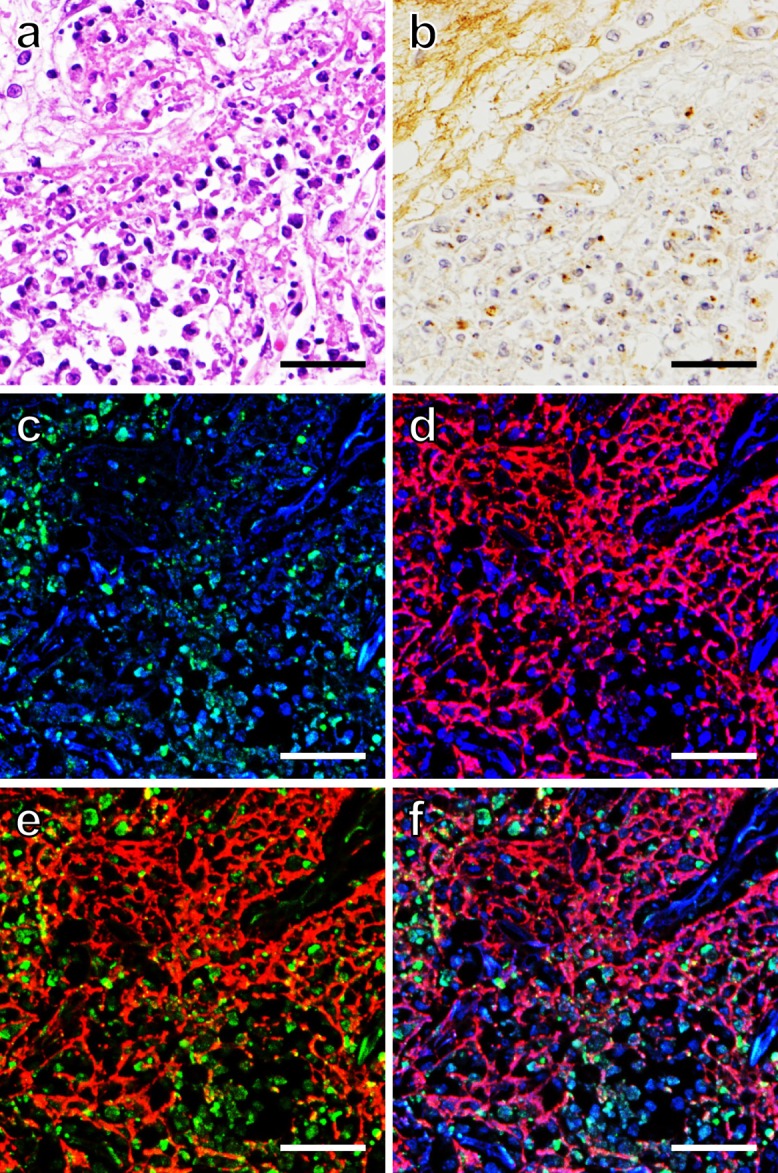
Localization of LF, FGG and DAPI (DNA) in a paraffin-embedded lung tissue of legionnaire’s pneumonia. Consecutive sections demonstrate HE-stained microscopic features (a), immunoperoxidase staining for Legionella pneumophila, serotype 1 (b), and CLSM images (c–f) of immunofluorescence staining for LF (green) and FGG (red) and DAPI-induced DNA fluorescence (blue). Dual fluorescence illustrates LF and DAPI (c), FGG and DAPI (d) and LF and FGG (e). Triple fluorescence is shown in (f). The brown-stained pathogens are observed in the cytoplasm of neutrophils. Fibrin meshwork is shown not only in HE staining (eosinophilic fibrils) but also with FGG red fluorescence. The cytoplasm and nuclei of neutrophils stain green and blue, respectively. The “dotted” (fine granular) light microscopic localization pattern of LF and DAPI shown here is different from the common NETs-related fibrillary structure. Bars=50 μm.
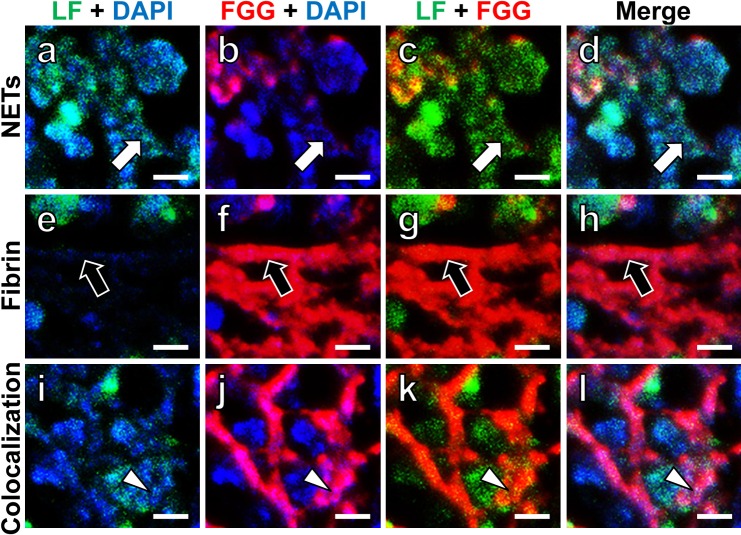
Next, high-magnification CLSM observation was performed to examine structural correlations among LF, FGG and DAPI. As shown by white arrows in Figure 2 (a–d), green-colored LF and blue-stained DNA were colocalized on the dotted structure, representing NETs. Red-colored fibrin fibrils (FGG immunoreactivity) forming thick fibrillar structures are indicated by black arrows in Figure 2 (e–h). As illustrated by arrowheads in Figure 2 (i–l), LF and DAPI (DNA) double-positive dotted signals were also seen on the FGG-positive fibrin fibrils. Bacillary entrapment was not observed at the site of evaluation.
Fig. 2.
High-powered fluorescent CLSM images of LF, FGG and DAPI (DNA) in NETs and fibrin fibrils. High-powered CLSM images of NETs (a–d), fibrin fibrils (e–h) and the colocalization of NETs and fibrin fibrils (i–l) are shown. NETs are stained with both LF (green) and DAPI (blue), and fibrin fibrils are stained with FGG (red). Two-color merged images are illustrated in (a–c, e–g, i–k). Three-color merged images are seen in (d, h, l). White arrows indicate dotted NETs. Black arrows demonstrate fibrin fibrils. White arrowheads indicate the site of colocalization of NETs and fibrin fibrils: fine NETs-related dots are observed on the fibrin fibrils. Bars=5 μm.
Correlative light and electron microscopic observation
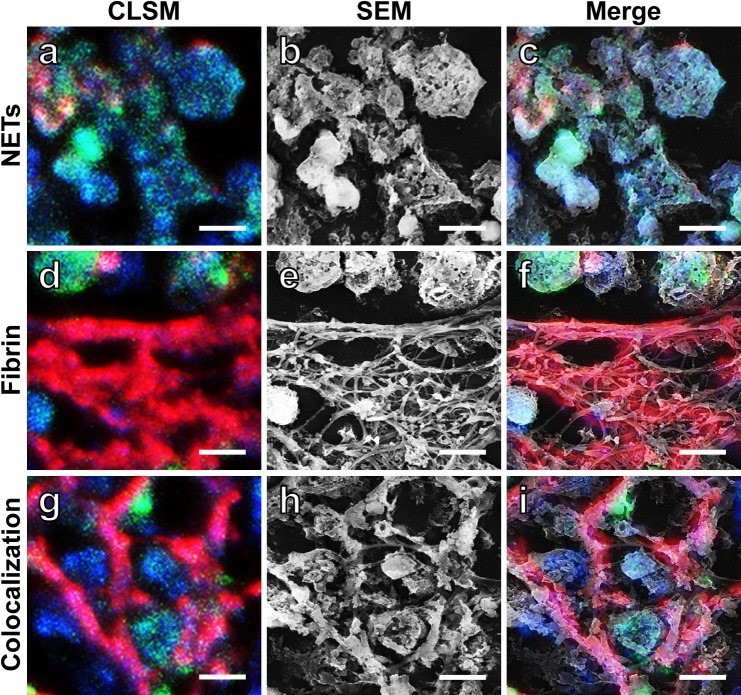
High magnification CLSM observation clearly distinguished NETs-related structures from fibrin fibrils. SEM observation expectedly visualizes the ultrastructure of the fibrils. To evaluate the fine structure of NETs and fibrin fibrils, the very same area of the section was observed with both methods. Figure 3 (a, d, g) demonstrates high magnification CLSM features of NETs (Fig. 3a), fibrin (Fig. 3d) and the colocalization of NETs and fibrin (Fig. 3g). The corresponding SEM features are shown in Figure 3 (b, e, h). Figure 3 (c, f, i) illustrates merged images of CLSM and SEM. It is evident that NETs appeared as clusters of globular materials (Fig. 3a–c), and that fibrin fibrils seen by CLSM are composed of a cluster of filamentous components (Fig. 3d–f). At the site of colocalization of NETs and fibrin, fibrin filaments were covered with NETs-related globular materials (Fig. 3g–i).
Fig. 3.
Correlation between CLSM and SEM images of NETs and fibrin fibrils. CLSM images (a, d, g) and SEM images (b, e, h) of NETs (a, b), fibrin fibrils (d, e) and the colocalization of NETs and fibrin fibrils (g, h) are shown. NETs are dually stained for LF (green) and DAPI (blue), while fibrin fibrils are stained for FGG (red). Merged features of the CLSM and SEM images are illustrated in (c, f, i). NETs are observed here as a cluster of globular materials (a–c). Note that thick fibrin fibrils are composed of a cluster of smooth-surfaced filaments (d–f). At the site of colocalization, fibrin filaments are attached with globular materials (g–i). Bars=5 μm.
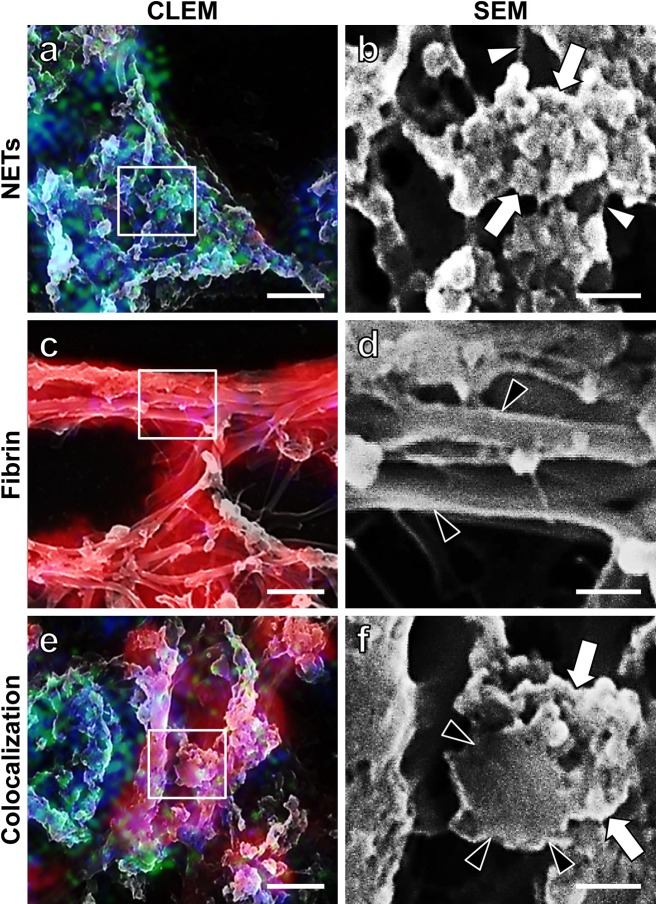
The ultrastructure of NETs and fibrin fibrils was further analyzed by using the merged (CLEM) images of CLSM and SEM. Again, we recognized three topographic patterns, NETs, fibrin and the colocalization of both (Fig. 4a, c, e). To examine the detailed ultrastructure of NETs and fibrin fibrils, the squared areas in the CLEM images (Fig. 4a, c, e) were observed by SEM at the highest magnification (Fig. 4b, d, f). As illustrated in Figure 4b, NETs (LF +, FGG −, DAPI +) were composed of fine filaments (white arrowheads) and masses of globular materials (white arrows). The fine filaments were DAPI-reactive but without LF labeling. Figure 4d illustrates that fibrin fibrils (LF −, FGG +, DAPI −) were solely composed of a cluster of smooth surfaced, thicker filaments (black arrowheads). Figure 4f, sampled from the area showing the colocalization of NETs and fibrin, demonstrates 1) a transverse section of a single fibrin filament (black arrowheads indicate marginal portions of the fibrin filament) and 2) masses of globular materials with features similar to Figure 4b, clung onto the fibrin filament (white arrows).
Fig. 4.
Highly magnified SEM images of NETs and fibrin fibrils, in relation to CLEM images. CLEM images illustrate NETs (a), fibrin fibrils (c) and the colocalization of NETs and fibrin fibrils (e). The squared areas in the CLEM images (a, c, e) are observed by SEM at the maximal magnification (b, d, f). White arrowheads in (b) indicate NETs filaments, and globular materials (white arrows) are clustered on the fine filaments. Black arrowheads in (d) demonstrate individual smooth-surfaced fibrin filaments. A transverse section of a single fibrin filament is indicated by black arrowheads in (f). NETs-related globular materials on the fibrin filament are shown by white arrows in (f). Bars=2 μm (a, c, e) and 0.5 μm (b, d, f).
On light microscopic CLSM images, the diameter of LF and DAPI-colocalized dots (representing NETs-related structures) was 2.5±0.6 μm. The FGG-positive fibrils (representing a component of fibrin meshwork) measured 1.7±0.4 μm in diameter. On ultrastructural CLEM images, the width of DAPI-positive filaments (representing NETs filaments) and FGG-positive filaments (representing fibrin filaments), as well as the diameter of LF-positive globular materials (as an element of LF and DAPI-colocalized dots), were evaluated. There was significant size difference between DAPI-positive filaments and FGG-positive filaments: DAPI-positive NETs filaments measured 26±12 nm, while FGG-positive fibrin filaments measured 295±53 nm (p<0.0001). The LF-positive globular materials were 58±8 nm in diameter at the ultrastructural level.
IV. Discussion
NETs represent an extracellular spider’s web-like structure of neutrophilic DNA stretches dotted with granule-derived antimicrobial proteins, including lactoferrin (LF), functionally preventing microbial invasions [3, 26, 28]. The pathophysiological significance of NETs has been reported in a variety of diseased conditions such as acute appendicitis, dysentery, preeclampsia, necrotizing fasciitis and pneumonia [1, 3, 4, 9]. In these acute inflammatory lesions, fibrin meshwork commonly deposited together with neutrophilic infiltration [5, 31], and NETs forming a web-like network were also demonstrated by SEM [20, 27].
We previously evaluated differences in the fibrillar structures of NETs and fibrin in formalin-fixed, paraffin-embedded sections of inflammatory lesions [20, 23]. LF functioned a good immunohistochemical marker of NETs, while fibrin was demonstrated by FGG immunostaining. In the light microscopic analysis, fibrils were categorized into three types: thin, thick and clustered thick [23]. Thin fibrils belonged to NETs and thick fibrils were composed of either mixed NETs and fibrin or fibrin alone, while clustered thick fibrils were solely composed of fibrin [23]. In the electron microscopic analysis using an autopsied lung tissue of legionnaire’s pneumonia, NETs were composed of fine filaments and globular materials attached onto the fibrin fibrils [20]. The smooth-surfaced fibrin filaments were much thicker than the NETs filaments [20]. A discrepancy to be solved is that the diameter of the fibrils at the light microscopic level appeared much larger than that seen at the electron microscopic level [20].
CLEM has successfully been utilized in a variety of morphologic studies. For example, Sindbis virus exit pathway [19] and macrophage uptake of cylindrical microparticles [25] were investigated by CLEM. These studies were performed without causing damages to the ultrastructure of the cell and tissue. In the present CLEM analysis using a routinely prepared autopsy lung tissue (legionnaire’s pneumonia), the same area of the inflammatory lesion was simultaneously observed with both CLSM and SEM [18]. It is of note that CLSM did not damage the SEM features. Both types of microscopy should be used at their full capabilities [15, 18, 32]. The results were consistent with those of our previous studies [20, 23]. NETs (LF +, FGG −, DAPI +) were composed of fine filaments, 26±12 nm in diameter, and masses of globular materials. Fibrin filaments (LF −, FGG +, DAPI −), 295±53 nm in diameter, were much thicker than the NETs filaments.
The CLEM analysis explained the discrepancy between the light and electron microscopic observations regarding the diameter of the fibrils and the shape of the meshwork structure. Fibrin, seen as a thick (1.7 μm-sized) fibril at the light microscopic level, consisted ultrastructurally of a cluster of fine filamentous skeletons. NETs-related dots, measuring 2.5±0.6 μm, seen at the light microscopic level were ultrastructurally composed of clustered globular materials, 58±8 nm in diameter, on the fine filamentous structure. These sizes of the filaments and globules were consistent with those of the previous reports [3, 16, 17, 30]. It should be of note that light microscopically, NETs can present as a form of clustered dots, but not in the form of spider’s web-like framework. The importance of the recognition of this variant of NETs should be emphasized, particularly when NETs are evaluated under pathologic conditions. Poor development of spider’s web-like NETs may be related to the intracellular growth features of the pathogen, Legionella pneumophila [12]. In fact, bacteria, the target of NETs functions, were not entrapped in the extracellular space of the lung where we evaluated. As described in our previous study using immunoperoxidase visualization of LF and FGG [20], LF and FGG double-positive meshwork structures were clearly demonstrated in the same pneumonia sample, where accumulation of LF-positive globules on the FGG-positive fibrin filament was accelerated. The appearance of the “dotted” NETs may be dependent upon the balance of neutrophilic LF release and fibrin meshwork formation.
The present study is the first report demonstrating, by means of LF, FGG and DAPI as probes, the ultrastructural features of NETs and fibrin fibrils in an inflammatory lesion by CLEM, combining CLSM and SEM. It should be emphasized that the CLEM analysis was applicable to routinely prepared formalin-fixed, paraffin-embedded sections of autopsy materials and that the ultrastructural localization of specific proteins was successfully demonstrated even after heating treatment. We sincerely hope that the CLEM analysis can be expanded to morpho-functional studies of NETs and fibrin fibrils in archival pathology materials under varied inflammatory conditions.
V. Competing Interest Statement
We have no conflict of interest to be claimed.
VI. Acknowledgments
We are grateful to Senior Assistant Prof. Gen Niimi, Ph.D. and Assistant Prof. Tomihiko Ide, Ph.D., Division of Electron Microscopy, Institute of Joint Research, Fujita Health University, Toyoake, for their technical advice. We are also grateful to Prof. Ken-ichi Inada, M.D., Ph.D. and Assistant Prof. Kouhei Sakurai, Ph.D., Department of Diagnostic Pathology, Banbuntane-Houtokukai Hospital, Fujita Health University School of Medicine, Nagoya, for their helpful discussion. Ms. Yukika Hasegawa, Ms. Sayaka Takeuchi, Ms. Mika Maeshima, and Ms. Chikayo Yashiro, Department of Pathology, Fujita Health University School of Medicine, Toyoake, are cordially acknowledged for their positive cooperation in our research activity. This work was supported by a Research Grant from Fujita Health University, 2015–2016 (no specific grant number given).
VII. References
- 1.Beiter K., Wartha F., Albiger B., Normark S., Zychlinsky A. and Henriques-Normark B. (2006) An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr. Biol. 16; 401–407. [DOI] [PubMed] [Google Scholar]
- 2.Benedetti L., Sogne E., Rodighiero S., Marchesi D., Milani P. and Francolini M. (2014) Customized patterned substrates for highly versatile correlative light-scanning electron microscopy. Sci. Rep. 4; 7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D. S., Weinrauch Y. and Zychlinsky A. (2004) Neutrophil extracellular traps kill bacteria. Science 303; 1532–1535. [DOI] [PubMed] [Google Scholar]
- 4.Buchanan J. T., Simpson A. J., Aziz R. K., Liu G. Y., Kristian S. A., Kotb M., Feramisco J. and Nizet V. (2006) DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr. Biol. 16; 396–400. [DOI] [PubMed] [Google Scholar]
- 5.Chung C. L., Chen Y. C. and Chang S. C. (2003) Effect of repeated thoracenteses on fluid characteristics, cytokines, and fibrinolytic activity in malignant pleural effusion. Chest 123; 1188–1195. [DOI] [PubMed] [Google Scholar]
- 6.Cortese K., Diaspro A. and Tacchetti C. (2009) Advanced correlative light/electron microscopy: current methods and new developments using Tokuyasu cryosections. J. Histochem. Cytochem. 57; 1103–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Boer P., Hoogenboom J. P. and Giepmans B. N. (2015) Correlated light and electron microscopy: ultrastructure lights up! Nat. Methods 12; 503–513. [DOI] [PubMed] [Google Scholar]
- 8.Giepmans B. N., Adams S. R., Ellisman M. H. and Tsien R. Y. (2006) The fluorescent toolbox for assessing protein location and function. Science 312; 217–224. [DOI] [PubMed] [Google Scholar]
- 9.Gupta A. K., Hasler P., Holzgreve W., Gebhardt S. and Hahn S. (2005) Induction of neutrophil extracellular DNA lattices by placental microparticles and IL-8 and their presence in preeclampsia. Hum. Immunol. 66; 1146–1154. [DOI] [PubMed] [Google Scholar]
- 10.Hell S. W. (2007) Far-field optical nanoscopy. Science 316; 1153–1158. [DOI] [PubMed] [Google Scholar]
- 11.Hellstrom K., Vihinen H., Kallio K., Jokitalo E. and Ahola T. (2015) Correlative light and electron microscopy enables viral replication studies at the ultrastructural level. Methods 90; 49–56. [DOI] [PubMed] [Google Scholar]
- 12.Jules M. and Buchrieser C. (2007) Legionella pneumophila adaptation to intracellular life and the host response: clues from genomics and transcriptomics. FEBS Lett. 581; 2829–2838. [DOI] [PubMed] [Google Scholar]
- 13.Kanyshkova T. G., Semenov D. V., Buneva V. N. and Nevinsky G. A. (1999) Human milk lactoferrin binds two DNA molecules with different affinities. FEBS Lett. 451; 235–237. [DOI] [PubMed] [Google Scholar]
- 14.Kinnula V. L., Soini Y., Kvist-Makela K., Savolainen E. R. and Koistinen P. (2002) Antioxidant defense mechanisms in human neutrophils. Antioxid. Redox Signal. 4; 27–34. [DOI] [PubMed] [Google Scholar]
- 15.Kopek B. G., Shtengel G., Xu C. S., Clayton D. A. and Hess H. F. (2012) Correlative 3D superresolution fluorescence and electron microscopy reveal the relationship of mitochondrial nucleoids to membranes. Proc. Natl. Acad. Sci. U S A 109; 6136–6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krautgartner W. D., Klappacher M., Hannig M., Obermayer A., Hartl D., Marcos V. and Vitkov L. (2010) Fibrin mimics neutrophil extracellular traps in SEM. Ultrastruct. Pathol. 34; 226–231. [DOI] [PubMed] [Google Scholar]
- 17.Liu F. C., Chuang Y. H., Tsai Y. F. and Yu H. P. (2014) Role of neutrophil extracellular traps following injury. Shock 41; 491–498. [DOI] [PubMed] [Google Scholar]
- 18.Liv N., Zonnevylle A. C., Narvaez A. C., Effting A. P., Voorneveld P. W., Lucas M. S., Hardwick J. C., Wepf R. A., Kruit P. and Hoogenboom J. P. (2013) Simultaneous correlative scanning electron and high-NA fluorescence microscopy. PLoS One 8; e55707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez M. G., Snapp E. L., Perumal G. S., Macaluso F. P. and Kielian M. (2014) Imaging the alphavirus exit pathway. J. Virol. 88; 6922–6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Onouchi T., Shiogama K., Matsui T., Mizutani Y., Sakurai K., Inada K. and Tsutsumi Y. (2016) Visualization of neutrophil extracellular traps and fibrin meshwork in human fibrinopurulent inflammatory lesions: II. Ultrastructural study. Acta Histochem. Cytochem. 49; 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rigby K. M. and DeLeo F. R. (2012) Neutrophils in innate host defense against Staphylococcus aureus infections. Semin. Immunopathol. 34; 237–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rilla K. and Koistinen A. (2015) Correlative light and electron microscopy reveals the HAS3-induced dorsal plasma membrane ruffles. Int. J. Cell Biol. 2015; 769163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shiogama K., Onouchi T., Mizutani Y., Sakurai K., Inada K. and Tsutsumi Y. (2016) Visualization of neutrophil extracellular traps and fibrin meshwork in human fibrinopurulent inflammatory lesions: I. Light microscopic study. Acta Histochem. Cytochem. 49; 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sjollema K. A., Schnell U., Kuipers J., Kalicharan R. and Giepmans B. N. (2012) Correlated light microscopy and electron microscopy. Methods Cell Biol. 111; 157–173. [DOI] [PubMed] [Google Scholar]
- 25.Tscheka C., Hittinger M., Lehr C. M., Schneider-Daum N. and Schneider M. (2015) Macrophage uptake of cylindrical microparticles investigated with correlative microscopy. Eur. J. Pharm. Biopharm. 95; 151–155. [DOI] [PubMed] [Google Scholar]
- 26.Urban C. F., Reichard U., Brinkmann V. and Zychlinsky A. (2006) Neutrophil extracellular traps capture and kill Candida albicans yeast and hyphal forms. Cell. Microbiol. 8; 668–676. [DOI] [PubMed] [Google Scholar]
- 27.Veklich Y., Francis C. W., White J. and Weisel J. W. (1998) Structural studies of fibrinolysis by electron microscopy. Blood 92; 4721–4729. [PubMed] [Google Scholar]
- 28.Vitkov L., Klappacher M., Hannig M. and Krautgartner W. D. (2009) Extracellular neutrophil traps in periodontitis. J. Periodont. Res. 44; 664–672. [DOI] [PubMed] [Google Scholar]
- 29.Vogel H. J. (2012) Lactoferrin, a bird’s eye view. Biochem. Cell Biol. 90; 233–244. [DOI] [PubMed] [Google Scholar]
- 30.Vorobjeva N. V. and Pinegin B. V. (2014) Neutrophil extracellular traps: mechanisms of formation and role in health and disease. Biochemistry 79; 1286–1296. [DOI] [PubMed] [Google Scholar]
- 31.Wait M. A., Sharma S., Hohn J. and Dal Nogare A. (1997) A randomized trial of empyema therapy. Chest 111; 1548–1551. [DOI] [PubMed] [Google Scholar]
- 32.Watanabe S., Punge A., Hollopeter G., Willig K. I., Hobson R. J., Davis M. W., Hell S. W. and Jorgensen E. M. (2011) Protein localization in electron micrographs using fluorescence nanoscopy. Nat. Methods 8; 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]