MicroRNA-21 (miR-21) is thought to promote liver regeneration after partial hepatectomy (PHx), but it also facilitates fibrosis in liver and other tissues. In ethanol-fed rats, hepatocyte proliferation after PHx is suppressed, miR-21 expression is enhanced, and hepatic stellate cell (HSC) activation markers are upregulated. Inhibition of miR-21 restored cell cycle progression and prevented upregulation of HSC activation markers after PHx in ethanol-fed rats, suggesting that a miR-21-promoted profibrotic state of HSCs contributes to inhibition of hepatocyte proliferation.
Keywords: microRNA antisense oligonucleotide, chronic alcohol treatment, gene expression pattern analysis, stellate cell activation
Abstract
Liver regeneration is a clinically significant tissue repair process that is suppressed by chronic alcohol intake through poorly understood mechanisms. Recently, microRNA-21 (miR-21) has been suggested to serve as a crucial microRNA (miRNA) regulator driving hepatocyte proliferation after partial hepatectomy (PHx) in mice. However, we reported recently that miR-21 is significantly upregulated in ethanol-fed rats 24 h after PHx, despite inhibition of cell proliferation, suggesting a more complex role for this miRNA. Here, we investigate how inhibition of miR-21 in vivo affects the early phase of liver regeneration in ethanol-fed rats. Chronically ethanol-fed rats and pair-fed control animals were treated with AM21, a mixed locked nucleic acid-DNA analog antisense to miR-21 that inhibited miR-21 in vivo to undetectable levels. Liver regeneration after PHx was followed by cell proliferation marker and gene expression analysis, miRNA profiling, and cell signaling pathway analysis. Although liver regeneration was not significantly impaired by AM21 in chow-fed rats, AM21 treatment in ethanol-fed animals completely restored regeneration and enhanced PHx-induced hepatocyte proliferation to levels comparable to those of untreated or chow-fed animals. In addition, a marked deposition of α-smooth muscle actin, a marker of stellate cell activation, which was evident in ethanol-treated animals after PHx, was effectively suppressed by AM21 treatment. Gene expression analysis further indicated that suppression of stellate cell-specific profibrogenic profiles and the Notch signaling contributed to AM21-mediated rescue from deficient hepatocyte proliferation in ethanol-fed animals. Our results indicate that the impact of miR-21 balances proproliferative effects with antiproliferative profibrogenic actions in regulating distinctive regenerative responses in normal vs. disease conditions.
NEW & NOTEWORTHY
MicroRNA-21 (miR-21) is thought to promote liver regeneration after partial hepatectomy (PHx), but it also facilitates fibrosis in liver and other tissues. In ethanol-fed rats, hepatocyte proliferation after PHx is suppressed, miR-21 expression is enhanced, and hepatic stellate cell (HSC) activation markers are upregulated. Inhibition of miR-21 restored cell cycle progression and prevented upregulation of HSC activation markers after PHx in ethanol-fed rats, suggesting that a miR-21-promoted profibrotic state of HSCs contributes to inhibition of hepatocyte proliferation.
the adult liver has a unique capacity to repair tissue damage by allowing fully differentiated, quiescent hepatocytes to reenter the cell cycle and undergo proliferation while maintaining liver-specific functions. In the experimental model of 70% partial hepatectomy (PHx) in the rat, the left lateral and medial (LLM) lobes are surgically removed to initiate a synchronized cell cycle entry of hepatocytes in the remnant tissue (15), driven in part by early stress signals (4, 12) and the release of proinflammatory cytokines (39) and reorganization of extracellular matrix components (27) to initiate a G0-G1 transition (priming) within 4–6 h. By 24 h after PHx, the majority of hepatocytes are in the S phase, as detected by bromodeoxyuridine (BrdU) incorporation and expression of S phase markers, followed at ∼48 h by replication of nonparenchymal cells (NPCs), with recovery of liver mass within 1–2 wk (11, 18). Liver regeneration is clinically important in tissue resection procedures to remove tumors and in live-donor liver transplantation. Liver regeneration is suppressed and delayed by chronic alcohol intake, evident in an almost-complete absence of the peak of the S phase at 24 h, with some recovery at later times (6, 41). The defects in liver regeneration may contribute to alcoholic liver disease.
How ethanol treatment affects the regeneration response is not well understood. Early cellular stress responses and cytokine-mediated hepatocyte-NPC interactions that initiate the coordinated and integrated repair response may be the targets. Previous studies from our group and others showed that adaptation to chronic ethanol intake affects the cytokine profile driving the transition from quiescence to the cell cycle and alters gene expression dynamics and transcription factor-binding profiles after PHx (2, 3, 41). How adaptation to ethanol elicits these changes remains unresolved.
Recently, we reported that several microRNAs (miRNAs) show dynamic responses after PHx (7, 8), with multiple miRNAs exhibiting different responses to PHx in ethanol-fed rats and pair-fed controls (8). Of particular interest was the differential response of miRNA-21 (miR-21). This miRNA is thought to be proproliferative, is highly overexpressed in hepatocellular carcinoma and other cancers, and is upregulated in rodent liver after PHx (19). In a recent study, partial suppression of miR-21 after PHx in mice inhibited cell proliferation in remnant livers, presumably by targeting cyclin D1 expression through the Rho-related GTP-binding protein (RhoB)/Akt/mammalian target of rapamycin (mTOR) pathway (28). We were surprised to find that miR-21 was significantly elevated in livers from ethanol-fed rats at 24 h after PHx relative to controls, despite near-complete suppression of cell proliferation. A comprehensive target analysis indicated that although traditional proproliferative miR-21 targets were not engaged in the remnant livers of ethanol-fed animals, several antiproliferative miR-21 targets were (7). Thus, under these conditions, the miR-21 increase is not associated with a proproliferative profile, and its target binding is context-dependent.
Here, we analyzed the role of miR-21 in cell proliferation during liver regeneration by treating animals with AM21, a miR-21-specific inhibitory mixed locked nucleic acid (LNA)-DNA analog complementary to the 5′ end of miR-21.
METHODS
Animals.
All animal protocols were approved by the Institutional Animal Care and Use Committee.
Male Sprague-Dawley rats (Charles River, Wilmington, MA) were fed an ethanol-containing liquid diet (36% of calories from ethanol, 11% from carbohydrate, 35% from fat, and 18% from protein; Bio-Serv, Frenchtown, NJ) for 5 wk; for littermate pair-fed controls, ethanol calories were replaced with maltose-dextran (22). Male Sprague-Dawley rats (Harlan, Indianapolis, IN) with ad libitum access to standard chow and water also were used as dietary controls. Animals were maintained on a 12:12-h light-dark cycle. PHx was performed under isoflurane anesthesia by removal of the LLM lobe through surgical ligation (4, 15). For sham surgical controls, the liver was externalized and gently palpated to mimic the surgical stress of the PHx procedure. Surgeries were performed between 8 and 11 AM to avoid circadian effects. Where indicated, BrdU was injected (100 mg/kg ip) 1.5 h before the animals were euthanized. Remnant liver (PHx sample) was harvested at 24, 48, or 80 h after surgery. Samples were freeze-clamped (29) and stored at −80°C, except for a small formalin-fixed section, which was used for histology. LLM tissue was retained at the time of surgery and used as within-animal control to diminish the effect of biological variation. To inhibit miR-21, rats were injected with AM21 (7.5 mg/kg ip), an LNA-modified oligonucleotide antisense to miR-21 (Exiqon, Vedbaek, Denmark), in 1 ml of saline. AM21 has a sequence 5′-TCAGTCTGATAAGCT-3′, with LNA nucleotides underlined, and contains a phosphorothioate backbone. AM21 was administered twice, at 72 h prior to PHx and immediately following PHx, for a total dose of 15 mg/kg. Controls were treated with saline (1 ml) or scrambled oligonucleotide (7.5 mg/kg per injection in 1 ml of saline) or left untreated.
RNA isolation.
An animal tissue RNA purification kit (Norgen Biotek, Thorold, ON, Canada) was used to isolate total RNA from frozen liver tissue. RNA concentration was determined using a spectrophotometer (ND-1000, NanoDrop, Wilmington, DE).
miR-21 expression.
miR-21 was measured by TaqMan RT-quantitative PCR (qPCR) miRNA assay (Applied Biosystems, Foster City, CA) according to the manufacturer's recommendations. Briefly, 10 ng of total RNA were reverse-transcribed, and 1 μl of RT product was amplified in triplicate using TaqMan Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems) on a sequence detection system (Prism 7000, Applied Biosystems). Relative expression was determined with the cycle threshold (ΔΔCt) method; miR-21 expression was normalized to small nucleolar RNA expression.
High-throughput qPCR.
High-throughput gene and miRNA expression analysis was performed following BioMark (Fluidigm, South San Francisco, CA) qPCR protocols 8 and 27. For gene expression analysis, cDNA was synthesized from total RNA (1.2 μg) using the EasyScript Plus cDNA synthesis kit (Applied Biological Materials, Richmond, BC, Canada). Primers (see Supplemental Table S1 in Supplemental Material for this article available online at the Journal website) were designed using Universal Probe Library Assay Design Center software (Roche Applied Science, https://www.roche-applied-science.com/sis/rtpcr/upl/index.jsp). cDNA (100 ng) was preamplified for 12 cycles using TaqMan PreAmp Master Mix (Applied Biosystems). For miRNA expression analysis, total RNA (300 ng) was reverse-transcribed using a pool of TaqMan RT-qPCR miRNA assay (Applied Biosystems) RT primers, and product was preamplified for 15 cycles using TaqMan PreAmp Master Mix and Megaplex Rodent Pool A PreAmp primers (Life Technologies). All qPCRs were performed using BioMark Dynamic Arrays (Fluidigm) with 40 amplification cycles (15 s at 95°C, 5 s at 70°C, 60 s at 60°C). Relative gene expression was determined by the −ΔΔCt method. The modified geNorm algorithm (40) in R was used to identify the set of most stable reference genes and miRNAs. miR-103, miR-16, and miR-145 were selected for miRNA expression normalization. Idh3B, Mrpl16, and Ubqln1 were used as reference genes.
Microarray gene expression data analysis.
Gene expression profiling was performed using Affymetrix Ra-Gene-2.0-ST arrays following the manufacturer's protocol. Raw expression data were logarithmically transformed and normalized across all samples using the robust multiarray-averaging algorithm; normalized data are available at the Gene Expression Omnibus (GEO) database (accession no. GSE67242). Data were filtered for genes with expression levels at log intensity >5 in at least one sample, yielding a total of 11,325 probe sets. These were filtered further for genes with 1) 1.5-fold PHx-induced up- or downregulation in the AM21-treated or untreated condition in the ethanol-fed group and 2) 1.5-fold AM21-induced change in response to PHx compared with the untreated condition in the ethanol-fed group.
Fold changes in response to PHx in AM21-treated and untreated conditions were computed for the two dietary groups (ethanol-fed and control), yielding a total of four ratios. We also computed two additional ratios representing the change in response to PHx due to AM21 in each dietary group, i.e., PHx-to-LLM ratio change in AM21-treated compared with untreated animals. These ratios were discretized as −1, 0, and +1 based on a threshold (≥1.5-fold up- or downregulation). Genes were categorized into patterns on the basis of whether AM21 treatment caused an upregulation or a downregulation of the response to PHx compared with the untreated condition in the ethanol-fed group.
Gene ontology (GO) terms enrichment analysis in AM21-responsive gene pattern groups was performed using the GOrilla tool (10); all genes with expression levels at a log intensity >5 in at least one sample were used as a background list.
Protein analysis.
Whole cell lysates were prepared from frozen liver tissue (∼50 mg) using RIPA buffer (Sigma-Aldrich, St. Louis, MO) containing cOmplete protease inhibitor cocktail (Roche Diagnostics, Basel, Switzerland) and phosphatase inhibitor (Sigma-Aldrich). Protein concentration was determined by bicinchoninic acid assay with an albumin standard on a Synergy HT microplate reader (BioTek Instruments, Winooski, VT). Equal amounts of protein were resolved by 10% Bis-Tris gel electrophoresis, transferred to nitrocellulose membranes using the iBlot dry blot system (Invitrogen), and detected via chemiluminescence. A list of antibodies is shown in Table 1. Blots were imaged on a Kodak Image Station 440 (Carestream Health, Rochester, NY). Protein levels were quantified by densitometry using Carestream Molecular Imaging software.
Table 1.
Antibodies used for Western blotting and immunohistochemistry
| Target | Catalog No. | Manufacturer | Source |
|---|---|---|---|
| Western blotting | |||
| Cyclin D1 | RM-9104-S1 | Thermo Scientific (Waltham, MA) | Rabbit |
| PTEN | sc-9145 | Santa Cruz Biotechnology (Dallas, TX) | Rabbit |
| p21 | sc-397 | Santa Cruz Biotechnology | Rabbit |
| PCNA | 2586 | Cell Signaling Technology (Danvers, MA) | Mouse |
| 4EBP1 | 9644 | Cell Signaling Technology | Rabbit |
| p4EBP1 | 9459 | Cell Signaling Technology | Mouse |
| Akt | 9272 | Cell Signaling Technology | Mouse |
| pAkt | 4051 | Cell Signaling Technology | Mouse |
| GAPDH | 2118 | Upstate Biotechnology (Lake Placid, NY) | Rabbit |
| Anti-rabbit | 31460 | Thermo Scientific | Goat |
| Anti-mouse | 31430 | Thermo Scientific | Goat |
| Immunohistochemistry | |||
| Anti-BrdU | OBT0030 | Accurate Chemical and Scientific (Westbury, NY) | Rat |
| Anti-Ki-67 | Ab16667 | AbCam (Cambridge, MA) | Rabbit |
| Anti-αSMA | Ab7817 | AbCam | Mouse |
| Anti-rat | AAR10B | AbD Serotech (Raleigh, NC) | Sheep |
| Anti-rabbit | BA-1000 | Vector Laboratories (Burlingame, CA) | Goat |
| Anti-mouse-Alexa Fluor 488 | A11001 | ThermoFisher Scientific | Goat |
PTEN, phosphatase and tensin homolog; PCNA, proliferating cell nuclear antigen; 4EBP1, 4E-binding protein-1; p4EBP1, phosphorylated 4EBP1; pAkt, phosphorylated Akt; BrdU, bromodeoxyuridine; αSMA, α−smooth muscle actin.
Immunohistochemistry.
Liver tissue was fixed overnight in 10% neutral buffered formalin followed by 70% ethanol, embedded in paraffin, and sectioned. For BrdU staining, sections were processed using the Vectastain ABC kit (Vector Laboratories, Burlingame, CA) according to the manufacturer's instructions. Diaminobenzidine (ImmPact DAB peroxidase substrate, Vector Laboratories) was used as detection agent (see Table 1 for a list of antibodies). Slides were counterstained with Harris hematoxylin. For each sample, three random fields were selected, total and positive cells were counted, and the mean percentage of labeled nuclei was calculated per sample. For α-smooth muscle actin (αSMA) staining, sections were probed using anti-αSMA antibody, which was detected using anti-mouse Alexa Fluor 488 secondary antibody (Life Technologies), and nuclei were counterstained with 4′,6-diaminido-2-phenylindole. αSMA staining was semiquantitatively scored by a modification of the method described by Schmitt-Graff et al. (35) as follows: 0 (no staining or <3% staining), 1 (3–33% staining), 2 (34–66% staining), and 3 (>66% staining). The periportal (zone 1), intermediate (zone 2), and perivenular (zone 3) regions of the liver lobule were scored separately. The sum of the average score for each zone was used to obtain the final “αSMA score” (range 0–9). At least five liver acini were evaluated in each sample (n = 4 for 80 h and n = 3 for 24 and 48 h).
Triglyceride assay.
Liver triglyceride content was analyzed by saponification of frozen liver tissue in ethanolic KOH and neutralized with MgCl2 as described by Salmon and Flatt (33), and assay for glycerol was performed using the Triglyceride LiquiColor Test Kit (Stanbio Laboratory, Boerne, TX) according to the manufacturer's instruction.
Statistics.
Power analysis conducted using G*POWER software (http://www.gpower.hhu.de/en.html) indicated >90% sensitivity for three biological replicates at a type I error of 5% when a ≥2-fold change and <20% SD of the fold change was expected, as was the case in our results. Statistical significance of differential expression was determined by ANOVA. Analysis of differential expression between PHx and LLM samples was paired by animal. Analysis of all other observations was unpaired. Statistical significance between cumulative distributions of predicted miR-21 target vs. nontarget gene expression was determined with a two-sided Kolmogorov-Smirnov test. The statistical significance of the difference in the number of predicted miR-21 targets between up- and downregulated groups (see Fig. 4B) was evaluated using Fisher's exact test. P values were calculated on the basis of a hypergeometric distribution that considered the random chance of observing at least as many predicted miR-21 targets as in the upregulated group, with the overall differentially regulated gene set considered the background set (i.e., union of up- and downregulated genes). For all measurements, P < 0.05 was considered significant.
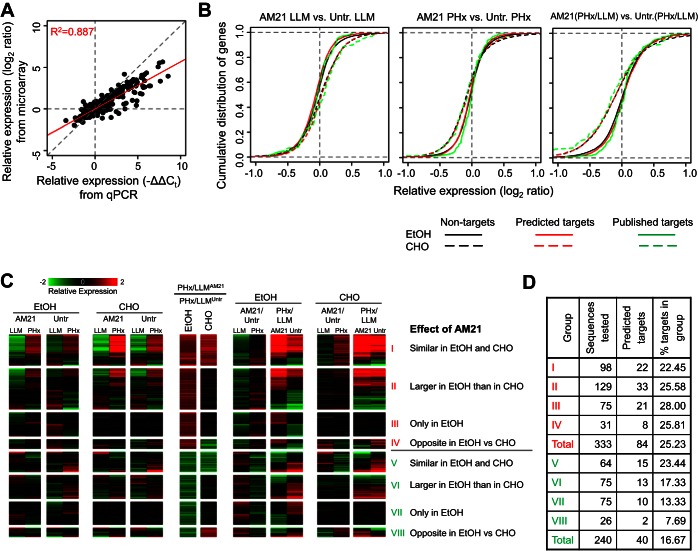
Fig. 4.
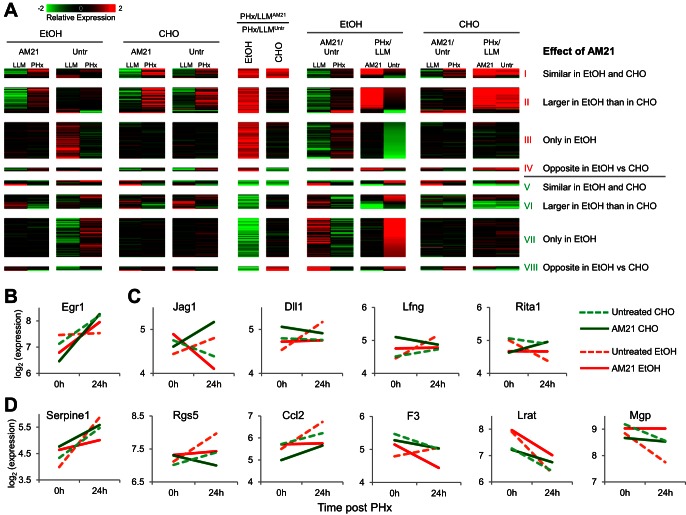
Effects of AM21 treatment on global gene expression patterns. A: expression changes measured in pooled samples using microarrays show a strong correlation with averaged expression changes of individual samples (n = 3, 904 points of comparison) measured by qPCR. Red diagonal line shows trend. B: cumulative distribution plots of differential expression of predicted (red curve) and published (green curve) miR-21 targets compared with differential expression of nontargets (black curve). Solid curves, ethanol diet; dashed curves, control carbohydrate diet. Untr, untreated. C: expression patterns of genes that showed an AM21-dependent PHx response in the ethanol-fed group. Genes were classified into 8 groups depending on the direction of the AM21 effect on the PHx response, as well as the similarity of this effect between the ethanol-fed and control groups. D: distribution of predicted miR-21 targets in 8 expression patterns of AM21-responsive genes.
RESULTS
miR-21 is effectively inhibited in vivo by AM21.
To inhibit miR-21 in vivo, rats were treated with a mixed LNA-DNA oligonucleotide complementary to the 5′ end of miR-21 (AM21) at 72 h before PHx followed by a second AM21 injection at the time of the surgery. Measurement of available miR-21 in the LLM samples at the time of PHx by qPCR demonstrates >97% inhibition of miR-21 (Fig. 1A). In AM21-treated animals, miR-21 was also undetectable in other tissues (heart, skeletal muscle, adipose tissue, and intestine), but not in the brain (data not shown). Saline and scrambled oligonucleotide injections did not affect miR-21 levels in the LLM tissue, nor did they increase miR-21 in the remnant liver after PHx (Fig. 1A). AM21 treatment had no detectable effect on liver histology (Fig. 1B) or triglyceride accumulation in the regenerating liver (data not shown) during this exposure period.
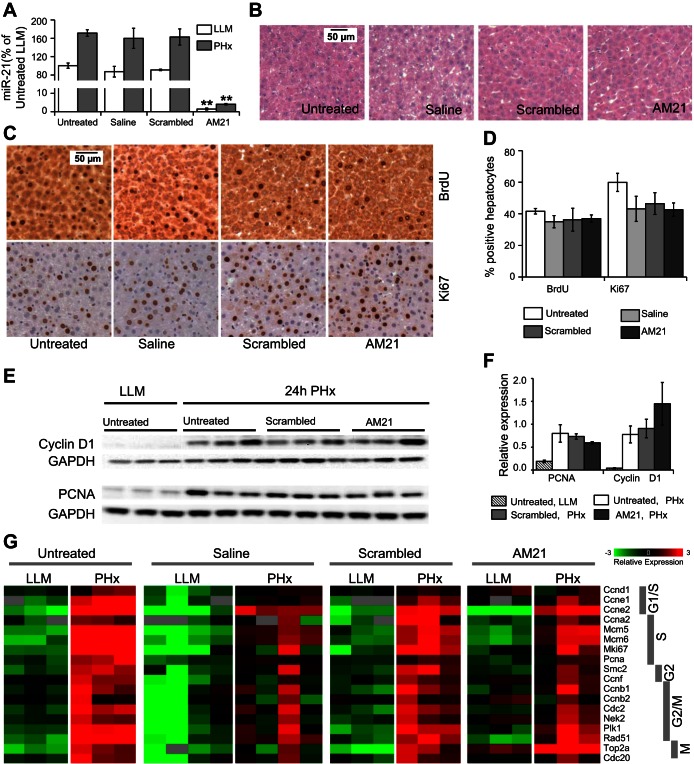
Fig. 1.
AM21 treatment does not affect cell cycle progression in regenerating liver of chow-fed rats. A: hepatic microRNA-21 (miR-21) is inhibited >97% by AM21 treatment. Control scrambled oligonucleotide and saline injections do not affect miR-21 expression levels in left lateral and medial (LLM) lobes or after 70% partial hepatectomy (PHx). miR-21 expression levels in rat liver were measured by quantitative PCR (qPCR). Relative expression levels compared with the mean of untreated rat LLM samples are shown. Values are means ± SE (n = 3). **P < 0.01 vs. corresponding untreated control. B: histology of hematoxylin-eosin-stained sections of AM21-treated LLM samples appears normal compared with untreated, saline, and scrambled oligonucleotide controls. C and D: bromodeoxyuridine (BrdU) incorporation and expression of cell cycle marker Ki-67 at the peak of the S phase, at 24 h after PHx, does not differ significantly between AM21-treated and control livers. E and F: cyclin D1 and proliferating cell nuclear antigen (PCNA) expression at 24 h after PHx are not altered by inhibition of miR-21. G: expression levels of cell cycle-related genes were measured by high-throughput qPCR and are presented as mean-centered −ΔCt. No significant difference in PHx-induced increase was observed between AM21-treated rats and any control groups.
Inhibition of miR-21 does not affect cell cycle progression during liver regeneration in chow-fed rats.
To examine the requirement for miR-21 in activating DNA synthesis in chow-fed rats, BrdU (100 mg/kg) was administered to AM21-treated and control animals 1.5 h before the remnant liver was harvested at 24 h after PHx, the peak of the S phase. No significant difference in BrdU incorporation was detected between AM21-treated and control groups (Fig. 1, C and D). Also, expression of the S phase marker Ki-67 was not affected by miR-21 inhibition (Fig. 1, C and D). Similarly, cyclin D1 and proliferating cell nuclear antigen (PCNA) protein levels at 24 h after PHx were unaffected by AM21 treatment (Fig. 1, E and F). In further experiments, we performed high-throughput qPCR to simultaneously measure expression levels of several cell cycle progression indicators (see Supplemental Data File S1). We did not observe any consistent negative effects of AM21 treatment on the expression of factors required for progression through the cell cycle at 24 h after PHx compared with untreated or scrambled oligonucleotide-treated animals. Saline-treated animals showed a variable expression profile, although BrdU incorporation and Ki-67 expression were similar to those in untreated or scrambled oligonucleotide-treated tissues (Fig. 1, C and D). Cell cycle-related genes were similarly increased in remnant livers from AM21-treated and control groups at 24 h after PHx (Fig. 1G). Thus these data indicate that miR-21 is not required for cell cycle progression from the G1 to the M phase during liver regeneration in chow-fed rats.
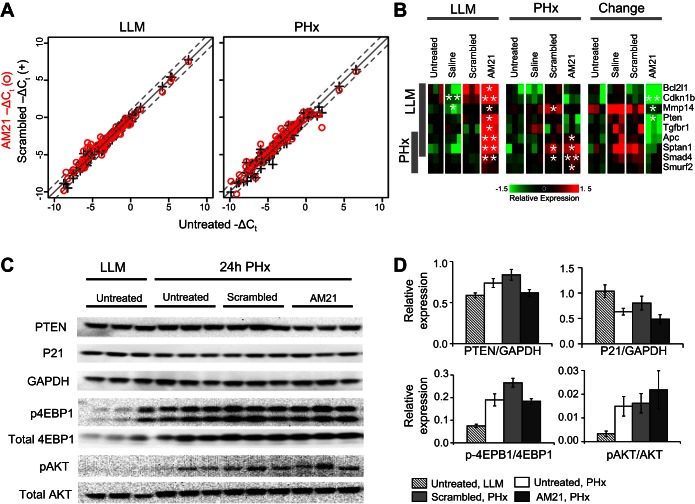
miR-21 inhibition has little effect on expression of miR-21 targets in chow-fed rats.
Using high-throughput qPCR, we also analyzed the effect of AM21 treatment on expression of 82 putative miR-21 targets reported in miRWalk (see Supplemental Data File S1). Despite complete inhibition of miR-21, the majority of these putative targets remained unchanged in LLM tissue or after PHx (Fig. 2A). Only a small subset of miR-21 targets analyzed was responsive to AM21 treatment (Fig. 2B). Well-recognized miR-21 targets (APC, Pten, and Cdkn1b), as well as several members of the Smad/TGFβ signaling pathway (Smad4, Smurf2, and Tgfbr1) were upregulated in response to miR-21 inhibition. However, protein levels of phosphatase and tensin homolog (PTEN) and p21 were not significantly affected by miR-21 inhibition (Fig. 2, C and D). Inhibition of miR-21 was reported to upregulate RhoB expression in regenerating mouse liver, thereby causing inhibition of the Akt/mTOR/4E-binding protein (4EBP) pathway to suppress cyclin D1 expression (28). Neither significant RhoB upregulation (see Supplemental Data File S1) nor a decrease in Akt or 4EBP phosphorylation (Fig. 2, C and D) could be detected in miR-21-inhibited rat livers after PHx compared with controls. These findings demonstrate that miR-21 is not required for the onset and progression of liver regeneration after PHx in the rat. Furthermore, the mediators of miR-21 effects that are reportedly upregulated after PHx in mice upon inhibition of miR-21 do not appear to show similar effects in rats.
Fig. 2.
Effect of AM21 treatment on expression of miR-21 targets. A: scatter plots comparing expression levels of 82 putative miR-21 targets in AM21-treated, scrambled oligonucleotide-treated, and untreated samples. Expression levels were measured using high-throughput qPCR. B: a subset of putative miR-21 targets responsive to AM21 treatment. Results are shown as mean-centered −ΔCt. *P < 0.05 and **P < 0.01 vs. corresponding untreated control. C: Western blots showing expression levels of putative miR-21 targets and Akt/mTOR signaling pathway proteins. D: relative protein levels [phosphatase and tensin homolog (PTEN), p21, phosphorylated 4E-binding protein (p-4EBP), and phosphorylated Akt (p-Akt)] compared with GAPDH or corresponding total protein. Values are means ± SE (n = 3).
AM21 treatment overcomes ethanol-induced suppression of liver regeneration.
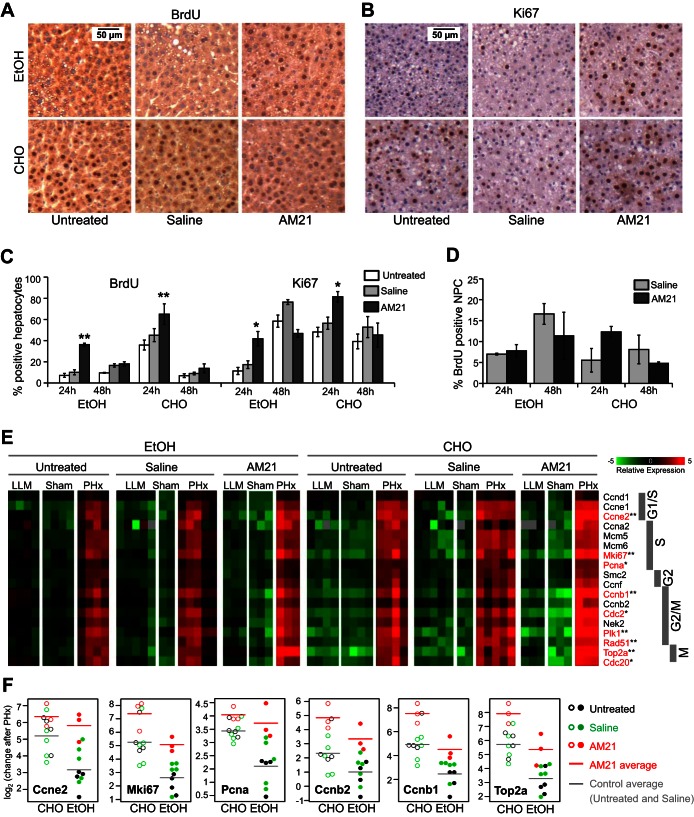
At the peak of DNA synthesis 24 h after PHx, 30–50% of hepatocytes show BrdU incorporation in remnant livers. However, chronic ethanol treatment substantially suppresses cell proliferation. Less than 10% of hepatocytes incorporated BrdU at 24 h after PHx, and BrdU incorporation remained low even at 48 h. Remarkably, AM21 treatment significantly increased BrdU incorporation in hepatocytes from ethanol-fed animals to levels comparable to untreated controls at 24 h after PHx (Fig. 3, A and C). Ki-67 expression was also increased in AM21-treated ethanol-fed rats at 24 h after PHx (Fig. 3, B and C). DNA synthesis in NPCs was not significantly affected by AM21 treatment within 48 h after PHx (Fig. 3D).
Fig. 3.
AM21 treatment recovers cell proliferation after PHx in ethanol-fed rats. A–C: AM21 treatment increased BrdU incorporation in hepatocytes of ethanol (EtOH)-fed animals to levels comparable to untreated carbohydrate (CHO)-fed controls. Expression of Ki-67 was also increased in AM21-treated rats. Values are means ± SE (n = 3). *P < 0.05 and **P < 0.01 vs. corresponding untreated control. D: AM21 treatment did not significantly affect BrdU incorporation in nonparenchymal cells (NPCs) within 48 h after PHx. E: expression level of cell cycle-related genes measured by qPCR and shown as mean-centered −ΔCt. Genes showing significantly lower upregulation after PHx in untreated ethanol-fed rats than untreated pair-fed controls are shown in red. *P < 0.05 and **P < 0.01. F: a subset of cell cycle-related genes showed significantly higher upregulation after PHx in AM21-treated ethanol-fed rats than untreated controls (P < 0.05).
Gene expression analysis confirmed the delay in cell cycle progression in ethanol-treated rats (see Supplemental Data File S1). Cell cycle genes from the G1 to the M phase were significantly upregulated in carbohydrate diet-fed controls at 24 h after PHx. Cell cycle genes were also upregulated in ethanol-treated animals after PHx, but changes in the majority of these genes, including cyclins E2 and B1 and PCNA, were significantly lower than in pair-fed controls (Fig. 3E). Sham surgery had no effects on the expression of cell cycle genes in any treatment group. As in chow-fed animals, inhibition of miR-21 by AM21 pretreatment did not cause downregulation of cell cycle-related genes (Fig. 3E). By contrast, AM21 treatment significantly enhanced the upregulation of several cell cycle genes, particularly in ethanol-fed animals, where the expression of Ki-67, PCNA, cyclins E2, B1, and B2, and topoisomerase 2 was increased to levels similar to those in control animals (Fig. 3F).
AM21 induces broad gene expression changes in ethanol-fed and control diet groups.
Microarray analysis was used to assess global effects of AM21 treatment on gene expression in regenerating rat livers. Pooled samples from three animals were analyzed using microarrays and compared for validation with the mean expression changes of individual samples assayed by high-throughput qPCR (Fig. 4A). Comparison of 904 data points for these two platforms showed strong correlation, with a Pearson's product-moment correlation coefficient of 0.887 (P < 2.2 × 10−16).
We assayed global gene expression changes in response to PHx and assessed how AM21 treatment modified this response in ethanol-fed and control groups. AM21-induced gene expression changes are complex, and most of those changes do not show profiles compatible with direct regulation by miR-21. AM21 treatment did not result in a statistically significant positive shift of the cumulated distribution of genes with predicted miR-21 binding sites or published miR-21 targets compared with nontargets (Fig. 4B).
For a more detailed analysis, we filtered the gene expression results to focus on AM21-mediated changes after PHx in the ethanol-fed group, yielding a total of 591 genes. We analyzed these results using a pattern analysis focusing on identifying groups of genes with similar directionality of change due to AM21 treatment. This yielded eight patterns, with four distinct cases each for AM21-mediated up- and downregulation in response to PHx (Fig. 4C; see Supplemental Data File S2). In pattern I, AM21 caused similar upregulation of PHx responses in ethanol-fed and control groups. This set includes both up- and downregulated genes. In the upregulated genes, AM21 amplified the response to PHx; in the downregulated genes, AM21 attenuated the downregulation response. Pattern II is characterized by a larger upregulatory AM21-induced shift in the ethanol-fed than control group. In pattern III, a shift toward upregulation occurred only in the ethanol-fed group. In pattern IV, a divergent effect was characterized by a shift toward upregulation in the ethanol-fed group but further attenuation for the same genes in the control group. Patterns V–VIII correspond to AM21-mediated shifts toward downregulation and follow a grouping similar to that described above. Patterns I and II contain several genes corresponding to cell cycle and cell proliferation, indicating that a key component of the AM21 effect in the ethanol-fed group is a shift in the response of cell cycle genes toward upregulation after PHx. The distribution of potential miR-21 targets was different among genes that showed a differential response to PHx only in untreated ethanol-fed rats compared with pair-fed controls and recovered normal expression patterns after AM21 treatment. There were significantly more potential miR-21 targets in the set of genes that is upregulated by AM21 treatment (25%) than in the downregulated set (16%, P = 0.0087; Fig. 4D).
Notably, the global gene expression data indicated that AM21 enhanced expression of proproliferative genes after PHx not only in ethanol-fed rats, but also in animals that received the control liquid diet. This phenomenon was also evident in the BrdU incorporation analysis shown in Fig. 3C, where >60% of cells were labeled in the AM21-treated animals receiving the control diet at 24 h after PHx. Global gene expression analysis confirmed the proproliferative effects of AM21 in regenerating liver in liquid diet-fed animals. GO analysis of the genes that showed higher upregulation after PHx in AM21-treated rats than corresponding untreated controls identified multiple cell cycle-related GO categories as being significantly enriched in both ethanol- and pair-fed rats (see Supplemental Table S2). The observation that BrdU incorporation was significantly enhanced by AM21 treatment in the control liquid diet group, but not in the chow-fed animals, suggests that miR-21 may also contribute to suppression of the proproliferative microenvironment in this group. It remains to be determined whether this is a consequence of particular dietary component(s) (or lack thereof) in the liquid control diet or by more general physiological parameters, such as the restricted food intake in the pair-fed control liquid diet group relative to the ad libitum chow-fed animals.
Pattern analysis identifies key nonparenchymal processes normalized by AM21.
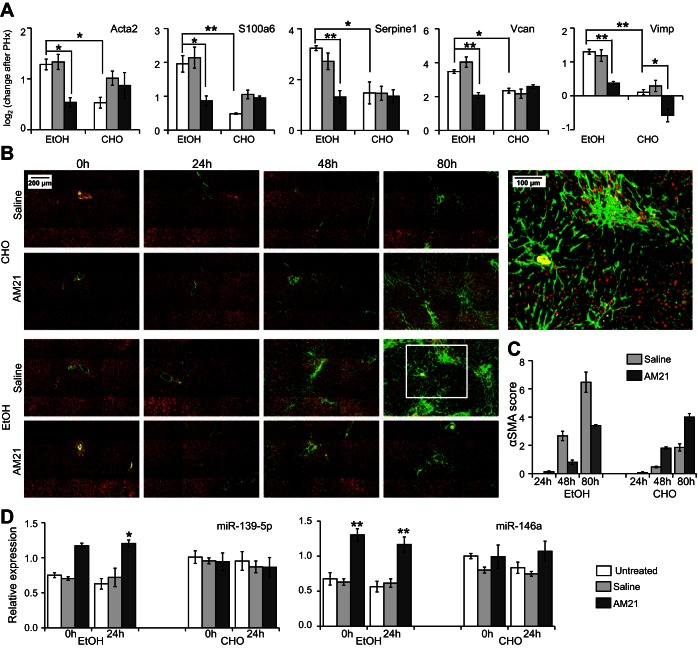
We analyzed the set of 591 genes with differential PHx response to AM21 treatment in the ethanol-fed group to identify those genes that showed a similar PHx response in the control untreated and AM21-treated groups and in the ethanol-fed group after AM21 treatment, i.e., the genes that showed a distinct PHx response only in the untreated ethanol-fed group that were normalized by AM21 treatment (referred to as the “AM21-renormalized set”). This filtering yielded 139 genes (see Supplemental Data File S2), largely derived from patterns III and VII, containing genes that were responsive only in the ethanol-fed group (Fig. 5A). A closer analysis of this set pointed to several key genes that were normalized by AM21 for their PHx response in the ethanol-fed group. Notably, Egr1 expression was unresponsive to PHx only in the untreated ethanol-fed group, and AM21 treatment recovered its upregulation (Fig. 5B). Early growth response 1 (EGR1) is a critical regulator of regeneration with upregulated expression and activity by 24 h in liver regeneration after CCl4-induced liver damage (30). EGR1 also contributes to increased proinflammatory responses in Kupffer cells after chronic ethanol exposure (31). In agreement with a role for EGR1 in regulating cell cycle progression, EGR1 knockout mice exhibit a delayed regenerative response after PHx and in a CCl4-induced liver injury model (21, 30). Our analysis also identified components of the Notch signaling pathway that were differentially responsive to AM21 treatment in the ethanol-fed group. Both ligands Jag1 and Dll1 and the positive regulator Lfng were upregulated, whereas the negative regulator Rita1 was downregulated, in the untreated group (Fig. 5C). AM21 treatment abrogated or reversed these changes to normalize the ethanol-fed group to response patterns similar to those in control groups. We also identified several genes associated with the hepatic stellate cell (HSC) activation state in the AM21-renormalized set. For instance, Serpine1 (plasminogen activator inhibitor 1), Rgs5, Ccl2, and F3 were significantly upregulated in the untreated ethanol-fed group, and these responses were attenuated after AM21 treatment (Fig. 5D). Lrat and Mgp were downregulated in both untreated ethanol-fed and untreated carbohydrate diet conditions. These genes have been reported to be downregulated during HSC activation (42), and AM21 treatment attenuated or abrogated this downregulation (Fig. 5D). To confirm the effects of AM21 treatment on HSC activation suggested by microarray data, we measured qPCR expression levels of several HSC activation-related genes that were too low to be detected by microarrays (see Supplemental Data File S1); inhibition of miR-21 suppressed upregulation of Acta2, S100a6, Serpine1, Vcan, and Vimp in ethanol-fed animals (Fig. 6A), supporting a role for HSC activation in ethanol inhibition of regeneration. In accordance with the changes in Acta2 expression, protein levels of αSMA significantly increased in the livers of ethanol-fed rats by 48 h and were markedly more pronounced at 80 h after PHx. Remarkably, this increase in αSMA protein was also prevented by AM21 treatment (Fig. 6B). However, we did not detect evidence of fibrosis in any of the regenerating tissues, as indicated by a lack of Sirius red staining, and there was no significant increase in the expression of type I collagen in the remnant liver of ethanol-fed animals (data not shown).
Fig. 5.
Patterns of gene expression normalized by AM21 treatment in the ethanol-fed group. A: expression patterns of select genes in C that showed a distinct PHx response only in the untreated ethanol-fed group. The pattern groups are same as those in C. Select genes are highlighted in B–D. B: upregulation of Egr1 was missing in the untreated ethanol-fed group and was restored to control levels after AM21 treatment. C: AM21 attenuated upregulation of Notch signaling components and prevented downregulation of Rita1, an inhibitor of Notch signaling. D: AM21 attenuates or prevents upregulation of genes corresponding to an activated hepatic stellate cell (HSC) phenotype (Serpine1, Rgs5, Ccl2, and F3). AM21 attenuated downregulation of genes that are typically at higher expression levels in quiescent than activated HSCs (Lrat and Mgp).
Fig. 6.
Inhibition of miR-21 suppressed upregulation of HSC activation-related genes and miRNAs in ethanol-fed rats after PHx. A: gene expression level measured by high-throughput qPCR presented as −ΔΔCt to corresponding LLM lobes. Values are means ± SE (n ≥ 3). *P < 0.05; **P < 0.01. B and C: α-smooth muscle actin (αSMA, green) staining was semiquantitatively scored by a modification of the method described by Schmitt-Graff et al. (35): 0 (no staining or <3% staining), 1 (3–33% staining), 2 (34–66% staining), and 3 (>66% staining). Periportal (zone 1), intermediate (zone 2), and perivenular (zone 3) regions of the liver lobule were scored separately. Sums of average scores for each zone were used to obtain the final “αSMA score” (range = 0–9). At least 5 liver acini were evaluated in each sample. Values are means ± SE (n = 4 for 80 h and n = 3 for 24 and 48 h). Nuclei stained red with 4′,6-diaminido-2-phenylindole. D: AM21 treatment upregulated expression of miR-146a and miR-139-5p in ethanol-fed animals. Expression level was measured by high-throughput qPCR and normalized to untreated control LLM lobes. Values are means ± SE (n = 3). *P < 0.05 and **P < 0.01 vs. corresponding untreated control.
The data shown above cannot distinguish direct and indirect effects of miR-21 inhibition. To assess whether effects of miR-21 inhibition could be mediated by changes in other hepatic miRNAs, we measured levels of 35 miRNAs in AM21-treated samples. miR-21 was the only miRNA consistently suppressed across all AM21-treated sample groups. miR-146a and miR-139-5p were significantly upregulated in AM21-treated ethanol-fed animals (Fig. 6C). Thus the effects of miR-21 inhibition may reflect the impact of a network of interacting miRNAs that collectively contribute to HSC activation.
DISCUSSION
In this study we used AM21, a LNA-DNA oligonucleotide antisense to the 5′ end of miR-21, to inhibit miR-21 in vivo. AM21 treatment was very effective in binding miR-21 in both chow-fed and liquid diet pair-fed animals, decreasing detectable levels of miR-21 by >97% in the liver. Of the 35 miRNAs analyzed, only miR-21 was consistently suppressed across all AM21-treated sample groups (see Supplemental Data File S1). Surprisingly, almost complete inhibition of miR-21 had relatively little effect on the expression levels of putative miR-21 targets. Only 12 genes of 82 targets tested were responsive to AM21 treatment, and these included such well-characterized miR-21 targets as APC, Pten, Cdkn1b, and several members of the Smad/TGFβ signaling pathway (Smad4, Smurf2, and Tgfbr1). miRNAs can exert their effects on target genes by degradation of their mRNA target or by inhibition of translation (38). We cannot exclude effects on target expression at the protein level without expression changes being evident in the transcripts due to such translational effects of miR-21. However, protein levels of PTEN and p21 were not affected by miR-21 inhibition in our model (Fig. 2C). It has also been suggested that LNA-DNA antisense oligonucleotide inhibitors such as AM21 may themselves act as novel miRNA mimics that could directly affect gene expression patterns (16). However, only a small fraction of analyzed miR-21 targets have potential binding sites for the AM21 sequence (of 82 putative miR-21 targets, 14 genes have a predicted AM21 binding site), and the possibility of direct inhibition of expression of these genes by AM21 acting as a miRNA mimic cannot explain the lack of response in these miR-21 target genes. Thus the modest effect of AM21 treatment on expression of well-characterized miR-21 targets more likely can be explained by adjustments of the miRNA regulatory network to prolonged inhibition of miR-21. These observations are compatible with other studies that indicate that biological consequences of miRNA expression are often context-dependent (36).
miR-21 is generally associated with highly proliferative states: it is highly overexpressed in the majority of cancer types (19) and has been demonstrated to be an oncomiR (25). In agreement with a proproliferative role, overexpression of miR-21 in cultured cells, including primary hepatocytes in culture, results in increased proliferation (1). Several groups, including our own, reported increased levels of miR-21 during the first round of hepatocyte proliferation after PHx (1, 7, 8, 23, 37). miR-21 has been suggested to contribute to cell cycle progression during mouse liver regeneration through its effects on cyclin D1 levels by direct targeting of RhoB and regulation of Akt/mTOR signaling (28, 37). However, in our current study almost complete inhibition of miR-21 did not have negative effects on cell cycle progression in the regenerating liver of chow-fed rats. AM21-treated rat livers did not show higher RhoB expression or decreased Akt or 4EBP phosphorylation compared with control tissue. The impact of miR-21 inhibition may be different in an acute surge suppression model as applied by Ng et al. (28) compared with complete inhibition over a longer period of time. Alternatively, the disparate outcome of miR-21 inhibition observed in the mouse (28) and rat (this study) may be due to the difference in timing of cell cycle progression between these two species. In the regenerating rat liver, the peak of DNA synthesis occurs at 24 h after surgery. Depending on the strain of the mice, the peak of the S phase occurs at 36–42 h after PHx (11). Despite different timing of cell cycle events, the temporal expression profile of miR-21 is the same in regenerating livers of both rats and mice, occurring at 12–24 h after PHx (1, 7). Our previous studies demonstrated that upregulation of hepatic miR-21 expression in chronic ethanol-fed animals, which have a defective liver regeneration response, is ∼50% higher than in their corresponding pair-fed controls. This overexpression of miR-21 may be a consequence of an ineffectual progression through the cell cycle, reflecting a compensatory response of the tissue. However, an equal increase in miR-21 expression after sham operation and after PHx indicates that miR-21 does not drive liver regeneration to the S phase in the rat (7). Together, these results suggest that the role of miR-21 in regeneration of the normal resected rat liver is not adequately described by its proproliferative effect.
By contrast, AM21 treatment significantly enhanced the DNA synthesis and expression levels of multiple cell cycle-related genes in chronic ethanol-fed animals subjected to PHx, bringing them to levels comparable to those of untreated pair-fed controls.
Although suppression of liver regeneration by chronic ethanol treatment has been widely reported (5, 6, 9, 41), the underlying mechanism(s) remains largely unknown. A detailed study of the impact of chronic ethanol feeding on the transcriptional dynamics after PHx was recently published by our group (20). Using a newly developed comparative pattern analysis, we demonstrated significant differences between control and ethanol-fed animals in the response to PHx at the time of hepatocyte priming (6 h) and at the peak of the S phase (24 h), time points when a surge of gene expression occurs, indicative of a major restructuring of liver homeostasis. The data reported in that study indicate that the adaptation to chronic ethanol intake, while having only a modest impact on the gene expression profile prior to PHx, markedly affects the acute response capacity to the PHx surgery. The present study complements the previous work by exploring the role of miR-21 in directing the response of the liver to the acute challenge of PHx and elucidating how adaptation to chronic ethanol intake alters this response capacity.
The target(s) of miR-21 that contributes to suppression of the regeneration response in ethanol-treated animals remains to be unequivocally identified. Our data do not indicate that inhibition of miR-21 alters the capacity for ethanol metabolism, as reflected in the expression profile of relevant genes (see Supplemental Table S2). Several key regulators of cell cycle progression are potential direct targets of miR-21. The target prediction program RNA22 predicts miR-21 binding sequences in the 5′-untranslated region of cyclin E2; hence, more robust upregulation of this gene in the regenerating livers of AM21-treated than control rats may be a direct consequence of miR-21 inhibition. Cyclin D1 has a predicted miR-21 binding site in its 3′-untranslated region. However, this gene did not respond to AM21 treatment. In agreement with the positive effects of AM21 treatment on cell cycle progression described above, inhibition of miR-21 also restored upregulation of Egr1 in the livers of chronic ethanol-fed rats to levels comparable to those in untreated pair-fed controls. This proproliferative transcription factor is required for normal progression of liver regeneration; Egr1 knockout mice show significant delay in cell cycle progression after PHx and in a CCl4-induced model of liver damage (21, 30).
Furthermore, miR-21 may play an important role in hepatic fibrogenesis. Elevated hepatic miR-21 levels were reported in an animal model of liver cirrhosis and in human patients, while downregulation of miR-21 inhibited primary HSC activation (44). Our data suggest that inhibition of miR-21 prevents an HSC activation switch toward a profibrogenic phenotype, which we observed in livers of untreated chronic ethanol-fed rats after PHx (20). The marked upregulation of such HSC activation-related genes as Acta2, S100a6, Vcan, Vimp, Serpine1, Rgs5, Ccl2, and F3, as well as the increased expression of αSMA protein, particularly at 48 and 80 h in the ethanol-fed animals, was effectively attenuated by AM21 treatment. Consistent with these results, the downregulation of the molecular signature for the quiescent state of HSC (expression of Lrat and Mgp) is also reversed by miR-21 inhibition. It should be emphasized, however, that the upregulation of markers for HSC activation in the ethanol-fed animals after PHx was not associated with a detectable increase in expression of type I collagen or deposition of extracellular matrix, suggesting that the αSMA protein expression reflected only a partial HSC activation. Further characterization of the HSC activation state and the mechanism by which it is induced in response to miR-21 is the subject of ongoing studies.
Inhibition of miR-21 also affected the expression levels of several members of the Notch signaling pathway. There is no conclusive evidence that Notch signaling in hepatocytes is required for normal progression of liver regeneration after PHx, but it may regulate interactions between different liver cell types involved in the repair process. Misregulation of Notch signaling is implicated in liver fibrosis by HSC activation (34) or modulation of inflammatory responses (13).
Inhibition of miR-21 was associated with a significant upregulation of miR-139-5p and miR-146a in ethanol-fed animals. Anticorrelative expression profiles of miR-139-5p and miR-21 have been observed in breast cancer patients (32). miR-139-5p directly targets Notch1 (26, 43). Although we did not observe significant changes in Notch1 levels in AM21-treated samples, cell type-specific effects on its expression are possible and, by suppression of Notch signaling in HSC upregulation of miR-139-5p, may contribute to attenuation of profibrotic tendencies in livers from ethanol-fed animals. Also, several studies reported anticorrelative expression of miR-21 and miR-146a during HSC activation in vitro (24, 44), as well as in hepatocellular carcinoma (17). miR-146a also suppresses the response of HSCs to TGFβ through a direct effect on Smad4 expression (14). Figure 7 illustrates schematically how multiple stress signaling pathways (NF-κB, Stat3, TGFβ, and Notch) may play a role in integrating the actions of these miRNAs into an interacting network that can target HSC activation.
Fig. 7.
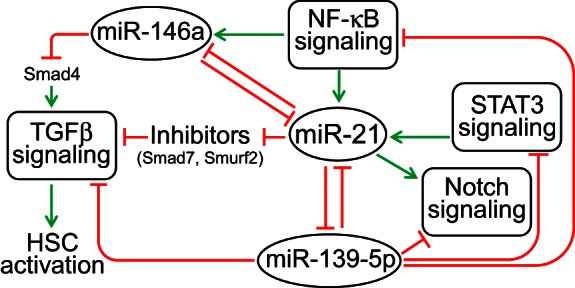
miR-21-centered miRNA regulatory network in the regenerating liver.
In summary, the findings reported in this study suggest that AM21 treatment may promote the regenerative process in chronic ethanol-fed animals at least in part by reversing antiproliferative profibrogenic processes driven by the shifting balance of HSC functional states. Figure 8 illustrates our current understanding of how adaptation to ethanol intake may affect the cell state transitions that accompany the regeneration process, with a specific focus on the interactions between hepatocytes and HSCs, and how miR-21 inhibition by AM21 (or corresponding changes in other interacting miRNAs) can shift the balance of these processes. This scheme emphasizes the point that such context-dependent effects of AM21 treatment are not restricted to hepatocytes themselves but may involve populations of NPCs that shape hepatocyte regenerative potential.
Fig. 8.
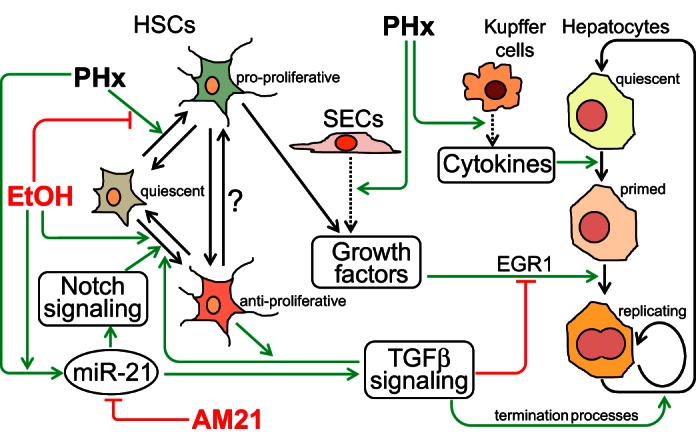
Summary of cell-cell interaction network through which AM21 affects ethanol suppression of liver regeneration after PHx. The scheme illustrates how enhanced upregulation of miR-21 in the liver of chronic ethanol-fed rats after PHx suppresses regeneration by promoting the antiproliferative phenotype of HSCs and TGFβ signaling. Inhibition of miR-21 by AM21 rescues hepatocyte proliferation by restoring the balance of pro- and antiproliferative cell phenotypes. SECs, sinusoidal endothelial cells.
GRANTS
This work was supported by National Institute of Alcohol Abuse and Alcoholism (NIAAA) Grants R01 AA-018873 and R21 AA-022417 to J. B. Hoek and R. Vadigepalli. E. Juskeviciute and R. P. Dippold were supported by training fellowships through NIAAA Grant T32 AA-007463.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
E.J., R.P.D., R.V., and J.B.H. developed the concept and designed the research; E.J., R.P.D., A.N.A., and A.S. performed the experiments; E.J., R.P.D., A.N.A., and R.V. analyzed the data; E.J., R.P.D., A.N.A., R.V., and J.B.H. interpreted the results of the experiments; E.J., R.P.D., A.N.A., and R.V. prepared the figures; E.J. and R.P.D. drafted the manuscript; R.V. and J.B.H. edited and revised the manuscript; R.V. and J.B.H. approved the final version of the manuscript.
Supplementary Material
REFERENCES
- 1.Castro RE, Ferreira DM, Zhang X, Borralho PM, Sarver AL, Zeng Y, Steer CJ, Kren BT, Rodrigues CM. Identification of microRNAs during rat liver regeneration after partial hepatectomy and modulation by ursodeoxycholic acid. Am J Physiol Gastrointest Liver Physiol 299: G887–G897, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cook D, Patra B, Kuttippurathu L, Hoek J, Vadigepalli R. A novel, dynamic pattern-based analysis of NF-κB binding during the priming phase of liver regeneration reveals switch-like functional regulation of target genes. Front Physiol 6: 189, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Correnti JM, Cook D, Aksamitiene E, Swarup A, Ogunnaike B, Vadigepalli R, Hoek JB. Adiponectin fine-tuning of liver regeneration dynamics revealed through cellular network modelling. J Physiol 593: 365–383, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crumm S, Cofan M, Juskeviciute E, Hoek JB. Adenine nucleotide changes in the remnant liver: an early signal for regeneration after partial hepatectomy. Hepatology 48: 898–908, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diehl AM, Chacon M, Wagner P. The effect of chronic ethanol feeding on ornithine decarboxylase activity and liver regeneration. Hepatology 8: 237–242, 1988. [DOI] [PubMed] [Google Scholar]
- 6.Diehl AM, Thorgeirsson SS, Steer CJ. Ethanol inhibits liver regeneration in rats without reducing transcripts of key protooncogenes. Gastroenterology 99: 1105–1112, 1990. [DOI] [PubMed] [Google Scholar]
- 7.Dippold RP, Vadigepalli R, Gonye GE, Hoek JB. Chronic ethanol feeding enhances miR-21 induction during liver regeneration while inhibiting proliferation in rats. Am J Physiol Gastrointest Liver Physiol 303: G733–G743, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dippold RP, Vadigepalli R, Gonye GE, Patra B, Hoek JB. Chronic ethanol feeding alters miRNA expression dynamics during liver regeneration. Alcohol Clin Exp Res 37: E59–E69, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duguay L, Coutu D, Hetu C, Joly JG. Inhibition of liver regeneration by chronic alcohol administration. Gut 23: 8–13, 1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fausto N. Liver regeneration. J Hepatol 32: 19–31, 2000. [DOI] [PubMed] [Google Scholar]
- 12.Garcin I, Tordjmann T. Calcium signalling and liver regeneration. Int J Hepatol 2012: 630670, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geisler F, Strazzabosco M. Emerging roles of Notch signaling in liver disease. Hepatology 61: 382–392, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Y, Huang C, Sun X, Long XR, Lv XW, Li J. MicroRNA-146a modulates TGF-β1-induced hepatic stellate cell proliferation by targeting SMAD4. Cell Signal 24: 1923–1930, 2012. [DOI] [PubMed] [Google Scholar]
- 15.Higgins GM, Anderson RM. Experimental pathology of the liver. Restoration of the liver of the white rat following partial surgical removal. Arch Pathol 12: 186–202, 1931. [Google Scholar]
- 16.Jin YY, Andrade J, Wickstrom E. Non-specific blocking of miR-17-5p guide strand in triple negative breast cancer cells by amplifying passenger strand activity. PLos One 10: e0142574, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karakatsanis A, Papaconstantinou I, Gazouli M, Lyberopoulou A, Polymeneas G, Voros D. Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog 52: 297–303, 2013. [DOI] [PubMed] [Google Scholar]
- 18.Koniaris LG, McKillop IH, Schwartz SI, Zimmers TA. Liver regeneration. J Am Coll Surg 197: 634–659, 2003. [DOI] [PubMed] [Google Scholar]
- 19.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med 13: 39–53, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuttippurathu L, Juskeviciute E, Dippold R, Hoek J, Vadigepalli R. A novel comparative pattern analysis approach identifies chronic alcohol mediated dysregulation of transcriptomic dynamics during liver regeneration. BMC Genomics 17: 260, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao Y, Shikapwashya ON, Shteyer E, Dieckgraefe BK, Hruz PW, Rudnick DA. Delayed hepatocellular mitotic progression and impaired liver regeneration in early growth response-1-deficient mice. J Biol Chem 279: 43107–43116, 2004. [DOI] [PubMed] [Google Scholar]
- 22.Lieber CS, DeCarli LM. The feeding of alcohol in liquid diets: two decades of applications and 1982 update. Alcohol Clin Exp Res 6: 523–531, 1982. [DOI] [PubMed] [Google Scholar]
- 23.Marquez RT, Wendlandt E, Galle CS, Keck K, McCaffrey AP. MicroRNA-21 is upregulated during the proliferative phase of liver regeneration, targets Pellino-1, and inhibits NF-κB signaling. Am J Physiol Gastrointest Liver Physiol 298: G535–G541, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maubach G, Lim MC, Chen J, Yang H, Zhuo L. miRNA studies in in vitro and in vivo activated hepatic stellate cells. World J Gastroenterol 17: 2748–2773, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 467: 86–90, 2010. [DOI] [PubMed] [Google Scholar]
- 26.Mi L, Chen Y, Zheng X, Li Y, Zhang Q, Mo D, Yang G. MicroRNA-139-5p suppresses 3T3-L1 preadipocyte differentiation through Notch and IRS1/PI3K/Akt insulin signaling pathways. J Cell Biochem 116: 1195–1204, 2015. [DOI] [PubMed] [Google Scholar]
- 27.Michalopoulos GK. Liver regeneration. J Cell Physiol 213: 286–300, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng R, Song G, Roll GR, Frandsen NM, Willenbring H. A microRNA-21 surge facilitates rapid cyclin D1 translation and cell cycle progression in mouse liver regeneration. J Clin Invest 122: 1097–1108, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palladino GW, Wood JJ, Proctor HJ. Modified freeze clamp technique for tissue assay. J Surg Res 28: 188–190, 1980. [DOI] [PubMed] [Google Scholar]
- 30.Pritchard MT, Malinak RN, Nagy LE. Early growth response (EGR)-1 is required for timely cell-cycle entry and progression in hepatocytes after acute carbon tetrachloride exposure in mice. Am J Physiol Gastrointest Liver Physiol 300: G1124–G1131, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pritchard MT, Nagy LE. Ethanol-induced liver injury: potential roles for egr-1. Alcohol Clin Exp Res 29: 146S–150S, 2005. [DOI] [PubMed] [Google Scholar]
- 32.Rask L, Balslev E, Sokilde R, Hogdall E, Flyger H, Eriksen J, Litman T. Differential expression of miR-139, miR-486 and miR-21 in breast cancer patients sub-classified according to lymph node status. Cell Oncol 37: 215–227, 2014. [DOI] [PubMed] [Google Scholar]
- 33.Salmon DM, Flatt JP. Effect of dietary fat content on the incidence of obesity among ad libitum fed mice. Int J Obes 9: 443–449, 1985. [PubMed] [Google Scholar]
- 34.Sawitza I, Kordes C, Reister S, Haussinger D. The niche of stellate cells within rat liver. Hepatology 50: 1617–1624, 2009. [DOI] [PubMed] [Google Scholar]
- 35.Schmitt-Graff A, Kruger S, Bochard F, Gabbiani G, Denk H. Modulation of α-smooth muscle actin and desmin expression in perisinusoidal cells of normal and diseased human livers. Am J Pathol 138: 1233–1242, 1991. [PMC free article] [PubMed] [Google Scholar]
- 36.Shields BB, Pecot CV, Gao H, McMillan E, Potts M, Nagel C, Purinton S, Wang Y, Ivan C, Kim HS, Borkowski RJ, Khan S, Rodriguez-Aguayo C, Lopez-Berestein G, Lea J, Gazdar A, Baggerly KA, Sood AK, White MA. A genome-scale screen reveals context-dependent ovarian cancer sensitivity to miRNA overexpression. Mol Syst Biol 11: 842, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song G, Sharma AD, Roll GR, Ng R, Lee AY, Blelloch RH, Frandsen NM, Willenbring H. MicroRNAs control hepatocyte proliferation during liver regeneration. Hepatology 51: 1735–1743, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szabo G, Satishchandran A. MicroRNAs in alcoholic liver disease. Semin Liver Dis 35: 36–42, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taub R. Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol 5: 836–847, 2004. [DOI] [PubMed] [Google Scholar]
- 40.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: 18, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang SQ, Lin HZ, Yin M, Albrecht JH, Diehl AM. Effects of chronic ethanol consumption on cytokine regulation of liver regeneration. Am J Physiol Gastrointest Liver Physiol 275: G696–G704, 1998. [DOI] [PubMed] [Google Scholar]
- 42.Yi SH, Zhang Y, Tang D, Zhu L. Mechanical force and tensile strain activated hepatic stellate cells and inhibited retinol metabolism. Biotechnol Lett 37: 1141–1152, 2015. [DOI] [PubMed] [Google Scholar]
- 43.Zhang L, Dong Y, Zhu N, Tsoi H, Zhao Z, Wu CW, Wang K, Zheng S, Ng SS, Chan FK, Sung JJ, Yu J. microRNA-139-5p exerts tumor suppressor function by targeting NOTCH1 in colorectal cancer. Mol Cancer 13: 124, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao J, Tang N, Wu K, Dai W, Ye C, Shi J, Zhang J, Ning B, Zeng X, Lin Y. MiR-21 simultaneously regulates ERK1 signaling in HSC activation and hepatocyte EMT in hepatic fibrosis. PLos One 9: e108005, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.