Summary
Effective immunity requires a complex network of cellular and humoral components that interact with each other and are influenced by different environmental and host factors. We used a systems biology approach to comprehensively assess the impact of environmental and genetic factors on immune cell populations in peripheral blood, including associations with immunoglobulin concentrations, from ∼500 healthy volunteers from the Human Functional Genomics Project. Genetic heritability estimation showed that variations in T cell numbers are more strongly driven by genetic factors, while B cell counts are more environmentally influenced. Quantitative trait loci (QTL) mapping identified eight independent genomic loci associated with leukocyte count variation, including four associations with T and B cell subtypes. The QTLs identified were enriched among genome-wide association study (GWAS) SNPs reported to increase susceptibility to immune-mediated diseases. Our systems approach provides insights into cellular and humoral immune trait variability in humans.
Graphical Abstract
Highlights
-
•
Understanding inter-individual variation of immune cells and immunoglobulin levels
-
•
Season and gender influence B cell subpopulation abundance
-
•
Identification of genetic loci that might regulate B cell levels in blood
-
•
Cell count QTLs overlap with risk SNPs for (auto)immune/inflammatory disease
As part of the Human Functional Genomics Project, this study by Aguirre-Gamboa et al. maps the contribution of genetics and non-heritable factors onto immune-cell counts and immunoglobulin levels. They find that season and gender influence the abundance of most of B cell subpopulations.
Introduction
Blood is a complex tissue consisting of a very specialized network of circulating immune cells and soluble factors that are the morphological substrate of the human immune response. Among immune cells, the monocyte, neutrophil, and natural killer (NK) compartments are essential for first-line, innate immune responses, while T cells, B cells, and the latter’s cognate immunoglobulin ([Ig] antibody) repertoire are essential for effective adaptive immune response to a wide variety of pathogens. Dysregulated immune cell or Ig numbers and/or functions can lead to an increased susceptibility to infections or to immune-mediated inflammatory disorders such as autoimmune diseases or allergy (Cho and Feldman, 2015, Tangye et al., 2012).
Both genetic and non-genetic factors may contribute to variations in the number and function of human immune cells, as well as the concentration of soluble mediators, resulting in considerable heterogeneity in individual immune responses. Recent cohort-based studies have highlighted the effect of both genetic (Brodin et al., 2015, Orrù et al., 2013, Roederer et al., 2015) and non-genetic factors, including cohabitation, chronic infection, aging, and microbiome (Carr et al., 2016, Roederer et al., 2015, Shaw et al., 2013) on the variation of human immune cell levels. However, a comprehensive analysis characterizing the interrelationship between different immune cell types (innate and adaptive) and Ig levels in freshly drawn (non-frozen) human blood as well as the effect of genetic and non-genetic factors on the variation in these immune traits has been lacking.
The Human Functional Genomics Project (HFGP) is an initiative comprising several cohorts of healthy individuals and patients that aims to identify the factors responsible for the variability of immune responses in health and disease (http://www.humanfunctionalgenomics.org). While three other studies accompanying this present study describe environmental (ter Horst et al., 2016), genetic (Li et al., 2016), and host microbiome (Schirmer et al., 2016) factors that affect pathogen-induced peripheral blood cytokine responses, this study is a comprehensive assessment of the impact of environmental and genetic host factors on circulating cell populations, focusing on both T cells and B cells and including associations of B cells with Ig concentrations. Our results provide a full picture of humoral immunity, as seen in serum Igs, and its interrelationship with immune cell levels.
We analyzed the determinants of variation in T and B cell counts and Ig levels by testing the association between immune traits and non-heritable factors such as age, gender, and season. We estimated the genetic heritability of different immune cells and show that the variation in T cell counts is predominantly (37%) explained by genetic factors, which is in contrast to B cell counts, which are more strongly influenced by the environment. We also tested the effect of genome-wide genetic variation on cell-level variation by using cell-count quantitative trait loci (ccQTL) mapping and identified eight independent genomic loci associated with lymphocyte counts, four of which have not been described before, and with four cell subsets that have not been characterized in previous studies. We also performed an integrative genomics analysis by using RNA-sequencing (RNA-seq) data from blood samples of 628 healthy individuals to identify putative causal genes, including long non-coding RNAs, at ccQTLs that may regulate cell counts. Lastly, we show that the genetics behind ccQTLs partially overlap with the previously described genetics of immune-mediated/related disease.
Results
Correlations of Cellular and Humoral Immune Compartments Highlight Factors that Drive Inter-individual Variation
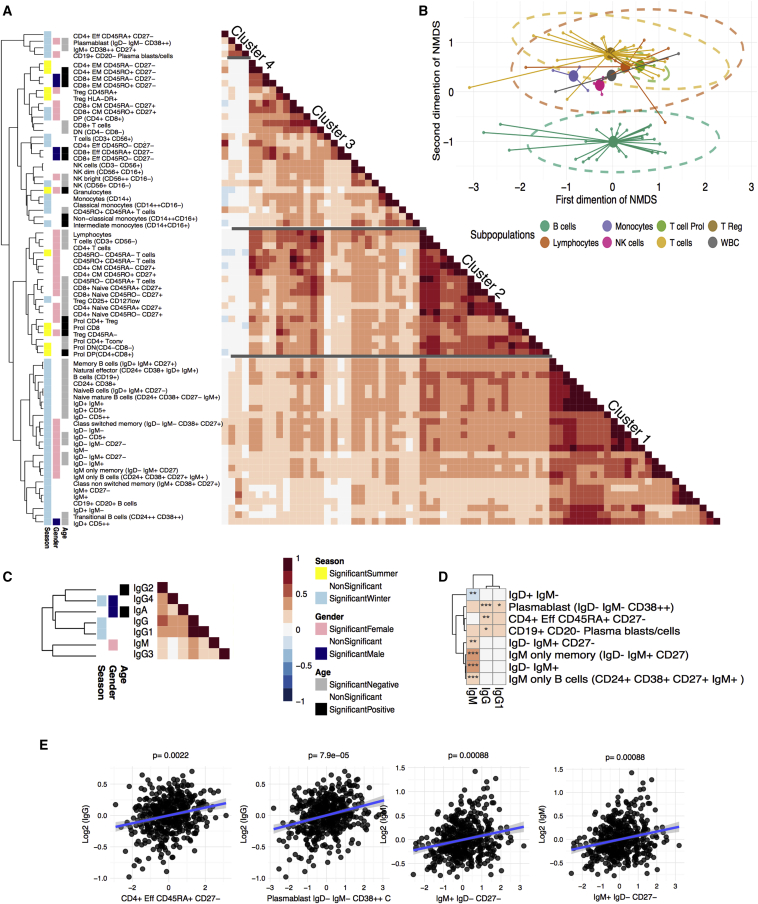
Both the cellular and humoral arms of our immune system are crucial for an effective immune response. However, information on the interrelationship between the cellular and humoral components is scarce. To analyze the underlying patterns of the variation within these immune components at the population level, we performed unsupervised hierarchical clustering within our measured immune cell populations and within Ig levels, after correcting for age, sex, and season effects. For immune cells, we identified four clusters of biological relevance (Figure 1A) in which subpopulations of B cells, T cells, and myeloid immune cells clustered into clusters 1, 2, and 3, respectively. Cluster 4 contains plasma cells and their precursors, as well as plasmablasts, with both groups clustering separately from the B cell cluster (cluster 1). A subpopulation of CD4+CD45RA+CD27− effector T cells was also present in cluster 4. These observations suggest that plasma cells and CD4+CD45RA+CD27− terminally differentiated effector T cells are co-regulated by similar factors. Moreover, using a nonmetric multi-dimensional scaling approach, we revealed, in a data-driven way, a separation between B cells and the other immune subpopulations at the second dimension (Figure 1B). This suggests that B cells might also be co-regulated independently of the other immune subsets.
Figure 1.
Interrelationship between Immune-Associated Cell Subpopulations and Immunoglobulin Levels in the General Population
(A) Unsupervised hierarchical clustering of the correlation within cell subpopulations.
(B) A two-dimensional representation of the correlations between each cell type by non-metric multidimensional scale analysis. Small circles represent individual cell types. Large circles represent the calculated centroid of the grouped cell types.
(C) Unsupervised clustering of immunoglobulin levels. The color code next to the dendogram represents any significant association of cell count with age, gender, or season.
(D) Heatmap of Spearman correlation coefficients between each independent cell subpopulation and immunoglobulin levels. Stars indicate significance of the correlation after FDR correction. ∗p ≤ 0.05, ∗∗p ≤ 0.005, ∗∗∗p ≤ 0.0005.
(E) Examples of cell subpopulations that are significantly associated with immunoglobulin levels. Regression line are included for visualization purposes.
The clustering patterns of Ig (sub)classes formed two major clusters, one containing IgM and IgG3 and the other containing IgG, IgG1, IgG4, and IgA (Figure 1C). For the IgM and IgG3 cluster, there is biological evidence associating these two humoral components. They are known to have the strongest complement binding capacity, a function that is required for optimal protection against (intracellular) pathogens (Schroeder and Cavacini, 2010). Interestingly, the regulation of both IgM and IgG3 appears to be controlled by the cytokines interleukin (IL)-4 and transforming growth factor β (TGF-β), indicating functional homogeneity under similar regulatory control (Brüggemann et al., 1987, Coffman et al., 1989, McIntyre et al., 1993, Snapper and Paul, 1987).
Having established the hierarchical clustering of immune cell populations and Ig levels, we analyzed the association between immune cell counts and Ig levels by using Spearman correlation (Figure 1). Out of 511 possible relations, nine significant correlations (false discovery rate [FDR] ≤ 0.05) were identified between Ig subclass and immune cell populations (Figure 1C). CD4+ effector T cells (CD45RA+ CD27−), which cluster with the plasma cells and plasmablasts (cluster 4), show a significant correlation with IgG levels (r = 0.2, p = 8.5e−6) (Figures 1D and 1E). This correlation may partly reflect the connection between these cell types in humans, where effective recall of antibody responses is dependent on T-cell-dependent memory B cell generation (Kurosaki et al., 2015). A significant correlation was also observed between IgM-only B cell levels and IgM serum levels (r = 0.24, p = 6.3e−8), and a negative was correlation observed between IgM serum levels and IgD+IgM− B cells (r = −0.2, p = 1.0e−8) (Figures 1C and 1D; Table S1). This correlation between IgM-only B cells in peripheral blood and IgM in serum suggests that high levels of IgM-only B cells predict higher levels of plasma cells in tissue. These results stress the importance of identifying the key factors driving the underlying inter-individual variation in the immune system.
Effect of Age, Gender, and Season on the Inter-individual Variation of Cellular and Humoral Immune Components
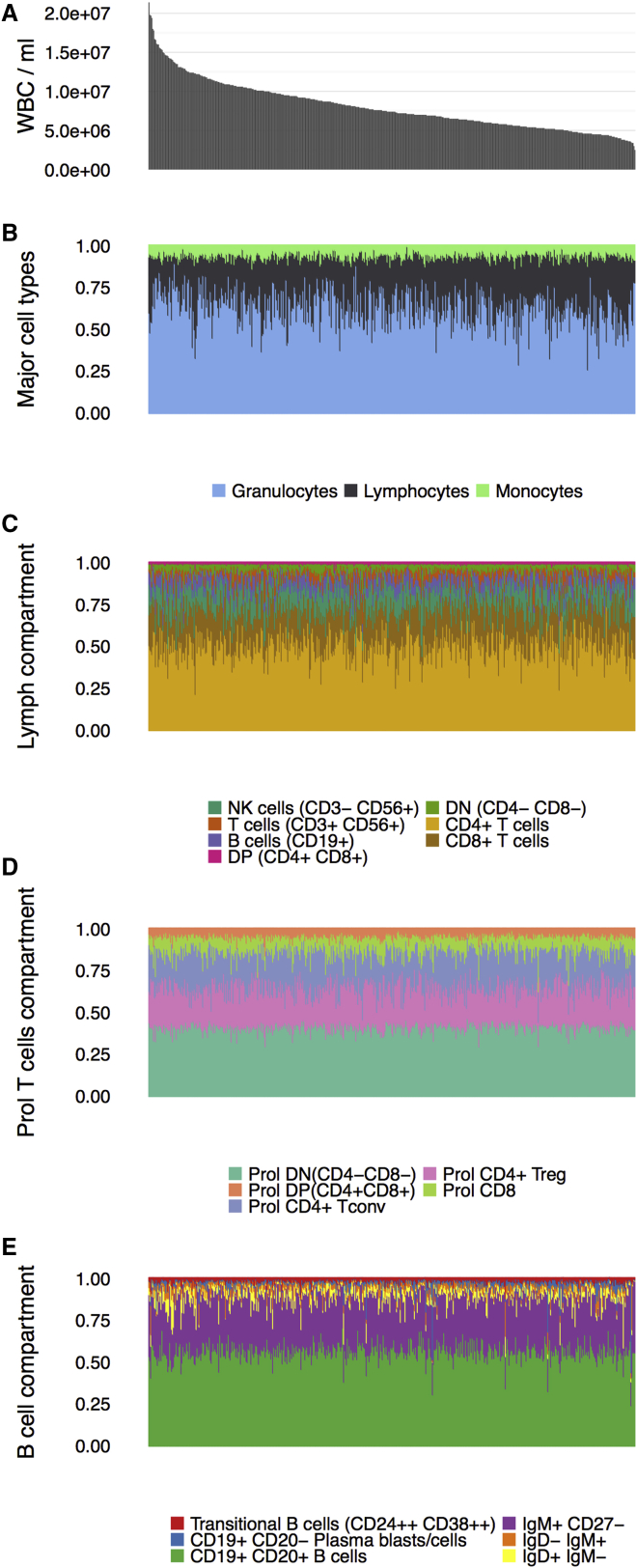
We investigated the distribution of immune cell counts and subset frequencies among ∼500 individuals in our cohort (500 Functional Genomics Project cohort [500FG] from the HGFP). We observed substantial variation in total white blood cell (WBC) counts (Figure 2A) and the levels of the lymphoid and myeloid cell populations (Figures 2B and 2E) between individuals. We then systematically tested the association of this variation with age, gender, and season.
Figure 2.
Variation of Cell Levels and Composition in the Dutch General Population
(A) Peripheral-blood white blood cell counts per ml blood (y axis) in 516 individuals (500FG cohort) (x axis).
(B–E) Relative cell proportions (y axis) of monocytes, lymphocytes and neutrophils (B), the lymphoid subpopulations (C), proliferating T cell subsets (D), and B cell subsets (E). Samples are presented in the exact same order in each figure.
Age Is Associated with Reduced Lymphoid but Increased Myeloid Cell Levels
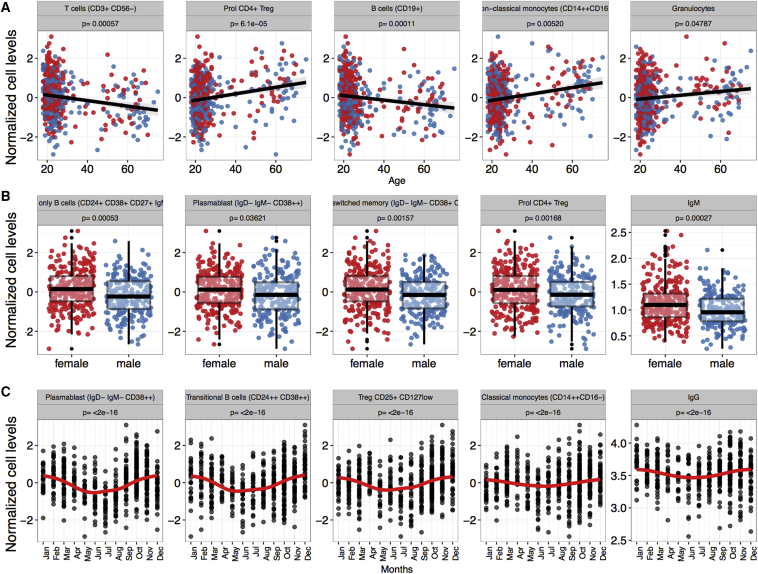
Aging plays a major role in shaping the immune profile (LeMaoult et al., 1997, Shaw et al., 2013, Solana et al., 2006). Using Spearman correlation, we observed consistent correlation with age (64% of the cell subpopulations studied are significantly correlated), both negative and positive. Aging was significantly associated (FDR ≤ 0.05, corrected for 73 tests) with a decrease in lymphoid immune cell levels (naive T cells, B cell subsets) and with a concomitant increase in myeloid immune cell levels of granulocytes, pro-inflammatory non-conventional monocytes (CD14++CD16+), and intermediate monocytes (CD14+CD16+) and levels of proliferating CD4+ regulatory T cells (Tregs) (Figure 3A; Table S2). To show the robustness of age effect on immune traits, we used a resampling. We randomly selected 90% of all the samples and tested for age effect on immune traits. We iterated this 100 times and observed that 91% of traits showed consistent results when compared with the original full dataset in more than 70% of the sampling iterations (Figure S1). We also compared the variation within cell counts in younger subjects (lower quartile of age distribution in the 500FG cohort; median age = 19 years) versus older subjects (upper quartile; median age = 65 years). We observed significant differences (p ≤ 0.05) in the variations of CD4+ (CD45RA−CD27+) effector T cell, NK cell (CD56+CD16−), and CD3+CD56+ T cell subpopulations (Figure S2A). Upon testing of associations between age and Ig levels, only IgG2 and IgA levels showed a significant positive correlation age (FDR ≤ 0.05, corrected for seven tests). These observations support the hypothesis that immune response shifts class in elderly individuals with de novo infections, with a restricted adaptive response being replaced by an innate type of immunity (Le Garff-Tavernier et al., 2010, Hazeldine et al., 2012, LeMaoult et al., 1997, Solana et al., 2006).
Figure 3.
Age, Gender, and Season Are Modulators of the Immune Traits
Examples of significant associations (FDR ≤ 0.05) between age (A), gender (B), or season (C) and cell counts or immunoglobulin levels.
Gender Is Associated with Different B Cell Subsets and Ig Levels
We observed a significant increase (FDR ≤ 0.05) in mature B cell subsets, IgM-only B cells, plasmablast B cells, proliferating and memory (CD45RA−) Treg cells, NK cell subsets, and IgM serum levels in women as compared to men (Figure 3B; Table S2). The significant association between higher levels of IgM-only B cells (p = 0.0005) and increased serum IgM levels (p = 0.0002) in women highlights the functional link between the cell type and its product (Amadori et al., 1995). By using the resampling approach, we observe that 87% of traits show consistent results when compared with the original full dataset in more than 70% of the iterations (Figure S1). In men, we observed an increased level of effector and effector memory T cells (Figure S2C) and a reduced level of IgG4 and IgA with nominal p values < 0.01 (see also ter Horst et al.)
Because we observed a significant effect of gender on different B cell and Ig levels, we investigated whether this effect was due to a difference in gender-associated hormone levels. We first tested whether the immune cell counts correlated with hormone levels in the 500FG cohort, but found no statistically significant correlation (Figure S2B). As expected, we observed lower testosterone concentrations in women than in men (Figure S2C). Although testosterone has been shown to inhibit Ig levels of human peripheral-blood mononuclear cells in vitro (Kanda et al., 1996), our analysis indicates that higher testosterone levels in women are significantly associated with increased IgG levels. Moreover, we observed a significant association of hydroxyprogesterone with IgG levels in women (Figure S2C). Hydroxyprogesterone levels vary with menstrual cycle, being highest in the luteal phase and lowest prior to ovulation. In men, this hormone showed less variation in serum levels.
Seasonal Variation Affects Both Cellular and Humoral Responses
We found a consistent seasonal effect on immune cell subpopulations, with 67% of the measured cell types showing a significant association with season (FDR ≤ 0.05). B cell subsets were the most consistently affected, with all B cell subpopulations showing significantly higher levels in winter. Treg, NK(T), and classical monocytes (CD14++CD16−) were also significantly higher in winter, while granulocytes, proliferating CD8+ T cells and CD4+ effector memory cells showed a higher peak during the summer months (Figure 3C; Table S2). IgG, IgG1, and IgG4 levels were also higher in winter, with nominal p values < 0.01 (see also ter Horst et al., 2016). By using the resampling approach, we observed that 94% of traits show consistent results when compared with the original full dataset in more than 70% of the iterations (Figure S1). Altogether, these results point to an important role for environmental factors that vary with season (e.g., allergies and viral infections) in the regulation of the magnitude of both the cellular and the humoral immune response (Dopico et al., 2015).
Genetic Factors Explain a Large Proportion of the Variation in Immune Traits
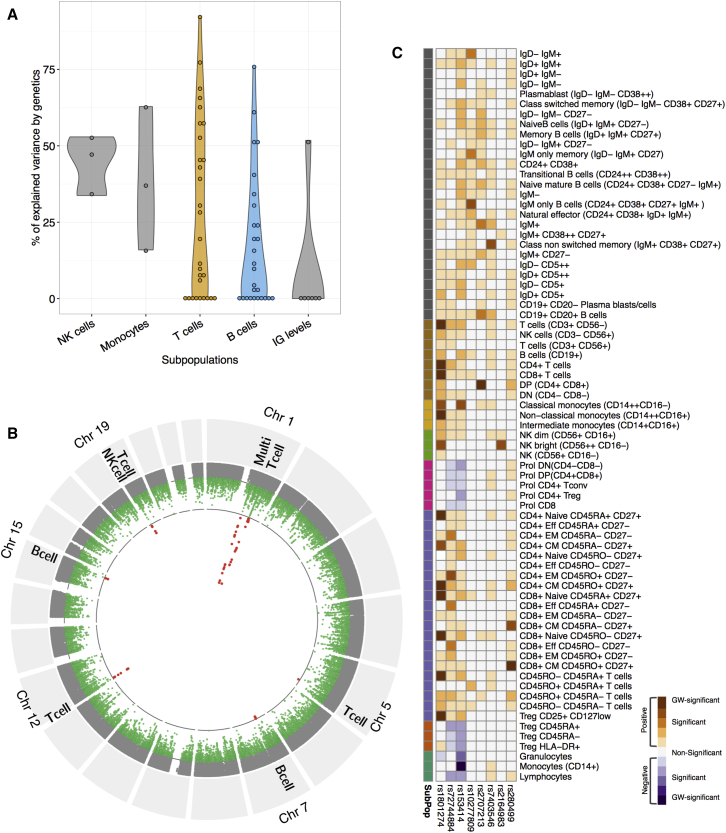
We observed that cell counts show high variability across individuals and that this variation could be partially ascribed to age-, gender-, or season-related factors. To further explore this inter-individual variation, we estimated the proportion of variance explained by genome-wide SNPs for each of 73 independent cell types after controlling for age, gender, and seasonal variation. As shown in Figure 4A and Figure S3, the majority of immune cell population variation is explained by non-heritable rather than heritable influences. The proportion of immune cell variation that was explained by genetics varies for each cell subpopulation. It was significantly higher for the 29 T cell immune traits as compared to the 27 B cell immune traits (median of 30% versus 18%, respectively; Student’s t test, p ≤ 0.05). Effector memory and effector CD4+ and CD8+ and CD4+ Tregs were also strongly influenced by genetic factors (Figure S3). The seemingly interdependent IgD+IgM+ and IgD+IgM− B cell populations showed completely opposing heritability estimates (Figure S3), likely reflecting the heterogeneity of the IgD+IgM+ population, which consists of both T-cell-dependent naive CD27− B cells and presumed T-cell-independent CD27+ memory B cells (Weller et al., 2004). Within the innate leucocytes, more than 50% of the variance in transitional monocytes (CD14+ CD16+), NK cells (CD3−CD56+), and NK-bright cells (CD56++CD16−) was explained by genetic variation. There is little contribution of genetics to the variation of granulocyte levels. Notably, 50% (± 20%) of the variance in IgM can be explained using genotype information. For the remaining Igs, we did not identify any contribution of genetics to the variance (Figure S3).
Figure 4.
The Genetics of Cell Counts and Immunoglobulin Level Variation in a General Population
(A) Violin plot representing the distribution of the percentage of variance explained by genetics for the immune traits. A total of 29 T cell subsets versus 27 B cell subsets were analyzed (mean percentages of variance explained by genetics of 29.5 versus 17.7, respectively; Student’s t test, p ≤ 0.05).
(B) Combined Manhattan plot of all cell types. Red dots mark genome-wide significant associations (p ≤ 5e−10). Immune cell types with the strongest association are indicated.
(C) Overview of the association of multiple genomic loci (ccQLTs) and immune cell types. Darkest colors indicate genome-wide significant ccQTLs, while divergence represents the direction of ccQLT effect.
Mapping of QTLs in the 500FG Cohort Identifies Eight Cell Count QTLs
To identify the genetic variants determining cell counts and Ig levels, we mapped ccQTL and Ig level QTLs (IgQTLs) using genome-wide SNP genotype data. After controlling for the effect of age, gender, and season, we identified eight independent genome-wide significant ccQTLs specific for three cell types: T cells (five ccQTLs), B cells (two ccQTLs), and NK cells (one ccQTL) (Figures 4B and 4C; Table 1 and Table S3). Four of these ccQTLs have been reported before (Table 1, Figures 4A–4D), providing validation for our analytical approach (Orrù et al., 2013, Roederer et al., 2015). The other four ccQTLs have not previously been associated to immune traits. One of these B cell ccQTL SNPs was also associated to Ig levels, although not at genome-wide significance (rs62433089, p < 5e−8) (Figure S4F). The higher numbers of T cell ccQTLs compared to B cell ccQTLs, when combined with our finding that a greater proportion of the variance in T cells (but not B cells) can be explained by genetics, would suggest a stronger genetic component for T cell immunity when compared to B cells. Furthermore, we also found that the IgG1 level is suggestively associated with a B-cell-specific ccQTL (rs10277809, p ≤ 0.001), implying a shared regulation of B cell and certain Ig levels in blood.
Table 1.
List of Eight Independent Genome-wide Significant Cell Count QTLs
| SNP | Chr. | Base-Pair Position | ccQTL p Valuea | Cell Type Name | No.b | Type | Replicated in | Candidate Genec | Functional Annotation | Disease SNPs |
|---|---|---|---|---|---|---|---|---|---|---|
| rs1801274 | 1 | 161479745 | 5.60e−25 | CD4+ naive CD45RO- CD27+ | 20 | 16/T cells |
Roederer et al., 2015 Orrù et al., 2013 |
FCGR2Ad HSPA7d |
missense | KD, UC, SLE, IBD |
| rs72744884 | 2 | 241782823 | 2.20e−9 | CD4+ EM CD45RO+ CD27− | 1 | T cell | – | KIF1A | – | – |
| rs153414 | 5 | 153748732 | 3.60e−8 | CD4+ T cells | 4 | T cell | Roederer et al., 2015 | GALNT10d | intronic | – |
| rs10277809 | 7 | 44948953 | 2.80e−8 | IgM-only B cells (CD24+ CD38+ CD27+ IgM+) | 2 | B cell | – |
RP4-647J21d MYO1G ZMIZ2d |
– | – |
| rs2707213 | 12 | 6899181 | 1.30e−9 | DP (CD4+ CD8+) |
3 | T cell | Orrù et al., 2013 | CD4d | intronic | – |
| rs7403546 | 15 | 87871288 | 2.30e−8 | class non-switched memory (IgM+ CD38+ CD27+) |
1 | B cell | – | AGBL1 | – | – |
| rs2164983 | 19 | 8789381 | 2.70e−8 | NK bright (CD56++ CD16−) |
2 | NK | Roederer et al., 2015 | ACTL9 | – | AD |
| rs280499 | 19 | 10489606 | 5.70e−9 | CD8+ CM CD45RO+ CD27+ | 2 | T cell | – | PDE4Ad | – | MS, CD, T1D, RA, UC, IBDe |
Abbreviations are as follows: EM, effector memory; KD, Kawasaki disease; UC, ulcerative colitis; SLE, systemic lupus erythematosus; IBD, inflammatory bowel disease; AD, atopic dermatitis; MS, multiple sclerosis; CD, Crohn’s disease; T1D, type 1 diabetes; RA, rheumatoid arthritis; JIA, juvenile idiopathic arthritis.
p value from a linear regression model after correcting for age, gender, and month of collection.
The number of additional cell subpopulations showing a nominal p value ≤ 1 × 10−6 at this SNP.
Predicted candidate genes based on eQTL analysis and/or close proximity with the ccQTL.
Genes with significant cis-eQTL based on ∼600 RNA-seq samples from peripheral blood.
Overlapping with ImmunoBase curated regions.
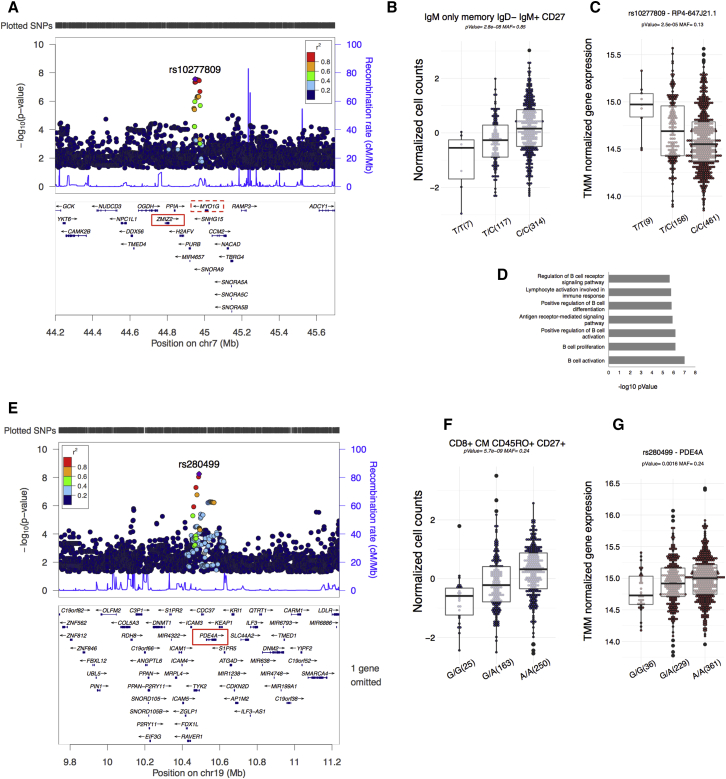
The MYO1B Locus on Chromosome 7 Is Associated with B Cell Levels
We found a B-cell-specific ccQTL (rs10277809, chromosome 7) (Figures 4B and 4C; Table 1) that showed a genome-wide significant association with three B cell subpopulations (CD24dim CD38dim, IgM+-only, and IgM-only memory IgD− IgM+ CD27+ B cells) (Figures 5A and 5B). To explore the biological role of the MYO1B locus, we mapped expression QTLs (eQTLs) by using RNA-seq data from peripheral-blood cells of 629 healthy individuals from the Lifelines Deep (LLDeep) cohort (Tigchelaar et al., 2015). We observed that SNP rs10277809 affects the expression levels of both lncRNA RP4-647J21 and the MYO1G protein-coding gene (Figure 5C). This further supports our finding that this ccQTL is associated with the abundance of peripheral B cell subsets in human peripheral blood. Co-expression analysis and pathway predictions using over 10,000 RNA-seq samples collected from public databases (Fehrmann et al., 2015) show a significant enrichment of B-cell-related functions for both MYO1G and RP4-647J21 (Figure 5D).
Figure 5.
ccQTLs Associated with B and T Cell Subpopulations in Healthy Volunteers
(A) Locus zoom plot showing a B-cell-specific ccQTL in chromosome 7. Red boxes in the gene area denote a significant eQTL effect (nominal p value ≤ 0.05) using ∼600 RNA-seq samples from an independent Dutch LLDeep cohort.
(B) Box-plot of the top associated B cell subpopulation (IgM-only memory IgD− IgM+ CD27) with the genotype.
(C) eQTL box-plot of the lncRNA RP4-647J2.1, which shows a high co-expression pattern with MYO1G, dotted red box in (A).
(D) Gene ontology enrichment analysis of co-expression genes using publically available RNA-seq data (∼10,000) indicates that candidate gene RP4-647J21 is involved in the regulation of B cell activation.
(E) Locus zoom plot showing a T-cell-specific ccQTL in chromosome 19. Red box marks the gene with a significant eQTL effect using the LLDeep cohort RNA-seq data (∼600 samples).
(F) ccQTL boxplot of the top associated T cell subpopulation (CD8+ CM CD45RO+CD27+).
(G) Box-plot of cis-eQTL of PDE4A using the LLDeep cohort RNA-seq data.
PDE4A Locus on Chromosome 19 Affects T Cell Levels
We found a T-cell-specific ccQTL, rs280499 on chromosome 19, that (Figures 4B and 4C; Table 1) particularly associated with CD8+ CM CD45RO+ CD27+ cells (Figures 5E and 5F). We then mapped cis-eQTLs for SNP rs280499 and found its effect on expression levels of PDE4A (Figure 5G). PDE4A encodes the protein phosphodiesterase 4A and has been implicated in T cell differentiation (Peter et al., 2007). PDE4A hydrolyses cyclic AMP, which modulates a variety of cellular responses to extracellular stimuli, including regulating lymphocyte proliferation and the biosynthesis of IL-2. Because PDE4A plays a role in inflammatory processes, it is therapeutically targeted in the treatment of a number of immune-mediated diseases (Mazur et al., 2015).
Shared Genetics between Immune Traits and Immune-Mediated Diseases
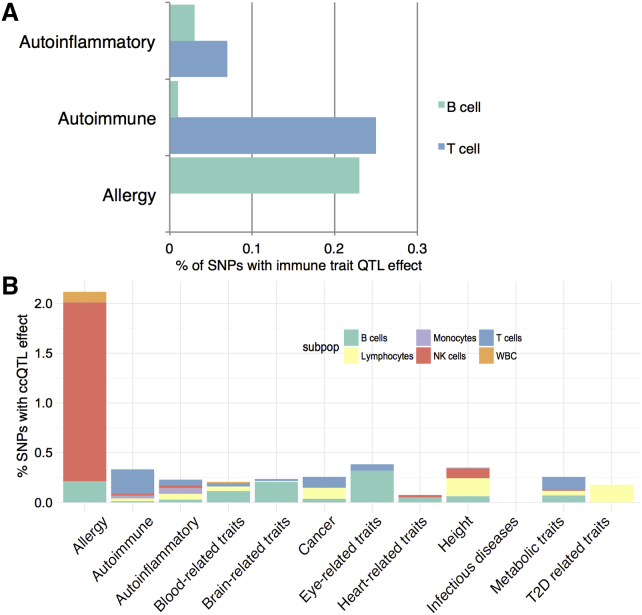
Three out of our eight ccQTLs have been previously associated with immune-mediated diseases (Table 1). In particular, rs1801274, which is a ccQTL for multiple cell types, is associated with several auto-immune diseases (Table 1; Figure S4A), including ulcerative colitis and Kawasaki disease, and has also been replicated in previous studies (Orrù et al., 2013, Roederer et al., 2015). On chromosome 19, the SNP rs2164983 associated to NK cells (Table 1) has been previously reported to be a risk factor for atopic dermatitis (Paternoster et al., 2011). Furthermore, ccQTL rs280499 overlaps with ImmunoBase regions associated with immune-mediated diseases such as multiple sclerosis and rheumatoid arthritis (https://immunobase.org/studies/). In addition, we make use of ccQTLs and IgQTLs at a suggestive significance threshold (p < 1e−5) and genome-wide association study (GWAS) catalog SNPs known to influence susceptibility to various diseases (Figure 6). Interestingly, SNPs that affect T cells levels are also enriched for SNPs associated to auto-inmmune and inflammatory diseases. In contrast, ccQTLs that affect B cells are enriched for SNPs associated with allergy-related diseases (Figure 6).
Figure 6.
Association of ccQTLs with Disease
(A) The percentage of auto-inflammatory-disease-, autoimmune-disease-, and allergy-associated SNPs with B cell and T cell count QTLs (p ≤ 1E-05).
(B) The percentage of disease-associated SNPs with cell count QTLs (p ≤ 1E−05).
Discussion
The HFGP project was initiated to better understand the variation of the immune landscape of human beings and to identify targets for personalized treatment interventions. To explore the determinants of variation in T and B lymphocytes and Ig levels, we tested the association between these immune traits and both heritable factors and non-heritable factors, such as age, gender, and seasonality, in the HFGP 500FG cohort of healthy volunteers.
The abundance of circulating T cells appears to be influenced more by genetics than the numbers of circulating B cells. This hypothesis is based on our observation that a higher percentage of variation is explained by genetics for T cells (∼30%) than for B cells (<∼18%) and on our identification of five T cell ccQTLs versus only two B cell ccQTLs. Most B cell subsets (and Ig levels) consistently showed seasonality effects, peaking during winter, suggesting that environmental factors might be more important in driving B cell count variation. This hypothesis is supported by results of multi-dimensional scaling analysis, revealing a separation between B cells and other immune cell subpopulations.
Despite the impact of environmental cues on B cell counts, B cell function is still affected by genetics. Moreover, only one type of Ig showed a significant genetic component to its variation: ∼50% of the proportion of variance in IgM levels was explained by genetics, while none of the other Igs we measured showed any genetic component. We also identified an IgM-specific QTL but didn’t find QTLs for any of the other Igs that we investigated. Both the IgM QTL and the ccQTL associated to IgM-only B cells, and this may be representative for that part of the B cell response that has innate-like features, such as the production of natural antibodies by dedicated B cell types. In contrast, the adaptive B cell response, featuring receptor editing and affinity maturation, might be under more stringent environmental control, as previously reported in a study of the seasonal pathogen influenza (Baumgarth et al., 1999).
Non-genetic factors such as age and gender have extensively been associated with changes in immune profiles. Fluctuating gender-associated hormone levels and the accumulation of environmental factors, such as an increasing infection burden with age, both leave a strong imprint on the nature and dynamics of the immune response (LeMaoult et al., 1997, Shaw et al., 2013, Solana et al., 2006). Notably, our results appear to support the hypothesis that aging is associated with an overall decrease in lymphoid immune cell levels and an increase in myeloid cell types, as well as increased Treg activity. This suggests that immune response type and regulation is altered toward a more innate-type of immunity with age, as previously reported (Le Garff-Tavernier et al., 2010, Hazeldine et al., 2012). In our current study, we replicate a number of previously reported age-related changes in the human immune system, such as depletion of naive B cells and T cells and a concomitant increase of memory B and T cells (LeMaoult et al., 1997, Shaw et al., 2013, Solana et al., 2006). We also identify age-related changes in specific cell subsets, such as monocyte subclasses, granulocytes, and proliferating T cell populations, that were not reported before.
With regard to gender, we see overall higher immune cell counts and Ig levels for women, with the notable exception of effector/memory T cells, which are more abundant in men. The significant correlation we observed between the higher levels of IgM-only B cells and increased serum levels of IgM in women could be explained by the functional link between these cell types and overall serum Ig levels in humans (Amadori et al., 1995). The enhanced antibody responses found in women upon vaccination fits this profile (Butterworth et al., 1967, Rowley and Mackay, 1969), as does the previously established positive correlation between estrogens and IgM and IgG levels (Kanda and Tamaki, 1999).
The generation of heterogeneous human memory T cell subsets, and how they develop upon activation of naive T cells, is a subject of intense research (Farber et al., 2014). Two developmental models have been proposed. Either (1) memory T cells arise directly from effector cells or (2) naive cells develop directly into memory cells without effector stage transition (Restifo and Gattinoni, 2013). In our unsupervised approach to study the inter-relationship between cell types, we observed that naive and central memory T cells co-cluster within the T cell cluster, while effector and effector memory T cells co-cluster with innate effector cells. Although we weren’t able to decipher the developmental route of these T cell maturation stages, this differential clustering of more quiescent naive and central memory T cells versus innate effector-like effector and effector memory T cells suggests clustering based on function. Meanwhile, the cluster composed of plasmablast B cells also grouped the T helper cytokine (Th2) subpopulation, and these two subpopulations of immune cells have previously been functionally linked given that they are increased in patients with IgG4-related disease (Akiyama et al., 2015). Unfortunately, we weren’t able to find any significant association between IgG4 levels and plasmablast or Th2 T cells within the general population.
The generation and isotype switching of Ig-producing plasma cells can be mediated in either a T-cell-dependent or a T-cell-independent fashion. We found that CD4+ effector T cells (CD27− CD45RA+) show a strong association with IgG levels, implying a functional link between these cell types in humans, which is in line with the finding that effective recall of antibody responses requires the generation of memory B cells controlled by T cell subsets (Kurosaki et al., 2015). We found a significant positive correlation between IgM-only B cell counts and IgM serum levels and a negative correlation between IgM serum levels and IgD+IgM− B cells. IgM-only peripheral-blood lymphocytes are non-activated resting B cells that resemble classical, class-switched memory B cells and express higher levels of mRNA than naive B cells (Klein et al., 1997). Whether this increase in transcription contributes to higher serum Ig levels is still unclear.
The proportion of variance explained by genetics per subpopulation can be quite variable. NK cells display the highest percentage of variance explained by genetics. A similar positive impact of genetics on NK cells was described previously (Roederer et al., 2015). With respect to the effector memory and effector T cells, high levels of variance explained by genetics are in agreement with recent findings in twins (Brodin et al., 2015). We also observed a genetic contribution to Treg counts, which is in contrast to the study by Brodin et al. (2015). For the majority of B cell subsets, with the exception of IgD+ IgM− and transitional B cells, the variance in cell counts explained by genetics was low (median < 18%). This result could suggest that B cell immunity is more susceptible to environmental cues, which is further exemplified by a prominent seasonal effect on both B cell counts and Ig levels. Additionally, in a recent vaccination cohort study, it was discovered that the inter- and intra-individual variations in immune response before and after vaccination can be influenced by age and gender, which also corroborates our findings (Frasca et al., 2012, Tsang et al., 2014).
Identifying ccQTLs associated with genomic regions of relevance to disease provides insight into disease etiology. We identified eight ccQTLs, four of which were not reported before. Those specific B cell subpopulations (which had a ccQTL effect) have not been studied before in the context of the general population. A multi-omics approach combining cell count data, genomics, and transcriptomics was applied to identify the functional and clinical relevance of the ccQTLs. Given the comprehensive analysis of B cell subpopulations, we identified eQTL-effects on MYO1G expression and on the expression of a neighboring lncRNA. MYO1G has previously been implicated in B cell biology and blood cell numbers in a mouse model (Maravillas-Montero et al., 2014). Together, these results suggest the involvement of MYO1G in the active regulation of B cell levels in humans. The lncRNA might or may not be involved in regulation of MYO1G expression (Quinn and Chang, 2016). Furthermore, we identified a T-cell-specific ccQTL in the PDE4A locus that modulates its expression. The fact that PDE4A is a common therapeutic target for immune-mediated diseases (Mazur et al., 2015) further supports an immune-associated role for our ccQTLs.
There are some drawbacks to the approach that we have used for our current study. A limitation of our and similar studies (Brodin et al., 2015, Carr et al., 2016, Orrù et al., 2013, Roederer et al., 2015) is that circulating immune cells are used and do not represent the full landscape of human immunity. Strong differences in immune cell composition have been reported between human peripheral blood, bone marrow, spleen, and lymph nodes (Peters et al., 2013). However, obtaining samples of lymphoid organs in a cohort of healthy individuals is not feasible for ethical and practical reasons. Moreover, given the sample size of our study, the standard error on the calculation of percentage of explained variance by genetics per trait can be substantial (Yang et al., 2010). Finally, in this study we were unable to set a discovery-replication scheme for the immune trait QTL mapping due to the limited sample size. Despite these drawbacks, we were able to identify a differential contribution of genetic versus environmental factors on lymphocyte subpopulations, we confirmed previously reported ccQTLs, and we identified ccQTLs for B cell and T cell subpopulations.
In conclusion, we assessed the influence of genetics, age, gender, and seasonality on cell count variation, Ig levels, and their interrelationship in healthy volunteers participating in the HFGP. Our findings indicate that T cell immunity has a stronger genetic imprint than B cell immunity, while the latter might be driven by environmental factors. We also found eight genome-wide significant loci associated to cell levels, four of which were not reported previously. Moreover, we were able to link immune cell count QTLs to GWAS SNPs associated with immune-mediated diseases. Within the HFGP 500FG cohort, three complementary studies focus on a broader understanding of the variability in human cytokine responses. ter Horst et al. (2016) identified host and environmental factors that contribute to variation of cytokine responses, while Li et al. (2016) and Schirmer et al. (2016) mapped 17 new genetic variants and microbiome factors, respectively, that explain variability of cytokine responses (Li et al., 2016, Schirmer et al., 2016). Like immune cell counts and Ig levels, cytokine responses were influenced by age and gender (ter Horst et al., 2016), and cytokine responses also revealed annual seasonal dependencies (ter Horst et al., 2016). Together, these different HFGP 500FG studies provide important resources for understanding the human immune response. Future studies using the HFGP cohorts will focus on assessing the effect of other factors (e.g., microbiome, infection, and immune-mediated inflammatory disease) on the variation of immune cell counts and function. These studies will contribute to the goal of precision medicine in infections and inflammation by allowing for more accurate predictions of disease status and better treatment efficacy.
Experimental Procedures
Ethics Statement
The HFGP study was approved by the ethical committee of Radboud University Nijmegen (no. 42561.091.12). Experiments were conducted according to the principles expressed in the Declaration of Helsinki. Samples of venous blood were drawn after informed consent was obtained.
Population Cohorts
The study was performed in a cohort of 516 healthy individuals of Western-European ancestry from the HFGP (500FG; for inclusion criteria and further description see http://www.humanfunctionalgenomics.org).
Analysis of Immune Cell Composition and Humoral Components in a Healthy Dutch Population
We measured myeloid and lymphoid immune cell levels by 10-color flow cytometry (Table S4) and serum Ig (sIg) concentrations by fluorescence enzyme immunoassay (Immunocap) in 516 Dutch individuals of Western-European descent, aged 18 to 75 years, recruited over the years 2013–2014 as part of the 500FG study within the HFGP (http://www.humanfunctionalgenomics.org). We focused on a set of 73 manually annotated immune cell subpopulations and seven different classes of Igs (Figure S5). To minimize biological variability, cells were processed immediately after blood sampling and typically analyzed within 2–3 hr. Cell populations were gated manually (see Supplemental Experimental Procedures for details).
Flow Cytometry and Data Analysis
Cells were analyzed within 2–3 hr after sample collection on a 10-color Navios flow cytometer (Beckman Coulter) equipped with three solid-state lasers (488 nm, 638 nm, and 405 nm). Calibration of the machine was performed once a week, and little adjustment to the machine setting had to be made during the inclusion period of the study. Data were then analyzed using Kaluza software version 1.3 (Beckman Coulter). The hierarchical gating strategy is illustrated in Figures S6 and S7. See Supplemental Experimental Procedures for details on cell processing, reagents, gating, and analysis.
Serum Ig and Hormone Levels
Genotyping, Quality Control, and Imputation
Volunteers from the 500FG cohort were genotyped using the Illumina Human OmniExpress Exome-8 v1.0 SNP chip. The genotype was called with Opticall 0.7.0 using the default settings, excluding samples with a call rate ≤ 0.99. Variants with Hardy-Weinberg equilibrium (HWE) ≤ 0.0001, call rate ≤ 0.99, and minor-allele frequency (MAF) ≤ 0.001 were also filtered out. Ethnic outliers were identified by multi-dimensional scaling plots of samples merged with 1000 Genome data and excluded from further analysis. A total of 482 samples and 518,980 variants passed quality control. For further imputation of this dataset, we aligned the strands and variant identifiers to the reference Genome of the Netherlands (GoNL) dataset using Genotype Harmonizer. The phasing was performed with SHAPEIT2 version 2 with the GoNL as a reference panel. Finally, the data were imputed using IMPUTE2 with the GoNL as the reference panel. Only imputed variants with a quality score ≥ 0.8 were used for further cell count quantitative loci mapping.
Statistical Analysis
All statistical analysis were performed using the statistical programming language R (R Core Team, 2012). Cell counts were normalized using an inverse rank transformation (IRT) algorithm, shown in Table S5. Ig levels were normalized using a log2 transformation. To properly ascertain cell count correlations, we first corrected the normalized cell counts for age, gender, and seasonal effects using a linear model. Associations were then calculated using the normalized and corrected cell counts via Spearman correlation analysis and clustered using these coefficients as distance by an unsupervised hierarchical clustering approach. The same methodology was applied to calculate the association between cell counts and Igs. Significance was declared after multiple testing correction (FDR ≤ 0.05) (Benjamini and Hochberg, 1995). The Euclidean distances used on the multi-dimensional scaling between cell types were obtained based on the Spearman coefficients described above (Venables and Ripley, 2002).
See Supplemental Experimental Procedures for details regarding statistical analysis of the association of cell counts or Ig levels with age, gender, and season.
Cell Count and Ig QTL Mapping
For 442 individuals, absolute cell count data and genotype information was available. For 407 individuals, Ig levels and genotype data were available. We calculated parental and grandparental percentages, which are defined as the percentage of a certain cell type within the subpopulation of cells from which it was isolated. This was performed for cell counts of all measured cell types because it has been shown that these percentages tend to reduce inter-experimental noise and therefore increase statistical power for QTL mapping (Orrù et al., 2013). Absolute cell counts and percentages were transformed by IRT (Orrù et al., 2013). Ig levels were normalized using a log2 transformation. We then corrected the IRT cell counts and log2 Ig values using a linear model correcting for age, gender, and month of sample collection. Lastly, QTL mapping was performed using a linear model as implemented in the Matrix-eQTL R package (Shabalin, 2012), where we associated immune traits to genotype information. A p value < 5e−6 was considered to be genome-wide significant.
Genome-wide Significant cis-eQTL Analysis
We used the LLDeep cohort (Tigchelaar et al., 2015), composed of 627 healthy Dutch volunteers, to test for possible eQTL effects of the ccQLTs. For LLDeep, both gene expression data (obtained through RNA-seq) and genotype information are available. We mapped cis-eQTLs for each identified ccQTL within a 1 Mb window. For this, we fitted a linear model using TMM-normalized (Robinson et al., 2010) expression data to the genotype information. Given that the number of tests depended on the ccQTL genomic location for each independent locus, a threshold of FDR (≤0.05) was used, depending on the number of tests performed in that specific window.
Estimation of Cell Count and Ig Level Heritability
To estimate the proportion of variance explained by genetics, we used a linear mixed model implemented in the GCTA tool (Yang et al., 2010). We applied it to each of the cell counts and percentages and to Ig levels using the complete set of genetic variants quantified in our cohort. The immune traits were pre-processed as described for QTL mapping using IRT cell counts and log2 Ig values corrected for age, sex, and month of sample collection. Given the relatively small sample size, the confidence intervals for heritability estimation can be wide (Zaitlen and Kraft, 2012).
Raw Flow Cytometry Data
The accession number for the raw flow cytometry data and analyzed data files are available upon request to the authors (http://hfgp.bbmri.nl). A pipeline is available regarding further collaborations and access to additional data and samples.
Author Contributions
M.G.N. and C.W. coordinated the recruitment of the cohorts. H.J.P.M.K., I.J., Y.L., V.K., M.G.N., and C.W. conceived and directed the study with input from all authors. R.A.-G., Y.L., I.J., and H.J.P.M.K. analyzed and interpreted the data. M.A.S. and L.F. provided the computational framework for the study. P.C.M.U., R.G.M., E.v.R., B.v.C., M.O., S.S., M.J., R.J.X., M.Z., A.E.v.H., F.S., and R.T.N. contributed to the data collection. R.A.-G., Y.L., H.J.P.M.K., I.J., V.K., S.W., and M.G.N. wrote the manuscript with input from all other authors. M.G.N., L.A.B.J., C.W., H.J.P.M.K., and I.J. acquired funding.
Acknowledgments
The authors thank all volunteers from the 500FG cohort of the HFGP for participation in the study. We thank Jackie Senior and Kate Mc Intyre for editorial assistance. The HFGP is supported by a European Research Council (ERC) Consolidator grant (3310372) and an IN-CONTROL CVON grant (CVON2012-03) to M.G.N., an ERC Advanced Grant (FP/2007-2013/ERC grant 2012-322698) and a Spinoza Prize (NWO SPI 92-266) to C.W., a Dutch Digestive Diseases Foundation (MLDS) grant (WO11-30) to C.W. and V.K., a European Union Seventh Framework Program (EU FP7) grant (TANDEM; HEALTH-F3-2012-305279) to C.W. and V.K., a Netherlands Organization for Scientific Research (NWO) VENI grant (863.13.011) to Y.L., a CONACYT-I2T2 scholarship (382117) to R.A.G., and a scholarship from Brazil’s Science Without Borders program (11920/13-0) to P.C.M.U. This study made use of data generated by the Genome of the Netherlands project funded by NWO (grant no. 184021007), which was made available as a Rainbow Project of BBMRI-NL.
Published: November 3, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.10.053.
Contributor Information
Yang Li, Email: y.li01@umcg.nl.
Hans J.P.M. Koenen, Email: hans.koenen@radboudumc.nl.
Supplemental Information
References
- Akiyama M., Suzuki K., Yamaoka K., Yasuoka H., Takeshita M., Kaneko Y., Kondo H., Kassai Y., Miyazaki T., Morita R. Number of Circulating Follicular Helper 2 T Cells Correlates With IgG4 and Interleukin-4 Levels and Plasmablast Numbers in IgG4-Related Disease. Arthritis Rheumatol. 2015;67:2476–2481. doi: 10.1002/art.39209. [DOI] [PubMed] [Google Scholar]
- Amadori A., Zamarchi R., De Silvestro G., Forza G., Cavatton G., Danieli G.A., Clementi M., Chieco-Bianchi L. Genetic control of the CD4/CD8 T-cell ratio in humans. Nat. Med. 1995;1:1279–1283. doi: 10.1038/nm1295-1279. [DOI] [PubMed] [Google Scholar]
- Baumgarth N., Herman O.C., Jager G.C., Brown L., Herzenberg L.A., Herzenberg L.A. Innate and acquired humoral immunities to influenza virus are mediated by distinct arms of the immune system. Proc. Natl. Acad. Sci. USA. 1999;96:2250–2255. doi: 10.1073/pnas.96.5.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J.R. Stat. Soc. 1995;57:289–300. [Google Scholar]
- Brodin P., Jojic V., Gao T., Bhattacharya S., Angel C.J.L., Furman D., Shen-Orr S., Dekker C.L., Swan G.E., Butte A.J. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160:37–47. doi: 10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüggemann M., Williams G.T., Bindon C.I., Clark M.R., Walker M.R., Jefferis R., Waldmann H., Neuberger M.S. Comparison of the effector functions of human immunoglobulins using a matched set of chimeric antibodies. J. Exp. Med. 1987;166:1351–1361. doi: 10.1084/jem.166.5.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterworth M., McClellan B., Allansmith M. Influence of sex in immunoglobulin levels. Nature. 1967;214:1224–1225. doi: 10.1038/2141224a0. [DOI] [PubMed] [Google Scholar]
- Carr E.J., Dooley J., Garcia-Perez J.E., Lagou V., Lee J.C., Wouters C., Meyts I., Goris A., Boeckxstaens G., Linterman M.A. The cellular composition of the human immune system is shaped by age and cohabitation. Nat. Immunol. 2016;17:461–468. doi: 10.1038/ni.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J.H., Feldman M. Heterogeneity of autoimmune diseases: pathophysiologic insights from genetics and implications for new therapies. Nat. Med. 2015;21:730–738. doi: 10.1038/nm.3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman R.L., Lebman D.A., Shrader B. Transforming growth factor beta specifically enhances IgA production by lipopolysaccharide-stimulated murine B lymphocytes. J. Exp. Med. 1989;170:1039–1044. doi: 10.1084/jem.170.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2012). R: A language and environment for statistical computing. http://R-project.org/.
- Dopico X.C., Evangelou M., Ferreira R.C., Guo H., Pekalski M.L., Smyth D.J., Cooper N., Burren O.S., Fulford A.J., Hennig B.J. Widespread seasonal gene expression reveals annual differences in human immunity and physiology. Nat. Commun. 2015;6:7000. doi: 10.1038/ncomms8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber D.L., Yudanin N.A., Restifo N.P. Human memory T cells: generation, compartmentalization and homeostasis. Nat. Rev. Immunol. 2014;14:24–35. doi: 10.1038/nri3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrmann R.S.N., Karjalainen J.M., Krajewska M., Westra H.-J., Maloney D., Simeonov A., Pers T.H., Hirschhorn J.N., Jansen R.C., Schultes E.A. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat. Genet. 2015;47:115–125. doi: 10.1038/ng.3173. [DOI] [PubMed] [Google Scholar]
- Frasca D., Diaz A., Romero M., Phillips M., Mendez N.V., Landin A.M., Blomberg B.B. Unique biomarkers for B-cell function predict the serum response to pandemic H1N1 influenza vaccine. Int. Immunol. 2012;24:175–182. doi: 10.1093/intimm/dxr123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazeldine J., Hampson P., Lord J.M. Reduced release and binding of perforin at the immunological synapse underlies the age-related decline in natural killer cell cytotoxicity. Aging Cell. 2012;11:751–759. doi: 10.1111/j.1474-9726.2012.00839.x. [DOI] [PubMed] [Google Scholar]
- Kanda N., Tamaki K. Estrogen enhances immunoglobulin production by human PBMCs. J. Allergy Clin. Immunol. 1999;103:282–288. doi: 10.1016/s0091-6749(99)70503-8. [DOI] [PubMed] [Google Scholar]
- Kanda N., Tsuchida T., Tamaki K. Testosterone inhibits immunoglobulin production by human peripheral blood mononuclear cells. Clin. Exp. Immunol. 1996;106:410–415. doi: 10.1046/j.1365-2249.1996.d01-842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U., Küppers R., Rajewsky K. Evidence for a large compartment of IgM-expressing memory B cells in humans. Blood. 1997;89:1288–1298. [PubMed] [Google Scholar]
- Kurosaki T., Kometani K., Ise W. Memory B cells. Nat. Rev. Immunol. 2015;15:149–159. doi: 10.1038/nri3802. [DOI] [PubMed] [Google Scholar]
- Le Garff-Tavernier M., Béziat V., Decocq J., Siguret V., Gandjbakhch F., Pautas E., Debré P., Merle-Beral H., Vieillard V. Human NK cells display major phenotypic and functional changes over the life span. Aging Cell. 2010;9:527–535. doi: 10.1111/j.1474-9726.2010.00584.x. [DOI] [PubMed] [Google Scholar]
- LeMaoult J., Szabo P., Weksler M.E. Effect of age on humoral immunity, selection of the B-cell repertoire and B-cell development. Immunol. Rev. 1997;160:115–126. doi: 10.1111/j.1600-065x.1997.tb01032.x. [DOI] [PubMed] [Google Scholar]
- Li Y., Oosting M., Smeekens S., Jaeger M., Aguirre-Gamboa R., Le K.T.T., Deelen P., Ricaño-Ponce I., Schoffelen T., Jansen A.F.M. A functional genomics approach to understand variation in cytokine production in humans. Cell. 2016;167:1099–1110. doi: 10.1016/j.cell.2016.10.017. [DOI] [PubMed] [Google Scholar]
- Maravillas-Montero J.L., López-Ortega O., Patiño-López G., Santos-Argumedo L. Myosin 1g regulates cytoskeleton plasticity, cell migration, exocytosis, and endocytosis in B lymphocytes. Eur. J. Immunol. 2014;44:877–886. doi: 10.1002/eji.201343873. [DOI] [PubMed] [Google Scholar]
- Mazur M., Karczewski J., Lodyga M., Żaba R., Adamski Z. Inhibitors of phosphodiesterase 4 (PDE 4): A new therapeutic option in the treatment of psoriasis vulgaris and psoriatic arthritis. J. Dermatolog. Treat. 2015;26:326–328. doi: 10.3109/09546634.2014.991267. [DOI] [PubMed] [Google Scholar]
- McIntyre T.M., Klinman D.R., Rothman P., Lugo M., Dasch J.R., Mond J.J., Snapper C.M. Transforming growth factor beta 1 selectivity stimulates immunoglobulin G2b secretion by lipopolysaccharide-activated murine B cells. J. Exp. Med. 1993;177:1031–1037. doi: 10.1084/jem.177.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrù V., Steri M., Sole G., Sidore C., Virdis F., Dei M., Lai S., Zoledziewska M., Busonero F., Mulas A. Genetic variants regulating immune cell levels in health and disease. Cell. 2013;155:242–256. doi: 10.1016/j.cell.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paternoster L., Standl M., Chen C.-M., Ramasamy A., Bønnelykke K., Duijts L., Ferreira M.A., Alves A.C., Thyssen J.P., Albrecht E., Australian Asthma Genetics Consortium (AAGC) Genetics of Overweight Young Adults (GOYA) Consortium. EArly Genetics & Lifecourse Epidemiology (EAGLE) Consortium Meta-analysis of genome-wide association studies identifies three new risk loci for atopic dermatitis. Nat. Genet. 2011;44:187–192. doi: 10.1038/ng.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter D., Jin S.L.C., Conti M., Hatzelmann A., Zitt C. Differential expression and function of phosphodiesterase 4 (PDE4) subtypes in human primary CD4+ T cells: predominant role of PDE4D. J. Immunol. 2007;178:4820–4831. doi: 10.4049/jimmunol.178.8.4820. [DOI] [PubMed] [Google Scholar]
- Peters J.H., Koenen H.J.P.M., Fasse E., Tijssen H.J., Ijzermans J.N.M., Groenen P.J.T.A., Schaap N.P.M., Kwekkeboom J., Joosten I. Human secondary lymphoid organs typically contain polyclonally-activated proliferating regulatory T cells. Blood. 2013;122:2213–2223. doi: 10.1182/blood-2013-03-489443. [DOI] [PubMed] [Google Scholar]
- Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- Restifo N.P., Gattinoni L. Lineage relationship of effector and memory T cells. Curr. Opin. Immunol. 2013;25:556–563. doi: 10.1016/j.coi.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roederer M., Quaye L., Mangino M., Beddall M.H., Mahnke Y., Chattopadhyay P., Tosi I., Napolitano L., Terranova Barberio M., Menni C. The genetic architecture of the human immune system: a bioresource for autoimmunity and disease pathogenesis. Cell. 2015;161:387–403. doi: 10.1016/j.cell.2015.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley M.J., Mackay I.R. Measurement of antibody-producing capacity in man. I. The normal response to flagellin from Salmonella adelaide. Clin. Exp. Immunol. 1969;5:407–418. [PMC free article] [PubMed] [Google Scholar]
- Schirmer M., Smeekens S.P., Vlamakis H., Jaeger M., Oosting M., Franzosa E.A., Jansen T., Jacobs L., Bonder M.J., Kurilshikov A., Fu J. Linking the gut microbiome to inflammatory cytokine production capacity. Cell. 2016;167:1125–1136. doi: 10.1016/j.cell.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder H.W., Jr., Cavacini L. Structure and function of immunoglobulins. J. Allergy Clin. Immunol. 2010;125(2, Suppl 2):S41–S52. doi: 10.1016/j.jaci.2009.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalin A.A. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A.C., Goldstein D.R., Montgomery R.R. Age-dependent dysregulation of innate immunity. Nat. Rev. Immunol. 2013;13:875–887. doi: 10.1038/nri3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapper C.M., Paul W.E. Interferon-gamma and B cell stimulatory factor-1 reciprocally regulate Ig isotype production. Science. 1987;236:944–947. doi: 10.1126/science.3107127. [DOI] [PubMed] [Google Scholar]
- Solana R., Pawelec G., Tarazona R. Aging and innate immunity. Immunity. 2006;24:491–494. doi: 10.1016/j.immuni.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Tangye S.G., Deenick E.K., Palendira U., Ma C.S. T cell-B cell interactions in primary immunodeficiencies. Ann. N Y Acad. Sci. 2012;1250:1–13. doi: 10.1111/j.1749-6632.2011.06361.x. [DOI] [PubMed] [Google Scholar]
- ter Horst R., Jaeger M., Smeekens S.P., Oosting M., Swertz M.A., Li Y., Kumar V., Diavatopoulos D.A., Jansen A.F.M., Lemmers H. Host and environmental factors influencing individual human cytokine responses. Cell. 2016;167:1111–1124. doi: 10.1016/j.cell.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigchelaar E.F., Zhernakova A., Dekens J.A.M., Hermes G., Baranska A., Mujagic Z., Swertz M.A., Muñoz A.M., Deelen P., Cénit M.C. Cohort profile: LifeLines DEEP, a prospective, general population cohort study in the northern Netherlands: study design and baseline characteristics. BMJ Open. 2015;5:e006772. doi: 10.1136/bmjopen-2014-006772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J.S., Schwartzberg P.L., Kotliarov Y., Biancotto A., Xie Z., Germain R.N., Wang E., Olnes M.J., Narayanan M., Golding H., Baylor HIPC Center. CHI Consortium Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014;157:499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables W.N., Ripley B.D. Springer; New York: 2002. Modern Applied Statistics with S. [Google Scholar]
- Weller S., Braun M.C., Tan B.K., Rosenwald A., Cordier C., Conley M.E., Plebani A., Kumararatne D.S., Bonnet D., Tournilhac O. Human blood IgM “memory” B cells are circulating splenic marginal zone B cells harboring a prediversified immunoglobulin repertoire. Blood. 2004;104:3647–3654. doi: 10.1182/blood-2004-01-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy B.P., Gordon S., Henders A.K., Nyholt D.R., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitlen N., Kraft P. Heritability in the genome-wide association era. Hum. Genet. 2012;131:1655–1664. doi: 10.1007/s00439-012-1199-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.