Figure 2.
High-Frequency Contribution of Identified FS Interneurons to the Trough of Spindle Oscillations during Natural Sleep
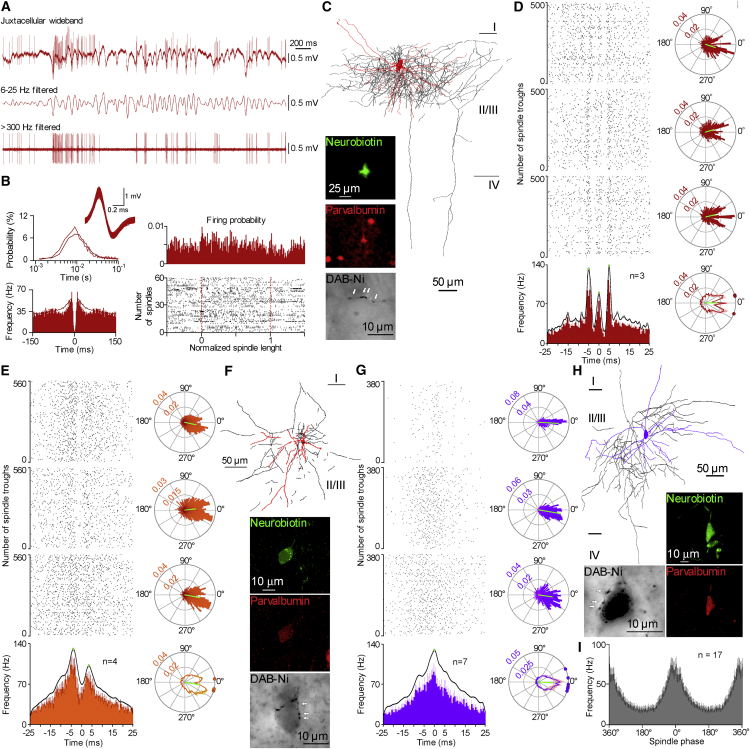
(A) Juxtacellularly recorded activity of an identified spindle ripple cell (top, wideband; middle, filtered for spindles; bottom, filtered for spikes).
(B) Normalized interspike interval distribution (top left, burgundy for overall, red for spindles; inset, time course of single spikes) and autocorrelogram (bottom left, burgundy for overall, red for spindles), and firing distribution of the cell during spindles of normalized length (right).
(C) Top: reconstruction of the dendritic (red) and axonal (black) arborization of the spindle ripple cell. Insets: the neurobiotin-labeled cell expressed parvalbumin and showed features of a basket cell with DAB-Ni containing axonal terminals (arrows) decorating an unlabeled soma.
(D) Top left: peri-trough raster plots of firing of individual spindle ripple cells (top three panels; the top panel shows the cell presented in A–C) and average firing frequency distribution (bottom left; bin width, 0.78 ms) with three peaks of activity centered around the spindle trough shown at 0 ms determined by kernel smoothing and peak detection. Data are based on successive spindle troughs recorded in SWS. Right: cumulative circular plots of firing probability in single cycles of spindles for the three cells (trough at 0°) and their average circular plot of firing distribution (bottom) with vectors of individual cells at the perimeter. Error bars represent SEM.
(E–H) Spindle high-gamma cells (E and F) and simple spindle trough cells (G and H) are presented along the same logic as spindle ripple cells in (D) and (C). Error bars in (E) and (G) represent SEM.
(I) Average firing probability of all spindle-trough-related FS cells tuned to the trough (0°) of spindle cycles, including spindle ripple, spindle high-gamma, and simple spindle trough cells. Error bars represent SEM.