Abstract
The identification of snails of the genus Biomphalaria can be done using morphological characteristics which depends on the size of the snails and skill and knowledge of researcher. These methods sometimes are not adequate for identification of species. The PCR-RFLP, using the ITS region of the rDNA, has been used to identify Brazilian species of the genus Biomphalaria. Nevertheless, there is a lack of information about snails from other Latin American countries. In addition, some snails may be infected by Schistosoma mansoni and when submitted to PCR-RFLP they show molecular profiles different from those previously standardized for the other mollusc species. In this work the molecular profiles of 15 species and the subspecies were established by PCR-RFLP of ITS-rDNA with the enzyme DdeI. Moreover, the molecular profiles of host species, B. glabrata, B. straminea, B. tenagophila, and B. prona, infected by S. mansoni were also established. The molluscs were dissected to permit morphological identification. These results contribute to a correct identification of snails of the genus Biomphalaria and detection of these snails infected by S. mansoni.
1. Introduction
Despite therapeutic advances in the last decade, Schistosomiasis remains one of the most prevalent parasitic diseases worldwide and endemic in 76 countries and territories [1]. In Africa and Neotropical Region there are species of the genus Biomphalaria (Gastropoda: Planorbidae) which are intermediate hosts of Schistosoma mansoni Sambon, 1907. In Latin America 24 species and one subspecies were registered (Table 1), four of them can be found naturally infected by S. mansoni, whereas six were found to be susceptible in the laboratory.
Table 1.
Molluscs of the genus Biomphalaria present in Latin America.
| Species | Geographical distribution | aSusceptibility to Schistosoma mansoni |
|---|---|---|
| Biomphalaria amazonica, Paraense 1966 | Brazil, Bolivia, Colombia | EI |
|
| ||
| Biomphalaria andecola (Orbigny, 1835) | Bolivia, Peru, Chile | NI |
|
| ||
| Biomphalaria cousini Paraense, 1966 | Brazil, Ecuador | EI |
|
| ||
| Biomphalaria edisoni (Estrada et al., 2006) | Colombia | NI |
|
| ||
| Biomphalaria equatoria (Cousin, 1887) | Ecuador | NI |
|
| ||
| Biomphalaria glabrata (Say, 1818) | Antigua, Brazil, Curacao, Dominica, Guadeloupe, French Guiana, Haiti, Saint Kitts and Nevis, Martinique, Montserrat, Puerto Rico, Dominican Republic, Saint Lucia, Suriname, Venezuela | S |
|
| ||
| Biomphalaria havanensis (Pfeiffer, 1839) | Haiti, Mexico, Puerto Rico, Cuba, Venezuela | EI |
|
| ||
| Biomphalaria helophila (Orbigny, 1835) | Peru, Cuba, Costa Rica, Guatemala, Belize, Haiti, Mexico, Saint Thomas, El Salvador, Dominican Republic, Puerto Rico, Barbados, Nicaragua | EI |
|
| ||
| Biomphalaria intermedia (Paraense & Deslandes, 1962) | Brazil, Argentine | NS |
|
| ||
| Biomphalaria kuhniana (Clessin, 1883) | Suriname, Brazil, Venezuela, Panama, Colombia | NS |
|
| ||
| Biomphalaria nicaraguana (Morelet, 1851) | Nicaragua | NI |
|
| ||
| Biomphalaria occidentalis Paraense, 1981 | Brazil, Paraguay, Argentine | NS |
|
| ||
| Biomphalaria oligoza Paraense, 1974 | Bolivia, Brazil, Argentine | EI |
|
| ||
| Biomphalaria orbignyi Paraense, 1975 | Argentine, Uruguay | EI |
|
| ||
| Biomphalaria obstructa (Morelet, 1849) | Mexico, Puerto Rico, Guatemala, El Salvador, Belize, Cuba | NS |
|
| ||
| Biomphalaria pallida (Adams, 1846) | Jamaica, Cuba | NI |
|
| ||
| Biomphalaria peregrina (Orbigny, 1835) | Ecuador, Bolivia, Chile, Brazil, Paraguay, Peru, Uruguay, Argentine, Colombia | EI |
|
| ||
| Biomphalaria prona (Martens, 1873) | Venezuela | S |
|
| ||
| Biomphalaria schrammi (Crosse, 1864) | French Guiana, Guadeloupe, Brazil | NS |
|
| ||
| Biomphalaria sericea (Dunker, 1848) | Ecuador | EI |
|
| ||
| Biomphalaria straminea (Dunker, 1848) | Venezuela, Suriname, French Guiana, Guyana, Peru, Brazil, Paraguay, Argentine, Dominica, Grenada, Guadeloupe, Martinique, Dominican Republic, Trinidad, Uruguay, Costa Rica | S |
|
| ||
| Biomphalaria subprona (Martens, 1899) | Mexico, Guatemala | NI |
|
| ||
| Biomphalaria tenagophila (Orbigny, 1835) | Argentine, Paraguay, Uruguay, Brazil, Peru, Bolivia | S |
|
| ||
| Biomphalaria tenagophila guaibensis Paraense, 1984 | Brazil, Uruguay, Paraguay, Argentine | NS |
|
| ||
| Biomphalaria trigyra (Philippi, 1869) | Peru, Ecuador | NS |
a: susceptible = S; not susceptible = NS; experimental infection = EI; not information = NI.
The classical identification of snails of the genus Biomphalaria is based on morphological characteristics of the shell and the reproductive system [2]. However, this approach is complicated in cases of inadequate fixation or interspecific similarity. The Polymerase Chain Reaction and Restriction Fragment Length Polymorphism (PCR-RFLP), directed to the internal transcribed spacer (ITS) region of the rDNA gene, has been used with success to resolve these cases. The molecular profile of Brazilian species of the genus Biomphalaria using this method has been established [3]. Thus, the specific profile of all these species together will be useful to facilitate interspecific identification in the genus Biomphalaria. Besides, the specific identification could be done by comparing the sequences between closely related species [4–6], as well as using the morphology associated with the species-specific PCR [7, 8].
Furthermore, the snails which were collected in the field may be infected with S. mansoni during the prepatent period, and when they are submitted to the molecular identification, their DNA is simultaneously amplified with the DNA from the parasite. In this case the molecular profile differs from the profile established for the snail alone.
The aim of the present work is to present the previously species-specific profiles established by PCR-RFLP of ITS-rDNA with DdeI and to establish the profiles for B. glabrata, B. tenagophila, B. straminea, and B. prona infected by S. mansoni.
2. Material and Methods
2.1. Samples
Of the 24 species registered for Latin America, the Medical Malacological Collection (Fiocruz-CMM) has fifteen species and a subspecies: B. glabrata, B. tenagophila, B. occidentalis, B. schrammi, B. oligoza, B. peregrina, B. intermedia, B. straminea, B. kuhniana, B. amazonica, B. cousini, B. prona, B. edisoni, B. havanensis, B. orbignyi, and B. tenagophila guaibensis. The molluscs were dissected to permit morphological identification. DNA of specimens of the Fiocruz-CMM collection was cryopreserved.
Biomphalaria glabrata, B. tenagophila, and B. straminea molluscs and AL, SJ, and LE strains of S. mansoni used in this study were maintained and raised in the “Lobato Paraense” Mollusc Rearing of René Rachou Research Center, CPqRR/Fiocruz, in Belo Horizonte, MG, Brazil. The LE strain was isolated, in 1968, from a patient residing in Belo Horizonte, MG (Brazil). The SJ strain was isolated, in 1975, from naturally infected snails from São José dos Campos, São Paulo (Brazil). The AL strain was isolated in 1980 from B. glabrata that originated from Alagoas state (Brazil). To obtain specimens of B. glabrata, B. tenagophila, and B. straminea shedding S. mansoni cercariae, experimental infection with LE, SJ, and AL strains, respectively, was carried out [9]. However, there was no population of B. prona in the “Lobato Paraense” Mollusc Rearing, so the DNA of the snails and the parasite (LE strain) was mixed and amplified together to obtain the profile of this infected species. DNA of adult worms of S. mansoni was used for control of amplification.
Cercaria macrogranulosa, Cercaria caratinguensis, and Cercaria ocellifera were obtained from field snails Biomphalaria.
2.2. Molecular Techniques
2.2.1. DNA Extraction and PCR-RFLP Assay
Total DNA from B. glabrata, B. tenagophila, and B. straminea infected by S. mansoni, B. prona, adult worms of S. mansoni and C. macrogranulosa, C. caratinguensis, and C. ocellifera were extracted using Wizard Genomic Purification Kit (Promega, Madison, USA) with some modifications. The DNA of all samples was used as template in the PCR-RFLP assay. The entire ITS was amplified using the primers ETTS2 (5′ TAACAAGGTTTCCGTAGGTGAA 3′) and ETTS1 (5′ TGCTTAAGTTCAGCGGGT 3′) anchored, respectively, in the conserved extremities of the 18S and 28S ribosomal genes [10]. The PCR amplification was undertaken in a volume of 10 μL consisting of 1–10 ng template DNA, 10 mM Tris-HCl, pH 8.5, 200 μM of each DNTP, 1.5 mM MgCl2, 0.5 U of Taq DNA polymerase, and 50 mM KCl, together with 1.0 pmol of each primer. The reactions were covered with a drop of mineral oil and subjected to the following thermal cycling program: initial denaturation step for 3 min at 95°C, and then 32 cycles with annealing at 54°C for 1 min, extension at 72°C for 2 min, denaturation at 95°C for 45 sec, and a final extension step at 72°C for 5 min. A negative control (no template DNA) was included in all experiments. Three microliters of the amplification products were visualized on silver stained 6% polyacrylamide gels to check the quality of amplification. The remaining 7 μL was mixed with water, and DdeI (10–12 units) enzyme was added, together with 1.0 μL of the respective enzyme buffer. The digestion was performed for 3.5 h at 37°C and at 80°C for 20 min for enzyme denaturation and the digestion products were evaluated on silver stained 6% polyacrylamide gels [3].
3. Results and Discussion
The PCR amplification resulted in a product of approximately 1200 pb for Biomphalaria, one of 800 pb for S. mansoni, and both fragments for infected molluscs (data not shown). The RFLP profiles obtained by digesting rDNA ITS with DdeI in Figure 1 allow the following: (1) to identify noninfected B. glabrata, B. tenagophila, B. straminea, and B. prona, by observation of species-specific fragments (Lanes 2, 3, 4, and 5); (2) to establish the species-specific profile of S. mansoni (Lanes 6 and 11); and (3) to detect by the presence of overlapping species-specific fragments the infection by S. mansoni in B. glabrata, B. tenagophila, B. straminea, and B. prona (Lanes 7, 8, 9, and 10).
Figure 1.
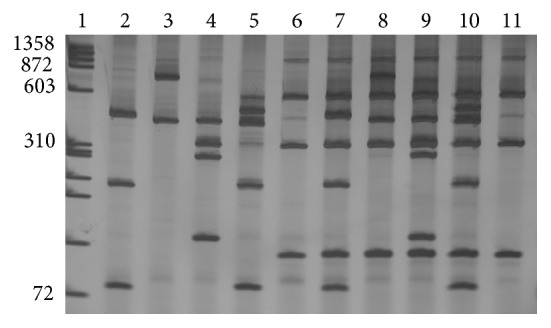
6% silver stained polyacrylamide gel showing restriction profiles obtained by digestion of the ITS region of DNA ribosomal with DdeI. Lane 1: molecular size markers Phi X 174/HaeIII; Lane 2: Biomphalaria glabrata; Lane 3: Biomphalaria tenagophila; Lane 4: Biomphalaria straminea; Lane 5: Biomphalaria prona; Lane 6: adult worm of Schistosoma mansoni; Lane 7: B. glabrata infected by S. mansoni; Lane 8: B. tenagophila infected by S. mansoni; Lane 9: B. straminea infected by S. mansoni; Lane 10: DNA of B. prona with DNA of S. mansoni; Lane 11: adult worm of S. mansoni. The numbers on the left of the gel represent the value in base pairs (bp).
All 15 species and the subspecies of Biomphalaria were dissected and their identification confirmed by analysis of specific diagnostic characters established for each species. In association with morphological identification, the profile of PCR-RFLP was established for these species and is shown in Figure 2.
Figure 2.
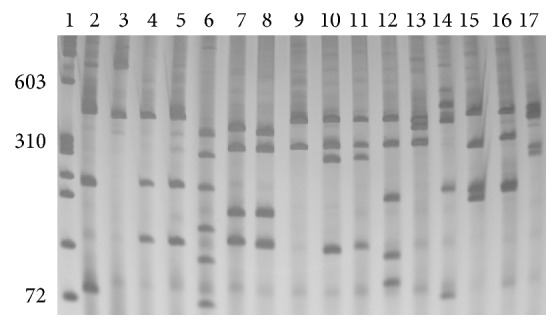
6% silver stained polyacrylamide gel showing restriction profiles obtained by digestion of the ITS region of DNA ribosomal with DdeI. Lane 1: molecular size markers Phi X 174/HaeIII; Lane 2: Biomphalaria glabrata; Lane 3: Biomphalaria tenagophila; Lane 4: Biomphalaria tenagophila guaibensis; Lane 5: Biomphalaria occidentalis; Lane 6: Biomphalaria schrammi; Lane 7: Biomphalaria oligoza; Lane 8: Biomphalaria peregrina; Lane 9: Biomphalaria intermedia; Lane 10: Biomphalaria straminea; Lane 11: Biomphalaria kuhniana; Lane 12: Biomphalaria amazonica; Lane 13: Biomphalaria cousini; Lane 14: Biomphalaria prona; Lane 15: Biomphalaria edisoni; Lane 16: Biomphalaria havanensis; Lane 17: Biomphalaria orbignyi. The numbers on the left of the gel represent the value in base pairs (bp).
Studies that incorporate morphological and molecular techniques in taxonomic analysis can generate data that allow a better interpretation and understanding of the biological diversity of the organisms under study. In fact, both the molecular and morphological taxonomy, if properly applied, successfully achieve the same goal [11]. In previous studies, the diagnosis of S. mansoni in molluscs has been performed using the LS-PCR [12], the conventional PCR assays for amplification of the Sm1–7 repeated sequence [13], and Loop-Mediated Isothermal Amplification [14] and otherwise the most frequent technique used to the identification of Biomphalaria is the PCR-RFLP.
This study has demonstrated the usefulness of the PCR-RFLP technique in the diagnosis of infection by S. mansoni in molluscs concurrently with identification of the four intermediate hosts, B. glabrata, B. tenagophila, B. straminea, and B. prona (Figure 1). In addition it was possible to identify a unique profile for the cercariae of S. mansoni, C. macrogranulosa, C. caratinguensis, and C. ocellifera, obtained from snails Biomphalaria collected in the field, after amplification of the ITS region of the rDNA digestion individually with the enzymes DdeI, AluI, HaeIII, RsaI, and HpaII (data not published).
Thus, this molecular biology technique has great utility for generating new knowledge about the taxonomy of molluscs of the genus Biomphalaria. Further, from the genetic analysis of various species of Schistosoma and Biomphalaria, it was observed that intraspecific genetic polymorphism of the parasite is limited while in the mollusc, it is very pronounced, showing the higher relevance of molluscan genetics over parasite genetics in determining the epidemiology of the disease [15]. For example, in adult B. glabrata, resistance to S. mansoni has been shown to be a dominant single-gene trait that is inherited by Mendelian genetics. In contrast, in juveniles, the genetics of resistance has been shown to involve 5 to 6 genes each with multiple alleles [16]. Additionally, Ittiprasert and Knight report reversing the resistance phenotype of resistant BS-90 B. glabrata by applying stress in the form of a mild heat pulse before they were exposed to S. mansoni, rendering these snails susceptible [17].
Acknowledgments
This work was partially supported by grants from FAPEMIG; CNPq (304121/2014-2); and Fiocruz. The authors would like to thank the “Lobato Paraense” Mollusc Rearing, the Medical Malacological Collection (Fiocruz-CMM) of René Rachou Research Center to the support for this research, and the Program for Technological Development in Tools for Health-PDTIS/Fiocruz for use of its facilities.
Competing Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Engels D., Chitsulo L., Montresor A., Savioli L. The global epidemiological situation of schistosomiasis and new approaches to control and research. Acta Tropica. 2002;82(2):139–146. doi: 10.1016/S0001-706X(02)00045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paraense W. L. Estado atual da sistemática dos planorbídeos brasileiros. Arquivos do Museu Nacional. 1975;55:105–128. [Google Scholar]
- 3.Vidigal T. H. D. A., Caldeira R. L., Simpson A. J. G., Carvalho O. S. Further studies on the molecular systematics of Biomphalaria snails from Brazil. Memorias do Instituto Oswaldo Cruz. 2000;95(1-2):57–66. doi: 10.1590/s0074-02762000000100009. [DOI] [PubMed] [Google Scholar]
- 4.Pointier J.-P., DeJong R. J., Tchuem Tchuenté L. A., Kristensen T. K., Loker E. S. A neotropical snail host of Schistosoma mansoni introduced into Africa and consequences for the Schistosomiasis transmission: Biomphalaria tenagophila in Kinshasa (Democratic Republic of Congo) Acta Tropica. 2005;93(2):191–199. doi: 10.1016/j.actatropica.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Dejong R. J., Morgan J. A. T., Lobato Paraense W., et al. Evolutionary relationships and biogeography of Biomphalaria (Gastropoda: Planorbidae) with implications regarding its role as host of the human bloodfluke, Schistosoma mansoni . Molecular Biology and Evolution. 2001;18(12):2225–2239. doi: 10.1093/oxfordjournals.molbev.a003769. [DOI] [PubMed] [Google Scholar]
- 6.Pointier J. P., Paraense W. L., Dejong R. J., Loker E. S., Bargues M. D., Mas-Coma S. A potential snail host of schistosomiasis in Bolivia: Biomphalaria amazonica Paraense, 1966. Memorias do Instituto Oswaldo Cruz. 2002;97(6):793–796. doi: 10.1590/s0074-02762002000600007. [DOI] [PubMed] [Google Scholar]
- 7.Lotfy W. M., DeJong R. J., Black B. S., Loker E. S. Specific identification of Egyptian Biomphalaria species and possible hybrids using the polymerase chain reaction based on nuclear and mitochondrial loci. Molecular and Cellular Probes. 2005;19(1):21–25. doi: 10.1016/j.mcp.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Abou-El-Naga I. F., El-Nassery S. M. F., Allam S. R., Shaat E. A., Mady R. F. M. Biomphalaria species in Alexandria water channels. Parasitology International. 2011;60(3):247–254. doi: 10.1016/j.parint.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 9.Jannotti-Passos L. K., Caldeira R. L., Carvalho O. S. Técnicas utilizadas no estudo dos moluscos do gênero Biomphalaria e na manutenção do ciclo de Schistosoma mansoni . In: Carvalho O. S., Coelho P. M. Z., Lenzi H. L., editors. Schistosoma Mansoni e Esquistossomose: Uma Visão Multidisciplinar. Rio de Janeiro, Brazil: 2008. pp. 531–544. [Google Scholar]
- 10.Kane R. A., Rollinson D. Repetitive sequences in the ribosomal DNA internal transcribed spacer of Schistosoma haematobium, Schistosoma intercalatum and Schistosoma mattheei . Molecular and Biochemical Parasitology. 1994;63(1):153–156. doi: 10.1016/0166-6851(94)90018-3. [DOI] [PubMed] [Google Scholar]
- 11.Moritz C., Hillis D. M. Molecular systematics: context and controversies. In: Hillis D. M., Moritz C., Mable B. K., editors. Molecular Systematics. Sunderland, Mass, USA: Sinauer Associates; 1996. pp. 11–16. [Google Scholar]
- 12.Jannotti-Passos L. K., Vidigal T. H. D. A., Dias-Neto E., et al. PCR amplification of the mitochondrial DNA minisatellite region to detect Schistosoma mansoni infection in Biomphalaria glabrata snails. Journal of Parasitology. 1997;83(3):395–399. doi: 10.2307/3284400. [DOI] [PubMed] [Google Scholar]
- 13.Hamburger J., He N., Xu Y. X., Ramzy R. M., Jourdane J., Ruppel A. A polymerase chain reaction assay for detecting snails infected with Bilharzia parasites (Schistosoma mansoni) from very early prepatency. American Journal of Tropical Medicine and Hygiene. 1998;59(6):872–876. doi: 10.4269/ajtmh.1998.59.872. [DOI] [PubMed] [Google Scholar]
- 14.Abbasi I., King C. H., Muchiri E. M., Hamburger J. Detection of Schistosoma mansoni and Schistosoma haematobium DNA by loop-mediated isothermal amplification: identification of infected snails from early prepatency. The American Journal of Tropical Medicine and Hygiene. 2010;83(2):427–432. doi: 10.4269/ajtmh.2010.09-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simpson A. J., Dias Neto E., Vidigal T. H., Pena H. B., Carvalho O. S., Pena S. D. DNA polymorphism of schistosomes and their snail hosts. Memórias do Instituto Oswaldo Cruz. 1995;90(2):211–213. doi: 10.1590/s0074-02761995000200014. [DOI] [PubMed] [Google Scholar]
- 16.Richards C. S., Knight M., Lewis F. A. Genetics of Biomphalaria glabrata and its effect on the outcome of Schistosoma mansoni infection. Parasitology Today. 1992;8(5):171–174. doi: 10.1016/0169-4758(92)90015-t. [DOI] [PubMed] [Google Scholar]
- 17.Ittiprasert W., Knight M. Reversing the resistance phenotype of the Biomphalaria glabrata snail host Schistosoma mansoni infection by temperature modulation. PLoS Pathogens. 2012;8(4) doi: 10.1371/journal.ppat.1002677.e1002677 [DOI] [PMC free article] [PubMed] [Google Scholar]


