Abstract
The genus Arcobacter includes species considered emerging food and waterborne pathogens. Despite Arcobacter has been linked to the presence of faecal pollution, few studies have investigated its prevalence in wastewater, and the only isolated species were Arcobacter butzleri and Arcobacter cryaerophilus. This study aimed to establish the prevalence of Arcobacter spp. at a WWTP using in parallel two culturing methods (direct plating and culturing after enrichment) and a direct detection by m-PCR. In addition, the genetic diversity of the isolates was established using the ERIC-PCR genotyping method. Most of the wastewater samples (96.7%) were positive for Arcobacter and a high genetic diversity was observed among the 651 investigated isolates that belonged to 424 different ERIC genotypes. However, only few strains persisted at different dates or sampling points. The use of direct plating in parallel with culturing after enrichment allowed recovering the species A. butzleri, A. cryaerophilus, Arcobacter thereius, Arcobacter defluvii, Arcobacter skirrowii, Arcobacter ellisii, Arcobacter cloacae, and Arcobacter nitrofigilis, most of them isolated for the first time from wastewater. The predominant species was A. butzleri, however, by direct plating predominated A. cryaerophilus. Therefore, the overall predominance of A. butzleri was a bias associated with the use of enrichment.
1. Introduction
The genus Arcobacter is included together with Campylobacter and Helicobacter in the family Campylobacteraceae, and all of these genera include species that might be pathogenic to humans and animals [1, 2]. Arcobacter butzleri is the fourth most common Campylobacter-like organism isolated from the stool of human patients with diarrhoea in two independent studies carried out in France [3] and Belgium [4]. Furthermore, in a recent study, Arcobacter species was the fourth most common pathogen group isolated from faecal samples from persons with acute enteric disease [5]. It has been demonstrated that the presence of Arcobacter in water correlates with the presence of faecal pollution [2]. In this sense, Arcobacter has been recovered in three outbreaks in which drinking water was contaminated with sewage ([2] and references therein). Food products, especially meat, shellfish, and milk, have also been found contaminated with bacteria of this genus, mainly with A. butzleri [2, 6]. Considering this, the International Commission on Microbiological Specifications for Foods has defined A. butzleri as a serious hazard to human health [6], and it has been identified as an important zoonotic agent to humans and animals ([2] and references therein).
Disposal of sewage is a critical issue in modern cities that normally deliver it to receiving waters after treatment at wastewater treatment plants (WWTPs). This treatment is aimed at reducing degradable organic matter under controlled conditions before it is discharged into natural bodies of water [7]. However, conventional primary and secondary treatments per se (without disinfection steps) do not eliminate the pathogens present in the water and as a result WWTP outflows contain a lot of microbes that are potentially pathogenic to humans and animals.
The presence of Arcobacter in water, including sewage from WWTPs, has been reported in several studies [2, 8–13]. In those studies, Arcobacter spp. were isolated in 40% to 100% of the samples studied, using different culture media and protocols, and were found in 66% to 100% of the samples when direct detection by molecular techniques was used [2, 10–14]. Three studies have investigated the presence of Arcobacter in WWTPs after using different treatments [8–10]. Despite different results being obtained all the studies showed the presence of these bacteria at all points of the treatment, including the water outflow. Furthermore, using pyrosequencing of the V6 hypervariable region of 16S rRNA gene, Arcobacter were found to be one of the predominant taxa in WWTPs in Milwaukee (USA) in contrast to their scarcity in surface waters [15]. In fact, considering those results, Arcobacter spp. were selected as “sewer signature microbes” together with Acinetobacter and Trichococcus (the most common taxa in sewage) in the detection of sewage contamination of surface waters [16].
Studies on wastewater samples using conventional culture protocols that included an enrichment step in a selective broth found that A. butzleri was more predominant than A. cryaerophilus [2, 8–12]. However, it has been suggested that growth of some Arcobacter species may be enhanced in the enrichment broth, which can mask other species, leading to a bias in the estimation of the diversity [17]. On the other hand, the best atmosphere of incubation (aerobic or microaerophilic) for arcobacters has not yet been determined, and half of the studies have used aerobic conditions [2]. Furthermore, only one study so far has compared the effect of both atmosphere incubation conditions on the recovery of Arcobacter, but it did not reach clear conclusions [11].
The genetic diversity of Arcobacter in sewage has seldom been studied and methods used include random amplification of polymorphic DNA [12] and enterobacterial repetitive intergenic consensus (ERIC-PCR) [18]. Results showed a wide range of genotypes, as happens in samples from other environments [2].
The objective of this survey is to establish the prevalence and genetic diversity of Arcobacter spp. in a WWTP using two culturing approaches (direct plating and culturing after enrichment) and comparing the recovery under aerobic or microaerophilic conditions, using direct detection by m-PCR in parallel.
2. Materials and Methods
2.1. Samples and Water Processing
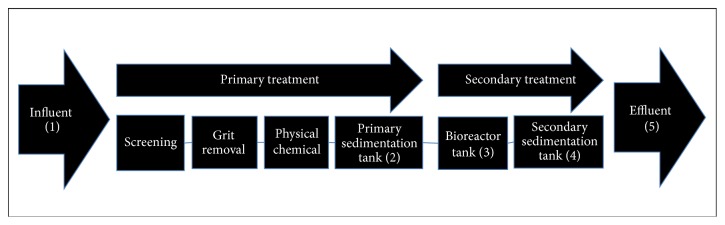
The wastewater samples were collected on six sampling dates (April, June, and October 2009 and May, June, and September 2011) from five sampling points (Figure 1) at the WWTP in Reus, Spain, that produced nondisinfected secondary treated wastewater. The sampling points were located in the influent and effluent water in the primary and secondary sedimentation tanks and in the bioreactor tank. Samples were collected into 2-litre sterile polypropylene bottles, which were then chilled in ice during transport. Microbiological assays began on the same day as sampling.
Figure 1.
Simplified scheme of the wastewater treatment plant indicating the five sampling points (1 to 5).
From each water sample 200 mL was filtered through a 0.45 μm membrane filter (47 mm diameter) in duplicate (Millipore Corp., Bedford, MA, USA). Then the filters were rolled and one of them was introduced into a tube containing 1 mL distilled water and vigorously mixed by vortexing. The other filter was then introduced into a tube containing 9 mL of Arcobacter-CAT broth (Arcobacter-enrichment broth supplemented with the CAT antibiotic supplement, i.e., Cefoperazone, amphotericin B and teicoplanin, and oxoid, Basingstoke, UK) and incubated aerobically (30°C, 48 to 72 h).
2.2. Molecular Detection
For molecular detection, 400 μL of water from the 2 tubes was centrifuged at 12,000 rpm min−1 and the obtained pellet was resuspended and washed 3 times with Milli-Q sterile water. Afterwards, DNA was extracted by using the InstaGene™ DNA Purification Matrix (Bio-Rad Laboratories, Hercules, CA), and direct detection of Arcobacter was carried out using the primers and conditions of the m-PCR designed by Houf et al. [19] for the detection of A. butzleri, A. cryaerophilus, and A. skirrowii. The procedure included initial denaturation for 2 min at 94°C followed by 32 cycles of denaturation for 45 sec at 94°C, annealing for 45 sec at 61°C, and chain extension for 60 sec at 72°C and a final extension for 5 min at 72°C. The PCRs were carried out in a reaction mixture containing 1 μL of DNA extract, 1.5 U of Taq DNA polymerase (Invitrogen, Carlsbad, CA), 0.2 mM of each deoxyribonucleotide triphosphate, 50 pmol of each primer set (Invitrogen, Carlsbad, CA), and 1.3 mM of MgCl2.
For molecular detection, 400 μL of water from the 2 tubes was centrifuged at 12,000 rpm min−1 and the obtained pellet was resuspended and washed 3 times with Milli-Q sterile water. Afterwards, DNA was extracted by using the InstaGene DNA Purification Matrix (Bio-Rad Laboratories, Hercules, CA), and direct detection of Arcobacter was carried out using the primers and conditions of the m-PCR designed by Houf et al. [19] for the detection of A. butzleri, A. cryaerophilus, and A. skirrowii. The procedure included initial denaturation for 2 min at 94°C followed by 32 cycles of denaturation for 45 sec at 94°C, annealing for 45 sec at 61°C, and chain extension for 60 sec at 72°C and a final extension for 5 min at 72°C. The PCRs were carried out in a reaction mixture containing 1 μL of DNA extract, 1.5 U of Taq DNA polymerase (Invitrogen, Carlsbad, CA), 0.2 mM of each deoxyribonucleotide triphosphate, 50 pmol of each primer set (Invitrogen, Carlsbad, CA), and 1.3 mM of MgCl2.
2.3. Culturing Procedure
For the direct detection by culturing, 200 μL of water from the nonenriched tube was transferred onto the surface of a 0.45 μm membrane filter (47 mm diameter), placed on blood agar medium (Trypticase Soy Agar, oxoid, Basingstoke, UK, supplemented with 5% sheep blood), and allowed to filter passively under ambient conditions for 30 min [5]. For culturing after enrichment, 200 μL of incubated Arcobacter-CAT broth (enrichment broth) was transferred onto the surface of a 0.45 μm membrane filter (47 mm diameter), placed on blood agar medium, and allowed to filter as described above. The filters were then removed and the plates were incubated (30°C, 48 to 72 h) under aerobic conditions.
For samples collected in 2011, direct and postenrichment culturing was also processed in duplicate in order to allow the parallel incubation under aerobic and microaerophilic conditions. The microaerophilic conditions (oxygen, 6% to 16%; carbon dioxide, 2% to 10%; and nitrogen, 80%) were generated by using GasPak EZ campy container system sachets (Becton Dickinson, Sparks, MD, USA).
2.4. Arcobacter Isolation, Genotyping, and Identification
From each positive sample, eight small, colourless or beige to off-white, translucent colonies were picked, streaked to purity, and confirmed as presumptive arcobacters on the basis of their response to phenotypic tests (i.e., gram negative slightly curved rods that were positive for oxidase and motility). All isolates were genotyped using the ERIC-PCR technique, using the Houf et al. [17] protocol for Arcobacter. DNA was extracted as described above and the concentration of each DNA template was determined using the GeneQuant pro (Amersham Biosciences, Cambridge, England) at A260 and adjusted to 25 ng mL−1. Gel images were saved as TIFF files, normalized with the 100 bp DNA Ladder (Invitrogen), and further analysed by Bionumerics software, version 6.5 (Applied Maths, Belgium). Patterns with one or more different bands were considered different genotypes [17].
All strains (1 representative of each genotype) were finally identified using in parallel two molecular identification methods, the m-PCR described above for the direct detection [19] and the 16S rDNA-RFLP [1, 20]. When identification was not possible with these methods or discordant results were obtained, the rpoB and/or 16S rRNA genes were sequenced using primers and conditions previously described [1].
2.5. Counting of Arcobacter
Direct counting of Arcobacter was carried out from all wastewater samples collected in 2011 as previously described [21, 22]. In brief, water samples were tenfold diluted in 0.1% peptone water (oxoid, Basingstoke, UK), from 100 to 10−8 and then 100 μL of each tenfold dilution was inoculated onto Arcobacter selective isolation agar plate (24 g litre−1 Arcobacter broth, oxoid, Basingstoke, UK; 12 g litre−1, Agar Technical No. 3, oxoid, Basingstoke, UK; supplemented with 100 mg litre−1 5-fluorouracil, 100 mg litre−1 cycloheximide, 10 mg litre−1, amphotericin B, 16 mg litre−1 cefoperazone, 32 mg litre−1 novobiocin, and 64 mg litre−1 trimethoprim, Sigma, USA) [23]. All plates were then incubated for 48 h at 30°C under microaerophilic conditions. After incubation, plates were checked for typical bluish colonies using Henry transillumination and the colony forming units (CFU) were counted and then informed as CFU mL−1 [22, 23]. The tenfold dilutions prepared in peptone water were used to enumerate Arcobacter using the MPN method. Briefly, each of the 5 dilutions (1, 0.1, 0.001, 0.0001, and 0.00001 mL of the original sample) was inoculated in 5 tubes containing Arcobacter-CAT broth for the MPN calculation as previously described by Levican [1]. The broths were incubated for 48 h under aerobic conditions and then 100 μL of each tube was inoculated by passive filtration onto 5% sheep blood agar plates and incubated under the same conditions. The MPN of Arcobacter in 100 mL was estimated from the obtained combination of positive tubes using the MPN CALCULATOR Software (Curiale M, 2004 available from http://www.i2workout.com/mcuriale/mpn/).
In order to confirm that typical colonies obtained by direct counting or from the MPN positive tubes belonged to Arcobacter spp., a representative number per plate was randomly selected to be identified by the molecular methods described above, that is, m-PCR [19] and 16S rDNA-RFLP [1, 20].
2.6. Statistical Analyses
The proportions obtained using different methods were compared using the Z test and a P value < 0.05 was considered as statistically significant.
3. Results and Discussion
3.1. Prevalence and Diversity of Arcobacter Species
Arcobacter spp. were recovered from 29 of the 30 samples (96.7%), from which 651 isolates recovered by culturing were confirmed to belong to the genus Arcobacter (Table 1). Those isolates were genotyped with ERIC-PCR and their patterns indicated that they belonged to 424 different genotypes, so the global genetic diversity was 65.1% (Table 1). In previous studies that used different culture media and protocols, the prevalence of Arcobacter spp. from wastewater samples ranged from 40% to 100% [10–14, 18]. When a genotyping method was applied, a high genetic diversity was observed. For example, Collado et al. [18] report that 90.2% of the isolates belonged to different ERIC-PCR genotypes, while González et al. [12] found that all of their isolates belonged to different RAPD-PCR genotypes. This high genetic diversity has previously been explained by possible multiple sources of contamination and/or as a consequence of genomic rearrangement [12, 18].
Table 1.
Number of isolates and strains of the identified Arcobacter species found from wastewater using the two molecular methods (m-PCRand 16S rRNA-RFLP) in parallel.
| Species | Number of isolates (%) | Number of strains (%) | % genetic diversity | Molecular identification |
|---|---|---|---|---|
| m-PCRa/16S rRNA-RFLPb | ||||
| A. butzleri (Ab) | 354 (54.4) | 247 (58.3) | 69.8% | Ab/Ab |
| A. cryaerophilus (Ac) | 226 (34.7) | 150 (35.4) | 66.4% | Ac/Ac |
| A. thereius (At) | 37 (5.7) | 9 (2.1) | 24.3% | Ac/Ab |
| A. skirrowii (As) | 16 (2.5) | 5 (1.2) | 31.3% | As/As |
| A. defluviic (Ad) | 12 (1.8) | 8 (1.9) | 66.7% | ~230 bp/Ad |
| A. ellisii (Ae) | 3 (0.5) | 2 (0.5) | 66.7% | Ac/Ae |
| A. cloacaed (Aclo) | 2 (0.3) | 2 (0.5) | 100% | Ac/Aclo |
| A. nitrofigilis (An) | 1 (0.2) | 1 (0.2) | 100% | As/An |
|
| ||||
| Total | 651 | 424 | 65.1% | |
The isolates were genotyped with ERIC-PCR to determine the ones that showed the same ERIC-pattern and therefore belonged to the same strain. aHouf et al. [19] and bFigueras et al. [20]. Results of the RFLP were verified by sequencing the rpoB gene. cNew species recognized on the basis of the new RFLP pattern described by Figueras et al [20]. dNew species recognized on the basis of the new RFLP pattern and described by Levican et al. [21].
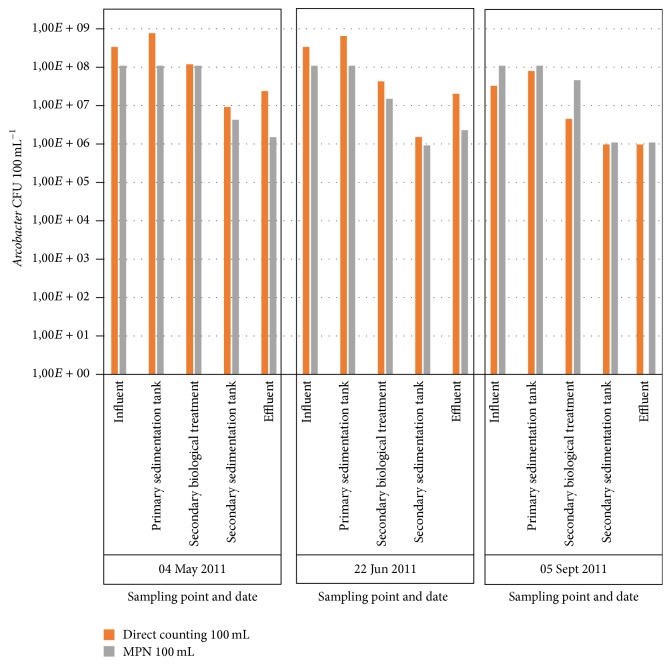
Arcobacter spp. were isolated from all sampling points, with the exception of only one sample taken at the water outflow (Table 2). The amount of arcobacters showed a decrease of at least 2 logarithms from the influent to the effluent of the WWTP and both enumeration methods showed similar results (Figure 2). The densities of Arcobacter found in the effluent water in our study are similar to the results shown in another study performed from the same WWTP [21, 24]. No seasonal variation was observed among results (Figure 2).
Table 2.
Arcobacter species detected according to the method at the 5 sampling points in the wastewater treatment plant on the 6 different sampling occasionsa.
| Species detected | m-PCR | Culturing method | |||
|---|---|---|---|---|---|
| Direct | Postenrichment | Direct | Postenrichment | ||
| A. butzleri (Ab) | 19 (100) | 15 (100) | 22 (78.6) | 24 (82.8) | |
| A. cryaerophilus (Ac) | 10 (52.6) | 9 (60) | 27 (96.4) | 16 (55.2) | |
| A. defluvii (Ad) | 0 | 0 | 2 (7.1) | 3 (10.3) | |
| A. nitrofigilis (Anit) | 0 | 0 | 1 (3.6) | 0 | |
| A. cloacae (Aclo) | 0 | 0 | 0 | 2 (6.9) | |
| A. skirrowii (As) | 0 | 0 | 1 (3.6) | 2 (6.9) | |
| A. thereius (At) | 0 | 0 | 3 (10.7) | 4 (13.8) | |
| A. ellisii (Ae) | 0 | 0 | 2 (7.1) | 0 | |
|
| |||||
| Sampling month | Sampling point | m-PCR | Culturing method | ||
| Direct | Postenrichment | Direct | Postenrichment | ||
|
| |||||
| April 2009 | Influent water | Ac+Ab | ND | Ac+Anit | Ac+Ad+Aclo |
| Primary sedimentation tank | Ac+Ab | ND | Ac+Ab+Ad | Ac+Ab | |
| Secondary bioreactor tank | Negative | ND | Ac+Ad | Ac+Ad | |
| Secondary sedimentation tank | Negative | ND | Negative | Ab+Ad | |
| Effluent water | Negative | ND | Negative | Negative | |
|
| |||||
| June 2009 | Influent water | Negative | ND | Ac+Ab | Ab |
| Primary sedimentation tank | Ab | ND | Ac+Ab | Ab | |
| Secondary bioreactor tank | Negative | ND | Ac+Ab | Ab | |
| Secondary sedimentation tank | Negative | ND | Ac | Ac+Ab | |
| Effluent water | Negative | ND | Ac+Ab | Ab | |
|
| |||||
| October 2009 | Influent water | Ac+Ab | ND | Ac | Ac+Ab |
| Primary sedimentation tank | Ac+Ab | ND | Ac+Ab | Ac+Ab | |
| Secondary bioreactor tank | Negative | ND | Ac | Ab | |
| Secondary sedimentation tank | Ab | ND | Ac+Ab | Ac+Ab | |
| Effluent water | Negative | ND | Ac+Ab | Ab | |
|
| |||||
| May 2011 | Influent water | Ab | Ab+Ac | Ac+At | Ab+Ac+At |
| Primary sedimentation tank | Ab+Ac | Ab+Ac | Ab+Ac+At | Ac+At | |
| Secondary bioreactor tank | Ab | Ab+Ac | Ab+Ac+As | Ac+As | |
| Secondary sedimentation tank | Ab | Ab+Ac | Ab+Ac | Ab+Ac | |
| Effluent water | Ab | Ab+Ac | Ab+Ac | Ab+Ac | |
|
| |||||
| June 2011 | Influent water | Ab+Ac | Ab | Ab+Ac+Ae | Ab+As |
| Primary sedimentation tank | Ab+Ac | Ab | Ab+Ac | Ab+Ac | |
| Secondary bioreactor tank | Ab | Ab | Ab+Ac+At | At | |
| Secondary sedimentation tank | Ab | Ab | Ab+Ac | Ab+Ac+At | |
| Effluent water | Ab | Ab | Ab+Ac | Ab | |
|
| |||||
| September 2011 | Influent water | Ab+Ac | Ab+Ac | Ab+Ac+Ae | Ab+Aclo |
| Primary sedimentation tank | Ab+Ac | Ab+Ac | Ab+Ac | Ab | |
| Secondary bioreactor tank | Ab+Ac | Ab+Ac | Ab+Ac | Ab+Ac | |
| Secondary sedimentation tank | Negative | Ab | Ab | Ab | |
| Effluent water | Negative | Ab+Ac | Ab+Ac | Ab+Ac | |
|
| |||||
| Total No. positive samples (%) | 19/30 (63.3) | 15/15 (100) | 28/30 (86.7) | 29/30 (86.7) | |
aThe identified species are only mentioned once, independently of the number of strains obtained from each specific sample.
m-PCR: multiplex PCR Houf et al., 2000 [19]
ND: not done.
Figure 2.
Amount of Arcobacter found in the different sampling points by date of sampling.
On the other hand, when the 424 genotypes were analysed with the Bionumerics software, only 4 of them (0.9%) were coincidentally recovered from different sampling points at the same time or on different samplings days (data not shown). This indicates that most of the Arcobacter strains do not persist over the time in the WWTP. As in previous studies [8, 10], Arcobacter was present at all sampling points suggesting that conventional wastewater treatment is not able to completely remove these bacteria.
A total of 8 Arcobacter spp. were recovered in this study among the 424 strains, the most prevalent being A. butzleri and A. cryaerophilus, which together accounted for 94.8% (n = 402) of strains. Both species showed a similar genetic diversity (69.8% and 66.4%, resp.; Table 1). In a previous study in river water impacted by sewage effluents [16], A. cryaerophilus had a slightly wider diversity (95.2%) than A. butzleri (90.2%). The remaining 22 strains (5.2%) belonged to 6 species (Table 1); two of them were new species recovered for the first time from these samples and were described elsewhere, that is, A. defluvii [10] and A. cloacae [1, 21]. To our knowledge this is the first time that the other 3 species, A. nitrofigilis, A. thereius, and A. ellisii, have been isolated from sewage. The species A. thereius had been isolated previously from animal faeces and porcine abortions [22, 25] and has very recently been reported from the faeces of patients with diarrhoea in Belgium [5]. However, A. nitrofigilis has so far only been genetically identified from mussels [2] since its description from the roots of a salt marsh plant [26] but A. ellisii has never again been isolated since its description from shellfish [27]. It is noticeable that the 8 Arcobacter spp. recovered in the present study have also been recovered from mussels in a recent study [28]. In this sense, wastewater may be the source of contamination of seawater with these bacteria in the shellfish harvesting area from where they can be concentrated in mussels by filtering. Therefore, our results also support the previous suggestion that potential pathogenic arcobacters enter seawater with sewage-polluted fresh water [2]. The two molecular identification methods used in this study [17, 18] showed the same results for 402 of the 424 strains (94.8%), 247 of them (58.3%) being A. butzleri, 150 (35.4%) being A. cryaerophilus, and 5 (1.2%) being A. skirrowii (Table 1). The other 22 strains (5.2%) gave different results with the two methods and their identity was confirmed by sequencing the rpoB and 16S rRNA genes (Table 1). Several of the available detection and identification methods for Arcobacter spp. have failed to recognize all known species or have confused them with the most common ones [29]. In this regard, the recognition of such a high number of different species in our study confirms the previous suggestion that the known diversity of Arcobacter spp. in different environments will become more precise as reliable identification methods are applied [29].
3.2. Detection Using m-PCR and Culturing Methods
Of the 30 samples studied, 28 (86.7%) were positive by direct plating, 29 (93.3%) by postenrichment, and 19 (63.3%) by direct m-PCR, and as indicated above only one sample taken from the WWTP outflow was negative by all three methods (Table 2). However, 15 of the 15 samples (100%) tested by m-PCR after enrichment were positive (Table 2). Therefore, comparing those methods, direct detection by m-PCR [19] performed worse (Table 2) than the other two. Previous studies that have investigated Arcobacter in wastewater by the same m-PCR detection method from the enrichment broth reported the same number of positive samples as by culturing [2, 11] or a higher number of positive samples by m-PCR (100%) than by culturing (45.5%) [12]. The bad performance of direct detection by m-PCR from the samples studied could be explained by different factors that were not controlled for in the present study, that is, the presence of inhibitors and the low concentration of the Arcobacter spp. in relation to the sensitivity of the m-PCR method for the detection of the different species. It is clear that the enrichment step improves growth, which might increase the level of target cells and thus the percentage of detection. Despite that, it has been demonstrated by Ho et al. [30] that the detection of the different species by m-PCR is biased when applied after the enrichment step. Regarding that, the latter authors suggested that the species that grow faster in enrichment are more easily detected. However, that study did not determine whether this behaviour was due to the different concentrations of the bacteria cells in the mixtures that could be under the detection limit of the method established by Houf et al. [19] at 103 cfu mL−1.
Regarding the species detected when using this m-PCR method [19], some of them might be underestimated because the method was specifically created to detect only A. butzleri, A. cryaerophilus, and A. skirrowii and we know that A. cloacae, for example, produces the same amplicon expected for A. cryaerophilus (257 bp) and A. defluvii a very similar one (~230 bp) [2] while A. nitrofigilis produces the amplicon expected for A. skirrowii (625 bp) [2, 29].
When comparing the performance of different incubation conditions, similar results were observed; that is, 45.7% of the strains were recovered under aerobic conditions and 45.4% under microaerophilic conditions (Table 3). In both cases, the predominant species were A. butzleri followed by A. cryaerophilus, with no significant difference between results. As commented in Introduction, although about half of the existing studies used aerobic conditions for incubation ([2] and references therein) this is a poorly explored aspect and the only study to assess this gave inconclusive results [11]. Based on the results obtained in our study, the use of microaerophilic conditions seems not to be justified, considering that aerobic conditions yielded almost the same results, and is cheaper and easier to carry out.
Table 3.
Number of Arcobacter spp. strains recovered depending on aerobic (A) and microaerophilic (MA) incubation conditions.
| Speciesa | Total recovered (%) | Only A (%) | Only MA (%) | A & MA (%) |
|---|---|---|---|---|
| A. butzleri | 170 (60.7) | 75 (58.6) | 83 (65.4) | 12 (48.0) |
| A. cryaerophilus | 94 (33.6) | 50 (39.1) | 39 (30.7) | 5 (20.0) |
| A. thereius | 9 (3.2) | 2 (1.6) | 2 (1.6) | 5 (20.0) |
| A. skirrowii | 4 (1.4) | 0 | 1 (0.8) | 3 (12.0) |
| A. ellisii | 2 (0.7) | 1 (0.8) | 1 (0.8) | 0 |
| A. cloacae | 1 (0.4) | 0 | 1 (0.8) | 0 |
|
| ||||
| 280 | 128 (45.7%) | 127 (45.4%) | 25 (8.9%) | |
aThe species A. defluvii and A. nitrofigilis do not appear at the table because they were isolated in 2009 where only aerobic incubation conditions were employed.
In relation to the comparative performance of culturing methods and independently of the incubation conditions (data not shown), direct plating obtained a higher, although not significant, number of strains (n = 218) than postenrichment (n = 189) as shown in Table 4. The predominant species isolated by each method were different (Table 4); that is, the most abundant species recovered under direct plating conditions were A. cryaerophilus (50%) and A. butzleri (46.8%). However, A. butzleri was the most frequently isolated under postenrichment culturing conditions (74.1%) followed by far by A. cryaerophilus (19%; Table 4). The species A. thereius, A. skirrowii, and A. defluvii were isolated by both methods, whereas A. nitrofigilis and A. ellisii were recovered only by direct plating and A. cloacae only by postenrichment (Tables 2 and 4). Contrary to that, previous studies in wastewater have shown lower species diversity with A. butzleri and/or A. cryaerophilus being the only species recovered [2, 8, 9, 11, 12, 18]. This lower diversity has probably been originated as a result of the lower number of isolates investigated and the applied methodology including only an enrichment step but no direct plating. In fact, if only culturing after enrichment was carried out in our study, A. butzleri would be 3.9 times more prevalent than A. cryaerophilus (140 versus 36 strains, Table 4). Nevertheless, the true proportion of both species determined by direct culturing was 0.94 (102 of A. butzleri versus 109 strains of A. cryaerophilus). The former 3.9 proportion is in agreement with 4 times more prevalence of A. butzleri (248 strains) than A. cryaerophilus (60 strains) reported in a previous study in which samples were cultured using the same enrichment procedure but no direct plating [18]. A previous study on Arcobacter in broiler carcasses from Belgium compared the diversity of strains obtained by direct plating and by postenrichment in parallel and found that A. butzleri was 5.4 times (49 versus 9 strains) more prevalent than A. cryaerophilus by postenrichment culturing, while both species showed more similar proportion (42 A. butzleri versus 31 A. cryaerophilus, i.e., 1.4 : 1) when they were recovered by direct plating [16]. Consequently, those authors recommend the use of the two methods in parallel in order to enhance the diversity of species recovered. Another study from the same country performed by De Smet et al. [22] compared the recovered isolates from pig faeces using again the two methods. Regarding the species diversity, they found more isolates of A. skirrowii and A. thereius by direct plating than by postenrichment and more of A. butzleri and A. trophiarum by postenrichment than by direct plating [22]. Those authors hypothesized that the predominance of one species over another is due to the isolation procedure and medium used to recover the species rather than to its higher occurrence in the samples [17, 22]. More evidence of the influence of the culturing method applied has recently been provided by Fisher et al. [31], when studying the Arcobacter populations in wastewater from different cities in the United States and from the city of Reus (Spain) using a metagenomic analysis targeting the V4V5 regions of 16S rRNA gene. Those authors found that the predominant oligotypes matched with A. cryaerophilus while A. butzleri was only the eleventh most abundant oligotype [31]. Interestingly, they also report a correlation between the abundance of some Arcobacter oligotypes and water temperature. Another study on the Arcobacter diversity in shellfish and seawater observed that adding a 2.5% NaCl to the Arcobacter-CAT enrichment broth and subculturing in marine agar produced a significant increase on the recovered number of species (11 known species and 7 new candidate species) more than with the conventional method (7 known species and 2 new candidate species) [32]. Those authors recommended this new protocol for the isolation of Arcobacter from marine and brackish environments in order to avoid underestimation of the number of species [32]. They also stated that this simple modification of the culture shows a big influence on the community of species recovered. This finding is considered especially relevant in this metagenomics era, when it is not clear to what extent the differences observed between culturing and nonculturing methods are influenced by the culture media and conditions applied [32]. Therefore, future studies are warranted to assess the effect on Arcobacter isolation when using different media or conditions such as the incubation temperature. In this regard, the observed high prevalence and genetic diversity of Arcobacter spp. from wastewater confirm that this is an important reservoir for bacteria of this genus and could be a good matrix for testing different isolation protocols for the recovery of these bacteria.
Table 4.
Number of strains (%) of the different Arcobacter spp. recovered by direct plating (DP) and postenrichment (PE).
| Species | Total recovered (%) | Only by DP (%) | Only by PE (%) | DP& PE (%) |
|---|---|---|---|---|
| A. butzleri | 247 (58.3) | 102 (46.8) | 140 (74.1) | 5 (29.4) |
| A. cryaerophilus | 150 (35.4) | 109 (50.0) | 36 (19.0) | 5 (29.4) |
| A. thereius | 9 (2.1) | 2 (0.9) | 4 (2.1) | 3 (17.6) |
| A. skirrowii | 5 (1.2) | 1 (0.5) | 3 (1.6) | 1 (5.9) |
| A. defluvii | 8 (1.9) | 1 (0.5) | 4 (2.1) | 3 (17.6) |
| A. ellisii | 2 (0.5) | 2 (0.9) | 0 | 0 |
| A. cloacae | 2 (0.5) | 0 | 2 (1.1) | 0 |
| A. nitrofigilis | 1 (0.2) | 1 (0.5) | 0 | 0 |
|
| ||||
| 424 | 218 (51.4%) | 189 (44.6%) | 17 (4.0%) | |
Acknowledgments
This work was supported by funds from the European Commission for the Aquavalens Project (KBBE.2012.2.5-01) and projects with references AGL2011-30461-C02-02 and JPIW2013-095-C03-03 from MINECO (Spain). Arturo Levican is indebted to Universitat Rovira i Virgili for a doctoral grant and to CONICYT, Chile, for financial support through Becas Chile. The authors would like to thank Jaume Cabré at the WWTP in Reus, Spain, for his help in providing them with the access to sampling points.
Disclosure
The authors are solely responsible for the content of this publication; it does not represent the opinion of the European Commission.
Competing Interests
The authors declare that there is no conflict of interest regarding the publication of this article.
References
- 1.Levican A. Sanitary importance of Arcobacter [Ph.D. thesis] Reus, Spain: Department of Basic Health Sciences, Universitat Rovira i Virgili; 2013. http://www.tesisenred.net/bitstream/handle/10803/125666/A_Levican_Phd_thesis.pdf?sequence=1. [Google Scholar]
- 2.Collado L., Figueras M. J. Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter . Clinical Microbiology Reviews. 2011;24(1):174–192. doi: 10.1128/cmr.00034-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prouzet-Mauléon V., Labadi L., Bouges N., Ménard A., Mégraud F. Arcobacter butzleri: underestimated enteropathogen. Emerging Infectious Diseases. 2006;12(2):307–309. doi: 10.3201/eid1202.050570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vandenberg O., Dediste A., Houf K., et al. Arcobacter species in humans. Emerging Infectious Diseases. 2004;10(10):1863–1867. doi: 10.3201/eid1010.040241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van den Abeele A.-M., Vogelaers D., Van Hende J., Houf K. Prevalence of Arcobacter species among humans, Belgium, 2008–2013. Emerging Infectious Diseases. 2014;20(10):1731–1734. doi: 10.3201/eid2010.140433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.ICMSF. International Commission on Microbiological Specifications for Foods. New York, NY, USA: Kluwer Academic/Plenum; 2002. Microorganisms in foods 7—microbiological testing in food safety management. [Google Scholar]
- 7.WHO. Guidelines for Safe Recreational Water Environments. Vol. 1. Geneva, Switzerland: World Health Organization; 2003. (Coastal and Fresh Waters). [Google Scholar]
- 8.Stampi S., Varoli O., Zanetti F., De Luca G. Arcobacter cryaerophilus and thermophilic campylobacters in a sewage treatment plant in Italy: two secondary treatments compared. Epidemiology and Infection. 1993;110(3):633–639. doi: 10.1017/s0950268800051050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stampi S., De Luca G., Varoli O., Zanetti F. Occurrence, removal and seasonal variation of thermophilic campylobacters and Arcobacter in sewage sludge. Zentralblatt für Hygiene und Umweltmedizin. 1999;202(1):19–27. [PubMed] [Google Scholar]
- 10.Moreno Y., Botella S., Alonso J. L., Ferrús M. A., Hernández M., Hernández J. Specific detection of Arcobacter and Campylobacter strains in water and sewage by PCR and fluorescent in situ hybridization. Applied and Environmental Microbiology. 2003;69(2):1181–1186. doi: 10.1128/aem.69.2.1181-1186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.González A., Botella S., Montes R. M., Moreno Y., Ferrús M. A. Direct detection and identification of Arcobacter species by multiplex PCR in chicken and wastewater samples from Spain. Journal of Food Protection. 2007;70(2):341–347. doi: 10.4315/0362-028x-70.2.341. [DOI] [PubMed] [Google Scholar]
- 12.González A., Suski J., Ferrús M. A. Rapid and accurate detection of arcobacter contamination in commercial chicken products and wastewater samples by real-time polymerase chain reaction. Foodborne Pathogens and Disease. 2010;7(3):327–338. doi: 10.1089/fpd.2009.0368. [DOI] [PubMed] [Google Scholar]
- 13.Merga J. Y., Royden A., Pandey A. K., Williams N. J. Arcobacter spp. isolated from untreated domestic effluent. Letters in Applied Microbiology. 2014;59(1):122–126. doi: 10.1111/lam.12256. [DOI] [PubMed] [Google Scholar]
- 14.Šilha D., Šilhová-Hrušková L., Vytřasová J. Modified isolation method of Arcobacter spp. from different environmental and food samples. Folia Microbiologica. 2015;60(6):515–521. doi: 10.1007/s12223-015-0395-x. [DOI] [PubMed] [Google Scholar]
- 15.McLellan S. L., Huse S. M., Mueller-Spitz S. R., Andreishcheva E. N., Sogin M. L. Diversity and population structure of sewage-derived microorganisms in wastewater treatment plant influent. Environmental Microbiology. 2010;12(2):378–392. doi: 10.1111/j.1462-2920.2009.02075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newton R. J., Bootsma M. J., Morrison H. G., Sogin M. L., McLellan S. L. A microbial signature approach to identify fecal pollution in the waters off an urbanized coast of lake Michigan. Microbial Ecology. 2013;65(4):1011–1023. doi: 10.1007/s00248-013-0200-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Houf K., De Zutter L., Van Hoof J., Vandamme P. Assessment of the genetic diversity among Arcobacters isolated from poultry products by using two PCR-based typing methods. Applied and Environmental Microbiology. 2002;68(5):2172–2178. doi: 10.1128/aem.68.5.2172-2178.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collado L., Kasimir G., Perez U., et al. Occurrence and diversity of Arcobacter spp. along the Llobregat River catchment, at sewage effluents and in a drinking water treatment plant. Water Research. 2010;44(12):3696–3702. doi: 10.1016/j.watres.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Houf K., Tutenel A., De Zutter L., Van Hoof J., Vandamme P. Development of a multiplex PCR assay for the simultaneous detection and identification of Arcobacter butzleri, Arcobacter cryaerophilus and Arcobacter skirrowii . FEMS Microbiology Letters. 2000;193(1):89–94. doi: 10.1111/j.1574-6968.2000.tb09407.x. [DOI] [PubMed] [Google Scholar]
- 20.Figueras M. J., Levican A., Collado L. Updated 16S rRNA-RFLP method for the identification of all currently characterised Arcobacter spp. BMC Microbiology. 2012;12, article 292 doi: 10.1186/1471-2180-12-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levican A., Collado L., Figueras M. J. Arcobacter cloacae sp. nov. and Arcobacter suis sp. nov., two new species isolated from food and sewage. Systematic and Applied Microbiology. 2013;36(1):22–27. doi: 10.1016/j.syapm.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 22.De Smet S., De Zutter L., Debruyne L., Vangroenweghe F., Vandamme P., Houf K. Arcobacter population dynamics in pigs on farrow-to-finish farms. Applied and Environmental Microbiology. 2011;77(5):1732–1738. doi: 10.1128/aem.02409-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houf K., Devriese L. A., De Zutter L., Van Hoof J., Vandamme P. Development of a new protocol for the isolation and quantification of Arcobacter species from poultry products. International Journal of Food Microbiology. 2001;71(2-3):189–196. doi: 10.1016/s0168-1605(01)00605-5. [DOI] [PubMed] [Google Scholar]
- 24.Fernandez-Cassi X., Silvera C., Cervero-Aragó S., et al. Evaluation of the microbiological quality of reclaimed water produced from a lagooning system. Environmental Science and Pollution Research. 2016;23(16):16816–16833. doi: 10.1007/s11356-016-6812-0. [DOI] [PubMed] [Google Scholar]
- 25.Houf K., On S. L. W., Coenye T., Debruyne L., De Smet S., Vandamme P. Arcobacter thereius sp. nov., isolated from pigs and ducks. International Journal of Systematic and Evolutionary Microbiology. 2009;59(10):2599–2604. doi: 10.1099/ijs.0.006650-0. [DOI] [PubMed] [Google Scholar]
- 26.McClung C. R., Patriquin D. G., Davis R. E. Campylobacter nitrofigilis sp. nov., a Nitrogen-Fixing Bacterium Associated with Roots of Spartina alterniflora Loisel. International Journal of Systematic Bacteriology. 1983;33(3):605–612. doi: 10.1099/00207713-33-3-605. [DOI] [Google Scholar]
- 27.Figueras M. J., Levican A., Collado L., Inza M. I., Yustes C. Arcobacter ellisii sp. nov., isolated from mussels. Systematic and Applied Microbiology. 2011;34(6):414–418. doi: 10.1016/j.syapm.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Levican A., Collado L., Yustes C., Aguilar C., Figueras M. J. Higher water temperature and incubation under aerobic and microaerobic conditions increase the recovery and diversity of Arcobacter spp. from shellfish. Applied and Environmental Microbiology. 2014;80(1):385–391. doi: 10.1128/aem.03014-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levican A., Figueras M. J. Performance of five molecular methods for monitoring Arcobacter spp. BMC Microbiology. 2013;13(1, article 220) doi: 10.1186/1471-2180-13-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho H. T. K., Lipman L. J. A., Gaastra W. Arcobacter, what is known and unknown about a potential foodborne zoonotic agent! Veterinary Microbiology. 2006;115(1–3):1–13. doi: 10.1016/j.vetmic.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 31.Fisher J. C., Levican A., Figueras M. J., McLellan S. L. Population dynamics and ecology of Arcobacter in sewage. Frontiers in Microbiology. 2014;5, article 525 doi: 10.3389/fmicb.2014.00525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salas-Massó N., Andree K. B., Furones M. D., Figueras M. J. Enhanced recovery of Arcobacter spp. using NaCl in culture media and re-assessment of the traits of Arcobacter marinus and Arcobacter halophilus isolated from marine water and shellfish. Science of The Total Environment. 2016;566-567:1355–1361. doi: 10.1016/j.scitotenv.2016.05.197. [DOI] [PubMed] [Google Scholar]