Abstract
The demand for improved pertussis vaccines is urgent due to the resurgence of whooping cough. A deeper understanding of the mode of action of pertussis vaccines is required to achieve this improvement. The vaccine-induced effects of a candidate outer membrane vesicle vaccine (omvPV) and a classical protective but reactogenic whole cell vaccine (wPV) were comprehensively compared in mice. The comparison revealed essential qualitative and quantitative differences with respect to immunogenicity and adverse effects for these vaccines. Both vaccines stimulated a mixed systemic Th1/Th2/Th17 response. Remarkably, omvPV evoked higher IgG levels, lower systemic pro-inflammatory cytokine responses and enhanced splenic gene expression than wPV. The omvPV-induced transcriptome revealed gene signatures of the IFN-signaling pathway, anti-inflammatory signatures that attenuate LPS responses, anti-inflammatory metabolic signatures, and IgG responses. Upon intranasal challenge, both immunized groups were equally efficient in clearing Bordetella pertussis from the lungs. This study importantly shows that immunization with omvPV provides a milder inflammatory responses but with equal protection to bacterial colonization and induction of protective antibody and Th1/Th17 type immune responses compared to wPV. These results emphasize the potential of omvPV as a safe and effective next-generation pertussis vaccine.
The efficacy of pertussis vaccines on the market, whole-cell pertussis vaccine (wPV) and acellular pertussis vaccine (aPV), is under scrutiny because of a whooping cough resurgence despite high vaccination coverage1,2. The current view on immunity to Bordetella pertussis (B. pertussis) is that T-helper 1 and 17 (Th1/Th17) responses3,4 and specific antibody responses are preferred for protection. wPV induce a predominant Th1/Th17 response4,5,6 and a broad systemic antibody repertoire7, but are associated with mild adverse effects8,9. The suspected correlation between wPV and serious acute neurological illness in children10,11 has finally led to the call for safer pertussis vaccines. This resulted in the introduction of better-defined aPVs in many countries. aPVs evoke high IgG1 antibody titers and mainly a Th2 response, which provides protection against disease, however, with a relatively short duration. Recent findings for aPV suggest waning immunity in children12 and impaired prevention of transmission in baboons13. It is thought that aPV induce suboptimal antibody7 and T-cell responses14. These drawbacks call again for improved pertussis vaccines.
Outer membrane vesicles from B. pertussis (omvPV) are a potential vaccine candidate. The protection in mice induced by omvPV is comparable to that of aPV, based on lung colonization data after B. pertussis challenge15,16. Nevertheless, the omvPV elicits a broader humoral immunity and T-cell response compared to aPV7,17. To match the high short-term efficacy and good safety profile of current aPVs, a more comprehensive insight into omvPV-induced responses is required to unravel the type of immunity and assist in future vaccine registration. To that end, an unbiased and detailed systems biology approach is desirable. The application of systems biology in vaccine research has provided a better understanding of immune mechanisms and has been useful for prediction of vaccine efficacy based on correlating biomarkers both for yellow fever and influenza18,19,20. Moreover, systems biology can serve to compare molecular signatures induced by distinct vaccines21,22 and gain insight into vaccine safety23. Previously we used a systems approach in mice to study B. pertussis infection-induced responses24.
Here, we implemented systems vaccinology to investigate the potency of omvPV in mice. As benchmark, wPV was included for its relevant immune responses. Importantly also markers for vaccine safety with respect to pro- and anti-inflammatory cytokine secretion and splenic transcriptome were compared. Finally, the recall of protective immune responses in omvPV versus wPV immunized mice after B. pertussis challenge was compared and related to responses in non-protected mice.
Methods
Vaccines and challenge culture
OmvPV from B. pertussis B1917 were produced as previously described25. For preparation of wPV, B. pertussis B1917 was heat-inactivated (30 min, 56 °C) in PBS. Both omvPV and wPV were diluted in PBS to a final concentration of 4 μg total protein per immunization dose (300 μl). Vaccine characterization included particle size, protein composition, and LPS and DNA content7. For the challenge culture, stock suspension of B. pertussis strain B1917 was diluted in Verweij medium (BBio, Bilthoven, The Netherlands) to a final concentration of 5 × 106 colony-forming units (cfu)/ml.
Animal experiment
An independent ethical committee of the Institute for Translational Vaccinology (Intravacc) approved the animal experiment with identifier 201200073. Animal handling in this study was carried out in accordance with relevant Dutch national legislation, including the 1997 Dutch Act on Animal Experimentation. 8-week old female BALB/c mice (Harlan, The Netherlands) were immunized subcutaneously on day 0 (left groin) and day 28 (right groin) with 4 μg total protein of either omvPV or wPV resulting in omvPV-immunized mice (omvPV-mice) and wPV-immunized mice (wPV-mice). Mice were challenged intranasally under anesthesia (isoflurane/oxygen) with 2 × 105 cfu B. pertussis B1917 in 40 μL Verweij medium on day 56. Non-immunized mice (N.I.-mice) were used as a control.
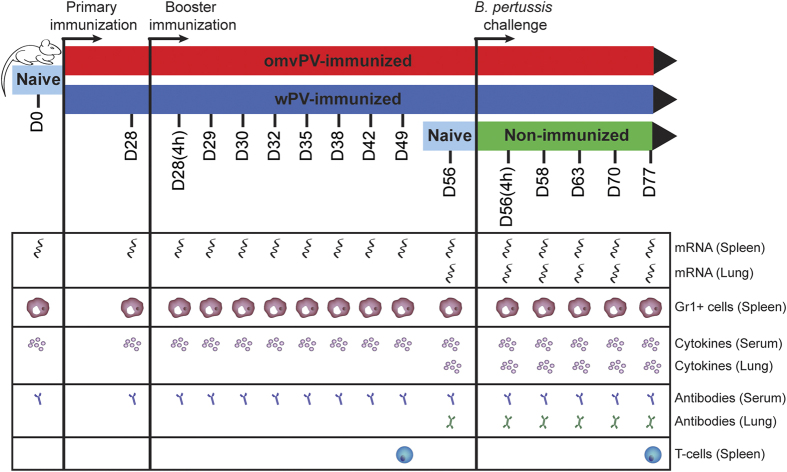
For the determination of gene expression in spleen, cytokine responses and antibody responses, mice (n = 4) were sacrificed on day 28 after primary immunization. In addition, mice (n = 4) were euthanized after booster immunization on day 28 + 4 hours (day 28(4 h)), and day 29, 30, 32, 35, 38, 42, 49 and 56. Finally, mice (n = 4 per group) were sacrificed 4 hours after challenge and on day 58, 63, 70, and 77 to measure bacterial load in the respiratory tract, antibody responses and cytokine responses. For the investigation of Th subsets, mice (n = 4) were sacrificed on day 49 and 77. Naive mice (n = 4) were included as additional control group on each time point. Mice were bled under anesthesia (isoflurane/oxygen) by orbital bleeding and sacrificed by cervical dislocation for further sample collection. An overview of the study design is depicted in Fig. 1.
Figure 1. Study design of the systems approach on omvPV- and wPV-induced responses.
BALB/c mice were subcutaneously immunized with 4 μg omvPV (red) or wPV (blue) on day 0 and day 28. Subsequently, the vaccine-induced responses of both vaccines were characterized over a period of 56 days at 10 different time points. Additionally, an intranasal B. pertussis challenge (2 × 105 cfu/mouse) was performed on day 56 in immunized groups and in non-immunized mice (green). Both vaccine- and infection-induced responses were characterized at a transcriptomic, proteomic and cellular level on given time points as depicted.
Sample collection and preparation
For investigation of cytokine and antibody responses in serum, whole blood from each mouse was collected in a blood collection tube (MiniCollect 0.8 ml Z Serum Sep GOLD, Greiner Bio-One, Austria). After coagulation (10 min. at room temperature) and centrifugation (10 min., 3000 g), sera were taken and stored at −80 °C. For the colonization assay, the right lung lobe was placed in 900 ml Verweij medium at room temperature. To determine pulmonary cytokine and antibody responses, the lung lysates used for colonization assays were filtered (Millex GV Filter unit 0.22 μm, Millipore) and stored at −80 °C. For characterization of pulmonary gene expression, the left lung lobe was placed in 1 ml RNAlater (Qiagen), incubated overnight at 4 °C, and stored at −80 °C. In addition, the spleen was excised and divided in two equal parts. For gene expression analysis, one piece was placed in 1 ml RNAlater (Qiagen), incubated overnight at 4 °C and stored at −80 °C. For detection of Gr1+ cells, the other piece was placed in in 5 ml RPMI-1640 medium (Gibco) supplemented with 10% FCS (Hyclone), 100 units penicillin, 100 units streptomycin, and 2.92 mg/ml L-glutamine (Invitrogen), hereafter called RPMI complete medium, and kept on ice. For the analysis of Th responses, the whole spleen was placed in 5 ml of RPMI medium and kept on ice. Splenocytes were isolated by homogenization of spleens using a 70-μm cell strainer (BD Falcon, BD Biosciences) in RPMI complete medium.
Colonization assay
The numbers of cfu in lung tissue were determined as previously described24.
RNA isolation and microarray analysis
RNA isolation from lung and spleen tissue and a RNA quality check (concentration and integrity) were performed as described before24. Microarray analysis was performed on RNA concentrates from spleen tissue of each omvPV-mice and wPV-mice (n = 3) sacrificed at 16 time points: D0, D28, D28(4 h), D29, D30, D31, D32, D35, D42, D49, D56, D56(4 h), D58, D63, D70 and D77 (Fig. 1). For microarray analysis on lung tissue, RNA samples were analyzed obtained from individual mice sacrificed at time points: D56, D56(4 h), D58, D63, D70 and D77. RNA samples extracted from lung and spleen tissue of N.I.-mice were pooled (n = 3) for according time points described before. Amplification, labeling and hybridization of RNA samples for either lung or spleen tissue to microarray chips (NimbleGen 126135 k Mus musculus, Roche, Germany) was carried out at the Microarray Department of the University of Amsterdam, The Netherlands, as described previously24.
Data analysis of gene expression
Quality control and normalization of raw microarray data was performed as described before24. To identify differentially expressed genes (DEGs) between experimental groups (naive and various time points post immunization or challenge) an ANOVA was applied. The induction or repression of individual genes was expressed as fold ratio (FR) by comparing mean gene expression levels of experimental groups to the naive mice. Average normalized gene expression levels contain data of three mice per immunized group. The data of challenged non-immunized mice are individual samples of pooled RNA of three mice. The dataset of challenged non-immunized mice (n = 3) of our previous study24 was incorporated in the analysis to increase the statistical power. Probes were considered differentially expressed if they met the following two criteria: (i) a p-value <0.001 (ANOVA), which corresponds to a Benjamini-Hochberg False discovery rate (FDR)26 of <5%; and (ii) an absolute FR ≥1.5 (experimental groups compared to naive mice) for at least one time point. If multiple probes corresponding to the same gene were differentially expressed, their data were averaged to remove redundancy for further analysis. GeneMaths XT (Applied Maths, St-Martens-Latem, Belgium) was used to visualize differences in gene expression in heatmaps and perform the hierarchical clustering based on Euclidean distance (linear scaling) with UPGMA. Genes were arranged according to similar expression patterns in time at which genes exceeded the FR cut-off of 1.5. To facilitate visual interpretation of heatmaps, only induction (red) and repression (green) of gene expression levels with fold ratios ≥1.5 are visualized, therefore presenting fold ratios ≤1.5 as naive level (black). Functional enrichment with an over-representation analysis (ORA) based on Gene Ontology Biological Processes (GO-BP) and Kyoto Encyclopedia of Genes and Genomes (KEGG) by using DAVID and the detection of cell-specific or tissue-specific gene expression based on BioGPS datasets was performed as described previously24. Involvement of type I and II IFN-signaling pathway was performed by using the Interferome database (http://www.interferome.org/interferome/home.jspx)27. Additional text mining on gene function was performed in PubMed.
Multiplex immunoassay (MIA) and ELISA for antibody response
Levels of pulmonary IgA and serum IgM, IgA, total IgG and IgG subclasses (IgG1, IgG2a, IgG2b and IgG3) specific for B. pertussis antigens pertussis toxin (Ptx), filamentous hemagglutinin (FHA), pertactin (Prn), combined fimbria type 2 and 3 antigens (Fim2/3) and outer membrane vesicles B1917 (OMV B1917) were determined as described previously24 and data are presented in fluorescent intensity units (F.I.). Whole-cell B. pertussis ELISA to quantify total serum IgG was performed as described before24.
Immunoproteomic profiling of serum IgG antibodies
Antigen specificity of B. pertussis-specific serum IgG antibody responses were determined by SDS-PAGE and subsequent Western blotting as described before7.
Gr1+ cells in the spleen
The percentage of Gr1+ cells in the spleen was determined with flow cytometry as described before24.
Isolation and in vitro restimulation of splenocytes
After treatment with erythrocyte lysis buffer, splenocytes were cultured in 24-well plates at 6 × 106cells/well for 7 days at 37 °C in a humidified atmosphere containing 5% CO2 in IMDM medium (Gibco) supplemented with 8% FCS, 100 units penicillin, 100 units streptomycin, 2.92 mg/ml L-glutamine, and 20 μM β-mercaptoethanol (Sigma). The cells were either left unstimulated (medium control) or stimulated with 1 μg/ml Prn, 1 μg/ml Ptx, 1 μg/ml FHA, 5 μg/ml OMV, or 5 μg/ml heat-killed B. pertussis. On day 7, supernatant was collected for cytokine analysis.
Cytokine profiling using multiplex technology
Concentrations (pg/ml) of 32 cytokines (Eotaxin, G-CSF, GM-CSF, IFNγ, IL-10, IL-12 (p40), IL-12 (p70), IL-13, IL-15, IL-17A, IL- 1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-9, IP-10, KC, LIF, LIX, M-CSF, MCP-1, MIG, MIP-1α, MIP-1β, MIP-2, RANTES, TNFα, VEGF) present in serum and lung lysates were determined using a MIA (Milliplex MAP Mouse Cytokine/Chemokine - Premixed 32 Plex; Merck KGaA, Darmstadt, Germany) following the manufacturer’s protocol.
The concentration of various Th cytokines (IL-4, IL-5, IL-10, IL-13, IL-17A, TNFα, and IFNγ) was determined in culture supernatant using a Milliplex mouse cytokine 7-plex luminex kit (Millipore), according to the manufacturer’s protocol. Measurements and data analysis were performed with Bio-Plex 200, using Bio-PlexManager software (version 5.0, Bio-Rad Laboratories). Results were corrected for the background (medium control) and cytokine concentrations were calculated and provided in pg/ml.
Statistical analysis
Data of the antibody, cytokine, and colonization assays were log-transformed after which a t-test was performed. p-values ≤ 0.05 were considered to indicate significant differences.
Results
OmvPV provides equal protection as wPV against B. pertussis challenge
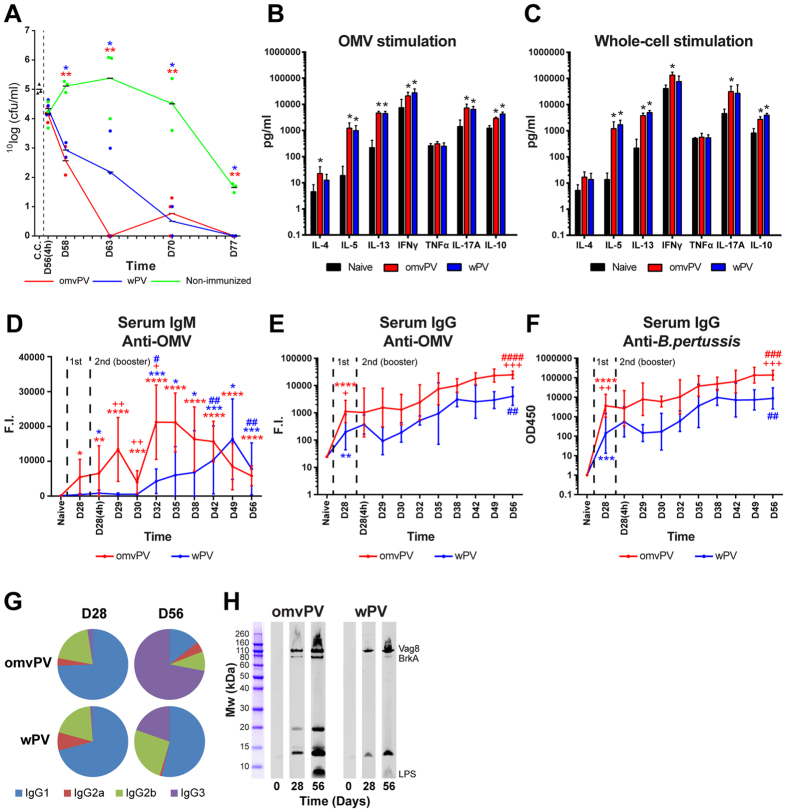
The composition of both omvPV and wPV, in terms of proteins, DNA, and LPS7, was determined before mice were immunized twice with a four-week interval (Fig. 1). Vaccine efficacy was assessed by determining lung colonization upon an intranasal B. pertussis challenge (Fig. 2A). N.I.-mice showed extensive colonization and lungs were not cleared at day 77 (21 days post challenge) as observed before24, whereas lungs of immunized mice were cleared faster. The clearance of lungs was not significantly different between omvPV-mice and wPV-mice, although the lungs of omvPV-mice tended to be cleared slightly faster than of wPV-mice.
Figure 2. Lung colonization and vaccine-induced adaptive responses in omvPV- and wPV-immunized mice.
(A) The number of B. pertussis cfu in 1 ml challenge culture (c.c.) was confirmed before challenge. Subsequently, the cfu/ml in the lungs of mice was determined 4 hours and 2, 7, 14, and 21 days after challenge (D56(4 h)-D77). * and **p ≤ 0.05 and p ≤ 0.01 for immunized group vs. non-immunized group. (B,C) Splenocytes were obtained post booster immunization (day 49) of mice that were naive (black), omvPV-immunized (red), or wPV-immunized (blue). Cytokine concentrations were determined in the culture supernatants after 7 days of stimulation with (B) 5 μg/ml OMVs or (C) 5 μg/ml B. pertussis whole-cells. Results for each mouse are corrected for medium stimulation. Data presented as mean ± SD (n = 4). *p ≤ 0.05 for immunized group vs. naive group. (D–H) The kinetics of serum (D) anti-OMV IgM, (E) anti-OMV IgG and (F) anti-B. pertussis IgG formation was determined for a period of 56 days after omvPV and wPV immunization. Data are presented as mean ± SD (n = 4). For (D) *, **, *** and ****p ≤ 0.05, p ≤ 0.01, p ≤ 0.001 and p ≤ 0.0001 for day 28–56 vs. naive, # and ##p ≤ 0.05 and p ≤ 0.01, for D28(4 h)-D56 vs. D28, + and ++p ≤ 0.05 and p ≤ 0.01 for omvPV-mice vs. wPV-mice. For (E,F) *, **, *** and ****p ≤ 0.05, p ≤ 0.01, p ≤ 0.001 and p ≤ 0.0001 for naive vs. D28, #, ##, ### and ####p ≤ 0.05, p ≤ 0.01, p ≤ 0.001 and p ≤ 0.0001 for D28 vs. D56, + and ++p ≤ 0.05 and p ≤ 0.01 for omvPV-mice vs. wPV-mice. The significance in (E) and (F) between D28(4 h)-D49 is not depicted. (G) Subclass distribution was determined at day 56 in serum of mice immunized with omvPV or wPV by calculating the ratio of the level for each subclass from the sum of the levels all subclasses. (H) To determine antigen-specificity, the immunoproteomic profiles of serum antibodies (pooled sera, n = 4) following primary immunization (day 28) and booster immunization (day 56) were obtained. The band positions of Vag8, BrkA and LPS antigens are indicated as identified previously7.
Immunization with omvPV and wPV induces a mixed Th1/Th2/Th17 response
Stimulation of splenocytes with B. pertussis OMVs or whole-cells resulted in enhanced production of IFNγ (Th1), IL-5 and IL-13 (Th2), IL-17A (Th17) and IL-10 in omvPV-mice and wPV-mice, compared to naive mice (Fig. 2B,C). TNFα was not observed, while IL-4 (Th2) was only detected after OMV stimulation in omvPV-mice. Hardly any cytokines were produced upon stimulation of splenocytes with the purified antigens Ptx, FHA and Prn, except for IL-17A after FHA stimulation in wPV-mice and IL-5 after Prn stimulation in omvPV-mice (Supplementary Fig. S1A–C, left panels).
OmvPV induces higher serum IgG antibody responses
OMV-specific IgM was induced by omvPV and at a lower rate by wPV with highest peaks at day 32 and day 49, respectively (Fig. 2D). Moderate IgG levels were detected 28 days after the primary immunization with either omvPV or wPV, using an anti-OMV MIA and a whole-cell ELISA (Fig. 2E,F). Booster immunization strongly increased IgG production in omvPV-mice and wPV-mice, but omvPV induced higher IgG levels than wPV. No IgG or IgM antibodies directed against purified antigens (Ptx, FHA, Prn, Fim2/3) were found (data not shown). The IgG subclass distribution on day 28 was dominated by IgG1 for both omvPV and wPV (Fig. 2G). Booster immunizations with wPV and to a much larger extent with omvPV stimulated IgG3 antibody production as is shown on day 56 (Fig. 2G). Most of these IgG3 antibodies were directed against LPS7. Confirmed here, higher levels of anti-LPS IgG antibodies were detected in omvPV-mice compared to wPV-mice (Fig. 2H). Moreover, these anti-LPS IgG antibodies were observed solely on day 56, indicating that these antibodies were elicited by the booster immunization. Additionally, both vaccines mainly induced anti-Vag8 and anti-BrkA antibodies (Fig. 2H).
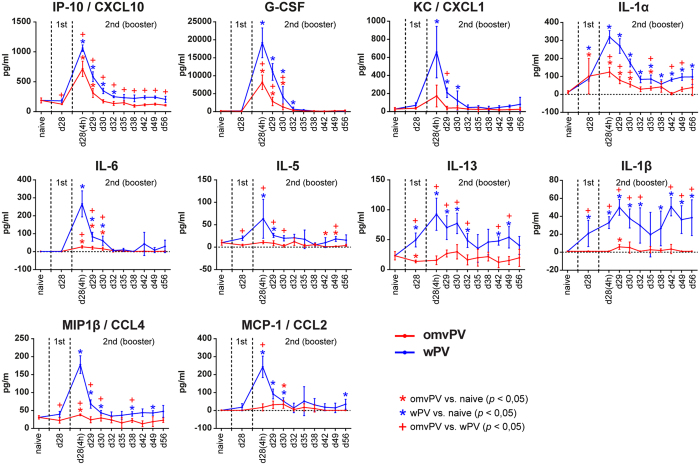
Less pro-inflammatory cytokine induction in serum by omvPV compared to wPV
Vaccine-induced cytokine responses in serum occurred mainly 4 hours after booster immunization (day 28(4 h)). Concentrations of CCL4, CXCL10, G-CSF, IL-1α, and IL-6 were significantly enhanced in omvPV-mice and wPV-mice compared to levels before immunization (Fig. 3). In omvPV-mice, all these cytokine concentrations were significantly lower than in wPV-mice. Moreover, CCL2, CXCL1, IL-1β, IL-5, and IL-13 concentrations were only significantly elicited in wPV-mice.
Figure 3. Serum cytokine secretion following omvPV or wPV immunization.
Concentrations of 32 cytokines were determined in serum at multiple time points after immunization with omvPV or wPV. Levels of CXCL10, G-CSF, CXCL1, IL-1α, IL-6, IL-5, IL-13, IL-1β, CCL4, and CCL2 were significantly altered and are depicted here. The different stages of primary immunization and booster immunization are depicted in the panels. Data represent mean ± SD (n = 4). *p ≤ 0.05 for immunized group vs. naive, +p ≤ 0.05 for omvPV group vs. wPV group.
Enhanced anti-inflammatory transcriptomic profiles in the spleen after omvPV immunization
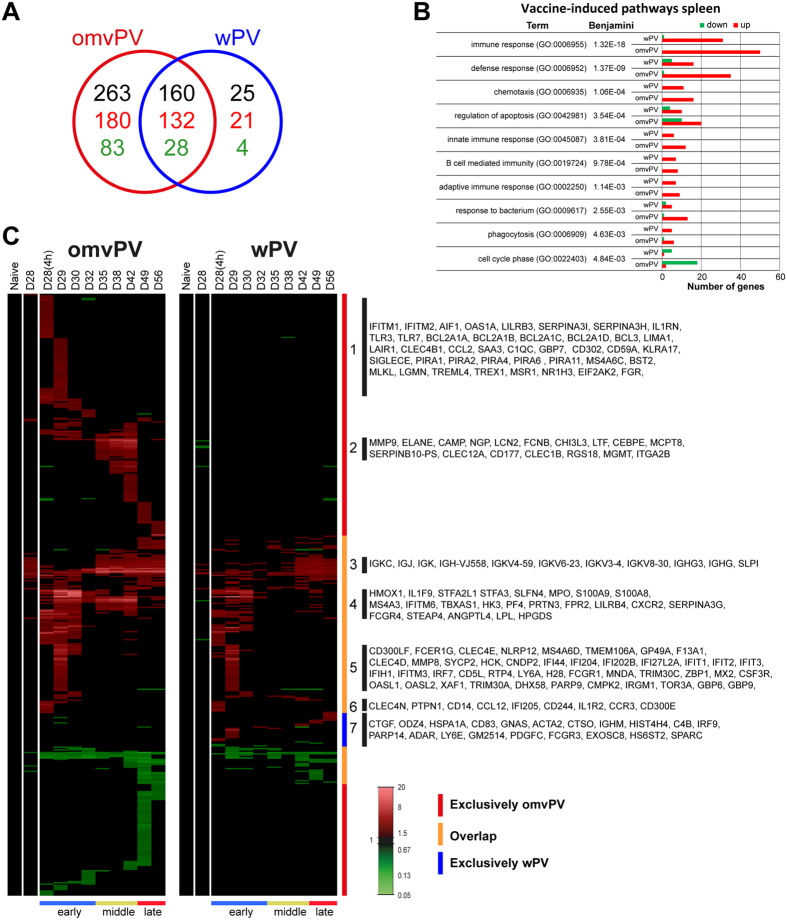
In total, 423 and 185 differentially expressed genes (DEGs) were identified in the spleen compared to naive mice (day 0) (p-value ≤ 0.001, FR ≥1.5) in omvPV-mice and wPV-mice, respectively (Fig. 4A). Of these DEGs, 160 (132 upregulated, 28 downregulated) overlapped between both immunized groups. Additionally, 263 DEGs (180 upregulated, 83 downregulated) were exclusively detected in omvPV-mice, whereas 25 DEGs (21 upregulated, 4 downregulated) were only found in wPV-mice.
Figure 4. Transcriptomic profiles in the spleen following omvPV and wPV booster immunization.
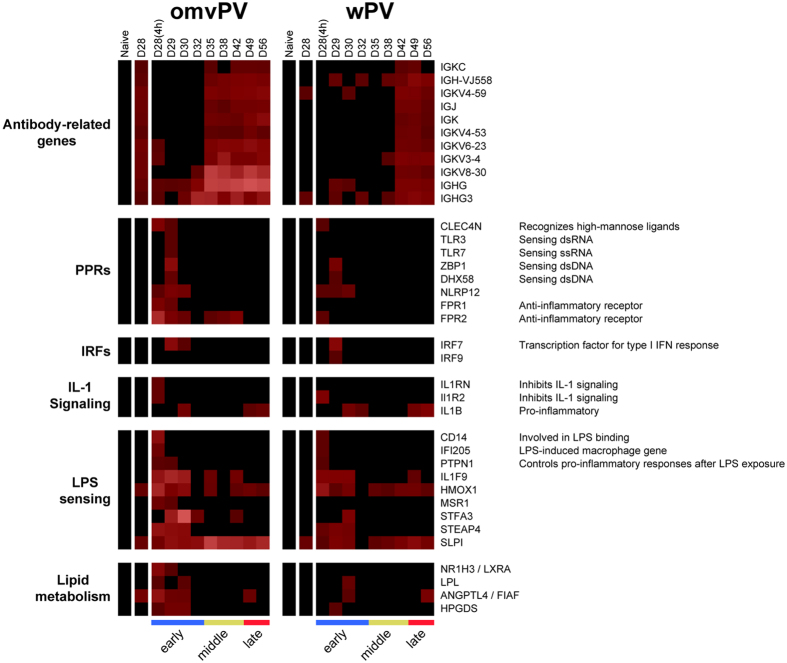
(A) Fold changes of DEGs were calculated compared to naive mice (FR ≥1.5, p-value ≤ 0.001). In total, 448 DEGs were found divided over both vaccine responses, as depicted in a Venn-diagram with total number of genes (black) and both upregulated (red) and downregulated genes (green). (B) Overrepresentation analysis on all 448 genes revealed the involvement of specific GO-BP terms and KEGG pathways with corresponding upregulated (red) and downregulated (green) genes. (C) All differentially upregulated (red) and downregulated (green) genes are portrayed as heatmap (mean of n = 4). Genes not surpassing a FR of 1.5 are shown as basal level (black) at this time point. Gene clustering is based on up/downregulation, time of involvement, and presence in the immunization group. The overlap and the exclusive presence of DEGs in either the omvPV or wPV groups are further depicted next to the heatmap. Booster immunization-induced responses were divided in three phases: early phase (D28(4 h)-D32), middle phase (D35-D42) and late phase (D49-D56) as calculated in Supplementary Fig. S2A. Selections of genes co-expressed in seven (1–7) areas of the heatmap are shown.
Overrepresentation analysis (ORA) gained insight into molecular pathways activated by immunization and revealed enrichment of 77 gene ontology biological pathways (GO-BP) terms and 2 KEGG pathways (Benjamini ≤0.05) in the dataset of omvPV-mice and/or wPV-mice. The immunity-related pathways are shown in Fig. 4B. The terms Immune response, Defense response, and Cell cycle contained more genes upregulated in omvPV-mice than in wPV-mice. Both groups had an equal number of genes related to Phagocytosis and B-cell mediated immunity (Fig. 4B).
Genes were clustered based on (i) co-expression over time and (ii) overlap between both immunized groups (Fig. 4C). Additionally, the vaccine-induced response could be divided in three phases based on hierarchical clustering (Supplementary Fig. S2A): early phase (D28(4 h)-D32), middle phase (D35–D42) and late phase (D49–D56) (Fig. 4C). By gene clustering, 7 groups were identified and reviewed in-depth using text mining. The results are described in the following paragraphs.
Group 1 consists of DEGs that were exclusively expressed in the early phase of the omvPV-induced response. This group comprised genes related to pathogen recognition, such as Fpr1 and genes related to Toll-like receptor (TLR) 3 (Tlr3, Mlkl) and TLR7 (Tlr7, Treml4, Lgmn, Trex1) mediated signaling. Trex1 limits pro-inflammatory signals following TLR7 activation in macrophages28, whereas Siglece represses TLR-signaling in general29. Moreover, IL-1 receptor antagonist (Il1rn) and members of the Bcl-family, (Bcl2a1a, Bcl2a1b, Bcl2a1c, Bcl2a1d, Bcl3), paired-Ig-like receptors (Pira1, Pira2, Pira4, Pira6, Pira11, Lilrb3), and IFN-induced transmembrane proteins (Ifitm1, Ifitm2) were detected. Genes related to processing of LPS included Bst2, Eif2ak2, essential for LPS-induced iNOS production30, and Msr1 that inhibits LPS-stimulated IL-6 secretion31.
Group 2 genes were exclusively upregulated in the middle phase of the omvPV-induced response. This group contained genes related to myeloid cells (Mcpt8, Clec12a, Chi3l3) and more specifically neutrophils (Cd177, Cebpe, Clec1b, Elane, Lcn2, Ltf, Ngp).
Group 3 includes antibody-related genes, such as Igkc, Igj, Igk, and Ighg, which showed higher expression in omvPV-mice than in wPV-mice (Fig. 5A), in line with the increased antibody levels (Fig. 2D–F).
Figure 5. Selected splenic transcriptome profiles following omvPV and wPV booster immunization.
The splenic transcriptome profiles of genes related to antibodies, pathogen recognition receptors (PRRs), interferon regulatory factors (IRFs), IL-1 signaling, LPS sensing, and lipid metabolism are depicted.
Group 4 genes were upregulated in the early phase of immunization in omvPV-mice, but were not or hardly upregulated in wPV-mice. Most of these genes were also differentially expressed in the middle phase. This group contained the anti-inflammatory Fpr232 and Steap4 that decreases inflammatory effects by repressing IL-6 production33. Moreover, genes involved in LPS responsiveness were upregulated, such as Slpi that suppresses responses to LPS34, Hmox1, Stfa3, Stfa2l1, and Angptl4 that are induced by LPS in the acute phase response35. Notably, Angptl4 has multiple functions, including inflammation and lipid metabolism36. Genes involved in metabolism (Hk3, Tbxas1) and more specifically the lipid metabolism (Angptl4, Lpl, Hpgds) were found in this group. Furthermore, this group comprised Pf4/Cxcl4, myeloid cell specific genes (Lilrb4, Ms4a3, Serpina3g), danger-associated molecular patterns (DAMPs; S100a9, S100a8), IFN-induced genes (Slfn4, Ifitm6) and Fcgr4 that binds IgG2a and IgG2b. Moreover, neutrophil-related genes (Mpo, Prtn3, Cxcr2, Il1f9) were detected. Notably, IL-1F9 promotes Th1 formation by binding IL-36R on T-cells37.
Group 5 contains genes that were expressed by both vaccines on day 29. Some of these genes are involved in the type I IFN-signaling pathway. This included Irf7, Oasl1, that inhibits IRF738, and other IFN-induced proteins (Ifi44, Ifi204, Ifi202b, Ifi27l2a, Ifit1, Ifit2, Ifit3, Ifih1, Ifitm3, Irgm1, H28, Mnda, Oasl2, Tor3a, Lgals3bp, Mx2). Moreover, two genes were detected that encode pathogen recognition receptors (PRRs) sensing cytosolic DNA: DAI (Zbp1)39 and LPG2 (Dhx58)40. Furthermore, genes encoding Fc receptors (Fcgr1, Fcer1g), a NOD-like receptor and neutrophil migration marker41 (Nlrp12), C-type lectins (Clec4d, Clec4e), and other membrane markers (Ly6a, Ms4a6d, Cd300lf, Gp49a) were identified. Moreover, differential gene expression was detected for two genes encoding tripartite-motif proteins (Trim30a, Trim30c) of which TRIM30α inhibits TLR4-mediated NF-kappaB activation42.
Group 6 genes were upregulated on day 28(4 h) by both vaccines. This group included an LPS-induced macrophage gene (Ifi205), the inflammatory chemokine Ccl12, the interleukin 1 receptor, type II (Il1r2), and Ptpn1 that controls pro-inflammatory responses after LPS exposure43. Moreover, genes encoding membrane markers were detected (Cd244, Cd300e, Ccr3) of which some are related to pathogen-recognition, such as dectin-2 (Clec4n) that recognizes high-mannose ligands, and Cd14 involved in LPS binding.
Group 7 genes were merely differentially expressed in the wPV-mice. This group included genes on immune cells such as C4b, Cd83 an important activation marker on B-cell, T-cell, and dendritic cell populations44, Fcgr3, Ighm, Irf9, Ly6e, and Parp14. PARP-14 promotes Th2 and Th17 cell formation and steers the antibody subclass distribution45,46.
Based on this text mining, sets of genes involved in antibody formation, PRRs, interferon regulatory factors (IRFs), IL-1 signaling, LPS sensing and lipid metabolism were found (Fig. 5). Subsequently, the dataset was matched with the Interferome database to identify which vaccine-induced genes were involved in the type I and type II IFN-signaling pathways (Supplementary Fig. S3). In total, 91 genes were related to type I IFN, 22 to type II IFN, and 60 genes to both pathways. The majority of IFN-related genes of both type I and II were induced by omvPV, whereas only 13 genes were found exclusively in wPV-mice. Moreover, the type II IFN-signaling pathway was mainly involved in the early phase. The type I IFN-signaling pathway genes were also mainly found in the early phase, but also partly in later phases (Supplementary Fig. S3).
Higher neutrophil responses elicited by omvPV compared to wPV
Transcriptome comparison with BioGPS databases particularly revealed involvement of MAC+GR1+ granulocytes (neutrophils) in vaccine-induced immune responses (Supplementary Fig. S4A). These 46 genes were all detected in omvPV-mice and mainly activated during the early and middle phase of the vaccine-induced response (Supplementary Fig. S4B). In contrast, only 16 of these 46 genes were upregulated in wPV-mice. Flow cytometry confirmed that the percentage of Gr1+ cells in the spleen was significantly increased by omvPV and wPV immunization between days 29–42 (Supplementary Fig. S4C). In line with the enhanced gene expression, a significant higher number of Gr1+ cells was detected in the middle phase (day 42) in omvPV-mice compared to wPV-mice.
Immunized mice showed reduced serum and pulmonary cytokine responses following B. pertussis challenge
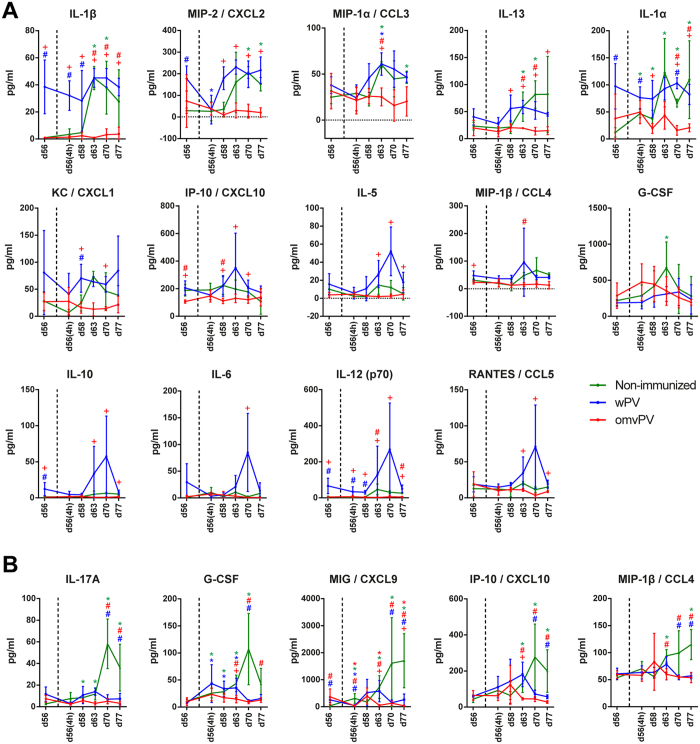
Subsequent to the investigation of vaccine-induced responses, we examined the immune responses after B. pertussis challenge (Fig. 1). First, concentrations of 32 cytokines were determined in serum and lung lysate (Fig. 6A,B). In sera of N.I.-mice, IL-1α, IL-1β, CXCL2, CCL3, IL-13, and G-CSF were significantly increased mainly between days 63–77. Notably, no significant serum cytokine secretion was observed in omvPV-mice (Fig. 6A). Serum concentrations of IL-1α, IL-1β, IL-5, IL-6, IL-10, IL-12(p70), IL-13, CCL3, CCL4, CCL5, CXCL1, CXCL2, and CXCL10 were significantly higher in wPV-mice compared to omvPV-mice, mainly on day 63. However, the B. pertussis challenge did not lead to significant induction of cytokine levels compared to day 56 in wPV-mice, except for CCL3 (Fig. 6A). In the lungs, IL-17A, G-CSF, CXCL9, CCL4, and CXCL10 were significantly increased in N.I.-mice, between day 63–77 whereas these cytokines were not elevated in either of the immunized groups, except for G-CSF in wPV-mice between day 56(4 h)-63 (Fig. 6B).
Figure 6. Serum and pulmonary cytokine secretion following B. pertussis challenge in mice immunized with omvPV or wPV and non-immunized mice.
(A,B) The concentrations of 32 cytokines were determined prior to and at multiple time points post challenge with B. pertussis in non-immunized mice (green) or mice immunized with omvPV (red) or wPV (blue). Only cytokines that were significantly altered in (A) serum and (B) lung lysate are depicted. Data are presented as mean ± SD for immunized groups (n = 4) and non-immunized groups (n = 3). *p ≤ 0.05 for challenged groups vs. day 56, #p ≤ 0.05 for immunized groups vs. non-immunized group, +p ≤ 0.05 for omvPV group vs. wPV group.
Reduced transcriptomic profiles in the spleen of immunized mice following B. pertussis challenge
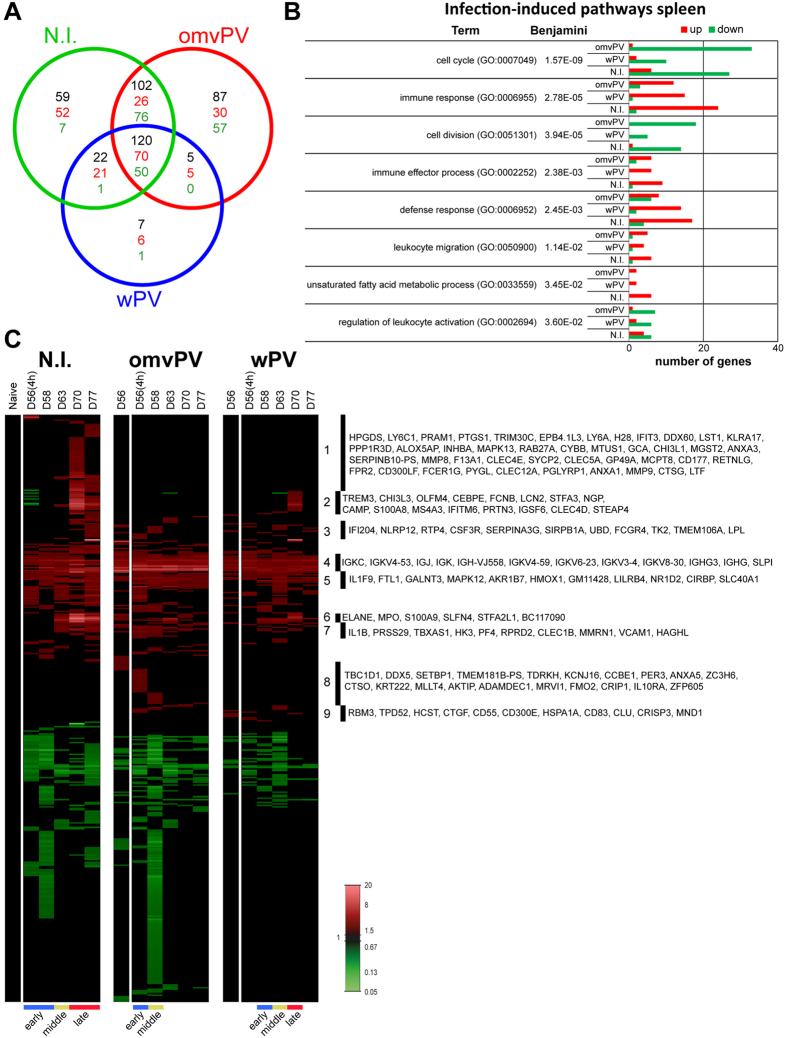
In total, 402 DEGs (p-value ≤ 0.001, FR ≥1.5) were detected in the spleen of immunized mice or N.I.-mice after B. pertussis challenge (Fig. 7A). This analysis demonstrated less intense gene expression profiles in omvPV-mice and wPV-mice than in N.I.-mice following the challenge with B. pertussis. These responses were even lower in omvPV-mice compared to wPV-mice. 87, 7, and 59 DEGs were exclusively detected in the challenged omvPV-mice, wPV-mice, and N.I.-mice, respectively. 120 DEGs (70 upregulated, 50 downregulated) were found in all three groups (Fig. 7A). The ORA on infection-induced responses in the spleen revealed enrichment of 24 GO-BP terms and 2 KEGG pathways (Benjamini ≤0.05) in omvPV-mice, wPV-mice, and N.I.-mice (Fig. 7B). In omvPV-mice, several downregulated genes are involved in Cell cycle (33 genes) and Cell division (18 genes). The terms Immune response and Defense response comprised a smaller number of genes in omvPV-mice than in wPV-mice and N.I.-mice. Genes were clustered based on (i) co-expression over time and (ii) overlap between both experimental groups (Fig. 7C). Hierarchical clustering on the three challenged groups indicated that infection-induced responses were distinct in immunized mice compared to the challenged N.I.-mice (Supplementary Fig. S2B). The infection-induced response of N.I.-mice was divided in three phases: early phase (D56(4 h)-D58), middle phase (D63), and late phase (D70–D77). In omvPV-mice, the response was faster compared to N.I.-mice. In wPV-mice, the response was slightly slower compared to omvPV (Fig. 7C and Supplementary Fig. S2B). A selection of genes involved in nine (1–9) areas is depicted in the heatmap. The involvement of neutrophils was more pronounced in challenged N.I.-mice as we detected 38 genes commonly expressed in MAC+GR1+ granulocytes compared to only 10 and 22 of these genes in omvPV-mice and wPV-mice, respectively (Supplementary Fig. S4D). Flow cytometry analysis confirmed a significantly increased number of Gr1+ cells, indicative for neutrophils, in N.I.-mice on day 70 (Supplementary Fig. S4E). This Gr1+ cell fraction in immunized mice was significantly lower between days 70–77 compared to N.I.-mice.
Figure 7. Transcriptomic profiles in the spleen following B. pertussis challenge in omvPV-, wPV-, and non-immunized mice.
(A) Fold changes in expression and significant gene expression were calculated compared to naive mice (FR ≥1.5, p-value ≤ 0.001). In total, 402 DEGs were found divided over the three groups in a Venn-diagram with total number of genes (black), upregulated genes (red), and downregulated genes (green). (B) Overrepresentation analysis on all 402 genes revealed the involvement of specific GO-BP terms and KEGG pathways with corresponding upregulated (red) and downregulated (green) genes. (C) All differentially upregulated (red) and downregulated (green) genes are portrayed as heatmap (mean of n = 3 for immunized groups, n = 1 (pool of 3 mice for non-immunized group)). Genes not surpassing a FR of 1.5 are shown as basal level (black). Gene clustering is based on up/downregulation, time of involvement, and presence in different groups. Infection-induced responses were divided in phases according to the hierarchical clustering calculated in Supplementary Fig. S2B. Selections of genes in nine (1–9) areas of the heatmap are shown.
B. pertussis challenge has less impact on gene expression in the lungs of omvPV-mice than in wPV-mice and N.I.-mice
In total, 3269 DEGs (p-value ≤ 0.001, FR ≥1.5) were detected in lungs of immunized mice or N.I.-mice following B. pertussis challenge (Supplementary Fig. S5A). The pulmonary transcriptome in omvPV-mice on day 56 prior to challenge differs a lot from the naive basal gene expression level. Compared to day 56, the B. pertussis challenge induced less change in pulmonary gene expression in omvPV-mice as compared to wPV-mice and N.I.-mice. Exclusive expression of 578, 156, and 90 DEGs was detected in challenged omvPV-mice, wPV-mice, and N.I.-mice, respectively. In total, 1702 DEGs (695 upregulated, 1007 downregulated) were found in all three groups (Supplementary Fig. S5A). Genes were clustered based on (i) co-expression over time and (ii) overlap between both experimental groups (Supplementary Fig. S5B). Hierarchical clustering of the pulmonary transcriptome of the three challenged groups demonstrated a distinct infection-induced response in immunized mice compared to challenged N.I.-mice (Fig. S2C). The infection-induced response of N.I.-mice was divided in three phases: early phase (D56(4 h)-D58), middle phase (D63 and D77) and late phase (D70), as seen before24. In omvPV-mice, the response was less diverse compared to day 56 and divided in two phases; early phase (D56(4 h)) and middle phase (D70). In wPV-mice, the response compared to day 56 was more diverse and developed slower than in omvPV-mice with an early phase (D63–D70) and middle phase (D77) (Supplementay Figs S2C and S5B).
Adaptive recall responses in omvPV-mice and wPV-mice following B. pertussis challenge
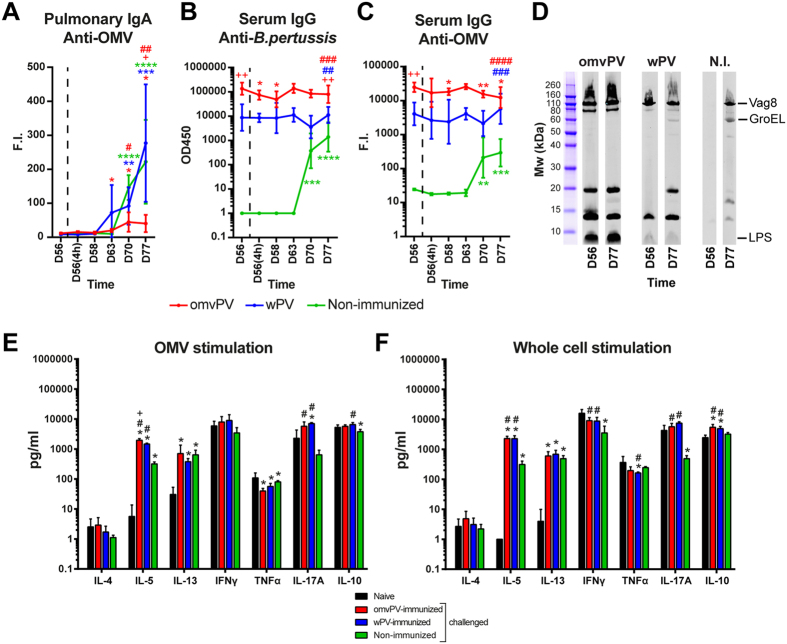
Prior to the intranasal B. pertussis challenge (day 56), no vaccine-induced anti-OMV IgA was detected in the lungs of omvPV-mice and wPV-mice. However, similar amounts of anti-OMV IgA were detected after the challenge of wPV-mice and N.I.-mice on day 63–77, but no anti-OMV IgA was found in omvPV-mice (Fig. 8A). Serum IgG levels specific for pertussis OMVs or whole-cells were unaltered in wPV-mice and slightly decreased in omvPV-mice after the challenge, whereas IgG levels in N.I.-mice were rising between day 63–77 (Fig. 8B,C). The IgG antibody specificity did not change in omvPV-mice after challenge, except for the disappearance of the 15 kDa band (Fig. 8D). The challenge of wPV-mice stimulated antibody formation against GroEL (60 kDa) and an unknown antigen of 20 kDa.
Figure 8. Humoral and cellular adaptive recall responses following B. pertussis challenge in omvPV- and wPV-immunized mice as compared to control mice before and after challenge.
(A–C) Kinetics of (A) pulmonary anti-OMV IgA, (B) serum anti-B. pertussis IgG, and (C) serum anti-OMV IgG levels were determined for a period of 21 days following intranasal challenge in immunized mice (n = 4) and non-immunized mice (N.I.) (n = 3). Data are presented as mean ± SD. Statistical significance of differences in (A), (B) and (C): *, **, *** and ****p ≤ 0.05, p ≤ 0.01, p ≤ 0.001 and p ≤ 0.0001 for vs. D56 per group, + and ++p ≤ 0.05 and p ≤ 0.01 for omvPV vs. wPV, #, ##, ### and ####p ≤ 0.05, p ≤ 0.01, p ≤ 0.001 and p ≤ 0.0001 for immunized groups vs. non-immunized group. (D) Immunoproteomic profiles of serum IgG antibodies (pooled sera, n = 4) before (D56) and 21 days post challenge (D77) were obtained on a B. pertussis lysate. (E,F) Cytokine concentrations were determined in the culture supernatants after 7 day stimulation with (E) 5 μg/ml OMVs, or (F) 5 μg/ml B. pertussis whole-cells. Splenocytes were obtained from naive mice (black) and post-challenge (D77) of mice that were omvPV- (red), wPV- (blue) or non-immunized (green). Results for each mouse are corrected for medium stimulation. Data in (E) and (F) presented as mean ± SD (n = 4). Statistical significance of differences in (E) and (F) *p ≤ 0.05 for challenged groups vs. naive, +p ≤ 0.05 for omvPV group vs. wPV group, #p ≤ 0.05 for immunized group vs. non-immunized group.
Regarding post challenge Th-responses, the stimulation of splenocytes with OMVs revealed altered Th1 (TNFα), Th2 (IL-5, IL-13), and Th17 (IL-17A) cytokines in challenged omvPV-mice, wPV-mice, and N.I.-mice compared to naive mice (Fig. 8E,F and Supplementary Fig. S1). IL-5 and IL-17A levels were higher in immunized mice compared to N.I.-mice (Fig. 8E). Stimulation with B. pertussis whole-cells led to altered levels of Th1 (IFNγ), Th2 (IL-5, IL-13) and Th17 (IL-17A) cytokines in omvPV-mice, wPV-mice, and N.I.-mice compared to naive mice. These levels were overall higher in immunized mice compared to N.I.-mice. Moreover, IL-10 production was only observed in immunized mice (Fig. 8F). IFNγ and TNFα were both high in naive control mice after stimulation with B. pertussis OMVs and whole-cells, most likely caused by LPS. Nevertheless, omvPV-mice and wPV-mice showed a similar result post challenge (day 77) as after immunization (day 49). Prn stimulation resulted in increased IL-13 and IL-17A concentrations in all challenged mice compared to naive control mice (Supplementary Fig. S1A, right panel). These IL-17A levels were significantly higher in immunized mice compared to N.I.-mice. Furthermore, significantly higher IL-5 production was observed in omvPV-mice and N.I.-mice than in wPV-mice. After Ptx stimulation, IL-17A production was exclusively detected in wPV-mice (Supplementary Fig. S1B, right panel). IL-5, IL-13 and IL-17A production was evoked by FHA stimulation in immunized and N.I.-mice. Moreover, enhancement of IFNγ, TNFα, and IL-10 was detected in wPV-mice (Supplementary Fig. S1C, right panel). Overall enhanced Th1/Th2/Th17 ex vivo recall responses were detected in challenged omvPV-mice and wPV-mice compared to challenged N.I.-mice.
Discussion
Introduction of an improved next-generation pertussis vaccine requires in-depth knowledge on vaccine-induced responses in comparison to current pertussis vaccines especially in terms of efficacy and safety. Parenteral injection of classical whole-cell pertussis vaccine (wPV) has been related to adverse effects8,9 and suspected of being correlated with serious acute neurological illness in children10,11 mainly caused by circulating pro-inflammatory cytokines such as IL-1β, TNFα, and IL-611,47. The adverse effects have been key reasons for the development of aPVs, which have a better safety profile. Our current in vivo study demonstrated that immunization with an OMV-based pertussis vaccine (omvPV) in comparison to wPV elicited reduced concentrations of serum pro-inflammatory cytokines (IL-1α, IL-1β, and IL-6), chemokines (CXCL1 and CXCL10), and G-CSF. Especially, reduced IL-1β secretion by omvPV may be beneficial, because IL-1β is related to acute neurological illness11. Although data obtained in mice may not be representative for humans, it was previously demonstrated with a human whole-blood stimulation assay that omvPV induces less IL-6 compared to wPV17, which is in line with the mouse data presented here. Other cytokines that were detected in the current in vivo mouse model could be investigated in a similar way or in future human clinical trials with omvPV. Activation of Toll-like receptors (TLRs)48 by pathogen-associated molecular patterns (PAMPs), like lipopolysaccharide (LPS) and bacterial DNA, result in the secretion of these cytokines. Previously we showed that omvPV contain less TLR-activating LPS (1.0 μg versus 1.5 μg) and DNA (0.2 μg versus 1.2 μg) than wPV7. Lower LPS and DNA concentrations in omvPV may induce less TLR4 and TLR9 activation and subsequently contribute to attenuated pro-inflammatory cytokine response by omvPV compared to wPV. Adsorption of LPS to aluminum hydroxide causes less pro-inflammatory cytokine production49, which demonstrates that free LPS might be more pyrogenic than bound LPS. OmvPV may contain less free LPS compared to wPV thus resulting in lower cytokine induction.
Unexpectedly, in light of the attenuated pro-inflammatory cytokine signatures, omvPV immunization induced a larger number of genes than wPV. Yet triggering of anti-inflammatory responses, diminished PRR responses, and expression of modifiers that dampen inflammatory LPS responses was more prominent in omvPV-mice than in wPV-mice and likely explains why omvPV elicited lower amounts of pro-inflammatory cytokines. The omvPV-activated genes encoding anti-inflammatory PRRs FPR1 and FPR2 that bind many different signal peptides of bacteria50 and are important for rapid neutrophil recruitment51. Furthermore, the involvement of omvPV-induced genes encoding FPR2, SPLI, STEAP4, PTPN1, and MSR1, which are attenuators of LPS responses31,32,33,34,43, may assist in repressing LPS-induced pro-inflammation. The more prominent anti-inflammatory response following omvPV immunization was strengthened by activation of genes encoding proteins with a dual function in lipid metabolism and anti-inflammation, namely ANGPTL435,36, HPGD2S52, and LXRα. These genes may have been activated by lipid mediators that were released during the omvPV-induced response, which needs to be further investigated.
Only omvPV immunization enhances gene expression of TLR3 and TLR7, which are intracellular sensors for viral and bacterial nucleotides. Activation of TLRs leads to induction of IFN-signaling pathways. For instance, IRF7, a factor downstream of TLR7 and important mediator of type I IFN-signaling53, was induced by both vaccines. Overall, a large number of genes of the type I IFN-signaling and type II IFN-signaling pathway were induced by both vaccines but were more profound in the omvPV response. Type I IFN-signaling can affect both innate and adaptive immune cells54 and with respect to CD4+ T-cells, the pathway is important for the regulation of Th1 and Th17 responses55. Whether there are more TLR3 and TLR7 ligands present in omvPV compared to wPV remains to be investigated.
In addition, TLR4 activation by LPS can direct the cellular responses towards Th1 and Th17 responses5,6. Thus, it is important to evaluate whether the reduced TLR4-signaling by omvPV does not affect its ability to induce B. pertussis specific Th1 and Th17 responses. Cytokine profiles of stimulated splenocytes revealed that omvPV and wPV both induce mixed systemic Th1/Th2/Th17 responses, in contrast to the Th2-biased aPV-induced response4,14. This indicates that despite lower LPS concentration and TLR4-signaling, omvPV still induced a similar Th response as wPV. This may suggest that the LPS concentration in omvPV is sufficient for the induction of Th1/Th17 responses or that other signaling pathways, such as via TLR2 through lipoproteins56,57, also promote Th17 responses. Induction of both types of IFN-signaling pathways by omvPV and wPV may contribute to these Th1/Th17 responses that are thought to be important for protection3,4,24.
The omvPV and wPV used in this study contain low concentrations of aPV components (Prn, FHA, and Ptx). except for FHA in wPV7, which also resulted in low antibody and T-cell responses specific for these antigens following immunization. However, high antibody levels against other B. pertussis-specific antigens such as BrkA, Vag8 and LPS, were detected. These antibodies were especially formed after booster immunization and overall higher in omvPV-mice. The omvPV also contains higher concentrations of BrkA and Vag87. As Prn-deficient B. pertussis strains are increasingly circulating, especially in aPV-immunized populations, it has been hypothesized that this might be the result of pathogen adaptation58,59. Therefore, the broad humoral response conferred by both omvPV and wPV might limit the risk of driving B. pertussis adaptation caused by vaccine-induced selection pressure. Remarkably, despite lower LPS concentrations in omvPV, the booster omvPV immunization induced higher anti-LPS IgG3 antibody levels than wPV7. This indicates that the LPS in omvPV was processed in a different way by the immune system during the first immunization compared to booster immunization. Since anti-LPS IgG3 antibodies are mostly produced in a T-cell independent manner60, this may indicate that omvPV evokes a more efficient T-cell independent B-cell response than wPV. Furthermore, mucosal B. pertussis-specific IgA induction in the lungs was absent after omvPV and wPV immunization, as expected. Mucosal IgA is produced after natural infection24 and contributes most likely to a faster lung clearance during B. pertussis challenge. Presumably, direct involvement of the respiratory tract through mucosal vaccine administration may result in more effective immunity.
Immunization with omvPV or wPV enabled rapid clearance of B. pertussis from the lungs after challenge when compared to N.I.-mice. Whereas no significant differences in lung clearance were observed between both immunized groups, the lungs of omvPV-mice tended to be cleared slightly faster perhaps because of the higher serum IgG levels. Previously, intraperitoneal immunization with alum-adjuvated omvPV and wPV derived from a B. pertussis Tohama strain also demonstrated equal protection15. The B. pertussis challenge elucidated the pre-existing immunity in omvPV-mice and wPV-mice. The immune responses of challenged immunized mice were characterized by (i) adaptive recall responses, (ii) a change in pulmonary environment by reduced cytokine secretion and transcriptome expression, (iii) altered systemic cytokine responses, and (iv) reduced splenic transcriptome expression in comparison to challenged N.I.-mice. The presence of high serum IgG levels and strong recall of Th1/Th2/Th17 responses enabled a rapid clearance in both immunized groups. In contrast to vaccine-induced responses, the challenge-induced responses revealed specific T-cell activation against purified proteins FHA, Prn and Ptx as a result of prolonged exposure to these antigens after B. pertussis challenge24. Furthermore, immunized mice responded less vigorously to an infection with respect to transcriptome, cytokine responses and number of splenic neutrophils. In line with our previous study24, N.I.-mice showed the largest changes in gene expression in the lungs 14 days after challenge, whereas these responses were overall lower in omvPV-mice than wPV-mice. Overall, B. pertussis challenge-induced responses in wPV-mice resembled those of N.I.-mice more than those of omvPV-mice, which may indicate that the B. pertussis challenge was controlled more effectively in omvPV-mice than wPV-mice. For instance, challenged omvPV-mice revealed no presence of antigen-specific IgA in the lungs nor IL-17A secretion after ex vivo Ptx stimulation of splenocytes, while these responses appeared in both wPV-mice and N.I.-mice. Therefore, the B. pertussis exposure in wPV-mice may have been longer than in omvPV-mice as this Ptx exposure only occurs during B. pertussis colonization, since Ptx is absent in both omvPV and wPV used in this study7.
In summary, we demonstrated that in comparison to a classical wPV, omvPV confers equal, if not more rapid protection with higher IgG levels and a comparable Th1/Th2/Th17 response. Additionally, the systems approach provided detailed insight into the molecular signatures of the vaccine as well as challenge-induced responses. Importantly, the inflammatory responses elicited by omvPV were milder than those by wPV, as reflected by reduced levels of pro-inflammatory cytokines. Probably, inflammatory responses are attenuated by the enhanced anti-inflammatory responses, i.e. ANGPTL4, FPR2, PTPN1, SPLI, and STEAP4. Therefore, it is tempting to speculate that the decreased inflammatory responses induced by omvPV reflect a better safety profile. In conclusion, our collective findings emphasize the potential of omvPV as a third-generation pertussis vaccine.
Additional Information
How to cite this article: Raeven, R. H. M. et al. Bordetella pertussis outer membrane vesicle vaccine confers equal efficacy in mice with milder inflammatory responses compared to a whole-cell vaccine. Sci. Rep. 6, 38240; doi: 10.1038/srep38240 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The authors are grateful to Tim Bindels of Intravacc for the production of the omvPV. The authors thank employees of the Animal Research Centre (ARC) of Intravacc for the performance of animal experiments, and employees of the Microarray Department (MAD) of the University of Amsterdam for the performance of the microarray analyses.
Footnotes
Author Contributions R.H.M.R., G.F.A.K. and B.M. conceived the experiments, R.H.M.R., J.B., L.M., W.T., and K.H. conducted the experiments, and R.H.M.R., J.B., J.L.A.P., L.M., W.T., and K.H. analyzed the results. R.H.M.R. and B.M. wrote the main manuscript. J.B., J.L.A.P., L.M., W.T., K.H., E.R., W.J., C.A.C.M.E., W.G.H.H. and G.F.A.K. reviewed the manuscript.
References
- Cherry J. D. Epidemic pertussis in 2012–the resurgence of a vaccine-preventable disease. The New England journal of medicine 367, 785–787, doi: 10.1056/NEJMp1209051 (2012). [DOI] [PubMed] [Google Scholar]
- Tan T. et al. Pertussis Across the Globe: Recent Epidemiologic Trends From 2000 to 2013. The Pediatric infectious disease journal 34, e222–232, doi: 10.1097/INF.0000000000000795 (2015). [DOI] [PubMed] [Google Scholar]
- Warfel J. M. & Merkel T. J. Bordetella pertussis infection induces a mucosal IL-17 response and long-lived Th17 and Th1 immune memory cells in nonhuman primates. Mucosal immunology 6, 787–796, doi: 10.1038/mi.2012.117 (2013). [DOI] [PubMed] [Google Scholar]
- Ross P. J. et al. Relative contribution of Th1 and Th17 cells in adaptive immunity to Bordetella pertussis: towards the rational design of an improved acellular pertussis vaccine. PLoS pathogens 9, e1003264, doi: 10.1371/journal.ppat.1003264 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins S. C., Jarnicki A. G., Lavelle E. C. & Mills K. H. TLR4 mediates vaccine-induced protective cellular immunity to Bordetella pertussis: role of IL-17-producing T cells. J Immunol 177, 7980–7989 (2006). [DOI] [PubMed] [Google Scholar]
- Banus S. et al. The role of Toll-like receptor-4 in pertussis vaccine-induced immunity. BMC immunology 9, 21, doi: 10.1186/1471-2172-9-21 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeven R. H. M. et al. Immunoproteomic Profiling of Bordetella pertussis Outer Membrane Vesicle Vaccine Reveals Broad and Balanced Humoral Immunogenicity. Journal of proteome research 14, 2929–2942, doi: 10.1021/acs.jproteome.5b00258 (2015). [DOI] [PubMed] [Google Scholar]
- Jefferson T., Rudin M. & DiPietrantonj C. Systematic review of the effects of pertussis vaccines in children. Vaccine 21, 2003–2014 (2003). [DOI] [PubMed] [Google Scholar]
- David S., Vermeer-de Bondt P. E. & van der Maas N. A. Reactogenicity of infant whole cell pertussis combination vaccine compared with acellular pertussis vaccines with or without simultaneous pneumococcal vaccine in the Netherlands. Vaccine 26, 5883–5887, doi: 10.1016/j.vaccine.2008.07.105 (2008). [DOI] [PubMed] [Google Scholar]
- Miller D. L., Ross E. M., Alderslade R., Bellman M. H. & Rawson N. S. Pertussis immunisation and serious acute neurological illness in children. British medical journal 282, 1595–1599 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong M. E., Loscher C. E., Lynch M. A. & Mills K. H. IL-1beta-dependent neurological effects of the whole cell pertussis vaccine: a role for IL-1-associated signalling components in vaccine reactogenicity. Journal of neuroimmunology 136, 25–33 (2003). [DOI] [PubMed] [Google Scholar]
- Klein N. P., Bartlett J., Rowhani-Rahbar A., Fireman B. & Baxter R. Waning protection after fifth dose of acellular pertussis vaccine in children. The New England journal of medicine 367, 1012–1019, doi: 10.1056/NEJMoa1200850 (2012). [DOI] [PubMed] [Google Scholar]
- Warfel J. M., Zimmerman L. I. & Merkel T. J. Acellular pertussis vaccines protect against disease but fail to prevent infection and transmission in a nonhuman primate model. Proceedings of the National Academy of Sciences of the United States of America 111, 787–792, doi: 10.1073/pnas.1314688110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelman J. et al. Modulation of the CD4(+) T cell response after acellular pertussis vaccination in the presence of TLR4 ligation. Vaccine 33, 1483–1491, doi: 10.1016/j.vaccine.2015.01.063 (2015). [DOI] [PubMed] [Google Scholar]
- Roberts R. et al. Outer membrane vesicles as acellular vaccine against pertussis. Vaccine 26, 4639–4646, doi: 10.1016/j.vaccine.2008.07.004 (2008). [DOI] [PubMed] [Google Scholar]
- Gaillard M. E. et al. Acellular pertussis vaccine based on outer membrane vesicles capable of conferring both long-lasting immunity and protection against different strain genotypes. Vaccine 32, 931–937, doi: 10.1016/j.vaccine.2013.12.048 (2014). [DOI] [PubMed] [Google Scholar]
- Bottero D. et al. Characterization of the immune response induced by pertussis OMVs-based vaccine. Vaccine 34, 3303–3309, doi: 10.1016/j.vaccine.2016.04.079 (2016). [DOI] [PubMed] [Google Scholar]
- Nakaya H. I. et al. Systems biology of vaccination for seasonal influenza in humans. Nature immunology 12, 786–795, doi: 10.1038/ni.2067 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman D. et al. Apoptosis and other immune biomarkers predict influenza vaccine responsiveness. Molecular systems biology 9, 659, doi: 10.1038/msb.2013.15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querec T. D. et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nature immunology 10, 116–125, doi: 10.1038/ni.1688 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermoser G. et al. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity 38, 831–844, doi: 10.1016/j.immuni.2012.12.008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nature immunology 15, 195–204, doi: 10.1038/ni.2789 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami T. et al. System vaccinology for the evaluation of influenza vaccine safety by multiplex gene detection of novel biomarkers in a preclinical study and batch release test. PloS one 9, e101835, doi: 10.1371/journal.pone.0101835 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeven R. H. M. et al. Molecular Signatures of the Evolving Immune Response in Mice following a Bordetella pertussis Infection. PloS one 9, e104548, doi: 10.1371/journal.pone.0104548 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollinger W. D. et al. Design and evaluation in mice of a broadly protective meningococcal group B native outer membrane vesicle vaccine. Vaccine 28, 5057–5067, doi: 10.1016/j.vaccine.2010.05.006 (2010). [DOI] [PubMed] [Google Scholar]
- Benjamini Y. & Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society 57, 289–300. (1995). [Google Scholar]
- Samarajiwa S. A., Forster S., Auchettl K. & Hertzog P. J. INTERFEROME: the database of interferon regulated genes. Nucleic acids research 37, D852–857, doi: 10.1093/nar/gkn732 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Lopes S. et al. The exonuclease Trex1 restrains macrophage proinflammatory activation. J Immunol 191, 6128–6135, doi: 10.4049/jimmunol.1301603 (2013). [DOI] [PubMed] [Google Scholar]
- Chen G. Y. et al. Broad and direct interaction between TLR and Siglec families of pattern recognition receptors and its regulation by Neu1. eLife 3, e04066, doi: 10.7554/eLife.04066 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Franchi L. & Nunez G. The protein kinase PKR is critical for LPS-induced iNOS production but dispensable for inflammasome activation in macrophages. European journal of immunology 43, 1147–1152, doi: 10.1002/eji.201243187 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jozefowski S., Biedron R., Srottek M. & Chadzinska M. & Marcinkiewicz, J. The class A scavenger receptor SR-A/CD204 and the class B scavenger receptor CD36 regulate immune functions of macrophages differently. Innate immunity 20, 826–847, doi: 10.1177/1753425913510960 (2014). [DOI] [PubMed] [Google Scholar]
- Dufton N. et al. Anti-inflammatory role of the murine formyl-peptide receptor 2: ligand-specific effects on leukocyte responses and experimental inflammation. J Immunol 184, 2611–2619, doi: 10.4049/jimmunol.0903526 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A. et al. Murine tumor necrosis factor alpha-induced adipose-related protein (tumor necrosis factor alpha-induced protein 9) deficiency leads to arthritis via interleukin-6 overproduction with enhanced NF-kappaB, STAT-3 signaling, and dysregulated apoptosis of macrophages. Arthritis and rheumatism 64, 3877–3885, doi: 10.1002/art.34666 (2012). [DOI] [PubMed] [Google Scholar]
- Jin F. Y., Nathan C., Radzioch D. & Ding A. Secretory leukocyte protease inhibitor: a macrophage product induced by and antagonistic to bacterial lipopolysaccharide. Cell 88, 417–426 (1997). [DOI] [PubMed] [Google Scholar]
- Lu B., Moser A., Shigenaga J. K., Grunfeld C. & Feingold K. R. The acute phase response stimulates the expression of angiopoietin like protein 4. Biochemical and biophysical research communications 391, 1737–1741, doi: 10.1016/j.bbrc.2009.12.145 (2010). [DOI] [PubMed] [Google Scholar]
- Zhu P., Goh Y. Y., Chin H. F., Kersten S. & Tan N. S. Angiopoietin-like 4: a decade of research. Bioscience reports 32, 211–219, doi: 10.1042/BSR20110102 (2012). [DOI] [PubMed] [Google Scholar]
- Vigne S. et al. IL-36R ligands are potent regulators of dendritic and T cells. Blood 118, 5813–5823, doi: 10.1182/blood-2011-05-356873 (2011). [DOI] [PubMed] [Google Scholar]
- Lee M. S., Kim B., Oh G. T. & Kim Y. J. OASL1 inhibits translation of the type I interferon-regulating transcription factor IRF7. Nature immunology 14, 346–355, doi: 10.1038/ni.2535 (2013). [DOI] [PubMed] [Google Scholar]
- Takaoka A. et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448, 501–505, doi: 10.1038/nature06013 (2007). [DOI] [PubMed] [Google Scholar]
- Pollpeter D., Komuro A., Barber G. N. & Horvath C. M. Impaired cellular responses to cytosolic DNA or infection with Listeria monocytogenes and vaccinia virus in the absence of the murine LGP2 protein. PloS one 6, e18842, doi: 10.1371/journal.pone.0018842 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur J. C. et al. Cutting edge: NLRP12 controls dendritic and myeloid cell migration to affect contact hypersensitivity. J Immunol 185, 4515–4519, doi: 10.4049/jimmunol.1002227 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M. et al. TRIM30 alpha negatively regulates TLR-mediated NF-kappa B activation by targeting TAB2 and TAB3 for degradation. Nature immunology 9, 369–377, doi: 10.1038/ni1577 (2008). [DOI] [PubMed] [Google Scholar]
- Traves P. G. et al. Pivotal role of protein tyrosine phosphatase 1B (PTP1B) in the macrophage response to pro-inflammatory and anti-inflammatory challenge. Cell death & disease 5, e1125, doi: 10.1038/cddis.2014.90 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechmann M., Shuman N., Wakeham A. & Mak T. W. The CD83 reporter mouse elucidates the activity of the CD83 promoter in B, T, and dendritic cell populations in vivo. Proceedings of the National Academy of Sciences of the United States of America 105, 11887–11892, doi: 10.1073/pnas.0806335105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley J. P. et al. PARP-14 binds specific DNA sequences to promote Th2 cell gene expression. PloS one 8, e83127, doi: 10.1371/journal.pone.0083127 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. H. et al. B cell-intrinsic and -extrinsic regulation of antibody responses by PARP14, an intracellular (ADP-ribosyl)transferase. J Immunol 191, 3169–3178, doi: 10.4049/jimmunol.1301106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loscher C. E., Donnelly S., McBennett S., Lynch M. A. & Mills K. H. Proinflammatory cytokines in the adverse systemic and neurologic effects associated with parenteral injection of a whole cell pertussis vaccine. Annals of the New York Academy of Sciences 856, 274–277 (1998). [DOI] [PubMed] [Google Scholar]
- O’Neill L. A. How Toll-like receptors signal: what we know and what we don’t know. Current opinion in immunology 18, 3–9, doi: 10.1016/j.coi.2005.11.012 (2006). [DOI] [PubMed] [Google Scholar]
- Shi Y., HogenEsch H., Regnier F. E. & Hem S. L. Detoxification of endotoxin by aluminum hydroxide adjuvant. Vaccine 19, 1747–1752 (2001). [DOI] [PubMed] [Google Scholar]
- Bufe B. et al. Recognition of bacterial signal peptides by mammalian formyl peptide receptors: a new mechanism for sensing pathogens. The Journal of biological chemistry 290, 7369–7387, doi: 10.1074/jbc.M114.626747 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H. et al. Ligand recognition and activation of formyl peptide receptors in neutrophils. Journal of leukocyte biology 79, 247–256, doi: 10.1189/jlb.0905498 (2006). [DOI] [PubMed] [Google Scholar]
- Rajakariar R. et al. Hematopoietic prostaglandin D2 synthase controls the onset and resolution of acute inflammation through PGD2 and 15-deoxyDelta12 14 PGJ2. Proceedings of the National Academy of Sciences of the United States of America 104, 20979–20984, doi: 10.1073/pnas.0707394104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M. et al. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS letters 441, 106–110 (1998). [DOI] [PubMed] [Google Scholar]
- McNab F., Mayer-Barber K., Sher A., Wack A. & O’Garra A. Type I interferons in infectious disease. Nature reviews. Immunology 15, 87–103, doi: 10.1038/nri3787 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber J. P. & Farrar J. D. Regulation of effector and memory T-cell functions by type I interferon. Immunology 132, 466–474, doi: 10.1111/j.1365-2567.2011.03412.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds J. M. et al. Toll-like receptor 2 signaling in CD4(+) T lymphocytes promotes T helper 17 responses and regulates the pathogenesis of autoimmune disease. Immunity 32, 692–702, doi: 10.1016/j.immuni.2010.04.010 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne A. et al. A novel TLR2 agonist from Bordetella pertussis is a potent adjuvant that promotes protective immunity with an acellular pertussis vaccine. Mucosal immunology 8, 607–617, doi: 10.1038/mi.2014.93 (2015). [DOI] [PubMed] [Google Scholar]
- Martin S. W. et al. Pertactin-negative Bordetella pertussis strains: evidence for a possible selective advantage. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 60, 223–227, doi: 10.1093/cid/ciu788 (2015). [DOI] [PubMed] [Google Scholar]
- Lam C. et al. Rapid increase in pertactin-deficient Bordetella pertussis isolates, Australia. Emerging infectious diseases 20, 626–633, doi: 10.3201/eid2004.131478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana F. J., Solomon A., Cohen I. R. & Nussbaum G. Induction of IgG3 to LPS via Toll-like receptor 4 co-stimulation. PloS one 3, e3509, doi: 10.1371/journal.pone.0003509 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.