Clinical case
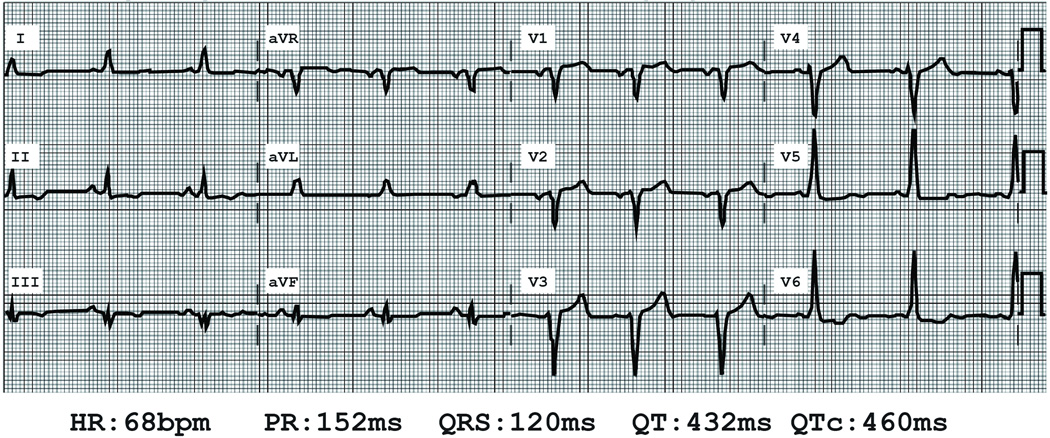
A 59 y/o female with a personal and family history of dilated cardiomyopathy (DCM) was referred by her cardiologist for genetic testing and counseling. The patient was initially diagnosed with DCM after presenting with dyspnea and fatigue. Her symptoms also included mild bilateral pedal edema but she denied orthopnea, paroxysmal nocturnal dyspnea, palpitations, pre-syncope, or syncope. An EKG showed sinus rhythm and a left bundle branch (LBBB) morphology (Figure 1). She underwent a gadolinium enhanced cardiac MRI that showed an enlarged left ventricle (internal diameter at end diastole and systole were 6.5 cm and 5.4 cm, respectively), and the left ventricular ejection fraction was ~25%. Abnormal septal wall motion consistent with a bundle branch block was noted. All other walls of the left ventricle were diffusely hypokinetic. Mild mid-myocardial hyperenhancement suggestive of idiopathic dilated cardiomyopathy was present. The RV was normal in size and function. There were no valvular abnormalities. Left heart catheterization showed no significant epicardial coronary artery disease (CAD). She was treated with a beta-adrenergic antagonist, an angiotensinogen inhibitor, a mineralocorticoid antagonist and a loop diuretic. Her symptoms improved; however, the left ventricle remained enlarged and had poor systolic function with an ejection fraction of ~30%. She subsequently underwent biventricular ICD implantation.
Figure 1.
Electrocardiogram of proband. Heart rate (HR) in beats per minute (bpm), P-R interval (PR) in milliseconds (ms), QRS duration (QRS) in ms, Q-T interval (QT) in ms and corrected Q-T interval using Bazett's formula (QTc) in ms are shown.
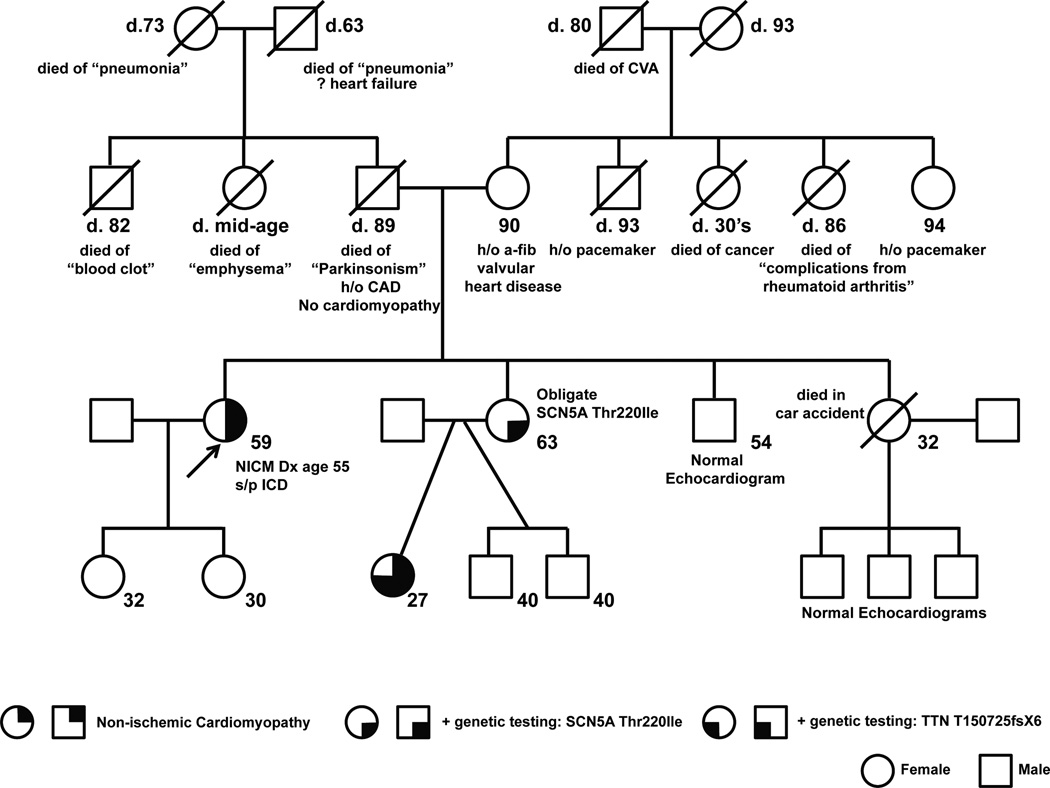
At the time of her initial diagnosis, the patient’s family history was notable for a 86 y/o mother who had atrial fibrillation and valvular heart disease; a father who died at age of 89 with CAD; a 59 y/o sister who had hypertension; a 50 y/o brother who had no known medical illnesses; and a sister who died in an automobile accident at age 32 (Figure 2). She also had two daughters, ages 32 and 30, who had no known medical illnesses. Of note, at the time of her diagnosis, our patient had no other family members who were reported to have DCM, although none had been screened prior to our patient’s diagnosis.
Figure 2.
Pedigree. Ages and clinical data are shown. Arrow denotes the proband.
Genetic test results
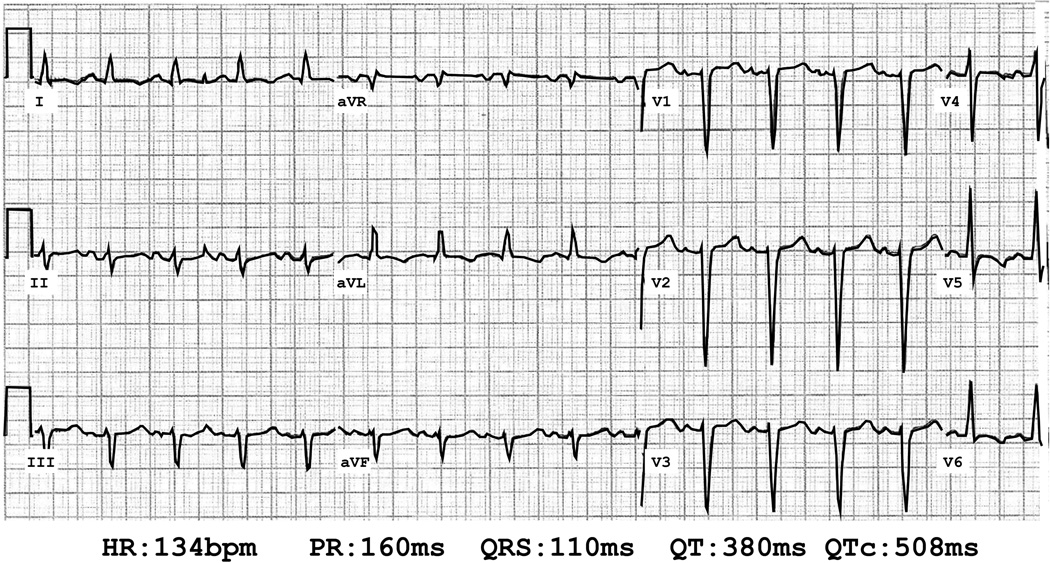
Four years after the patient’s initial diagnosis of DCM, her 27 y/o niece who lived out of state developed clinical left sided congestive heart failure and DCM. Her echocardiogram revealed moderate LV enlargement, severe functional mitral regurgitation and severe LV systolic dysfunction with EF <20%. ECG had incomplete LBBB, left axis deviation and diffuse T wave abnormalities (figure 3). She was treated with a beta-adrenergic antagonist, an angiotensinogen inhibitor, a mineralocorticoid antagonist and a loop diuretic. Her symptoms resolved and at 9 months her LV size, systolic dysfunction and functional mitral regurgitation normalized.
Figure 3.
Electrocardiogram of proband’s niece. Heart rate (HR) in beats per minute (bpm), P-R interval (PR) in milliseconds (ms), QRS duration (QRS) in ms, Q-T interval (QT) in ms and corrected Q-T interval using Bazett's formula (QTc) in ms are shown.
Her niece underwent genetic testing that revealed a missense mutation (Thr220Ile) in the type V alpha subunit of the voltage gated sodium channel (SCN5A). Variants in SCN5A have been shown to impair the sodium ion channel and associated with dilated cardiomyopathy with and without conduction disease, Brugada syndrome, and other arrhythmias.1–6 The SCN5A Thr220Ile variant (dbSNP ID rs45620037) has reported allele frequencies of 0.1% (allele count of 78 out of 22106) in an aggregate population and 0.7% (allele count of 29 out of 4160) in the Non-Finnish European population according to the Exome Aggregation Consortium (ExAC) (http://exac.broadinstitute.org/variant/3-38655278-G-A).7 Probands that carry the SCN5A Thr220Ile variant have been reported to carry additional potentially pathogenic variants. Additionally, ClinVar, an archive of reports of relationships among medically important variants and phenotypes, described conflicting interpretation of the pathogenicity of SCN5A Thr220Ile among seven submissions – two likely benign, three pathogenic, and two of uncertain significance (http://www.ncbi.nlm.nih.gov/clinvar/variation/9396/#clinical-assertions). Therefore, the contribution of the SCN5A Thr220Ile variant to the phenotype was unclear, suggesting that other genetic variants likely were contributing to the niece’s presentation. A more comprehensive panel was obtained and a second variant was identified (Thr15072SerfsX6) in the Titin (TTN) gene. In light of her niece’s test results, the patient was referred to our Adult Cardiovascular Genetics Clinic. After genetic counseling, the patient underwent targeted genetic testing that identified the SCN5A Thr220Ile variant; however, she did not harbor the TTN Thr15072SerfsX6 variant. The niece’s mother (Proband’s sister) had a history of hypertension and was asymptomatic. Cardiac evaluation revealed an ECG with left axis deviation. Echocardiography showed normal LV dimensions.
Genetic testing in DCM
Genetic testing utilizing Sanger or next generation DNA sequencing for familial DCM is clinically available. DCM-specific commercial panels comprehensively assay from 13 to over 30 genes depending on the panel; comprehensive cardiomyopathy panels that are not specific to DCM can range to over 70 genes.8, 9 The sensitivity (i.e. the likelihood of identifying a genetic variant using these panels in DCM) of testing remains about the same, however, ranging from 25% in DCM without conduction disease to as high as 40% in DCM with conduction disease.
Among patients who present with unexplained cardiomyopathy, ~50% are attributed to idiopathic DCM.10 A familial component has been estimated to occur in up to 40% of these idiopathic DCM cases.8 The identification of an inheritance pattern in multiple affected individuals in a family supports further genetic testing. The most important initial step in evaluating a patient who may have familial DCM is to obtain a careful family history for at least three generations with a goal of elucidating relatives with histories of early onset heart failure, syncope, sudden cardiac death without coronary involvement, and other muscular disease. The most common inheritance pattern observed with DCM with or without conduction disease is autosomal dominant, however, this pattern may be masked by variable clinical expressivity, reduced penetrance, and heterogeneous genetic involvement. The presence of other affected family members (e.g.; a pedigree consistent with familial DCM) may be the best predictor of informative genetic testing.
Interpreting Variants of Uncertain Significance
Variant interpretation, i.e. classifying genetic changes identified from sequencing as pathogenic, likely pathogenic, or variants of uncertain significance (VUS), can vary at each laboratory offering such testing. This clinical scenario demonstrates several important and complex issues pertaining to the evaluation of a genetic cause of DCM. The SCN5A Thr220Ile variant replaces the polar threonine amino acid with an isoleucine amino acid in the transmembrane region of the sodium channel and has been associated with DCM. It was considered a pathogenic variant by the initial testing laboratory and a VUS by the second testing laboratory. This difference in assignment of genetic variants by testing laboratories highlights a challenge to clinicians. As mentioned, segregation analyses of variants may provide some insight into the potential pathogenicity of a VUS. Additionally, the field of human genetics is rapidly evolving and variants that were initially classified as VUS may be re-classified as pathogenic in light of advances in knowledge.11–13 In cases of well-documented pathogenic variants, the clinician should consider predictive genetic testing in other family members. Importantly, casade screening of VUS is not recommended.
The families that have been studied with the SCN5A Thr220Ile variant have been complex and a clear causative role has not been established with certainty; thus, there is room for differences in interpretation of this variant. This is highlighted by the conflicting interpretation of the pathogenicity of SCN5A Thr220Ile listed in the ClinVar database.
Titin is an essential component of the sarcomere and mutations in TTN have been implicated in several diseases including limb-girdle muscular dystrophy, Salih myopathy, tibial muscular dystrophy, hereditary myopathy with early respiratory failure, and DCM.14, 15 TTN Thr15072SerfsX6 is a truncating variant; changes that cause truncation of TTN have been implicated in ~25% of familial cases of DCM and in ~18% of sporadic cases of DCM. Conversely, TTN truncating variants have been found in approximately 3% of healthy controls.14 The TTN Thr15072SerfsX6 is a novel genetic change that has not previously been published, but was classified as a pathogenic variant by the testing laboratory.
The proband did not carry the TTN Thr15072SerfsX6 suggesting that this variant may represent a VUS, underscoring the complexities of variant interpretation. The designation of pathological variants, either de novo or inherited, requires additional supportive data. In the de novo setting, negative clinical data in both patients and the absence of the variant (with confirmed paternity) is required. If the variant is presumed not to be de novo, then her mother or father has a 50% chance of passing the variant to the proband. The mother was evaluated clinically and did not have DCM. However, the father has not been evaluated and we recommended clinical screening.
The use of multiple laboratories highlights the differences in available testing panels. The interpretation of variants can be complex; our patient’s SCN5A variant was “downgraded” to a VUS by the second laboratory. Given our patient’s clinical history and lack of carriage of the TTN variant along with evidence in the literature, one interpretation is that the SCN5A Thr220Ile variant is responsible for the DCM presentation in our patient. Alternatively, an unidentified variant in a gene other than SCN5A could explain the clinical presentation. Moreover, TTN Thr15072SerfsX6 and SCN5A Thr220Ile potentially contribute to the niece’s presentation, which is supported by her DCM presentation at a younger age as compared to our patient. These issues highlight the importance of cardiovascular genetics – namely the careful phenotyping of all at risk relatives, identifying as many variants as possible, and testing unaffected relatives to understand the potential segregation of variants.
The Importance of formal evaluation of relatives
Guidelines for genetic testing in familial DCM have been established.8 Genetic testing provides important information that guides the care of patients and relatives. First, genetic test results have the potential to assist in risk stratification, prognosis, and management decisions. For example, identification of LMNA (A-type nuclear lamin) cardiomyopathy has significant implications regarding decisions concerning pacemaker or defibrillator therapy. Second, genetic testing can identify familial disease that may be missed even with careful documentation of family history. A careful family history for at least three generations is an important initial step. Combined genetic, family history, and clinical evaluation is optimal. Importantly, given the limitations and inaccuracies of reported family history, objective evaluation can be quite valuable. Clinical screening for DCM in asymptomatic first-degree relatives is recommended, i.e. with echocardiography for assessment of left ventricular function and size. Clinical screening for DCM should be considered in asymptomatic first-degree relative every 3–5 years if no causal variant is identified in the family or every 1–3 years in adults if a causal variant is present. Additionally, clinical screening is necessary at any time signs or symptoms appear. Referrals to centers that are expert in evaluation, genetic counseling, and genetic testing should be considered when there is concern for familial DCM. When evaluating several family members concurrently for potential genetic testing, testing should be considered for the single most clearly affected family member to facilitate screening, and genetic counseling is recommended for all patients and families who have DCM. Third, genetic test results can assist in the segregation analyses of variants and analysis of the potential pathogenicity of VUS’s.
Our patient’s symptoms have improved and she remains active with NYHA class I-II symptoms. Variants in SCN5A have been associated with congenital and drug-induced long QT syndrome and, therefore, she was cautioned about the use of medication that could potentially prolong her QT interval and lead to life threatening arrhythmias (http://www.brugadadrugs.org/drug-lists).1, 2, 16 The patient’s genetic testing results also had implications for other family members. As a result of genetic testing and counseling, our patient’s 30 y/o daughter was evaluated in our Adult Cardiovascular Genetics Clinic and underwent genetic screening that showed that she did not inherit the SCN5A Thr220Ile variant as mentioned above. The other daughter is considering genetic testing. Analysis of the pedigree suggests that our patient’s sister (her niece’s mother) is an obligate carrier of the SCN5A variant, although she did not pursue clinical or genetic testing herself.
Conclusion
This clinical case highlights some of the complexity and issues involved with genetic testing in DCM. The understanding of the genetic basis of DCM is rapidly evolving, requires iterative re-evaluation of VUS identified during genetic testing, and interpreting VUS in light of new functional data from ongoing investigations to correlate genotype to phenotype. Regardless, the field of cardiovascular genetics is rapidly evolving and identification of patients who may have familial DCM is critical to improving the health and lives of patients and families. Genetic testing for familial DCM should be considered and if appropriate and desired by the patient, referral to a clinic with expertise in this disease, and capabilities for genetic counseling should be pursued.
Footnotes
Conflict of Interest Disclosures: None
References
- 1.Benson DW, Wang DW, Dyment M, Knilans TK, Fish FA, Strieper MJ, et al. Congenital sick sinus syndrome caused by recessive mutations in the cardiac sodium channel gene (SCN5A) J Clin Invest. 2003;112:1019–1028. doi: 10.1172/JCI18062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapplinger JD, Tester DJ, Alders M, Benito B, Berthet M, Brugada J, et al. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm. 2010;7:33–46. doi: 10.1016/j.hrthm.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McNair WP, Ku L, Taylor MR, Fain PR, Dao D, Wolfel E, et al. SCN5A mutation associated with dilated cardiomyopathy, conduction disorder, and arrhythmia. Circulation. 2004;110:2163–2167. doi: 10.1161/01.CIR.0000144458.58660.BB. [DOI] [PubMed] [Google Scholar]
- 4.McNair WP, Sinagra G, Taylor MR, Di Lenarda A, Ferguson DA, Salcedo EE, et al. SCN5A mutations associate with arrhythmic dilated cardiomyopathy and commonly localize to the voltage-sensing mechanism. J Am Coll Cardiol. 2011;57:2160–2168. doi: 10.1016/j.jacc.2010.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olesen MS, Yuan L, Liang B, Holst AG, Nielsen N, Nielsen JB, et al. High prevalence of long QT syndrome-associated SCN5A variants in patients with early-onset lone atrial fibrillation. Circ Cardiovasc Genet. 2012;5:450–459. doi: 10.1161/CIRCGENETICS.111.962597. [DOI] [PubMed] [Google Scholar]
- 6.Olson TM, Michels VV, Ballew JD, Reyna SP, Karst ML, Herron KJ, et al. Sodium channel mutations and susceptibility to heart failure and atrial fibrillation. JAMA. 2005;293:447–454. doi: 10.1001/jama.293.4.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Exome Aggregation Consortium (ExAC) Cambridge, MA: [[July, 2015]]. (URL: http://exac.broadinstitute.org) [Google Scholar]
- 8.Hershberger RE, Lindenfeld J, Mestroni L, Seidman CE, Taylor MR, Towbin JA, et al. Genetic evaluation of cardiomyopathy--a Heart Failure Society of America practice guideline. J Card Fail. 2009;15:83–97. doi: 10.1016/j.cardfail.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Hershberger RE, Morales A, Siegfried JD. Clinical and genetic issues in dilated cardiomyopathy: a review for genetics professionals. Genet Med. 2010;12:655–667. doi: 10.1097/GIM.0b013e3181f2481f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felker GM, Thompson RE, Hare JM, Hruban RH, Clemetson DE, Howard DL, et al. Underlying causes and long-term survival in patients with initially unexplained cardiomyopathy. N Engl J Med. 2000;342:1077–1084. doi: 10.1056/NEJM200004133421502. [DOI] [PubMed] [Google Scholar]
- 11.Andreasen C, Nielsen JB, Refsgaard L, Holst AG, Christensen AH, Andreasen L, et al. New population-based exome data are questioning the pathogenicity of previously cardiomyopathy-associated genetic variants. Eur J Hum Genet. 2013;21:918–928. doi: 10.1038/ejhg.2012.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herman DS, Lam L, Taylor MR, Wang L, Teekakirikul P, Christodoulou D, et al. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norton N, Li D, Rampersaud E, Morales A, Martin ER, Zuchner S, Guo S, et al. Exome sequencing and genome-wide linkage analysis in 17 families illustrate the complex contribution of TTN truncating variants to dilated cardiomyopathy. Circ Cardiovasc Genet. 2013;6:144–153. doi: 10.1161/CIRCGENETICS.111.000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Postema PG, Wolpert C, Amin AS, Probst V, Borggrefe M, Roden DM, et al. Drugs and Brugada syndrome patients: review of the literature, recommendations, and an up-to-date website ( www.brugadadrugs.org) Heart Rhythm. 2009;6:1335–1341. doi: 10.1016/j.hrthm.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]