Abstract
Cisplatin, one of the most widely used anticancer drugs, crosslinks DNA and ultimately induces cell death. However, the genomic pattern of cisplatin–DNA adducts has remained unknown owing to the lack of a reliable and sensitive genome-wide method. Herein we present “cisplatin-seq” to identify genome-wide cisplatin crosslinking sites at base resolution. Cisplatin-seq reveals that mitochondrial DNA is a preferred target of cisplatin. For nuclear genomes, cisplatin–DNA adducts are enriched within promoters and regions harboring transcription termination sites. While the density of GG dinucleotides determines the initial crosslinking of cisplatin, binding of proteins to the genome largely contributes to the accumulative pattern of cisplatin–DNA adducts.
Keywords: cisplatin, DNA damage, DNA structures, high-throughput sequencing, medicinal chemistry
Cisplatin plays a central role in cancer chemotherapy.[1] It is highly effective in treating a variety of solid tumors, including testicular, ovarian, cervical, head and neck, lung, and colorectal cancer.[2] Although cisplatin can bind to proteins, RNA, membrane phospholipids, microfilaments, and thiol-containing peptides,[3] DNA is generally considered as its major biological target.[2] The platinum atom of cisplatin can form covalent bonds to the N7 positions of purine bases, resulting in about 65% cis-[Pt(NH3)2{d(GpG)}] (“cis-GG”), approximately 25% cis-[Pt(NH3)2{d(ApG)}] (“cis-AG”) 1,2-intrastrand adducts, and about 5–10% 1,3-intrastrand adducts (“cis-GNG”).[4] Compared to the 1,2-intrastrand adducts, 1,3-intrastrand adducts are more readily excised in vitro by the nucleotide excision repair (NER) machinery.[5] A small percentage of monofunctional adducts and interstrand crosslinks also exist.[4] Transplatin, the clinically ineffective isomer of cisplatin, mainly forms 1,3-intrastrand and interstrand crosslinks; it is unable to form 1,2-intrastrand cis-GG or cis-AG adducts owing to stereochemical constraints.[6] Collectively, these findings suggest that the 1,2-intrastand adducts may be important to the anticancer activity of cisplatin.[3a]
Cisplatin crosslinking distorts DNA duplex structures, which can be recognized by various classes of proteins.[2] One well-studied example is the high mobility group (HMG) box protein HMGB1,[7] which is an abundant and highly conserved non-histone chromosomal protein. As a non-sequence-specific DNA binding protein, HMGB1 regulates many cellular processes, including transcription, replication, recombination, and chromatin remodeling.[8] It consists of a highly acidic C-terminal tail and two tandem HMG boxes (domain A and domain B). While each domain as well as the full-length protein can bind to cisplatin-modified DNA, domain A interacts more strongly with cisplatin-modified DNA than domain B.[2] Instead of directly recognizing the platinum modification, domain A binds to the cisplatin-induced widened minor groove of the highly distorted DNA duplex.[9]
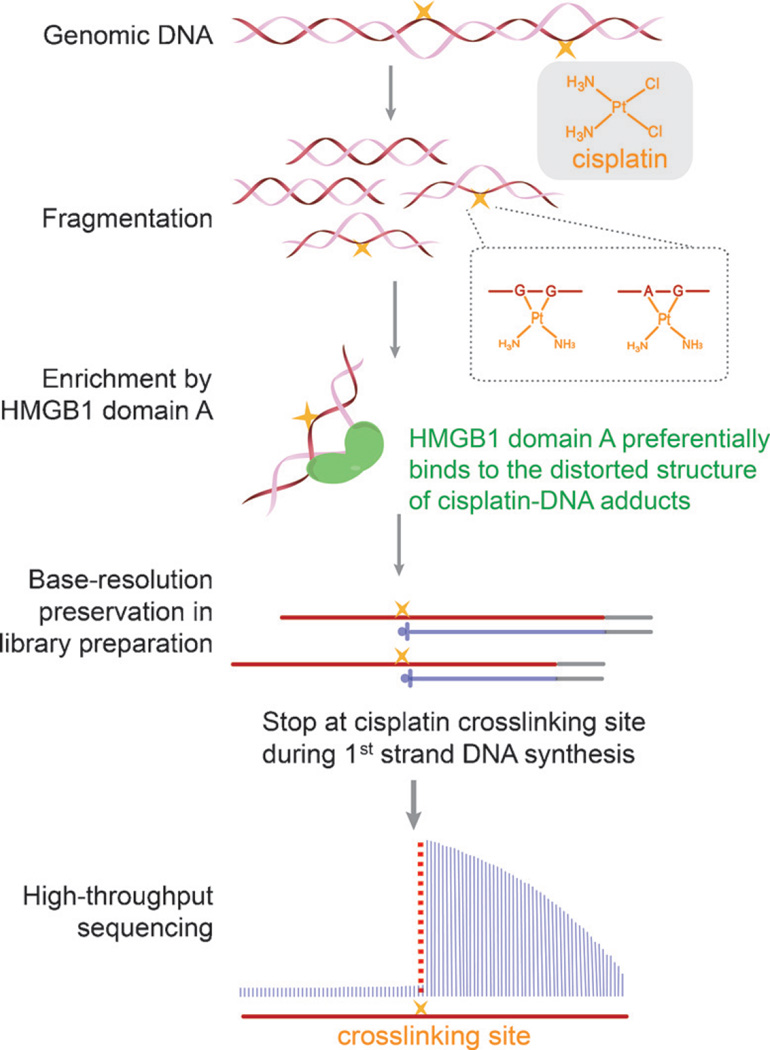
To uncover the genomic distribution of cisplatin–DNA adducts, we developed “cisplatin-seq” to identify cisplatin crosslinking sites in the human genome with base resolution. Although previous studies have reported cisplatin adducts in several specific genomic regions using primer extension and PCR,[10] a genome-wide method to detect cisplatin–DNA adducts is lacking. In this study, we took advantage of the preferential binding of HMGB1 domain A to distorted DNA structures to selectively enrich cisplatin-modified DNA for high-throughput sequencing. Owing to the ability of cisplatin–DNA adducts to stall DNA synthesis,[11] cisplatin crosslinking sites could be identified at base resolution throughout the genome (Scheme 1).
Scheme 1.
Workflow of cisplatin-seq. Following DNA fragmentation, cisplatin-modified DNA fragments are enriched by domain A of HMGB1. Cisplatin–DNA adducts cause primer extension (first strand DNA synthesis) to stop at the sites of cisplatin crosslinking, which gives base-resolution information of cisplatin crosslinking sites.
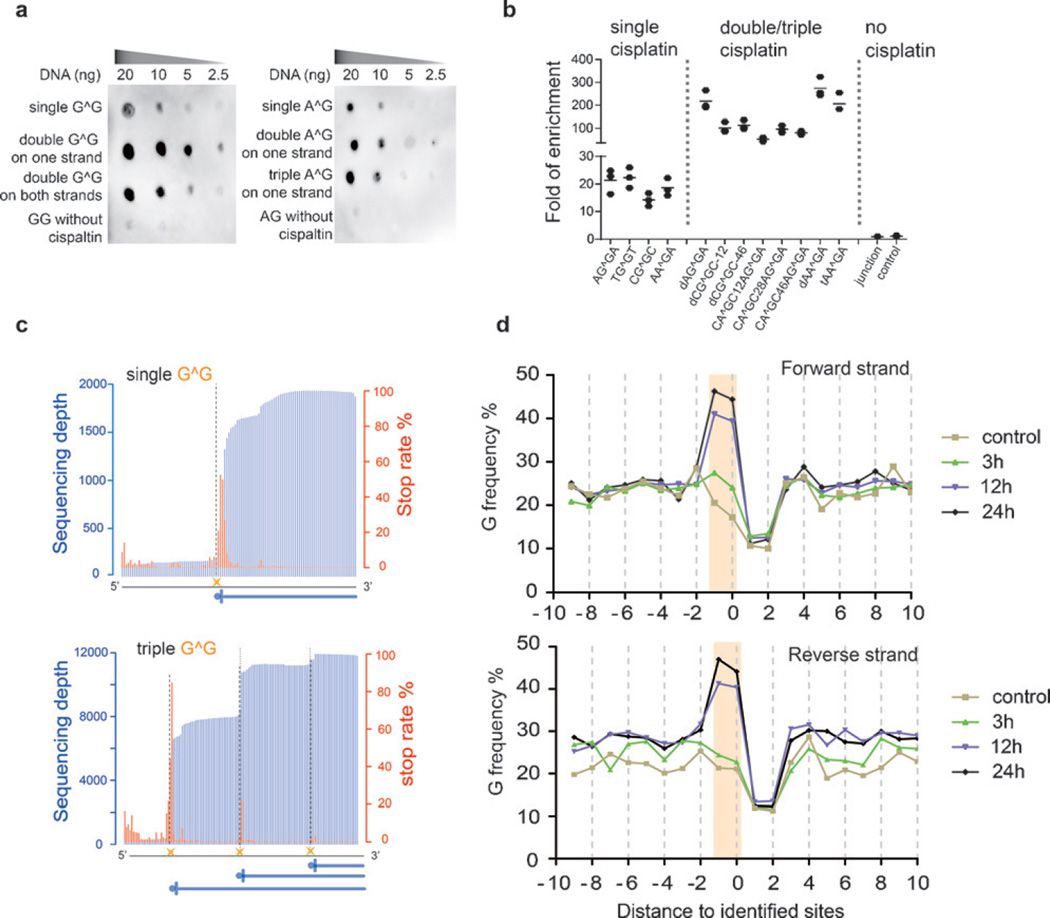
To identify a suitable construct of HMGB1 to enrich cisplatin-modified DNA, we first expressed and purified a series of protein constructs comprising different truncations and mutations to wild-type HMGB1 (see the Supporting Information, Figure S3a, b), and compared their specificities as well as affinities to cisplatin-modified model DNA sequences. Each model sequence contains one or more site-specific, fully modified cisplatin–DNA adducts (Figures S1 and S2). Dot blot analyses showed that among all of the HMGB1 constructs tested, domain A demonstrates the highest specificity and affinity to cisplatin-modified DNA (Figure S3c), which is consistent with results from previous gel shift assays.[2] Owing to its ability to recognize the distorted duplex structure instead of the crosslinking sites, domain A efficiently recognizes both cis-GG and cis-AG adducts (Figure 1a), which cannot be achieved with a commercial antibody (Figure S4). Compared to sequences with a single adduct, domain A exhibited a higher binding affinity to sequences with multiple adducts, independent of the relative positions of the platinum modifications (whether they are on different DNA strands or separated by different distances; Figure 1a and Figure S5). We then optimized the conditions to selectively pull down cisplatin-modified DNA (Figure S6). Model sequences with a single cisplatin were enriched by about 20 fold, and sequences containing two or three cisplatin modifications were enriched by about 50–300 fold (Figure 1b). Importantly, no enrichment was observed under our optimized conditions for a control sequence with a four-way junction structure,[12] suggesting the specificity of cisplatin-seq.
Figure 1.
Cisplatin-seq reliably detects cisplatin–DNA adducts in model DNA and the human genome. a) Dot blot analysis showed that HMGB1 domain A specifically binds to cisplatin-modified model DNA sequences (“G^G” denotes cis-GG 1,2-intrastrand adducts; all sequences in this Figure have two adenosines flanking the G^G and A^G adducts). b) Enrichment of cisplatin-modified model DNA sequences after HMGB1 domain A pull-down experiments (n = 3). For the sequence named “dCG^GC-12”, “d” stands for “double” cis-GG adducts, and “12” means a 12 bp distance between two cis-GG adducts. Similarly, in other sequences, “t” stands for “triple” and the number represents the distance between two nearby cisplatin adducts; “junction” represents a sequence with a four-way junction structure, which was used as a control. c) The stop rate information in model sequences was used to detect cisplatin crosslinking sites at base resolution. The blue vertical lines are sequencing depths whereas the red lines are calculated stop rates (see the Supporting Information). The dashed lines represent the cisplatin sites. The sequences in this Figure have two cytidines flanking the G^G adducts. d) Frequency of guanine nucleosides spanning the identified sites for treatment for 3 h, 12 h, and 24 h. “Position 0” corresponds to sites of high stop rates.
We then subjected the model sequences to high-throughput sequencing. To preserve the base-resolution information of cisplatin crosslinking, we utilized a modified library preparation procedure in which the second adaptor was ligated after the synthesis of the first DNA strand (see the Experimental Section in the Supporting Information). Indeed, sequencing reads were truncated at the sites of cisplatin crosslinking (Figure 1c and Figure S7); moreover, for model sequences that contained multiple platinum modifications on one strand, consecutive stops within sequencing reads were observed (Figure 1c). Thus cisplatin-seq is able to identify platinum modifications at base resolution even when they are clustered within short distances.
Having validated our method in model sequences, we next applied cisplatin-seq to cisplatin-modified genomic DNA from Hela cells. We first confirmed the capability of cisplatin-seq to effectively enrich cisplatin-modified genomic DNA using a commercial anti-cisplatin antibody in a dot blot assay (Figure S8). To identify genome-wide cisplatin–DNA adducts at base resolution, we adopted a stringent bioinformatics procedure: We performed peak calling and calculated the stop rate for each nucleotide throughout the genome; only sites that had high stop rates and were also located within cisplatin peaks were considered to be cisplatin crosslinking sites. 1782, 3917, and 3281 sites were identified from cells treated with cisplatin for 3 h, 12 h, and 24 h, respectively. In fact, most of the sites with high stop rates (ca. 75%) were found to be present within the cisplatin peaks, and these sites were used for subsequent analysis.
We then calculated the frequency of guanosine spanning the identified sites (defined as “position 0”); within the 20 nt window, only position −1 and position 0 demonstrated dramatically increased frequencies of guanosine upon prolonged cisplatin treatment (Figure 1d). This observation is consistent with the fact that cis-GG is a major product of cisplatin crosslinking. In addition, the amount of AG dinucleotides also increased (Figure S9). These results demonstrate that cisplatin-seq reliably detects cisplatin–DNA adducts in the whole genome.
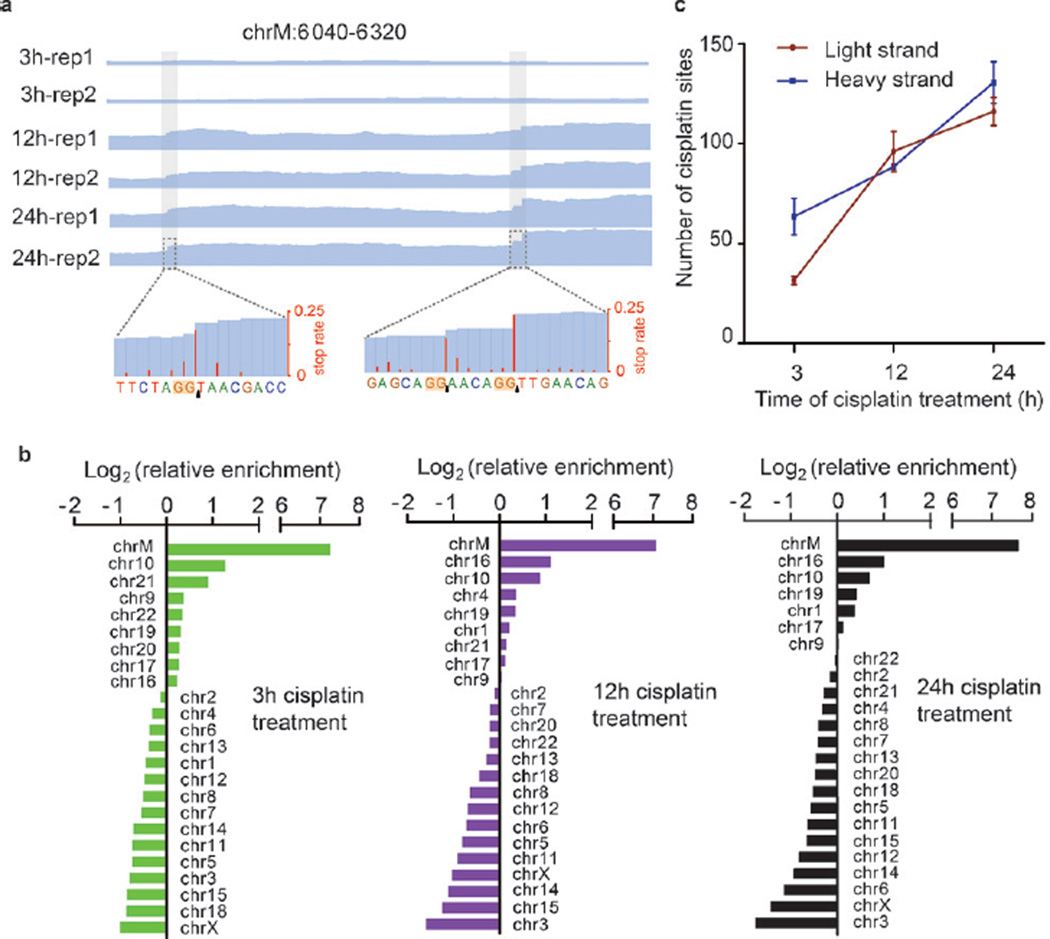
We next analyzed the distribution of cisplatin crosslinking sites in the human genome; one typical example is shown in Figure 2a. Mitochondrial DNA, which is devoid of histone proteins or NER, was found to be a major target of cisplatin (Figure 2b and Figure S10a). Initially, fewer cisplatin–DNA adducts were found in the light strand of mitochondrial DNA, which carries more genes than the heavy strand; after extended cisplatin treatment, similar numbers of cisplatin crosslinking sites were found in the two strands (Figure 2c and Figure S10b).
Figure 2.
Mitochondrial DNA is a preferred target of cisplatin. a) IGV views of representative cisplatin sites. Blue lines are sequencing depths; the enrichment of regions containing cisplatin modifications also gradually increased from 3 h to 12 h and 24 h. In the zoom-in view, both single and consecutive cisplatin crosslinking sites can be identified. b) Relative enrichment of cisplatin sites on each chromosome. Enrichments were normalized by the sequencing coverage of each chromosome in the “input” sample. c) The number of cisplatin sites in light and heavy strands of mitochondrial DNA after 3 h, 12 h, or 24 h of cisplatin treatment.
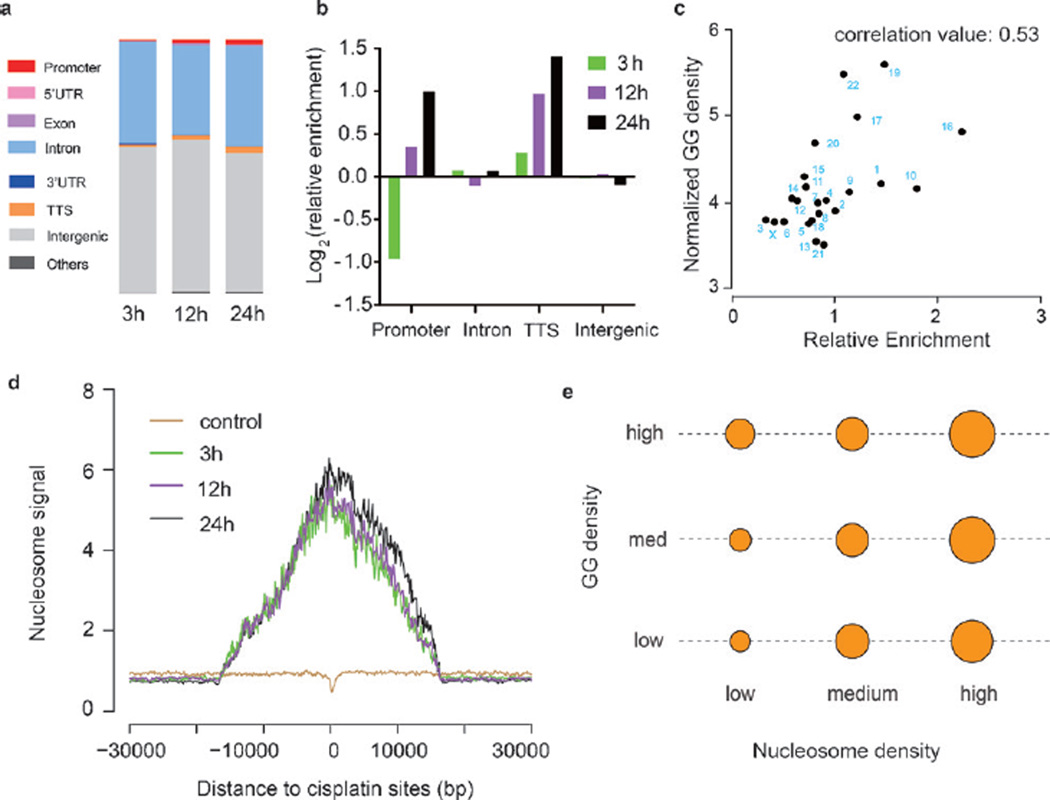
For nuclear DNA, cisplatin crosslinking displays an uneven distribution (Figure 3a,b). Cisplatin is enriched within promoters and regions near transcription termination sites (TTSs). The GG dinucleotide densities (denoted as “GG density”) of cisplatin-modified promoters and TTS regions were found to be higher than that of other promoters and TTS regions (Figure S11a, b), which is consistent with the fact that GG dinucleotides are the major target of cisplatin crosslinking. Moreover, higher genomic GG densities were also observed in the regions surrounding cisplatin modifications (Figure S11c). Furthermore, the enrichment of cisplatin sites for different nuclear chromosomes demonstrates a positive correlation with their GG densities (Figure 3c). Therefore, the density of GG dinucleotides positively impacts the genomic pattern of cisplatin crosslinking.
Figure 3.
Distribution of cisplatin–DNA adducts on nuclear DNA. a) Overall distribution of cisplatin sites in the human genome after cisplatin treatment for 3 h, 12 h, or 24 h. b) Relative enrichment of cisplatin crosslinking sites in promoter, intron, TTS, and intergenic regions. c) The enrichment of cisplatin sites on nuclear chromosomes positively correlates with their GG densities (Pearson correlation test, two-sided, correlation value: 0.53, p = 0.009). d) Nucleosome sequencing signals across the cisplatin sites. e) The enrichment of cisplatin–DNA adducts increases with elevated nucleosome density regardless of the GG densities. The area of each circle corresponds to the log value of the cisplatin density. For each column and row, the nucleosome and GG density are fixed, respectively.
While the density of GG dinucleotides may influence the initial crosslinking of cisplatin, other factors including the chromatin states have been proposed to affect the accumulative pattern of cisplatin crosslinking.[13] We first calculated the chromatin immunoprecipitation sequencing (ChIP-Seq) signals of several histone modification markers (including H3K4me3, H3K27ac, and H3k27me3) around cisplatin crosslinking sites; however, no noticeable differences were found between the treated and untreated samples (Figure S12). Interestingly, cisplatin was found to preferentially crosslink genomic regions with high nucleosome signals (Figure 3d). Given that nucleosome signals are also correlated with the GG density (Figure S13), we further separated the impact of these two factors on the cisplatin distribution (Figure 3e). The strength of the nucleosome signal significantly influences the distribution of cisplatin no matter whether the GG density is low, medium, or high (Figure 3e), suggesting that histone binding contributes to the cisplatin accumulation independent of the GG density. We then examined whether the binding of non-histone proteins could also influence the accumulation of cisplatin. Utilizing ChIP-seq data of three distinct types of DNA-binding proteins (Pol II, EZH2, and CTCF) and one histone variant (H2AZ) available from the ENCODE database, we found that cisplatin–DNA adducts tend to accumulate in regions with concentrated binding of all of these proteins (Figure S14). Thus the binding of DNA-binding proteins will influence the accumulation of cisplatin within the corresponding genomic regions. In fact, previous reports have shown that protein binding limits the DNA accessibility to the NER machinery and hence impairs the NER activity.[14] Collectively, we concluded that after the preferential cisplatin targeting of regions with high GG density, shielding of cisplatin–DNA adducts by protein binding largely affects the accumulative pattern of cisplatin modification in the human genome.
In conclusion, the cisplatin-seq approach provides the first genome-wide profile of cisplatin–DNA adducts at base resolution. Our results are genome-wide evidence that mitochondrial DNA is a major target of cisplatin. Whereas the GG dinucleotide density determines the initial cisplatin crosslinking, binding of proteins to the genome largely contributes to the accumulation of cisplatin–DNA adducts. Cisplatin-seq may also be applied to profile cisplatin–DNA adducts for different cisplatin dosages, when cisplatin is used in combination therapies with other drugs,[1a] or in cisplatin-resistant cancer cells.[15]
Supplementary Material
Acknowledgments
This work was supported by the National Basic Research Foundation of China (2016YFC0900301 to C.Y.), the National Natural Science Foundation of China (21472009 and 21522201 to C.Y.), and the US National Institutes of Health (R01 HG006827 to C.H.). C.H. is supported by the Howard Hughes Medical Institute. This work was also supported by the Beijing Advanced Innovation Centre for Genomics at Peking University.
Footnotes
Supporting information for this article can be found under: http://dx.doi.org/10.1002/anie.201607380. Sequencing data have been deposited into the Gene Expression Omnibus (GEO) under the accession number GSE84637.
Contributor Information
Xiaoting Shu, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Department of Chemical Biology and Synthetic and Functional Biomolecules Center, College of Chemistry and Molecular Engineering and Peking-Tsinghua Center for Life Sciences, Peking University, Beijing 100871 (China); Academy for Advanced Interdisciplinary Studies, Peking University, Beijing 100871 (China).
Xushen Xiong, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Department of Chemical Biology and Synthetic and Functional Biomolecules Center, College of Chemistry and Molecular Engineering and Peking-Tsinghua Center for Life Sciences, Peking University, Beijing 100871 (China); Academy for Advanced Interdisciplinary Studies, Peking University, Beijing 100871 (China).
Jinghui Song, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Department of Chemical Biology and Synthetic and Functional Biomolecules Center, College of Chemistry and Molecular Engineering and Peking-Tsinghua Center for Life Sciences, Peking University, Beijing 100871 (China).
Chuan He, Department of Chemistry, Department of Biochemistry and Molecular Biology and Institute for Biophysical Dynamics, Howard Hughes Medical Institute, The University of Chicago, 929 East 57th Street, Chicago, IL 60637 (USA); Department of Chemical Biology and Synthetic and Functional Biomolecules Center, College of Chemistry and Molecular Engineering, Beijing Advanced Innovation Center for Genomics, Peking University, Beijing 100871 (China).
Chengqi Yi, State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Department of Chemical Biology and Synthetic and Functional Biomolecules Center, College of Chemistry and Molecular Engineering and Peking-Tsinghua Center for Life Sciences, Peking University, Beijing 100871 (China).
References
- 1.a) Wang D, Lippard SJ. Nat. Rev. Drug Discovery. 2005;4:307. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]; b) Muggia FM, Bonetti A, Hoeschele JD, Rozencweig M, Howell SB. J. Clin. Oncol. 2015;33:4219. doi: 10.1200/JCO.2015.60.7481. [DOI] [PubMed] [Google Scholar]; c) Fuertes MA, Alonso C, Perez JM. Chem. Rev. 2003;103:645. doi: 10.1021/cr020010d. [DOI] [PubMed] [Google Scholar]
- 2.Jung Y, Lippard SJ. Chem. Rev. 2007;107:1387. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 3.a) Jamieson ER, Lippard SJ. Chem. Rev. 1999;99:2467. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]; b) Chapman EG, Hostetter AA, Osborn MF, Miller AL, DeRose VJ. Met. Ions Life Sci. 2011;9:347. doi: 10.1039/9781849732512-00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Fichtinger-Schepman AM, van der Veer JL, den Hartog JH, Lohman PH, Reedijk J. Biochemistry. 1985;24:707. doi: 10.1021/bi00324a025. [DOI] [PubMed] [Google Scholar]; b) Eastman A. Biochemistry. 1986;25:3912. doi: 10.1021/bi00361a026. [DOI] [PubMed] [Google Scholar]
- 5.a) Huang JC, Zamble DB, Reardon JT, Lippard SJ, Sancar A. Proc. Natl. Acad. Sci. USA. 1994;91:10394. doi: 10.1073/pnas.91.22.10394. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zamble DB, Mu D, Reardon JT, Sancar A, Lippard SJ. Biochemistry. 1996;35:10004. doi: 10.1021/bi960453+. [DOI] [PubMed] [Google Scholar]
- 6.Lepre CA, Strothkamp KG, Lippard SJ. Biochemistry. 1987;26:5651. doi: 10.1021/bi00392a011. [DOI] [PubMed] [Google Scholar]
- 7.Pil PM, Lippard SJ. Science. 1992;256:234. doi: 10.1126/science.1566071. [DOI] [PubMed] [Google Scholar]
- 8.Stros M. Biochim. Biophys. Acta Gene Regul. Mech. 2010;1799:101. doi: 10.1016/j.bbagrm.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Ohndorf UM, Rould MA, He Q, Pabo CO, Lippard SJ. Nature. 1999;399:708. doi: 10.1038/21460. [DOI] [PubMed] [Google Scholar]
- 10.a) Grimaldi KA, McAdam SR, Souhami RL, Hartley JA. Nucleic Acids Res. 1994;22:2311. doi: 10.1093/nar/22.12.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Murray V, Motyka H, England PR, Wickham G, Lee HH, Denny WA, McFadyen WD. Biochemistry. 1992;31:11812. doi: 10.1021/bi00162a020. [DOI] [PubMed] [Google Scholar]; c) Burstyn JN, Heiger-Bernays WJ, Cohen SM, Lippard SJ. Nucleic Acids Res. 2000;28:4237. doi: 10.1093/nar/28.21.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Davies NP, Hardman LC, Murray V. Nucleic Acids Res. 2000;28:2954. doi: 10.1093/nar/28.15.2954. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) McGurk CJ, McHugh PJ, Tilby MJ, Grimaldi KA, Hartley JA. Methods Enzymol. 2001;340:358. doi: 10.1016/s0076-6879(01)40431-9. [DOI] [PubMed] [Google Scholar]
- 11.a) Comess KM, Burstyn JN, Essigmann JM, Lippard SJ. Biochemistry. 1992;31:3975. doi: 10.1021/bi00131a013. [DOI] [PubMed] [Google Scholar]; b) Malina J, Novakova O, Vojtiskova M, Natile G, Brabec V. Biophys. J. 2007;93:3950. doi: 10.1529/biophysj.107.116996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Webb M, Thomas JO. J. Mol. Biol. 1999;294:373. doi: 10.1006/jmbi.1999.3150. [DOI] [PubMed] [Google Scholar]
- 13.a) Han C, Srivastava AK, Cui T, Wang QE, Wani AA. Carcinogenesis. 2016;37:129. doi: 10.1093/carcin/bgv247. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Powell JR, Bennett MR, Evans KE, Yu S, Webster RM, Waters R, Skinner N, Reed SH. Sci. Rep. 2015;5:7975. doi: 10.1038/srep07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Wang D, Hara R, Singh G, Sancar A, Lippard SJ. Biochemistry. 2003;42:6747. doi: 10.1021/bi034264k. [DOI] [PubMed] [Google Scholar]; b) Hara R, Mo J, Sancar A. Mol. Cell. Biol. 2000;20:9173. doi: 10.1128/mcb.20.24.9173-9181.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Sabarinathan R, Mularoni L, Deu-Pons J, Gonzalez-Perez A, Lopez-Bigas N. Nature. 2016;532:264. doi: 10.1038/nature17661. [DOI] [PubMed] [Google Scholar]; d) Perera D, Poulos RC, Shah A, Beck D, Pimanda JE, Wong JW. Nature. 2016;532:259. doi: 10.1038/nature17437. [DOI] [PubMed] [Google Scholar]
- 15.a) Brabec V, Kasparkova J. Drug Resist. Updates. 2005;8:131. doi: 10.1016/j.drup.2005.04.006. [DOI] [PubMed] [Google Scholar]; b) Andrews PA, Howell SB. Cancer Cells. 1990;2:35. [PubMed] [Google Scholar]; c) Reed E. Cancer Treat. Rev. 1998;24:331. doi: 10.1016/s0305-7372(98)90056-1. [DOI] [PubMed] [Google Scholar]; d) Kang TH, Sancar A. Cell Cycle. 2009;8:1665. doi: 10.4161/cc.8.11.8707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.