Abstract
Regulation of deubiquitinating enzyme (DUB) activity is an essential step for proper function of cellular ubiquitin signals. UAF1 is a WD40 repeat protein, which binds and activates three important DUBs, USP1, USP12 and USP46. Here, we report the crystal structure of the USP12-Ub/UAF1 complex at a resolution of 2.8 Å and of UAF1 at 2.3 Å. In the complex we find two potential sites for UAF1 binding, analogous to what was seen in a USP46/UAF1 complex. In line with these observed dual binding states, we show here that USP12/UAF1 complex has 1:2 stoichiometry in solution, with a two-step binding at 4 nM and 325 nM respectively. Mutagenesis studies show that the fingers sub-domain of USP12 interacts with UAF1 to form the high affinity interface. Our activation studies confirm that the high affinity binding is important for activation while the second UAF1 binding does not affect activation. Nevertheless, we show that this two step binding is conserved in the well-studied USP12 paralog, USP1. Our results highlight the interfaces essential for regulation of USP12 activity and show a conserved second binding of UAF1 which could be important for regulatory functions independent of USP12 activity.
Keywords: Deubiquitinating enzyme, Allosteric regulation, Protein structure, Small angle X-ray scattering (SAXS), Surface Plasmon Resonance (SPR), Enzyme catalysis
1. Introduction
Ubiquitination of proteins is a reversible post-translational modification that is critical for almost any cellular process. The control of these crucial pathways lies in the precise regulation of ubiquitinating and deubiquitinating enzymes (DUBs). DUBs are carefully regulated intra-cellular peptidases that cleave ubiquitin from target substrates. There are approximately 90 DUBs in the human genome, in 5 different families (Clague et al., 2013, Komander et al., 2009). The most abundant are the ubiquitin specific proteases (USPs) with 60 members that share a conserved USP catalytic domain, in which the ubiquitin core is held by the “fingers” while the catalytic centre lies between the “palm” and “thumb” subdomains. Despite this common catalytic core, the USP family members have many different modes of regulation (Sahtoe and Sixma, 2015).
Such regulation can take place in different ways and at different sites. USPs are regulated by changes in the catalytic domain, where the catalytic triad may be misarranged (Hu et al., 2002), via additional domains within the protein itself (Clerici et al., 2014, Faesen et al., 2011a), via post translational modifications (Nicassio et al., 2007), or by sub-cellular localization (Row et al., 2007). An interesting form of regulation is seen in a small sub-family of USPs, that includes USP1, USP12 and USP46. These three proteins are activated by complex formation with a WD40 repeat protein called UAF1 (USP1 associated factor, also known as WDR48) that leads to increased catalytic turnover for these enzymes (Cohn et al., 2007).
USP12 and USP46 are small proteins (370 and 366 residues respectively) with a highly conserved catalytic domain and high sequence similarity (88% identity). The related paralog USP1 (31% identity) is much larger (785 residues), due to additional inserts within its catalytic domain. The USP12/UAF1 and USP46/UAF1 complex can be further activated by binding to a second WD repeat protein, WDR20. This hyper-activation is not observed for the USP1/UAF1 complex (Kee et al., 2010). The lack of hyper-activation in USP1 could be due to the presence of the long inserts which might prevent it from interacting with WDR20. All three enzymes have low intrinsic activity in isolation and binding to UAF1 leads to activation which was shown to be due to an increase in kcat with no drastic change in the KM (Cohn et al., 2007, Faesen et al., 2011b, Villamil et al., 2012a). The exclusive kcat activation is unusual for DUBs as most intermolecular activators (except GMPS) affect substrate binding (Faesen et al., 2011b).
The UAF1 regulation of this subfamily of DUBs has attracted considerable attention due to the biological importance of the enzymes involved. USP1 is important in DNA repair, where it acts on mono-ubiquitinated FANCD2 and PCNA in DNA cross-link repair and DNA-damage avoidance pathways (Huang et al., 2006, Nijman et al., 2005). It is also found to deubiquitinate the ID family of transcriptional regulators (Williams et al., 2011). Due to these important functions, USP1 is considered a major possible drug target (Liang et al., 2014). USP46 plays important roles in neurobiology, as a small deletion mutation in USP46 leads to neurological effects in mice, including anxiety and changes in learning and memory (Imai et al., 2013, Zhang et al., 2011). The molecular basis for these effects is not yet clear. For USP12 several possible roles have been described. It is involved in stabilizing the Akt phosphatases resulting in decreased levels of pAkt (Gangula and Maddika, 2013). It has also been reported that USP12 and USP46 deubiquitinate histone H2A and H2B thereby playing a role in Xenopus development (Joo et al., 2011). Recently USP12 was shown to stabilize the T-cell receptor complex at the cell surface by deubiquitinating TCR adaptor proteins LAT and Trat1 (Jahan et al., 2016).
Several studies have tried to uncover the detailed mechanism of USP1/12/46 activation by UAF1 and also the interfaces involved in the formation of this complex. It was suggested that UAF1 binding modulates the active site conformation of USP1 resulting in a productive catalytic triad and also that phosphorylation of Ser313 is necessary for its interaction (Villamil et al., 2012a, Villamil et al., 2012b). Other studies have shown that the regions in and around the fingers domain of USP1 might be necessary for UAF1 binding (Olazabal-Herrero et al., 2015). Recently the crystal structure of the USP46-Ub and its complex with UAF1 were determined. In these structures two possible interfaces for UAF1 binding and activation were identified. The authors used mutational analysis to show that Interface 1 is critical for UAF1 binding and activation (Yin et al., 2015).
Here we present the structure of USP12-Ub/UAF1580 complex which was solved at a resolution of 2.8 Å and compare it to the UAF1 structure alone. Intriguingly, we find that these structures resemble the USP46/UAF1 complex structure (Yin et al., 2015), including the presence of the second binding site. We then show that in solution USP12 can bind to a second molecule UAF1, but with lower affinity. We confirm for USP12 that Interface 1 at the fingers site is the high affinity interface while the second low affinity interface could be at the backside of the ubiquitin binding cleft. Moreover, mutagenesis studies suggest that the first binding event at the fingers is responsible for activation while the second binding does not influence activity.
2. Materials and methods
2.1. Plasmids and cloning
Human USP12, USP46, USP1 and UAF1 constructs were obtained from Martin Cohn. The USPs were cloned into pFastbac-HTb vector and UAF1 was cloned into pFastbac1 (N-terminal Strep tag) for insect cell expression. The sequence verified insert containing pFastbac vectors were transformed into DH10Bac cells for bacmid preparation. The recombinant bacmid was used for transfection of Spodoptera frugiperda (sf9) insect cells to produce the recombinant baculovirus. USP12WT (24–370) and USP46 (8–366) were also cloned into the pGEX and pET bacterial expression vectors of the NKI LIC suite, respectively (Luna-Vargas et al., 2011). All USP12 mutants were made using the QuikChange site directed mutagenesis method on pGEX USP12 bacterial construct.
2.2. Protein expression and purification
N-terminal His-tagged USP1 (21–785) and USP12FL were expressed using Baculovirus expression in sf9 insect cells. After 72 h of infection the cells were harvested in 20 mM Tris (pH 8.0) + 200 mM NaCl + 2 mM TCEP + protease inhibitor (lysis buffer) and lysed by sonication. The lysed cells were spun down (21,000 rpm for 1 h) and the lysate was loaded on a His-affinity column (GE, USA). The column was washed with lysis buffer supplemented with 50 mM Imidazole (pH 8.0) followed by elution with lysis buffer supplemented with 500 mM imidazole. The His-USP1 elution fractions were concentrated and loaded on a size exclusion chromatography column (Superdex 200, GE, USA) equilibrated in GF buffer (20 mM HEPES (pH 7.5) + 150 mM NaCl + 2 mM DTT) following which the protein fractions were concentrated up to 4.5 mg/ml and stored in −80 °C. The USP12FL and USP46FL fractions were incubated with TEV protease and dialyzed overnight in lysis buffer without imidazole. The dialysed protein sample was then loaded on a His-affinity column where the protein was obtained in the wash fractions. The protein containing fractions were then concentrated and loaded on a size exclusion chromatography column (Superdex 200, GE, USA) equilibrated in GF buffer following which protein fractions were concentrated up to 6 mg/ml and stored in −80 °C.
N-terminal GST-tagged USP12WT and mutants were expressed in E. coli. The cells were induced with 0.2 mM IPTG at 18 °C for 10–12 h followed by which they were harvested in lysis buffer and lysed by sonication. The lysate was loaded on a GST-affinity column and washed with lysis buffer followed by elution with lysis buffer supplemented with 20 mM Glutathione. The protein containing fractions were then concentrated and loaded on a size exclusion chromatography column (Superdex 200, GE, USA) equilibrated in GF buffer following which protein fractions were concentrated up to 15 mg/ml and stored in −80 °C.
N-terminal His-tagged USP46 (8–366) was expressed in E. coli. The cells were induced with 0.5 mM IPTG at 20 °C for ∼16 h followed by which they were harvested in 20 mM Tris (pH 7.5), 150 mM NaCl, 10 mM Imidazole (pH 8.0) and 5 mM beta-mercaptoethanol (lysis buffer) and lysed by sonication. The lysate was loaded on a Ni-affinity column and washed with lysis buffer supplemented with 50 mM imidazole followed by elution with lysis buffer supplemented with 500 mM imidazole. USP46 was incubated with TEV protease and dialysed overnight in lysis buffer without imidazole. The dialysed protein sample was then loaded on a His-affinity column where the protein was obtained in the wash fractions. USP46 was loaded on a size exclusion chromatography column (Superdex 200, GE, USA) equilibrated with 20 mM Tris (pH 7.5), 150 mM NaCl and 2 mM DTT.
UAF1FL and UAF1580 were also purified from sf9 insect cells and were lysed in similar conditions as the USP purifications. The UAF1 lysates were loaded on a Strep-affinity column (IBA life sciences) and washed with 50 mM Tris (pH 8.0) + 150 mM NaCl + 2 mM TCEP (wash buffer). The strep-UAF1 protein was then eluted using wash buffer supplemented with 2.5 mM Desthiobiotin. The protein containing fractions were concentrated and loaded on a size exclusion chromatography column (Superdex 200, GE, USA) equilibrated in GF buffer. The fractions corresponding to the peak were collected. UAF1 samples were treated with 10 mM iodoacetamide to prevent background DUB activity due to minute amounts of co-purified insect cell DUBs. Following iodoacetamide treatment the protein fractions were re-purified using size exclusion chromatography (Superdex 200, GE, USA). The protein fractions were then concentrated up to 10 mg/ml and stored in −80 °C.
The USP/UAF1 complexes were co-expressed in baculovirus infected sf9 insect cells. USP/UAF1 complexes were purified by first carrying out a His-affinity purification followed by Strep-tag (IBA life Sciences) affinity purification as described above. The protein fractions were then concentrated and loaded on a size exclusion chromatography (Superdex S200, GE, USA) equilibrated in GF buffer. The protein containing fractions were concentrated up to 5 mg/ml and stored at -80 °C.
Covalent complexes of USP46 and USP12FL/UAF1580 with Ub-VME and Ub-PRG respectively were generated under reducing conditions at 4 °C for 12–16 h. The USP46 reaction was very slow and only 50% of the enzyme reacted to Ub-VME whereas approximately 80% of the USP12FL/UAF1580 complex reacted to the Ub-PRG probe. The reacted USPs were then purified from the excess probe by using a size exclusion chromatography column (Superdex 200, GE, USA) equilibrated in GF buffer. The purified USP-Ub complex was then concentrated and stored at -80 °C.
2.3. Crystallization and structure determination
USP12 and UAF1 crystals were obtained by sitting drop vapour diffusion experiment at 20 °C using equal ratio of protein and mother liquor solution. UAF1580 was setup for crystallization at 8 mg/ml and crystals were obtained in 20% PEG3350, 200 mM Tri-Sodium Citrate, Bis-Tris Propane pH 6.5. USP12FL-Ub-PRG/UAF1580 was setup at 5 mg/ml and crystals were obtained in 3.2% PEG4000, 0.1 mM MMT pH 6.5 and 0.1 mM TCEP. The crystals were cryo-protected by brief washing in mother liquor solution with 30% glycerol prior to flash freezing in liquid nitrogen. Diffraction data were collected at the Swiss Light Source beamline PXIII at 100 K.
USP46-Ub-VME at 8 mg/ml was set up for crystallization by sitting drop vapour diffusion experiment at 20 °C. Crystals were obtained in 0.96 M Sodium Citrate pH 7.5 and 0.1 mM ZnCl2 with protein: precipitant ratio 1.5: 1. The crystals were cryo-protected by brief washing in mother liquor solution with 20% glycerol prior to flash freezing in liquid nitrogen. Diffraction data were collected at the European Synchrotron Radiation Facility (ESRF) beamline ID14-1 at 100 K.
Crystallographic data were processed with XDS (Kabsch, 2010) or iMOSFLM (Battye et al., 2011) and scaled using XSCALE (Kabsch, 2010) or Aimless from the CCP4 suite (Winn et al., 2011). The USP46-Ub-VME structure was solved by molecular replacement using the USP2-Ub model (PDB 2IBI) in Phaser (McCoy et al., 2007) followed by automated model building using ARP/WARP (Langer et al., 2008). The UAF1 (9–580) structure was solved by molecular replacement (PDB-1VYH) in Phaser (McCoy et al., 2007) followed by automated model building using ARP/Warp (Langer et al., 2008). The USP12FL-Ub/UAF1580 structure was solved by molecular replacement using the USP46, UAF1580 and ubiquitin structures as search models in Phaser. All structures were refined by Phenix (Adams et al., 2010), autoBUSTER (Smart et al., 2012), Refmac (Murshudov et al., 1997), PDB_REDO (Joosten et al., 2014) and models were built using COOT (Emsley et al., 2010). Interface analysis was performed with PISA (Krissinel and Henrick, 2007). All structure figures were generated using PyMOL (Schrödinger, LLC).
2.4. Size exclusion chromatography Multi Angle Laser Light Scattering (SEC-MALLS)
The SEC-MALLS experiments were performed using the miniDawn Tristar light scattering detector (Wyatt technologies, USA) in line with size exclusion chromatography. After equilibration of the Superdex 200 10/300 GL and the Superose 6 increase 10/300 GL (GE, USA) in buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl, 2 mM DTT, the USP12FL-Ub/UAF1580 (8 μM) and the USP1/UAF1FL complex (6 μM) were loaded on the Superdex 200 10/300 GL while the USP12FL-Ub/UAF1FL sample (42 μM) was loaded on the Superose 6 increase 10/300 GL (GE, USA). Molecular weight estimation was done by using the refractive index signal as measure of the concentration with the Astra software (Wyatt Technologies, USA).
2.5. Size exclusion chromatography – small angle X-ray scattering (SEC-SAXS)
USP12FL-Ub/UAF1FL purified from insect cells was concentrated up to 38 μM. The buffer used for the size exclusion chromatography was also used for SEC-SAXS experiment on the BM29 SAXS beamline at the ESRF. Following equilibration, 30 μl of the purified USP12FL-Ub/UAF1FL was loaded on the Superdex 200 5/150 GL (GE, USA) and 1500 successive 1 s frames were collected through the protein elution peak. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank (derived from the buffer run) was subtracted. Data were analysed using the ATSAS software package (Svergun et al., 2001). A moving set of 10 or 20 frames was analysed across the high-intensity peak to estimate molecular weight from various methods (DATPOROD (Konarev et al., 2006), excluded volume from DAMMIF model (Franke and Svergun, 2009) and SAXS MoW2 (Fischer et al., 2010)) and fit the data to the crystallographic structures in CRYSOL (Svergun et al., 1995). Frames prior to 670 and frames latter than 910 were considered too noisy or weak for analysis. OLIGOMER (Konarev et al., 2003) was used to perform a fit to the experimental curve from different possible models of the complex and determine volume fractions of each component. Several OLIGOMER runs were performed using the 2:1 complex, both 1:1 complexes and UAF1 alone as models in different combinations.
2.6. Ub-rhodamine enzymatic assays
Enzymatic activity was followed as release of fluorescent rhodamine from the quenched Ub-Rho substrate, providing a direct readout for DUB activity. The fluorescent intensity was measured using Pherastar plate reader (BMG LABTECH GmbH, Germany). Activity of USP12WT at varying concentrations of UAF1FL (20–1250 nM) was tested at 100 nM of USP12WT and at a single substrate concentration of 1 μM. Activity was quantified by calculating the initial rates and plotted as a function of UAF1FL concentration. 100 nM of enzyme was used against different concentrations of the minimal substrate (32–0.25 μM) for the Michaelis Menten analysis. The initial rates were then plotted against substrate concentration and fitted with a Michaelis Menten model using non-linear regression in GraphPad Prism 6 software (GraphPad Software Inc, USA).
2.7. Binding assays
Surface Plasmon Resonance experiments were carried out to test binding of His-USP1 and Gst-USP12WT to UAF1FL in the Biacore T200 system (GE, USA). The SPR buffer for Gst-USP12WT and mutants consisted of 20 mM HEPES 7.5, 150 mM NaCl, 20% glycerol, 0.05% Tween-20, 2 mM TCEP (Tris (2-carboxyethyl) phosphine hydrochloride) and 1 mg/ml dextran. The SPR buffer for His-USP1 had 300 mM NaCl to neutralize any unspecific interaction with Ni on the chip surface and the rest of the components were identical. The Gst-USP12 was immobilized via goat anti-GST antibody that was pre immobilized to the CM5 chip using amine coupling. The His-USP1 was directly immobilized to the surface of NTA chip via His-tag. The binding experiments were carried out in the single cycle kinetics mode with 5 sequential injection of UAF1FL in each cycle. An initial experiment with 5 injections from 1 nM to 10 μM and a more detailed assay with 15 injections from 0.56 nM to 2.56 μM were carried out. Data from a reference flow cell, run in parallel with an empty chip (in case of NTA immobilization) or with GST (in case of anti-GST immobilization) were subtracted from the signal using the Biacore T200 Evaluation Software. Final analysis and the figures were done using GraphPad Prism 6 (GraphPad Software Inc, USA).
3. Results
3.1. Crystal structures of USP46, UAF1580 and USP12FL-Ub/UAF1580 complex
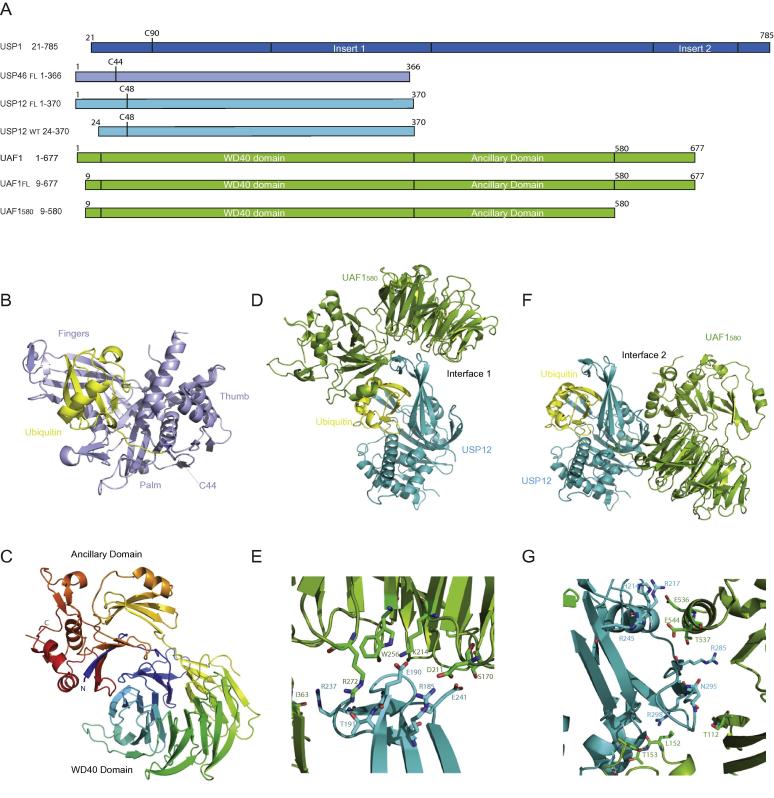
To study how the regulator UAF1 modulates the activity of USPs, we purified the human USP46, human USP12 and several constructs of the human UAF1. We refer to UAF1 (9–580) as UAF1580 and to UAF1 (9–677) as UAF1FL (Fig. 1a). By reaction of USP46 and USP12 with suicide inhibitors of ubiquitin, Ub-VME and Ub-PRG respectively, we could create covalently linked USP-Ub complexes, which were used for crystallization attempts.
Fig. 1.
Crystal structure analysis shows two possible binding options for USP12. A) Constructs used for the present study. B) Crystal structure of USP46-Ub-VME in cartoon representation. C) Crystal structure of UAF1580 coloured in a rainbow representation. D) Crystal structure of USP12FL-Ub/UAF1580 showing UAF1580 bound to Interface 1. E) Zoomed Interface 1 between UAF1580 and USP12FL-Ub showing the interface residues. F) Crystal structure of USP12FL-Ub/ UAF1580 showing UAF1580 bound to Interface 2. G) Zoomed Interface 2 between UAF1580 and USP12FL-Ub showing the interface residues. (Colours in panel B, D–G: Ub in yellow, other proteins as in A).
We generated well diffracting crystals for USP46-Ub (1.85 Å), USP12FL-Ub/UAF1580 (2.8 Å) and UAF1580 (2.3 Å). All three crystal structures were solved by molecular replacement where the covalent USP2-Ub complex model (2IBI) was used for the USP46-Ub structure and the protein PAF-AH (1VYH) was used as a model for UAF1580. The resulting structures of USP46-Ub and UAF1580 were used as models for the USP12FL-Ub/UAF1580 complex. All three structures were refined to acceptable R-factors and good geometry (Table.1).
Table 1.
Crystallography details.
| Data collection | |||
|---|---|---|---|
| USP46-UbVME | UAF1580 | USP12-UbPRG/UAF1580 | |
| Wavelength (Å) | 0.93 | 0.92 | 1.00 |
| Resolution (Å) | 28.41–1.85 (1.90–1.85) | 48.97–2.30 (2.35–2.30) | 47.41–2.79 (2.92–2.79) |
| Space Group | C 2 2 21 | P 2 21 21 | I 2 2 2 |
| Unit Cell a, b, c (Å) | 91.95 104.66 135.31 | 73.30 131.60 148.67 | 103.68 152.82 182.93 |
| CC1/2 | 0.999 (0.720)∗ | 0.999 (0.585) | 0.995 (0.638) |
| Rmerge | 0.12 (0.84) | 0.05 (1.0) | 0.08 (0.72) |
| Rpim | 0.04 (0.84) | 0.06 (0.57) | |
| I/σI | 17.68 (2.0) | 16.1 (1.4) | 10.4 (1.6) |
| Completeness (%) | 98.8 (99.7) | 99.6 (95.9) | 99.3 (94.8) |
| Redundancy | 3.56 (3.3) | 4.5 (4.1) | 4.5 (4.3) |
| Refinement | |||
| Unique Reflections (nr) | 55,291 | 64,580 | 36,148 |
| Atoms total (nr) | 3442 | 8586 | 7290 |
| Protein atoms (nr) | 3192 | 8195 | 7266 |
| Solvent atoms (nr) | 250 | 391 | 24 |
| B-factors | 31.9 | 58.3 | 72.4 |
| TLS groups | 4 | 4 | 4 |
| Rwork/Rfree (%) | 16.6/19.4 | 18.5/22.4 | 20.8/25.9 |
| Rmsd bond lengths (Å) | 0.017 | 0.007 | 0.014 |
| Rmsd bond angles (°) | 1.758 | 1.235 | 1.642 |
| Ramachandran (%) Preferred/outliers | 97.89/0 | 97.27/0.30 | 95.5/0.34 |
| Molprobity score | 1.01 | 0.82 | 1.77 |
| PDB code | 5L8H | 5L8E | 5L8W |
High resolution shell in parentheses; (∗ – Reprocessed with aimless).
The crystal structure of USP46-Ub reveals the canonical USP domain with a catalytic centre in the catalytically proficient state (Fig. 1b). The crystallization contained only 50% of the Ub-bound form, but the crystal was fully modified, indicating a preferential crystallization of the ubiquitin-bound form. This structure is in good agreement with the recently published structure of USP46 ((5CVM), RMSd on 308 Cα: 0.3 Å). The structure of UAF1580 forms a 7 bladed β-propeller with an ancillary domain (Fig. 1c), although WD40 domain predictions obtained from the protein sequence predicted 8 blades. The first N-terminal residues (13–24) are present in the ancillary domain and the following residues from 25 to 360 form the 7 bladed β-propeller. The predicted eighth blade (361–400) forms the expected 4-stranded sheet but is located in the ancillary domain where it sits on top of a β sheet in which the N-terminal residues form an integral part. The UAF1580 structure is very similar to the recently published structure of UAF1 ((5CVL), RMSd on 517 Cα: 1.2 Å).
In the crystal lattice of the USP12FL-Ub/UAF1580 complex, the asymmetric unit contains one molecule of USP12 and one molecule of UAF1. However, USP12 makes two possible interfaces with UAF1 in these crystals, one on the fingers sub-domain (Interface 1) (Fig. 1d, e) and the other on the backside of the ubiquitin binding cleft (Interface 2) (Fig. 1f, g). The interactions in Interface 1 are dominated by hydrophilic and charged interactions, with 794 Å2 and 698 Å2 buried surface area on USP12 and UAF1580 respectively (Fig. 1e). The Interface 2 is mostly hydrophobic and buries a surface area of 1030 Å2 and 1111 Å2 on USP12 and UAF1580 respectively (Fig. 1g). Intriguingly the same crystal packing was seen for the USP46/UAF1 complex (PDB code: 5CVN) that was published during the refinement of our structures (Yin et al., 2015). In that publication the authors confirmed by mutational analysis that Interface 1 is the relevant interface for activation. USP46/UAF1 was crystallized in the same space group, with similar cell dimensions, which is not surprising since USP12 and USP46 have 88% sequence similarity, although neither interface is fully conserved. A second crystal form for USP46/UAF1FL was determined in a different crystal lattice (C2), with different packing, but even in this crystal lattice, the USP still has maintained both interfaces (PDB code: 5CVO). The striking similarity and conservation of these two USP/UAF1 interfaces among three different crystal structures suggested that this was not due to a crystal packing artefact and further analysis of the second interface would be worthwhile (Supp. Fig. 1a).
A superposition of our USP46 structure on USP12 in complex with UAF1580 revealed no significant conformational change in the USP moiety (RMSd on 309 Cα: 0.9 Å). When the UAF1 moieties are compared by superposition of the UAF1580 onto the USP12FL-Ub/UAF1580 complex, no conformational change is observed in UAF1580 close to Interface 1 (Supp. Fig. 1b), whereas a large shift is observed in the extended loop of blade 3, the region where UAF1580 forms Interface 2 (Supp. Fig. 1c). This conformational change due to interaction of UAF1580 at Interface 2 is also observed in the USP46/UAF1 structure upon UAF1 binding (Yin et al., 2015). Since the conformational change as well as the presence of two UAF1 interfaces seems to be conserved between USP46 and USP12, we wondered what stoichiometry the USP12/UAF1 complex has in solution.
3.2. The USP12/UAF1 complex has 1:2 stoichiometry
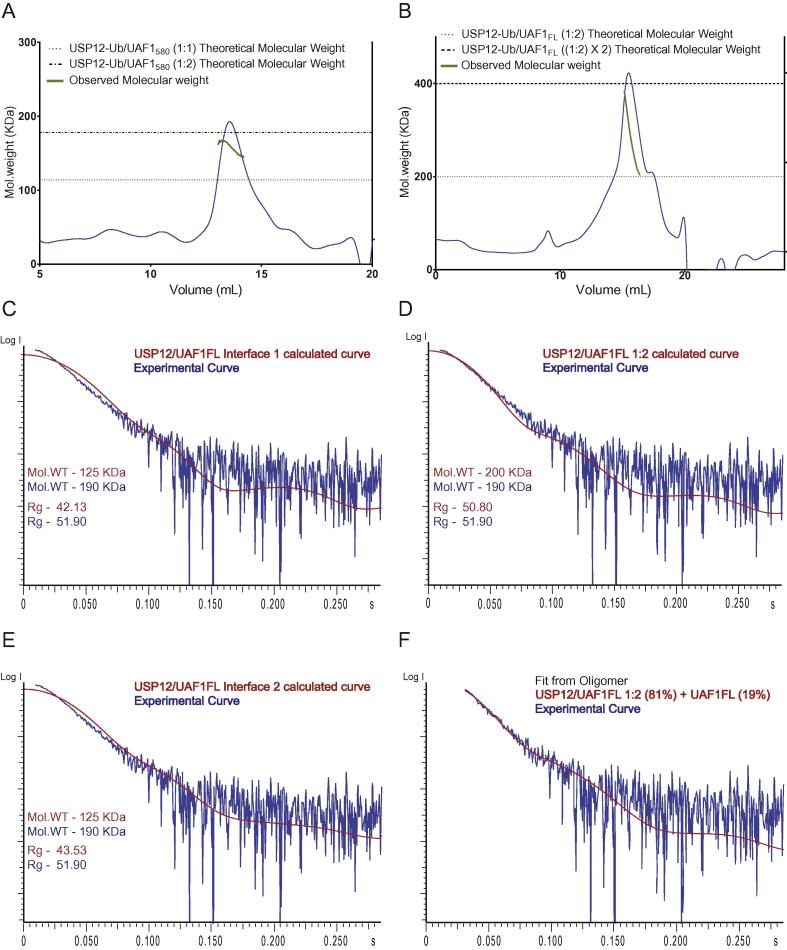
To study relative amounts of USP12 and UAF1 in solution we carried out size exclusion chromatography coupled with multi-angle laser scattering (SEC-MALLS) of the purified USP12FL-Ub/UAF1580 complex which was used for obtaining the crystal structure. The molecular weight obtained from the MALLS experiment was approximately 166 KDa which is much closer to the calculated mass for 1:2 complex (180 KDa) than for a 1:1 complex (115 KDa). The calculated molecular weight was not constant within the peak, which suggests a dynamic equilibrium between the 1:2 and 1:1 complexes (Fig. 2a).
Fig. 2.
USP12/UAF1 has 1:2 stoichiometry. A) SEC-MALLS analysis of USP12FL-Ub/UAF1580 (8 μM) shows a molecular weight corresponding to a 1:2 complex. B) SEC-MALLS analysis of USP12FL-Ub/UAF1FL at high concentration (42 μM) shows a dynamic equilibrium between high molecular weight species and the 1:2 complex in agreement with the SAXS data. C–E) Experimental SEC-SAXS curve of USP12FL-Ub/UAF1FL fits better with the scattering curve based on the structure of the 1:2 complex as compared to the 1:1 complex at either of the two interfaces. F) OLIGOMER fit of the Experimental SEC-SAXS curve of USP12FL-Ub/UAF1FL.
Next, we carried out small angle X-ray scattering (SAXS) experiments coupled to size exclusion chromatography. At the high concentration necessary for SAXS experiments higher order species are observed (Fig. 2b). The purified complex of USP12FL with covalently bound Ub-PRG and UAF1FL was used for these experiments. Frames corresponding to the different parts of the peak were averaged and analysed separately (Supp. Fig. 1e). Molecular weight estimation of the data (898–909) from the latter half of the peak was consistent, when carried out using different methods (Porod volume (Konarev et al., 2006), excluded volume from DAMMIF model (Franke and Svergun, 2009), and SAXS MoW2 (Fischer et al., 2010)), where we observed that USP12FL-Ub/UAF1FL has a molecular weight of 190 KDa corresponding to a 1:2 stoichiometry of the complex. Additionally, Porod analysis of the frames in the early half of the peak corresponded to a molecular weight of 390 KDa indicating formation of high molecular weight species which fits well with the MALLS data (Fig. 2b).
Moreover, we compared the experimental SAXS profile of frames (898–909) with calculated scattering curves based on the crystal structure either with one UAF1FL bound (in either of the two sites), or with two UAF1FL molecules bound. We obtained a much worse fit with the 1:1 profiles (Fig. 2c, e) compared to the possible 1:2 profile (Fig. 2d). To further analyse these data, we tested whether the OLIGOMER algorithm would change the fit. Other possible options did not improve the fit, but when testing for the 1:2 profile, we found an improved fit (Fig. 2f) with volume fractions of 81% for the 1:2 complex and a smaller fraction for UAF1FL (19%). In principle such a fraction was expected to be a mixture of the 1:1 complex and UAF1FL. It is possible that the computational analysis selects against this complex mixture when fractions are small.
3.3. UAF1 binds USP12 in two distinct steps with different affinities
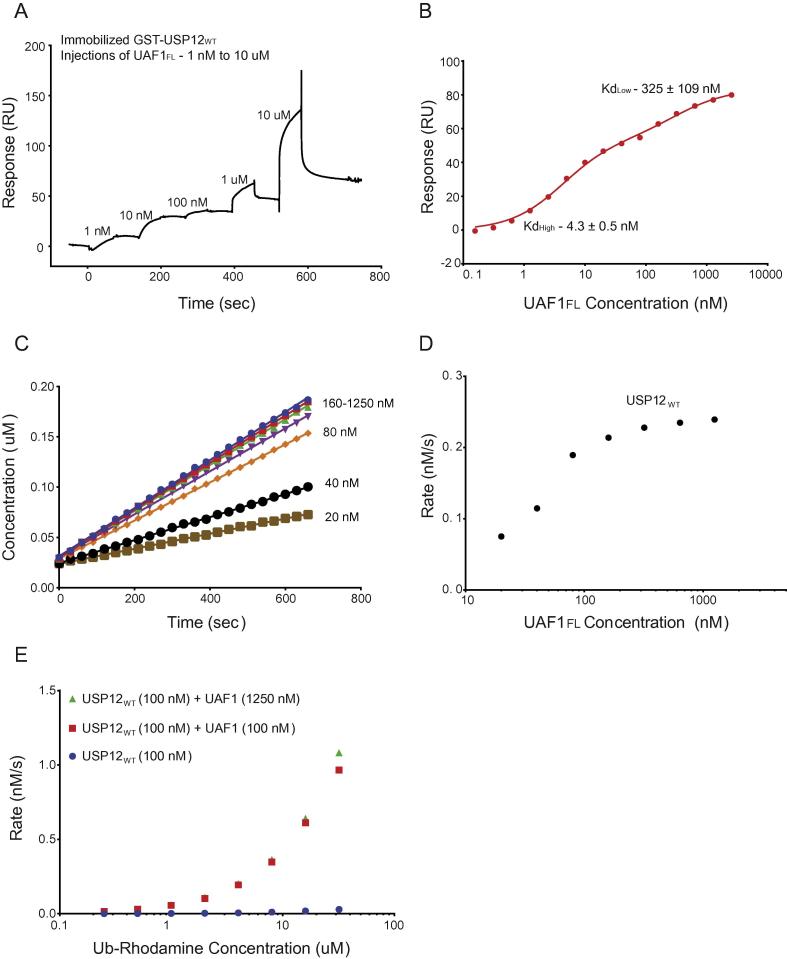
We investigated if the two binding events indicative of the 1:2 stoichiometry could be observed in an in vitro binding experiment. Therefore, we analysed the binding of UAF1FL to GST-USP12WT by Surface Plasmon Resonance (SPR) and indeed observed two distinct binding events (Fig. 3a). We found a high-affinity binding (Kd = 4 nM) with an extremely low off-rate, which saturated at 100 nM. Moreover, when we added higher concentrations of UAF1FL, a second binding event could be observed (Kd = 325 nM) (Fig. 3b) with faster binding and dissociation. In conclusion, we observed two binding events for UAF1FL binding to USP12WT with different binding characteristics.
Fig. 3.
UAF1 binds USP12 in two steps with different affinities. A) Qualitative SPR analysis of five successive injections of UAF1FL on immobilized USP12WT, raw data show how initial injections show slow kinetics, and binding at higher concentrations displays fast off-rates. B) Fitting quantitative SPR analysis of UAF1FL to USP12WT, obtained from 15 successive injections of UAF1FL. Curve is fit to a two-step binding model with KD as indicated. C) Cleavage of Ub-Rhodamine by USP12WT at different concentrations of UAF1FL(10 nM–1250 nM). D) Initial rates of USP12WT plotted as a function of UAF1FL concentration shows the importance of high affinity site for activation. E) Michaelis Menten analysis of USP12WT (100 nM) alone and in two different concentrations of UAF1FL (100 nM, 1250 nM) shows that the second binding of UAF1 leads to no further activation.
We tested which of these events contributed to activation. We performed an enzymatic assay against the minimal substrate ubiquitin rhodamine (Ub-Rho), as a function of activator concentration. We could see a clear activation that correlates with the high-affinity binding site. In contrast, the second binding event does not affect the activation status, as no further activation is observed when the UAF1FL binds the second site (Fig. 3c, d). We then performed a kinetic analysis of USP12 activity, either at equal concentration to UAF1 (1:1) or when an excess of UAF1 is present (Fig. 3e). The Michaelis Menten parameters Kcat and KM did not change with higher UAF1 concentration, confirming that only Interface 1 is important for UAF1 mediated USP12 activation (Table 2).
Table 2.
Michaelis Menten analysis of USP12WT with UAF1.
| Protein | Kcat (×10−2s−1) | KM (μM) | Kcat/KM (M−1 s−1) |
|---|---|---|---|
| USP12WT (100 nM) | 0.076 | 50 | 0.15 × 102 |
| USP12WT + UAF1 (100 nM) | 2.6 | 50 | 5.20 × 102 |
| USP12WT + UAF1 (1250 nM) | 3.3 | 64 | 5.15 × 102 |
3.4. The Fingers sub-domain in USP12 is crucial for binding and activation by UAF1
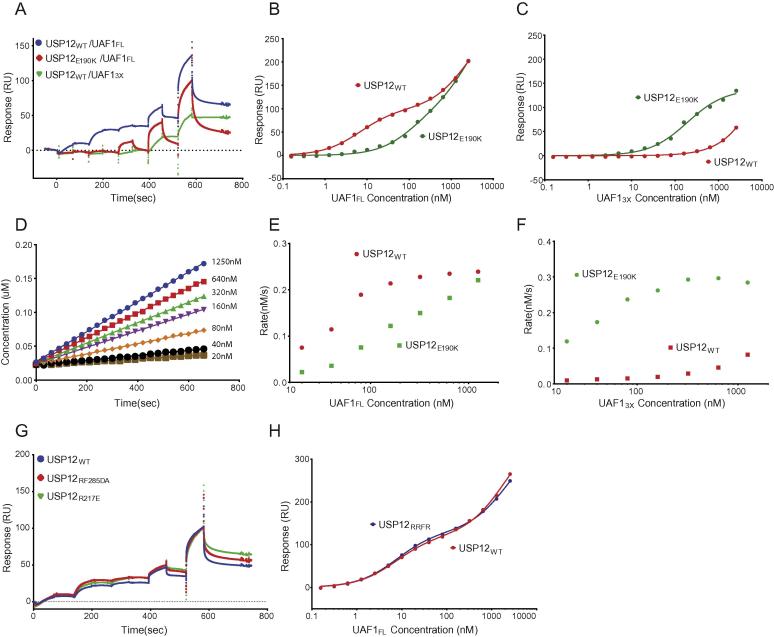
We validated the role of Interface 1 by making a series of mutations. In line with their role in the USP46/UAF1 interface (Yin et al., 2015), a triple mutant (UAF13X = K214E + W256A + R272D) on UAF1 and a reciprocal mutant (E190K) on USP12 interfered with high affinity binding (Fig. 4a, b). The high affinity binding could be partially rescued by combining the USP12E190K with the UAF13X mutant, in a similar fashion to what was observed for USP46 and UAF1 binding (Fig. 4c) (Yin et al., 2015). Additionally, the binding of the USP12E190K to the low affinity site remained unchanged. On comparing the binding characteristics of these mutants with USP12WT, we noted that the UAF13X mutant only binds at very high concentrations and does not release easily, while the USP12E190K mutant binds with a fast release (Fig. 4a). We also made a series of mutations at Interface 2 on USP12WT, by either reversing charges (R217E or R285D) or changing the hydrophobic interface (F287A) but none of them could disrupt binding of UAF1 to this site (Fig. 4g, h).
Fig. 4.
Interface 1 is the high affinity site and is responsible for activation by UAF1. A) Comparing Qualitative SPR analysis of five successive injections of UAF1FL and UAF13X on immobilized USP12WT and USP12E190K shows differences in binding characteristics. B) Semi-quantitative SPR analysis of UAF1FL to USP12WT and USP12E190K mutant highlights weaker binding of the mutant to the high affinity interface. C) Semi-quantitative SPR analysis of UAF13X to USP12WT and USP12E190K, curves show disruption of binding in USP12WT which is rescued by the USP12E190K mutant. D) Cleavage of Ub-Rhodamine by USP12E190K at different concentrations of UAF1FL shows loss in activation by UAF1FL. E) Comparison of initial rates of USP12WT and USP12E190K at different UAF1FL concentrations shows that when high affinity interaction is lost the activity is impaired. F) Comparison of initial rates of USP12WT and USP12E190K at different concentrations of UAF13X shows the rescue of activation by the USP12E190K mutant. G) Comparing Qualitative SPR analysis of five successive injections of UAF1FL on immobilized USP12WT and USP12 interface 2 mutants shows no significant change in binding characteristics. H) Semi-quantitative SPR analysis of UAF1FL to Interface 2 mutant USP12RRFR and USP12WT shows identical binding.
We then carried out an in vitro Ub-Rho assay to analyse the effects of these mutations on USP12 activation and compare it with previously published findings for USP46 (Yin et al., 2015). Wild type UAF1 was unable to activate USP12E190K to similar levels as compared to USP12WT (Fig. 4d, e) and similarly, the UAF13X mutant did no longer activate. However, as shown previously for USP46/UAF1 (Yin et al., 2015), the combination of these complementary mutants rescues the activation (Fig. 4f), highlighting the importance of the fingers sub domain in USP12 and USP46 (Yin et al., 2015) activation by UAF1.
3.5. The two-step binding is conserved in USP1
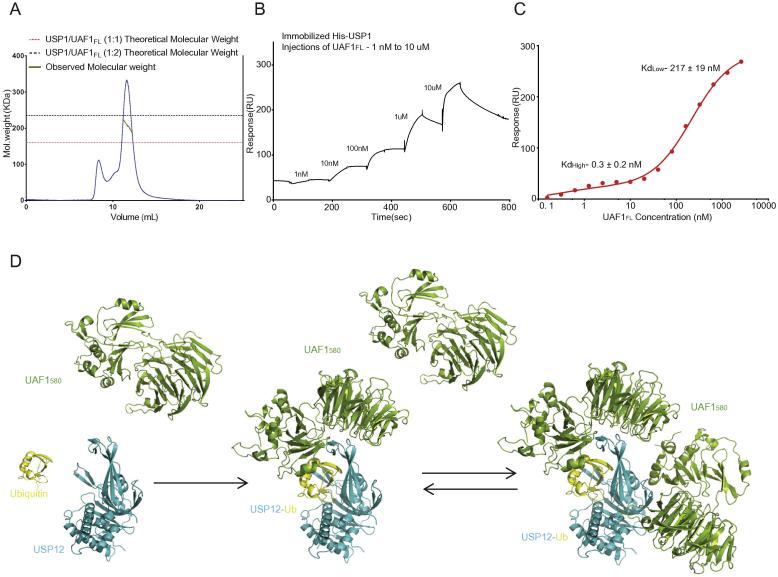
Some aspects of USP1 regulation are different from the USP12 and USP46, as USP1 is a much larger enzyme, and it cannot be hyper-activated by WDR20. We therefore wondered whether the 1:2 stoichiometry is also conserved in this important DNA repair enzyme. We carried out SEC-MALLS of the purified USP1/UAF1 complex and the molecular weight obtained was approximately 220 KDa which closely corresponds to the 1:2 complex (235 KDa) rather than a 1:1 complex (160 KDa). Similar to the USP12/UAF1 MALLS data (Fig. 2a), the molecular weight was not constant within the peak suggesting a dynamic equilibrium between the two states of the complex (Fig. 5a). We then performed the UAF1 binding assay with His-USP1 on the SPR chip (Fig. 5b, c). Again, we observed a two-step binding, and although the high RU values for the second step suggests an additional unexplained background process, the fitting identified similar affinities for USP1 to those observed for USP12. Thus from our SPR result in addition to the MALLS data, we could confirm the presence of this unusual two-step binding in USP1.
Fig. 5.
The two step UAF1 binding is conserved in USP1. A) SEC-MALLS analysis of USP1/UAF1FL (6 μM) shows a molecular weight corresponding to the 1:2 complex. B) Qualitative SPR analysis shows that USP1 has conserved the two-step binding to UAF1FL, with similar kinetics. C) Fitting of quantitative SPR analysis of UAF1FL to USP1 obtained from 15 successive injections of UAF1FL to quantify the KD values. Curve is fit to a two-step binding model with affinities as indicated. D) UAF1 binds USP12 at the fingers sub-domain which activates USP12 and then UAF1 also binds on the backside of the ubiquitin binding cleft.
4. Discussion and conclusions
Here we present a series of structures that contribute to the understanding of regulation of DUBs by non-substrate partners. In our analysis we identified two possible binding sites for UAF1 on USP12-Ub (Fig. 5d), that are conserved from USP46. The two UAF1 binding sites are distant from the catalytic centre and binding of UAF1 did not induce significant rearrangements in the USP structure. Thus it is very hard to envision from these structures how USP12 is activated by UAF1, especially when the activation is due to an increase in kcat and not KM. It is possible that UAF1 binding stabilizes the flexible USP12 “fingers”, transforming it into a catalytically proficient state and this may not be visible since we compare it to a ubiquitin-bound USP structure. Therefore an apo-structure of USP12 could shed more light on this issue.
The binding analysis shows that Interface 1 provides the high affinity interaction that is responsible for the activation of the USP. We measure a binding affinity in the order of 4 nM with extremely slow dissociation. The affinity is less tight than the 0.1 nM observed for USP46 by Yin et al., 2015, most likely due to differences in the experimental setup. Their lowest concentration of UAF1 used was far above the reported KD (sevenfold), and the KD was estimated based on very slow dissociation constant derived from the global kinetic fit. Overall, the structure of USP12-Ub/UAF1580 and the UAF1 mediated activation through Interface 1 bears close resemblance to the published data on USP46 (Yin et al., 2015), again highlighting the high degree of conservation between these USPs. The Interface 1 has a relatively small surface area, with a predominance of charged interactions. For such a small interface, even the observed affinity of 4 nM is rather surprising since such strong binding between two proteins is rarely mediated by interfaces having less than 1000 Å2 buried surface area (Chen et al., 2013). However, previous reports have suggested the importance of the fingers domain for UAF1 binding and activation (Olazabal-Herrero et al., 2015). It is still unclear how S313 phosphorylation which is present in the long insert of USP1 could play a role in UAF1 binding (Villamil et al., 2012b), as the (shorter) corresponding region on USP12 is not involved in UAF1 binding for either of the interfaces.
In the USP12/UAF1580 complex we observe a conserved second interface (Interface 2) at the backside of the ubiquitin binding cleft. The buried surface area of this interface is larger than for Interface 1, and the contact is more hydrophobic, indicating that its properties are very different. Our binding data confirm the presence of a secondary interaction, with an affinity of ∼300 nM and fast kinetics. Unfortunately we were unable to validate this interface by mutational analysis. It is possible that this is due to the large surface area involved in this binding. Additionally the surface shape complementarity between the extended loop in USP12 and the relatively concave architecture of ancillary domain and β-propeller in the UAF1 molecule could contribute to the interaction primarily by backbone interactions.
The two binding interfaces on UAF1 seem independent of each other as they reside in distant parts of the protein. Thus mutating residues on Interface 1 is unlikely to affect binding to Interface 2 or vice versa. Our binding studies with the UAF13X mutant showed binding to the high affinity site at very high concentrations and no binding to the low affinity site. This could mean that UAF1 can only bind the low affinity site once the high affinity site has been fully occupied. It is possible that high overall flexibility in USP12 in the unbound state prevents binding to the low affinity site, thereby making the high affinity binding mandatory for binding the second UAF1.
During revision of this manuscript, a structure for the USP12/UAF1/WDR20 complex was published (Li et al., 2016). In this structure WDR20 binds to a number of structural elements at the palm domain of USP12 with an affinity of 7 nM. Interestingly, the binding interface for WDR20 partially overlaps with the second binding site of UAF1 on USP12 (Supp. Fig. 1d), but the contacts are very different, explaining how a F287A/V279D mutation can disrupt WDR20 binding whereas F287A does not affect the UAF1 interface, which is dominated by backbone interactions in this area. The partial overlap of the binding interfaces and the high affinity of WDR20 suggests that WDR20 binding to USP12/UAF1 can either prevent binding of the second UAF1 molecule or it can actively compete out UAF1 if it is bound to the USP12/UAF1 complex. The partial overlap of this interface could hint towards a possible regulatory role for the second UAF1 binding.
The second binding of UAF1 to USP12 does not seem to play a role in the activation of USP12 on a minimal substrate. Meanwhile the effect of the second UAF1 binding on the catalytic activity of USP12 against one of its natural substrates remains to be tested and this could yield more insight on the possible function of this regulatory event. In any case, the conservation of the 1:2 stoichiometry and the two step binding of UAF1 in this small family of USPs highlights the importance of the second binding and implies that the binding could be important for functions which are independent of activation on a minimal substrate.
All available data (Cohn et al., 2009, Sowa et al., 2009) and the high affinity with slow off-rates indicate that USP12/UAF1 form a constitutive complex in cells. In contrast, the fast kinetics of the second binding site indicates that it could play a role in regulation. This is in line with the recent quantitative analysis of protein abundance in HeLa cells that place the concentration of USP12 and UAF1 at 2 and 77 nM, respectively (Hein et al., 2015). At that concentration the first interface would be fully saturated, and a significant fraction of the USPs would start to bind a second molecule. This means that the second binding could potentially play an important regulatory role.
Conflict of Interest
The authors declare that there is no actual or perceived conflict of interest on the part of any author.
Author contributions
SD designed, performed and analysed biochemical experiments, expressed UAF1 constructs, performed UAF1 and USP12/UAF1 structural analysis and wrote the paper. MC set up expression and purification protocols and determined the USP46 structure. AF designed, performed and analysed the SPR experiments. WJvD provided technical assistance. TKS conceived and coordinated the study and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgements
We thank Michael Uckelmann and Robbert Kim for critical reading of the manuscript and other members in the lab for their helpful suggestions. We thank Martin Cohn for the USP1, USP12, USP46 and UAF1 constructs, Huib Ovaa and Farid El Oualid for providing us with the Ub-PRG, Ub-VME and Ub-Rhodamine. We thank Tassos Perrakis for discussion of SAXS data and Tatjana Heidebrecht for technical assistance. We thank beamline scientists at beam lines SLS PXIII, ESRF ID14-1 and ESRF BM29 where diffraction and SAXS data were collected. The authors acknowledge funding from the Dutch Cancer Society KWF, projects NKI-2014-6858 and NKI 2008-4014, ERC advanced grant, Ubiquitin Balance (249997) and CGC.nl.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jsb.2016.09.011.
Appendix A. Supplementary data
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C., Richardson J.S., Terwilliger T.C., Zwart P.H. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battye T.G.G., Kontogiannis L., Johnson O., Powell H.R., Leslie A.G.W. IMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Sawyer N., Regan L. Protein-protein interactions: general trends in the relationship between binding affinity and interfacial buried surface area. Protein Sci. 2013;22:510–515. doi: 10.1002/pro.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague M.J., Barsukov I., Coulson J.M., Liu H., Rigden D.J., Urbé S. Deubiquitylases from genes to organism. Physiol. Rev. 2013;93:1289–1315. doi: 10.1152/physrev.00002.2013. [DOI] [PubMed] [Google Scholar]
- Clerici M., Luna-Vargas M.P.a., Faesen A.C., Sixma T.K. The DUSP-Ubl domain of USP4 enhances its catalytic efficiency by promoting ubiquitin exchange. Nat. Commun. 2014;5:5399. doi: 10.1038/ncomms6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn M.A., Kee Y., Haas W., Gygi S.P., D’Andrea A.D. UAF1 is a subunit of multiple deubiquitinating enzyme complexes. J. Biol. Chem. 2009;284:5343–5351. doi: 10.1074/jbc.M808430200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn M.A., Kowal P., Yang K., Haas W., Huang T.T., Gygi S.P., D’Andrea A.D. A UAF1-containing multisubunit protein complex regulates the fanconi anemia pathway. Mol. Cell. 2007;28:786–797. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faesen A.C., Dirac A.M.G., Shanmugham A., Ovaa H., Perrakis A., Sixma T.K. Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Mol. Cell. 2011;44:147–159. doi: 10.1016/j.molcel.2011.06.034. [DOI] [PubMed] [Google Scholar]
- Faesen A.C., Luna-Vargas M.P.A., Geurink P.P., Clerici M., Merkx R., Van Dijk W.J., Hameed D.S., El Oualid F., Ovaa H., Sixma T.K. The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chem. Biol. 2011;18:1550–1561. doi: 10.1016/j.chembiol.2011.10.017. [DOI] [PubMed] [Google Scholar]
- Fischer H., De Oliveira Neto M., Napolitano H.B., Polikarpov I., Craievich A.F. Determination of the molecular weight of proteins in solution from a single small-angle X-ray scattering measurement on a relative scale. J. Appl. Crystallogr. 2010;43:101–109. [Google Scholar]
- Franke D., Svergun D.I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangula N.R., Maddika S. WD repeat protein WDR48 in complex with deubiquitinase USP12 suppresses akt-dependent cell survival signaling by stabilizing ph domain leucine-rich repeat protein phosphatase 1 (PHLPP1) J. Biol. Chem. 2013;288:34545–34554. doi: 10.1074/jbc.M113.503383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein M.Y., Hubner N.C., Poser I., Cox J., Nagaraj N., Toyoda Y., Gak I.A., Weisswange I., Mansfeld J., Buchholz F., Hyman A.A., Mann M. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- Hu M., Li P., Li M., Li W., Yao T., Wu J.W., Gu W., Cohen R.E., Shi Y. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- Huang T.T., Nijman S.M.B., Mirchandani K.D., Galardy P.J., Cohn M.A., Haas W., Gygi S.P., Ploegh H.L., Bernards R., D’Andrea A.D. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- Imai S., Kano M., Nonoyama K., Ebihara S. Behavioral characteristics of ubiquitin-specific peptidase 46-deficient mice. PLoS One. 2013;8:339. doi: 10.1371/journal.pone.0058566. 10.1371/journal.pone.0058566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan A.S., Lestra M., Swee L.K., Fan Y., Lamers M.M., Tafesse F.G., Theile C.S., Spooner E., Bruzzone R., Ploegh H.L., Sanyal S. Usp12 stabilizes the T-cell receptor complex at the cell surface during signaling. Proc. Natl. Acad. Sci. 2016;113:201521763. doi: 10.1073/pnas.1521763113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo H.Y., Jones A., Yang C., Zhai L., Smith A.D., IV, Zhang Z., Chandrasekharan M.B., Sun Z.W., Renfrow M.B., Wang Y., Chang C., Wang H. Regulation of histone H2A and H2B deubiquitination and xenopus development by USP12 and USP46. J. Biol. Chem. 2011;286:7190–7201. doi: 10.1074/jbc.M110.158311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten R.P., Long F., Murshudov G.N., Perrakis A. The PDB_REDO server for macromolecular structure model optimization. IUCrJ. 2014;1:213–220. doi: 10.1107/S2052252514009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y., Yang K., Cohn M.A., Haas W., Gygi S.P., D’Andrea A.D. WDR20 regulates activity of the USP12.UAF1 deubiquitinating enzyme complex. J. Biol. Chem. 2010;285:11252–11257. doi: 10.1074/jbc.M109.095141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D., Clague M.J., Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Konarev P.V., Petoukhov M.V., Volkov V.V., Svergun D.I. ATSAS 2.1, a program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2006;39:277–286. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarev P.V., Volkov V.V., Sokolova A.V., Koch M.H.J., Svergun D.I. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Langer G., Cohen S.X., Lamzin V.S., Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Lim K.S., Kim H., Hinds T.R., Jo U., Mao H., Weller C.E., Sun J., Chatterjee C., D’Andrea A.D., Zheng N. Allosteric activation of ubiquitin-specific proteases by β-propeller proteins UAF1 and WDR20. Mol. Cell. 2016;1–12 doi: 10.1016/j.molcel.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q., Dexheimer T.S., Zhang P., Rosenthal A.S., Villamil M.a, You C., Zhang Q., Chen J., Ott C.a, Sun H., Luci D.K., Yuan B., Simeonov A., Jadhav A., Xiao H., Wang Y., Maloney D.J., Zhuang Z. A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nat. Chem. Biol. 2014;10:298–304. doi: 10.1038/nchembio.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna-Vargas M.P.A., Christodoulou E., Alfieri A., van Dijk W.J., Stadnik M., Hibbert R.G., Sahtoe D.D., Clerici M., Marco V.De., Littler D., Celie P.H.N., Sixma T.K., Perrakis A. Enabling high-throughput ligation-independent cloning and protein expression for the family of ubiquitin specific proteases. J. Struct. Biol. 2011;175:113–119. doi: 10.1016/j.jsb.2011.03.017. [DOI] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997 doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nicassio F., Corrado N., Vissers J.H.A., Areces L.B., Bergink S., Marteijn J.A., Geverts B., Houtsmuller A.B., Vermeulen W., Di Fiore P.P., Citterio E. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr. Biol. 2007;17:1972–1977. doi: 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- Nijman S.M.B., Huang T.T., Dirac A.M.G., Brummelkamp T.R., Kerkhoven R.M., D’Andrea A.D., Bernards R. The deubiquitinating enzyme USP1 regulates the fanconi anemia pathway. Mol. Cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Olazabal-Herrero A., García-Santisteban I., Rodríguez J.A. Structure-function analysis of USP1: insights into the role of Ser313 phosphorylation site and the effect of cancer-associated mutations on autocleavage. Mol. Cancer. 2015;14:33. doi: 10.1186/s12943-015-0311-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Row P.E., Liu H., Hayes S., Welchman R., Charalabous P., Hofmann K., Clague M.J., Sanderson C.M., Urbé S. The MIT domain of UBPY constitutes a CHMP binding and endosomal localization signal required for efficient epidermal growth factor receptor degradation. J. Biol. Chem. 2007;282:30929–30937. doi: 10.1074/jbc.M704009200. [DOI] [PubMed] [Google Scholar]
- Sahtoe D.D., Sixma T.K. Layers of DUB regulation. Trends Biochem. Sci. 2015;40:456–467. doi: 10.1016/j.tibs.2015.05.002. [DOI] [PubMed] [Google Scholar]
- Smart O.S., Womack T.O., Flensburg C., Keller P., Paciorek W., Sharff A., Vonrhein C., Bricogne G. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012;68:368–380. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa M.E., Bennett E.J., Gygi S.P., Harper J.W. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun D., Barberato C., Koch M.H. CRYSOL – a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- Svergun D.I., Petoukhov M.V., Koch M.H. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villamil M.A., Chen J., Liang Q., Zhuang Z. A noncanonical cysteine protease USP1 is activated through active site modulation by USP1-associated factor 1. Biochemistry. 2012;51:2829–2839. doi: 10.1021/bi3000512. [DOI] [PubMed] [Google Scholar]
- Villamil M.A., Liang Q., Chen J., Choi Y.S., Hou S., Lee K.H., Zhuang Z. Serine phosphorylation is critical for the activation of ubiquitin-specific protease 1 and its interaction with WD40-repeat protein UAF1. Biochemistry. 2012;51:9112–9113. doi: 10.1021/bi300845s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.A., Maecker H.L., French D.M., Liu J., Gregg A., Silverstein L.B., Cao T.C., Carano R.A.D., Dixit V.M. USP1 deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell. 2011;146:918–930. doi: 10.1016/j.cell.2011.07.040. [DOI] [PubMed] [Google Scholar]
- Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G.W., McCoy A., McNicholas S.J., Murshudov G.N., Pannu N.S., Potterton E.A., Powell H.R., Read R.J., Vagin A., Wilson K.S. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Schoeffler A.J., Wickliffe K., Newton K., Starovasnik M.A., Dueber E.C., Harris S.F. Structural insights into WD-repeat 48 activation of ubiquitin-specific protease 46. Structure. 2015;23:2043–2054. doi: 10.1016/j.str.2015.08.010. [DOI] [PubMed] [Google Scholar]
- Zhang W., Tian Q.B., Li Q.K., Wang J.M., Wang C.N., Liu T., Liu D.W., Wang M.W. Lysine 92 amino acid residue of USP46, a gene associated with “behavioral despair” in mice, influences the deubiquitinating enzyme activity. PLoS One. 2011;6:2–7. doi: 10.1371/journal.pone.0026297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.