Abstract
Genome-wide studies are increasingly becoming a must, especially for complex diseases such as cancer where multiple genes and diverse molecular mechanisms are known to be involved in genes’ function alteration. In this review, we report our latest genomic and epigenomic findings in African-American colorectal cancer patients. This population suffers a higher burden of the disease and most investigators in this field are looking for the underlying genetic and epigenetic targets that might be responsible for this disparity. We here report genome-wide copy number variations, single nucleotide mutations and DNA methylation findings that might be specific to this population.
Keywords: Colorectal cancer, Genomics, Epigenomics, Exome sequencing, DNA methylation, Single nucleotide mutations, Copy number
Introduction
Several studies have investigated the molecular mechanisms leading to colorectal cancer (CRC), which is the third most common cancer in the US [1]. Many of the CRC cases arise from adenomas, in a process described as adenoma-carcinoma sequence [2]. The initiation and progression of CRC are associated with alterations in the function of oncogenes and tumor suppressor genes (TSG). These changes might occur genetically or epigenetically. Indeed, three major mechanisms that affect genes’ function in CRC have been described as CpG island methylation phenotype (CIMP) where global DNA methylation occurs, microsatellite instability (MSI) where uncorrected mismatch repairs generated during DNA replication at nucleotides repeat sequences (microsatellites) leads to their instability; and chromosomal instability (CIN) where deletions, amplifications and translocations of chromosomal fragments lead to global genome alterations [2]. Most times, these mechanisms overlap within the same tumor [2]. In MSI tumors, which occur in about 15% of CRC cases, DNA mismatch repair genes are mutated or methylated which leads to tumors with an MSI phenotype [3]. In contrast, the CIN phenotype is characterized by global genomic rearrangements of chromosomal fragments [4]. The accumulation of chromosomal gains and/or losses throughout the progression from high-grade adenomas to invasive carcinomas have been established in CIN CRC tumors [5]. CIN results from specific mutations or gene rearrangements as a consequence of structural defects involving centromeres, telomere erosion, microtubule dysfunction, chromosome breakage and failure of cell cycle checkpoints [5]. MSI and CIN tumors have been considered as being mutually exclusive, and it has been speculated that MSI tumors have stable diploid karyotypes [6,7]. Nonetheless, recent studies have found that MSI and CIN can occur in the same tumor [8,9]. Trautmann et al. reported that at least 50% of MSI-H tumors have some degree of simultaneous chromosomal alterations [10]. Even though evidence for some degree of CIN could be seen in the majority of MSI-H tumors, patterns of concrete gains and losses between MSI-H and MSS tumors are still poorly understood.
One of the most important epigenetic events is DNA methylation that is thought to occur during the early stages of colon oncogenic transformation. Hence, molecular studies intended for the precise identification of CRC methylation markers may provide useful insights for a better understanding of CRC initiation and progression [11–13].
DNA methylation of some genes and their significance in CRC have been well documented [11–13]. Several genes’ hypermethylation, including MLH1, p16INK4a, TIMP3 as well as other tumor suppressor genes were studied by qualitative and quantitative approaches such as qPCR, MSP (methylation specific PCR), and pyrosequencing [13]. The abnormal methylation of CpG islands that are within gene promoters is a well-studied epigenetic event, which can lead to the transcriptional silencing of the corresponding tumor suppressor genes (TSGs) in CRC tumors. Irrespective of the biological significances of methylation-induced silencing of TSGs, this epigenetic variation establishes molecular signatures that can serve as promising specific biomarkers for early detection of CRC. Such markers need to be validated using genome-wide studies. Additionally, patterns of DNA methylation can be practical for a more precise molecular classification of neoplastic changes, as well as for the prediction of therapeutical responsiveness and prognosis. As our current understanding of the role of genetic and epigenetic alterations in colon carcinogenesis improves, better targeted therapies for CRC might be developed in the population as a whole and high-risk populations such as African Americans in particular [11–13].
Here, we review the outcome of genomic and epigenomic studies performed in African Americans, a population that carries a higher burden of CRC that needs to be addressed through targeted and comprehensive studies.
Array Comparative Genomic Hybridization (aCGH) Findings
In many of our previous studies we reported on the role of MSI, methylation of specific CRC genes such as p16 and MLH1, and mutation of known genes such as KRAS and BRAF from African American CRC patients. Our studies were influential in illuminating some of the precise genetic and epigenetic alterations that happen in this population. We have also capitalized on our previous work and conducted a whole genome analysis of copy number aberrations in African American patients with CRC to find primary targets of amplification and deletion. More significantly, we performed a phylogenetic clustering of the tumors based on aCGH data to establish the sequential order of the detected aberrations.
The analyzed samples displayed that 17% (5 out of 30 tested tumors) had the MSI-H phenotype (MSI-H tumors display instability at 2 or more of the tested markers). This MSI-H rate rests at a higher rate than that documented in the general population [14]. We also detected 4 tumors that were MSI-L (tumors with instability at no more than 1 of the tested markers) while the remaining 21 were MSS (tumors that were stable for all tested markers). The average copy number variations (CNVs) was 25.46 per tumor. The MSI-H tumors indicated a lower rate of 19.0 CNV aberrations while the non-MSI-H tumors had a higher rate of 26.7 CNVs. This quantitative CNV difference supports the concept that MSI-L tumors are different than MSI-H tumors and that they are primarily driven by chromosomal instability [15].
Chromosomes 3, 5, 7 and 8 were the most frequently altered in our study population. Several publications reported that these chromosomes carry genes of relevance to cancer in general and colon cancer in particular. Deletion, mutation or methylation of MLH1, a DNA mismatch repair gene on chromosome 3, leads to the MSI-H phenotype [16]. PPM1L, which is another CRC gene on chromosome 3, was shown to have CNV in APC-negative familial adenomatous polyposis CRC [17]. Chromosome 5 harbors the APC gene, another critical CRC gene that has a crucial role in the early steps of colon carcinogenesis in sporadic CRC and hereditary FAP syndrome. Chromosome 5 displayed 41 aberrations equally distributed among deletions and amplifications (20/21) [18]. Caretaker gene PMS2 [19], a DNA mismatch repair gene, and TSG PIK3CG are on chromosome 7 [20]. Chromosome 8 was the one that showed the most aberrations (25 amplifications/23 deletions). This chromosome is known as a hotspot for CRC tumor progression [21].
An additional chromosome with striking patterns of aberrations was chromosome X, which contained 24 aberrations (14 amplifications/20 deletions). Chromosome X was especially shown to be amplified in male patients. This chromosome has been described as the carrier of tumor suppressor genes [22]. Only 4 out of 15 female CRC patients displayed amplification for chromosome X in comparison to 10/15 male patients. Similar findings have also been observed in Japanese male CRC patients [23].
After comparing our altered genes to Sjoblom et al. cancer genes’ list (Table 1) [24], we found that most of these genes were also altered in our cohort, with 10 genes being primarily deleted, and 19 were shown to be preferentially amplified. TP53 was equally amplified and deleted in our set of samples. P53 (TP53) is a tumor suppressor gene and it has been known to be altered through deletion leading to a loss of function [25]. SMAD2 and SMAD4 were the most deleted genes in this cohort (16 out of 30 samples).
Table 1.
Comparison of African American aCGH data with cancer genes’ list from Sjoblom et al. [24].
| African Americans (n=30) | |||
|---|---|---|---|
| Gene | Chromosome | Amplified (%) | Deleted (%) |
| ABCA1 | 9q31.1 | 1(3) | 1(3) |
| ACSL5 | 10q25 | 1(3) | 5(16) |
| ADAM29 | 4q34 | 0(0) | 8(26) |
| ADAMTS15 | 11q25 | 3(10) | 3(10) |
| ADAMTS18 | 16q23 | 11(36) | 1(3) |
| ADAMTSL3 | 15q25.2 | 1(3) | 6(20) |
| APC | 5q22 | 2(6) | 8(26) |
| C10orf137 | 10q26.1 | 1(3) | 4(13) |
| C15orf2 | 15q11 | 0(0) | 7(23) |
| CD109 | 6q13 | 8(26) | 1(3) |
| CD248 | 11q13 | 11(36) | 3(10) |
| CD46(MCP) | 1q32 | 4(13) | 3(10) |
| CHL1 | 3p26.1 | 2(6) | 1(3) |
| CNTN4 | 3p26 | 4(13) | 1(3) |
| CSMD3 | 8q23.3 | 14(46) | 2(6) |
| EPHA3 | 3p11.2 | 3(10) | 1(3) |
| EPHB6 | 7q34 | 18(60) | 0(0) |
| ERCC6 | 10q11.2 | 3(10) | 2(6) |
| ERGIC3(SBDCAG84) | 20q12 | 20(66) | 0(0) |
| EVL | 14q32.2 | 4(13) | 5(16) |
| EXOC4(SEC8L1) | 7q31 | 18(60) | 0(0) |
| EYA4 | 6q23 | 4(13) | 2(6) |
| FBXW7 | 4q31.3 | 0(0) | 8(26) |
| GALNS | 16q24.3 | 14(46) | 1(3) |
| GNAS | 20q13.3 | 19(63) | 0(0) |
| GUCY1A2 | 11q22 | 3(10) | 6(20) |
| HAPLN1 | 5q14.3 | 1(3) | 9(30) |
| HIST1H1B | 6p22 | 11(36) | 0(0) |
| KCNQ5 | 6q14 | 9(30) | 1(3) |
| KIAA1409 | 14q32.1 | 5(16) | 2(6) |
| KRAS | 12p12.1 | 8(26) | 1(3) |
| KRT73(K6IRS3) | 12q13.3 | 11(36) | 0(0) |
| LGR6 | 1q32.1 | 4(13) | 3(10) |
| LMO7 | 13q22.2 | 17(56) | 1(3) |
| LRP2 | 2q31 | 6(20) | 2(6) |
| MAP2 | 2q34–35 | 6(20) | 2(6) |
| ACTL9 | 19p13.2 | ||
| MKRN3 | 15q11 | 0(0) | 5(16) |
| MLL3 | 7q36.1 | 16(53) | 0(0) |
| MMP2 | 16q12–13 | 15(50) | 1(3) |
| NF1 | 17q11.2 | 12 (40) | 8(26) |
| OBSCN | 1q42.1 | 4(13) | 3(10) |
| P2RX7 | 12q24 | 8(26) | 1(3) |
| P2RY14 | 3q25 | 6(20) | 1(3) |
| PHIP | 6q14 | 8(26) | 1(3) |
| PKHD1 | 6p12.2 | 5(16) | 1(3) |
| PKNOX1 | 21q22.3 | 7(23) | 7(23) |
| PRKD1 | 14q11 | 2(6) | 6(20) |
| PTPRD | 9p23–24 | 1(3) | 1(3) |
| PTPRU | 1p35 | 6(20) | 6(20) |
| RET | 10q11.2 | 0 | 2(6) |
| RUNX1T1 | 8q22 | 14 (46) | 2(6) |
| SCN3B | 11q23.3 | 3(10) | 3(10) |
| SFRS6 | 20q13.1 | 19(63) | 0 |
| SLC29A1 | 6p21 | 11(36) | 2(6) |
| SLC44A4(C6orf29) | 6p21.3 | 11(36) | 2(6) |
| SMAD2 | 18q21.1 | 0 | 16 (53) |
| SMAD3 | 15q22.3 | 1(3) | 6(20) |
| SMAD4 | 18q21.1 | 0 | 16 (53) |
| SYNE1 | 6q25 | 4(13) | 2(6) |
| TBX22 | Xq21.1 | 12 (40) | 4(13) |
| TCF7L2 | 10q25.3 | 1(3) | 5(16) |
| TGFBR2 | 3p22 | 4(13) | 1(3) |
| TP53 | 17p13.1 | 10 (33) | 10 (33) |
| TTLL3 | 3p25.3 | 5(16) | 2(6) |
| UHRF2 | 9p24.1 | 1(3) | 1(3) |
| UQCRC2 | 16p12 | 10 (33) | 1(3) |
| ZNF442 | 19p13.2 | 12 (40) | 7(23) |
This alteration profile is in line with the known TSG status of these genes in many types of cancers [26]. Neurofibromin (NF1) was also lost in many samples of our cohort, has been known to act as a TSG in colon through the Ras pathway [27]. FBXW7, a component of the SCF (Skp1/Cullin/F-box protein) E3 ubiquitin ligase complex, acts as a TSG in many tissues and targets multiple transcriptional activators and proto-oncogenes for ubiquitin-mediated degradation. The gene FBXW7, which was deleted in several samples, influences murine intestinal homeostasis, targeting Notch and Jun pathways’ [28].
With regards to the amplified genes, CD248 (TEM-1) amplification in 11 samples might be justified by its established role in tumor angiogenesis [29]. Although EPHB6 is amplified in our cohort, its function is known to be a metastasis suppressor in non-small cell lung cancer, suggesting that it might have a different function in colon tissue and/or in non-metastatic tumors [30]. The MMP2 gene was amplified in our study population. The use of MMP1/2 inhibitors was shown to promote cell invasion in CRC cell lines [31]. GNAS has been shown to be activated through amplification primarily in ovarian cancer as well as through activating mutations in colorectal cancer [32,33]. GNAS was revealed to act through the activation of Wnt and ERK1/2 MAPK pathways as was shown in mice models of colorectal cancer [33]. This gene was primarily amplified in our study population. LMO7 gene, also amplified in our samples, is known to mediate cell-specific activation of Rho-MRTF_SRF pathway, where it plays a significant role in breast cancer cells migration (Table 2) [34].
Table 2.
Comparison of African American a CGH data with those from caucasian patients from Lassmann et al. [72].
| Caucasians | African Americans (n=30) |
||||
|---|---|---|---|---|---|
| Gene | Chromosome | Amplified (%) |
Deleted (%) |
Amplified (%) |
Deleted (%) |
| THRB | 3p24.3 | 32 | 13 | 3 | |
| RAF1 | 3p25 | 14 | 20 | 6 | |
| RFC2 | 7q11.2 | 36 | 60 | 3 | |
| CYLN2 | 7q11.23 | 36 | |||
| MET | 7q31 | 23 | 60 | 0 | |
| LPL | 8p22 | 23 | 10 | 40 | |
| E2F5 | 8q22-q21.3 | 36 | 53 | 0 | |
| LPL | 8p22 | 23 | |||
| EXT1 | 8q24.11–q24.13 | 32 | 46 | 6 | |
| MYC | 8q24.12–q24.13 | 36 | 46 | 0 | |
| EGR2 | 10q21.3 | 23 | 10 | 13 | |
| DMBT1 | 10q25.3–q26.1 | 23 | 3 | 16 | |
| LRRC32 | 11q13.5 | 32 | 10 | 13 | |
| ATM | 11q22.3 | 27 | 10 | 16 | |
| INS | 11p tel | 32 | 26 | 6 | |
| BRCA2 | 13q12–q13 | 36 | 60 | 0 | |
| RB1 | 13q14 | 41 | 53 | 0 | |
| MAP2K5 | 15q23 | 32 | 3 | 20 | |
| SP6 | 17ptel | 23 | 30 | 30 | |
| TOP3A | 17p11.2 | 10 | 33 | ||
| LLGL1 | 17p12–17p11.2 | 36 | 10 | 33 | |
| FLII | 17p12–17p11.2 | 23 | 10 | 33 | |
| HIC1 | 17p13.3 | 32 | 20 | 30 | |
| CTDP1 | 18q tel | 45 | 0 | 40 | |
| LAMA3 | 18q11.2 | 14 | 0 | 40 | |
| BCL2 | 18q21.3 | 23 | 0 | 40 | |
| DCC | 18q21.3 | 32 | 18 | 0 | 50 |
| TPD52L2 | 20qtel | 27 | 76 | 3 | |
| TOP1 | 20q12–q13.1 | 32 | 56 | 0 | |
| TNFRSF6B | 20q13 | 32 | 53 | 3 | |
| NCOA3 | 20q13 | 32 | 63 | 0 | |
| AURKA | 20q13 | 36 | 56 | 0 | |
| CSE1L | 20q13 | 27 | 63 | 0 | |
| MYBL2 | 20q13.1 | 32 | 63 | 0 | |
| PTPN1 | 20q13.1–q13.2 | 23 | 63 | 0 | |
| CYP24A1 | 20q13.2 | 36 | 63 | 0 | |
| ZNF217 | 20q13.2 | 32 | 63 | 0 | |
| PRPF6 | 20q13.3 | 27 | 60 | 3 | |
| PCNT | 21qtel | 18 | 16 | 20 | |
| XIST | Xq13.2 | 36 | 33 | 13 | |
| STS | Xp22.3 | 23 | 23 | 13 | |
| KAL1 | Xp22.3 | 36 | 30 | 13 | |
While the above discussion centered on genes already known to be oncogenes or TSGs based on prior studies, most of the detected aberrations affected genes without an annotated role in cancer in general and colorectal cancer in particular. We used the global aCGH data in a phylogenetic clustering analysis of the tumors to establish the analyzed tumors relatedness and possible correlations with clinical, pathological or molecular features. These analyses revealed that gender, age and tumor location do not have an impact on the nature of the chromosomal aberrations. The most striking discovery from our parsimony analysis was the clustering of 80% MSI-H tumors in the generated cladogram, away from MSI-L and MSS tumors. These MSI-H tumors had fewer aberrations (<15) compared to MSI-L and MSS tumors (>25). The parsimony phylogenetic analysis implies that this difference is not only quantitative but also qualitative. This is in agreement with Trautmann et al. findings regarding the difference in nature of chromosomal alterations in MSI vs. MSS tumors [10]. CGH array data are very informative. Though, due to many of the chromosomal aberrations spanning large genomic areas and affecting many genes at once, it is difficult at present to assign weight and value to genes within a given aberration. To distinguish between passenger and driver genes within an aberration, CGH experiments need to be added to genome, exome, and/or transcriptome sequencing data to establish genetic variations within the amplified/deleted chromosomal fragments and their impact on gene expression and function [35].
Whole Exome Sequencing of Colorectal Tumors
Whole exome sequencing is likely to lead to the discovery of specific driver mutations. Exponentially growing genomic databases of many specimens across different racial/ethnic groups has the potential to allow the detection of disease-causing and/or race-specific driving mutations in neoplastic lesions [36–38].
We examined alterations which happen in major colorectal cancer pathways and associated effectors using whole exome sequencing (WES) in 12 pairs of African American CRC tumors along with their matched normal showed that WNT and EGFR signaling pathways were the most altered, highlighting their significance in colorectal carcinogenesis, regardless of sex, age or race of the patients [39–41]. Our results defined a total of 52 genes that have been recently cited by Vogelstein et al. [39]. It is noteworthy that many of the identified mutations, especially in the APC gene are novel and different when compared to previously reported mutations.
Copy number variations (CNVs) for the analyzed samples were generated from the exome data using R software. No copy number variations at the APC locus was detected for the analyzed samples. This is consistent with the fact that APC gene is a target of single nucleotide variants (SNVs) rather than CNV alterations [39]. This noticeably illustrates the intricacies between different genomic alterations mechanisms in the path to cancer (Figure 1).
Figure 1. Circular arrangement of all alterations in tumors and normal pairs.
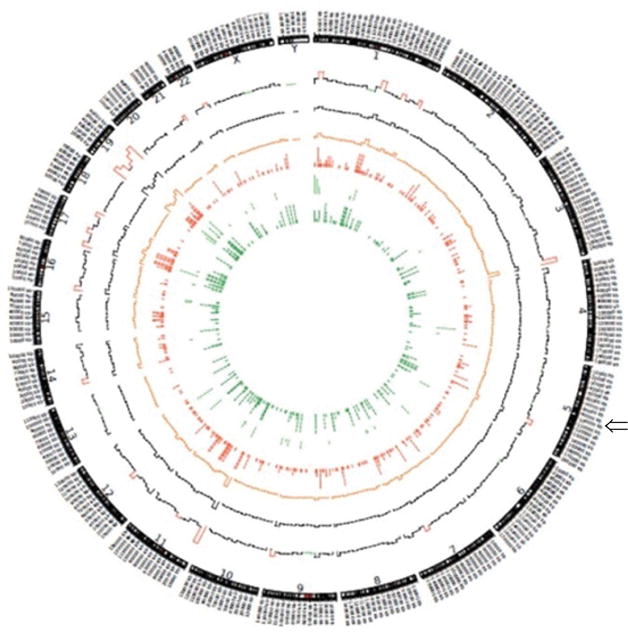
Circos Plot, circular arrangement of all alterations in tumors and normal pairs using Circos version 0.63 (http://circos.ca/). From the outside to the inside of the plot, 1) chromosome, 2) number of total mutations in 10 Mb (red implies >1000 combined mutations in 12 samples in 10 Mb. Green implies <100), 3) histogram of number of synonymous mutations in 10 Mb (Black), 4) histogram of number of missense mutations in 10 Mb (Orange), 5) tiled plot of nonsense mutations (Red) (we used tiled plot since the number of mutations are not very high), 6) tiled plot of non-stop mutations (Green), 7) tiled plot of frameshift insertions and deletions (Dark Green), Arrow show APC loci in chromosome 5.
The number of variants was different among samples. Two samples had more than 7 variants per 106 bases while ten had less than 3 variants per 106 bases. The MSI status of the analyzed samples was established to assess whether the high level of variants within the samples correlates with the MSI-H status. To our surprise, the 3 MSI-H tumors were not part of the hyper-mutated tumor samples [14,22,42–45].
Among the CRC genes established by the cancer genome atlas database (TCGA) [40], the most frequently mutated genes were APC, CACNA1C, DNAH17, FCRL5, KRAS, OBSCN, PR1L1, TELO2, ZNF568, and SRMS. Beyond identifying new non-synonymous mutations and candidate CRC genes, our study determined that we are far from having a complete catalog of CRC genes, with many specific genes at clinically significant frequencies within individual CRC tumors still awaiting detection and experimental validation. The number of such genes is still increasing steeply with the number of analyzed samples and tumor types, which should reflect the diversity of the general population.
Two non-synonymous SNVs were found in tumor suppressor TP53. KRAS gene displayed 4 non-synonymous and 1 non-frameshift deletion. MSH3 and MSH6 had 2 and 1 non-synonymous variants, respectively. MSH3 and MSH6 mutations are in line with a high rate of MSI-H in African American CRC patients. Indeed, we have previously reported an 18% rate of MSI-H in urban African American CRC patients that was primarily assigned to another DNA MMR gene methylation (MLH1). Our exome data shows that SNVs targeting other DNA MMR genes do likely participate in the higher MSI rate in this population [14,46,47].
Our results have shown 1 frameshift deletion in AXIN2 and 2 non-synonymous alterations in ATM gene. We found 2 non-synonymous variants in PIK3CA gene. Three non-synonymous variants were detected in CACNA1C that belongs to the MAPK/Ca channel pathway [48].
In the APC gene, we detected 1 missense mutation, 5 frameshift deletions, 5 stopgains, 2 frameshift insertions, and 1 non-frameshift deletion. APC protein binds to many other proteins including β-catenin which interacts with LEF-1/TCF DNA-binding proteins, which in turn activates Wnt target genes through a unique C-terminal activation domain [49,50]. APC is also recruited to Wnt target genes upon activation of the pathway and regulates the periodic exchange of β-catenin and TLE1/Gro co-repressor complexes [42–44].
Functional annotation of the mutated genes showed several pathways including Wnt, cell adhesion, and ubiquitin-mediated proteolysis pathways were altered genetically in the early stage of colorectal tumorigenesis. Therefore, the functional biological effects of these mutations especially in APC are reflected in the CRC carcinogenicity. Annotation and functional analysis of the novel mutations is likely to establish changes in the activity of APC protein, due to their location in exons 5 and 15.
Also, many of the novel SNVs such as APC4664, APC3418, and APC3862, are situated in exon 15, which is the APC exon most highly associated with CRC risk. These mutations may contribute to major biological alterations of this protein with clinical and pathological implications in this population. These mutations are novel and as such their frequencies are not known in African Americans and other patients. It will be useful to establish such frequencies for personalized therapeutical management approaches and also to assess whether these mutations have any specific racial specificity [51].
There have been other relatively large genome-wide or exome sequencing studies [39–41] on CRC, but only one of them specifically targeted African American patients using WES. This recent publication by Guda et al. on novel recurrently mutated genes in African Americans with CRC provided a set of novel genes that seem to be primarily targeted by mutation in African Americans [52]. They specifically discussed EPHA6 and FLCN, the top genes in their 20 genes’ panel, identified in microsatellite stable (MSS) and low mutation rates CRC tumors. Their approach was focused on detecting genes with differential mutation rates between African Americans and Caucasians while our approach’s focus was to look for distinct mutations within known cancer pathways. There are limited exome studies in African Americans with CRC. As such, it is a challenging task to predict the frequency and incidence of any mutation, be it in a novel or well-known cancer gene. Both of our studies combined, account only for 41 whole exome sequenced African American CRC tumors. Therefore, more exome studies are needed in this population to have a meaningful evaluation of mutations’ weight and to assess cohorts’ variations in the context of CRC disease and disparity, nationwide.
When we checked Guda et al.’s newly identified genes and mutations in our samples, we found 7 somatic protein-changing mutations common between the two cohorts [53]. These mutations occurred in 5 genes, including APC and KRAS. These mutations were found in 6/12 of our samples and 12/29 samples in Guda et al.’s dataset. We have 9 somatic protein-changing mutations in 6 genes (particularly WDR87) from their 20 genes panel, but none observed in their two top genes: FLCN and EPHA6. That result is not surprising because the complete mutation frequency of these two genes reported in their dataset is low (2.91% and 5.83%, respectively). Considering all of the mutations observed for the samples, we have 459 mutations in 18 out of the 20 genes with WDR87 as the dominantly targeted gene. Accordingly, these 20 genes mutational profile in our cohort gives more credibility to their findings as to the potential race specific nature of these genes as targets of mutation in African Americans with CRC. If supplemented with our approach of looking for novel, highly pathogenic mutations within known cancer pathways and genes, the outcome would likely allow a better definition and understanding of race-specific cancer genes and mutations.
Methylome Study of Colorectal Lesions from African Americans
Several approaches were used to establish DNA methylation status in cancer specimens. Qualitative (MSP: methylation specific PCR) and quantitative (qPCR, methylight, pyrosequencing) techniques have been widely used. These techniques, however, were still used at the gene-specific level with a pre-knowledge of the methylated target gene. Our understanding of epigenetic studies at the genomic level have become possible thanks to advances in high-throughput technologies, such as next-generation sequencing and methylation microarrays. We recently published a microarray methylome study that exemplified a genome-wide methylation-based clustering of various genes and loci in CRC neoplasia specimens as a function of CRC status [54]. We also recently used the reduced representation bisulfite sequencing (RRBS), to analyze methylome profiles in various colon tissues and validated a subset of highly methylated targets in an independent cohort of colorectal specimens.
RRBS uses sodium bisulfite modified DNA that is digested with specific restriction endonucleases to enrich for GC-rich regions allowing a focused (reduced representation) sequencing of primarily CpG rich genomic regions. The application of this technique in the context of our study led to the identification of several potential methylation markers. Indeed, we noted a gradual hypomethylation of LINE (long interspersed nuclear elements) sequences during the progression of colorectal carcinogenesis. Cancer samples showed less methylation in LINE sequences than adenoma samples. The same was true for adenoma samples that also had lower methylation of these repeat elements than normal colon samples. Interestingly, this was specific for LINE, but not SINE (short interspersed nuclear elements) sequences that displayed similar methylation pattern across the analyzed tissues. These repeat sequences were known as “junk DNA”. However, recent studies, primarily from the ENCODE project [53–57], have revealed that these sequences play major roles in DNA expression and regulation. Indeed, Hur et al. reported that LINE-1 hypomethylation led to the activation of multiple oncogenes in CRC, hence providing a mechanistic explanation for one of the biological roles of LINE elements’ hypomethylation in colon cancer [58]. These findings are of interest because they reflect the way methylation alterations facilitate the recruitment of oncogenes in the carcinogenic pathway while most of the focus in methylation studies was on tumor suppressor genes. The expression of some oncogenes seems to be unleashed by hypomethylation of regulatory regions embedded within LINE sequences.
As for methylated promotor regions, data analysis led to the identification of specific CpG sites that were located within the following genes; ATXN7L1, BMP3, EID3, GAS7, GPR75 and TNFAIP2 with the following number of identified CpG sites; 2, 7, 15, 1, 24 and 1, respectively. Validation experiments allowed 4 of these genes to be successfully confirmed as potential methylation markers.
EID3 acts as a repressor of nuclear receptor-dependent transcription possibly by interfering with CREBBP-dependent co-activation [59,60]. EID3 may function as a co-inhibitor of other CREBBP/EP300-dependent transcription factors, and its methylation/silencing likely affects several genes that promote cell proliferation and division [61,62]. Ingenuity pathway analysis (IPA) positioned this gene within the Sept9 proteins network. Sept9, a CRC stool methylation marker is a member of this network. The association of EID3 with such a well-recognized biomarker is a confirmation of the RRBS based data credibility.
IPA analysis also placed two other validated genes, BMP3, and GAS7, in TGFβ1 and insulin pathways. GPR75 was not shown to have been located on this same network, probably, due to lack of published related functional data. Indeed, other GPR proteins were within TGFβ1 network. The co-positioning of these methylated markers in the same pathway along with insulin and TGFβ1 is important as it puts colon neoplasia in the large picture of diabetes-associated molecules such as insulin and TGFβ1. African Americans are indeed well known to have a higher incidence and predisposition to diabetes [63,64]. Increased insulin production is very common during the neoplastic transformation [65]. This is perhaps associated to the hormone signaling pathway, which has many elements in common with several growth factors’ signaling pathways such as TGFβ1. Higher levels of this growth factor have been reported in the serum of several types of cancer patients. Also, it has been reported that insulin signaling directly interacts with that of the Ras oncogene [66]. It is worth noting that BMP3 methylation in stools has been found to be linked to CRC in IBD patients [67]. There are however very few publications that are available for these 4 markers, which attest to their novelty as CRC methylation markers [68,69].
There are some genes (NDRG, Sept9) of which the methylation status has already been established in other populations such as Caucasians. However, for the 4 methylation genes (EID3, BMP3, GAS7, and GPR75) we report in African American CRC patients, there have been no other documented investigation except for GAS7, which is a Korean publication reported recently [69]. For the other three genes as stated above, our report is the first in the framework of CRC. GAS7 hypermethylation has recently been shown in CRC Korean patients, but no functional studies have been reported [69]. In another study in breast cancer, it has been shown that BMP3 and GAS7 have major epigenetic differences that affect gene expression, which can play a significant role in the induction and maintenance of different phenotypes [70].
These 4 novel methylation gene targets (BMP3, EID3, GAS7, and GPR75) need to be validated in a larger African American cohort as well as in other populations, and their function need to be analyzed to dissect further their correlation to other known pathways, especially the Insulin/TGFβ1.
As for the TNFAIP2, this gene’ methylation was not validated and it was found to be hypomethylated in the validation set. The hypomethylation status is more in tune with its involvement in invasion and metastasis. [71]
In a study we recently published, we described a series of genes that were cited in many studies as methylation targets in colorectal cancer patients but still not cited in any CIMP genes’ list [54]. Of this list of 15 genes, 4 were methylated in this RRBS dataset. These genes-SFRP1, 3OST2, LAMA1, and KCNQ5- might be eligible now to make the CIMP genes’ list since our current RRBS data reflect their methylation status in the analyzed samples [72].
Conclusion
Integrative genome-wide studies to elucidate CRC driver genes
Our data above reflect the need for comprehensive genome-wide studies that should integrate copy number variations, along with methylome, and exome studies to delineate the intricacies of genomic and epigenomic mechanisms that trigger and promote colon oncogenic transformation []. It is worth noting however that the few studies with limited number of samples have been performed so far. This constitutes a major limitation in the field of cancer research in general, and colorectal cancer in particular in minority populations. As such, there has to be more research with large sample sizes to further dissect the role of each gene altering mechanism and to define specific driver genes in a given tumor. Such findings will be instrumental in determining the type of therapy that is most suitable for a particular patient and in predicting prognosis based on genome-wide established profiles.
Acknowledgments
The authors acknowledge support from NIH-RCMI grant# 5G12MD007597.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, et al. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 3.Markowitz S. DNA repair defects inactivate tumor suppressor genes and induce hereditary and sporadic colon cancers. J Clin Oncol. 2000;18:75S–80S. [PubMed] [Google Scholar]
- 4.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 5.Ried T, Knutzen R, Steinbeck R, Blegen H, Schrock E, et al. Comparative genomic hybridization reveals a specific pattern of chromosomal gains and losses during the genesis of colorectal tumors. Genes Chromosomes Cancer. 1996;15:234–245. doi: 10.1002/(SICI)1098-2264(199604)15:4<234::AID-GCC5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 6.Aaltonen LA, Peltomaki P, Leach FS, Sistonen P, Pylkkanen L, et al. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 7.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 8.Goel A, Arnold CN, Niedzwiecki D, Chang DK, Ricciardiello L, et al. Characterization of sporadic colon cancer by patterns of genomic instability. Cancer Res. 2003;63:1608–1614. [PubMed] [Google Scholar]
- 9.Tang R, Changchien CR, Wu MC, Fan CW, Liu KW, et al. Colorectal cancer without high microsatellite instability and chromosomal instability: an alternative genetic pathway to human colorectal cancer. Carcinogenesis. 2004;25:841–846. doi: 10.1093/carcin/bgh074. [DOI] [PubMed] [Google Scholar]
- 10.Trautmann K, Terdiman JP, French AJ, Roydasgupta R, Sein N, et al. Chromosomal instability in microsatellite-unstable and stable colon cancer. Clin Cancer Res. 2006;12:6379–6385. doi: 10.1158/1078-0432.CCR-06-1248. [DOI] [PubMed] [Google Scholar]
- 11.Ashktorab H, Daremipouran M, Goel A, Varma S, Leavitt R, et al. DNA methylome profiling identifies novel methylated genes in African American patients with colorectal neoplasia. Epigenetics. 2014;9:503–512. doi: 10.4161/epi.27644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashktorab H, Rahi H, Wansley D, Varma S, Shokrani B, et al. Toward a comprehensive and systematic methylome signature in colorectal cancers. Epigenetics. 2013;8:807–815. doi: 10.4161/epi.25497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fatemi M, Paul TA, Brodeur GM, Shokrani B, Brim H, et al. Epigenetic silencing of CHD5, a novel tumor-suppressor gene, occurs in early colorectal cancer stages. Cancer. 2014;120:172–180. doi: 10.1002/cncr.28316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brim H, Mokarram P, Naghibalhossaini F, Saberi-Firoozi M, Al-Mandhari M, et al. Impact of BRAF, MLH1 on the incidence of microsatellite instability high colorectal cancer in populations based study. Mol Cancer. 2008;7:68. doi: 10.1186/1476-4598-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carethers JM. One Colon Lumen but Two Organs. Gastroenterology. 2011;141:411–412. doi: 10.1053/j.gastro.2011.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hitchins MP, Lin VA, Buckle A, Cheong K, Halani N, et al. Epigenetic inactivation of a cluster of genes flanking MLH1 in microsatellite-unstable colorectal cancer. Cancer Res. 2007;67:9107–9116. doi: 10.1158/0008-5472.CAN-07-0869. [DOI] [PubMed] [Google Scholar]
- 17.Thean LF, Loi C, Ho KS, Koh PK, Eu KW, et al. Genome-wide scan identifies a copy number variable region at 3q26 that regulates PPM1L in APC mutation-negative familial colorectal cancer patients. Genes Chromosomes Cancer. 2010;49:99–106. doi: 10.1002/gcc.20724. [DOI] [PubMed] [Google Scholar]
- 18.Arnold CN, Goel A, Niedzwiecki D, Dowell JM, Wasserman L, et al. APC promoter hypermethylation contributes to the loss of APC expression in colorectal cancers with allelic loss on 5q. Cancer Biol Ther. 2004;3:960–964. doi: 10.4161/cbt.3.10.1113. [DOI] [PubMed] [Google Scholar]
- 19.Will O, Carvajal-Carmona LG, Gorman P, Howarth KM, Jones AM, et al. Homozygous PMS2 deletion causes a severe colorectal cancer and multiple adenoma phenotype without extraintestinal cancer. Gastroenterology. 2007;132:527–530. doi: 10.1053/j.gastro.2006.11.043. [DOI] [PubMed] [Google Scholar]
- 20.Kratz CP, Emerling BM, Bonifas J, Wang W, Green ED, et al. Genomic structure of the PIK3CG gene on chromosome band 7q22 and evaluation as a candidate myeloid tumor suppressor. Blood. 2002;99:372–374. doi: 10.1182/blood.v99.1.372. [DOI] [PubMed] [Google Scholar]
- 21.Hutter CM, Slattery ML, Duggan DJ, Muehling J, Curtin K, et al. Characterization of the association between 8q24 and colon cancer: gene-environment exploration and meta-analysis. BMC Cancer. 2010;10:670. doi: 10.1186/1471-2407-10-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashktorab H, Schaffer AA, Daremipouran M, Smoot DT, Lee E, et al. Distinct genetic alterations in colorectal cancer. PLoS One. 2010;5:e8879. doi: 10.1371/journal.pone.0008879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Unotoro J, Kamiyama H, Ishido Y, Yaginuma Y, Kasamaki S, et al. Analysis of the relationship between sex and chromosomal aberrations in colorectal cancer by comparative genomic hybridization. J Int Med Res. 2006;34:397–405. doi: 10.1177/147323000603400409. [DOI] [PubMed] [Google Scholar]
- 24.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 25.Copelli SB, Mazzeo C, Gimenez A, Casco C, Meiss R, et al. Molecular analysis of p53 tumor-suppressor gene and microsatellites in preneoplastic and neoplastic lesions of the colon and esophagus. Oncol Rep. 2001;8:923–929. doi: 10.3892/or.8.4.923. [DOI] [PubMed] [Google Scholar]
- 26.Maliekal TT, Antony ML, Nair A, Paulmurugan R, Karunagaran D. Loss of expression, and mutations of Smad 2 and Smad 4 in human cervical cancer. Oncogene. 2003;22:4889–4897. doi: 10.1038/sj.onc.1206806. [DOI] [PubMed] [Google Scholar]
- 27.Cacev T, Radosevic S, Spaventi R, Pavelic K, Kapitanovic S. NF1 gene loss of heterozygosity and expression analysis in sporadic colon cancer. Gut. 2005;54:1129–1135. doi: 10.1136/gut.2004.053348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Babaei-Jadidi R, Li N, Saadeddin A, Spencer-Dene B, Jandke A, et al. FBXW7 influences murine intestinal homeostasis and cancer, targeting Notch, Jun, and DEK for degradation. J Exp Med. 2011;208:295–312. doi: 10.1084/jem.20100830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rmali KA, Puntis MC, Jiang WG. Prognostic values of tumor endothelial markers in patients with colorectal cancer. World J Gastroenterol. 2005;11:1283–1286. doi: 10.3748/wjg.v11.i9.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu J, Bulk E, Ji P, Hascher A, Tang M, et al. The EPHB6 receptor tyrosine kinase is a metastasis suppressor that is frequently silenced by promoter DNA hypermethylation in non-small cell lung cancer. Clin Cancer Res. 2010;16:2275–2283. doi: 10.1158/1078-0432.CCR-09-2000. [DOI] [PubMed] [Google Scholar]
- 31.Wong JC, Chan SK, Schaeffer DF, Sagaert X, Lim HJ, et al. Absence of MMP2 Expression Correlates with Poor Clinical Outcomes in Rectal Cancer, and Is Distinct from MMP1-Related Outcomes in Colon Cancer. Clin Cancer Res. 2011;17:4167–4176. doi: 10.1158/1078-0432.CCR-10-1224. [DOI] [PubMed] [Google Scholar]
- 32.Tominaga E, Tsuda H, Arao T, Nishimura S, Takano M, et al. Amplification of GNAS may be an independent, qualitative, and reproducible biomarker to predict progression-free survival in epithelial ovarian cancer. Gynecol Oncol. 2010;118:160–166. doi: 10.1016/j.ygyno.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 33.Wilson CH, McIntyre RE, Arends MJ, Adams DJ. The activating mutation R201C in GNAS promotes intestinal tumourigenesis in Apc(Min/+) mice through activation of Wnt and ERK1/2 MAPK pathways. Oncogene. 2010;29:4567–4575. doi: 10.1038/onc.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu Q, Guo C, Li Y, Aronow BJ, Zhang J. LMO7 Mediates Cell-Specific Activation of Rho-MRTF-SRF Pathway and Plays an Important Role in Breast Cancer Cell Migration. Mol Cell Biol. 2011;31:3223–3240. doi: 10.1128/MCB.01365-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abu-Asab MS, Chaouchi M, Alesci S, Galli S, Laassri M, et al. Biomarkers in the age of omics: time for a systems biology approach. OMICS. 2011;15:105–112. doi: 10.1089/omi.2010.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 37.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamborero D, Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Kandoth C, et al. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci Rep. 2013;3:2650. doi: 10.1038/srep02650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Network CGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chevillard-Briet M, Quaranta M, Grezy A, Mattera L, Courilleau C, et al. Interplay between chromatin-modifying enzymes controls colon cancer progression through Wnt signaling. Hum Mol Genet. 2014;23:2120–2131. doi: 10.1093/hmg/ddt604. [DOI] [PubMed] [Google Scholar]
- 43.Ashktorab H, Belgrave K, Hosseinkhah F, Brim H, Nouraie M, et al. Global histone H4 acetylation and HDAC2 expression in colon adenoma and carcinoma. Dig Dis Sci. 2009;54:2109–2117. doi: 10.1007/s10620-008-0601-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kume K, Iizumi Y, Shimada M, Ito Y, Kishi T, et al. Role of N-end rule ubiquitin ligases UBR1 and UBR2 in regulating the leucine-mTOR signaling pathway. Genes Cells. 2010;15:339–349. doi: 10.1111/j.1365-2443.2010.01385.x. [DOI] [PubMed] [Google Scholar]
- 45.Ashktorab H, Smoot DT, Farzanmehr H, Fidelia-Lambert M, Momen B, et al. Clinicopathological features and microsatellite instability (MSI) in colorectal cancers from African Americans. Int J Cancer. 2005;116:914–919. doi: 10.1002/ijc.21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brim H, Hasszadeh H, Schaffer AA, Yunis A, Razjouyan H, et al. Genomic Aberrations in an African American Colorectal Cancer Cohort Reveals a MSI-Specific Profile and Chromosome X Amplification in Male Patients. PLoS One. 2012;7:e40392. doi: 10.1371/journal.pone.0040392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar K, Brim H, Mokarram P, Naghibalhossaini F, Saberi-Firoozi M, et al. Distinct BRAF (V600E) and KRAS mutations in high microsatellite instability sporadic colorectal cancer in African Americans. Clin Cancer Res. 2009;15:1155–1161. doi: 10.1158/1078-0432.CCR-08-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choi K, Chen J, Mitra S, Sarna SK. Impaired integrity of DNA after recovery from inflammation causes persistent dysfunction of colonic smooth muscle. Gastroenterology. 2011;141:1293–1301. doi: 10.1053/j.gastro.2011.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 50.Valenta T, Hausmann G, Basler K. The many faces and functions of beta-catenin. EMBO J. 2012;31:2714–2736. doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ashktorab H, Daremipouran M, Devaney J, Varma S, Rahi H, et al. Identification of novel mutations by exome sequencing in African American colorectal cancer patients. Cancer. 2014;121:34–42. doi: 10.1002/cncr.28922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guda K, Veigl ML, Varadan V, Nosrati A, Ravi L, et al. Novel recurrently mutated genes in African American colon cancers. Proc Natl Acad Sci USA. 2015;112:1149–1154. doi: 10.1073/pnas.1417064112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Birney E. The making of ENCODE: Lessons for big-data projects. Nature. 2012;489:49–51. doi: 10.1038/489049a. [DOI] [PubMed] [Google Scholar]
- 54.Ecker JR, Bickmore WA, Barroso I, Pritchard JK, Gilad Y, et al. Genomics: ENCODE explained. Nature. 2012;489:52–55. doi: 10.1038/489052a. [DOI] [PubMed] [Google Scholar]
- 55.Gerstein M. Genomics: ENCODE leads the way on big data. Nature. 2012;489:208. doi: 10.1038/489208b. [DOI] [PubMed] [Google Scholar]
- 56.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maher B. ENCODE: The human encyclopaedia. Nature. 2012;489:46–48. doi: 10.1038/489046a. [DOI] [PubMed] [Google Scholar]
- 58.Hur K, Cejas P, Feliu J, Moreno-Rubio J, Burgos E, et al. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut. 2013;63:635–646. doi: 10.1136/gutjnl-2012-304219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bavner A, Matthews J, Sanyal S, Gustafsson JA, Treuter E. EID3 is a novel EID family member and an inhibitor of CBP-dependent co-activation. Nucleic Acids Res. 2005;33:3561–3569. doi: 10.1093/nar/gki667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guerineau M, Kriz Z, Kozakova L, Bednarova K, Janos P, et al. Analysis of the Nse3/MAGE-binding domain of the Nse4/EID family proteins. PLoS One. 2012;7:e35813. doi: 10.1371/journal.pone.0035813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carmona FJ, Azuara D, Berenguer-Llergo A, Fernandez AF, Biondo S, et al. DNA methylation biomarkers for noninvasive diagnosis of colorectal cancer. Cancer Prev Res (Phila) 2013;6:656–665. doi: 10.1158/1940-6207.CAPR-12-0501. [DOI] [PubMed] [Google Scholar]
- 62.Church TR, Wandell M, Lofton-Day C, Mongin SJ, Burger M, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2013;63:317–325. doi: 10.1136/gutjnl-2012-304149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCormack S, Grant SF. Genetics of obesity and type 2 diabetes in African Americans. J Obes. 2013;2013:396416. doi: 10.1155/2013/396416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ricci-Cabello I, Ruiz-Perez I, Nevot-Cordero A, Rodriguez-Barranco M, Sordo L, et al. Health care interventions to improve the quality of diabetes care in African Americans: a systematic review and meta-analysis. Diabetes Care. 2013;36:760–768. doi: 10.2337/dc12-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Giovannucci E, Harlan DM, Archer MC, Bergenstal RM, Gapstur SM, et al. Diabetes and cancer: a consensus report. Diabetes Care. 2010;33:1674–1685. doi: 10.2337/dc10-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kisiel JB, Yab TC, Nazer Hussain FT, Taylor WR, Garrity-Park MM, et al. Stool DNA testing for the detection of colorectal neoplasia in patients with inflammatory bowel disease. Aliment Pharmacol Ther. 2013;37:546–554. doi: 10.1111/apt.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cai C, Ashktorab H, Pang X, Zhao Y, Sha W, et al. MicroRNA-211 expression promotes colorectal cancer cell growth in vitro and in vivo by targeting tumor suppressor CHD5. PLoS One. 2012;7:e29750. doi: 10.1371/journal.pone.0029750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mokarram P, Kumar K, Brim H, Naghibalhossaini F, Saberi-firoozi M, et al. Distinct high-profile methylated genes in colorectal cancer. PLoS One. 2009;4:e7012. doi: 10.1371/journal.pone.0007012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim YH, Lee HC, Kim SY, Yeom YI, Ryu KJ, et al. Epigenomic analysis of aberrantly methylated genes in colorectal cancer identifies genes commonly affected by epigenetic alterations. Ann Surg Oncol. 2011;18:2338–2347. doi: 10.1245/s10434-011-1573-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ronneberg JA, Fleischer T, Solvang HK, Nordgard SH, Edvardsen H, et al. Methylation profiling with a panel of cancer related genes: association with estrogen receptor, TP53 mutation status and expression subtypes in sporadic breast cancer. Mol Oncol. 2011;5:61–76. doi: 10.1016/j.molonc.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen LC, Chen CC, Liang Y, Tsang NM, Chang YS, et al. A novel role for TNFAIP2: its correlation with invasion and metastasis in nasopharyngeal carcinoma. Mod Pathol. 2011;24:175–184. doi: 10.1038/modpathol.2010.193. [DOI] [PubMed] [Google Scholar]
- 72.Lassmann S, Weis R, Makowiec F, Roth J, Danciu M, et al. Array CGH identifies distinct DNA copy number profiles of oncogenes and tumor suppressor genes in chromosomal- and microsatellite-unstable sporadic colorectal carcinomas. J Mol Med (Berl) 2007;85:293–304. doi: 10.1007/s00109-006-0126-5. [DOI] [PubMed] [Google Scholar]
- 73.Ashktorab H, Varma S, Brim H. Next-generation sequencing in African Americans with colorectal cancer. Proc Natl Acad Sci USA. 2015;112:E2852. doi: 10.1073/pnas.1503760112. [DOI] [PMC free article] [PubMed] [Google Scholar]


