Abstract
Interferon (IFN) regulates immune responses and antitumor activity. Arginine–glycine–aspartic acid (RGD) peptides can specifically bind to integrin αvβ3, a transmembrane receptor that is highly expressed on the surface of various cancer cells. In this study, we expressed recombinant RGD-IFN-α2a-core fusion proteins and assessed their antitumor activity in vitro. Two RGD-IFN-α2a-core fusion proteins and a negative control protein were expressed in vitro. These two RGD-IFN-α2a-core fusion proteins could bind the tumor cell surface specifically and did not bind to normal cells. RGD-IFN-α2a-core fusion protein treatment of tumor cells significantly reduced cell viability and induced apoptosis in a dose-dependent manner. At the ‘mRNA’ level, both proteins could upregulate CASP3 expression. These data indicate that both laboratory-engineered RGD-IFN-α2a-core fusion proteins could bind the surface of tumor cells and induce apoptosis in vitro. Further studies will investigate the in-vivo antitumor activities of the RGD-IFN-α2a-core fusion proteins.
Keywords: antitumor activity, arginine–glycine–aspartic acid peptides, cancer-targeting therapy, interferon-α2a
Introduction
Despite recent advancements in early detection, prevention, and treatment, cancer remains one of the most devastating diseases endangering the health and quality of life of humans worldwide. To date, surgery remains the only effective curative option to treat most cancers, although radiotherapy and chemotherapy can sometimes effectively control tumor progression and prolong human life 1. However, in addition to destroying cancer cells, radiotherapy and chemotherapy considerably damage normal cells 2. Therefore, cancer treatment research is shifting to specifically targeted molecular therapies that are capable of targeting tumor cells and sparing normal cells.
The integrin family is a class of membrane receptors that mediates cell–cell or cell–extracellular matrix interactions. Integrin consists of two subunits, α and β, and different heterodimers of these two subunits form different integrins 3. Integrin plays essential regulatory roles in cell adhesion, proliferation, differentiation, and apoptosis. Integrin αvβ3 is an important integrin 4. Previous studies have indicated that integrin αvβ3 is specifically overexpressed in activated endothelial cells and tumor cells, but is not expressed, or is rarely expressed, in the vast majority of mature endothelial cells and normal cells 5. Therefore, integrin αvβ3 is a promising target of molecular cancer therapy. Indeed, arginine–glycine–aspartic acid (RGD) can specifically bind to integrin αvβ3 6 and provides a useful candidate for specific targeting and delivery of drugs or other antitumor agents.
In this study, we selected interferon-α2a (IFN-α2a), which functions as an innate immune response against viral infection and various human tumors and is produced by natural killer cells, B lymphocytes and T lymphocytes, macrophages, fibroblasts, and endothelial cells 7. IFN is used widely as an antitumor therapy in medical oncology clinics. It has a complex antitumor mechanism that primarily acts by inhibiting tumor cell proliferation and oncogene expression, and by inducing apoptosis through its role in immune regulation, antiangiogenesis, and inhibition of metastasis. In this study, we constructed expression vectors that fused two different RGD-IFN-α2a-core cDNA clones and in-vitro expressed these fusion proteins using a prokaryotic expression system before purifying them using a glutathione S-transferase (GST) purification method. Finally, we assessed the antitumor activity of both RGD-IFN-α2a-core fusion proteins in vitro.
Materials and methods
Materials
Restriction endonuclease enzymes (EcoRI, BamHI and SalI), DNA polymerase Pfu, DNA marker, gel extraction kits, T4 DNA ligase, the PureLink RNA kit, and the cDNA SuperScript First Strand Synthesis kits were purchased from TaKaRa (Shiga, Japan). pGEX-4T-1, Escherichia coli DH5α, and E. coli Rosseta cells were maintained in our laboratory. Glutathione agarose was purchased from Thermo Fisher Scientific Inc. (Waltham, Massachusetts, USA). Anti-IFN-α2a antibody was purchased from Gene Tex Inc. (Irvine, California, USA). Horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody and DAB color development kits were obtained from Beyotime Biotechnology Company (Hangzhou, China). Reverse transcriptase PCR kits, RNA extraction kits, Lipofectamine 2000, Dulbecco’s modified Eagle’s medium (DMEM), and fetal bovine serum were purchased from Invitrogen (Life Technologies, Grand Island, New York, USA). West Pico ECL reagent was purchased from Pierce Biotechnology Inc. (Rockford, Illinois, USA). Penicillin G and streptomycin were purchased from Shanghai Biotechnology Company (Shanghai, China). PCR primers were synthesized by Shanghai Sangon Biotechnology Company (Shanghai, China).
Cell cultures and culture
Human non-small-cell lung cancer cell line A549, human colon cancer cell line SW480, and normal human lung epithelial cell line BEAS-2B were cultured in DMEM supplemented with 10% fetal bovine serum, 100 U/ml penicillin G, and 100 μg/ml streptomycin at 37°C in a 5% CO2 incubator.
Expression and purification of the fusion protein GST-RGD-IFN-α2a-core
To construct the RGD-IFN-α2a-core fusion protein expression vector, we prepared the pGEX-4T-core plasmid to harbor the RGD and IFN-α2a cDNA using PCR from the T-hepatitis C virus (T-HCV) core as a template and the following primers: 5′-TAGAATTCATGAGCACGAATCCTAAACCTCA-3′ and 5′-GGGGCGGCCGCTTAGGCTGAAGCGGGCACAGTCA-3′. RGD-IFN-α2a, RGD-IFN-α2a(300), and IFN-α2a fragments were generated using PCR from pET28a-RGD-IFN-α2a with the following primers: 5′-TAGGATCCATGTGTCGTGGCGATTGT-3′ and 5′-TAGAATTCTCATTCCTTACTTCTTAAACTTTCTTGCAAG-3′; 5′-AGGATCCATGGCCCTGTCCTTTTCTTTACT-3′ and 5′-TAGAATTCGGCTGAAGCGGGCACAGT-3′; 5′-TAGGATCCATGTGTCGTGGCGATTGT-3′ and 5′-TAGAATTCTCATTCCTTACTTCTTAAACTTTCTTGCAAG-3′, respectively. pGEX-4T-RGD-IFN-α2a-core, pGEX-4T-IFN-α2a-core, and pGEX-4T-RGD-IFN-α2a(300)-core were constructed using these PCR fragments and the recombinant plasmids were verified by restriction endonuclease digestion and DNA sequencing. After confirmation, these plasmids were named GST-RGD-IFN-α2a(300)-core (Q1), GST-RGD-IFN-α2a-core (Q2), and GST-IFN-α2a-core [nitrate reductase (NR)]. The completed and confirmed vectors were transfected into E. coli for in-vitro expression of the fusion proteins. The fusion proteins were then purified by affinity chromatography/glutathione agarose according to the manufacturer’s instructions.
Flow cytometry
The cells were fixed with 80% methanol (5 min) and then permeabilized with 0.1% PBS-Tween for 20 min. The cells were then incubated in 1× PBS/10% normal goat serum/0.3 mol/l glycine to block nonspecific protein–protein interactions, followed by the 27.1 (VNR-1) antibodies (2 µg/1×106 cells; Abcam, Cambridge, Massachusetts, USA) for 30 min at 22°C. The secondary antibody used was DyLight 488 goat anti-mouse IgG (H+L) (ab96879) at a 1/500 dilution for 30 min at 22°C. Acquisition of less than 10 000 events was analyzed using a Becton Dickinson FACSCalibur (BD Biosciences, San Diego, California, USA).
Immunofluorescence
Immunofluorescence staining was used to visualize fusion protein expression in tumor cells. Briefly, we plated tumor cells on glass coverslips and grew them at 37°C in a 5% CO2 incubator for 12 h. Next, the cells were fixed with 2% paraformaldehyde, permeabilized with 0.2% Triton X-100, blocked with 5% BSA in PBS, incubated with an anti-HCV core monoclonal antibody, and visualized by incubating the cells with Cy3-labeled goat anti-mouse IgG. Stained cells were viewed using an Olympus Fluoview FV1000 confocal laser-scanning microscope (Olympus, Tokyo, Japan).
Fusion protein-binding assay
Briefly, 3×104 cells were plated into 96-well plates precoated with 0, 0.1, 1, and 10 nmol/l of Q1, Q2, or NR protein and grown at 37°C under 5% CO2 for 1 h. Cells were then washed three times with incomplete DMEM and fixed with 4% paraformaldehyde for 10 min. The cells that were bound to the precoated plates were visualized by incubating with 0.5% crystal violet dye, viewed under an inverted microscope, and counted.
Cell viability MTT assay
Cell viability 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays were performed according to the protocol outlined in Mosmann 8, with slight modifications. Cells were seeded in 96-well plates at a density of 6×103 cells/well and incubated in a CO2 incubator at 37°C overnight. The next day, the recombinant fusion proteins were added to each well at three different concentrations and cells were further cultured for up to 5 days. At the end of each experiment, 20 µl of MTT solution (5 mg/ml) was added to each well and the cells were incubated for an additional 3 h. Afterwards, the solution was discarded and 100 µl of dimethyl sulfoxide was added to each well to solubilize the crystals. The plates were then measured using an ELISA plate reader (BioTek Instruments, Winooski, Vermont, USA) at 570 nm. Each fusion protein concentration was assayed in five parallel wells and the experiments were repeated three times. The data were then plotted as time points on the x-axis and A570 optical density numbers on the y-axis.
Hoechst 33342 staining
Hoechst 33342 staining was performed to detect altered nuclear morphology in A549 and SW480 cells after Q1, Q2, or NR treatment. Cells were plated on coverslips and treated with different concentrations of the three fusion proteins for 1 day and, after treatment, the cells were stained with 10 µmol/l Hoechst 33342 solution for 15 min at 37°C. After staining, cells were washed three times with PBS, then viewed and counted under a fluorescence microscope with standard excitation filters (Nikon, Tokyo, Japan). The excitation wavelength was 346 nm and the emission wavelength was 460 nm.
Annexin V/FITC flow cytometric assay
The flow cytometric assay was carried out using the annexin V Kit (BD Biosciences Pharmingen, San Diego, California, USA). Cells were seeded overnight in six-well plates at a density of 1.0×105 cells/well and treated with 10 nmol/l recombinant protein (Q1, Q2, or NR) for 72 h. After treatment, cells were harvested, pelleted by centrifugation, immediately resuspended in binding buffer, and subsequently stained with 5 µl FITC annexin V or 5 µl propidium iodide according to the kit instructions. The mixture was placed on ice in the dark and analyzed using the FACS system (Becton Dickinson, San Jose, California, USA).
Protein extraction and western blot
After treatment, cells were washed with ice-cold PBS and harvested in SDS-polyacrylamide gel electrophoresis loading buffer. Twenty micrograms of total protein sample was separated on a 12% sodium dodecyl sulfate polyacrylamide gel by electrophoresis and transferred to polyvinylidene fluoride membranes (Millipore, Billerica, Massachusetts, USA). For western blot, the membrane was blocked in 5% milk in PBS for 1 h at room temperature. The expression levels of caspase-3 (Cell Signaling Technology, Danvers, Massachusetts, USA), activated caspase-3 (Cell Signaling Technology), and tubulin (Cell Signaling Technology) were detected by incubating the membrane with the appropriate antibodies, followed by HRP-conjugated secondary antibody (Abcam, Cambridge, Massachusetts, USA) for 1 h at room temperature before being developed with West Pico ECL reagent (Pierce, Rockford, Illinois, USA).
Statistical analysis
All data were summarized as mean±SD and analyzed statistically using SPSS software, version 16 (SPSS Inc., Chicago, Illinois, USA) using Student’s t-test. P values of up to 0.05 were considered statistically significant.
Results
Cloning, expression, and purification of fusion proteins
We assessed the effects of two RGD-IFN-α2a-core fusion proteins on tumor cell viability and apoptosis. First, we cloned, expressed, and purified RGD-IFN-α2a, RGD-IFN-α2a(300), and IFN-α2a fusion proteins. Successful cloning was confirmed by digesting the expression vectors with BamHI and EcoRI (Fig. 1), and further confirmed by DNA sequencing. The verified fusion proteins were expressed in E. coli Rosseta and purified using glutathione agarose affinity resin under native conditions. The isolated proteins were then resolved by 10% SDS-polyacrylamide gel electrophoresis at 61 kDa for Q1 and NR and 70 kDa for Q2 (Fig. 1).
Fig. 1.
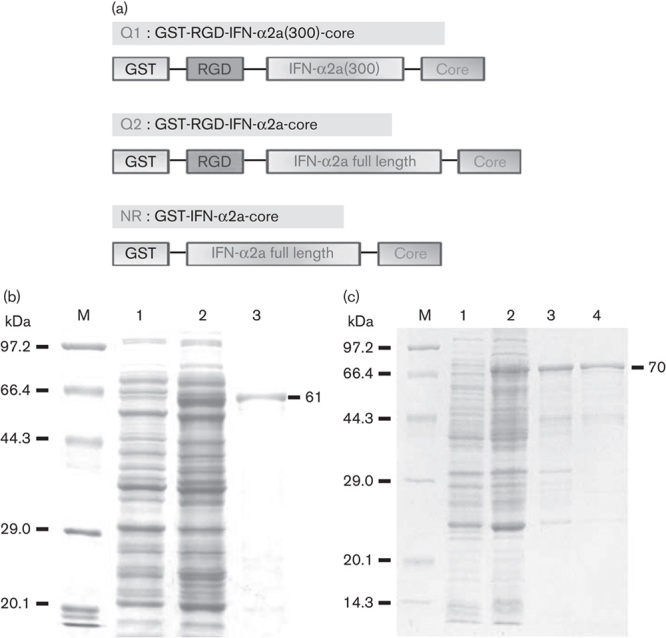
Cloning, expression, and purification of GST and RGD-IFN-α2a-core fusion proteins. (a) Schematic diagram of pGEX-4T-RGD-IFN-α2a(300)-core (Q1), pGEX-4T-RGD-IFN-α2a-core (Q2), and pGEX-4T-IFN-α2a (NR). (b) Purified Q1 protein. M: protein marker; 1: pGEX-4T-RGD-IFN-α2a(300)-core without being induced; 2: pGEX-4T-RGD-IFN-α2a(300)-core induced by IPTG; 3: purified Q1 protein. (c) Purified Q2 and NR proteins. M: protein marker; 1: pGEX-4T-RGD-IFN-α2a-core without being induced; 2: pGEX-4T-RGD-IFN-α2a-core induced by IPTG; 3: purified Q2 protein; 4: purified NR protein. GST, glutathionine S-transferase; IFN, interferon; NR, nitrate reductase; RGD, arginine–glycine–aspartic acid.
Tumor cell-binding specificity of RGD-IFN-α2a-core fusion proteins
Because αvβ3 expression is usually very high on the cancer cell surface, our RGD constructs were specifically designed to bind integrin αvβ3. Therefore, we first assessed whether the recombinant RGD-IFN-α2a-core proteins could bind to tumor cells. As expected, the recombinant RGD-IFN-α2a-core protein specifically bound to the human non-small-cell lung cancer cell line A549 and human colon cancer cell line SW480, but did not bind to the normal human lung epithelial BEAS-2B cell line (Fig. 2). The binding affinities of Q1 and Q2 were similar and dose dependent (between 1 and 10 nmol/l), but NR was not (Fig. 2). After protein concentrations reached 10 nmol/l, the binding affinities of both Q1 and Q2 reached their maximal capacity (Fig. 2). The Q1 cells’ binding number of 10 nmol/l in A549 was almost five times as much as 1 nmol/l and the Q1 cells’ binding number of 10 nmol/l in SW480 was almost six times as much as 1 nmol/l. The results for Q2 were the same as Q1 (P<0.01). Immunofluorescence staining further confirmed and showed that both Q1 and Q2 specifically bound to the membranes of A549 and SW480 cells, but not to the membranes of BEAS-2B cells (Fig. 3).
Fig. 2.
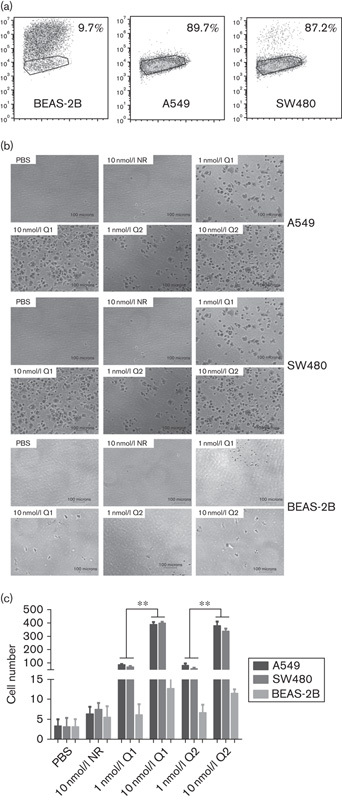
RGD-IFN-α2a-core fusion protein A549 and SW480 cancer cell surface-binding specificity analyzed in-vitro by the cell adhesion assay. (a) Flow cytometric analysis of αvβ3 integrin expression in different cell lines. (b) Cells were seeded into PBS-precoated, NR-precoated, Q1-precoated, or Q2-precoated 96-well plates and incubated for 1 h. After incubation, unbound cells were washed away with PBS and bound cells were fixed and stained with 0.5% crystal violet dye, viewed under an inverted microscope, and quantified. (c) Quantification of experiment B. The data are expressed as means±SEM from at least three independent experiments. **P<0.01 compared with the 0.1 nmol/l group. IFN, interferon; NR, nitrate reductase; RGD, arginine–glycine–aspartic acid.
Fig. 3.
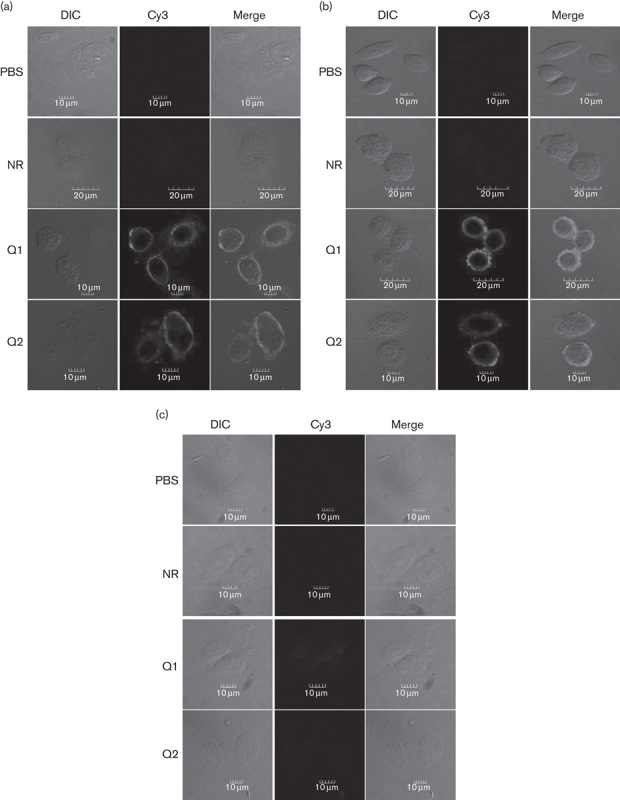
RGD-IFN-α2a-core fusion protein A549 and SW480 cancer cell surface-binding specificity analyzed in-vitro by immunofluorescence staining. Cells were seeded onto coverslips and treated with 10 nmol/l of NR, Q1, or Q2 for 12 h. The cells were then subjected to immunofluorescence staining with an anti-HCV core monoclonal antibody. (a) A549 cells, (b) SW480 cells, and (c) BEAS-2B cells. Cy, cyanine; DIC, differential interference contrast; HCV, hepatitis C virus; IFN, interferon; NR, nitrate reductase; RGD, arginine–glycine–aspartic acid.
RGD-IFN-α2a-core fusion proteins decrease tumor cell viability and increase apoptosis
We assessed the effects of the RGD-IFN-α2a-core fusion proteins on tumor cell viability and apoptosis. The MTT assay showed that both Q1 and Q2 reduced A549 and SW480 cell viability in a dose-dependent manner (Fig. 4) and that 10 nmol/l NR inhibited A549 and SW480 cells after 3 days of treatment. The antiproliferation effect of Q1 is more potent in A549 than in SW480 (Fig. 4).
Fig. 4.
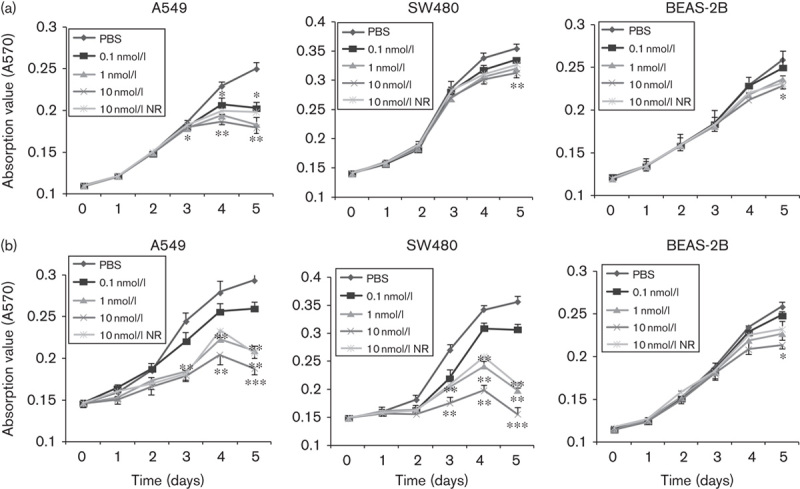
Effect of RGD-IFN-α2a-core fusion proteins on tumor cell viability. Cells were seeded into 96-well plates and treated with different concentrations of PBS, NR, Q1, or Q2 for up to 5 days and then subjected to cell viability MTT assays. (a) Q1 and (b) Q2. *P<0.05, **P<0.01 and ***P<0.001 compared with the negative control (PBS group) or the NR group. IFN, interferon; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; NR, nitrate reductase; RGD, arginine–glycine–aspartic acid.
We also observed Q1 and Q2 induced morphological changes in A549 and SW480 cells. Hoechst 33342 staining showed that, after Q1 or Q2 treatment, cell nuclei became agglutinated and smaller and the chromatin was partially condensed into small spherical or crescent shapes, or even apoptotic bodies (Fig. 5a). Flow cytometric apoptosis assays showed that Q1 and Q2 induced apoptosis in tumor cells (Fig. 5b and c). Caspase-3 activation was induced after 48 h of treatment with 10 nmol/l of either Q1 or Q2 (Fig. 5d and f).
Fig. 5.
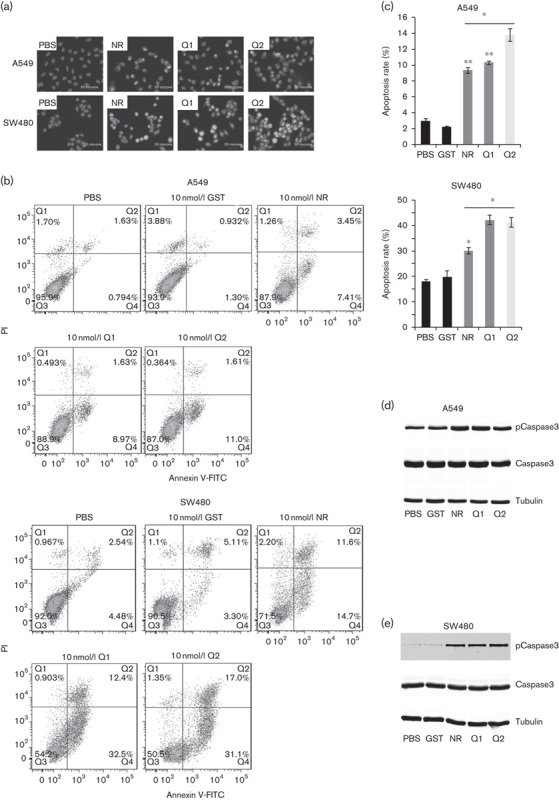
Effects of RGD-IFN-α2a-core fusion proteins on tumor cell apoptosis induction. (a) Hoechst 33342 staining. Cells were seeded onto coverslips and treated with 10 nmol/l of NR, Q1, or Q2 for 24 h and then subjected to Hoechst 33342 staining. (a–d) A549 cells treated with PBS, NR, Q1, or Q2; (d, e) SW480 cells treated with PBS, NR, Q1, or Q2. (b) Flow cytometric assay. The cells described above were subjected to flow cytometric assay after the treatments described above. (c) Summarized data of B. The data are expressed as means±SEM from at least three independent experiments. *P<0.05 and **P<0.01 compared with the control. (d) Western blot analysis of caspase-3 levels in A549 cell lines. Duplicated cells were subjected to western blot analysis. The values of PBS-treated, GST-treated, IFN-α2a-treated, Q1-treated, and Q2-treated tumor cells were expressed as means±SEM (n=5). (e) Western blot analysis of caspase-3 levels in SW480 cell lines. Duplicated cells were subjected to western blot analysis. The values of PBS-treated, GST-treated, NR-treated, Q1-treated, and Q2-treated tumor cells were expressed as means±SEM (n=5). GST, glutathionine S-transferase; IFN, interferon; NR, nitrate reductase; PI, propidium iodide; RGD, arginine–glycine–aspartic acid.
Discussion
The antitumor activity of IFN is well documented in the literature 7,9. In the current study, we constructed recombinant RGD-IFN-α2a-core fusion proteins to assess their antitumor activity against human cancer cells in vitro. Our data showed that the recombinant RGD-IFN-α2a-core fusion proteins specifically bound to tumor cells, but did not bind to normal cells. Recombinant RGD-IFN-α2a-core fusion protein treatment reduced the viability of non-small-cell lung cancer cells, and induced apoptosis. Future studies are needed to assess their in-vivo effects against human cancer.
IFN is produced by leukocytes and serves as an innate immune response against viral or other pathogenic infections, such as bacteria, parasites, or even tumor cells 7,10 11. In the human body, IFN production induces certain infection-related symptoms, including fever, muscle pain, and other flu-like symptoms, and triggers the immune system to eradicate pathogens 11. IFN also limits viral spread by increasing p53 activity and kills virus-infected cells by promoting apoptosis 12. IFN binds to specific cellular receptors and activates signal transducer and activator of transcription complexes. Signal transducer and activator of transcriptions are a family of transcription factors that regulate the expression of certain immune system genes. IFN is used widely in oncology clinics as an antitumor agent in individual or as combination therapy against human cancers 7,9. Our current study further confirmed the antitumor induction of apoptosis by IFN in human non-small-cell lung cancers.
We chose to conjugate IFN to a small molecule or peptide that could specifically bring IFN to tumor cells and destroy them. We utilized the RGD peptide, which is a class of short-chain peptides that can specially bind integrin αvβ3 on the surface of tumor cells. Previous studies confirmed that this RGD peptide could specifically bind integrin αvβ3 and inhibit its activity 13,14. All five αV integrins, two β1 integrins (α5 and α8), and IIb3 share or recognize the same ligands containing an RGD tripeptide active site. Crystal structures of αVβ3 and αIIbβ3 binding to RGD ligands showed identical atomic structure and interactions 15. RGD binds to an interface between the α and β subunits, the R residue fits a cleft in a propeller module in the subunit, and the D coordinates cation binding to a Von Willebrand factor A domain in the β subunit. The ligands which are recognized by the RGD binding integrins are the most promiscuous in the integrins family, with β3 integrins in particular binding to a large number of extracellular matrix and soluble vascular ligands 16. Furthermore, integrin is a transmembrane receptor that bridges cell–cell or cell–extracellular matrix interactions 17,18, and in turn integrin plays important roles in cellular adhesion, proliferation, apoptosis, and differentiation, as well as in tumor transformation, invasion, and metastasis 6. In our current study, we verified that the RGD-IFN-α2a-core fusion proteins specifically bound to the surface of SW480 and A549 cells, but did not bind to BEAS-2B human normal lung epithelial cells. Previous studies have reported integrin αvβ3 overexpression in various human cancer cells, including osteosarcoma, lung, breast, prostate, bladder, glioblastoma, invasive melanoma, and also the neovascular endothelial cells of tumor tissues 19. Thus, overexpression of integrin αvβ3 provides a novel target for personalized targeting therapy in a wide variety human cancer types.
A previous study showed that an HCV core and a peptide RGD could assemble into virus-like particles in a baculovirus expression system 20. To evaluate the effect of fusion proteins on tumor cell proliferation and apoptosis, we constructed recombinant plasmids with RGD, different length fragments of IFN-α2a and an HCV core, and purified the RGD-IFN-α2a-core fusion proteins Q1, Q2, and NR. These experiments were successful and we could generate a stable and functional fusion protein.
Our current study was just a proof-of-principle, and showed the antitumor activity of this recombinant RGD-IFN-α2a-core fusion protein in vitro. However, much more work remains to be done. Future studies are needed to assess the following: (a) the in-vivo antitumor activity of the RGD-IFN-α2a-core fusion proteins against human cancers; (b) the stability of the RGD-IFN-α2a-core fusion proteins in the human body; (c) the immune response of the human body to the RGD-IFN-α2a-core fusion proteins; and (d) the means to deliver the RGD-IFN-α2a-core fusion proteins to cancer patients, whether viral, plasmid vector, or even direct administration of the proteins.
Acknowledgements
This work was supported in part by grants from the Natural Science Foundation of Zhejiang Province (#LY15C070003 and #LY13H100003) and the Natural Science Foundation of China (#30900344).
X.L. conceived and designed the experiments; Z.W., Q.J., X.K., and L.Z. performed the experiments; X.L., Z.W., Q.J., X.K., and J.Y. analyzed the data; J.G. and Y.L. contributed the materials and analysis tools; X.L. and L.H. wrote the manuscript. All authors read and approved the final manuscript.
Conflicts of interest
There are no conflicts of interest.
Footnotes
Zhengwei Wen and Qunying Jia contributed equally to the writing of this article.
References
- 1.Huang SH, O’Sullivan B. Oral cancer: current role of radiotherapy and chemotherapy. Med Oral Patol Oral Cir Bucal 2013; 18:e233–e240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hensing TA. Clinical evaluation and staging of patients who have lung cancer. Hematol Oncol Clin North Am 2005; 19:219–235. [DOI] [PubMed] [Google Scholar]
- 3.Huveneers S, Truong H, Danen HJ. Integrins: signaling, disease, and therapy. Int J Radiat Biol 2007; 83:743–751. [DOI] [PubMed] [Google Scholar]
- 4.Garanger E, Boturyn D, Dumy P. Tumor targeting with RGD peptide ligands-design of new molecular conjugates for imaging and therapy of cancers. Anticancer Agents Med Chem 2007; 7:552–558. [DOI] [PubMed] [Google Scholar]
- 5.Von Wallbrunn A, Höltke C, Zühlsdorf M, Heindel W, Schäfers M, Bremer C. In vivo imaging of integrin αVβ3 expression using fluorescence-mediated tomography. Eur J Nucl Med Mol Imaging 2007; 34:745–754. [DOI] [PubMed] [Google Scholar]
- 6.Humphries JD, Byron A, Humphries MJ. Integrin ligands at a glance. J Cell Sci 2006; 119 (Pt 19):3901–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldstein D, Laszlo J. The role of interferon in cancer therapy: a current perspective. CA Cancer J Clin 1988; 38:258–277. [DOI] [PubMed] [Google Scholar]
- 8.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 1983; 65:55–63. [DOI] [PubMed] [Google Scholar]
- 9.Hauschild A, Gogas H, Tarhini A, Middleton MR, Testori A, Dréno B, Kirkwood JM. Practical guidelines for the management of interferon-alpha-2b side effects in patients receiving adjuvant treatment for melanoma: expert opinion. Cancer 2008; 112:982–994. [DOI] [PubMed] [Google Scholar]
- 10.Isaacs A, Lindenmann J, Valentine RC, Isaacs A, Lindenmann J, Valentine RC, et al. Virus interference. II. Some properties of interferon. Proc R Soc Lond B Biol Sci 1957; 147:268–273.13465721 [Google Scholar]
- 11.Fensterl V, Sen GC. Interferons and viral infections. Biofactors 2009; 35:14–20. [DOI] [PubMed] [Google Scholar]
- 12.Takaoka A, Hayakawa S, Yanai H, Stoiber D, Negishi H, Kikuchi H, et al. Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature 2003; 424:516–523. [DOI] [PubMed] [Google Scholar]
- 13.Moreno-Manzano V, Lucio-Cazana J, Konta T, Nakayama K, Kitamura M. Enhancement of TNF-alpha-induced apoptosis by immobilized arginine–glycine–aspartate: involvement of a tyrosine kinase-dependent, MAP kinase-independent mechanism. Biochem Biophys Res Commun 2000; 277:293–298. [DOI] [PubMed] [Google Scholar]
- 14.Davis ME, Chen ZG, Shin DM. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nat Rev Drug Discov 2008; 7:771–782. [DOI] [PubMed] [Google Scholar]
- 15.Xiao T, Takagi J, Coller BS, Wang JH, Springer TA. Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature 2004; 432:59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong JP, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin αVβ3 in complex with an Arg–Gly–Asp ligand. Science 2002; 296:151–155. [DOI] [PubMed] [Google Scholar]
- 17.Humphries MJ. Integrin structure. Biochem Soc Trans 2000; 28:311–339. [PubMed] [Google Scholar]
- 18.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell 2002; 110:673–687. [DOI] [PubMed] [Google Scholar]
- 19.Buckley CD, Pilling D, Henriquez NV, Parsonage G, Threlfall K, Scheel-Toellner D, et al. RGD peptides induce apoptosis by direct caspase-3 activation. Nature 1999; 397:534–539. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Xu X, Jin A, Jia Q, Zhou H, Kang S, et al. Self-assembled HCV core virus-like particles targeted and inhibited tumor cell migration and invasion. Nanoscale Res Lett 2013; 8:401. [DOI] [PMC free article] [PubMed] [Google Scholar]


