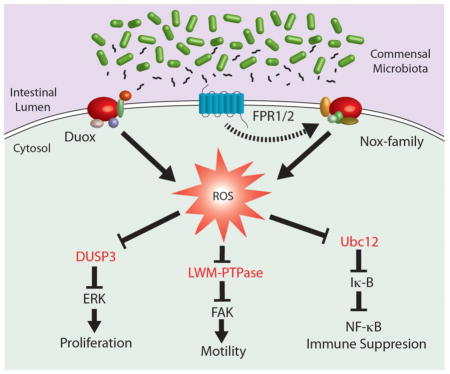
Abstract
The microbiota that inhabits the mammalian intestine can influence a range of physiological functions, including the modulation of immune responses, enhancement epithelial barrier function, and the stimulation of cell proliferation. While the mechanisms by which commensal prokaryotes stimulate immune signaling networks are well-characterized, less is known about the mechanistic control over homeostatic pathways within tissues. Recent reports by our research group have demonstrated that contact between the gut epithelia and some groups of enteric commensal bacteria prompts the rapid generation of reactive oxygen species (ROS) within host cells. Whereas the bacterial-induced production of ROS in phagocytes in response to ligand binding to Formyl Peptide Receptors (FPRs) and ensuing activation of NADPH oxidase 2 (Nox2) is a well-defined mechanism, ROS generated by other cell types such as intestinal epithelia in response to microbial signals via FPRs and the NADPH oxidase 1 (Nox1) is less appreciated. Importantly, enzymatically generated ROS have been shown to function as second messengers in many signal transduction pathways via the transient oxidative activity on sensor proteins bearing oxidant-sensitive thiol groups. Examples of redox sensitive proteins include tyrosine phosphatases that serve as regulators of MAPK pathways, focal adhesion kinase, as well as components involved NF-kB activation. Here, we review the leading edge discoveries gleaned from investigations that focus on microbial-induced generation of ROS and their functional effects on host physiology. These studies identify the functional molecular elements and mechanistic events that mediate the established effects of the normal microbiota on intestinal physiology.
Keywords: Probiotics, Nox enzymes, Reactive oxygen species, Formyl peptide receptors
Graphical Abstract
The intestinal physiology and eukaryotic-prokaryotic interactions
Symbiotic host-microbe communications has evolved in virtually every metazoan, with the human gut microbial population an example of increasingly documented medical significance [1]. In utero, the mammalian gut is sterile. The microbial colonization progression begins during birth, culminating in a diverse and stable community, though typically, microbial composition between individuals varies [2]. Recent advancements in high-throughput sequencing and bioinformatic methodologies have profoundly enhanced our knowledge of the diversity of the microbial population within the gut, revealing that the bulk of the microbial population are represented by Bacteroidetes and Firmicutes [3–6]. Particular taxa of the gut microbial population may be free-living in the luminal fecal stream thereby occupying a planktonic niche, or may be adherent to the gut mucous layer or to epithelial cells of the mucosa.
Microbes in the intestine thrive in the nutrient rich environment, with certain taxa contributing favorable influences to the host that include, but not limited to the production of short chain fatty acids and enhanced energy extraction from foodstuffs, competitive exclusion of pathogenic microorganisms, and priming of innate and adaptive immune system responses, and influences on bone homeostasis [6, 7]. In addition, investigations utilizing germ-free mice have established a function for the gut microbiome and metabolic regulation [8]. The gut microbiome has also been shown to positively influence homeostasis of the intestinal mucosa by enhancing barrier function, as well as epithelial cell proliferation and survival [9–15]. For instance, villi of the small intestine of the germ-free mice have impaired angiogenesis [16] and have slower turnover rates of epithelial cells [17]. Mono-association of germ-free mice with a gut symbiont (Bacteriodes thetaiotaomicron) evoked a vigorous host transcriptional response, showing that the host can actively sense, perceive, and respond to the presence of commensal of symbiotic within the lumen [18]. These data demonstrate the existence of an active dynamic association between host cell and microbes inhabiting the gut. Nevertheless, it is also illustrates that anomalies in the quality and diversity of the gut microbiome (“dysbiosis”) may be sufficient by itself to aggravate intestinal inflammation as seen in Inflammatory bowel diseases (IBD). In addition, changes in the diversity of the gut microbiome have been linked with infectious disease such as pseudomembranous colitis, in systemic immune disorders such as multiple sclerosis, in allergic diseases such as celiac disease and asthma, and in the onset of diabetes and obesity in metabolic syndromes in adults [19–21].
Over the past few years, investigations have focused on exploiting the positive influences of certain taxa of the gut microbiome by supplementing the indigenous microbiome with purified cultures of symbiotic bacteria. This tactic, called ‘probiotics’, has been successful in suppressing inflammation, strengthening barrier function, facilitating tissue repair in response to injury in the intestine, thus offering a therapeutic approach to ameliorate disorders of the intestinal tract [22]. Altogether, mounting evidence have empirically demonstrated that the gut microbiome favorably influences intestinal physiology. However, there is a gap in our knowledge pertaining to an understanding of how the intestinal cells sense symbiotic bacteria, and mechanistically modulate gut physiology. This manuscript will review recent discoveries that have identified a redox based response within cells, that is emerging as a critically conserved element of host cell and symbiotic microbe interactions.
Epithelial perception and monitoring of the microbiota
The intestinal mucosa encompasses the outward facing epithelial cells, the structural components of the underlying lamina propria, and the immune cells residing in sub-epithelial compartments, which together form a functional barrier preventing the luminal contents from entering systemic compartments. The gut luminal contents are physically separated from the host interior by a thin layer of mucus that overlays a monolayer of columnar epithelial cells. Within epithelial cells are apical surface factors that function in the uptake of nutrients while at the same time, existing in intimate contact with the luminal contents. This active process exists against a background of the intestinal epithelium continually renewing itself in a progression involving asymmetrical proliferation of stem cells, ensuing differentiation, migration and eventual programed apoptosis and shedding at the villous apex - a homeostatic cycle that occurs over 5–7 days in humans.
The intestine may become damaged following exposure to a pathogenic and/or immunologic insults. While overcoming and resolving damage to tissues, the gut mucosa must also maintain the beneficial relationship with the symbiotic taxa of the gut microbiota [23]. To succeed in this management, cells of the intestinal mucosa have dedicated sentinel elements for monitoring bacteria. Examples of such sentinel elements are Toll-like receptors (TLRs) and related Nod-like receptors (NLRs), collectively termed “pattern recognition receptors” (PRRs), that bind to motifs known as “microbe associated molecular patterns” (MAMPs) that are ubiquitously present across the bacterial phylogenetic domain [24]. Sensing of MAMPs by TLRs activate innate immune signaling cascades such as the MAPK and NF-kB pathways [25–28], which are by and large considered pro-inflammatory, although at lower ‘tonic’ levels of activation, have been connected to mechanisms of normal gut homeostasis [29, 30].
A distinct group of PRRs are the formylated peptide receptors (FPR), which are G-protein-linked seven membrane pass receptors originally discovered on the surface of professional phagocytes. There are three structurally-related FPRs in humans, known as FPR1, FPR2 and FPR3 [31]. Functionally, FPRs sense and bind to peptides that harbor a bacterial specific N-formyl group of which an example is N-formyl methionyl-leucyl-phenylalanine (fMLF). FPR1 binds to fMLF with particularly high-affinity with an ED50 in the nanomolar range, whereas FPR2 binds with lower affinity in the micromolar range. Furthermore, agonists generated by eukaryotic cells such as AnxA1, LXA4 and SAA, or mitochondrially derived translation products can also stimulate FPRs [32]. The importance of these receptors is highlighted by the observation that FPR1-deficient mice have augmented susceptibility to pathogens, suggesting that FPR1 is functions in supporting processes of acute inflammation [33]. In addition, mitochondrial derived formyl peptides are recognized as being the source of the agonists in non-infectious tissue injury (“sterile inflammation”). Here, mitochondria, which are prokaryotic endosymbionts, and as such, retain the prokaryotic translation machinery, including the bacterial-specific use of N-formyl-methionine capping of nascent transcripts. This means that formylated peptides are present inside cells within mitochondria. In a pathological context such as tissue necrosis (not apoptosis), cellular contents are released extracellularly and can be perceived by other cells and phagocytes as “danger associated molecular patterns” or DAMPS [34]. Once the agonist binds, FPRs are phosphorylated and glycosylated, which initiates interaction with pertussis toxin-sensitive Gi proteins [35–38]. Ensuring cell signaling cascades involves MAPK and phosphatidylinositol 3-kinase (PI3K) pathway activation, which together with small GTPases initiate phagocytic functions such as stimulation of actin dynamics and chemotaxis, and the activation of ROS generation by NADPH oxidase enzyme (Nox2) in a process known as the respiratory burst [39–41]. Indeed, it was the known function of agonist binding to FPRs eventuating in ROS generation in phagocytes that proved to be the rationale and the springboard for assessing the function of generated ROS in epithelial cells following in host-symbiotic bacterial interactions. This notion was substantiated with the discovery of functional FPRs located on the apical surface of the intestinal epithelial cells, suggesting that FPRs function in an similar physiological process in the intestinal mucosa [42]. Indeed, in epithelial cells, it was shown that MAPK pathway signaling is activated by fMLF by an FPR-dependent mechanism, and that fMLF binding to FPR also induced the generation of ROS in the epithelial cells [43].
The generation of physiological levels of reactive oxygen species (ROS)
Reactive oxygen species (ROS) generated at high levels by professional phagocytes mediate the capacity of these cells to kill bacteria. Agonist binding to FPRs in phagocytes initiate the respiratory burst, which involves extensive enzymatic production of superoxide within the vacuole harboring phagocytosed microbe. ROS generation in phagocytes is catalyzed by a multi component and membrane-bound NADPH oxidase enzyme called Nox2 (formerly designated gp120phox). Nox2 is basally a dimer of gp91phox and gp22phox [44], and considering the potential harmfulness of elevated superoxide levels to surrounding tissue, explicably, ROS generation is firmly controlled by the consecutive recruitment of separate subunits of the Nox2 enzyme. The function of Nox2 in vivo is demonstrated by the discovery that Nox2 null mice develop chronic granulomatous disease (CGD), which is a disorder in patients where phagocytes are unable to produce ROS and are thus highly susceptible to repeated pyogenic infections.
Nox2/gp120phox was first protein of the NADPH oxidase group or “Nox’es” to be discovered. Importantly, Nox enzymes are also functional in non-phagocytic cell types and tissues, with the discovery that Nox1 and Duox2 are strongly expressed in enterocytes, which are in close proximity to the gut luminal bacterial content being relevant to this review [45–48]. Generally, the Nox enzymes functional within non-phagocytic cells have analogous, but not identical subunit regulation and assembly to Nox2 in phagocytes. For example, Nox1 requires Rac-GTPase-initiated cascades for its enzymatic activity, whereas Duox2 activity is calcium dependent. Nox1 catalyzed generation of ROS within epithelial cells is hypothesized to function in regulating several signal transduction pathways, which was detected following the stimulation of Nox1 function by growth factors, hormones, and cytokines in a wide range of cells and tissues [45, 47]. Indeed, Nox enzymes orthologs catalyze ROS production in a breadth of multi cellular organisms [49–52]. In plants, ROS generated Nox enzymes regulates the transition from proliferation to differentiation in root tips [53]. In flies, Duox-generated ROS in gut epithelia functions in the control of the diversity of the intestinal microbiome [54–56], which implies a role for ROS generation in epithelial cells in host cell and microbe interactions. However, by and large, the functions of Duox in the epithelia are less studied, but have been implicated more in anti-microbial functions that in signaling events. Pertaining to the catalytic activity of Nox1 and Duox2, both enzymes are involved in trans-membrane generation of superoxide generation which is generally thought to be rapidly dismutated to H2O2. Thereafter, H2O2 may be transported back across the plasma membrane, likely via aquaporins, for cytoplasmic signaling functions [57–59].
Redox signaling and the oxidation of reactive cysteine residues
Non-radical ROS, such as H2O2, that are generated by Nox enzymes in non-phagocytic tissues, are well documented as functioning in a variety of signaling pathways within a variety of cell types [60]. The diverse biological outcomes induced by ROS depends on the specific species generated, the as well as the duration, and the subcellular locations of generation [47, 52]. Because they are extremely short-lived, ROS have slight functional radii, which also means they can precisely discriminate their influence. Indeed, some receptors physically interact with Nox enzymes, apparently to bound the influences of the generated ROS to the vicinity of target proteins [61].
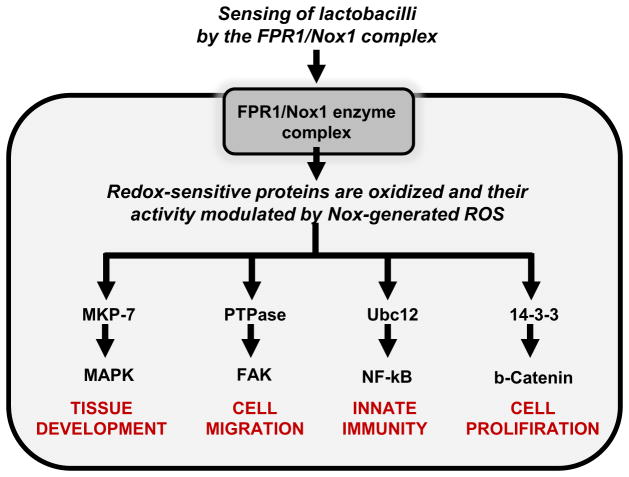
The mechanism through which ROS modulate cell signaling is through their capacity to oxidize certain reactive cysteine residues within enzymes controlling the activation of cell signaling pathways [62–64]. Proteins that harbor reactive cysteine residues can therefore function redox sensors and transducers of signaling initiated by elevated ROS. Moreover, reversible oxidation of cysteine residues allows for graded response to intracellular ROS concentrations, meaning that the cell can sense and respond to fluctuating levels of ROS. At the molecular level, the vast majority of cysteine residues are protonated at physiological pH (Cys-SH) (pKa ~8.5) and are unreactive at physiological pH. It is only a few cysteine residues that have ROS-sensitive properties, which due to them being charged at a low pKa, and thus extant as thiolate anions (Cys-S-) at physiological pH. Low pKa cysteine residues are highly reactive and are easily oxidized by ROS such as H2O2 [64]. Mass spectrometry analysis has revealed that redox-sensitive thiolates are present in a limited subset of enzymes. Examples include protein tyrosine phosphatases (PTP) [65], the lipid phosphatase (PTEN) [62, 66], MAP kinase phosphatases (MAPKP) such as DUSP3 [63], and low-molecular-weight protein tyrosine phosphatases (LMW-PTP) [67], in enzymes involved in the sumoylation and neddylation reactions, and well as in oxidant sensors such as Keap1, which control of overall redox balance of the cell. (Figure 1).
Figure 1.
Host signaling events controlled by symbiotic bacterial-induced cellular ROS generation. Symbiotic bacteria residing in the gut lumen stimulate intestinal tissue homeostatic events via the reversible activation of cellular redox signaling processes. The gut microbiota generate formylated peptides that are sensed by formyl peptide receptors (FPRs) situated on the apical surface of epithelial cells. Receptor binding initiates a singling cascade that trigger the NADPH oxidase Nox1 to catalyze localized ROS generation, which then oxidizes critical cysteine residues and the regulatory influence of redox sensor proteins including the Nedd8 ligase, Ubc12, DUSP3, and LMW-PTPase. Ensuing signaling processes influence gut physiology by including stem cell proliferation, epithelial cell motility, and dampening of innate immune responses.
Symbiotic bacterial-induced ROS generation in intestinal epithelial cells
Symbiotic bacterial-induced ROS generated through catalytic activities of Nox enzymes have been detected in numerous forms of multicellular organisms, extending from social amoebae, to florae, and to humans. Thus it is thought that ROS signaling represents an evolutionary ancient form of host cell and microbe cross-talk [49, 54, 68, 69]. Of late, our research group and others reported that contact of certain symbiotic taxa with host epithelial cells can induce the generation of rapid, non-pathogenic levels of ROS, in a process that requires the catalytic activity of Nox1 in host cells. Furthermore, contact of intestinal epithelial cells with bacteria of the lactobacilli taxon induced increased oxidation of proteins that function as soluble redox sinks, including thioredoxin and glutathione, and result in the induction of the transcription of redox-stimulated modulons such as the Nrf2 pathway activity. These observations point to an active and dynamic response to increased levels of cellular ROS. Furthermore, specific bacterial contact with epithelial cells have regulatory effects on cytoskeletal dynamics and on host immune activity [14, 43, 70]. Of these taxa, diverse commensal bacteria elicit distinctly varying capacities of inducing cellular ROS generation, with lactobacilli particularly powerful inducers of ROS generation, although the majority of bacteria assayed exhibited some degree of ability to induce ROS generation in host epithelial cells. Lactobacilli have been shown to harbor membrane components or secreted factors that trigger cellular responses. For example, it has been demonstrated that soluble factors produced by Lactobacillus rhamnosus GG mediate beneficial effects in in vivo injury models [71]. In addition, lactobacilli that stimulate ROS may also have enhanced ability to penetrate the mucus layer, or have enhanced adhesive properties to come into contact with cellular receptors such as FPRs or TLRs.
While levels of ROS produced by the action of non-phagocyte NADPH oxidases are indeed orders of magnitude less than the output of Nox2 in phagocytes, ROS production is not so vanishingly small that it is beyond the sensitivity of current biochemical assays. For example, ROS produced can be visualized by a variety of redox sensitive dyes in immune fluorescence. In our hands, the most faithful reporters of ROS generation in tissues are the ROSstarTM hydrocyanine probes [72]. These probes are specific for oxygen radicals, superoxide, and hydroxyl radicals and detect intracellular ROS with high sensitivity and specificity. The probes are cell-permeable and initially non-fluorescent. The probes become fluorescent upon oxidation. The probes are used to detect oxidative stress by fluorescence microscopy, flow cytometry, and can be quantified by microplate fluorometry. In addition, images captured by fluorescence microscope may be were quantified using image densitometry software.
Physiological outcomes of symbiotic bacteria-induced redox signaling
Effects on inflammatory signaling: Extensive reports have described the suppressive activity of lactobacilli on host inflammatory signaling pathways, having the net effect of dampening the ensuing innate immunity [71, 73–75]. For example, intestinal microbes can modulate gut inflammation, and probably additional cellular processes by modulating the ubiquitin-proteosome pathway [76]. Contact of lactobacilli with epithelial cells has the effect of inhibiting IκB ubiquitination by interfering with the IκB ubiquitin ligase, SCFβTrCP (Skp1, Cdc53/Cullin, F box receptor), thereby blocking activation of the pro-inflammatory NF-κB pathway [77–79]. Specifically, for activation of the SCFβTrCPcomplex, a covalent modification involving neddylation of the cullin-1 (Cul-1) regulatory subunit must occur. Oxidative signaling negatively regulates neddylation by the reversible inactivation of Ubc12, which is a redox sensitive Nedd8 ligase [70]. Contact of epithelial cells with lactobacilli induces rapid loss of the Nedd8 modification, thereby inhibiting SCF ubiquitin ligase function, and the suppression of NF-κB pathway activity [78]. Together, ROS generation in the gut epithelium modulates SCF ligase activity, and the NF-κB pathway by controlling the balance between neddylated and un-neddylated Cul-1. Interestingly, other pathways are known to be regulated by E3-SCFβTrCP, such as β-catenin, Snail, Twist and Hedgehog [80], pointing to further molecular influences that lactobacilli-induced generation of ROS could regulate various facets of intestinal physiology.
Effects on epithelial cell motility. As stated previously, germ-free mice exhibit impaired gut physiology including proliferation and wound healing rates of the epithelial layer, showing that the enteric luminal contents have potent influences on host cells. Furthermore, germ-free mice have impaired rates of epithelial cell migration, which is a process that is regulated by the exquisitely coordinated restructuring of the actin cytoskeleton at the advancing edge of the cell to specialized signaling nidus points called focal adhesions (FA) of the extracellular matrix. The dynamics of the FA assembly is controlled by an enzyme called focal adhesion kinase (FAK), which is a 125 kDa protein that is held in its inactive state under the dephosphorylating influences of the redox-sensitive tyrosine phosphatases LMW-PTPase and SHP-2. However, interactions between growth factors and integrins at the basement membrane triggers Nox1 catalyzed ROS production which oxidative inactivates PTPase, resulting in releasing FAK from its dephosphorylated state thereby initiating FA turnover and cell motility [81]. Concerning the influence of the gut microbe population on cell motility, we have shown that contact between intestinal epithelia with symbiotic bacteria such as some lactobacilli strains induces generation of ROS in these cells, particularly at the edges of in vivo colonic biopsy wounds, and at the leading edge of the migrating cells. Mechanistically, the ROS generated in response to cell contact with lactobacilli reversibly oxidizes low pKa cysteines within LMW-PTP and SHP-2, thereby activating FAK activity and FA dynamic assembly and disassembly at the migrating edge of the monolayer. These molecular mechanisms induce augmented epithelial migration rates as demonstrated in improved wound sealing of in vitro and in vivo injury models [66]. Additionally, we showed that fMLF triggers FPR-dependent ROS generation in epithelial cells. Importantly, in either FPR or Nox1 null mice, lactobacilli-induced ROS generation, ERK and FAK phosphorylation, and improved wound healing are abolished. Consequently, these are molecular evidence for events that following ROS production associated with FPR/Nox1 dependent ROS generation following lactobacilli-epithelial contact, which eventuate in faster epithelial motility. It was also demonstrated that ROS generated by Nox are necessary for the function of invadopodia, which are actin-rich membrane protrusions in cells. The study proposed that the invadopodia protein Tks5 is a part of the Nox complex, and showed that depletion of Tks5 levels reduces total ROS amounts in cells [82]. These data demonstrate a further mechanism by which the microbiota positively influences physiological processes within the gut mucosa and may also point to possible mechanisms by which probiotics exert their beneficial influence on epithelial barrier integrity.
Effects on epithelial growth and differentiation: The intestinal gut epithelium is the most dynamically renewing tissue in the adult body. The epithelium is a three dimensional architecture of invaginated crypts and projecting villi. At the base of the crypts reside multipotent stem cell niches that are the source of epithelial renewal and homeostasis. Stem cell undergo asymmetric division where the cell targeted for a differentiated fate migrates to a transient amplifying compartment where further signaling events eventuate in the specification of the cells as they absorptive enterocytes, mucus secreting goblet cells, or neuroendocrine epithelial cells. Germ-free mice have been shown to have slower rates of cell migration up the crypt-villus axis, while studies in germ-free Drosophila corroborate distinctly curbed proliferation rates of epithelial precursor cells, pointing to a role for the luminal microbiota in controlling the intestinal epithelial development and homeostasis [83, 84]. Importantly, ROS act as mediators of cell proliferation and differentiation in wide variety of unrelated systems such as in the growing plant root hair [53], to Drosophila hematopoiesis [85]. Our research group demonstrated lactobacilli-induced ROS generation leads to the activation of the pro-proliferation and developmental ERK pathway by a mechanism involving the redox inactivation of the ERK phosphatase DUSP6 [14, 43, 66]. Furthermore, Nox1 generation of ROS has been reported to regulate Wnt and Notch1 pathway signaling in the colon [86]. Indeed, the function of Nox1 as pivotal factor in cell fate through Wnt/beta-catenin and Notch1 signaling pathways was shown in Nox1-deficient mice which had elevated numbers of goblet cells as a result of PI3K/AKT/Wnt/beta-catenin and Notch1 signaling repression [86]. By extension, lactobacilli-induced and Nox dependent redox signaling may also function in gut development by similar mechanisms.
Cytoprotection by lactobacilli-activation of Keap1/Nrf2/ARE signaling
Another example of host signaling circuitry that responds to ROS generation in the cytoplasm of cells is the Keap1/Nrf2/ARE signaling module. Nrf2 (NF-E2-Related Factor 2) and its antagonist Keap1 (Kelch-like ECH-Associated Protein 1) are essential factors that mediate cytoprotection in response to xenobiotics [87]. The Keap1/Nrf2/ARE module is evolutionarily conserved across kingdoms with examples in Caenorhabditis elegans [88], D. melanogaster [89], zebrafish [90], and in mice [91]. Nrf2 activity is regulated by the binding action of its agonist and inhibitor, Keap1 [92]. Under non-stimulated situations of low cellular ROS levels, Keap1 attaches to Nrf2 and directing Nrf2 to Cullin-dependent E3 ubiquitin ligase proteosomal degradation. However, electrophilic stress in the cytoplasm, typically in situations of elevated ROS levels, results in the oxidation of low pKa cysteine residues within Keap1, thereby causing a conformational change in Keap1 structure that results in its disassociation from Nrf2. Nrf2 is therefore free to translocate the nuclear membrane where it associates with a conserved DNA sequence known as antioxidant response elements (ARE) which are situated in the promoters of a battery of cytoprotective factor genes [93]. Investigation of the result of bacterial-induced ROS generation on Nrf2 pathway signaling discovered that lactobacilli-induced, and Nox1 catalyzed generation of ROS triggered the transcription of Nrf2-responsive cytoprotective elements, and resulted in organismal cytoprotection against oxidative stress in Drosophila, and against radiological insult in mice [94]. Thus, the Nrf2 signaling pathway is yet another mechanism whereby host cells perceive and respond to bacterial contact, this time to activate cytoprotection within cells (Figure 2).
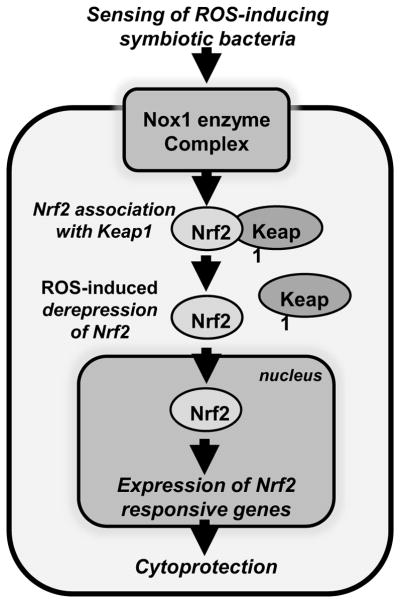
Figure 2.
Activation of the redox-sensitive Nrf2 cytoprotective pathway by lactobacilli-induced generation of cellular ROS. Under homeostatic conditions, Keap1 binds to Nrf2 and inhibits its nuclear translocation. Generation of lactobacilli-induced ROS catalyzed by Nox1 oxidizes cysteine residues within Keap1, resulting in conformation change and release of binding from Nrf2, allowing it to translocate into the nucleus. Nrf2 then binds to an anti-oxidant response DNA promoter element and induces the transcription of a battery of Nrf2-responsive genes including a plethora of cytoprotective factors. The Nrf2 responsive gene products protect macromolecules from oxidative damage thereby promoting cell survival and preserving tissue physiological integrity.
Because we established that lactobacilli induce Nrf2 signaling, this unlocks the prospect of recognizing mechanisms by which probiotic bacteria may elicit beneficial effects on disease states that involve Nrf2 pathway signaling. As stated previously, Nrf2 signaling has been widely investigated in relation to cytoprotection from xenobiotic stresses, whose presence in the cell induces the basal Nrf2 transcriptional regulon of several hundred genes [95]. Importantly, studies into Nrf2 function exposed a plethora of cellular processes other than cytoprotection which are regulated by its signaling. These include diabetes [96], cancer cell growth and chemoresistance [97–99], neurodegenerative diseases [100], redox homeostasis in the aging heart [101], as well as oxidative stress and inflammatory pathways [102]. Furthermore, ROS are generated as by products during inflammation in the gut epithelia, primarily due to respiratory burst activity from phagocytes described earlier in this review. At the site of injury, Nrf2-responsive elements defend stem cell populations and promote restitutive cell proliferation and migration [103]. Indeed, each of the listed examples of cellular processes that are influenced Nrf2, are by extension potentially modulated by lactobacilli (probiotic) stimulation of Nrf2 pathway that would augment cytoprotective and reparative responses.
Future Perspectives
Experimental evidence generated by our research group demonstrate that intestinal epithelial cells generation ROS in response to contact with symbiotic bacteria, by mechanisms involving receptors and enzymatic processes similar to those evolved in phagocytic cells to induce microbial death. Evidence from the Drosophila model point to the notion that ROS generation in the gut epithelia may represent an evolutionary conserved response to microbes [55]. In mice, ROS generated in epithelial cells in response to lactobacilli undoubtedly functions in a signaling cell signaling events through reversible redox inactivation of regulatory proteins [52, 65]. Importantly, leading edge proteomic methodology can be employed as a screening system to identify microbial-specific, oxidant-sensitive regulatory proteins [104]. Identification of the function of identified reactive cysteine residues within redox-sensitive proteins in vivo will be challenging future work.
The strength and duration of lactobacilli-ROS-mediated signaling may vary with quantitative and qualitative variations in bacterial populations, for example, as would occur as a result of antibiotic administration, probiotic use, dietary changes, or the acquisition of the microbiota following birth. The population density of the gut microbiota differs by several orders of magnitude along the course of the digestive tract, with the peak density and numbers occurring in the cecum. The significance of this is that various regions of the intestine are likely to undergo dissimilar levels of bacterial-induced ROS. Furthermore, the fact that specific bacterial taxa such as lactobacilli exhibit potent ROS-inducing activity ties with the idea that qualitative changes in this taxa can negatively influence host biology. Conceivably, the relative amounts of a particular microbial taxa in the intestinal lumen might result in specific physiological outcomes at the organism level. A comprehension of the association amongst microbes and cellular ROS generation will contribute towards for describing the “eubiotic” and the “dysbiotic” population diversity that lead to positive health or inflammatory disease, and certainly has implications for the characterization of new types of probiotics. In addition, lasting cellular adaptation to the intimate bacterial company, as occurs in the colon, or short term contact, as occurs in the small intestine may also have differential outcomes on redox biology.
Future studies should also consider the influence of tissue oxygen partial pressure (pO2) at the interface of the epithelium and lumen, and its potential modulating influence on ROS/Nox signaling. A non-invasive method of determining tissue pO2 by electron paramagnetic resonance (EPR) oximetry showed oxygen levels of less than 2% in the intestinal lumen, 8% in the small intestinal wall, and 3 % at the villus tip [105]. These conditions of physiological oxygenation were termed physioxia. However, we detected similar qualitative ROS generation along the crypt villus axis, including the apex, where tissue pO2 is lowest [13]. Very little is known about how pO2 in the gut influences Nox activity, although reports using in vitro cultured cells claim that mRNAs of nox enzymes are sensitive to oxygen tension [106]. Nevertheless, our research group did described mucosal tissue oxygen depletion in the context of active neutrophilic inflammation, presumably due to oxygen consumption by emigrated phagocytes [11]. It is possible that, in this context, oxygen depletion my limit ROS production.
Another subject of intense investigation in our laboratory at the moment is to determine the duration and persistence of lactobacilli-induced ROS generation. Our current hypothesis holds that this type of generation is acute and highly localized to sub-cellular organelles, and to specific cell types. The influence of ROS in the cytoplasm is undoubtedly short-lived due the presence of redox sinks such as glutathione-s-transferase (GSH) in the cytoplasm, and the activity of redox sensitive transcriptional pathways such as Nrf2, which serve to upregulate antioxidant gene products. In addition, Nox enzyme activity is presumably deactivated by currently unknown mechanisms. In all, we hypothesize that ROS signaling appears to be induced within minutes, with consequent modulation of protein activity and transcriptional responses in several hours, followed by a re-establishment of basal conditions. This pulsatile nature of ROS kinetics are also consistent with feeding cycles. Another variable to consider is the region of the gastrointestinal tract that ROS signaling is strongest. Areas of the gut, such as the upper small intestine, have vastly less microbial population than the colon. We have also demonstrated the importance of the mucus layer for limiting ROS activation, and that the loss of mucus seen in injury is associated with markedly increased ROS production [11]. Thus, future investigations must focus and consider the effects of local micro- and macro- environmental, temporal, bacterial taxa dependent, and disease related influences on microbial mediated ROS generation.
In conclusion, microbial contact-induced epithelial ROS generation is an extremely conserved phenomenon across phyla with several known, and expected physiological consequences. This mechanism is an universal and non-discriminating means by which bacterial communities can effect a variety of signaling and homeostatic processes in the host [79]. It is anticipated that a comprehensive understanding of this association will improve our knowledge pertaining to the function of the gut microbiota, and its dysregulation on gut physiology.
Highlights.
Bacterial contact with host epithelial cells can result in the enzymatic generation of reactive oxygen species and consequent redox signaling.
ROS is produced by mechanisms similar to the oxidant burst in phagocytes, involving formyl peptide receptors and NAPDH oxidases.
Distinct bacterial taxa have very different abilities to stimulate redox signaling
Redox signaling is involved in immune regulation, cytoprotection and control of cellular motility and proliferation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136(1):65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, Henrissat B, Coutinho PM, Minx P, Latreille P, Cordum H, Van Brunt A, Kim K, Fulton RS, Fulton LA, Clifton SW, Wilson RK, Knight RD, Gordon JI. Evolution of Symbiotic Bacteria in the Distal Human Intestine. PLoS Biol. 2007;5(7):e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449(7164):811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312(5778):1355–9. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-Bacterial Mutualism in the Human Intestine. Science. 2005;307(5717):1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 7.Li JY, Chassaing B, Tyagi AM, Vaccaro C, Luo T, Adams J, Darby TM, Weitzmann MN, Mulle JG, Gewirtz AT, Jones RM, Pacifici R. Sex steroid deficiency-associated bone loss is microbiota dependent and prevented by probiotics. J Clin Invest. 2016;126(6):2049–63. doi: 10.1172/JCI86062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Seminars in Immunology. 2007;19(2):59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Ismail AS, Hooper LV. Epithelial cells and their neighbors. IV. Bacterial contributions to intestinal epithelial barrier integrity. Am J Physiol Gastrointest Liver Physiol. 2005;289(5):G779–84. doi: 10.1152/ajpgi.00203.2005. [DOI] [PubMed] [Google Scholar]
- 10.Madsen K, Cornish A, Soper P, McKaigney C, Jijon H, Yachimec C, Doyle J, Jewell L, De Simone C. Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterology. 2001;121(3):580–91. doi: 10.1053/gast.2001.27224. [DOI] [PubMed] [Google Scholar]
- 11.Alam A, Leoni G, Quiros M, Wu H, Desai C, Nishio H, Jones RM, Nusrat A, Neish AS. The microenvironment of injured murine gut elicits a local pro-restitutive microbiota. Nat Microbiol. 2016;1:15021. doi: 10.1038/nmicrobiol.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alam A, Leoni G, Wentworth CC, Kwal JM, Wu H, Ardita CS, Swanson PA, Lambeth JD, Jones RM, Nusrat A, Neish AS. Redox signaling regulates commensal-mediated mucosal homeostasis and restitution and requires formyl peptide receptor 1. Mucosal Immunol. 2014;7(3):645–55. doi: 10.1038/mi.2013.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones RM, Luo L, Ardita CS, Richardson AN, Kwon YM, Mercante JW, Alam A, Gates CL, Wu H, Swanson PA, Lambeth JD, Denning PW, Neish AS. Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J. 2013;32(23):3017–28. doi: 10.1038/emboj.2013.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wentworth CC, Alam A, Jones RM, Nusrat A, Neish AS. Enteric commensal bacteria induce extracellular signal-regulated kinase pathway signaling via formyl peptide receptor-dependent redox modulation of dual specific phosphatase 3. J Biol Chem. 2011;286(44):38448–55. doi: 10.1074/jbc.M111.268938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones RM. The Influence of the Gut Microbiota on Host Physiology: In Pursuit of Mechanisms. Yale J Biol Med. 2016;89(3):285–297. [PMC free article] [PubMed] [Google Scholar]
- 16.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proceedings of the National Academy of Sciences. 2002;99(24):15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proceedings of the National Academy of Sciences. 2005;102(1):99–104. doi: 10.1073/pnas.0405979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hooper LV, Gordon JI. Commensal Host-Bacterial Relationships in the Gut. Science. 2001;292(5519):1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 19.Sartor RB. Microbial Influences in Inflammatory Bowel Diseases. Gastroenterology. 2008;134(2):577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 20.Noverr MC, Huffnagle GB. Does the microbiota regulate immune responses outside the gut? Trends Microbiol. 2004;12(12):562–8. doi: 10.1016/j.tim.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 21.Wills-Karp M, Santeliz J, Karp CL. The germless theory of allergic disease: revisiting the hygiene hypothesis. Nat Rev Immunol. 2001;1(1):69–75. doi: 10.1038/35095579. [DOI] [PubMed] [Google Scholar]
- 22.Hord NG. Eukaryotic-Microbiota Crosstalk: Potential Mechanisms for Health Benefits of Prebiotics and Probiotics. Annu Rev Nutr. 2008;28(15):1–17. doi: 10.1146/annurev.nutr.28.061807.155402. [DOI] [PubMed] [Google Scholar]
- 23.Sansonetti PJ. War and peace at mucosal surfaces. Nat Rev Immunol. 2004;4(12):953–64. doi: 10.1038/nri1499. [DOI] [PubMed] [Google Scholar]
- 24.Jones RM, Neish AS. Recognition of bacterial pathogens and mucosal immunity. Cell Microbiol. 2011;13(5):670–6. doi: 10.1111/j.1462-5822.2011.01579.x. [DOI] [PubMed] [Google Scholar]
- 25.Ishii KJ, Koyama S, Nakagawa A, Coban C, Akira S. Host innate immune receptors and beyond: making sense of microbial infections. Cell Host Microbe. 2008;3(6):352–63. doi: 10.1016/j.chom.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Jones RM, Sloane VM, Wu H, Luo L, Kumar A, Kumar MV, Gewirtz AT, Neish AS. Flagellin administration protects gut mucosal tissue from irradiation-induced apoptosis via MKP-7 activity. Gut. 2011;60(5):648–57. doi: 10.1136/gut.2010.223891. [DOI] [PubMed] [Google Scholar]
- 27.Zeng H, Wu H, Sloane V, Jones R, Yu Y, Lin P, Gewirtz AT, Neish AS. Flagellin/TLR5 responses in epithelia reveal intertwined activation of inflammatory and apoptotic pathways. Am J Physiol Gastrointest Liver Physiol. 2006;290(1):G96–G108. doi: 10.1152/ajpgi.00273.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vijay-Kumar M, Wu H, Jones R, Grant G, Babbin B, King TP, Kelly D, Gewirtz AT, Neish AS. Flagellin suppresses epithelial apoptosis and limits disease during enteric infection. Am J Pathol. 2006;169(5):1686–700. doi: 10.2353/ajpath.2006.060345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of Commensal Microflora by Toll-Like Receptors Is Required for Intestinal Homeostasis. Cell. 2004;118(2):229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 30.Lee J, Mo JH, Katakura K, Alkalay I, Rucker AN, Liu YT, Lee HK, Shen C, Cojocaru G, Shenouda S, Kagnoff M, Eckmann L, Ben-Neriah Y, Raz E. Maintenance of colonic homeostasis by distinctive apical TLR9 signalling in intestinal epithelial cells. Nat Cell Biol. 2006;8(12):1327–36. doi: 10.1038/ncb1500. [DOI] [PubMed] [Google Scholar]
- 31.Migeotte I, Communi D, Parmentier M. Formyl peptide receptors: a promiscuous subfamily of G protein-coupled receptors controlling immune responses. Cytokine Growth Factor Rev. 2006;17(6):501–19. doi: 10.1016/j.cytogfr.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 32.Le Y, Murphy PM, Wang JM. Formyl-peptide receptors revisited. Trends Immunol. 2002;23(11):541–8. doi: 10.1016/s1471-4906(02)02316-5. [DOI] [PubMed] [Google Scholar]
- 33.Gao JL, Lee EJ, Murphy PM. Impaired antibacterial host defense in mice lacking the N-formylpeptide receptor. J Exp Med. 1999;189(4):657–62. doi: 10.1084/jem.189.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, Hauser CJ. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464(7285):104–7. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ali H, Richardson RM, Tomhave ED, Didsbury JR, Snyderman R. Differences in phosphorylation of formylpeptide and C5a chemoattractant receptors correlate with differences in desensitization. J Biol Chem. 1993;268(32):24247–54. [PubMed] [Google Scholar]
- 36.Tardif M, Mery L, Brouchon L, Boulay F. Agonist-dependent phosphorylation of N-formylpeptide and activation peptide from the fifth component of C (C5a) chemoattractant receptors in differentiated HL60 cells. J Immunol. 1993;150(8 Pt 1):3534–45. [PubMed] [Google Scholar]
- 37.Bokoch GM, Gilman AG. Inhibition of receptor-mediated release of arachidonic acid by pertussis toxin. Cell. 1984;39(2 Pt 1):301–8. doi: 10.1016/0092-8674(84)90008-4. [DOI] [PubMed] [Google Scholar]
- 38.Lad PM, Olson CV, Smiley PA. Association of the N-formyl-Met-Leu-Phe receptor in human neutrophils with a GTP-binding protein sensitive to pertussis toxin. Proc Natl Acad Sci U S A. 1985;82(3):869–73. doi: 10.1073/pnas.82.3.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jesaitis AJ, Naemura JR, Sklar LA, Cochrane CG, Painter RG. Rapid modulation of N-formyl chemotactic peptide receptors on the surface of human granulocytes: formation of high-affinity ligand-receptor complexes in transient association with cytoskeleton. J Cell Biol. 1984;98(4):1378–87. doi: 10.1083/jcb.98.4.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sklar LA, Hyslop PA, Oades ZG, Omann GM, Jesaitis AJ, Painter RG, Cochrane CG. Signal transduction and ligand-receptor dynamics in the human neutrophil. Transient responses and occupancy-response relations at the formyl peptide receptor. J Biol Chem. 1985;260(21):11461–7. [PubMed] [Google Scholar]
- 41.Jesaitis AJ, Tolley JO, Painter RG, Sklar LA, Cochrane CG. Membrane-cytoskeleton interactions and the regulation of chemotactic peptide-induced activation of human granulocytes: the effects of dihydrocytochalasin B. J Cell Biochem. 1985;27(3):241–53. doi: 10.1002/jcb.240270306. [DOI] [PubMed] [Google Scholar]
- 42.Babbin BA, Jesaitis AJ, Ivanov AI, Kelly D, Laukoetter M, Nava P, Parkos CA, Nusrat A. Formyl peptide receptor-1 activation enhances intestinal epithelial cell restitution through phosphatidylinositol 3-kinase-dependent activation of Rac1 and Cdc42. J Immunol. 2007;179(12):8112–21. doi: 10.4049/jimmunol.179.12.8112. [DOI] [PubMed] [Google Scholar]
- 43.Wentworth CC, Jones RM, Kwon YM, Nusrat A, Neish AS. Commensal-epithelial signaling mediated via formyl peptide receptors. Am J Pathol. 2010;177(6):2782–90. doi: 10.2353/ajpath.2010.100529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng G, Lambeth JD. NOXO1, Regulation of Lipid Binding, Localization, and Activation of Nox1 by the Phox Homology (PX) Domain. J Biol Chem. 2004;279(6):4737–42. doi: 10.1074/jbc.M305968200. [DOI] [PubMed] [Google Scholar]
- 45.Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol. 2004;4(3):181–9. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- 46.Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87(1):245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 47.Ogier-Denis E, Mkaddem SB, Vandewalle A. NOX enzymes and Toll-like receptor signaling. Semin Immunopathol. 2008 doi: 10.1007/s00281-008-0120-9. [DOI] [PubMed] [Google Scholar]
- 48.Ardita CS, Mercante JW, Kwon YM, Luo L, Crawford ME, Powell DN, Jones RM, Neish AS. Epithelial adhesion mediated by pilin SpaC is required for Lactobacillus rhamnosus GG-induced cellular responses. Appl Environ Microbiol. 2014;80(16):5068–77. doi: 10.1128/AEM.01039-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kotchoni SO, Gachomo EW. The reactive oxygen species network pathways:an essential prerequisite for perception of pathogen attack and the acquired disease resistance in plants. J Biosci. 2006;31(3):389–404. doi: 10.1007/BF02704112. [DOI] [PubMed] [Google Scholar]
- 50.Pauly N, Pucciariello C, Mandon K, Innocenti G, Jamet A, Baudouin E, Herouart D, Frendo P, Puppo A. Reactive oxygen and nitrogen species and glutathione: key players in the legume-Rhizobium symbiosis. J Exp Bot. 2006;57(8):1769–1776. doi: 10.1093/jxb/erj184. [DOI] [PubMed] [Google Scholar]
- 51.Tanaka A, Christensen MJ, Takemoto D, Park P, Scott B. Reactive Oxygen Species Play a Role in Regulating a Fungus-Perennial Ryegrass Mutualistic Interaction. Plant Cell. 2006;18(4):1052–1066. doi: 10.1105/tpc.105.039263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Terada LS. Specificity in reactive oxidant signaling: think globally, act locally. J Cell Biol. 2006;174(5):615–23. doi: 10.1083/jcb.200605036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsukagoshi H, Busch W, Benfey PN. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell. 2010;143(4):606–16. doi: 10.1016/j.cell.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 54.Ha EM, Oh CT, Ryu JH, Bae YS, Kang SW, Jang IH, Brey PT, Lee WJ. An antioxidant system required for host protection against gut infection in Drosophila. Dev Cell. 2005;8(1):125–32. doi: 10.1016/j.devcel.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 55.Ha E-M, Oh C-T, Bae YS, Lee W-J. A Direct Role for Dual Oxidase in Drosophila Gut Immunity. Science. 2005;310(5749):847–850. doi: 10.1126/science.1117311. [DOI] [PubMed] [Google Scholar]
- 56.Luo L, Reedy AR, Jones RM. Detecting Reactive Oxygen Species Generation and Stem Cell Proliferation in the Drosophila Intestine. Methods Mol Biol. 2016;1422:103–13. doi: 10.1007/978-1-4939-3603-8_10. [DOI] [PubMed] [Google Scholar]
- 57.Jang JY, Rhee JY, Chung GC, Kang H. Aquaporin as a membrane transporter of hydrogen peroxide in plant response to stresses. Plant Signal Behav. 2012;7(9):1180–1. doi: 10.4161/psb.21178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Velde AAT, Pronk I, de Kort F, Stokkers PCF. Glutathione peroxidase 2 and aquaporin 8 as new markers for colonic inflammation in experimental colitis and inflammatory bowel diseases: an important role for H(2)O(2)? European Journal of Gastroenterology & Hepatology. 2008;20(6):555–560. doi: 10.1097/MEG.0b013e3282f45751. [DOI] [PubMed] [Google Scholar]
- 59.Laforenza U. Water channel proteins in the gastrointestinal tract. Molecular Aspects of Medicine. 2012;33(5–6):642–650. doi: 10.1016/j.mam.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 60.Hernández-García D, Wood CD, Castro-Obregón S, Covarrubias L. Reactive oxygen species: A radical role in development? Free Radical Biology and Medicine. 2010;49(2):130–143. doi: 10.1016/j.freeradbiomed.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 61.Karrasch T, Kim JS, Muhlbauer M, Magness ST, Jobin C. Gnotobiotic IL-10−/−;NF-kappa B(EGFP) mice reveal the critical role of TLR/NF-kappa B signaling in commensal bacteria-induced colitis. J Immunol. 2007;178(10):6522–32. doi: 10.4049/jimmunol.178.10.6522. [DOI] [PubMed] [Google Scholar]
- 62.Barford D. The role of cysteine residues as redox-sensitive regulatory switches. Curr Opin Struct Biol. 2004;14(6):679–86. doi: 10.1016/j.sbi.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 63.Kamata H, Honda S, Maeda S, Chang L, Hirata H, Karin M. Reactive oxygen species promote TNFalpha-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell. 2005;120(5):649–61. doi: 10.1016/j.cell.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 64.Rhee SG, Kang SW, Jeong W, Chang TS, Yang KS, Woo HA. Intracellular messenger function of hydrogen peroxide and its regulation by peroxiredoxins. Curr Opin Cell Biol. 2005;17(2):183–9. doi: 10.1016/j.ceb.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 65.Chiarugi P, Buricchi F. Protein tyrosine phosphorylation and reversible oxidation: two crosstalking posttranslation modifications. Antioxid Redox Signal. 2007;9(1):1–24. doi: 10.1089/ars.2007.9.1. [DOI] [PubMed] [Google Scholar]
- 66.Swanson PA, Kumar A, Samarin S, Vijay-Kumar M, Kundu K, Murthy N, Hansen J, Nusrat A, Neish AS. Enteric commensal bacteria potentiate epithelial restitution via reactive oxygen species-mediated inactivation of focal adhesion kinase phosphatases. Proceedings of the National Academy of Sciences. 2011;108(21):8803–8808. doi: 10.1073/pnas.1010042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tonks NK. Redox redux: revisiting PTPs and the control of cell signaling. Cell. 2005;121(5):667–70. doi: 10.1016/j.cell.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 68.Neish AS, Jones RM. Redox signaling mediates symbiosis between the gut microbiota and the intestine. Gut Microbes. 2014;5(2):250–3. doi: 10.4161/gmic.27917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jones RM, Mercante JW, Neish AS. Reactive oxygen production induced by the gut microbiota: pharmacotherapeutic implications. Curr Med Chem. 2012;19(10):1519–29. doi: 10.2174/092986712799828283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kumar A, Wu H, Collier-Hyams LS, Hansen JM, Li T, Yamoah K, Pan ZQ, Jones DP, Neish AS. Commensal bacteria modulate cullin-dependent signaling via generation of reactive oxygen species. Embo J. 2007;26(21):4457–66. doi: 10.1038/sj.emboj.7601867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yan F, Cao H, Cover TL, Whitehead R, Washington MK, Polk DB. Soluble Proteins Produced by Probiotic Bacteria Regulate Intestinal Epithelial Cell Survival and Growth. Gastroenterology. 2007;132(2):562–575. doi: 10.1053/j.gastro.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kundu K, Knight SF, Willett N, Lee S, Taylor WR, Murthy N. Hydrocyanines: A Class of Fluorescent Sensors That Can Image Reactive Oxygen Species in Cell Culture, Tissue, and In Vivo. Angewandte Chemie-International Edition. 2009;48(2):299–303. doi: 10.1002/anie.200804851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Menard S, Candalh C, Bambou JC, Terpend K, Cerf-Bensussan N, Heyman M. Lactic acid bacteria secrete metabolites retaining anti-inflammatory properties after intestinal transport. Gut. 2004;53(6):821–8. doi: 10.1136/gut.2003.026252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pena JA, Versalovic J. Lactobacillus rhamnosus GG decreases TNF-alpha production in lipopolysaccharide-activated murine macrophages by a contact-independent mechanism. Cell Microbiol. 2003;5(4):277–85. doi: 10.1046/j.1462-5822.2003.t01-1-00275.x. [DOI] [PubMed] [Google Scholar]
- 75.Madsen KL, Doyle JS, Jewell LD, Tavernini MM, Fedorak RN. Lactobacillus species prevents colitis in interleukin 10 gene-deficient mice. Gastroenterology. 1999;116(5):1107–14. doi: 10.1016/s0016-5085(99)70013-2. [DOI] [PubMed] [Google Scholar]
- 76.Petrof EO, Kojima K, Ropeleski MJ, Musch MW, Tao Y, De Simone C, Chang EB. Probiotics inhibit nuclear factor-kappaB and induce heat shock proteins in colonic epithelial cells through proteasome inhibition. Gastroenterology. 2004;127(5):1474–87. doi: 10.1053/j.gastro.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 77.Neish AS, Gewirtz AT, Zeng H, Young AN, Hobert ME, Karmali V, Rao AS, Madara JL. Prokaryotic Regulation of Epithelial Responses by Inhibition of Ikappa B-alpha Ubiquitination. Science. 2000;289(5484):1560–1563. doi: 10.1126/science.289.5484.1560. [DOI] [PubMed] [Google Scholar]
- 78.Collier-Hyams LS, Sloane V, Batten BC, Neish AS. Cutting Edge: Bacterial Modulation of Epithelial Signaling via Changes in Neddylation of Cullin-1. J Immunol. 2005;175(7):4194–4198. doi: 10.4049/jimmunol.175.7.4194. [DOI] [PubMed] [Google Scholar]
- 79.Lee WJ. Bacterial-modulated signaling pathways in gut homeostasis. Sci Signal. 2008;1(21):pe24. doi: 10.1126/stke.121pe24. [DOI] [PubMed] [Google Scholar]
- 80.Zhou BP, Hung MC. Wnt, hedgehog and snail: sister pathways that control by GSK-3beta and beta-Trcp in the regulation of metastasis. Cell Cycle. 2005;4(6):772–6. doi: 10.4161/cc.4.6.1744. [DOI] [PubMed] [Google Scholar]
- 81.Chiarugi P, Pani G, Giannoni E, Taddei L, Colavitti R, Raugei G, Symons M, Borrello S, Galeotti T, Ramponi G. Reactive oxygen species as essential mediators of cell adhesion: the oxidative inhibition of a FAK tyrosine phosphatase is required for cell adhesion. J Cell Biol. 2003;161(5):933–44. doi: 10.1083/jcb.200211118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Diaz B, Shani G, Pass I, Anderson D, Quintavalle M, Courtneidge SA. Tks5-Dependent, Nox-Mediated Generation of Reactive Oxygen Species Is Necessary for Invadopodia Formation. Science Signaling. 2009;2(88) doi: 10.1126/scisignal.2000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 2009;23(19):2333–44. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Buchon N, Broderick NA, Poidevin M, Pradervand S, Lemaitre B. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe. 2009;5(2):200–11. doi: 10.1016/j.chom.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 85.Owusu-Ansah E, Banerjee U. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature. 2009;461(7263):537–41. doi: 10.1038/nature08313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Coant N, Ben Mkaddem S, Pedruzzi E, Guichard C, Treton X, Ducroc R, Freund JN, Cazals-Hatem D, Bouhnik Y, Woerther PL, Skurnik D, Grodet A, Fay M, Biard D, Lesuffleur T, Deffert C, Moreau R, Groyer A, Krause KH, Daniel F, Ogier-Denis E. NADPH oxidase 1 modulates WNT and NOTCH1 signaling to control the fate of proliferative progenitor cells in the colon. Mol Cell Biol. 30(11):2636–50. doi: 10.1128/MCB.01194-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Venugopal R, Jaiswal AÄ. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1‚Äâgene. Proceedings of the National Academy of Sciences. 1996;93(25):14960–14965. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes & Development. 2003;17(15):1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sykiotis GP, Bohmann D. Keap1/Nrf2 Signaling Regulates Oxidative Stress Tolerance and Lifespan in Drosophila. Developmental Cell. 2008;14(1):76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kobayashi M, Itoh K, Suzuki T, Osanai H, Nishikawa K, Katoh Y, Takagi Y, Yamamoto M. Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system. Genes to Cells. 2002;7(8):807–820. doi: 10.1046/j.1365-2443.2002.00561.x. [DOI] [PubMed] [Google Scholar]
- 91.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y-i. An Nrf2/Small Maf Heterodimer Mediates the Induction of Phase II Detoxifying Enzyme Genes through Antioxidant Response Elements. Biochemical and Biophysical Research Communications. 1997;236(2):313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 92.Motohashi H, O’Connor T, Katsuoka F, Engel JD, Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294(1–2):1–12. doi: 10.1016/s0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 93.Kobayashi A, Kang M-I, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative Stress Sensor Keap1 Functions as an Adaptor for Cul3-Based E3 Ligase To Regulate Proteasomal Degradation of Nrf2. Mol Cell Biol. 2004;24(16):7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jones RM, Desai C, Darby TM, Luo L, Wolfarth AA, Scharer CD, Ardita CS, Reedy AR, Keebaugh ES, Neish AS. Lactobacilli Modulate Epithelial Cytoprotection through the Nrf2 Pathway. Cell Rep. 2015;12(8):1217–25. doi: 10.1016/j.celrep.2015.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Malhotra D, Portales-Casamar E, Singh A, Srivastava S, Arenillas D, Happel C, Shyr C, Wakabayashi N, Kensler TW, Wasserman WW, Biswal S. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Research. 2010;38(17):5718–5734. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jimenez-Osorio AS, Gonzalez-Reyes S, Pedraza-Chaverri J. Natural Nrf2 activators in diabetes. Clin Chim Acta. 2015;448:182–92. doi: 10.1016/j.cca.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 97.Furfaro AL, Traverso N, Domenicotti C, Piras S, Moretta L, Marinari UM, Pronzato MA, Nitti M. The Nrf2/HO-1 Axis in Cancer Cell Growth and Chemoresistance. Oxid Med Cell Longev. 2016;2016:1958174. doi: 10.1155/2016/1958174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ryoo IG, Lee SH, Kwak MK. Redox Modulating NRF2: A Potential Mediator of Cancer Stem Cell Resistance. Oxid Med Cell Longev. 2016;2016:2428153. doi: 10.1155/2016/2428153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Leinonen HM, Kansanen E, Polonen P, Heinaniemi M, Levonen AL. Dysregulation of the Keap1-Nrf2 pathway in cancer. Biochem Soc Trans. 2015;43(4):645–9. doi: 10.1042/BST20150048. [DOI] [PubMed] [Google Scholar]
- 100.Esteras N, Dinkova-Kostova AT, Abramov AY. Nrf2 activation in the treatment of neurodegenerative diseases: a focus on its role in mitochondrial bioenergetics and function. Biol Chem. 2016 doi: 10.1515/hsz-2015-0295. [DOI] [PubMed] [Google Scholar]
- 101.Silva-Palacios A, Konigsberg M, Zazueta C. Nrf2 signaling and redox homeostasis in the aging heart: A potential target to prevent cardiovascular diseases? Ageing Res Rev. 2016;26:81–95. doi: 10.1016/j.arr.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 102.Zhang C, Shu L, Kong AT. MicroRNAs: New players in cancer prevention targeting Nrf2, oxidative stress and inflammatory pathways. Curr Pharmacol Rep. 2015;1(1):21–30. doi: 10.1007/s40495-014-0013-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schäfer M, Dütsch S, auf dem Keller U, Navid F, Schwarz A, Johnson DA, Johnson JA, Werner S. Nrf2 establishes a glutathione-mediated gradient of UVB cytoprotection in the epidermis. Genes & Development. 2010;24(10):1045–1058. doi: 10.1101/gad.568810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sethuraman M, McComb ME, Huang H, Huang S, Heibeck T, Costello CE, Cohen RA. Isotope-coded affinity tag (ICAT) approach to redox proteomics: identification and quantitation of oxidant-sensitive cysteine thiols in complex protein mixtures. J Proteome Res. 2004;3(6):1228–33. doi: 10.1021/pr049887e. [DOI] [PubMed] [Google Scholar]
- 105.Fisher EM, Khan M, Salisbury R, Kuppusamy P. Noninvasive Monitoring of Small Intestinal Oxygen in a Rat Model of Chronic Mesenteric Ischemia. Cell Biochemistry and Biophysics. 2013;67(2):451–459. doi: 10.1007/s12013-013-9611-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fan J, Cai H, Tan WS. Role of the plasma membrane ROS-generating NADPH oxidase in CD34+ progenitor cells preservation by hypoxia. J Biotechnol. 2007;130(4):455–62. doi: 10.1016/j.jbiotec.2007.05.023. [DOI] [PubMed] [Google Scholar]