Abstract
Owing to their central roles in cellular signaling, construction, and biochemistry, protein-protein interactions (PPIs) and protein self-assembly have become a major focus of molecular design and synthetic biology. In order to circumvent the complexity of constructing extensive non-covalent interfaces, which are typically involved in natural PPIs and protein self-assembly, we have developed two design strategies, Metal-Directed Protein Self-Assembly (MDPSA) and Metal-Templated Interface Redesign (MeTIR). These strategies, inspired by both the proposed evolutionary roles of metals and their prevalence in natural PPIs, take advantage of the favorable properties of metal coordination (bonding strength, directionality, and reversibility) to guide protein self-assembly with minimal design and engineering. Using a small, monomeric protein (cytochrome cb562) as a model building block, we employed MDPSA and MeTIR to create a diverse array of functional supramolecular architectures which range from structurally tunable oligomers to metalloprotein complexes that can properly self-assemble in living cells into novel metalloenzymes. The design principles and strategies outlined herein should be readily applicable to other protein systems with the goal of creating new PPIs and protein assemblies with structures and functions not yet produced by natural evolution.
Keywords: Protein Design, Enzyme Design, Metal Coordination, Supramolecular Chemistry, Directed Evolution
1. Introduction
Proteins are nature's most versatile building blocks for the construction of supramolecular architectures which fulfill essential structural and catalytic roles in cellular metabolism, signaling, and biomaterial formation (Petsko, & Ringe, 2004). The protein-protein interactions (PPIs) that mediate the self-assembly of these protein architectures occur primarily through the formation of numerous non-covalent interfacial contacts (salt bridges, hydrophobic contacts, hydrogen bonds) that extend over large molecular surfaces (Conte, Chothia, & Janin, 1999; Fletcher, & Hamilton, 2006; Tsai, Lin, Wolfson, & Nussinov, 1996; Xu, Tsai, & Nussinov, 1997). There has been considerable progress in the de novo design of PPIs and protein assemblies through computationally guided approaches (Der, Machius, Miley, Mills, Szyperski, & Kuhlman, 2012; Gonen, DiMaio, Gonen, & Baker, 2015; Huang, Love, & Mayo, 2007; King, Bale, Sheffler, McNamara, Gonen, Gonen, et al., 2014; Mandell, & Kortemme, 2009; Mou, Huang, Hsu, Huang, & Mayo, 2015; Schreiber, & Fleishman, 2013). Yet, it still remains a formidable challenge to accurately capture the scope, specificity, and functionality of natural PPIs and protein assemblies purely through the design of non-covalent interactions.
Transition metal ions are powerful molecular templating/directing agents because of their ability to form strong bonds (relative to noncovalent interactions) which are highly directional and reversible (Rulíšek, & Vondrášek, 1998) (Harding, Nowicki, & Walkinshaw, 2010) Accordingly, installing chelating motifs on a protein provides a simple route to creating conditionally interactive protein surfaces without extensive design and engineering (Fig. 1). In fact, metal ions have been proposed to serve as templates to guide the formation of PPIs during the structural/functional evolution of some protein superfamilies. In such systems, it has been suggested that metals directed the formation of a simple aggregate species that subsequently evolved a more specialized binding interface around the metal ion (Liu, & Xu, 2002). For example, the vicinal oxygen chelate (VOC) superfamily, a diverse group of metalloproteins (Armstrong, 2000; Bergdoll, Eltis, Cameron, Dumas, & Bolin, 1998), as well as the quintessential oxygen transport proteins (hemocyanin, hemerythrin and hemoglobin) (Volbeda, & Hol, 1989) are thought to have originated from a single metal-bridged dimeric precursor, which underwent extensive rearrangement of the overall fold and metal-containing active site to give descendant proteins with diverse structures and functions. It is estimated that at least 4-5% of all structurally characterized protein oligomers contain a mid- to late-first row transition metal ion (Mn, Fe, Co, Ni, Cu and Zn) or a metallocofactor in an interface that serves an essential structural and/or functional role (Song, Sontz, Ambroggio, & Tezcan, 2014).
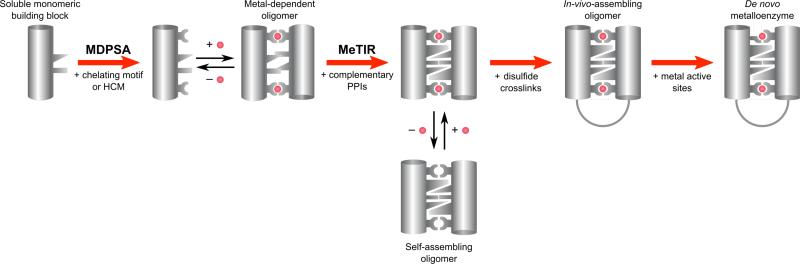
Figure 1.
Scheme showing the iterative engineering of a soluble, non-self-associating protein building block by MDPSA and MeTIR. By MDPSA, chelating motifs (e.g., bis-His clamps) can be incorporated into the protein surface, enabling the formation of metal-directed oligomers. By MeTIR, the nascent interfaces in these oligomers can be reinforced with the installation of complementary PPIs to generate self-assembling complexes that can form in the absence of templating metals. The installation of disulfide crosslinks increases scaffold robustness and the fidelity of self-assembly. These robust scaffolds can accommodate reactive metal sites to generate de novo designed metalloenzymes.
Taking this hypothetical evolutionary trajectory as a synthetic blueprint, we have implemented the advantageous properties of metal coordination (bonding strength, directionality, and reversibility) for developing a new strategy to design PPIs and self-assembled protein architectures: Metal-Directed Protein Self-Assembly (MDPSA). In proof-of-principle studies, MDPSA was applied to a small monomeric protein, cytochrome cb562 (cyt cb562), to generate discrete metal-mediated complexes whose oligomerization state and geometry were strictly controlled by metal coordination geometry (Salgado, Faraone-Mennella, & Tezcan, 2007; Salgado, Lewis, Faraone-Mennella, & Tezcan, 2008). The newly generated PPIs in these metal-mediated complexes provided a platform for the subsequent installation of favorable covalent and non-covalent interactions, an approach we have termed Metal-Templated Interface Redesign (MeTIR) (Salgado, Ambroggio, Brodin, Lewis, Kuhlman, & Tezcan, 2010). Together, the MDPSA and MeTIR approaches provide a straightforward, yet powerful route for the design of new metalloprotein scaffolds and metalloenzymes.
2. Metal Directed Protein Self-Assembly (MDPSA)
2.1 Choosing a protein building block for MDPSA
The choice of the protein building block is critical to successfully developing metal-mediated protein complexes through MDPSA. Ideally, a protein should possess the following characteristics:
Recombinantly expressible in high yields
Non-self-associating even at high concentrations (so as to eliminate non-specific aggregation)
Thermodynamically stable (so as to enable extensive surface modifications/mutations)
Uniform overall topology (so as to promote the formation of supramolecular assemblies with well-defined topologies and to facilitate crystallographic investigation)
In proof-of-principle studies, we used a model building block, cyt cb562 that fit all these criteria (Section 4.1). Cyt cb562 is a highly stable, monomeric electron-transfer protein containing a c-type heme cofactor. The protein adopts a classic 4-helix bundle fold with a rigid, cylindrical shape (Faraone-Mennella, Tezcan, Gray, & Winkler, 2006). Given such a simple uniform architecture, we could readily envision arranging multiple copies of this protein to rationally design larger assemblies mediated by strategically placed intermolecular interactions.
2.2 MDPSA using metal chelating motifs composed of natural amino acids
Once a suitable building block like cyt cb562 has been selected, a region on its surface can be chosen to implement MDPSA (Fig. 1). MDPSA takes advantage of the chelate effect through the installation of multidentate metal coordination motifs onto the protein surface. Such motifs can outcompete the many monodentate metal-binding functionalities present on the surface of a typical protein. α-helices are well suited for this purpose as their structural periodicity readily allows the pairing of histidine (His), aspartate (Asp), glutamate (Glu) and cysteine (Cys) residues at i and i+4 or i and i+3 positions to create bidentate motifs (clamps) with low-micromolar metal binding affinities (Arnold, & Haymore, 1991; Ghadiri, & Choi, 1990; Handel, Williams, & Degrado, 1993; Krantz, & Sosnick, 2001). In principle, installation of chelating motifs is possible on surface-facing β-strands through i and i+2 pairings of His, Asp, Glu and Cys side chains, or any two appropriately-oriented residues on a well-defined surface feature (Harding, 2004).
For installing metal binding motifs on a building block, protein-protein packing interactions in the crystal lattice can provide a good structural guide to select an appropriate region of the protein surface. In the case of cyt cb562, each protein monomer is paired in an antiparallel fashion with another monomer along the helix-helix face (residues 56-81) formed by Helix3 (Fig. 2). Given these extensive packing interactions (~775 Å2 of buried surface area), we envisioned incorporating bis-His clamps at both ends of Helix3 to facilitate metal-mediated oligomerization. Since residues 59 and 63 in one monomer oppose residues 73 and 77 of another, a pair of bis-His clamps was installed around the existing H63 with three additional mutations (W59H, D73H, K77H), affording Metal Binding Protein Construct 1 (MBPC1) (Fig. 2) (Salgado, et al., 2007).
Figure 2.
Application of MDPSA to the natively monomeric cyt cb562 (PDB ID: 2BC5), giving the MBPC1 metalloprotein scaffold. In the crystal lattice, cyt cb562 forms extensive packing interactions along Helix 3 (residues 56-81), which was chosen as the surface to install two bis-His clamps (H59/H63 & H73/H77). MBPC1 forms three distinct metal-bound oligomers dictated by the stereochemical preferences of the metal ions: a Ni-bound trimer (PDB ID: 3DE9), a Cu-bound dimer (PDB ID: 3DE8), and a Zn-bound tetramer (PDB ID: 2QLA). Metal ligands are shown as sticks and metal ions are shown as spheres. Close-up images show the metal coordination environments: Ni2+ is coordinated either by three H59/H63 (shown) or three H73/H77 bis-His clamps from three different protomers. Cu2+ is coordinated by H59, H63, H73' and H77' from two different protomers. Zn2+ is coordinated by H63, H73', H77' and D74” from three different protomers.
A central tenet of protein self-assembly is the reversibility of PPIs such that kinetic traps are avoided and desired thermodynamically favored architectures can be formed under ambient conditions. The fulfillment of this design feature in MDPSA requires the use of metal ions that strongly associate with amino acid residues, yet possess fast ligand exchange rates (i.e., are substitution-labile). These requirements are best met by late-first-row transition metal ions (Co2+, Ni2+, Cu2+, Zn2+) (Bertini, Gray, Stiefel, & Valentine, 2007; Frausto da Silva, & Williams, 2001; Lippard, & Berg, 1994). Although earth-alkaline metal ions such as Mg2+ and Ca2+ are labile, they have relatively weak affinities for proteinaceous functionalities and their coordination is largely limited to carboxylate and hydroxyl groups. In contrast, second- and third-row transition metal ions can tightly associate with protein surfaces, but possess very slow ligand exchange rates at room temperature (Bertini, et al., 2007; Lippard, et al., 1994).
A further advantage of late-first-row transition metal ions is their distinct preferences for coordination geometry (Ni2+, octahedral; Cu2+, tetragonal/square planar; Zn2+, tetrahedral) (Rulíšek, et al., 1998) (Harding, et al., 2010). Thus, they can also dictate the oligomerization state and oligomerization geometry of a protein building block. Indeed, MPBC1 self-assembles into a C3 symmetric trimer upon Ni2+ binding (via octahedral coordination by three bis-His clamps), a C2 symmetric dimer upon Cu2+ binding (via square planar coordination by two bis-His clamps), and a D2 symmetric tetramer upon Zn2+ binding (via tetrahedral coordination by a bis-His clamp from one protomer, D74 from a second protomer, and H63 from a third protomer (Fig. 2) (Salgado, et al., 2007; Salgado, Lewis, Mossin, Rheingold, & Tezcan, 2009). Notably, the Zn4:MBPC14 structure demonstrates the formation of the thermodynamically favored, albeit unforeseen, tetrameric architecture rather than the predicted Zn2:MBPC12 dimer mediated by tetrahedral coordination via two bis-His clamps. These studies indicate the following: 1) in terms of designing protein assemblies, metal coordination provides structural versatility and controllability that is difficult and perhaps impossible to match through the design of non-covalent interactions, 2) this versatility is achieved through minimal alterations of the protein building blocks, and 3) protein oligomers formed through MDPSA give rise to nascent protein-protein interfaces and internalized metal centers that provide a rich platform for further design efforts (Section 3).
2.3 MDPSA using synthetic metal-coordinating ligands
Precise placement of multiple amino acid side chains to form a chelating motif can be challenging, particularly in the case of proteins with irregular surface features. An alternative route for installing multidentate metal coordinating motifs on a protein surface is to employ non-natural metal chelating functionalities, such as the bidentate 8-hydroxyquinoline (quin) and 1,10-phenanthroline (phen), or the tridentate 2,2’:6’,2’’-terpyridine (terpy). These ligands have metal binding affinities many orders of magnitude higher than those of monodentate amino acid residues (Smith, Martell, & Motekaitis, 2004), and can be readily conjugated to surface-exposed cysteine residues through the use of their iodoacetamide or maleimide derivatives (Section 4.2). In the case of cyt cb562, a cysteine (C70) was installed near the middle of the solvent-exposed face of Helix3 to provide an appropriate attachment site for a metal chelating group (Fig. 3A). C70cyt cb562 was modified with iodoacetamide derivatives of quin, phen, or terpy, and the resulting adducts readily dimerized upon metal coordination (Fig. 3A-C) (Radford, Nguyen, & Tezcan, 2010).
Figure 3.
MDPSA via hybrid coordination motifs (HCMs). (A) C70cyt cb562 is labeled with 5-(iodoacetamido)-8-hydroxyquinoline to afford HQuin1. Upon the addition of Ni2+, HQuin1 forms the Ni:HQuin12 dimer (3L1M). (B) Ni2+ (green sphere) is coordinated by the two bidentate quin ligands, and the two H63 residues (purple sticks). (C) Chemical structures of the iodoacetamide-derivatized metal-chelating ligands (IA-phen, IA-quin, and IA-terpy).
These motifs are easily generalizable, since the only requirement is a surface-exposed cysteine residue. Nevertheless, the flexibility of the Cys-attachment site can give rise to unpredictability in the architecture of the resulting protein oligomer. To circumvent this potential problem, we envisioned using the Cys-attached phen or quin functionalities in tandem with a nearby His residue to form a tridentate hybrid coordination motif (HCM). In exploratory experiments with cyt cb562, this strategy was implemented by pairing C70-attached phen or quin with H63 in an i and i+7 arrangement on the Helix3 surface (Fig. 3A). The resulting tridentate HCM not only binds Co2+, Ni2+, Cu2+, Zn2+ with nanomolar to femtomolar affinities, but it also allows the formation of a discrete dimeric architecture upon Ni2+ binding in an octahedral geometry (Radford, Nguyen, Ditri, Figueroa, & Tezcan, 2010; Radford, et al., 2010) (Fig. 3A). HCMs could, in principle, be applied to any α-helical domain on a protein surface, provided that the helix spans at least two turns.
3. Metal-Templated Interface Redesign
3.1 Redesign of non-covalent interfaces with MeTIR
Metals are proposed to have been drivers of evolution in some protein superfamilies through the formation of metal-bridged protein/peptide aggregates (Armstrong, 2000; Bergdoll, et al., 1998; Liu, et al., 2002; Volbeda, et al., 1989). These ancestral oligomers subsequently developed stable and specific protein interfaces around the metal-nucleation sites, eventually leading to the emergence of modern proteins with evolved networks of bonding interactions to stabilize their tertiary or quaternary architectures. Whether or not such an evolutionary pathway was indeed operative, one can still use it as a blueprint for protein design in an approach we have termed Metal-Templated Interface Redesign (MeTIR) (Fig. 1). Self-assembled protein architectures obtained by MDPSA possess, by default, nascent PPIs between non-self-complementary surfaces surrounding the metal templates. These interfaces can be rendered self-complementary by replacing unfavorable amino acid contacts with favorable ones through rational design. In this way, MeTIR can furnish novel self-assembling proteins that form stable protein architectures with tunable metal binding sites in their interiors (see Sections 5.3, 5.4 and 5.5 for methods).
The Zn-directed MBPC1 tetramer, Zn4:MBPC14, was used as a model system for MeTIR (Salgado, et al., 2007). This tetramer features an extensive buried surface (>5000 Å2) between the MBPC1 protomers, distributed among three pairs of interconnected, C2 symmetric interfaces termed i1, i2, and i3 (PDB ID: 2QLA) (Fig. 4). We chose i1 (2 × 1080 Å2) as a promising candidate for initial redesign because, as the most extensive interface, it could accommodate a greater number of favorable contacts than i2 (2 × 870 Å2) or i3 (2 × 490 Å2). Additionally, i1 brings the protomers into closer proximity than i2, a prerequisite for forming favorable side-chain packing interactions. The redesign process was carried out by a computationally guided approach using Rosetta (Liu, & Kuhlman, 2006) (discussed in detail in Chapter XXX of this issue). In this process, the residues that coordinate the Zn2+ ions and the protein backbone were both held fixed as demanded by the templating strategy. Clusters of i1 residues were subjected to concerted mutation and optimization with respect to rotamer geometry and packing energy (Salgado, et al., 2010). This procedure led to the selection of six mutations (R34A, L38A, Q41W, K42S, D66W, and V69I), which were installed to convert MBPC1 into the variant RIDC1 (Rosetta Interface Designed Cytochrome 1) (Fig. 4).
Figure 4.
Application of MeTIR to the metal-directed Zn4:MBPC14 tetramer (PDB ID: 2QLA) to give Zn4:RIDC14 (PDB ID: 3HNI). Red axes denote the C2-symmetric interfaces i1, i2 and i3. The redesigned i1 residues are shown as cyan sticks. Metal-coordinating ligands are shown as gray sticks. Zn2+ ions are shown as gray spheres. Beneath are close-up views of the i1 PPIs between two MBPC1 or RIDC1 protomers within the Zn-bound tetramers.
As expected from the hydrophobicity of the engineered surface residues and determined by molecular docking simulations with RosettaDock (Gray, Moughon, Wang, Schueler-Furman, Kuhlman, Rohl, et al., 2003), the geometric specificity of i1 interactions is low. Even so, these interactions are strong enough to dimerize RIDC1 monomers in solution (KD = 25 μM) without metal (Salgado, et al., 2010). More importantly, in the presence of Zn2+, RIDC1 forms a tetramer (Zn4:RIDC14) that is isostructural with Zn4:MBPC14, but is considerably stabilized by the redesigned i1 interfaces (Fig. 4).
3.2 Disulfide crosslinks to enhance scaffold robustness
For an oligomeric protein complex with multiple interaction surfaces, such as the D2 symmetric Zn4:MBPC14 and Zn4:RIDC14 tetramers, the redesign of a single set of interfaces is insufficient to maintain the scaffold in the absence of bound metal. A second set of interfaces must be redesigned to hold the tetrameric assembly intact in the absence of the metal templates. However, computational redesign of non-covalent interactions is not only labor-intensive, but can be sub-optimal in interfaces that are too wide or too small. A straightforward alternative is to install disulfide bonds across these interfaces that–in combination with computationally redesigned interfaces– can lead to the formation of stable protein complexes in a metal-independent manner. In this regard, the unique properties of disulfide bonds must be emphasized: they are strong covalent bonds that are very specific (i.e., they only pair with themselves), yet they are reversible (Fass, 2012). Thus, in terms of protein self-assembly, disulfide bonds share many of the advantageous characteristics of metal coordination mentioned previously.
Cysteine residues can be installed at a pair of symmetry-related sites with the appropriate α-C separation and relative orientation to form disulfide bonds (Petersen, Jonson, & Petersen, 1999). In the case of Zn4:RIDC14, the T96/T96’ residue pairs across the i2 interfaces and the E81/E81’ across the i3 interfaces were both suitable for this purpose. Accordingly, we generated a single-cysteine (C96RIDC1) (Brodin, Medina-Morales, Ni, Salgado, Ambroggio, & Tezcan, 2010) and a double-cysteine (C81/C96RIDC1) (Medina-Morales, Perez, Brodin, & Tezcan, 2013) variant which properly self-assembled into stable metal-free tetramers (C96RIDC14 and C81/C96RIDC14). Moreover, these tetramers adopted the same structure as the parent tetramer Zn4:MBPC14 upon binding Zn2+. Importantly, the quadruply-disulfide-linked C81/C96RIDC14 scaffold is sufficiently preorganized to permit altering the inner-coordination sphere of the templating Zn ions without perturbing the quaternary structure (Fig. 5 A-C) (Medina-Morales, et al., 2013).
Figure 5.
(A) The quadruply-disulfide crosslinked Zn4:C81/C96RIDC14 (PDB ID: 4JEA). Red axes denote the C2-symmetric interfaces i1, i2 and i3. Disulfide crosslinks (B) C81/C81’ across i3 and (C) C96/C96’ across i2. (D) The artificial metallo-β-lactamase Zn8:G57/A104AB34 (PDB ID: 4U9E) bearing redesigned i1 contacts (cyan sticks), C96/C96’ disulfide crosslinks (green sticks), and sets of structural Zn2+ sites and catalytic Zn2+ sites (gray sticks). Zn2+ ions are shown as gray spheres, and the catalytic water molecules are shown as red spheres. Shown in purple is a natively well-structured loop which became highly mobile (residues 43-57, not observed crystallographically) as a result of directed evolution. (E) Surface representation of the structured regions of Zn8:G57/A104AB34 showing the proximity of the solvent-exposed catalytic Zn2+ site, with ligands colored in blue, to the mobile loop, shown in purple. (F) Close-up view of a catalytic Zn2+ site.
3.3 In vivo assembly and functional screening of metal-templated protein assemblies
A major goal in protein design (and synthetic biology) is to create artificial constructs that function in living systems. We envisioned that, through the combined action of non-covalent interface redesign and disulfide bond formation, self-assembly of artificial protein architectures could also occur in bacterial cells. The C96RIDC14 and C81/C96 RIDC14 assemblies were indeed found to properly self-assemble in vivo and bind Zn2+ ions with high selectivity (Medina-Morales, et al., 2013; Song, & Tezcan, 2014). To form disulfide bonds in vivo, the protein must be expressed in the oxidizing environment of the periplasm in Escherichia coli (Inaba, & Ito, 2008). Additionally, the periplasmic space exhibits comparatively relaxed metallostasis (Braymer, & Giedroc, 2014; Hu, Gunasekera, Spadafora, Bennett, & Crowder, 2008), which permits the incorporation of metal ions inside the cell (Medina-Morales, et al., 2013; Song, et al., 2014). Since proteins are synthesized in the cytoplasm, a genetically-encoded localization sequence is required for its translocation into the periplasm (Mergulhão, Summers, & Monteiro, 2005). In the case of cyt cb562, the signaling sequence was derived from Rhodopseudomonas palustris cytochrome c556 (Faraone-Mennella, et al., 2006; McGuirl, Lee, Lyubovitsky, Thanyakoop, Richards, Gray, et al., 2003). When expressed in E. coli, a periplasmic extraction procedure allows for the recovery of intact oligomers (e.g., Zn4:C81/C96 RIDC14) to examine in vivo assembly and Zn2+ content (Section 4.6).
A robust oligomeric protein scaffold that self-assembles in vivo is an ideal candidate for the de novo design of a functional, metalloenzyme. A catalytic metal site should be coordinatively unsaturated, such as a hydrolytic Zn center that coordinates and activates water (Alberts, Nadassy, & Wodak, 1998; Holm, Kennepohl, & Solomon, 1996). In C96RIDC14 and C81/C96RIDC14, replacing a coordinating ligand (D74) of the core (templating) Zn2+ ions with a non-coordinating alanine residue did not yield catalytically competent constructs, presumably because the Zn sites were inaccessible or did not possess a proper secondary sphere environment for substrate binding and activation (Medina-Morales, et al., 2013). Therefore, interfacial sites in the solvent-exposed peripheral regions of Zn4:C96RIDC14 were investigated as potential locations for installing catalytic Zn centers (Song, et al., 2014). Examination of i2 in C96RIDC14 revealed several three-residue permutations that could accommodate a tripodal Zn-coordination motif. One such permutation was the E86/H89/H100 triad on C96RIDC14. The installation of these residues gave the AB3 construct, which upon Zn-coordination yielded the tetrameric Zn8:AB34 with four core Zn sites and four potentially catalytic, peripheral Zn sites. Removal of an occluding lysine residue (K104) near the peripheral Zn sites resulted in the formation of tetrahedral Zn-H2O/OH− motifs reminiscent of natural metalloenzyme hydrolytic sites (Crowder, Spencer, & Vila, 2006).
In vitro enzymatic activity assays showed that Zn8:A104AB34 was indeed catalytically active in the hydrolysis of activated ester bonds (Song, et al., 2014). This observation, combined with the ability of A104AB34 to properly self-assemble in the periplasm of E. coli, raised the possibility of evolving the activity of this nascent, artificial enzyme in vivo (Jäckel, Kast, & Hilvert, 2008). Enzymatic hydrolysis of an antibiotic, such as ampicillin, could be coupled to cellular survival, providing a selection mechanism for directed evolution. The β-lactamase activity of Zn8:A104AB34 was sufficient to permit E. coli growth on media containing ampicillin, which allowed for screening of a library of active-site point mutants for improved enzymatic activity (Section 4.7). Library variants were identified and subsequently purified to quantify the enzymatic activities in vitro (Song, et al., 2014), which correlated with the in vivo survival frequencies of the variants. X-ray crystallographic studies indicated that best performing variant, Zn8:G57/A104AB34, possessed a highly mobile loop atop the catalytic Zn centers, which is a conserved feature of natural metallo-β-lactamases (Fig. 5 D-F) (Scrofani, Chung, Huntley, Benkovic, Wright, & Dyson, 1999; Tomatis, Fabiane, Simona, Carloni, Sutton, & Vila, 2008). Thus, by iterative protein design, cyt cb562 was converted into an in-vivo active β-lactamase while conserving nearly 90% sequence identity.
4. Methods
4.1 Cytochrome cb562 variant expression and purification
The gene encoding cyt cb562 was housed in a modified pET20b+ vector encoding the N-terminal periplasmic localization sequence of R. palustris cytochrome c556. Variants of the cyt cb562 gene were obtained by site-directed mutagenesis. Plasmids encoding cyt cb562 variants weretransformed into BL21(DE3) E. coli cells harboring the cytochrome c maturation cassette (ccm or pEC86) (Arslan, Schulz, Zufferey, Kunzler, & Thony-Meyer, 1998). Cells were plated onto selective LB/agar plates (34 μg/mL chloramphenicol and 100 μg/mL ampicillin) and the protein was expressed and purified as follows:
Transfer single colonies into three 5 mL of LB medium supplemented with 100 μg/mL ampicillin and 34 μg/mL chloramphenicol.
Shake cultures at 250 rpm, 37 °C until cultures are visibly turbid (OD600 > 0.6, 6-8 h)
Remove a 1 mL portion of each culture and pellet the cells by centrifugation (1 min, 10,000 × g). The color of the cell pellets provides a convenient indicator for the expression and folding of cyt cb562 through proper heme incorporation.
Transfer 75 μL of the culture which gave the most intensely red-colored pellet into 1 L of LB medium (typically 16 L total) supplemented with 100 μg/mL ampicillin and 34 μg/mL chloramphenicol.
Shake inoculated cell cultures at 250 rpm and 37 °C for 16-20 h. Protein expression occurs by auto-induction.
Harvest the cells by centrifugation (5,000 × g, 4 °C, 10 min), decant the supernatant and retain the red cell pellet.
Resuspend the cell pellet in 100 mL of a buffered solution (5 mM sodium acetate, pH 5) and freeze the suspension at −80 °C.
Thaw the cell suspension in a bath of water at room temperature, add ~100 mg lysozyme, and sonicate on ice.
Titrate the cell lysate with 40% (w/v) sodium hydroxide to pH 10 and then immediately with 50% (v/v) acetic acid to pH 5. Cream-colored precipitates will form in the red lysate.
Subject the lysate to centrifugation (10,000 × g, 4 °C, 20 min) and retain the clear, red supernatant. Repeat steps 9-10 to recover additional protein as needed.
Dilute the cleared lysate by 150 mL per 1 L cell culture, and apply to a CM Sepharose (GE Healthcare Life Sciences) cation exchange column equilibrated in a buffered solution (5 mM sodium acetate, pH 5). If the cyt cb562 variant of interest in unstable at pH 5, the purification can be carried out with a Q Sepharose (GE Healthcare Life Sciences) column equilibrated in 5 mM sodium phosphate, pH 8.
Elute the protein using a gradient of 0-1 M sodium chloride and recover eluate that is visibly red.
Concentrate the recovered protein sample using a DiaFlow concentrator (Amicon) fitted with a 3-kDa cutoff membrane to a final volume of 50 mL.
Dialyze the protein sample overnight at 4 °C against 4 L of buffered solution (10 mM sodium phosphate, pH 8) to remove residual salt.
Apply the dialyzed sample to a fast protein liquid chromatography (FPLC) workstation equipped with a Macroprep High Q-cartridge (BioRad) anion exchange column equilibrated with a buffered solution (10 mM sodium phosphate, pH 8).
Elute protein using a 0-0.5 M sodium chloride gradient.
Estimate protein purity spectrophotometrically and combine fractions with a Reinheitzahl (RZ) value (A415/A280) above 3.
Concentrate the combined fractions to a volume of <5 mL and apply the sample to an FPLC workstation equipped with a preparative-scale Superdex 75 size-exclusion column equilibrated in a buffered solution (20 mM TRIS, pH 7 and 150 mM sodium chloride).
Estimate protein purity spectrophotometrically and combine fractions with an RZ value (A415/A280) above 4.
Concentrate the combined fractions to a volume of ~500 μL and add >10-fold excess ethylenediaminetetraacetic acid (EDTA) to remove any residual bound metal ions.
Apply the protein sample to a desalting column pre-equilibrated as desired to remove the EDTA.
The protein concentration can be determined using the extinction coefficient of the Soret band of cyt cb562 (ε415 = 148,000 M−1 cm−1).
Flash freeze aliquots of the protein stock solutions at the desired concentration in liquid nitrogen for storage at −80 °C.
4.2 Iodoacetamide ligand synthesis and protein labeling
Below is a protocol for the synthesis of N-(8-hydroxyquinolin-5-yl)-2-iodoacetamide (IA-quin) for labeling C70cyt cb562 based on previously reported procedures (Radford, et al., 2010; Smith, et al., 2013). This procedure can be readily adapted for the synthesis of iodoacetamide-derivatized ligands and the Cys-labeling of proteins in general.
In a 50-mL round-bottom flask, combine 1.19 g (6.4 mmol) of iodoacetic acid dissolved in 10 mL ethyl acetate with 660 mg (3.2 mmol) of N,N’-dicyclohexylcarbodiimide (DCC) dissolved in 10 mL of ethyl acetate. A white precipitate should form immediately.
Stir at room temperature for 2 h protected from light using aluminum foil.
Remove and discard the white precipitate (dicyclohexylurea) by filtration.
Remove solvent in vacuo protected from light using aluminum foil to recover iodoacetic anhydride.
In a 25-mL round-bottom flask, combine 500 mg (2.1 mmol) of 5-amino-8-hydroxyquinolate dihydrochloric acid dissolved in 10 mL of acetonitrile with 1 mL (7 mmol) triethylamine.
Reflux the mixture at 80 °C for 2 h. This should afford a clear, dark solution. Allow this solution to cool to room temperature.
Dissolve the iodoacetic anhydride in 5 mL of acetonitrile and add dropwise to the reaction mixture.
Stir overnight at room temperature protected from light using aluminum foil.
Isolate the desired product by filtration and wash the retained solid with 5% (w/v) sodium bicarbonate in water.
Dry the retained solid in vacuo to afford the desired IA-quin. Store the sample protected from light.
A major side product of this reaction is N-(8-hydroxyquinolin-5-yl)-2-chloroacetamide (CA-quin), which would also react site-specifically with a cysteine residue to append the desired chelating motif, albeit much more slowly than IA-quin. The crude mixture of IA-quin is approximately 50% pure and forms in 80% yield as-synthesized. Though not necessary, conversion of the CA-quin to IA-quin can be achieved as follows:
Reflux the CA-quin with five molar equivalents of sodium iodide in acetone overnight.
Remove solvent in vacuo.
Resuspend the dried solid in water to remove excess sodium chloride and recover the solid by filtration.
Dry the retained solid in vacuo, affording IA-quin.
Once the iodoacetamide derivatives are prepared, labeling of surface-exposed cysteine residues can be carried out as follows:
Add a 10-fold molar excess of dithiothreitol (DTT) to a 5 mL solution of 0.3 mM cyt C70cb562 in 0.1 M TRIS, pH 7.75 and incubate for 30 min at room temperature reduce any disulfide bonds.
Transfer the cyt C70cb562 solution into a glass vial sealed with a gas-tight rubber septum.
Degas the cyt C70cb562 solution either on a Schlenk line using 3 × 10 cycles of vacuum/argon-fill or by purging with argon or nitrogen.
Transfer the protein sample into an anaerobic glove box (<10 ppm oxygen) under an inert (argon or nitrogen) atmosphere.
Dialyze the protein sample twice against 1 L of degassed 0.1 M TRIS, pH 7.75 in the anaerobic glove box to remove the DTT.
Recover the protein sample and place in a glass vial.
To the protein sample, add a 10-fold molar excess of an IA-quin stock dissolved in degassed dimethylformamide (DMF) dropwise over the course of one minute.
Incubate the reaction mixture at room temperature overnight, protected from light with aluminum foil.
Remove the protein sample from the glove box and dialyze it twice against 1 L of 10 mM sodium acetate, pH 5, supplemented with 1 mM EDTA to remove any trace metals from the sample.
Purify the crude labeled protein by applying it to an FPLC workstation equipped with an Uno-S cation-exchange column equilibrated in 10 mM sodium acetate, pH 5.
Elute protein using a 0-0.5 M sodium chloride gradient to isolate the quin-functionalized protein sample. Labeling efficiency was >60%.
Confirm the identity of the functionalized protein by MALDI or ESI-mass spectrometry.
4.3 Determining oligomeric state by sedimentation velocity experiments
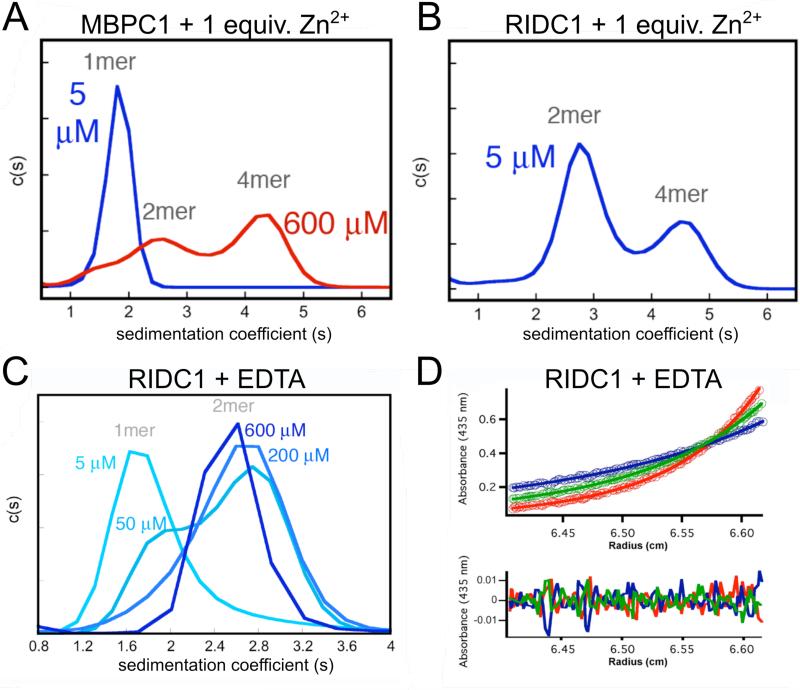
Protein oligomerization can be characterized in solution by sedimentation-velocity analytical ultracentrifugation (SV-AUC). An SV-AUC experiment requires an optical signal (absorbance or fluorescence) which can be used to monitor protein sedimentation. The heme cofactor of cyt cb562 provides a convenient optical handle (εmax at 415 nm=148,000 M−1 cm−1) that allows the AUC experiments to be conducted from very low (≥5 μM; near the Soret peak) to very high protein concentrations (>600 μM; near the shoulders of the Soret band) (Fig. 6A) (Faraone-Mennella, et al., 2006). Metal-induced protein oligomerization can be assessed under both metal-free and metal-loaded conditions (Fig. 6A-C). To assess metal-induced oligomerization, the protein sample is pre-incubated with a stoichiometric amount (one molar equivalent) of M2+ per coordination site. For substitution-labile metals, brief incubations (<1 hr at 4 °C) are sufficient, whereas longer incubations (~20 h at 4 °C) may be necessary for substitution-inert metals. The metal stock should be added slowly (~0.25 equivalents at a time) to prevent nonspecific protein aggregation. The choice of buffering agent is also an important consideration for examining protein oligomerization. Weakly chelating buffering agents, such as TRIS, will suppress nonspecific metal binding. For weak metal-binding events, a non-chelating buffering agent such as MOPS is more appropriate. Buffering agents that form insoluble metal salts, such as phosphates, should be avoided.
Figure 6.
SV-AUC profiles showing Zn-induced assembly of (A) MBPC1 and (B) RIDC1 as well as (C) RIDC1 in the absence of metal ions. (D) SE-AUC scans of metal-free RIDC1 (circles) globally fit to a monomer-dimer equilibrium model for centrifugation at 20,000 (red), 25,000 (green) and 30,000 (blue) rpm along with the residuals of the data-fitting. Adapted from (Salgado, Faraone-Mennella, & Tezcan, 2007).
Samples are sedimented by high-speed centrifugation at a fixed velocity. The sedimentation is monitored by changes in absorbance at different radii of rotation. To examine oligomerization by SV-AUC, the hydrodynamic behavior of the protein is accounted for by applying the Lamm equation (Correia, & Stafford, 2015). AUC-fitting programs can be used to model the sedimentation behavior of the protein. The range of AUC-fitting programs and procedures available are beyond the scope of this review, and are discussed elsewhere (Cole, Lary, Moody, & Laue, 2008; Lebowitz, Lewis, & Schuck, 2002). The procedure used for cyt cb562 variants is briefly described:
Load the protein sample into a two-sector sample cell with a slight excess of an appropriate buffer blank (e.g., 420 μL sample and 450 μL buffer blank).
Sediment the protein sample at 135,000 × g at 25 °C, monitoring continuously at the wavelength(s) of interest, typically for 500 scans over 20 h.
Load the sedimentation velocity scans into HETEROANALYSIS (http://biotech.uconn.edu/auf/) and use the match function to determine when sedimentation has ended.
Load the sedimentation velocity scans up to the point where sedimentation has ended into SEDFIT (Schuck, 2000).
Manually set the cell limits and data-fitting limits of the overlaid scans. Hold these fixed during the fitting procedure.
Estimate the partial specific volume in mL/g by taking the quotient of the protein volume and the molecular weight.
Estimate the viscosity and density of the buffer using SEDNTERP (http://sednterp.unh.edu/).
Using the appropriate partial specific volume, buffer viscosity, and buffer density, fit the data to a continuous distribution of molecular weight (c(M) model) or sedimentation coefficient (c(S) model).
Using the “Run” command in SEDFIT, set the baseline of the overlaid velocity scans.
Using the “Run” command in SEDFIT, subtract the time-invariant or radius-invariant noise of the overlaid velocity scans.
Fit the weight-averaged frictional coefficient (f/f0) of the protein using the “Fit” command in SEDFIT with the confidence set to 0. A value of 1.2-1.3 is typical for symmetric proteins.
Obtain the final distribution profile by setting the confidence to 0.95 and executing the “Run” command in SEDFIT.
Using this procedure, the peaks of the sedimentation profile can be assigned to protein oligomeric states. With a c(M) profile, this can be done from the reported molecular weights. With a c(S) profile, this can be done by comparing the sedimentation profile with those of related proteins, or by modeling the sedimentation coefficients using a program such as HYDROPRO (Ortega, Amoros, & Garcia de la Torre, 2011).
4.4 Measuring protein-protein affinities with sedimentation equilibrium experiments
Once the oligomerization behavior of a cyt cb562 construct has been examined across a range of protein concentrations and metal ions by SV-AUC (Fig. 6A-C), one can measure the dissociation constants (Kd) of the oligomers in the absence of metal, or the apparent dissociation constant (Kd,app) in the presence of metal by sedimentation-equilibrium AUC (SE-AUC) (Cole, et al., 2008) (Fig. 6D). In an SE-AUC experiment, samples are subjected to angular velocities that permit the counteracting forces of sedimentation and diffusion to equilibrate. These velocities are slower than what is used in a typical SV-AUC experiment. An important consideration is the time needed to achieve equilibrium. This can be done by manual inspection of overlaid scans, or through the scan match functionalities in programs like HETEROANALYSIS ((http://biotech.uconn.edu/auf/). Once equilibrium is established, the extent of sedimentation correlates with the molecular weight of the protein. A protein sample under a given set of conditions is sedimented at various speeds, and the resulting datasets are analyzed globally using programs such as SEDPHAT (Fig. 7) (Schuck, 2004). Non-interacting (e.g., monomer-only) and self-assembly (e.g., monomer-dimer) models can be considered to find the best model based on the goodness-of-fit statistics. In more complex cases, SV-AUC data can help to eliminate models that are difficult to rule out. For Zn-induced RIDC1 assembly, SV-AUC experiments suggested the trimer was not formed, and assembly was modeled as a monomer-dimer-tetramer equilibrium.
4.5 Crystallization of cyt cb562 variants
As discussed previously, crystallography is a powerful and necessary tool for assessing the outcome and progress of MDPSA/MeTIR, because both design approaches are intimately dependent on detailed structural knowledge at the atomic level. Crystals of cyt cb562 variants can be obtained by sitting-drop vapor diffusion. In each well of a 24-well crystal tray, 500 μL of a screening solution containing a precipitant, a buffering agent, and a salt (see Table 1 for details on screening solutions) is added to the reservoir. The sitting drop consists of 2 μL of ~3 mM of a cyt cb562 variant and 2 μL of the reservoir solution. Crystal formation and growth can be monitored on a timescale of days to weeks. The initial screens can used to determine fine-screening conditions, in which the precipitant concentration is systematically lowered (typically as low as half the initial concentration). Mature crystals were cryoprotected with either 20% glycerol or perfluoropolyether cryo oil (Hampton) and frozen in liquid nitrogen for X-ray crystallographic analysis. We have typically used synchrotron beam lines with tunable energies as this enables confirmation of metal identity via anomalous scattering. Diffraction data are typically indexed and integrated with Mosflm (Battye, Kontogiannis, Johnson, Powell, & Leslie, 2011) and scaled with SCALA (Winn, Ballard, Cowtan, Dodson, Emsley, Evans, et al., 2011). Crystallographic phases are readily determined through molecular replacement with either PHASER (McCoy, Grosse-Kunstleve, Adams, Winn, Storoni, & Read, 2007) or MOLREP (Vagin, & Teplyakov, 1998) using an appropriate cyt cb562 monomer as a search model. Structure determination and refinement are accomplished by rigid-body, thermal, and positional refinement with REFMAC (Murshudov, Vagin, & Dodson, 1997). COOT is used for manual model rebuilding and placement of water/ligands (Emsley, & Cowtan, 2004). The resultant models provide detailed structural information and serve as a guide for further rational design efforts.
Table 1.
Components of homemade crystallography screening solutions.
| Buffering Agent (100 mM) | Salt (200 mM) | Precipitant (% w/v) | |
|---|---|---|---|
| 100 mM BIS-TRIS pH 6.5 | CaCl2 | 45% MPD | 25% PEG 3350 |
| NaCI | 40% PEG 400 | 25% PEG 4000 | |
| 100 mM HEPES pH 7.5 | MgCl2 | 30 % PPG 400 | 25% PEG 5000 |
| 100 mM TRIS pH 8.5 | NH4OAc | 25% PEG 1500 | 25% PEG 6000 |
| (NH4)2SO4 | 25% PEG 2000 | 25% PEG 8000 | |
4.6 Periplasmic extraction of cytcb562 assemblies from E. coli
A periplasmic extraction procedure is necessary to prevent disulfide bond reduction during purification. This protocol, adapted from a previously-reported method (Nossal, & Heppel, 1966), allows for the recovery of intact cyt cb562 oligomers for further analysis:
Pellet liquid cell culture expressing the protein of interest by centrifugation (10 min, 5000 × g, 4 °C) in a tared vessel.
Decant the supernatant and note the cell pellet mass.
Wash cell pellet three times with an ice-cold low-salt buffered solution (10 mM TRIS, pH 7.3 with 30 mM sodium chloride).
Gently resuspend the cell pellet in 10 volumes (v/w) of 33 mM TRIS, pH 7.3.
Add 10 volumes (v/w) of 33 mM TRIS, pH 7.3 supplemented with 40 % (w/v) sucrose and 0.4 mM EDTA.
Gently shake the sample for 10 min at room temperature.
Pellet cells by centrifugation (10 min, 5000 × g, 4 °C)
Discard the supernatant and wash the cell pellet with ice-cold 33 mM TRIS, pH 7.3 supplemented with 40% sucrose.
Resuspend the cells in 20 volumes (v/w) of ice-cold 33 mM TRIS, pH 7.3 supplemented with 0.5 mM magnesium chloride to lyse the outer membrane by osmotic shock.
Gently swirl protein sample for 10 min at room temperature.
Pellet the cells by centrifugation (20 min, 5000 × g, 4 °C) and retain the supernatant as the periplasmic extract.
Immediately add excess iodoacetamide (5 mM) to prevent post-lytic formation of disulfide bonds.
Immediately apply the periplasmic extract to a High Q-cartridge column (BioRad) equilibrated in 10 mM sodium phosphate, pH 8 and elute the protein using a 0-0.5 M sodium chloride gradient.
Successful periplasmic extraction can be assessed by using UV-visible spectrophotometry to verify that the heme cofactor of cyt cb562 variant has remained oxidized, which can be noted by a shift in the Soret band peak (Fe3+-cyt cb562 ε415 = 148,000 M−1 cm−1; Fe2+-cyt cb562 ε421 = 162,000 M−1 cm−1). Protein samples purified by periplasmic extraction can be examined for oligomerization and disulfide bond formation by analytical size-exclusion chromatography and non-reducing SDS-PAGE, respectively. Samples can be assayed for metal content by inductively coupled plasma optical emission spectrometry (ICP-OES).
4.7 In vivo screening of cyt cb562 assemblies with β-lactamase activity
Screening for β-lactamase activity required the removal of the ampicillin-resistance gene from the pET20b(+) vector housing the C96RIDC1 gene, and replacement with the kanamycin resistance gene from the pET24 vector by molecular cloning (Song, et al., 2014). The resultant construct can be used to generate libraries for in vivo β-lactamase screening. The following protocol outlines saturation mutagenesis at a single site to achieve 95% library coverage:
Use available crystal structures to identify residues in the secondary coordination sphere of the peripheral metal sites.
Design a primer pair that contains a single NNK codon in the sense primer and a single MNN codon in the antisense primer to generate variants at the site of interest. N is any nucleotide, K is a guanine or thymine, and M is cytosine or adenosine.
Carry out saturation mutagenesis using the above primers via polymerase chain reaction (PCR). This can be accomplished with a commercially available product, such as the QuikChange Site Directed Mutagenesis Kit (Agilent).
Transform the library PCR reaction into XL-1 Blue cells and spread onto LB/agar plates containing 50 μg/mL kanamycin.
Swab >100 colonies to inoculate a single culture of 100 mL of LB medium containing 50 μg/mL kanamycin. The number of colonies needed to achieve the desired library coverage can be estimated using CASTER (http://www.kofo.mpg.de/en/research/biocatalysis).
Incubate the culture overnight at 37 °C, while shaking at 250 rpm.
Extract the plasmid library using a Qiagen Plasmid purification kit.
Confirm sequence randomization by DNA sequence analysis.
Transform the plasmid library into BL21(DE3) cells housing the ccm plasmid and spread onto LB/agar plates containing 50 μg/mL kanamycin and 30 μg/mL chloramphenicol.
Swab >100 colonies to inoculate a single culture of 5 mL LB medium supplemented with 50 μg/mL kanamycin, 30 μg/mL chloramphenicol, and 50 μM zinc chloride (to ensure formation of Zn-bound protein).
Incubate cultures at 37 °C for 20 h, shaking at 250 rpm.
Prepare a triplicate series of LB/agar screening plates that contain 50 μg/mL kanamycin, 30 μg/mL chloramphenicol, 50 μM zinc chloride, 25 μM IPTG, and varied ampicillin concentrations (0-1.6 μg/mL). Store plates overnight at 4 °C. Storage of the plates for longer periods of time is not recommended.
Incubate the LB/agar screening plates at 37 °C for 30-60 min to pre-warm and dry the media.
Spread 20 μL aliquots of the cell culture onto half of each LB/agar screening plate. Library culture should be spread onto half of each plate and C96RIDC1 culture spread onto the other half to detect background cell growth.
Select individual colonies from plates that exhibit no C96RIDC1 growth and separately inoculate each into 5 mL of LB medium containing 50 μg/mL of kanamycin and 30 μg/mL chloramphenicol.
Isolate β-lactamase plasmids individually and subject to DNA sequencing analysis as outlined in steps 6-8.
5. Conclusion
Inspired by both the evolutionary roles of metal ions and their utility in nature's proteome, MDPSA and MeTIR provide a straightforward means to design functional protein assemblies from simple building blocks. The utility of these approaches has been primarily demonstrated with a model protein, cyt cb562, although the design strategies and principles described here can be readily extended to other building blocks. While MDPSA and MeTIR were originally motivated by the desire to circumvent the design of extensive non-covalent interactions, these strategies are not meant to bypass computational protein design approaches, but rather to complement them. Indeed, it is only with the prudent combination of rational (chemical and computational) design approaches and laboratory evolution that it will be possible to engineer systems that surpass the structural, functional, and dynamic complexity of natural PPIs and protein complexes.
Acknowledgments
We thank Drs. Cedric Owens, Jonathan Rittle, and Woon Ju Song for their helpful comments. X-ray crystal structures were collected at the Stanford Synchrotron Radiation Laboratory, which is supported by the U.S. Department of Energy, Offices of Basic Energy Sciences and Biological and Environmental Research, as well as by the NIH. The work reported here was supported in part by the University of California, San Diego. F.A.T. was supported by a Hellman Faculty Scholar Award, a Beckman Young Investigator Award, the Arnold and Mabel Beckman Foundation, the Sloan Foundation, the National Science Foundation (CHE-0908115 and CHE-1306646 for Metal-Templated Interface Redesign), and the Department of Energy (DE-FG02-10ER46677 for Metal-Directed Protein Self-Assembly). L.A.C. was supported by the Molecular Biophysics Training Grant (NIH). R.H.S. was supported by the Chemistry-Biology Interface Training Program (NIH).
Abbreviations
- CA-quin
N-(8-hydroxyquinolin-5-yl)-2-chloroacetamide
- Ccm
cytochrome c maturation cassette
- Cyt
cb562 cytochrome cb562
- DCC
N,N’-dicyclohexylcarbodiimide
- DMF
N,N-dimethylformamide
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- ESI
electrospray ionization
- FPLC
fast protein liquid chromatography
- HCM
hybrid coordination motif
- IA-phen
2-iodo-N-(1,10-phenanthrolin-5-yl)acetamide
- IA-quin
N-(8-hydroxyquinolin-5-yl)-2-iodoacetamide
- IA-terpy
N-([2,2':6',2”-terpyridin]-4'-yl)-2-iodoacetamide
- ICP-OES
inductively coupled plasma optical emission spectrometry
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- LB
lysogeny broth
- MALDI
Matrix-Assisted Laser Desorption/Ionization
- MBPC1
Metal Binding Protein Construct 1
- MDPSA
Metal-Directed Protein Self-Assembly
- MeTIR
Metal-Templated Interface Redesign
- MOPS
3-morpholinopropane-1-sulfonic acid
- PCR
polymerase chain reaction
- PDB
Protein Data Bank
- Phen
1,10-phenanthroline
- PPI
protein-protein interaction
- Quin
8-hydroxyquinoline
- RIDC1
Rosetta Interface Designed Cytochrome 1
- RZ
Reinheitzahl
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SE-AUC
sedimentation equilibrium analytical ultracentrifugation
- SV-AUC
sedimentation velocity analytical ultracentrifugation
- Terpy
2,2’:6’2’’-terpyridine
- TRIS
2-amino-2-hydroxymethyl-propane-1,3-diol
- VOC
vicinal oxygen chelate
References
- 1.Alberts IL, Nadassy K, Wodak SJ. Analysis of zinc binding sites in protein crystal structures. Protein Science. 1998;7:1700–1716. doi: 10.1002/pro.5560070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong RN. Mechanistic diversity in a metalloenzyme superfamily. Biochemistry. 2000;39:13625–13632. doi: 10.1021/bi001814v. [DOI] [PubMed] [Google Scholar]
- 3.Arnold FH, Haymore BL. Engineered metal-binding proteins - purification to protein folding. Science. 1991;252:1796–1797. doi: 10.1126/science.1648261. [DOI] [PubMed] [Google Scholar]
- 4.Arslan E, Schulz H, Zufferey R, Kunzler P, Thony-Meyer L. Overproduction of the Bradyrhizobium japonicum c-type cytochrome subunits of the cbb3 oxidase in Escherichia coli. Biochemical Biophysical Research Communications. 1998;251:744–7. doi: 10.1006/bbrc.1998.9549. [DOI] [PubMed] [Google Scholar]
- 5.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallographica Section D Biological Crystallography. 2011;67:271–81. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergdoll M, Eltis LD, Cameron AD, Dumas P, Bolin JT. All in the family: Structural and evolutionary relationships among three modular proteins with diverse functions and variable assembly. Protein Science. 1998;7:1661–1670. doi: 10.1002/pro.5560070801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertini I, Gray HB, Stiefel EI, Valentine JS. Biological inorganic chemistry, structure & reactivity. University Science Books; Sausalito, CA: 2007. [Google Scholar]
- 8.Braymer JJ, Giedroc DP. Recent developments in copper and zinc homeostasis in bacterial pathogens. Current Opinion in Chemical Biology. 2014;19:59–66. doi: 10.1016/j.cbpa.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brodin JD, Medina-Morales A, Ni T, Salgado EN, Ambroggio XI, Tezcan FA. Evolution of metal selectivity in templated protein interfaces. Journal of the American Chemical Society. 2010;132:8610–8617. doi: 10.1021/ja910844n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole JL, Lary JW, Moody T, Laue TM. Analytical ultracentrifugation: sedimentation velocity and sedimentation equilibrium. Methods in Cell Biology. 2008;84:143–79. doi: 10.1016/S0091-679X(07)84006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conte LL, Chothia C, Janin J. The atomic structure of protein-prtein recognition sites. Journal of Molecular Biology. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 12.Correia JJ, Stafford WF. Sedimentation velocity: A classical perspective. Methods in Enzymology. 2015;562:49–80. doi: 10.1016/bs.mie.2015.06.042. [DOI] [PubMed] [Google Scholar]
- 13.Crowder MW, Spencer J, Vila AJ. Metallo-beta-lactamases: novel weaponry for antibiotic resistance in bacteria. Accounts of Chemical Research. 2006;39:721–8. doi: 10.1021/ar0400241. [DOI] [PubMed] [Google Scholar]
- 14.Der BS, Machius M, Miley MJ, Mills JL, Szyperski T, Kuhlman B. Metal-mediated affinity and orientation specificity in a computationally designed protein jomodimer. Journal of the American Chemical Society. 2012;134:375–385. doi: 10.1021/ja208015j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 16.Faraone-Mennella J, Tezcan FA, Gray HB, Winkler JR. Stability and folding kinetics of structurally characterized cytochrome cb562. Biochemistry. 2006;45:10504–10511. doi: 10.1021/bi060242x. [DOI] [PubMed] [Google Scholar]
- 17.Fass D. Disulfide bonding in protein biophysics. Annual Review of Biophysics. 2012;41:63–79. doi: 10.1146/annurev-biophys-050511-102321. [DOI] [PubMed] [Google Scholar]
- 18.Fletcher S, Hamilton AD. Targeting protein-protein interactions by rational design: mimicry of protein surfaces. Journal of the Royal Society Interface. 2006;3:215–233. doi: 10.1098/rsif.2006.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frausto da Silva JJR, Williams RJP. The biological chemistry of the elements. Oxford University Press; Oxford: 2001. [Google Scholar]
- 20.Ghadiri MR, Choi C. Secondary structure nucleation in peptides - transition-metal ion stabilized alpha-helices. Journal of the American Chemical Society. 1990;112:1630–1632. [Google Scholar]
- 21.Gonen S, DiMaio F, Gonen T, Baker D. Design of ordered two-dimensional arrays mediated by noncovalent protein-protein interfaces. Science. 2015;348:1365–1368. doi: 10.1126/science.aaa9897. [DOI] [PubMed] [Google Scholar]
- 22.Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, et al. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. Journal of Molecular Biology. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 23.Handel TM, Williams SA, Degrado WF. Metal-ion dependent modulation of the dynamics of a designed protein. Science. 1993;261:879–885. doi: 10.1126/science.8346440. [DOI] [PubMed] [Google Scholar]
- 24.Harding MM. The architecture of metal coordination groups in proteins. Acta Crystallographica D Biological Crystallography. 2004;60:849–59. doi: 10.1107/S0907444904004081. [DOI] [PubMed] [Google Scholar]
- 25.Harding MM, Nowicki MW, Walkinshaw MD. Metals in protein structures: A review of their principal features. Crystallography Reviews. 2010;16:247–302. [Google Scholar]
- 26.Holm RH, Kennepohl P, Solomon EI. Structural and functional aspects of metal sites in biology. Chemical Reviews. 1996;96:2239–2314. doi: 10.1021/cr9500390. [DOI] [PubMed] [Google Scholar]
- 27.Hu Z, Gunasekera TS, Spadafora L, Bennett B, Crowder MW. Metal content of metallo-β-lactamase L1 Is determined by the bioavailability of metal ions. Biochemistry. 2008;47:7947–7953. doi: 10.1021/bi8004768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang P-S, Love JJ, Mayo SL. A de novo designed protein–protein interface. Protein Science. 2007;16:2770–2774. doi: 10.1110/ps.073125207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inaba K, Ito K. Structure and mechanisms of the DsbB–DsbA disulfide bond generation machine. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2008;1783:520–529. doi: 10.1016/j.bbamcr.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 30.Jäckel C, Kast P, Hilvert D. Protein design by directed evolution. Annual Review of Biophysics. 2008;37:153–173. doi: 10.1146/annurev.biophys.37.032807.125832. [DOI] [PubMed] [Google Scholar]
- 31.King NP, Bale JB, Sheffler W, McNamara DE, Gonen S, Gonen T, et al. Accurate design of co-assembling multi-component protein nanomaterials. Nature. 2014;510:103–108. doi: 10.1038/nature13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krantz BA, Sosnick TR. Engineered metal binding sites map the heterogeneous folding landscape of a coiled coil. Nature Structural Biology. 2001;8:1042–1047. doi: 10.1038/nsb723. [DOI] [PubMed] [Google Scholar]
- 33.Lebowitz J, Lewis MS, Schuck P. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Science. 2002;11:2067–79. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lippard S, Berg J. Principles of bioinorganic chemistry. University Science Books; Mill Valley, CA: 1994. [Google Scholar]
- 35.Liu CL, Xu HB. The metal site as a template for the metalloprotein structure formation. Journal of Inorganic Biochemistry. 2002;88:77–86. doi: 10.1016/s0162-0134(01)00337-3. [DOI] [PubMed] [Google Scholar]
- 36.Liu Y, Kuhlman B. RosettaDesign server for protein design. Nucleic Acids Research. 2006;34:W235–238. doi: 10.1093/nar/gkl163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mandell DJ, Kortemme T. Computer-aided design of functional protein interactions. Nature Chemical Biology. 2009;5:797–807. doi: 10.1038/nchembio.251. [DOI] [PubMed] [Google Scholar]
- 38.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGuirl MA, Lee JC, Lyubovitsky JG, Thanyakoop C, Richards JH, Gray HB, et al. Cloning, heterologous expression, and characterization of recombinant class II cytochromes c from Rhodopseudomonas palustris. Biochimica Et Biophysica Acta-General Subjects. 2003;1619:23–28. doi: 10.1016/s0304-4165(02)00437-3. [DOI] [PubMed] [Google Scholar]
- 40.Medina-Morales A, Perez A, Brodin JD, Tezcan FA. In vitro and cellular self-assembly of a Zn-binding protein cryptand via templated disulfide bonds. Journal of the American Chemical Society. 2013;135:12013–12022. doi: 10.1021/ja405318d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mergulhão FJM, Summers DK, Monteiro GA. Recombinant protein secretion in Escherichia coli. Biotechnology Advances. 2005;23:177–202. doi: 10.1016/j.biotechadv.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 42.Mou Y, Huang PS, Hsu FC, Huang SJ, Mayo SL. Computational design and experimental verification of a symmetric protein homodimer. Proceedings of the National Academy Sciences USA. 2015;112:10714–9. doi: 10.1073/pnas.1505072112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica Section D Biological Crystallography. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 44.Nossal NG, Heppel LA. The release of enzymes by osmotic shock from Escherichia coli in exponential phase. Journal of Biological Chemistry. 1966;241:3055–3062. [PubMed] [Google Scholar]
- 45.Ortega A, Amoros D, Garcia de la Torre J. Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophysical Journal. 2011;101:892–898. doi: 10.1016/j.bpj.2011.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petersen MTN, Jonson PH, Petersen SB. Amino acid neighbours and detailed conformational analysis of cysteines in proteins. Protein Engineering. 1999;12:535–548. doi: 10.1093/protein/12.7.535. [DOI] [PubMed] [Google Scholar]
- 47.Petsko GA, Ringe D. Protein structure and function. New Science Press; London: 2004. [Google Scholar]
- 48.Radford RJ, Nguyen PC, Ditri TB, Figueroa JS, Tezcan FA. Controlled protein dimerization through hybrid coordination motifs. Inorganic Chemistry. 2010;49:4362–4369. doi: 10.1021/ic100534y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Radford RJ, Nguyen PC, Tezcan FA. Modular and versatile hybrid coordination motifs on alpha-helical protein surfaces. Inorganic Chemistry. 2010;2010:7106–7115. doi: 10.1021/ic100926g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rulíšek L. r., Vondrášek J. Coordination geometries of selected transition metal ions (Co2+, Ni2+, Cu2+, Zn2+, Cd2+, and Hg2+) in metalloproteins. Journal of Inorganic Biochemistry. 1998;71:115–127. doi: 10.1016/s0162-0134(98)10042-9. [DOI] [PubMed] [Google Scholar]
- 51.Salgado EN, Faraone-Mennella J, Tezcan FA. Controlling protein-protein interactions through metal coordination: Assembly of a 16-helix bundle protein. Journal of the American Chemical Society. 2007;129:13374–13375. doi: 10.1021/ja075261o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salgado EN, Lewis RA, Faraone-Mennella J, Tezcan FA. Metal-mediated self-assembly of protein superstructures: Influence of secondary interactions on protein oligomerization and aggregation. Journal of the American Chemical Society. 2008;130:6082–6084. doi: 10.1021/ja8012177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salgado EN, Lewis RA, Mossin S, Rheingold AL, Tezcan FA. Control of protein oligomerization symmetry by metal coordination: C2 and C3 symmetrical assemblies through CuII and NiII Coordination. Inorganic Chemistry. 2009;48:2726–2728. doi: 10.1021/ic9001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salgado EN, Ambroggio XI, Brodin JD, Lewis RA, Kuhlman B, Tezcan FA. Metal-templated design of protein interfaces. Proceedings of the National Academy Sciences USA. 2010;107:1827–1832. doi: 10.1073/pnas.0906852107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schreiber G, Fleishman SJ. Computational design of protein–protein interactions. Current Opinion in Structural Biology. 2013;23:903–910. doi: 10.1016/j.sbi.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 56.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophysical Journal. 2000;78:1606–19. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schuck P. A model for sedimentation in inhomogeneous media: dynamic density gradients from sedimenting co-solutes. Biophysical Chemistry. 2004;108:187–200. doi: 10.1016/j.bpc.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 58.Scrofani SD, Chung J, Huntley JJ, Benkovic SJ, Wright PE, Dyson HJ. NMR characterization of the metallo-beta-lactamase from Bacteroides fragilis and its interaction with a tight-binding inhibitor: Role of an active-site loop. Biochemistry. 1999;38:14507–14. doi: 10.1021/bi990986t. [DOI] [PubMed] [Google Scholar]
- 59.Smith RM, Martell AE, Motekaitis RJ. NIST critically selected stability constants of metal complexes. NIST standard reference database 46.6.0. 2004 [Google Scholar]
- 60.Song WJ, Sontz PA, Ambroggio XI, Tezcan FA. Metals in protein-protein interfaces. Annual Review of Biophysics. 2014;43:409–431. doi: 10.1146/annurev-biophys-051013-023038. [DOI] [PubMed] [Google Scholar]
- 61.Song WJ, Tezcan FA. A designed supramolecular protein assembly with in vivo enzymatic activity. Science. 2014;346:1525–1528. doi: 10.1126/science.1259680. [DOI] [PubMed] [Google Scholar]
- 62.Tomatis PE, Fabiane SM, Simona F, Carloni P, Sutton BJ, Vila AJ. Adaptive protein evolution grants organismal fitness by improving catalysis and flexibility. Proceedings of the National Academy Sciences USA. 2008;105:20605–20610. doi: 10.1073/pnas.0807989106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsai C-J, Lin SL, Wolfson HJ, Nussinov R. Protein-protein interfaces: Architectures and interactions in protein-protein interfaces and in protein cores - their similarities and differences. Critical Reviews in Biochemistry and Molecular Biology. 1996;31:127–152. doi: 10.3109/10409239609106582. [DOI] [PubMed] [Google Scholar]
- 64.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. Journal of Applied Crystallography|Journal of Applied Crystallography. 1998;30:1022–1025. [Google Scholar]
- 65.Volbeda A, Hol WGJ. Pseudo 2-fold symmetry in the copper-binding domain of arthropodan hemocyanins - Possible implications for the evolution of oxygen-transport proteins. Journal of Molecular Biology. 1989;206:531–546. doi: 10.1016/0022-2836(89)90499-3. [DOI] [PubMed] [Google Scholar]
- 66.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, et al. Overview of the CCP4 suite and current developments. Acta Crystallographica Section D-Biological Crystallography. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu D, Tsai CJ, Nussinov R. Hydrogen bonds and salt bridges across protein-protein interfaces. Protein Engineering. 1997;10:999–1012. doi: 10.1093/protein/10.9.999. [DOI] [PubMed] [Google Scholar]