Abstract
Mitochondria are highly dynamic organelles that undergo continuous cycles of fission and fusion to maintain essential cellular functions. An imbalance between these two processes can result in many pathophysiological outcomes. Dengue virus (DENV) interacts with cellular organelles, including mitochondria, to successfully replicate in cells. This study used live-cell imaging and found an increase in mitochondrial length and respiration during DENV infection. The level of mitochondrial fission protein, Dynamin-related protein 1 (Drp1), was decreased on mitochondria during DENV infection, as well as Drp1 phosphorylated on serine 616, which is important for mitochondrial fission. Viral proteins NS4b and NS3 were also associated with subcellular fractions of mitochondria. Induction of fission through uncoupling of mitochondria or overexpression of Drp1 wild-type and Drp1 with a phosphomimetic mutation (S616D) significantly reduced viral replication. These results demonstrate that DENV infection causes an imbalance in mitochondrial dynamics by inhibiting Drp1-triggered mitochondrial fission, which promotes viral replication.
Keywords: Dengue virus, Drp1, mitochondria, fusion, fission, live-cell imaging, dendritic cells
Introduction
DENV is a mosquito-borne human pathogen of global medical importance with an estimation of 390 million infections every year [1–3]. DENV causes an acute febrile illness that, in some patients, is associated with a life-threatening plasma leakage syndrome termed dengue hemorrhagic fever (DHF) [4]. The principal target cells for DENV infection are from the myeloid-derived lineage (monocytes, macrophages and dendritic cells), hepatocytes and endothelial cells [5–15]. Several studies indicate that higher viremia levels during DENV infection correlate with the severity of disease [16–19]. DENV infection is also often associated with liver dysfunction as a result of direct viral effect on hepatocytes and dysregulated immune response against the virus [20]. Both viral and host factors seem to play a role in disease pathogenesis, but little is known about DENV-induced mechanisms that trigger modulation of organelles during viral infection [17, 21, 22].
DENV is a member of the Flaviviridae family and its positive-sense single-stranded RNA genome of 10.7 kb encodes for 10 viral proteins. This limited coding capacity makes it fully reliant on the host machinery for its viral life cycle. Like many viruses, DENV needs to modulate host cell metabolism and organelles for its own replication, interfering with different cellular organelles [23–26].
Mitochondria are essential organelles involved in cellular energy production and sensing of metabolic homeostasis. They serve as an anti-viral signaling platform for MAVS signaling [27] and contribute to activation of apoptosis and cell death [28–30]. Recently, mitochondria were shown to release their DNA into the cytosol during stress to trigger a type I interferon response [31]. These processes indicate an important role of mitochondria in initiating responses against viruses.
Mitochondrial morphology depends on highly dynamic processes, fusion and fission [32]. Mitochondrial fission involves the oligomerization of Dynamin-related protein 1 (Drp1) around the outer mitochondrial membrane, constricting the mitochondrion to trigger its division [33, 34]. Localization of Drp1 is regulated by differential phosphorylation of serine residues at positions 616 and 637 (S616-P and S637-P, respectively). Phosphorylation at S637 is thought to retain Drp1 in the cytoplasm whereas phosphorylation at S616 induces Drp1 localization to the fission site on the mitochondrion [35]. The endoplasmic reticulum and actin polymerization play a role in Drp1-triggered mitochondrial fission by initiating constriction of mitochondria [36, 37]. On the other hand, mitofusin 1 (Mfn1) and Mfn2, together with optic atrophy protein 1 (Opa1) are the core components of the mitochondrial fusion machinery. Mitochondrial fission and fusion proteins are regulated by post-translational modifications, including phosphorylation as described above for Drp1 [38]. The ability of mitochondria to undergo fission and fusion, and to move in the cells is critical for their function, as defects in mitochondrial dynamics are implicated in many neurodegenerative diseases [39–41].
DENV infection has previously been reported to induce structural and functional changes in organelles in vitro, including mitochondria [42–45]. DENV protease NS2b3 was shown to partially cleave Mfn1 and Mfn2 resulting in degradation of the proteins, which attenuated interferon responses and cell death [46]. A recent report was published while this manuscript was being prepared that showed that DENV NS4b induced mitochondrial elongation by inhibition of Drp1 activation and limited activation of the interferon response in a liver cell line [47]. Although these reports signify the importance of mitochondria and mitochondrial proteins during viral infection, the underlying mechanisms that link mitochondria and its proteins to DENV replication in target cell lines and primary cells has not been well studied.
Using live-cell imaging of DENV-infected cell cultures, we observed that DENV infection induces significant mitochondrial elongation. Mitochondria from DENV-infected cells displayed increased respiration, which can lead to an increase in ATP production, when compared to uninfected cells, and viral proteins such as NS4b were preferentially localized in subcellular fractions of mitochondria. Characterization of fusion and fission proteins revealed a decrease in Drp1 on mitochondria in the Huh7 cell line and in monocyte-derived dendritic cells (DCs). Drp1 decrease was correlated with a reduction of Drp1 phosphorylation on S616 in the Huh7 cell line. Conversely, induction of mitochondrial fission or overexpression of wild-type (WT) Drp1 and Drp1 S616D inhibited DENV replication. These results demonstrate that DENV infection causes an imbalance in mitochondrial dynamics by inhibiting Drp1-triggered mitochondrial fission, which promotes viral replication.
Results
DENV infection induces mitochondrial elongation and reduces mitochondrial motility
We recently described a reporter system for live-cell imaging of DENV-infected cells based on the pNS4b5-eGFP plasmid, which expresses a GFP fusion protein that translocates from the cytoplasm to the nucleus in infected cells [48]. To examine the structure of mitochondria in cells infected with DENV, we transfected cells with the pNS4b5-eGFP reporter and subsequently stained cells with MitoTracker Deep Red, a fluorescent stain that accumulates in active mitochondria. Uninfected liver cell lines (Huh7, Fig. 1A; HepG2, Fig. S1A and Huh7.5, Fig. S1B) and an epithelial cell line (Vero, Fig. 1B) showed both fused and fragmented mitochondria. In contrast, DENV-infected cells showed mostly fused mitochondria at 48h post-infection in all cell lines. The mitochondrial network in DENV-infected cells was filamentous and interconnected. To confirm that the mitochondrial elongation was not due to transfection of the reporter, which contains the DENV protein NS4b, cells were transfected with pTagRFP-mito (Huh7.5 and Huh7 cells, Fig. S1D and S1E, respectively) and infected 24h later with DENV. At 48h post-infection, cells were stained for DENV antigen and analyzed by confocal microscopy. Similarly, we stained DENV-infected and uninfected cells with COX-IV antibody, a mitochondrial marker, and DENV antigen (HepG2 cells, Fig. S1C). As observed previously, all cell lines infected with DENV showed elongated mitochondria at 48h post-infection compared to uninfected cells. Quantitative measurement of the change in mitochondrial lengths using a macro developed in ImageJ (NIH) showed that median lengths of mitochondria increased at 24h and became statistically significant by 48h post-infection when compared to uninfected cells (Fig. 1B). Analysis of DENV-infected Huh7 cells by transmission electron microscopy, characterized by the presence of convoluted membranes (DENV, Fig. 1C and 1D) and invaginated vesicles (DENV, Fig. 1D and 1E) specifically induced by DENV as previously described [24], also showed an increase of mitochondrial lengths when compared to uninfected cells (Fig. 1C and 1D).
Fig. 1. DENV infection induces mitochondrial elongation.
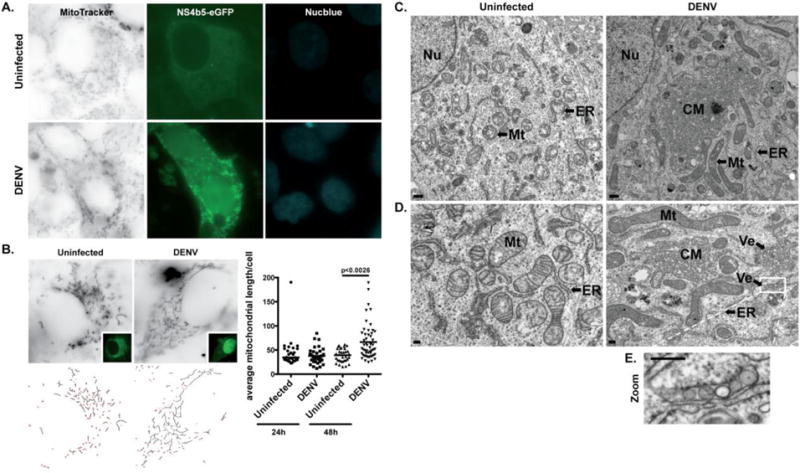
A. Immunofluorescence analysis of mitochondrial morphology in uninfected (cytoplasmic GFP) or DENV-infected (cytoplasmic and nuclear GFP) Huh7 cells. Cells were transfected with pNS4b5-eGFP reporter and infected with DENV (MOI of 0.1) for 48h. Cells were subsequently stained with MitoTracker Deep Red FM prior to imaging. Images were taken at 100× magnification and are representative of at least three independent experiments. B. Quantitative analysis of mitochondrial lengths in uninfected and infected Vero cells after 24h and 48h of infection. Vero cells transfected with pNS4b5-eGFP reporter were infected with DENV (MOI of 1) for 24h and 48h and mitochondria were stained with MitoTracker Deep Red FM. The average mitochondrial lengths for cells within each condition were calculated using a macro developed in ImageJ (NIH). Statistical analysis was done on the average mitochondrial lengths of each cell as described. The results reflect three independent experiments. C. and D. Ultra-thin sections TEM images of uninfected and DENV-infected (MOI of 1) resin-embedded Huh7 cells at different magnifications (C., ×7900, scale bar: 500 nm; D., ×19000, scale bar: 200 nm). Uninfected Huh7 cells exhibit typical ultrastructural morphology of non-infected cells. DENV-infected cells display elongated mitochondria (Mt), along with previously described convoluted membranes (CM) and invaginated vesicles (Ve) in proximity to the endoplasmic reticulum (ER) membranes. Nucleus (Nu) appears on lower magnification images (C.). E. Magnified image of box area illustrates invaginated vesicles (Ve) present in DENV-infected cells.
Defects in fusion and fission have been shown to reduce motility of mitochondria [49]. Therefore, we performed time-lapse imaging in Huh7 cells transfected with pTagRFP-mito and pNS4b5-eGFP and infected with DENV for 48h to evaluate the motility of mitochondria in cells during DENV infection, (Fig. S2, Movie S1 and S2). Prior to imaging mitochondria movements in cells, DENV infected and uninfected cells were identified by localization of GFP to the nucleus or cytoplasmic GFP only, respectively (Fig. S2). We observed not only an increase in mitochondrial lengths but also a reduction of mitochondrial movements in DENV-infected cells (Movie S2) compared to uninfected cells (Movie S1), indicating that mitochondrial motility is reduced in DENV-infected cells.
DENV NS4b and NS3 proteins are enriched in the mitochondrial fraction
Recently, HCV proteins C and NS4b were found to be associated with mitochondria [50–52]. Therefore, to assess whether DENV proteins interact with mitochondria, we analyzed the presence of DENV proteins in isolated mitochondria by western blot. We found that NS4b and NS3 were enriched in the mitochondrial fraction of DENV-infected Huh7 cells and DCs (Fig. 2A and 2B, respectively). Immunofluorescence analysis in DENV-infected Huh7 cells revealed that NS4b did not colocalize with mitochondria, but instead NS4b staining was mostly found juxtaposed to mitochondria, in dot-like structures colocalizing with the endoplasmic reticulum (ER) marker Sec61β (Fig. 2C). ER membranes have been reported to associate with mitochondria to form mitochondria-associated membranes (MAMs). To assess the presence of MAMs in the mitochondrial fractions, we analyzed the levels of FACL4, a MAM marker, by western blot. FACL4 was detected in the mitochondrial fractions of Huh7 cells and DCs (Fig. 2A and 2B), suggesting that DENV proteins interact with mitochondria through the MAMs. These results suggest that DENV may influence mitochondrial changes through viral protein interactions at the MAM-mitochondrial interface. We noticed the presence of additional bands of higher and lower molecular weight after FACL4 blotting. Whether they are a result of post-translational modifications or cross-reaction of FACL4 antibody with other isoforms of FACL is not clear. We also observed a decrease of the highest density band of FACL4 in the mitochondrial fraction of DENV-infected cells, particularly Huh7, whereas it is increased in the total cell lysate and cytosolic fraction, suggesting a decrease of ER-mitochondria contacts in DENV-infected cells.
Fig. 2. DENV NS4b and NS3 proteins are enriched in the mitochondrial fraction.
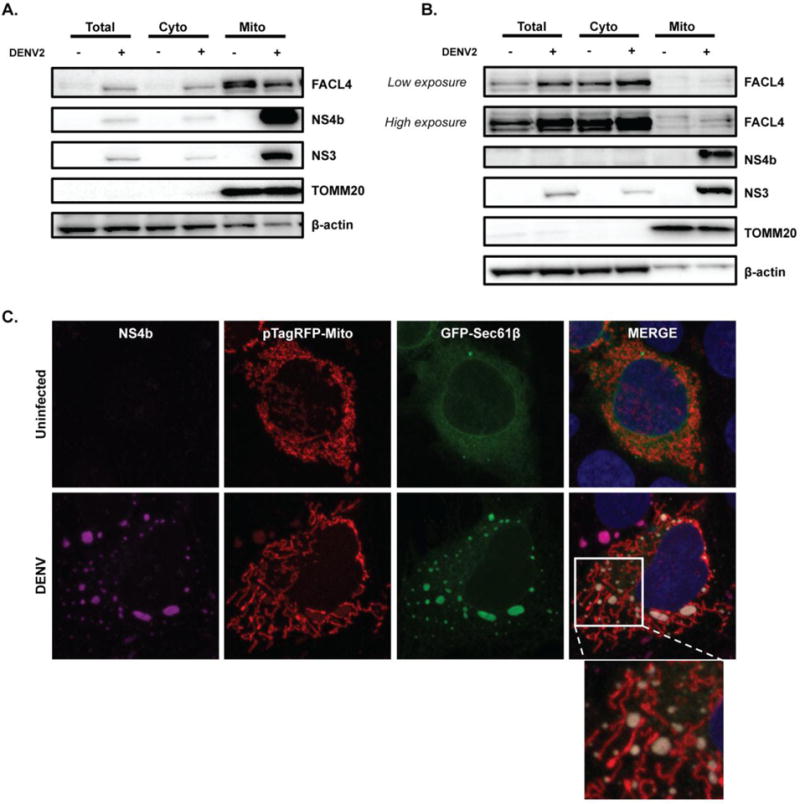
A. and B. Western blot analysis of NS4b and NS3 protein levels was performed in subcellular fractions from uninfected and DENV-infected Huh7 cells (A.) and DCs (B.). After 48h of infection, cells were fractionated to separate cytosolic and mitochondrial fractions, and compared to total cell lysates. TOMM20 was used as a loading and purity control for mitochondrial fraction. FACL4 antibody served as a marker for MAMs. β-actin antibody was used as a loading control. Western blots are representative of three independent experiments. C. Immunofluorescence analysis of NS4b protein localization in uninfected and DENV-infected Huh7 cells after 48h (MOI of 1). Cells were transfected with pTagRFP-mito and pAcGFP-Sec61β. Images were taken with confocal LSM800 at ×63×1.5 magnification.
DENV-infected cells display increased mitochondrial respiration
To assess if changes in mitochondrial morphology during DENV infection affect the function of mitochondria, the oxygen consumption rate (OCR), a measure of cellular respiration, was measured in uninfected and DENV-infected cells (Fig. 3A). Sequential addition of specific mitochondrial inhibitors to the cell culture medium was used to assess the function of each component of the respiratory chain (Fig. 3B). Basal levels of OCR in DENV-infected cells were increased compared to uninfected cells. In turn, mitochondrial respiration was also increased in DENV-infected cells. The addition of oligomycin, which inhibits F0/F1 ATPase (complex V), demonstrated that the potential for ATP production was significantly higher in DENV-infected cells, as compared with uninfected cells, but proton leak was unchanged. DENV-infected cells displayed increased respiration compared to uninfected cells. There was no significant difference in coupling efficiency between DENV-infected and uninfected cells. Maximal respiration was comparable between uninfected and DENV-infected cells, and similar to basal respiration in DENV-infected cells, suggesting that DENV increased mitochondrial respiration to their maximum. Accordingly, spare respiratory capacity was decreased in DENV-infected cells. These results indicate that elongated mitochondria in DENV-infected cells are functional and display increased respiration similar to maximal, resulting in more energy production when compared to uninfected cells.
Fig. 3. DENV-infected cells display increased mitochondrial respiration.
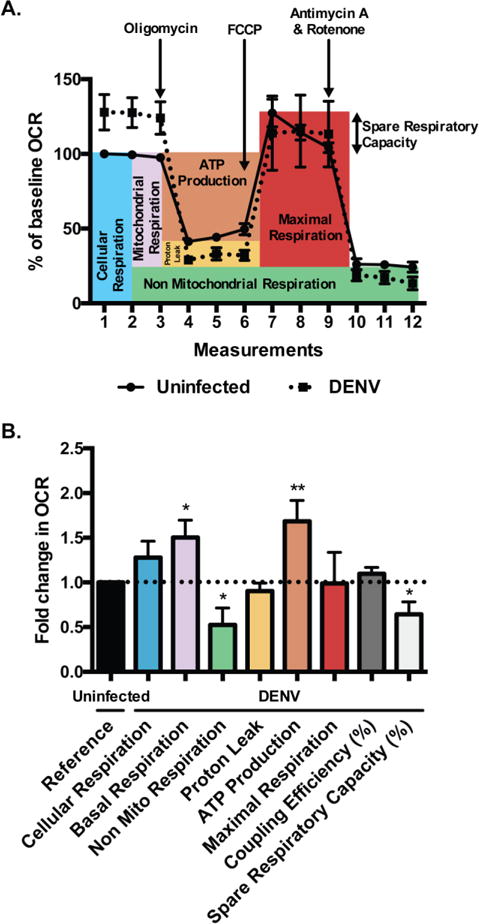
A. OCR in uninfected and DENV-infected Huh7 cells at 48h post-infection (MOI of 1) were measured in real time under basal conditions and in response to sequential injections of oligomycin (1 μM), FCCP (1 μM) and antimycin A/rotenone (0.5 μM) using XF24 Seahorse analyzer. Data are shown as percent of baseline OCR relative to first measurement from uninfected cells and represent mean +/− SEM of three independent experiments with at least five biological replicates per condition and measurement. B. Mitochondrial function parameters were analyzed according to Seahorse Bioscience instructions and represented as fold change in OCR relative to uninfected cells. Data are shown as mean +/− SD, analyzed with unpaired Student-t test.
DENV infection decreases the level of Drp1 and Mfn2 on mitochondria
Mitochondrial fusion requires fusion of the inner membrane of mitochondria by Opa1 and the outer membrane by Mfn1 and Mfn2 [53, 54]. Similarly, overexpression of Opa1 or mitofusins promote mitochondrial elongation. To analyze whether mitochondrial elongation seen during DENV infection was due to an increase in fusion events, mitochondrial and cytosolic fractions were isolated from DENV-infected and uninfected cells and fusion protein levels were assessed by western blot. Mfn1 and Opa1 protein levels in mitochondrial fractions from Huh7 cells and DCs were similar between uninfected and DENV-infected cells (Fig. 4A and 4B). By contrast, the level of Mfn2 protein on mitochondria was decreased in DENV-infected cells compared to uninfected cells. Immunofluorescence analysis also showed a decrease in Mfn2 fluorescence of DENV-infected cells, and no change in Mfn1 (Fig. S3A and S3B). These results indicate that elongated mitochondria are not due to an increase in fusion during DENV infection.
Fig. 4. DENV infection decreases the level of Drp1 and Mfn2 on mitochondria.
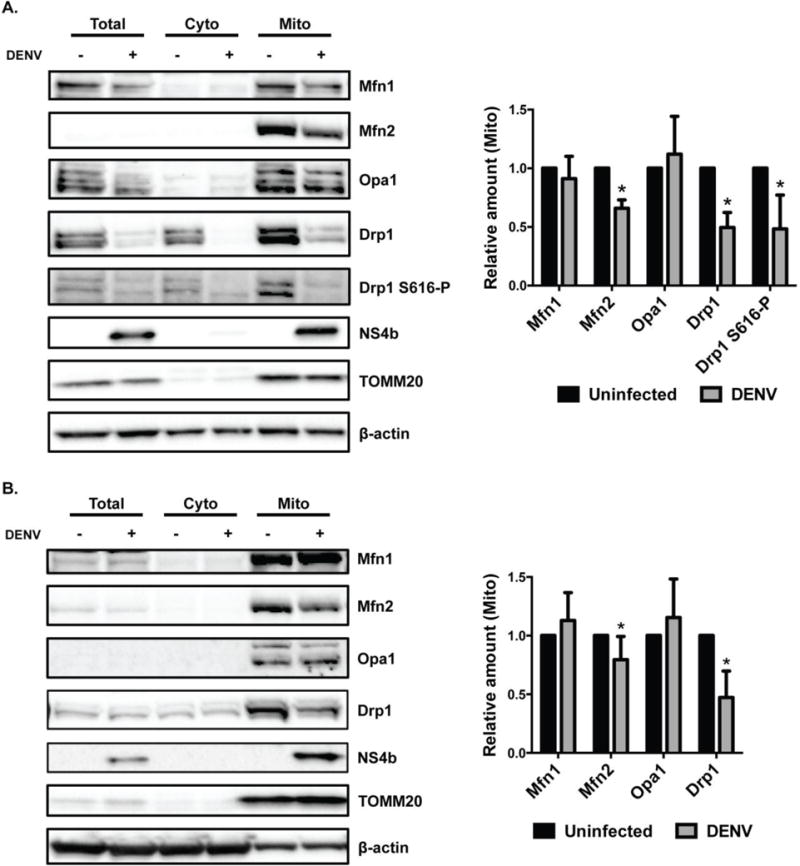
A. and B. DENV-infected and uninfected Huh7 cells (A.) and DCs (B.) were harvested at 48h post-infection (pi) and total, cytosolic and mitochondrial fractions were analyzed by western blot for Mfn1, Mfn2, Opa1, Drp1 and Drp1 S616-P protein levels. NS4b antibody served as a control for DENV infection. TOMM20 antibody was used as a loading and purity control for mitochondrial fraction. β-actin antibody was used as a loading control for all fractions. For quantification of protein levels in mitochondrial fraction, blots were evaluated after normalization to loading controls and expressed as mean +/− SD of at least three independent experiments. Statistical analysis was performed using unpaired Student-t test.
Mitochondrial elongation due to a defect in mitochondrial fission and decreased expression of Drp1 has previously been described [55, 56]. To investigate whether DENV-induced mitochondrial elongation was a result of impaired mitochondrial fission, we analyzed Drp1 and Drp1 S616-P protein levels from uninfected and DENV-infected Huh7 cells and DCs by western blot. The level of Drp1 in total cell lysates from DENV-infected Huh7 cells was decreased when compared to uninfected cells by western blot and confirmed in cells by immunofluorescence analysis (Fig. 4A and S4A); there was no change in Drp1 mRNA levels (Fig. S5A). We observed at higher magnification that the remaining Drp1 in DENV-infected Huh7 cells was less associated with mitochondria compared to uninfected cells (Fig. S5B). In contrast, Drp1 was not decreased in total cell lysates in DENV-infected DCs when compared to uninfected cell lysates by western blot (Fig. 4B).
Mitochondrial fractions from both DENV-infected Huh7 cells and DCs showed a significant decrease in Drp1 protein levels when compared to uninfected cells (Fig. 4A and 4B). Accordingly, the level of Drp1 S616-P was also markedly decreased in total cell lysate and in mitochondrial fraction of Huh7 cells (Fig. 4A). Immunofluorescence analysis of DENV-infected Huh7 cells showed a significant decrease in Drp1 S616-P (Fig. S4B). Drp1 S616-P also showed less colocalization with mitochondria in DENV-infected cells (Fig. S5C). In DCs, Drp1 S616-P was poorly detected by western blot (unpublished data). Overall, these results suggest that DENV infection inhibits Drp1-triggered mitochondrial fission by decreasing the levels of Drp1. Also, DENV infection reduced Drp1 S616-P protein on mitochondria in a cell type-specific manner.
Induction of mitochondrial fission inhibits DENV replication
Several approaches were used to investigate the effect of mitochondrial fission on DENV replication. DENV-infected cells treated with a reversible mitochondrial uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP) [57] for 6h (from 24h to 30h post-infection) displayed short mitochondria and viral titers in culture supernatants were decreased compared to untreated cells (Fig. 5A and 5B). Removal of CCCP from the media resulted in an increase in mitochondrial lengths and viral titers back to the level in untreated DENV-infected cell cultures, demonstrating a reversible effect on mitochondrial function rather than a toxic effect on cells. The intensity of DENV antigen staining also was significantly decreased in DENV-infected cells treated with CCCP and increased back to levels comparable to DENV-infected untreated cells when CCCP was removed (Fig 5B and S6). These results suggest that mitochondrial fission reduces the production of DENV proteins and infectious viral particles.
Fig. 5. CCCP-induced mitochondrial fragmentation or depletion of mitochondrial fusion protein Mfn2 affects DENV infection.
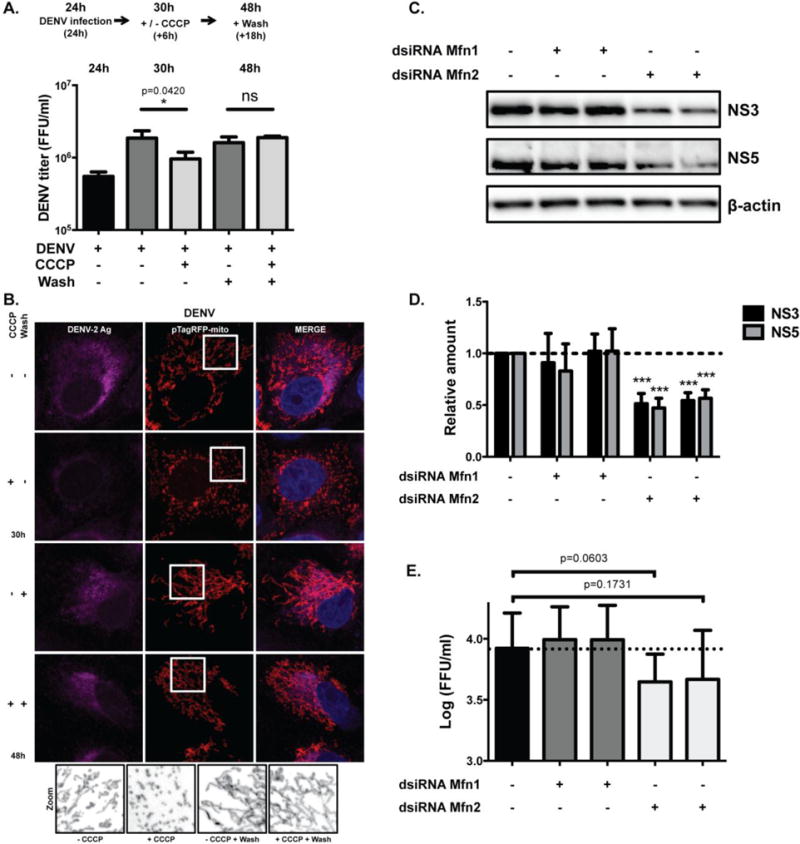
A. Supernatants from DENV infected Vero cells were collected after 24h of infection. Cells were treated with medium containing DMSO or CCCP 10 μM and supernatants were again collected after 6h post treatment (30h pi). To reverse CCCP treatment, DMSO and CCCP-treated cells were washed once with PBS and supernatant was collected (48h pi). DENV titers from collected supernatants were analyzed using an ELISpot assay. Data are representative of one out of two independent experiments, analyzed with unpaired Student-t test. Error bars represent SD of three biological replicates per condition. B. Immunofluorescence analysis of mitochondrial morphology of uninfected and DENV-infected Vero cells after indicated treatments. Cells were transfected with pTagRFP-mito, challenged with DENV (MOI of 1) and treated as indicated before being fixed and stained with antibody against DENV antigen. Images were taken with confocal LSM800 at ×63×1.5 magnification. Magnified images of boxed area show mitochondria stained with pTagRFP-mito for each condition. Images were converted to black and white using ImageJ. C. Huh7.5 cells were transfected with two independent sets of dsiRNAs against Mfn1 or Mfn2 (set (A), left and set (B), right) and infected with DENV (MOI of 0.1) for 24h. Western blots were probed with NS3 and NS5 antibodies and equal protein loading was assessed using an antibody against β-actin. Blot is representative of at least four independent experiments. D. For quantification of NS3 and NS5 protein levels, blots of four independent experiments were evaluated after normalization to β-actin and expressed as mean +/− SEM. Statistical analysis was performed using unpaired Student-t test. E. DENV titers from collected supernatants of Huh7.5 cells treated with dsiRNAs against Mfn1 or Mfn2 were analyzed using an ELISpot assay. DENV titers were transformed to log (FFU/ml) and displayed as mean +/− SEM. Statistical analysis was performed using paired t-test. Titers are representative of independent experiments (n=3).
Since CCCP decouples mitochondria and impairs ATP production, the impairment in viral production may be due to lower availability of energy in the cells. Therefore, we wanted to induce fission of mitochondria by knocking down the fusion proteins, Mfn1 and Mfn2 (Fig. S7A). siRNA knockdown of Mfn2 protein, but not Mfn1 protein, decreased the expression of DENV proteins NS3 and NS5 (Fig. 5C and 5D). Viral titers were reduced by at least 40% after 24h of infection when compared to controls and were on the threshold of statistical significance (p = 0.06 and 0.17, by paired t-test) (Fig. 5E).
To further examine the effect of mitochondrial fission on DENV infection, we knocked down endogenous Drp1 with siRNAs targeting the 3′ untranslated region (3′UTR) of Drp1 and transfected cells with plasmids expressing WT, phosphomimetic, or phosphodefective mutants of human Drp1 [55]. Endogenous Drp1 was observed as a double band and bands of higher molecular weight represent Drp1 WT or mutant constructs. In cells expressing Drp1 K38A there is an additional unidentified, intermediate-sized band (Fig. S7B). DENV-infected cells overexpressing WT Drp1 displayed a decrease in the levels of DENV NS3 and NS4b proteins by western blot analysis (Fig. 6A and 6B) and a reduction in viral titers (Fig. 6C) when compared to DENV-infected cells transfected with a control vector. Drp1 S616-P was reported to trigger mitochondrial fission [35] whereas S637-P results in a reduction in mitochondrial fission [58]. The overexpression of phosphomimetic Drp1 S616D caused a significant decrease in NS3 and NS4b levels (Fig 6A and 6B), as well as a decrease in virus production (Fig. 6C) when compared to the control, indicating that the ‘fission active’ form of Drp1 affects DENV protein expression and virus production. Overexpression of phosphodefective Drp1 S637A partially reduced virus titer, although the difference was not statistically significant. In contrast, the levels of DENV proteins and virus production were not affected by overexpression of phosphodefective Drp1 S616A or phosphomimetic Drp1 S637D, both of which are thought to be defective at inducing fission [55]. We further analyzed the effect of Drp1 GTPase activity on DENV infection by transfecting cells with a dominant negative form of Drp1 with defective GTPase activity (Drp1 K38A) [59]. Overexpression of Drp1 K38A increased levels of NS4b and NS3 proteins and viral titers when compared to cells expressing WT Drp1, indicating that GTPase activity of Drp1 is required for inhibition of DENV replication. Overall, our results indicate that inducing mitochondrial fission perturbs DENV production.
Fig. 6. Mitochondrial fission inhibits DENV replication.
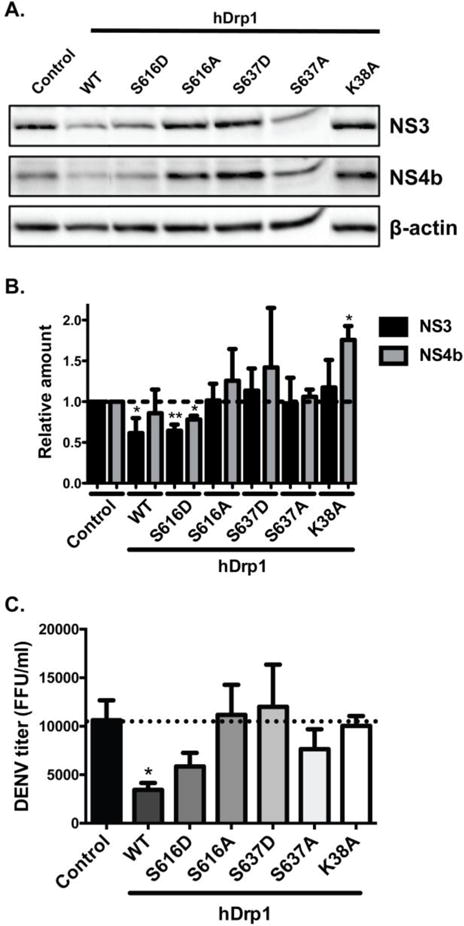
A. Huh7.5 cells were transfected with dsiRNA against the untranslated region of Drp1. Cells were transfected with plasmids expressing Drp1 wildtype, phosphomimetic mutants of human Drp1 (S616D and S637D), phosphodeficient mutants (S616A and S637A) and dominant negative protein Drp1 K38A and infected with DENV (MOI of 0.1) for 24h. Western blots were probed with antibodies directed against NS3 and NS4b proteins and equal protein loading was assessed using an antibody against β-actin. Blot is representative of at least three independent experiments. B. For quantification of protein levels, blots of three independent experiments were evaluated after normalization to β-actin and expressed as mean +/− SEM. Statistical analysis was performed using unpaired Student-t test. C. DENV titers from collected supernatants of Huh7.5 cells expressing indicated Drp1 proteins were analyzed using an ELISpot assay. Titers are representative of independent experiments (n=3).
Discussion
Virus-induced mitochondrial changes
The use of live-cell imaging of DENV-infected cells showed significant increase in mitochondrial elongation over time when compared to uninfected cells due to an inhibition of mitochondrial fission. Mitochondria are dynamic organelles that constantly undergo cycles of fusion and fission [32]. Many mitochondrial and cellular functions rely on mitochondria dynamics, suggesting that altering mitochondria dynamics could serve as a viral strategy to interfere with cellular signaling pathways. Several studies have reported that mitochondria dynamics can be altered during infection with pathogens such as viruses, bacteria and parasites, leading to changes in mitochondrial morphology [60–64]. Drp1 has also previously been shown to be targeted by viruses [60, 65]. Our results, together with those from Chatel-Chaix et al, show that inhibition of fission during DENV infection is due to a reduction in Drp1 protein levels on mitochondria. DENV NS4b is reported to trigger mitochondrial elongation, which was also observed with other DENV serotypes and Zika virus [47], and we also found a decrease of Drp S616-P after infection with two strains of Zika virus (unpublished data). In turn, overexpression of the ‘fission active’ form of Drp1 inhibits DENV titers and viral antigen suggesting that DENV-specific modulation of Drp1 promotes viral replication. Both liver cell line and DCs showed decrease of Drp1 (as well as Mfn2) on mitochondria during DENV infection. However, Drp1 S616-P was poorly detected in DCs compared to Huh7 cells by western blot. This result indicates that phosphorylation of S616 may be important in liver cells but not necessary in DCs. Phosphorylation state of other serines in Drp1 have been shown to regulate fission [35, 66] and these sites may be important for mitochondrial regulation in DCs. The advantage of using cellular fractionation in this study was the ability to define significant changes in the protein profile on mitochondrial fractions in both cell lines and DCs during DENV infection that, in some cases, would not have been identified using total cell lysates.
Drp1 is the master regulator of mitochondrial fission [55]. The current model suggests that ER encircles mitochondria at sites of fission, and ER-associated inverted formin 2 (INF2) then stimulates actin polymerization, providing the force required for partial constriction of the mitochondria, thereby facilitating the translocation of Drp1 to these pre-constriction contact sites in the outer mitochondrial membrane [36]. The ER is also the site of DENV replication and many DENV proteins remain embedded in the ER through their transmembrane domains [67]. Whether DENV prevents Drp1 binding to receptors on mitochondria or DENV affects formation of ER-mitochondria contacts to prevent Drp1-triggered mitochondrial fission remains to be evaluated.
We isolated mitochondrial fractions using magnetic beads coated with TOM22 antibody. Our results show that we not only pulled down mitochondria but also pulled down NS4b in DENV-infected mitochondrial fractions. Immunofluorescence did not reveal colocalization of mitochondria with NS4b but rather that they were in close proximity. Electron microscopy observations performed in our study indicate that DENV-induced convoluted membranes, which have previously been described as ER-derived membranes and contain non-structural proteins such as NS4b [68], are in close contact with elongated mitochondria. These results suggest that mitochondrial isolation using magnetic beads were also able to pull down mitochondrial-associated membranes. The decrease of FACL4 in mitochondrial fractions of DENV-infected cells suggests that interactions between the ER and mitochondria are reduced, which may also impair Drp1 recruitment to mitochondria to induce fission. On the other hand, we observed higher levels of FACL4 in DENV-infected cell lysates, which may be associated with changes in lipid metabolism during infection. There also remains the possibility that viral proteins in the ER membrane or MAMs may be located in regions where mitochondrial division is initiated and inhibit Drp1 binding to mitochondria-ER contact sites.
Mitochondrial fusion proteins during DENV infection
Our data revealed a decrease of Mfn2 protein on the mitochondria during DENV infection, whereas the level of Mfn1 protein was not significantly affected. Both proteins are anchored to the outer mitochondrial membrane and can form homotypic or heterotypic interactions that are both functional for mitochondrial fusion. Several studies have reported redundancy between the two proteins for mitochondrial fusion function [53, 69, 70]. Therefore, the decrease of Mfn2 on the mitochondria during DENV infection could be compensated by Mfn1 during fusion. One group recently reported that Mfn2, as well as Mfn1 protein, was cleaved by NS2b3 protease in A549 cells during DENV infection [46]. Cleavage could contribute to the decrease in Mfn2 levels observed in our experiments. However, the cleavage reported in A549 cells affected only a small fraction of the total Mfn2 protein [46], suggesting another mechanism is involved in the reduction of Mfn2. Mitochondrial fusion in A549 cells infected with DENV was reported to be Mfn1 mediated [46]. In contrast, our results in Huh7 cells and DCs show that mitochondrial elongation is due to inhibition of Drp1 and not due to Mfn1. The difference in findings from the previous study and our results may stem from the cell lines or cell types used for analysis. Uninfected A549 cells have been reported to display elongated mitochondria due to low expression levels of Drp1 [71, 72].
Although knockdown of Mfn2 indicates that it plays a role in DENV replication, Mfn2 did not play a role in mitochondrial elongation during DENV infection. Mfn2 has been reported to tether the ER to mitochondria, facilitating the transfer of calcium between the two organelles [73–75]. HIV-1 Vpr protein triggers the ubiquitinylation and turnover of Mfn2 by the CUL4 E3 ligase, leading to reduced ER-mitochondria contacts [76]. In contrast, other studies reported an increase of ER-mitochondria contacts in Mfn2 depleted cells compared to wild-type cells [77, 78]. It is possible that decreased levels of Mfn2 during DENV infection perturb the interaction between the ER and mitochondria, which could also contribute to a decrease in Drp1 recruitment to mitochondria during DENV infection. Accordingly, the level of MAM marker FACL4 was decreased in the mitochondria fraction of DENV-infected cells.
Using live-cell imaging, we also noted a decrease in mitochondrial motility within cells infected with DENV. As described mainly in neurons, mitochondrial transport occurs along microtubules and relies on the activity of the Miro, Milton, and kinesin heavy chain motor complex [79, 80]. Miro depletion releases the mitochondria from the motor complex, preventing mitochondrial movement. Interestingly, Mfn2 also interacts with both Miro and Milton proteins and is essential for efficient mitochondrial mobility [81, 82]. Whether Mfn2 depletion is involved in reduction of mitochondrial motility during DENV infection remains to be determined.
Mitochondrial elongation and function
Previous reports have established that elongated mitochondria are associated with increased oxidative phosphorylation and ATP production [83–85]. An interconnected mitochondrial network induced during DENV infection may serve to optimize mitochondrial function, increasing bioenergetic capacity of the cell as a pro-survival response against stress. Our results indicate that DENV-infected cells increase mitochondrial respiration associated with ATP production with little to no change in proton leak or maximal respiration. These findings suggest that DENV-induced elongation of mitochondria enhances mitochondrial respiration and energy production, which is in contrast with previous observations indicating mitochondrial dysfunction and alterations in cellular ATP balance in DENV-infected HepG2 cells [42]. These differences may stem from cell-type specificity.
Mitochondria hyperfusion has been described under conditions of stress such as starvation [84] or oxidative stress [86]. However, the induction of mitochondrial fragmentation with CCCP or overexpression of WT and S616-phosphorylated Drp1 reduce viral replication, suggesting that mitochondrial elongation is advantageous to DENV replication, and is not solely a consequence of metabolic stress. By contrast to DENV, HCV which is a strong inducer of oxidative stress [87] triggers mitochondrial fission via Drp1 overexpression [60], indicating that metabolic stress during virus infection is not always associated with mitochondrial elongation. Interestingly, DENV modulates the unfolded protein response in a time-dependent manner, preventing apoptosis and prolonging viral life cycle [88], as well as represses the formation of stress granules and processing (P) bodies [89], suggesting that DENV handles host cell stress responses. Whether DENV protein(s) specifically induce(s) mitochondrial elongation is under investigation.
In summary, this report demonstrates DENV-induced mitochondrial elongation through inhibition of the fission protein, Drp1. The mechanism of Drp1 inhibition such as protein degradation and/or inhibition of kinases involved in Drp1 phosphorylation will need to be determined. Similarly, whether DENV proteins play a direct role in Drp1 regulation at the ER-mitochondrial interface remains to be investigated.
Materials and Methods
Cell culture and virus infection
Vero, Huh7.5 (obtained from American Type Culture Collection (ATCC)) and Huh7 (a generous gift from Dr. Kate Fitzgerald) cells were maintained in Dulbecco’s modified minimal essential medium and HepG2 (ATCC) cells were maintained in Eagle’s Minimum Essential Medium, supplemented with 10% heat-inactivated fetal bovine serum (Sigma-Aldrich), 1% penicillin-streptomycin (Sigma-Aldrich), 1% non-essential amino acids (Lonza) and 1% L-Glutamine solution (Sigma-Aldrich). All cells were incubated in a humidified chamber at 37°C and 5% CO2. DENV-2 strain 16681 was originally obtained from Walter Reed Army Institute of Research and passaged in C6/36 cells (ATCC). Virus titers were determined by immunostained plaque assay on Vero cells [48, 90].
Antibodies and reagents
The following primary antibodies were used for immunofluorescence analysis or western blot: rabbit anti-Drp1 (ab180769, Abcam), anti-Drp1-P Ser616 (D9A1, Cell Signaling Technology), anti-Mfn1 (D6E2S, Cell Signaling Technology), anti-Mfn2 (D1E9, Cell Signaling Technology), anti-OPA1 (GTX48589, GeneTex), anti-TOMM20 (ab186734, Abcam), anti-beta-actin-HRP (ab20272, Abcam), anti-NS3 (GTX124252, GeneTex), anti-NS4b (GTX103349, GeneTex), anti-COX-IV (3E11, Cell Signaling Technology) and mouse anti-Dengue Virus Type II (MAB8702, EMD Millipore). Secondary antibodies were purchased from Thermo Fisher Scientific: Pierce anti-rabbit poly-HRP, anti-rabbit conjugated with Alexa Fluor® 647 or Alexa Fluor® 488 and anti-mouse conjugated with Alexa Fluor® 647. Carbonyl cyanide 3-chlorophenylhydrazone (CCCP, Sigma-Aldrich) was resuspended in medium and incubated at the concentration and time indicated in the figure legend.
Generation of DCs
Peripheral blood mononuclear cells (PBMCs) were isolated from buffy coat of healthy donors obtained from Oklahoma Blood Institute (Oklahoma City, OK) by density gradient centrifugation on Ficoll-Paque Premium (GE Healthcare). Monocytes were isolated from PBMCs by positive selection using CD14+ microbeads (Miltenyi Biotec). Monocytes were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated FBS (Sigma-Aldrich), 1% penicillin-streptomycin (Sigma-Aldrich), 50 ng/mL human interleukin-4 (IL-4) and 160 ng/mL human granulocyte-macrophage colony-stimulating factor (GM-CSF) for 5 days to generate monocyte-derived dendritic cells (DCs). Fresh medium with cytokines was added at day 3, and cells were infected with DENV at day 5. DCs were screened by flow cytometric analysis (MACSQuant, Miltenyi Biotec) for expression of CD11c (VioBlue), CD14 (VioGreen), HLA-DR (FITC), CD3 (PE), CD19 (PE), CD86 (PE-Vio770), CD83 (APC), CD1a (APC-Vio770) (Miltenyi Biotec).
Cell fractionation
Total cell lysates were prepared by solubilizing cells in lysis buffer (Miltenyi Biotec) supplemented with a protease and phosphatase inhibitor cocktail that consists of 1% Halt protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific), 5 mM EDTA (Thermo Fisher Scientific), 1% phosphatase inhibitor cocktail 3 (Sigma-Aldrich), and 0.1% pepstatin A (Research Products International Corp.). The cytosolic fraction was obtained from supernatant of total cell lysate centrifuged at 13,000 g for 10 min, 4°C. Mitochondria were isolated from total cell lysates using the Mitochondria Isolation Kit (Miltenyi Biotec), according to manufacturer’s instructions. Briefly, cells were lysed using the lysis buffer mentioned earlier and homogenized using a glass dounce tissue grinder (Sigma-Aldrich), with 250–300 strokes per sample, on ice. Anti-TOM22 magnetic beads were added to the cell lysates to label the mitochondria, followed by mitochondria isolation in the magnetic field of a MidiMACS separation unit. Isolated mitochondria were resuspended in RIPA buffer (150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl, pH=8.0) supplemented with a protease and phosphatase inhibitor cocktail.
Western Blot analysis
Cells were lysed in RIPA buffer freshly supplemented with a protease and phosphatase cocktail. Lysates were incubated on ice for 30 min and centrifuged at 10,000 rpm for 10 min at 4°C. Supernatants were used as total cell lysates. Protein concentrations were determined using Pierce BCA Protein Assay kit (Thermo Fisher Scientific) following manufacturer’s instructions and measured using an Envision plate reader (PerkinElmer). BOLT LDS Sample Buffer and Reducing Agent (Thermo Fisher Scientific) were added to cell lysates at a final concentration of 1x and samples were denatured at 70°C for 10 min. Proteins were separated on 4–12% or 8% BOLT Bis-Tris Plus gel (Thermo Fisher Scientific) and transferred onto nitrocellulose membranes using the Trans-Blot® Turbo™ RTA Mini Nitrocellulose Transfer kit (Bio-Rad). Membranes were blocked in 5% skim milk for 1h at room temperature (rt) followed by incubation with the appropriate primary antibody overnight at 4°C. Membranes were washed and incubated with the appropriate secondary antibody diluted for 1h at rt. For β-actin analysis, membranes were stained for 1 h with anti-β-actin-HRP. Detection was performed using Amersham ECL Select Western Blotting Detection Reagent (GE Healthcare Life Sciences) following manufacturer’s guidelines and images were captured with ChemiDoc™ XRS+ System (Bio-Rad). Detection and quantification of band intensities was performed using Image Lab 5.1 (Bio-Rad). Band intensities were normalized to loading control TOMM20 for mitochondrial fractions.
Plasmids, dsiRNA and transfections
The following plasmids were used in our study: pNS4b5-eGFP [48], pTagRFP-mito (Evrogen), pAcGFP-Sec61β (gift from Tom Rapoport, Addgene plasmid #15108), pEYFP-Drp1, pEYFP-Drp1 S616D, pEYFP-Drp1 S616A, pEYFP-Drp1 S637D, pEYFP-Drp1 S637A (Drp1 phosphomimetic and phosphodefective mutant plasmids were a gift from Dr. Luca Scorrano, University of Padua), pDrp1 K38A-CFP (gift from Dr. Gyorgy Hajnoczky, Thomas Jefferson University)). Transfection of cells with plasmids was performed using GeneJuice® Transfection Reagent (EMD Millipore), according to the manufacturer’s instructions.
Dicer substrate siRNAs (dsiRNAs) were purchased from Integrated DNA Technologies (IDT). Drp1, Mfn1 and Mfn2 dsiRNAs were designed with Custom RNAi Design Tool (IDT). Drp1 dsiRNA targets the 3′UTR region of human Drp1. DsiRNA sequences were as follows: Drp1 sense strand 5′-GCAGGAAUGCCUACAUUAAUUCCTA-3′ Drp1 anti-sense strand 5′-UAGGAAUUAAUGUAGGCAUUCCUGCUU-3′; Set (A) Mfn1 sense strand 5′-AGCGGAGACUUAGCAUAAUGGCAGA-3′ Mfn1 antisense strand 5′-UCUGCCAUUAUGCUAAGUCUCCGCUCC-3′; Set (B) Mfn1 sense strand 5′-AGAAUCCUAACAAUAGAGAUUGCTT-3′ Mfn1 antisense strand 5′-AAGCAAUCUCUAUUGUUAGGAUUCUUC-3′; Set (A) Mfn2 sense strand 5′-GCAUGGUACCAAGGAGUUAAGUUGA-3′ Mfn2 antisense strand 5′-UCAACUUAACUCCUUGGUACCAUGCUG-3′; Set (B) Mfn2 sense strand 5′-GGCUCAAGACUAUAAGCUGCGAATT-3′ Mfn2 antisense strand 5′-AAUUCGCAGCUUAUAGUCUUGAGCCAA-3′. A dsiRNA with no homology to any human gene was used as a negative control (DS NC1, IDT). Transfection of cells with dsiRNAs was performed at 5 nM final concentration using RiboJuice™ siRNA Transfection Reagent (EMD Millipore), according to the manufacturer’s instructions.
Immunofluorescence analysis
Cells were plated in 8-wells Nunc® Lab-Tek® II glass chamber slides (Sigma-Aldrich) and treated as indicated. Cells were fixed in 4% paraformaldehyde (Affymetrix) for 15 min at rt. Fixed cells were blocked and permeabilized in PBS with 0.1% Triton-X-100 and 10% normal goat serum (Cell Signaling Technology) for 30 min at rt. After blocking, cells were incubated with the appropriate primary antibody overnight at 4°C. Cells were washed and incubated with the appropriate secondary antibody for 1h 30min at rt. After washing, chambers were removed and slides mounted on coverslips with VectaShield (Vector Laboratories) mounting medium containing 6-diamino-2-phenylindole (DAPI) for detection of nuclei. Slides were analyzed on a LSM 800 confocal laser-scanning microscope (Zeiss) or Nikon C1si confocal microscope. Orthogonal projections were created from z-stacks, processed using median filter to reduce noise and improve image quality, and adjusted for brightness and contrast identically between each condition using ImageJ (NIH). The average mitochondrial lengths for cells within each condition were calculated using a macro developed in ImageJ (NIH). Briefly, the red channel image containing the MitoTracker Deep Red fluorescence was converted to 32 bit to keep image integrity and then to 16 bit and inverted. The image was further converted to black and white for analysis. The background was subtracted (rolling= 40 light) and Gaussian blur (sigma=0.90) was applied. The threshold was set (0, 235) and the image was converted to mask and then made binary. Skeletonization was applied to reduce pixel widths of captured mitochondria to a single pixel. Analyze particles was applied (size=0-infinity, circularity=0.00–1.00). The pixel lengths were determined for each particle captured from the image that corresponded to mitochondria. Statistical analysis was done on the average mitochondrial lengths of each cell using hierarchical linear regression models to adjust for the lack of independence of mitochondria within cells. Statistical analyses were performed utilizing JMP version 8 software (SAS institute).
Live cell imaging
Cells were plated in 8-wells Nunc™ Lab-Tek™ chambered coverglass (Thermo Fisher Scientific) and treated as indicated. Mitochondria were visualized in cells transfected with pTagRFP-mito (Evrogen) or after incubation with 200 nM MitoTracker® Deep Red FM (Thermo Fisher Scientific) for 15 min at 37°C. Nuclei were visualized using NucBlue® Live ReadyProbes® Reagent (Thermo Fisher Scientific) following manufacturer’s instructions. Images and time-lapse videos were captured on EVOS fluorescence microscope (Thermo Fisher Scientific) with 100× oil immersion objective. For time-lapse imaging, images were collected every 10 sec for 4 min and then processed identically between each condition to adjust brightness and contrast using ImageJ software (NIH).
Transmission Electron Microscopy
Huh7 cells were cultured on Permanox Lab-Tek chamber slides, fixed with 1.25% glutaraldehyde in 0.15M sodium cacodylate buffer at 4°C. Following fixation, cells were buffer rinsed and post-fixed in 1% osmium tetroxide. After distilled water rinses, samples were stained en bloc with 1% aqueous uranyl acetate for 30 min in the dark. Slides were rinsed and dehydrated through a graded ethanol series. Media chambers and gaskets were removed. Slides were covered with Epox 812 resin and placed over resin filled slide-duplicating molds and polymerized overnight at 60°C. The resin was separated from the slide, and cultures were examined by light microscopy to select regions of interest. Selected areas were cut out and mounted on blocks for sectioning. Ultra-thin sections (50nm) were prepared using a Reichert Ultracut S microtome (Leica Biosystems), retrieved onto 300 mesh copper grids, and contrasted with uranyl acetate and lead citrate stains. Sections were examined at 80 kV using a CM-10 electron microscope (FEI). Images were collected with a model 785 Erlangshen ES1000W CCD camera (Gatan).
qRT-PCR analysis
Total RNA extracted from cell pellets using the RNeasy Mini Kit (Qiagen) and the QIAcube. cDNA was synthetized from 250 ng of RNA with the RT2 First Strand Kit (Qiagen). Standard quantitative RT-PCR was performed with the RT2 SYBR® Green qPCR Mastermix (Qiagen) on a Bio-Rad CFX machine, and gene expression was normalized to the beta-actin expression level. Primer sets were designed using Primer-BLAST (NCBI) and purchased from Integrated DNA Technologies (IDT). Primer sequences were as follows: forward primer Drp1: 5′-CACCCGGAGACCTCTCATTC-3′, reverse primer Drp1: 5′-TTTACCCCATTCTTCTGCTTCCA-3′; forward primer beta-actin 5′-AGAGCTACGAGCTGCCTGAC-3′, reverse primer beta-actin 5′-AGTTTCGTGGATGCCACAGG-3′.
Mitochondrial function profile
Oxygen consumption rate (OCR), an indicator of mitochondrial respiration, was measured using a Seahorse Bioscience XF24 extracellular flux analyzer (Seahorse Bioscience). Huh7 cells were seeded in a XF24 microplate using complete growth medium and infected 1 day later with DENV (MOI 1) for 48h. The day before the assay, the cartridge sensor was hydrated overnight with 1mL Seahorse Bioscience XF24 Calibration Buffer at 37°C without CO2. On the day of the assay, the growth medium was replaced with XF24 Base Assay medium and cells were incubated at 37°C in a non-CO2 incubator for 1h. OCR values were monitored under basal condition and measured after sequential injection of oligomycin (1 μM), FCCP (1 μM), and antimycin A/rotenone (0.5 μM) using XF Cell Mito Stress kit (Seahorse Bioscience). Mitochondrial function parameters were analyzed according to Seahorse Bioscience instructions.
Supplementary Material
A. and B. Immunofluorescence analysis of mitochondrial morphology in uninfected (cytoplasmic GFP) or DENV-infected (cytoplasmic and nuclear GFP) HepG2 (A.) and Huh7.5 (B.) cells. Cells were transfected with pNS4b5-eGFP reporter and infected with DENV (MOI of 0.1) for 48h. Cells were subsequently stained with MitoTracker Deep Red FM prior to imaging. Images were converted to 8-bit and inverted for ImageJ analysis of mitochondrial lengths. Images were taken at 100× magnification and are representative of at least three independent experiments. C. Immunofluorescence analysis of mitochondrial morphology in HepG2 cells uninfected or infected with DENV (MOI of 1) for 48h. Cells were fixed and stained with antibodies against DENV antigen and COX-IV. Images were taken with Nikon Leica C1si confocal microscope ×120 magnification. D. and E. Immunofluorescence analysis of mitochondrial morphology in Huh7.5 (D.) and Huh7 (E.) cells transfected with pTagRFP-mito and uninfected or infected with DENV (MOI of 1) for 48h. Cells were fixed and stained with antibody against DENV antigen. Images were taken with confocal LSM800 at ×63×1.5 magnification and are representative of at least three independent experiments.
Immunofluorescence analysis of Huh7 cells transfected with pTagRFP-mito and pNS4b5-eGFP and uninfected or infected with DENV (MOI of 1) for 48h. Live cells were imaged with EVOS fluorescence microscope at ×100 magnification. Infected cells (cytoplasmic and nuclear GFP) were discriminated from uninfected cells (cytoplasmic GFP) and processed for time-lapse imaging (Movie S1 and S2). Time-lapse live cell imaging was performed on RFP channel to visualize mitochondria in uninfected cell (Movie S1) and infected cell (Movie S2). Time-lapse videos show images collected every 10 sec for 4 min and then processed using ImageJ software (NIH).
A. and B. Mfn1 and Mfn2 protein levels in uninfected or DENV-infected (MOI of 1) Huh7 cells were analyzed by immunofluorescence at 48h pi. Cells were fixed and stained with antibodies against Mfn1 (A.), Mfn2 (B.) and DENV antigen. Images were taken with confocal LSM800 at ×20 magnification.
A. and B. Drp1 and Drp1 S616-P levels in uninfected or DENV-infected (MOI of 0.1 or 1) Huh7.5 cells were analyzed by immunofluorescence at 48h pi. Cells were fixed and stained with antibodies against Drp1 (A.) or Drp1 S616-P (B.) protein and DENV antigen. Images were taken with confocal LSM800 at ×20 magnification. Quantification of Mean Fluorescence Intensity (MFI) in individual cells from image corresponding to Drp1 or Drp1 S616-P staining was performed using ImageJ and compared for statistical analysis using unpaired Student-t test.
A. qRT-PCR analysis of Drp1 gene expression was performed in uninfected and DENV-infected DCs after 8h, 12h and 24h of infection. Ct values were normalized to the housekeeping gene β-actin. Data are represented as mean +/− SD of three independent experiments (except 12h, n=2). Statistical analysis was done with unpaired Student-t test. B and C. Drp1 and Drp1 S616-P levels in uninfected or DENV-infected Huh7.5 cells were analyzed by immunofluorescence at 48h pi. Cells were transfected with pTagRFP-mito, challenged with DENV (MOI of 1) and after 48h of infection, cells were fixed and stained with antibodies against Drp1 (B.) or Drp1 S616-P (C.) and DENV antigen. Images were taken with confocal LSM800 at ×63×1.5 magnification.
DENV-2 antigen levels in uninfected or DENV-infected (MOI of 1) Vero cells, treated as indicated, were analyzed by immunofluorescence with confocal LSM800 at ×20 magnification. Cells were fixed and stained with antibody against DENV antigen.
A. Huh7.5 cells were transfected with dsiRNAs against Mfn1 or Mfn2 and infected with DENV (MOI of 0.1) for 24h. Western blots were probed with Mfn1 and Mfn2 antibodies and equal protein loading was assessed using an antibody against β-actin. Blot is representative of at least four independent experiments. B. Huh7.5 cells were transfected with a negative control dsiRNA or dsiRNA targeting the untranslated region of Drp1. Cells were transfected with plasmids expressing Drp1 wild-type or mutants and infected with DENV (MOI of 0.1) for 24h. Western blots were probed with antibodies directed against Drp1 protein and equal protein loading was assessed using an antibody against β-actin.
Movie S1. Time-lapse live cell imaging of mitochondria in uninfected Huh7 cell.
Movie S2. Time-lapse live cell imaging of mitochondria in DENV-infected Huh7 cell.
Highlights.
Mitochondrial length and respiration are increased during DENV infection.
DENV inhibits Drp1-triggered mitochondrial fission.
DENV titers are reduced by mitochondrial fragmentation, Drp1 WT and S616D expression.
Viral proteins NS4b and NS3 are associated with subcellular fractions of mitochondria.
Acknowledgments
We would like to thank Dr. Robbert Creton (Brown University) for his guidance in development of the ImageJ macro to capture mitochondrial lengths, Jennifer Friedman (Rhode Island Hospital) for help in the statistical analysis of mitochondria, Dr. Meenakshi Khare and Siraj Janoudi (University of Rhode Island) for technical assistance, Dr. Luca Scorrano (University of Padua) for generously providing the Drp1 phosphorylation mutant plasmids, Dr. David Guertin lab (UMass Medical School) for access to the Seahorse Bioanalyzer and Dr. Gyorgy Hajnoczky (Thomas Jefferson University) for providing the pDrp1 K38A-CFP plasmid. We also thank Dr. Kate Fitzgerald (UMass Medical School) and lab members for helpful discussions.
Funding information
This work was supported by National Institute of General Medical Sciences of the National Institutes of Health under grant number P20 GM104317 (COBRE) and P20 GM103430 (INBRE).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, et al. The global distribution and burden of dengue. Nature. 2013;496(7446):504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brady OJ, Gething PW, Bhatt S, Messina JP, Brownstein JS, Hoen AG, Moyes CL, Farlow AW, Scott TW, Hay SI. Refining the global spatial limits of dengue virus transmission by evidence-based consensus. PLoS Negl Trop Dis. 2012;6(8):e1760. doi: 10.1371/journal.pntd.0001760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simmons CP, Farrar JJ, Nguyen v V, Wills B. Dengue. N Engl J Med. 2012;366(15):1423–1432. doi: 10.1056/NEJMra1110265. [DOI] [PubMed] [Google Scholar]
- 4.Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control: New Edition. Geneva: 2009. [PubMed] [Google Scholar]
- 5.Aye KS, Charngkaew K, Win N, Wai KZ, Moe K, Punyadee N, Thiemmeca S, Suttitheptumrong A, Sukpanichnant S, Prida M, et al. Pathologic highlights of dengue hemorrhagic fever in 13 autopsy cases from Myanmar. Hum Pathol. 2014;45(6):1221–1233. doi: 10.1016/j.humpath.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 6.Balsitis SJ, Coloma J, Castro G, Alava A, Flores D, McKerrow JH, Beatty PR, Harris E. Tropism of dengue virus in mice and humans defined by viral nonstructural protein 3-specific immunostaining. Am J Trop Med Hyg. 2009;80(3):416–424. [PubMed] [Google Scholar]
- 7.Couvelard A, Marianneau P, Bedel C, Drouet MT, Vachon F, Henin D, Deubel V. Report of a fatal case of dengue infection with hepatitis: demonstration of dengue antigens in hepatocytes and liver apoptosis. Hum Pathol. 1999;30(9):1106–1110. doi: 10.1016/s0046-8177(99)90230-7. [DOI] [PubMed] [Google Scholar]
- 8.Durbin AP, Vargas MJ, Wanionek K, Hammond SN, Gordon A, Rocha C, Balmaseda A, Harris E. Phenotyping of peripheral blood mononuclear cells during acute dengue illness demonstrates infection and increased activation of monocytes in severe cases compared to classic dengue fever. Virology. 2008;376(2):429–435. doi: 10.1016/j.virol.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall WC, Crowell TP, Watts DM, Barros VL, Kruger H, Pinheiro F, Peters CJ. Demonstration of yellow fever and dengue antigens in formalin-fixed paraffin-embedded human liver by immunohistochemical analysis. Am J Trop Med Hyg. 1991;45(4):408–417. doi: 10.4269/ajtmh.1991.45.408. [DOI] [PubMed] [Google Scholar]
- 10.Huerre MR, Lan NT, Marianneau P, Hue NB, Khun H, Hung NT, Khen NT, Drouet MT, Huong VT, Ha DQ, et al. Liver histopathology and biological correlates in five cases of fatal dengue fever in Vietnamese children. Virchows Arch. 2001;438(2):107–115. doi: 10.1007/s004280000329. [DOI] [PubMed] [Google Scholar]
- 11.Jessie K, Fong MY, Devi S, Lam SK, Wong KT. Localization of dengue virus in naturally infected human tissues, by immunohistochemistry and in situ hybridization. J Infect Dis. 2004;189(8):1411–1418. doi: 10.1086/383043. [DOI] [PubMed] [Google Scholar]
- 12.Kou Z, Quinn M, Chen H, Rodrigo WW, Rose RC, Schlesinger JJ, Jin X. Monocytes, but not T or B cells, are the principal target cells for dengue virus (DV) infection among human peripheral blood mononuclear cells. J Med Virol. 2008;80(1):134–146. doi: 10.1002/jmv.21051. [DOI] [PubMed] [Google Scholar]
- 13.Povoa TF, Alves AM, Oliveira CA, Nuovo GJ, Chagas VL, Paes MV. The pathology of severe dengue in multiple organs of human fatal cases: histopathology, ultrastructure and virus replication. PLoS One. 2014;9(4):e83386. doi: 10.1371/journal.pone.0083386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmid MA, Diamond MS, Harris E. Dendritic cells in dengue virus infection: targets of virus replication and mediators of immunity. Front Immunol. 2014;5:647. doi: 10.3389/fimmu.2014.00647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suksanpaisan L, Cabrera-Hernandez A, Smith DR. Infection of human primary hepatocytes with dengue virus serotype 2. J Med Virol. 2007;79(3):300–307. doi: 10.1002/jmv.20798. [DOI] [PubMed] [Google Scholar]
- 16.Brandt WE, McCown JM, Gentry MK, Russell PK. Infection enhancement of dengue type 2 virus in the U-937 human monocyte cell line by antibodies to flavivirus cross-reactive determinants. Infect Immun. 1982;36(3):1036–1041. doi: 10.1128/iai.36.3.1036-1041.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halstead SB, Mahalingam S, Marovich MA, Ubol S, Mosser DM. Intrinsic antibody-dependent enhancement of microbial infection in macrophages: disease regulation by immune complexes. Lancet Infect Dis. 2010;10(10):712–722. doi: 10.1016/S1473-3099(10)70166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kliks SC, Nisalak A, Brandt WE, Wahl L, Burke DS. Antibody-dependent enhancement of dengue virus growth in human monocytes as a risk factor for dengue hemorrhagic fever. Am J Trop Med Hyg. 1989;40(4):444–451. doi: 10.4269/ajtmh.1989.40.444. [DOI] [PubMed] [Google Scholar]
- 19.Laoprasopwattana K, Libraty DH, Endy TP, Nisalak A, Chunsuttiwat S, Vaughn DW, Reed G, Ennis FA, Rothman AL, Green S. Dengue Virus (DV) enhancing antibody activity in preillness plasma does not predict subsequent disease severity or viremia in secondary DV infection. J Infect Dis. 2005;192(3):510–519. doi: 10.1086/431520. [DOI] [PubMed] [Google Scholar]
- 20.Samanta J, Sharma V. Dengue and its effects on liver. World J Clin Cases. 2015;3(2):125–131. doi: 10.12998/wjcc.v3.i2.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothman AL. Cellular immunology of sequential dengue virus infection and its role in disease pathogenesis. Curr Top Microbiol Immunol. 2010;338:83–98. doi: 10.1007/978-3-642-02215-9_7. [DOI] [PubMed] [Google Scholar]
- 22.Whitehorn J, Farrar J. Dengue. Br Med Bull. 2010;95:161–173. doi: 10.1093/bmb/ldq019. [DOI] [PubMed] [Google Scholar]
- 23.de Armas-Rillo L, Valera MS, Marrero-Hernandez S, Valenzuela-Fernandez A. Membrane dynamics associated with viral infection. Rev Med Virol. 2016;26(3):146–160. doi: 10.1002/rmv.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5(4):365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allonso D, Andrade IS, Conde JN, Coelho DR, Rocha DC, da Silva ML, Ventura GT, Silva EM, Mohana-Borges R. Dengue Virus NS1 Protein Modulates Cellular Energy Metabolism by Increasing Glyceraldehyde-3-Phosphate Dehydrogenase Activity. J Virol. 2015;89(23):11871–11883. doi: 10.1128/JVI.01342-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heaton NS, Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 2010;8(5):422–432. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tait SW, Green DR. Mitochondria and cell signalling. J Cell Sci. 2012;125(Pt 4):807–815. doi: 10.1242/jcs.099234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281(5381):1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 29.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A. 2005;102(49):17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24(5):633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 31.West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520(7548):553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27(2):433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, Nunnari J. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol. 2005;170(7):1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bui HT, Shaw JM. Dynamin assembly strategies and adaptor proteins in mitochondrial fission. Curr Biol. 2013;23(19):R891–899. doi: 10.1016/j.cub.2013.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem. 2007;282(15):11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- 36.Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334(6054):358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korobova F, Ramabhadran V, Higgs HN. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 2013;339(6118):464–467. doi: 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elgass K, Pakay J, Ryan MT, Palmer CS. Recent advances into the understanding of mitochondrial fission. Biochim Biophys Acta. 2013;1833(1):150–161. doi: 10.1016/j.bbamcr.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 39.Knott AB, Bossy-Wetzel E. Impairing the mitochondrial fission and fusion balance: a new mechanism of neurodegeneration. Ann N Y Acad Sci. 2008;1147:283–292. doi: 10.1196/annals.1427.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burte F, Carelli V, Chinnery PF, Yu-Wai-Man P. Disturbed mitochondrial dynamics and neurodegenerative disorders. Nat Rev Neurol. 2015;11(1):11–24. doi: 10.1038/nrneurol.2014.228. [DOI] [PubMed] [Google Scholar]
- 41.Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125(7):1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 42.El-Bacha T, Midlej V, Pereira da Silva AP, Silva da Costa L, Benchimol M, Galina A, Da Poian AT. Mitochondrial and bioenergetic dysfunction in human hepatic cells infected with dengue 2 virus. Biochim Biophys Acta. 2007;1772(10):1158–1166. doi: 10.1016/j.bbadis.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 43.Ho LJ, Wang JJ, Shaio MF, Kao CL, Chang DM, Han SW, Lai JH. Infection of human dendritic cells by dengue virus causes cell maturation and cytokine production. J Immunol. 2001;166(3):1499–1506. doi: 10.4049/jimmunol.166.3.1499. [DOI] [PubMed] [Google Scholar]
- 44.Mosquera JA, Hernandez JP, Valero N, Espina LM, Anez GJ. Ultrastructural studies on dengue virus type 2 infection of cultured human monocytes. Virol J. 2005;2:26. doi: 10.1186/1743-422X-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zargar S, Wani TA, Jain SK. Morphological changes in vero cells postinfection with dengue virus type-2. Microsc Res Tech. 2011;74(4):314–319. doi: 10.1002/jemt.20908. [DOI] [PubMed] [Google Scholar]
- 46.Yu CY, Liang JJ, Li JK, Lee YL, Chang BL, Su CI, Huang WJ, Lai MM, Lin YL. Dengue virus impairs mitochondrial fusion by cleaving mitofusins. PLoS Pathog. 2015;11(12):e1005350. doi: 10.1371/journal.ppat.1005350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chatel-Chaix L, Cortese M, Romero-Brey I, Bender S, Neufeldt CJ, Fischl W, Scaturro P, Schieber N, Schwab Y, Fischer B, et al. Dengue Virus Perturbs Mitochondrial Morphodynamics to Dampen Innate Immune Responses. Cell Host Microbe. 2016 doi: 10.1016/j.chom.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Medin CL, Valois S, Patkar CG, Rothman AL. A plasmid-based reporter system for live cell imaging of dengue virus infected cells. J Virol Methods. 2015;211:55–62. doi: 10.1016/j.jviromet.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen H, Chan DC. Mitochondrial dynamics–fusion, fission, movement, and mitophagy–in neurodegenerative diseases. Hum Mol Genet. 2009;18(R2):R169–176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okuda M, Li K, Beard MR, Showalter LA, Scholle F, Lemon SM, Weinman SA. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology. 2002;122(2):366–375. doi: 10.1053/gast.2002.30983. [DOI] [PubMed] [Google Scholar]
- 51.Schwer B, Ren S, Pietschmann T, Kartenbeck J, Kaehlcke K, Bartenschlager R, Yen TS, Ott M. Targeting of hepatitis C virus core protein to mitochondria through a novel C-terminal localization motif. J Virol. 2004;78(15):7958–7968. doi: 10.1128/JVI.78.15.7958-7968.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramage HR, Kumar GR, Verschueren E, Johnson JR, Von Dollen J, Johnson T, Newton B, Shah P, Horner J, Krogan NJ, et al. A combined proteomics/genomics approach links hepatitis C virus infection with nonsense-mediated mRNA decay. Mol Cell. 2015;57(2):329–340. doi: 10.1016/j.molcel.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160(2):189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cipolat S, Martins de Brito O, Dal Zilio B, Scorrano L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc Natl Acad Sci U S A. 2004;101(45):15927–15932. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cereghetti GM, Stangherlin A, Martins de Brito O, Chang CR, Blackstone C, Bernardi P, Scorrano L. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc Natl Acad Sci U S A. 2008;105(41):15803–15808. doi: 10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mai S, Klinkenberg M, Auburger G, Bereiter-Hahn J, Jendrach M. Decreased expression of Drp1 and Fis1 mediates mitochondrial elongation in senescent cells and enhances resistance to oxidative stress through PINK1. J Cell Sci. 2010;123(Pt 6):917–926. doi: 10.1242/jcs.059246. [DOI] [PubMed] [Google Scholar]
- 57.Ishihara N, Jofuku A, Eura Y, Mihara K. Regulation of mitochondrial morphology by membrane potential, and DRP1-dependent division and FZO1-dependent fusion reaction in mammalian cells. Biochem Biophys Res Commun. 2003;301(4):891–898. doi: 10.1016/s0006-291x(03)00050-0. [DOI] [PubMed] [Google Scholar]
- 58.Chang CR, Blackstone C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J Biol Chem. 2007;282(30):21583–21587. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- 59.Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, Smith CL, Youle RJ. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1(4):515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 60.Kim SJ, Syed GH, Khan M, Chiu WW, Sohail MA, Gish RG, Siddiqui A. Hepatitis C virus triggers mitochondrial fission and attenuates apoptosis to promote viral persistence. Proc Natl Acad Sci U S A. 2014;111(17):6413–6418. doi: 10.1073/pnas.1321114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stavru F, Palmer AE, Wang C, Youle RJ, Cossart P. Atypical mitochondrial fission upon bacterial infection. Proc Natl Acad Sci U S A. 2013;110(40):16003–16008. doi: 10.1073/pnas.1315784110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen TT, Wu LS, Hsu PW, Pang CY, Lee KM, Cheng PC, Peng SY. Mitochondrial dynamics in the mouse liver infected by Schistosoma mansoni. Acta Trop. 2015;148:13–23. doi: 10.1016/j.actatropica.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 63.Gorbunov NV, McDaniel DP, Zhai M, Liao PJ, Garrison BR, Kiang JG. Autophagy and mitochondrial remodelling in mouse mesenchymal stromal cells challenged with Staphylococcus epidermidis. J Cell Mol Med. 2015;19(5):1133–1150. doi: 10.1111/jcmm.12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lum M, Morona R. Dynamin-related protein Drp1 and mitochondria are important for Shigella flexneri infection. Int J Med Microbiol. 2014;304(5–6):530–541. doi: 10.1016/j.ijmm.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 65.Kim SJ, Khan M, Quan J, Till A, Subramani S, Siddiqui A. Hepatitis B virus disrupts mitochondrial dynamics: induces fission and mitophagy to attenuate apoptosis. PLoS Pathog. 2013;9(12):e1003722. doi: 10.1371/journal.ppat.1003722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yan J, Liu XH, Han MZ, Wang YM, Sun XL, Yu N, Li T, Su B, Chen ZY. Blockage of GSK3beta-mediated Drp1 phosphorylation provides neuroprotection in neuronal and mouse models of Alzheimer’s disease. Neurobiol Aging. 2015;36(1):211–227. doi: 10.1016/j.neurobiolaging.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 67.Murray CL, Jones CT, Rice CM. Architects of assembly: roles of Flaviviridae nonstructural proteins in virion morphogenesis. Nat Rev Microbiol. 2008;6(9):699–708. doi: 10.1038/nrmicro1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paul D, Bartenschlager R. Flaviviridae Replication Organelles: Oh, What a Tangled Web We Weave. Annu Rev Virol. 2015;2(1):289–310. doi: 10.1146/annurev-virology-100114-055007. [DOI] [PubMed] [Google Scholar]
- 69.Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280(28):26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- 70.Detmer SA, Chan DC. Complementation between mouse Mfn1 and Mfn2 protects mitochondrial fusion defects caused by CMT2A disease mutations. J Cell Biol. 2007;176(4):405–414. doi: 10.1083/jcb.200611080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rehman J, Zhang HJ, Toth PT, Zhang Y, Marsboom G, Hong Z, Salgia R, Husain AN, Wietholt C, Archer SL. Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. FASEB J. 2012;26(5):2175–2186. doi: 10.1096/fj.11-196543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thomas KJ, Jacobson MR. Defects in mitochondrial fission protein dynamin-related protein 1 are linked to apoptotic resistance and autophagy in a lung cancer model. PloS one. 2012;7(9):e45319. doi: 10.1371/journal.pone.0045319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rowland AA, Voeltz GK. Endoplasmic reticulum-mitochondria contacts: function of the junction. Nat Rev Mol Cell Biol. 2012;13(10):607–625. doi: 10.1038/nrm3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raturi A, Simmen T. Where the endoplasmic reticulum and the mitochondrion tie the knot: the mitochondria-associated membrane (MAM) Biochim Biophys Acta. 2013;1833(1):213–224. doi: 10.1016/j.bbamcr.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 75.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456(7222):605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 76.Huang CY, Chiang SF, Lin TY, Chiou SH, Chow KC. HIV-1 Vpr triggers mitochondrial destruction by impairing Mfn2-mediated ER-mitochondria interaction. PLoS One. 2012;7(3):e33657. doi: 10.1371/journal.pone.0033657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cosson P, Marchetti A, Ravazzola M, Orci L. Mitofusin-2 independent juxtaposition of endoplasmic reticulum and mitochondria: an ultrastructural study. PLoS One. 2012;7(9):e46293. doi: 10.1371/journal.pone.0046293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Filadi R, Greotti E, Turacchio G, Luini A, Pozzan T, Pizzo P. Mitofusin 2 ablation increases endoplasmic reticulum-mitochondria coupling. Proc Natl Acad Sci U S A. 2015;112(17):E2174–2181. doi: 10.1073/pnas.1504880112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saxton WM, Hollenbeck PJ. The axonal transport of mitochondria. J Cell Sci. 2012;125(Pt 9):2095–2104. doi: 10.1242/jcs.053850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schwarz TL. Mitochondrial trafficking in neurons. Cold Spring Harb Perspect Biol. 2013;5(6) doi: 10.1101/cshperspect.a011304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Misko A, Jiang S, Wegorzewska I, Milbrandt J, Baloh RH. Mitofusin 2 is necessary for transport of axonal mitochondria and interacts with the Miro/Milton complex. J Neurosci. 2010;30(12):4232–4240. doi: 10.1523/JNEUROSCI.6248-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baloh RH, Schmidt RE, Pestronk A, Milbrandt J. Altered axonal mitochondrial transport in the pathogenesis of Charcot-Marie-Tooth disease from mitofusin 2 mutations. J Neurosci. 2007;27(2):422–430. doi: 10.1523/JNEUROSCI.4798-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mitra K, Wunder C, Roysam B, Lin G, Lippincott-Schwartz J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc Natl Acad Sci U S A. 2009;106(29):11960–11965. doi: 10.1073/pnas.0904875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tondera D, Grandemange S, Jourdain A, Karbowski M, Mattenberger Y, Herzig S, Da Cruz S, Clerc P, Raschke I, Merkwirth C, et al. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 2009;28(11):1589–1600. doi: 10.1038/emboj.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ, Capaldi RA. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 2004;64(3):985–993. doi: 10.1158/0008-5472.can-03-1101. [DOI] [PubMed] [Google Scholar]
- 86.Redpath CJ, Bou Khalil M, Drozdzal G, Radisic M, McBride HM. Mitochondrial hyperfusion during oxidative stress is coupled to a dysregulation in calcium handling within a C2C12 cell model. PLoS One. 2013;8(7):e69165. doi: 10.1371/journal.pone.0069165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Paracha UZ, Fatima K, Alqahtani M, Chaudhary A, Abuzenadah A, Damanhouri G, Qadri I. Oxidative stress and hepatitis C virus. Virol J. 2013;10:251. doi: 10.1186/1743-422X-10-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pena J, Harris E. Dengue virus modulates the unfolded protein response in a time-dependent manner. J Biol Chem. 2011;286(16):14226–14236. doi: 10.1074/jbc.M111.222703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Emara MM, Brinton MA. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc Natl Acad Sci U S A. 2007;104(21):9041–9046. doi: 10.1073/pnas.0703348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu L, Wen K, Li J, Hu D, Huang Y, Qiu L, Cai J, Che X. Comparison of plaque- and enzyme-linked immunospot-based assays to measure the neutralizing activities of monoclonal antibodies specific to domain III of dengue virus envelope protein. Clin Vaccine Immunol. 2012;19(1):73–78. doi: 10.1128/CVI.05388-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. and B. Immunofluorescence analysis of mitochondrial morphology in uninfected (cytoplasmic GFP) or DENV-infected (cytoplasmic and nuclear GFP) HepG2 (A.) and Huh7.5 (B.) cells. Cells were transfected with pNS4b5-eGFP reporter and infected with DENV (MOI of 0.1) for 48h. Cells were subsequently stained with MitoTracker Deep Red FM prior to imaging. Images were converted to 8-bit and inverted for ImageJ analysis of mitochondrial lengths. Images were taken at 100× magnification and are representative of at least three independent experiments. C. Immunofluorescence analysis of mitochondrial morphology in HepG2 cells uninfected or infected with DENV (MOI of 1) for 48h. Cells were fixed and stained with antibodies against DENV antigen and COX-IV. Images were taken with Nikon Leica C1si confocal microscope ×120 magnification. D. and E. Immunofluorescence analysis of mitochondrial morphology in Huh7.5 (D.) and Huh7 (E.) cells transfected with pTagRFP-mito and uninfected or infected with DENV (MOI of 1) for 48h. Cells were fixed and stained with antibody against DENV antigen. Images were taken with confocal LSM800 at ×63×1.5 magnification and are representative of at least three independent experiments.
Immunofluorescence analysis of Huh7 cells transfected with pTagRFP-mito and pNS4b5-eGFP and uninfected or infected with DENV (MOI of 1) for 48h. Live cells were imaged with EVOS fluorescence microscope at ×100 magnification. Infected cells (cytoplasmic and nuclear GFP) were discriminated from uninfected cells (cytoplasmic GFP) and processed for time-lapse imaging (Movie S1 and S2). Time-lapse live cell imaging was performed on RFP channel to visualize mitochondria in uninfected cell (Movie S1) and infected cell (Movie S2). Time-lapse videos show images collected every 10 sec for 4 min and then processed using ImageJ software (NIH).
A. and B. Mfn1 and Mfn2 protein levels in uninfected or DENV-infected (MOI of 1) Huh7 cells were analyzed by immunofluorescence at 48h pi. Cells were fixed and stained with antibodies against Mfn1 (A.), Mfn2 (B.) and DENV antigen. Images were taken with confocal LSM800 at ×20 magnification.
A. and B. Drp1 and Drp1 S616-P levels in uninfected or DENV-infected (MOI of 0.1 or 1) Huh7.5 cells were analyzed by immunofluorescence at 48h pi. Cells were fixed and stained with antibodies against Drp1 (A.) or Drp1 S616-P (B.) protein and DENV antigen. Images were taken with confocal LSM800 at ×20 magnification. Quantification of Mean Fluorescence Intensity (MFI) in individual cells from image corresponding to Drp1 or Drp1 S616-P staining was performed using ImageJ and compared for statistical analysis using unpaired Student-t test.
A. qRT-PCR analysis of Drp1 gene expression was performed in uninfected and DENV-infected DCs after 8h, 12h and 24h of infection. Ct values were normalized to the housekeeping gene β-actin. Data are represented as mean +/− SD of three independent experiments (except 12h, n=2). Statistical analysis was done with unpaired Student-t test. B and C. Drp1 and Drp1 S616-P levels in uninfected or DENV-infected Huh7.5 cells were analyzed by immunofluorescence at 48h pi. Cells were transfected with pTagRFP-mito, challenged with DENV (MOI of 1) and after 48h of infection, cells were fixed and stained with antibodies against Drp1 (B.) or Drp1 S616-P (C.) and DENV antigen. Images were taken with confocal LSM800 at ×63×1.5 magnification.
DENV-2 antigen levels in uninfected or DENV-infected (MOI of 1) Vero cells, treated as indicated, were analyzed by immunofluorescence with confocal LSM800 at ×20 magnification. Cells were fixed and stained with antibody against DENV antigen.
A. Huh7.5 cells were transfected with dsiRNAs against Mfn1 or Mfn2 and infected with DENV (MOI of 0.1) for 24h. Western blots were probed with Mfn1 and Mfn2 antibodies and equal protein loading was assessed using an antibody against β-actin. Blot is representative of at least four independent experiments. B. Huh7.5 cells were transfected with a negative control dsiRNA or dsiRNA targeting the untranslated region of Drp1. Cells were transfected with plasmids expressing Drp1 wild-type or mutants and infected with DENV (MOI of 0.1) for 24h. Western blots were probed with antibodies directed against Drp1 protein and equal protein loading was assessed using an antibody against β-actin.
Movie S1. Time-lapse live cell imaging of mitochondria in uninfected Huh7 cell.
Movie S2. Time-lapse live cell imaging of mitochondria in DENV-infected Huh7 cell.


