Abstract
Prion diseases affect a wide range of mammal species and are caused by a misfolded self-propagating isoform (PrPSc) of the normal prion protein (PrPC). Distinct strains of prions exist and are operationally defined by differences in a heritable phenotype under controlled experimental transmission conditions. Prion strains can differ in incubation period, clinical signs of disease, tissue tropism, and host range. The mechanism by which a protein-only pathogen can encode strain diversity is only beginning to be understood. The prevailing hypothesis is that prion strain diversity is encoded by strain-specific conformations of PrPSc; however, strain-specific cellular cofactors have been identified in vitro that may also contribute to prion strain diversity. Although much progress has been made on understanding the etiological agent of prion disease, the relationship between the strain-specific properties of PrPSc and the resulting phenotype of disease in animals is poorly understood.
Distinct prion strains cause distinct disease phenotypes under controlled experimental conditions. But the molecular basis for prion strain variation is unclear; strain-specific conformations or cofactors may be involved.
PRIONS AND PRION DISEASES
Prion diseases are a group of transmissible neurodegenerative diseases that affect animals, including humans. The animal prion diseases include scrapie in sheep and goats, transmissible mink encephalopathy (TME) in ranch-raised mink, chronic wasting disease (CWD) in cervids, and bovine spongiform encephalopathy (BSE) (Cuillé and Chelle 1936; Zlotnik and Stamp 1961; Marsh and Hanson 1969; Williams and Young 1980, 1992; Wells 1987; Hope et al. 1988; Marsh et al. 1991; Liberski et al. 2009; Saunders et al. 2012). The human prion diseases can be acquired, inherited, or can occur sporadically, and include Creutzfeldt–Jakob disease (CJD), Gerstmann–Sträussler–Scheinker disease (GSS), fatal familial insomnia (FFI), and kuru (Alpers and Gajdusek 1965; Gibbs et al. 1968, 1980; Goldfarb et al. 1992; Tateishi et al. 1995). Prion diseases have long asymptomatic incubation periods that range from months to decades and are followed by a short symptomatic phase that is characterized by progressive cognitive and/or motor deficits (Dickinson and Outram 1979; Race et al. 2001). During the asymptomatic phase, prions can be detected in the central nervous system (CNS) and in extraneural locations (Hadlow et al. 1987). Currently, effective treatment for prion diseases is not available, and they are inevitably fatal.
The prion is composed primarily, if not entirely, of an abnormal isoform (PrPSc) of the host-encoded prion protein (PrPC) (Bolton et al. 1982; Prusiner 1982; Basler et al. 1986; Deleault et al. 2007; Wang et al. 2010). Prion propagation is thought to occur in a three-step process in which PrPSc first binds to PrPC, followed by a conformational conversion of PrPC to PrPSc. This conversion results in a change in the physical properties of PrPC, which includes an increase in β-pleated sheet content and decreased solubility in nondenaturing detergents (Caughey and Raymond 1991; Pan et al. 1993; Moore et al. 2011). Next, fragmentation of the growing PrPSc polymer results in the generation of new free ends for PrPC to bind to. In the absence of preexisting PrPSc, PrPC can spontaneously misfold into PrPSc that is enhanced by PrPC mutations, providing a molecular basis for sporadic and familial forms of human prion disease.
PRION STRAIN DEFINITION
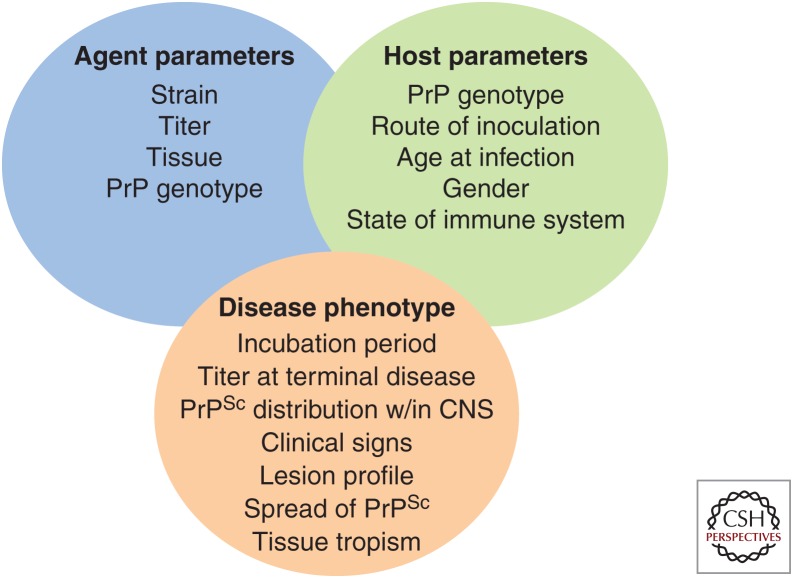
Prion strains are operationally defined as a phenotype of disease under a fixed set of agent and host parameters (Fig. 1). For example, agent parameters such as titer strongly influence the incubation period of disease (Marsh and Hanson 1978; Prusiner et al. 1981, 1982). Host parameters, including route of infection and PrP genotype, influence the incubation period and effective titer of disease (Dickinson et al. 1968, 1969; Dickinson and Fraser 1969; Dickinson and Meikle 1971; Kimberlin and Walker 1977, 1979, 1982, 1986; Bruce and Dickinson 1985). Under experimental conditions in which these parameters are precisely controlled, distinct phenotypes of disease correspond with prion strains (Fig. 1). Differences in the distribution and relative intensity of spongiform degeneration in select areas of the CNS are, at the current time, the most well-accepted criteria to distinguish strains (Fraser and Dickinson 1967, 1968). Not all disease phenotypes are unique to a given strain; for example, strains with distinct distributions of spongiform degeneration can have similar incubation periods and/or clinical signs of disease (Ayers et al. 2011). Importantly, the strain-specific phenotype is maintained on serial passage and is therefore heritable. Under passage conditions that are not precisely controlled, or when serial passage is not available, the term “prion isolate” may best reflect this uncertainty. A significant shortcoming of the operational definition of prion strains is that the degree to which the disease phenotype has to vary to be considered a unique strain is not agreed upon.
Figure 1.
Host–agent interactions in prion disease.
PRION STRAIN TROPISM
Prion strains can differ in the range of pathological changes observed in the CNS. The relative intensity of spongiform degeneration in specified regions of the CNS at terminal disease is the basis of the lesion profile (Fraser and Dickinson 1967). Strains of prions are defined by differences in the lesion profile. Although semiquantitative, the lesion profile is a robust and highly reproducible technique that has been used for more than 40 years. There is a wealth of existing lesion-profile data that is useful for comparison to newly described prions. The use of the lesion profile can also aid in establishing the etiology of disease, as in the case of transmission of BSE to humans, which results in the emergence of variant CJD (Bruce et al. 1997). Although the lesion profile is the defining characteristic of prion strains, it does not necessarily reflect strain-specific differences in neuronal tropism because neurons can harbor high levels of PrPSc at terminal disease yet fail to develop spongiform degeneration (van Keulen et al. 1995).
Differences in the regional deposition of PrPSc within the CNS are strain specific. Histoblot analysis of the brains from hamsters or transgenic mice expressing hamster PrPC infected with either the Sc237 or 139H strains of hamster-adapted scrapie have regional differences in the localization of PrPSc (Hecker et al. 1992; DeArmond et al. 1993). Immunohistochemistry can also reveal differences in the localization of PrPSc immunoreactivity between different strains and can provide additional information on the cellular distribution of PrPSc within neurons and glia (Jeffrey et al. 2001, 2003; Sisó et al. 2010; Ayers et al. 2011).
The tropism of prions outside the CNS is a determinant of prion strains. In natural prion disease, differences in the distribution of PrPSc in the spleen and lymph node are apparent between classical and atypical scrapie. Atypical scrapie is characterized by a relative lack of detectable PrPSc in these tissues compared with the classical form; however, lymphoreticular system (LRS) tissues from atypical scrapie cases still harbor infectivity (Benestad et al. 2003; Klingeborn et al. 2006; Andréoletti et al. 2011). A more striking example of tissue tropism is observed in hamsters infected with either the hyper (HY) or drowsy (DY) strains of TME. PrPSc, as well as infectivity, is widely distributed in HY TME–infected hamsters and is detected in the brain, LRS, skeletal muscle, nasal secretions, and blood (Bessen and Marsh 1992b; Marsh and Bessen 1994; Mulcahy et al. 2004; Bessen et al. 2010; Elder et al. 2013, 2015). In contrast, infectivity and PrPSc are limited to the CNS of DY TME–infected hamsters (Bartz et al. 2003, 2004; Bessen et al. 2009). Additionally, hamsters are not susceptible to extraneural infection with the DY TME agent, perhaps attributable to a failure to establish infection in secondary LRS tissues, which is an important component of extraneural prion pathogenesis (Dickinson and Fraser 1972; Race et al. 2000; McCulloch et al. 2011).
The mechanism responsible for PrPSc tropism is poorly understood. Ocular inoculation of prions results in a sequential spread of spongiform degeneration that is consistent with transport along neuroanatomical tracks (Fraser and Dickinson 1985; Scott and Fraser 1989). Consistent with this seminal observation, the temporal and spatial spread of PrPSc in the nervous system is consistent with spread along neuroanatomical tracks regardless of the initial site of inoculation (Beekes et al. 1998; Andréoletti et al. 2000; Beekes and McBride 2000; van Keulen et al. 2000; Bartz et al. 2003; Kincaid and Bartz 2007). Sciatic nerve inoculation results in direct neuronal spread of prions in rats, mice, and hamsters (Kimberlin et al. 1983; Bassant et al. 1986; Bartz et al. 2002). Detailed analysis of the temporal and spatial spread of PrPSc in the peripheral and central nervous system of hamsters inoculated in the sciatic nerve with either the HY or DY TME strains indicates that both these strains are retrogradely transported along the same four descending neuroanatomical pathways (Ayers et al. 2009). At terminal disease, strain-specific differences in PrPSc distribution in the CNS are observed; however, this could be overcome by altering the route of infection (Ayers et al. 2009). These data suggest that, once in the CNS, strain-specific differences in transport or the ability of neurons to support PrPSc formation do not exist, but rather strain-specific differences in neurodegeneration (i.e., clinical target areas) contribute to the observed differences in PrPSc deposition at terminal disease (Kimberlin et al. 1987a; Kimberlin and Walker 1988; Mirabile et al. 2014). Alternatively, recent evidence suggests that prion replication cofactors can alter strain properties in vitro (Deleault et al. 2012b; Miller et al. 2013; Supattapone 2014). It is possible, in vivo, that differences in the cellular distribution of strain-specific cofactors influence tropism. Less is known about the mechanism underlying tissue tropism outside the CNS. It is possible that strain-specific replication cofactors or inhibitors govern PrPSc formation; alternatively, strain-specific differences in the balance between PrPSc formation and clearance may influence tissue tropism (Choi and Priola 2013).
PRION STRAIN PROPERTIES
Prion strain diversity may be encoded by distinct conformations of PrPSc. The mere existence of prion strain diversity was used as evidence against the prion hypothesis (Bruce and Dickinson 1987). In the absence of a prion-specific nucleic acid genome, new mechanisms to encode strain diversity must exist. Studies of rodent-adapted prions were the first to indicate that differences in the biochemical properties of PrPSc correspond with different prion strains (Kascsak et al. 1987). Subsequent studies using the well-characterized HY and DY strains of TME were the first to show that PrPSc could have strain-specific differences in the proteinase K (PK) digestion site, relative PK resistance, and detergent solubility (Bessen and Marsh 1992a,b, 1994). Consistent with these findings, strain-specific differences in PrPSc migration properties of human prion isolates were preserved on transmission to transgenic mice expressing chimeric mouse–human PrPC (Telling et al. 1996). These observed differences in the PK cleavage site of PrPSc from several strains suggest that the conformation of PrPSc differs among prion strains. Structural studies of PrPSc using Fourier transform infrared (FTIR) spectroscopy indicate that strain-specific differences in the secondary structure of PrPSc exist (Caughey et al. 1998; Moore et al. 2011). Electron microscopy of PrPSc fibrils enriched from murine brain indicate that the diameter and twist periodicity could differentiate prion strains (Sim and Caughey 2009). The conformation-dependent immunoassay (CDI) measures changes in immunoreactivity of PrPSc under conditions of increasing denaturation compared with immunoreactivity of PrPC and can identify strain-specific differences in PrPSc conformation between several rodent prion strains (Safar et al. 1998). Additionally, the PrPSc conformational stability assay has identified strain-specific differences in PrPSc stability from a wide range of natural and synthetic prion strains (Peretz et al. 2001; Thackray et al. 2007; Colby et al. 2009; Ayers et al. 2011; Gonzalez-Montalban et al. 2011; Ghaemmaghami et al. 2013). Although these assays indicate strain-specific differences in PrPSc conformation, it is unclear what conformational differences in the structure of PrPSc (e.g., tertiary or quaternary) contribute to the measured differences. Strain-specific differences in the PrPSc aggregate state have been observed using sedimentation equilibrium but not sedimentation velocity ultracentrifugation, suggesting that strain-specific differences in PrPSc size occur (Tixador et al. 2010; Laferrière et al. 2013). Luminescent conjugated polythiophenes (LCPs) are a class of amyloid binding dyes whose emission wavelength spectra are influenced by the morphology of the bound ligand (Nilsson et al. 2006). When LCP is bound to the PrPSc of different prion strains, a strain-specific LCP emission wavelength is detected, which suggests strain-specific differences in the tertiary or quaternary structure of PrPSc (Sigurdson et al. 2007).
Cellular cofactors can influence the rate of PrPSc formation, and there is increasing evidence to suggest that cellular cofactors can also contribute to prion strain diversity. Phosphatidylethanolamine (PE) can enhance the formation of murine PrPSc in vitro (Deleault et al. 2012a). When three distinct prion strains were propagated using protein misfolding cyclic amplification (PMCA) that contained only PrPC and PE as the cofactor, the properties of each of the input strains changed. Interestingly, all three strains were altered into a single indistinguishable strain with unique properties in animals compared with the input strains as defined by incubation period, neuropathology, and the conformational stability of PrPSc. This convergence of strain properties that is induced by the presence of PE indicates that cofactors can modify strain properties in vitro that are maintained in vivo (Deleault et al. 2012b). Consistent with this finding is the observation that cofactors can initiate structural rearrangements of PrPC that may initiate or facilitate the formation of PrPSc (Miller et al. 2013). These seminal studies provide evidence that cellular cofactors can influence strain properties in vitro, but what remains to be determined is how, in vivo, these cellular cofactors are involved in regulating the strain-specific phenotype of disease.
Strain-specific differences in the accumulation of infectivity and PrPSc during the time course of disease are well documented (Marsh and Kimberlin 1975; Kimberlin and Walker 1979; Hecker et al. 1992; Beekes et al. 1996; van Keulen et al. 2000; Mulcahy and Bessen 2004; Kaatz et al. 2012). These studies suggest strain-specific differences in the rate of prion replication in various tissues from natural and experimental prion disease but are limited by the complexity of the animal host and the resulting alternative explanations. For example, strain-specific differences in the rate of transport to new areas of the tissue that support prion formation, the rate of prion clearance, and differences in cell death all can contribute to the overall titer of a tissue (Ayers et al. 2009; Choi and Priola 2013). In an attempt to overcome these weaknesses, the relative efficiency of PrPSc formation was calculated using PMCA, which only measures PrPSc formation and not clearance in a closed system (Shikiya et al. 2014). Using this technique, strain-specific differences in the efficiency of PrPSc formation have been observed (Makarava et al. 2010; Shikiya et al. 2010; Ayers et al. 2011; Gonzalez-Montalban et al. 2011). Interestingly, prion strains with short incubation periods have correspondingly higher efficiencies of PrPSc formation compared with strains with long incubation periods (Ayers et al. 2011). The structural underpinnings of these observations are unknown. The aggregate size of PrPSc has a strong influence on prion formation rates, with small PrPSc aggregates having higher specific activity compared with larger PrPSc aggregates (Silveira et al. 2005). Therefore, strain-specific differences in the distribution of PrPSc aggregates may influence the rate of PrPSc formation and the outcome of disease. In addition, sialation of PrP can influence the rate of PrPSc formation, and strain-specific differences in PrP sialation may contribute to the overall rate of PrPSc formation (Katorcha et al. 2014). Strain-specific differences in the requirement for RNA in the formation of PrPSc have been identified; however, it is unclear whether RNA contributes to the strain-specific properties of disease (Deleault et al. 2010; Piro et al. 2011; Saá et al. 2012; Gonzalez-Montalban et al. 2013).
Fragmentation of PrPSc is hypothesized to be required for PrPSc propagation. Although a PrPSc fragmentation assay does not currently exist, differences in the conformational stability of PrPSc in denaturants have been used as a surrogate marker of PrPSc fragmentation. Initial studies in murine prion strains have indicated that strains with shorter incubation periods have PrPSc with a lower conformational stability compared with PrPSc from long incubation period strains (Colby et al. 2009; Colby and Prusiner 2011; Ghaemmaghami et al. 2011). This finding suggests that PrPSc with low conformational stability can more readily fragment, facilitating a more rapid formation of PrPSc compared with PrPSc with correspondingly longer incubation periods. This interpretation is consistent with the elegant studies of yeast prions that show that the rate of [PSI+] strain propagation inversely corresponds to low conformational stability (i.e., thermal denaturation) (Tanaka et al. 2004). Subsequent studies examining a large panel of hamster-adapted prion strains came to the opposite conclusion—that short incubation period strains with a more efficient rate of PrPSc formation, as determined by PMCA, have high PrPSc conformational stabilities compared with long incubation period strains with relatively less efficient PrPSc formation and lower PrPSc conformational stabilities (Peretz et al. 2001; Ayers et al. 2011). These conflicting results have several possible interpretations. It is possible that overly high or low PrPSc conformational stabilities enhance fragmentation, resulting in higher rates of PrPSc formation and shorter incubation periods (Choi et al. 2014). It has been suggested that low PrPSc conformational stability corresponds with increased neuronal clearance in vivo; however, this hypothesis has not been supported by cell culture studies (Ayers et al. 2011; Choi et al. 2014). Finally, it is possible that the conformational stability assay may be measuring a property of PrPSc other than fragmentation.
The relationship between strain-specific PrPSc properties and the biological phenotype of disease is poorly understood. Although there is much evidence to support the hypothesis that prion strain properties are encoded by distinct conformations of PrPSc, definitive proof remains elusive. A major shortcoming to directly test this hypothesis is the lack of knowledge regarding the relationship between the different biochemical properties of PrPSc and how they translate into the phenotype of disease. Importantly, it is not well understood what portion, if any, of the identified strain-specific biochemical differences of PrPSc actually encode strain diversity or are a consequence of it (i.e., causation versus correlation).
EVIDENCE FOR PRION STRAINS IN NATURAL PRION DISEASE
The first evidence of prion strain variation was observed in small ruminants. Sheep scrapie brain pool 1 (SSBP/1) is made up of brain homogenate from three different sheep (one Cheviot sheep and two Cheviot/Border Leicester sheep) and was maintained by serial passage in sheep without apparent changes in the disease (Dickinson 1976). Interspecies transmission of SSBP/1 to goats resulted in the identification of two distinct disease syndromes, termed scratching and drowsy (Pattison and Millson 1961). Intraspecies transmission of the scratching and drowsy syndromes in goats maintained the clinical phenotype of disease for at least nine serial passages. More recently, an atypical form of sheep scrapie, also known as Nor98, has a different clinical presentation of disease and histopathological features compared with classical scrapie (Benestad et al. 2003). Importantly, atypical scrapie has unique PK-resistant PrPSc fragments and is more sensitive to PK digestion compared with classical forms of scrapie (Benestad et al. 2003; Klingeborn et al. 2006).
In cattle, three forms of BSE are recognized. Classical BSE reached epidemic proportions since its identification in 1995 and is responsible for the emergence of the variant form of CJD (Wells 1987; Bruce et al. 1997; Hill et al. 1997; Scott et al. 1999). With increased prion surveillance efforts in cattle, two additional forms of BSE have been identified. Bovine amyloidotic spongiform encephalopathy (BASE), also known as L-type BSE, is characterized by lower molecular weight migration of the PK-resistant fragment of PrPSc compared with classical BSE (Casalone et al. 2004; Buschmann et al. 2006). The regional distribution of PrPSc in the CNS from BASE is more rostral in the CNS compared with classical BSE (Casalone et al. 2004). Additionally, amyloid plaques are found in CNS tissue of BASE that are not observed in classical BSE (Casalone et al. 2004). In contrast to BASE, the PK-resistant fragment of PrPSc from H-type BSE migrates to a higher molecular weight compared with classical BSE. Both atypical forms of BSE are rare and are largely identified in aged cattle, which suggests they have a sporadic etiology (Brown et al. 2006).
Several types of CJD have been identified. Two major forms of PrPSc are found in CJD, types 1 and 2, which are characterized by migration of the unglycosylated PrPSc polypeptide at 21 and 19 kDa, respectively (Parchi et al. 1996). These differences result from PK cleavage of PrPSc at amino acid residue 82 in type 1 and residue 97 in type 2 (Parchi et al. 2000). These two subtypes can be further refined into six subtypes based on the PRNP codon 129 methionine/valine polymorphism. The subtypes of CJD correspond with specific clinical and pathological characteristics (Parchi et al. 1996, 1999), and may illuminate the etiology of CJD and suggest that some cases of sCJD may actually be attributable to an infectious etiology (Kobayashi et al. 2015).
Experimental transmission studies under controlled conditions firmly established the existence of prion strains in natural prion disease. Although the evidence for strains in the natural host species is strong, factors that are not controlled under field conditions could influence the outcome of disease (Fig. 1). As mentioned earlier in this section, the scratching and drowsy strains of scrapie are preserved following serial passage, indicating that strain diversity exists in natural disease. Experimental passage of SSBP/1 to mice results in the isolation of several distinct strains, suggesting that more than one strain of sheep scrapie is present in the inoculum (Thackray et al. 2011, 2012). Interspecies transmission, however, can lead to generation of new strains, complicating the interpretation of these studies (Kimberlin et al. 1987b; Bartz et al. 2000). To eliminate this variable, transmission of prions to transgenic mice expressing PrPC with the same amino acid sequence as the inoculum PrPSc can preserve strain properties (Scott et al. 1989); however, recent evidence suggests that this is not always the case (Crowell et al. 2015). Transmission of sCJD and FFI, which have 21 and 19 kDa deglycosylated PrPSc, respectively, to transgenic mice that express a chimeric human–mouse PrP gene results in the preservation of strain-specific biochemical properties of PrPSc, which indicates that these are bona fide prion strains (Telling et al. 1996). Similarly, experimental passage of H- and L-type BSE to transgenic mice expressing bovine PrPC result in the preservation of strain-specific migration of PrPSc, confirming that these are indeed distinct strains of BSE (Buschmann et al. 2006). Passage of sheep scrapie to transgenic mice expressing ovine PrPC results in the preservation of the distinctive strain properties and provides further evidence for the diversity of strains in sheep scrapie (Thackray et al. 2011, 2012).
In natural prion disease, more than one prion strain can be present in a single host. The coexistence of classical and atypical (Nor98) strains of scrapie is suggested by the presence of both classical and atypical PrPSc and pathology in a single sheep (Mazza et al. 2010). Passage of a single-sheep scrapie isolate to transgenic mice expressing ovine PrPC resulted in the isolation of multiple distinct strains, suggesting that the field isolate of scrapie contained more than one prion strain (Thackray et al. 2011). Evidence for the coexistence of prion strains in humans infected with CJD is based on the detection of both types 1 and 2 PrPSc in the same individual (Puoti et al. 1999). The relative percentage of CJD cases in which types 1 and 2 PrPSc coexist, however, is controversial (Polymenidou et al. 2005; Schoch et al. 2006; Notari et al. 2007). Experimentally, incomplete PK digestion of PrPSc may allow for an overestimation of the co-occurrence of types 1 and 2 PrPSc (Notari et al. 2004). Conversely, co-occurrence of types 1 and 2 PrPSc may be underestimated if a large panel of anti-PrP antibodies is not used or if tissue sampling from a limited number of brain regions occurs. Regardless, it is clear that in natural prion disease, mixtures of prion strains occur frequently. The effect of strain mixtures on the development of disease and transmission in cases of natural prion disease is unclear.
CONSEQUENCES OF PRION STRAIN MIXTURES
Selection of a strain from a mixture influences prion adaptation. Transmission of prions to a new species can result in extended incubation periods and lower attack rates (i.e., species barrier) compared with intraspecies transmission in the original host species (Pattison 1966). The species-barrier effect influences differences in the amino acid sequence of the donor and host PrPSc and PrPC (Bartz et al. 1994; Kocisko et al. 1995; Priola and Chesebro 1995; Browning et al. 2004). Subsequent serial passages in the new host species result in strain adaptation, which is characterized by a shortening and eventual stabilization of the incubation period (Kimberlin and Walker 1978). Adaptation is thought to occur via selection of the strain that is the most fit for the new host species from either a preexisting mixture of strains or from strains that are generated on interspecies transmission (Kimberlin and Walker 1978; Kimberlin et al. 1987b; Bartz et al. 2000). In both of these possibilities, the number and relative ratio of the strains in a mixture is thought to influence strain emergence.
An adapted strain that is stably passaged may contain a mixture of strains. Consistent with this hypothesis, changing the prion replication environment either in cell culture or PMCA can alter the properties of the strain, perhaps by selecting for a minor strain that was present in the starting inoculum (Li et al. 2010; Mahal et al. 2010; Gonzalez-Montalban et al. 2013; Makarava et al. 2013). This can also be accomplished by selection of prions that propagate in the presence of antiprion drugs that, once the drug is removed, revert back to the drug-sensitive phenotype (Li et al. 2010). These data suggest that prions act as a quasispecies. Viral quasispecies are defined as a population of similar but not identical viral particles that are a consequence of a high mutation rate of the viral genome (Domingo et al. 1978; Domingo 2000). Likewise, prion quasispecies are hypothesized to be a similar but not identical population of PrPSc conformations (Collinge and Clarke 2007; Li et al. 2010). Although intriguing, little is understood about the mutation rate of prions, and the distribution of PrPSc conformations is unknown (Kimberlin and Walker 1986; Bruce and Dickinson 1987; Kimberlin et al. 1987b). The implication of multiple strains existing as one mixture on the biology of prion disease is only beginning to be understood.
Prion strains can interfere with each other. This was first observed in mice when the long incubation period strain 22C was inoculated before superinfection with 22A. As the interval between inoculation with the blocking strain 22C and superinfection with 22A increased, 22C was able to extend the incubation period or completely block 22A from causing disease (Dickinson et al. 1972). Prion-strain interference can occur by either superinfection, as in the previous example, or during co-infection in which the short and long incubation period strains are inoculated simultaneously. Strain interference occurs between strains from mouse, hamster, and human prions, indicating that the phenomenon is not limited to one species (Manuelidis 1998; Baron and Biacabe 2001; Bartz et al. 2004; Schutt and Bartz 2008; Nilsson et al. 2010; Haldiman et al. 2013). Strain interference can occur following inoculation via either neuronal (intracerebral, sciatic nerve) (Dickinson et al. 1975; Shikiya et al. 2010) or non-neuronal (intraperitoneal, oral) (Dickinson et al. 1975) routes of inoculation. Overall, strain interference is a common property of prions.
The relative onset of replication of the blocking and superinfecting strain influences strain emergence. When the blocking and the superinfecting strains are co-infected, as the relative titer of the blocking strain to the superinfecting strain is increased, the blocking strain has an increased ability to interfere with the superinfecting strain (Dickinson et al. 1975). Interestingly, the titer of the blocking strain must be greater compared with the superinfecting strain for strain interference to occur in animals and in vitro using the PMCA strain interference model (Shikiya et al. 2010). In strain interference attributable to superinfection, extending the time between inoculation of the blocking strain and superinfecting strain increases the ability of the blocking strain to interfere with the superinfecting strain (Dickinson et al. 1972, 1975; Bartz et al. 2004, 2007; Shikiya et al. 2010). For strain interference to occur in both circumstances (i.e., co-infection or superinfection), the onset of replication of the slowly replicating long incubation period strain before the onset of replication of the faster replicating short incubation period strain is required (Bartz et al. 2007).
The blocking strain replication is required for strain interference to occur. Inactivation of the 22A strain by boiling, exposure to 12 M urea, or ionizing radiation eliminates the ability to interfere with the superinfected 22C strain, indicating that blocking strain replication and infectivity is necessary for strain interference (Kimberlin and Walker 1985). Mice inoculated with TME do not develop prion disease, and when the TME agent is used as the blocking strain, it does not extend the incubation period of 22A, 22C, 79A, 87A, 139A, or ME7 strains (Taylor et al. 1986). These data suggest that replication (and not simply site blocking) is required for strain interference to occur.
Strain interference requires that the blocking and superinfecting strains infect the same cells. Sciatic nerve inoculation of hamsters with a long incubation period strain (DY TME) before superinfection with a short incubation period strain (HY TME) leads to extension of the incubation period of complete blockage of HY TME (Bartz et al. 2004, 2007). In this system, ventral motor neurons (VMNs) ipsilateral to the side of inoculation are the first cells infected by both HY and DY TME, suggesting that VMNs are where strain interference occurs (Bartz et al. 2007; Ayers et al. 2009; Shikiya et al. 2010). Consistent with this hypothesis, superinfection of the sciatic nerve with HY TME opposite (i.e., contralateral) to the DY TME–infected nerve eliminates strain interference, and the animals develop disease with incubation periods similar to animals inoculated with HY TME alone (Bartz et al. 2007). This conclusively shows that both strains need to infect the same population of neurons (VMNs in this example) for the strain interference effect to occur. Finally, these data indicate that a soluble secreted factor does not produce the strain interference effect.
The mechanism of prion strain interference is not known. The replication site hypothesis predicts that prion strains compete for a limited number of prion replication sites in the host that are necessary for propagation of both the blocking and superinfection strains (Dickinson and Outram 1979). The replication site was hypothesized to be the gene product of Sinc (Dickinson and Outram 1979), which was subsequently identified as PrPC (Hunter et al. 1987; Westaway et al. 1987). Consistent with this hypothesis, PMCA strain interference studies indicate that the blocking strain DY TME does not convert all of the available PrPC to PrPSc, yet it is still able to interfere with the superinfecting strain HY TME (Shikiya et al. 2010). In animals, the blocking strain does not disrupt transport to or result in the death of cells required for superinfection replication, and the only observed change that corresponds with strain interference is the formation of PrPSc (Shikiya et al. 2010). These data suggest that the blocking strain is sequestering PrPC or preventing its use by the superinfecting strain (Shikiya et al. 2010). Recent evidence indicates that just before the onset of clinical disease, PrPC expression is downregulated in the CNS (Mays et al. 2014, 2015). This could provide another mechanism by which the blocking strain limits the number of replication sites; however, it remains to be established whether PrPC downregulation occurs in neurons at the early time points postinfection when the blocking strain is able to interfere with the superinfecting strain.
CONCLUSIONS
A significant challenge in prion biology is to identify the novel mechanisms that a protein-only infectious agent uses to accomplish complex biological tasks. Prion strains are operationally defined by differences in neuropathology when passaged under controlled experimental conditions. The current definition of prion strains is based on a fundamental lack of understanding of the molecular basis that encodes prion strain variation. It is unclear whether strain-specific differences in PrPSc conformation alone can encode strain variation or whether strain-specific cofactors are involved. It is unknown whether the biochemical properties of PrPSc that correspond with strain variation are causative or correlative. Further complicating the understanding of prion strains is recent evidence suggesting that strains are not static and unchanging but rather a dynamic mixture of many strains. Although much has been accomplished, further work is clearly needed to resolve these longstanding, important problems in prion biology.
ACKNOWLEDGMENTS
This article is dedicated to the late Dr. Richard Bessen, mentor, colleague, and friend.
Footnotes
Editor: Stanley B. Prusiner
Additional Perspectives on Prion Diseases available at www.perspectivesinmedicine.org
REFERENCES
- Alpers M, Gajdusek DC. 1965. Changing patterns of kuru: Epidemiological changes in the period of increasing contact of the Fore people with Western civilization. Am J Trop Med Hyg 14: 852–879. [DOI] [PubMed] [Google Scholar]
- Andréoletti O, Berthon P, Marc D, Sarradin P, Grosclaude J, van Keulen L, Schelcher F, Elsen J, Lantier F. 2000. Early accumulation of PrPSc in gut-associated lymphoid and nervous tissues of susceptible sheep from a Romanov flock with natural scrapie. J Gen Virol 81: 3115–3126. [DOI] [PubMed] [Google Scholar]
- Andréoletti O, Orge L, Benestad SL, Beringue V, Litaise C, Simon S, Le Dur A, Laude H, Simmons H, Lugan S, et al. 2011. Atypical/Nor98 scrapie infectivity in sheep peripheral tissues. PLoS Pathog 7: e1001285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayers J, Kincaid AE, Bartz JC. 2009. Prion strain targeting independent of strain-specific neuronal tropism. J Virol 83: 81–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayers JI, Schutt CR, Shikiya RA, Aguzzi A, Kincaid AE, Bartz JC. 2011. The strain-encoded relationship between PrP replication, stability and processing in neurons is predictive of the incubation period of disease. PLoS Pathog 7: e1001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron TGM, Biacabe AG. 2001. Molecular analysis of the abnormal prion protein during coinfection of mice by bovine spongiform encephalopathy and a scrapie agent. J Virol 75: 107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartz JC, McKenzie DI, Bessen RA, Marsh RF, Aiken JM. 1994. Transmissible mink encephalopathy species barrier effect between ferret and mink: PrP gene and protein analysis. J Gen Virol 75: 2947–2953. [DOI] [PubMed] [Google Scholar]
- Bartz JC, Bessen RA, McKenzie D, Marsh RF, Aiken JM. 2000. Adaptation and selection of prion protein strain conformations following interspecies transmission of transmissible mink encephalopathy. J Virol 74: 5542–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartz JC, Kincaid AE, Bessen RA. 2002. Retrograde transport of transmissible mink encephalopathy within descending motor tracts. J Virol 76: 5759–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartz JC, Kincaid AE, Bessen RA. 2003. Rapid prion neuroinvasion following tongue infection. J Virol 77: 583–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartz JC, Aiken JM, Bessen RA. 2004. Delay in onset of prion disease for the HY strain of transmissible mink encephalopathy as a result of prior peripheral inoculation with the replication-deficient DY strain. J Gen Virol 85: 265–273. [DOI] [PubMed] [Google Scholar]
- Bartz JC, Kramer ML, Sheehan MH, Hutter JAL, Ayers JI, Bessen RA, Kincaid AE. 2007. Prion interference is due to a reduction in strain-specific PrPSc levels. J Virol 81: 689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler K, Oesch B, Scott M, Westaway D, Walchli M, Groth DF, McKinley MP, Prusiner SB, Weissmann C. 1986. Scrapie and cellular PrP isoforms are encoded by the same chromosomal gene. Cell 46: 417–428. [DOI] [PubMed] [Google Scholar]
- Bassant M, Baron H, Gumpel M, Cathala F, Court L. 1986. Spread of scrapie agent to the central nervous system: Study of a rat model. Brain Res 383: 397–401. [DOI] [PubMed] [Google Scholar]
- Beekes M, McBride P. 2000. Early accumulation of pathological PrP in the enteric nervous system and gut-associated lymphoid tissue of hamsters orally infected with scrapie. Neurosci Lett 278: 181–184. [DOI] [PubMed] [Google Scholar]
- Beekes M, Baldauf E, Diringer H. 1996. Sequential appearance and accumulation of pathognomonic markers in the central nervous system of hamsters orally infected with scrapie. J Gen Virol 77: 1925–1934. [DOI] [PubMed] [Google Scholar]
- Beekes M, McBride PA, Baldauf E. 1998. Cerebral targeting indicates vagal spread of infection in hamsters fed with scrapie. J Gen Virol 79: 601–607. [DOI] [PubMed] [Google Scholar]
- Benestad SL, Sarradin P, Thu B, Schönheit J, Tranulis MA, Bratberg B. 2003. Cases of scrapie with unusual features in Norway and designation of a new type, Nor98. Vet Rec 153: 202–208. [DOI] [PubMed] [Google Scholar]
- Bessen R, Marsh R. 1992a. Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J Virol 66: 2096–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessen R, Marsh R. 1992b. Identification of two biologically distinct strains of transmissible mink encephalopathy in hamsters. J Gen Virol 73: 329–334. [DOI] [PubMed] [Google Scholar]
- Bessen RA, Marsh RF. 1994. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J Virol 68: 7859–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessen RA, Martinka S, Kelly J, Gonzalez D. 2009. Role of the lymphoreticular system in prion neuroinvasion from the oral and nasal mucosa. J Virol 83: 6435–6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessen RA, Shearin H, Martinka S, Boharski R, Lowe D, Wilham JM, Caughey B, Wiley JA. 2010. Prion shedding from olfactory neurons into nasal secretions. PLoS Pathog 6: e1000837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton D, McKinley M, Prusiner S. 1982. Identification of a protein that purifies with the scrapie prion. Science 218: 1309–1311. [DOI] [PubMed] [Google Scholar]
- Brown P, McShane LM, Zanusso G, Detwile L. 2006. On the question of sporadic or atypical bovine spongiform encephalopathy and Creutzfeldt–Jakob disease. Emerg Infect Dis 12: 1816–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Mason GL, Seward T, Green M, Eliason GAJ, Mathiason C, Miller MW, Williams ES, Hoover E, Telling GC. 2004. Transmission of prions from mule deer and elk with chronic wasting disease to transgenic mice expressing cervid PrP. J Virol 78: 13345–13350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce M, Dickinson A. 1985. Genetic control of amyloid plaque production and incubation period in scrapie-infected mice. J Neuropathol Exp Neurol 44: 285–294. [DOI] [PubMed] [Google Scholar]
- Bruce M, Dickinson A. 1987. Biological evidence that scrapie agent has an independent genome. J Gen Virol 68: 79–89. [DOI] [PubMed] [Google Scholar]
- Bruce ME, Will RG, Ironside JW, McConnell I, Drummond D, Suttie A, McCardle L, Chree A, Hope J, Birkett C, et al. 1997. Transmissions to mice indicate that “new variant” CJD is caused by the BSE agent. Nature 389: 498–501. [DOI] [PubMed] [Google Scholar]
- Buschmann A, Gretzschel A, Biacabe AG, Schiebel K, Corona C, Hoffmann C, Eiden M, Baron T, Casalone C, Groschup MH. 2006. Atypical BSE in Germany—Proof of transmissibility and biochemical characterization. Vet Microbiol 117: 103–116. [DOI] [PubMed] [Google Scholar]
- Casalone C, Zanusso G, Acutis P, Ferrari S, Capucci L, Tagliavini F, Monaco S, Caramelli M. 2004. Identification of a second bovine amyloidotic spongiform encephalopathy: Molecular similarities with sporadic Creutzfeldt–Jakob disease. Proc Natl Acad Sci 101: 3065–3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughey B, Raymond G. 1991. The scrapie-associated form of PrP is made from a cell surface precursor that is both protease- and phospholipase-sensitive. J Biol Chem 266: 18217–18223. [PubMed] [Google Scholar]
- Caughey B, Raymond GJ, Bessen RA. 1998. Strain-dependent differences in β-sheet conformations of abnormal prion protein. J Biol Chem 273: 32230–32235. [DOI] [PubMed] [Google Scholar]
- Choi YP, Priola SA. 2013. A specific population of abnormal prion protein aggregates is preferentially taken up by cells and disaggregated in a strain dependent manner. J Virol 87: 11552–11561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YP, Head MW, Ironside JW, Priola SA. 2014. Uptake and degradation of protease-sensitive and -resistant forms of abnormal human prion protein aggregates by human astrocytes. Am J Pathol 184: 3299–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby DW, Prusiner SB. 2011. De novo generation of prion strains. Nat Rev Microbiol 9: 771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby DW, Giles K, Legname G, Wille H, Baskakov IV, DeArmond SJ, Prusiner SB. 2009. Design and construction of diverse mammalian prion strains. Proc Natl Acad Sci 106: 20417–20422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge J, Clarke A. 2007. A general model of prion strains and their pathogenicity. Science 318: 930–936. [DOI] [PubMed] [Google Scholar]
- Crowell J, Hughson A, Caughey B, Bessen RA. 2015. Host determinants of prion strain diversity independent of prion protein genotype. J Virol 89: 10427–10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuillé J, Chelle PL. 1936. La maladie dite “tremblante du mouton” est-elle inoculable? [Is the disease known as “scrapie” inoculable?] C R Seances Acad Sci 203: 1552–1554. [Google Scholar]
- DeArmond S, Yang S, Lee A, Bowler R, Taraboulos A, Groth D, Prusiner S. 1993. Three scrapie prion isolates exhibit different accumulation patterns of the prion protein scrapie isoform. Proc Natl Acad Sci 90: 6449–6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleault NR, Harris BT, Rees JR, Supattapone S. 2007. Formation of native prions from minimal components in vitro. Proc Natl Acad Sci 104: 9741–9746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleault NR, Kascsak R, Geoghegan JC, Supattapone S. 2010. Species-dependent differences in cofactor utilization for formation of the protease-resistant prion protein in vitro. Biochemistry 49: 3928–3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleault NR, Piro JR, Walsh DJ, Wang F, Ma J, Geoghegan JC, Supattapone S. 2012a. Isolation of phosphatidylethanolamine as a solitary cofactor for prion formation in the absence of nucleic acids. Proc Natl Acad Sci 109: 8546–8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleault NR, Walsh DJ, Piro JR, Wang F, Wang X, Ma J, Rees JR, Supattapone S. 2012b. Cofactor molecules maintain infectious conformation and restrict strain properties in purified prions. Proc Natl Acad Sci 109: E1938–E1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson A. 1976. Scrapie in sheep and goats. Front Biol 44: 209–241. [PubMed] [Google Scholar]
- Dickinson A, Fraser H. 1969. Genetical control of the concentration of ME7 scrapie agent in mouse spleen. J Comp Pathol 79: 363–366. [DOI] [PubMed] [Google Scholar]
- Dickinson AG, Fraser H. 1972. Scrapie: Effect of Dh gene on incubation period of extraneurally injected agent. Heredity 29: 91–93. [DOI] [PubMed] [Google Scholar]
- Dickinson A, Meikle V. 1971. Host-genotype and agent effects in scrapie incubation: Change in allelic interaction with different strains of agent. Mol Gen Genet 112: 73–79. [DOI] [PubMed] [Google Scholar]
- Dickinson A, Outram G. 1979. The scrapie replication-site hypothesis and its implications for pathogenesis. In Slow transmissible diseases of the central nervous system, Vol. 2 (ed. Prusiner SB, Hadlow WJ), pp. 13–31. Academic, New York. [Google Scholar]
- Dickinson A, Meikle V, Fraser H. 1968. Identification of a gene which controls the incubation period of some strains of scrapie agent in mice. J Comp Pathol 78: 293–299. [DOI] [PubMed] [Google Scholar]
- Dickinson A, Meikle V, Fraser H. 1969. Genetical control of the concentration of ME7 scrapie agent in the brain of mice. J Comp Pathol 79: 15–22. [DOI] [PubMed] [Google Scholar]
- Dickinson A, Fraser H, Meikle V, Outram G. 1972. Competition between different scrapie agents in mice. Nat New Biol 237: 244–245. [DOI] [PubMed] [Google Scholar]
- Dickinson AG, Fraser H, McConnell I, Outram GW, Sales DI, Taylor DM. 1975. Extraneural competition between different scrapie agents leading to loss of infectivity. Nature 253: 556. [DOI] [PubMed] [Google Scholar]
- Domingo E. 2000. Viruses at the edge of adaptation. Virology 270: 251–253. [DOI] [PubMed] [Google Scholar]
- Domingo E, Sabo D, Taniguchi T, Weissmann C. 1978. Nucleotide sequence heterogeneity of an RNA phage population. Cell 13: 735–744. [DOI] [PubMed] [Google Scholar]
- Elder AM, Henderson DM, Nalls AV, Wilham JM, Caughey BW, Hoover EA, Kincaid AE, Bartz JC, Mathiason CK. 2013. In vitro detection of prionemia in TSE-infected cervids and hamsters. PLoS ONE 8: e80203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder AM, Henderson DM, Nalls AV, Hoover EA, Kincaid AE, Bartz JC, Mathiason CK. 2015. Immediate and ongoing detection of prions in the blood of hamsters and deer following oral, nasal, or blood inoculations. J Virol 89: 7421–7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser H, Dickinson A. 1967. Distribution of experimentally induced scrapie lesions in the brain. Nature 216: 1310–1311. [DOI] [PubMed] [Google Scholar]
- Fraser H, Dickinson A. 1968. The sequential development of the brain lesion of scrapie in three strains of mice. J Comp Pathol 78: 301–311. [DOI] [PubMed] [Google Scholar]
- Fraser H, Dickinson A. 1985. Targeting of scrapie lesions and spread of agent via the retino-tectal projection. Brain Res 346: 32–41. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Watts JC, Nguyen H-O, Hayashi S, Dearmond SJ, Prusiner SB. 2011. Conformational transformation and selection of synthetic prion strains. J Mol Biol 413: 527–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Colby DW, Nguyen HOB, Hayashi S, Oehler A, Dearmond SJ, Prusiner SB. 2013. Convergent replication of mouse synthetic prion strains. Am J Pathol 182: 866–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs C, Gajdusek D, Asher D, Alpers MP, Beck E, Daniel P, Matthews W. 1968. Creutzfeldt–Jakob disease (spongiform encephalopathy): Transmission to the chimpanzee. Science 161: 388–389. [DOI] [PubMed] [Google Scholar]
- Gibbs C, Amyx H, Bacote A, Masters C, Gajdusek D. 1980. Oral transmission of kuru, Creutzfeldt–Jakob disease, and scrapie to nonhuman primates. J Infect Dis 142: 205–208. [DOI] [PubMed] [Google Scholar]
- Goldfarb L, Petersen R, Tabaton M, Brown P, LeBlanc A, Montagna P, Cortelli P, Julien J, Vital C, Pendelbury W, et al. 1992. Fatal familial insomnia and familial Creutzfeldt–Jakob disease: Disease phenotype determined by a DNA polymorphism. Science 258: 806–808. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Montalban N, Makarava N, Savtchenko R, Baskakov IV. 2011. Relationship between conformational stability and amplification efficiency of prions. Biochemistry 50: 6815–6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Montalban N, Lee YJ, Makarava N, Savtchenko R, Baskakov IV. 2013. Changes in prion replication environment cause prion strain mutation. FASEB J 27: 3702–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadlow WJ, Race RE, Kennedy RC. 1987. Temporal distribution of transmissible mink encephalopathy virus in mink inoculated subcutaneously. J Virol 61: 3235–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldiman T, Kim C, Cohen Y, Chen W, Blevins J, Qing L, Cohen ML, Langeveld J, Telling GC, Kong Q, et al. 2013. Coexistence of distinct prion types enables conformational evolution of human PrPSc by competitive selection. J Biol Chem 288: 29846–29861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker R, Taraboulos A, Scott M, Pan KM, Yang SL, Torchia M, Jendroska K, Dearmond SJ, Prusiner SB. 1992. Replication of distinct scrapie prion isolates is region specific in brains of transgenic mice and hamsters. Genes Dev 6: 1213–1228. [DOI] [PubMed] [Google Scholar]
- Hill A, Desbruslais M, Joiner S, Sidle K, Gowland I, Collinge J, Doey L, Lantos P. 1997. The same prion strain causes vCJD and BSE. Nature 389: 448–450, 526. [DOI] [PubMed] [Google Scholar]
- Hope J, Reekie L, Hunter N, Multhaup G, Beyreuther K, White H, Scott A, Stack M, Dawson M, Wells G. 1988. Fibrils from brains of cows with new cattle disease contain scrapie-associated protein. Nature 336: 390–392. [DOI] [PubMed] [Google Scholar]
- Hunter N, Hope J, McConnell I, Dickinson AG. 1987. Linkage of the scrapie-associated fibril protein (PrP) gene and Sinc using congenic mice and restriction fragment length polymorphism analysis. J Gen Virol 68: 2711–2716. [DOI] [PubMed] [Google Scholar]
- Jeffrey M, Martin S, González L, Ryder S, Bellworthy S, Jackman R. 2001. Differential diagnosis of infections with the bovine spongiform encephalopathy (BSE) and scrapie agents in sheep. J Comp Pathol 125: 271–284. [DOI] [PubMed] [Google Scholar]
- Jeffrey M, Martin S, González L. 2003. Cell-associated variants of disease-specific prion protein immunolabelling are found in different sources of sheep transmissible spongiform encephalopathy. J Gen Virol 84: 1033–1045. [DOI] [PubMed] [Google Scholar]
- Kaatz M, Fast C, Ziegler U, Balkema-Buschmann A, Hammerschmidt B, Keller M, Oelschlegel A, McIntyre L, Groschup MH. 2012. Spread of classic BSE prions from the gut via the peripheral nervous system to the brain. Am J Pathol 181: 515–524. [DOI] [PubMed] [Google Scholar]
- Kascsak R, Rubenstein R, Merz P, Tonna-DeMasi M, Fersko R, Carp R, Wisniewski H, Diringer H. 1987. Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J Virol 61: 3688–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katorcha E, Makarava N, Savtchenko R, D’Azzo A, Baskakov IV. 2014. Sialylation of prion protein controls the rate of prion amplification, the cross-species barrier, the ratio of PrPSc glycoform and prion infectivity. PLoS Pathog 10: e1004366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberlin R, Walker C. 1977. Characteristics of a short incubation model of scrapie in the golden hamster. J Gen Virol 34: 295–304. [DOI] [PubMed] [Google Scholar]
- Kimberlin R, Walker C. 1978. Evidence that the transmission of one source of scrapie agent to hamsters involves separation of agent strains from a mixture. J Gen Virol 39: 487–496. [DOI] [PubMed] [Google Scholar]
- Kimberlin R, Walker C. 1979. Pathogenesis of mouse scrapie: Dynamics of agent replication in spleen, spinal cord and brain after infection by different routes. J Comp Pathol 89: 551–562. [DOI] [PubMed] [Google Scholar]
- Kimberlin R, Walker C. 1982. Pathogenesis of mouse scrapie: Patterns of agent replication in different parts of the CNS following intraperitoneal infection. J R Soc Med 75: 618–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberlin R, Walker C. 1985. Competition between strains of scrapie depends on the blocking agent being infectious. Intervirology 23: 74–81. [DOI] [PubMed] [Google Scholar]
- Kimberlin R, Walker C. 1986. Pathogenesis of scrapie (strain 263K) in hamsters infected intracerebrally, intraperitoneally or intraocularly. J Gen Virol 67: 255–263. [DOI] [PubMed] [Google Scholar]
- Kimberlin R, Walker C. 1988. Pathogenesis of experimental scrapie. Ciba Found Symp 135: 37–62. [DOI] [PubMed] [Google Scholar]
- Kimberlin R, Hall S, Walker C. 1983. Pathogenesis of mouse scrapie. Evidence for direct neural spread of infection to the CNS after injection of sciatic nerve. J Neurol Sci 61: 315–325. [DOI] [PubMed] [Google Scholar]
- Kimberlin R, Cole S, Walker C. 1987a. Pathogenesis of scrapie is faster when infection is intraspinal instead of intracerebral. Microb Pathog 2: 405–415. [DOI] [PubMed] [Google Scholar]
- Kimberlin R, Cole S, Walker C. 1987b. Temporary and permanent modifications to a single strain of mouse scrapie on transmission to rats and hamsters. J Gen Virol 68: 1875–1881. [DOI] [PubMed] [Google Scholar]
- Kincaid AE, Bartz JC. 2007. The nasal cavity is a route for prion infection in hamsters. J Virol 81: 4482–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingeborn M, Wik L, Simonsson M, Renström L, Ottinger T, Linné T. 2006. Characterization of proteinase K-resistant N-and C-terminally truncated PrP in Nor98 atypical scrapie. J Gen Virol 87: 1751. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Parchi P, Yamada M, Brown P, Saverioni D, Matsuura Y, Takeuchi A, Mohri S, Kitamoto T. 2015. Transmission properties of atypical Creutzfeldt–Jakob disease: A clue to disease etiology? J Virol 89: 3939–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocisko D, Priola S, Raymond G, Chesebro B, Lansbury P, Caughey B. 1995. Species specificity in the cell-free conversion of prion protein to protease-resistant forms: A model for the scrapie species barrier. Proc Natl Acad Sci 92: 3923–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laferrière F, Tixador P, Moudjou M, Chapuis J, Sibille P, Herzog L, Reine F, Jaumain E, Laude H, Rezaei H, et al. 2013. Quaternary structure of pathological prion protein as a determining factor of strain-specific prion replication dynamics. PLoS Pathog 9: e1003702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Browning S, Mahal SP, Oelschlegel AM, Weissmann C. 2010. Darwinian evolution of prions in cell culture. Science 327: 869–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberski PP, Sikorska B, Guiroy D, Bessen RA. 2009. Transmissible mink encephalopathy—Review of the etiology of a rare prion disease. Folia Neuropathol 47: 195–204. [PubMed] [Google Scholar]
- Mahal SP, Browning S, Li J, Suponitsky-Kroyter I, Weissmann C. 2010. Transfer of a prion strain to different hosts leads to emergence of strain variants. Proc Natl Acad Sci 107: 22653–22658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarava N, Kovács GG, Bocharova O, Savtchenko R, Alexeeva I, Budka H, Rohwer RG, Baskakov IV. 2010. Recombinant prion protein induces a new transmissible prion disease in wild-type animals. Acta Neuropathol 119: 177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarava N, Savtchenko R, Baskakov IV. 2013. Selective amplification of classical and atypical prions using modified protein misfolding cyclic amplification. J Biol Chem 288: 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis L. 1998. Vaccination with an attenuated Creutzfeldt–Jakob disease strain prevents expression of a virulent agent. Proc Natl Acad Sci 95: 2520–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh R, Bessen R. 1994. Physicochemical and biological characterizations of distinct strains of the transmissible mink encephalopathy agent. Philos Trans R Soc Lond B Biol Sci 343: 413–414. [DOI] [PubMed] [Google Scholar]
- Marsh RF, Hanson RP. 1969. Physical and chemical properties of the transmissible mink encephalopathy agent. J Virol 3: 176–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh R, Hanson R. 1978. The Syrian hamster as a model for the study of slow virus diseases caused by unconventional agents. Fed Proc 37: 2076–2078. [PubMed] [Google Scholar]
- Marsh R, Kimberlin R. 1975. Comparison of scrapie and transmissible mink encephalopathy in hamsters. II: Clinical signs, pathology, and pathogenesis. J Infect Dis 131: 104–110. [DOI] [PubMed] [Google Scholar]
- Marsh R, Bessen R, Lehmann S, Hartsough G. 1991. Epidemiological and experimental studies on a new incident of transmissible mink encephalopathy. J Gen Virol 72: 589–594. [DOI] [PubMed] [Google Scholar]
- Mays CE, Kim C, Haldiman T, van der Merwe J, Lau A, Yang J, Grams J, Di Bari MA, Nonno R, Telling GC, et al. 2014. Prion disease tempo determined by host-dependent substrate reduction. J Clin Invest 124: 847–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mays CE, van der Merwe J, Kim C, Haldiman T, McKenzie D, Safar JG, Westaway D. 2015. Prion infectivity plateaus and conversion to symptomatic disease originate from falling precursor levels and increased oligomeric PrPSc species. J Virol 89: 12418–12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza M, Iulini B, Vaccari G, Acutis PL, Martucci F, Esposito E, Peletto S, Barocci S, Chiappini B, Corona C, et al. 2010. Co-existence of classical scrapie and Nor98 in a sheep from an Italian outbreak. Res Vet Sci 88: 478–485. [DOI] [PubMed] [Google Scholar]
- McCulloch L, Brown KL, Bradford BM, Hopkins J, Bailey M, Rajewsky K, Manson JC, Mabbott NA. 2011. Follicular dendritic cell-specific prion protein (PrP) expression alone is sufficient to sustain prion infection in the spleen. PLoS Pathog 7: e1002402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, Wang DW, Wang F, Noble GP, Ma J, Woods VL, Li S, Supattapone S. 2013. Cofactor molecules induce structural transformation during infectious prion formation. Structure 21: 2061–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabile I, Jat PS, Brandner S, Collinge J. 2014. Identification of clinical target areas in the brainstem of prion infected mice. Neuropathol Appl Neurobiol 41: 613–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RA, Timmes AG, Wilmarth PA, Safronetz D, Priola SA. 2011. Identification and removal of proteins that co-purify with infectious prion protein improves the analysis of its secondary structure. Proteomics 11: 3853–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy ER, Bessen RA. 2004. Strain-specific kinetics of prion protein formation in vitro and in vivo. J Biol Chem 279: 1643–1649. [DOI] [PubMed] [Google Scholar]
- Mulcahy ER, Bartz JC, Kincaid AE, Bessen RA. 2004. Prion infection of skeletal muscle cells and papillae in the tongue. J Virol 78: 6792–6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson KP, Hammarström P, Ahlgren F, Herland A, Schnell EA, Lindgren M, Westermark GT, Inganäs O. 2006. Conjugated polyelectrolytes—Conformation-sensitive optical probes for staining and characterization of amyloid deposits. Chembiochem 7: 1096–1104. [DOI] [PubMed] [Google Scholar]
- Nilsson KP, Joshi-Barr S, Winson O, Sigurdson CJ. 2010. Prion strain interactions are highly selective. J Neurosci 30: 12094–12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notari S, Capellari S, Giese A, Westner I, Baruzzi A, Ghetti B, Gambetti P, Kretzschmar HA, Parchi P. 2004. Effects of different experimental conditions on the PrPSc core generated by protease digestion: Implications for strain typing and molecular classification of CJD. J Biol Chem 279: 16797–16804. [DOI] [PubMed] [Google Scholar]
- Notari S, Capellari S, Langeveld J, Giese A, Strammiello R, Gambetti P, Kretzschmar HA, Parchi P. 2007. A refined method for molecular typing reveals that co-occurrence of PrPSc types in Creutzfeldt–Jakob disease is not the rule. Lab Invest 87: 1103–1112. [DOI] [PubMed] [Google Scholar]
- Pan K, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick R, Cohen F, et al. 1993. Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci 90: 10962–10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchi P, Castellani R, Capellari S, Ghetti B, Young K, Chen S, Farlow M, Dickson D, Sima A, Trojanowski J, et al. 1996. Molecular basis of phenotypic variability in sporadic Creutzfeldt–Jakob disease. Ann Neurol 39: 767–778. [DOI] [PubMed] [Google Scholar]
- Parchi P, Giese A, Capellari S, Brown P, Schulz-Schaeffer W, Windl O, Zerr I, Budka H, Kopp N, Piccardo P, et al. 1999. Classification of sporadic Creutzfeldt–Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann Neurol 46: 224–233. [PubMed] [Google Scholar]
- Parchi P, Zou W, Wang W, Brown P, Capellari S, Ghetti B, Kopp N, Schulz-Schaeffer WJ, Kretzschmar HA, Head MW, et al. 2000. Genetic influence on the structural variations of the abnormal prion protein. Proc Natl Acad Sci 97: 10168–10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattison I. 1966. The relative susceptibility of sheep, goats and mice to two types of the goat scrapie agent. Res Vet Sci 7: 207–212. [PubMed] [Google Scholar]
- Pattison IH, Millson GC. 1961. Scrapie produced experimentally in goats with special reference to the clinical syndrome. J Comp Pathol 71: 101–109. [DOI] [PubMed] [Google Scholar]
- Peretz D, Scott MR, Groth D, Williamson RA, Burton DR, Cohen FE, Prusiner SB. 2001. Strain-specified relative conformational stability of the scrapie prion protein. Protein Sci 10: 854–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piro JR, Harris BT, Supattapone S. 2011. In situ photodegradation of incorporated polyanion does not alter prion infectivity. PLoS Pathog 7: e1002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Stoeck K, Glatzel M, Vey M, Bellon A, Aguzzi A. 2005. Coexistence of multiple PrPSc types in individuals with Creutzfeldt–Jakob disease. Lancet Neurol 4: 805–814. [DOI] [PubMed] [Google Scholar]
- Priola S, Chesebro B. 1995. A single hamster PrP amino acid blocks conversion to protease-resistant PrP in scrapie-infected mouse neuroblastoma cells. J Virol 69: 7754–7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner S. 1982. Novel proteinaceous infectious particles cause scrapie. Science 216: 136–144. [DOI] [PubMed] [Google Scholar]
- Prusiner S, Cochran S, Downey D, Groth D. 1981. Determination of scrapie agent titer from incubation period measurements in hamsters. Adv Exp Med Biol 134: 385–399. [DOI] [PubMed] [Google Scholar]
- Prusiner S, Cochran S, Groth D, Downey D, Bowman K, Martinez H. 1982. Measurement of the scrapie agent using an incubation time interval assay. Ann Neurol 11: 353–358. [DOI] [PubMed] [Google Scholar]
- Puoti G, Giaccone G, Rossi G, Canciani B, Bugiani O, Tagliavini F. 1999. Sporadic Creutzfeldt–Jakob disease: Co-occurrence of different types of PrPSc in the same brain. Neurology 53: 2173–2176. [DOI] [PubMed] [Google Scholar]
- Race R, Oldstone M, Chesebro B. 2000. Entry versus blockade of brain infection following oral or intraperitoneal scrapie administration: Role of prion protein expression in peripheral nerves and spleen. J Virol 74: 828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Race R, Raines A, Raymond G, Caughey B, Chesebro B. 2001. Long-term subclinical carrier state precedes scrapie replication and adaptation in a resistant species: Analogies to bovine spongiform encephalopathy and variant Creutzfeldt–Jakob disease in humans. J Virol 75: 10106–10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saá P, Sferrazza GF, Ottenberg G, Oelschlegel AM, Dorsey K, Lasmézas CI. 2012. Strain-specific role of RNAs in prion replication. J Virol 86: 10494–10504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen FE, Prusiner SB. 1998. Eight prion strains have PrPSc molecules with different conformations. Nat Med 4: 1157–1165. [DOI] [PubMed] [Google Scholar]
- Saunders SE, Bartelt-Hunt SL, Bartz JC. 2012. Occurrence, transmission, and zoonotic potential of chronic wasting disease. Emerg Infect Dis 18: 369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch G, Seeger H, Bogousslavsky J, Tolnay M, Janzer RC, Aguzzi A, Glatzel M. 2006. Analysis of prion strains by PrPSc profiling in sporadic Creutzfeldt–Jakob disease. PLoS Med 3: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutt CR, Bartz JC. 2008. Prion interference with multiple prion isolates. Prion 2: 61–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J, Fraser H. 1989. Transport and targeting of scrapie infectivity and pathology in the optic nerve projections following intraocular infection. Prog Clin Biol Res 317: 645–652. [PubMed] [Google Scholar]
- Scott M, Foster D, Mirenda C, Serban D, Coufal F, Walchli M, Torchia M, Groth D, Carlson G, DeArmond S, et al. 1989. Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell 59: 847–857. [DOI] [PubMed] [Google Scholar]
- Scott M, Will R, Ironside J, Nguyen H, Tremblay P, DeArmond S, Prusiner S. 1999. Compelling transgenetic evidence for transmission of bovine spongiform encephalopathy prions to humans. Proc Natl Acad Sci 96: 15137–15142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikiya RA, Ayers JI, Schutt CR, Kincaid AE, Bartz JC. 2010. Coinfecting prion strains compete for a limiting cellular resource. J Virol 84: 5706–5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikiya RA, Eckland TE, Young AJ, Bartz JC. 2014. Prion formation, but not clearance, is supported by protein misfolding cyclic amplification. Prion 8: 415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdson CJ, Nilsson KP, Hornemann S, Manco G, Polymenidou M, Schwarz P, Leclerc M, Hammarström P, Wüthrich K, Aguzzi A. 2007. Prion strain discrimination using luminescent conjugated polymers. Nat Methods 4: 1023–1030. [DOI] [PubMed] [Google Scholar]
- Silveira J, Raymond G, Hughson A, Race R, Sim V, Hayes S, Caughey B. 2005. The most infectious prion protein particles. Nature 437: 257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim VL, Caughey B. 2009. Ultrastructures and strain comparison of under-glycosylated scrapie prion fibrils. Neurobiol Aging 30: 2031–2042. [DOI] [PubMed] [Google Scholar]
- Sisó S, Jeffrey M, Martin S, Chianini F, Dagleish MP, González L. 2010. Characterization of strains of ovine transmissible spongiform encephalopathy with a short PrPd profiling method. J Comp Pathol 142: 300–310. [DOI] [PubMed] [Google Scholar]
- Supattapone S. 2014. Synthesis of high titer infectious prions with cofactor molecules. J Biol Chem 289: 19850–19854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. 2004. Conformational variations in an infectious protein determine prion strain differences. Nature 428: 323–328. [DOI] [PubMed] [Google Scholar]
- Tateishi J, Brown P, Kitamoto T, Hoque Z, Roos R, Wollman R, Cervenakova L, Gajdusek D. 1995. First experimental transmission of fatal familial insomnia. Nature 376: 434–435. [DOI] [PubMed] [Google Scholar]
- Taylor D, Dickinson A, Fraser H, Marsh R. 1986. Evidence that transmissible mink encephalopathy agent is biologically inactive in mice. Neuropathol Appl Neurobiol 12: 207–215. [DOI] [PubMed] [Google Scholar]
- Telling GC, Parchi P, DeArmond S, Cortelli P, Montagna P, Gabizon R, Mastrianni J, Lugaresi E, Gambetti P, Prusiner S. 1996. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science 274: 2079–2082. [DOI] [PubMed] [Google Scholar]
- Thackray AM, Hopkins L, Klein MA, Bujdoso R. 2007. Mouse-adapted ovine scrapie prion strains are characterized by different conformers of PrPSc. J Virol 81: 12119–12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thackray AM, Hopkins L, Lockey R, Spiropoulos J, Bujdoso R. 2011. Emergence of multiple prion strains from single isolates of ovine scrapie. J Gen Virol 92: 1482–1491. [DOI] [PubMed] [Google Scholar]
- Thackray AM, Lockey R, Beck KE, Spiropoulos J, Bujdoso R. 2012. Evidence for co-infection of ovine prion strains in classical scrapie isolates. J Comp Pathol 147: 316–329. [DOI] [PubMed] [Google Scholar]
- Tixador P, Herzog L, Reine F, Jaumain E, Chapuis J, Le Dur A, Laude H, Beringue V. 2010. The physical relationship between infectivity and prion protein aggregates is strain-dependent. PLoS Pathog 6: e1000859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Keulen L, Schreuder B, Meloen R, Poelen-van den Berg M, Mooij-Harkes G, Vromans M, Langeveld J. 1995. Immunohistochemical detection and localization of prion protein in brain tissue of sheep with natural scrapie. Vet Pathol 32: 299–308. [DOI] [PubMed] [Google Scholar]
- van Keulen L, Schreuder B, Vromans M, Langeveld J, Smits M. 2000. Pathogenesis of natural scrapie in sheep. Arch Virol Suppl 16: 57–71. [DOI] [PubMed] [Google Scholar]
- Wang F, Wang X, Yuan CG, Ma J. 2010. Generating a prion with bacterially expressed recombinant prion protein. Science 327: 1132–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells G. 1987. A novel progressive spongiform enchephalopathy in cattle. Vet Rec 121: 419–420. [DOI] [PubMed] [Google Scholar]
- Westaway D, Goodman P, Mirenda C, McKinley M, Carlson G, Prusiner S. 1987. Distinct prion proteins in short and long scrapie incubation period mice. Cell 51: 651–662. [DOI] [PubMed] [Google Scholar]
- Williams E, Young S. 1980. Chronic wasting disease of captive mule deer: A spongiform encephalopathy. J Wildl Dis 16: 89–98. [DOI] [PubMed] [Google Scholar]
- Williams E, Young S. 1992. Spongiform encephalopathies in Cervidae. Rev Sci Tech 11: 551–567. [DOI] [PubMed] [Google Scholar]
- Zlotnik I, Stamp JT. 1961. Scrapie disease of sheep. World Neurol 2: 895–907. [PubMed] [Google Scholar]