Abstract
The centrosome acts as a microtubule-organizing center (MTOC) from the G1 to G2 phases of the cell cycle; it can mature into a spindle pole during mitosis and/or transition into a cilium by elongating microtubules (MTs) from the basal body on cell differentiation or cell cycle arrest. New studies hint that the centrosome functions in more than MT organization. For instance, it has recently been shown that a specific substructure of the centrosome—the mother centriole appendages—are required for the recycling of endosomes back to the plasma membrane. This alone could have important implications for a renaissance in our understanding of the development of primary cilia, endosome recycling, and the immune response. Here, we review newly identified roles for the centrosome in directing membrane traffic, the immunological synapse, and the stress response.
The centrosome is involved in more than just microtubule organization. Newly identified roles for the centrosome include directing membrane traffic, forming immunological synapses, and sensing stress.
1. INTRODUCTION: CENTROSOMES—SMALL BUT NOT SIMPLE
Theodore Boveri published his seminal work in 1888, describing the origin of the centrosome from the sperm centriole after fertilization (Scheer 2014). This initial centriole will become the mother centriole and will duplicate to form a daughter centriole. Another round of duplication is required to make two mitotic spindle poles, each containing two centrioles. These two duplication cycles are required to form the first mitotic spindle, thus initiating the process of development and growth of an embryo.
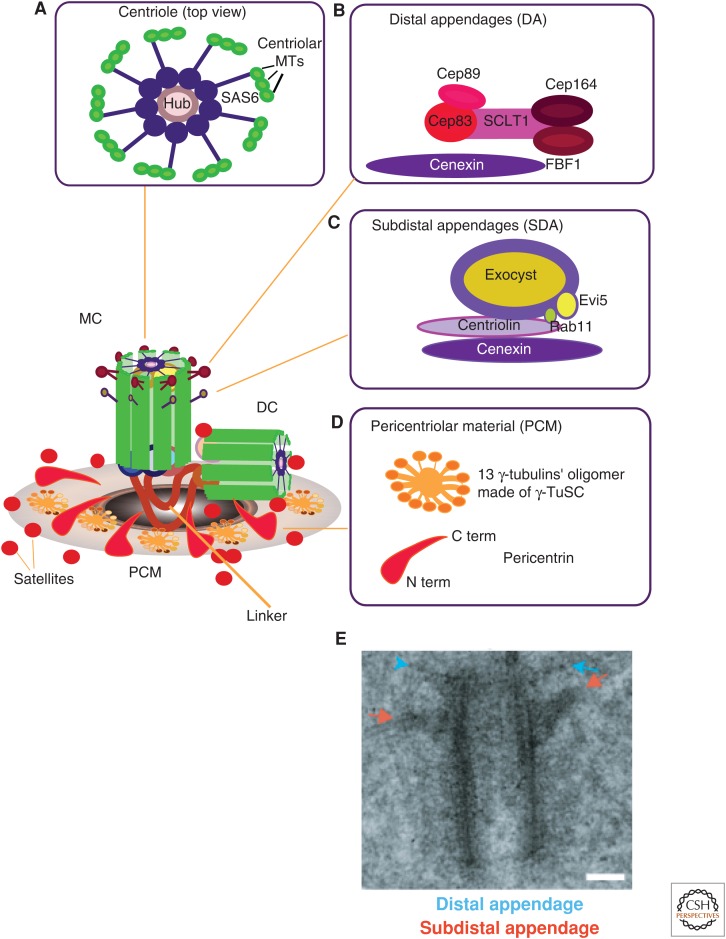
The centrosome has been visualized by light microscopy since the 1880s and, subsequently, by transmission electron microscopy (TEM), which revealed the two barrel-shaped centrioles surrounded by pericentriolar material (PCM). The centriole itself contains substructures such as a cartwheel-like structure and two sets of appendages at the distal end of the oldest centriole (a.k.a. “mother centriole”; Fig. 1). The cartwheel-like structure serves as a platform to assemble microtubule (MT) triplets arranged with ninefold symmetry. It is argued that the assembly of this structure de novo involves a complex of proteins (Kitagawa et al. 2011), which are regulated by a polo-like kinase—the serine/threonine-protein kinase PLK4 (centriole assembly has been reviewed elsewhere; Brito et al. 2012). However, the role of appendages in centrosome function remains more enigmatic. We know that the oldest centriole (the mother) is structurally distinct from the younger centriole (the “daughter”) in that it contains distal appendages and subdistal appendages (Fig. 1). The proteins that comprise these appendages are enriched specifically at the mother centriole, whereas the daughter centriole possesses its own specific molecular components (e.g., centrobin—a protein required to ensure the length of the newly formed centrioles; Zou et al. 2005; Gudi et al. 2015). The current proposed role for distal appendages is in docking the centrosome to the plasma membrane during the formation of cilia (Tanos et al. 2013), whereas the subdistal appendages are proposed to act in MT anchoring in interphase cells (Delgehyr et al. 2005). More recent evidence indicates that these appendage proteins are also important for ensuring symmetric division and membrane trafficking (see below).
Figure 1.
Centrosome substructures. A model highlighting the centrosome and its specific substructures, namely, (A) the centrioles, (B) distal appendages, (C) subdistal appendages, and (D) pericentriolar material (PCM). The centrosome comprises the mother centriole (MC), daughter centriole (DC), tethered together by “linker” structures, PCM, and satellites. The “linker” ensures centriole engagement and timely linker degradation that licenses centriole duplication during S phase. The protein composition of the “linker” remains elusive, but the involvement of pericentrin, Cep215, and Cep68 in centriole connections has recently been shown (Pagan et al. 2015). (A) Illustration showing the centriole barrel (in top view). Depicted is a central hub, a rod-shaped structure of SAS6 homodimers that form oligomers (Kitagawa et al. 2011; Guichard et al. 2013), from which nine spokes of SAS6 homodimers emanate, which each radiate toward a microtubule (MT) triplet. (B) Relative organization of molecular players forming the distal appendages (DAs), with Cep83 being in closest proximity to the centriole. (C) A model in which it is proposed that cenexin/Odf2 is responsible for the integrity of the subdistal appendages (SDAs) and for connecting them to the MC. (D) Organization of the PCM into 13 protofilament oligomers that contain γ-tubulin in the outermost layer; the carboxyl terminus (C term) of pericentrin is located close to the centriole barrel, whereas the amino terminus (A term) is oriented toward the outer lattice layer. (E) Electron micrograph of a mother centriole with two sets of appendages; DAs are highlighted by blue arrows and SDAs are highlighted by red arrows. Scale bar, 0.1 µm. (E, Reproduced from Hung et al. 2016, with permission from Elsevier.)
Another important structure of the centrosome is the PCM that surrounds the two centrioles. It was originally speculated that the PCM was a disorganized meshwork of proteins (as reviewed elsewhere; Mennella et al. 2014). With the advent of superresolution microscopy and deconvolution, the PCM appears to have a lattice-like organization (Dictenberg et al. 1998) and ring-like arrangements (Lawo et al. 2012). Superresolution microscopy also revealed molecular components of the PCM (e.g., pericentrin, γ-tubulin), which were detected at a resolution of <200 nm. These studies allowed the precise modeling of different PCM components within the lattice (Fu and Glover 2012; Lawo et al. 2012; Mennella et al. 2012; Sonnen et al. 2012). The first striking observation was that pericentrin is organized with its carboxyl terminus closer to the centriole wall, and the amino terminus of the protein extends toward the outer layer of the lattice (Mennella et al. 2012). γ-tubulin is found in a more peripheral layer. Pericentrin displays a structure similar to “arms” reaching across and, potentially, securing the lattice network (Lawo et al. 2012), arguing for its importance in lattice stability (Mennella et al. 2012). This topology might explain why γ-tubulin is lost so readily in cells lacking pericentrin (Zimmerman et al. 2004; Chen et al. 2014).
2. THE CONSTANTLY EVOLVING ROLE OF THE CENTROSOME THROUGHOUT THE CELL CYCLE
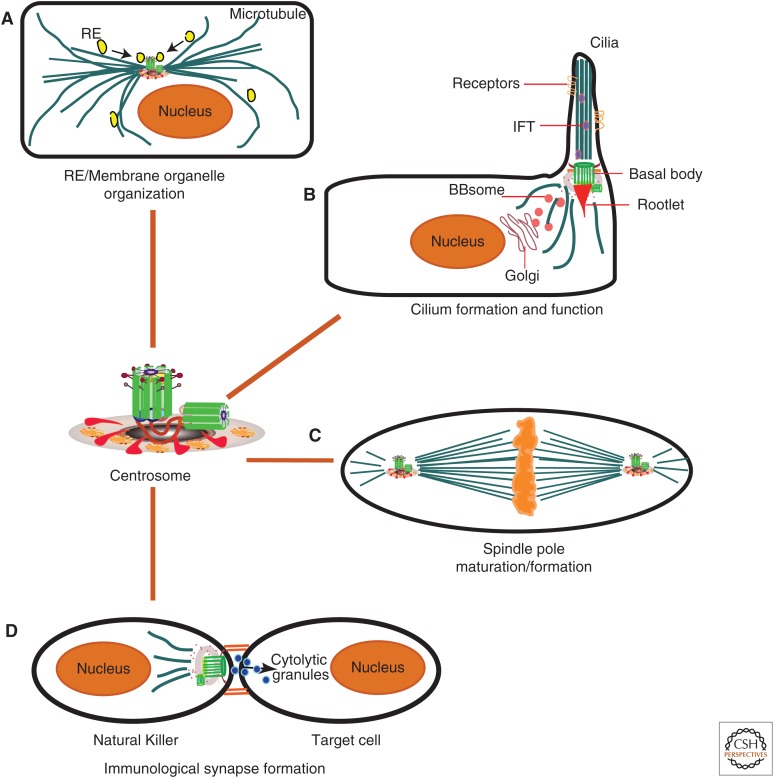
The dynamic, yet organized, centrosome provides a platform for multiple functions. As the cell enters mitosis, the ability of the centrosome to nucleate MTs increases, concomitant with recruitment of additional PCM and signaling components, thus transforming the once PCM-poor interphase centrosome into a mature PCM-rich spindle pole. Once division is completed and the cell commits to differentiate, the once-again PCM-poor centrosome can move toward the plasma membrane and function as a basal body for cilia formation (Fig. 2). Besides these classical functions of the centrosome, its recently appreciated roles in membrane trafficking and formation of immunological synapses (Fig. 2) direct a fast-burgeoning area of research, driven in part by the identification of more than 100 different centrosome proteins and an open-access centrosome proteome to peruse (Andersen et al. 2003; Jakobsen et al. 2011). However, the framework for how these proteins are organized, the dynamics of their localization to the centrosome throughout the cell cycle, and their organization have not been fully elucidated. With the ability to use live-cell imaging and superresolution microscopy, we envision that these questions will quickly be resolved.
Figure 2.
The various functions of the centrosome. The figure summarizes the main activities of the centrosome, including (A) organization of membrane organelles (e.g., recycling endosome); (B) cilium formation and function; (C) spindle pole maturation during mitosis; and (D) formation of an immunological synapse, in which a natural killer cell recognizes an infected cell. IFT, intraflagellar transport; RE, recycling endosome.
Proteins that make up the proteasome were one of many interesting centrosome candidates that have been identified (Badano et al. 2005; Wigley et al. 1999), but their specific subcentrosome localization is still unknown. In eukaryotes, proteasomes drive the selective degradation of protein substrates containing covalently linked ubiquitin chains. Although proteasomes are distributed throughout the cell, their localization at the centrosome argues for a specific biological function at this distinct subcellular site. For instance, studies linking the proteasome to the centrosome show a role for centrosome localization in neuronal function. Specifically, an E3-ubiquitin ligase linked to Parkinson’s disease is enriched at the centrosome (Zhao et al. 2003). These E3 ligases are enzymes that recognize targets for degradation by tagging them with ubiquitin. This work suggests that the centrosomal localization of this E3 ligase provides a subcellular site to specifically ubiquitinate and degrade protein aggregates that are crucially involved in the pathogenesis of Parkinson’s disease. A centrosomal localization of proteasome components was also proposed to be important for the development of dendrites. This was tested by developing a tool to inhibit proteasome function specifically at the centrosome (Puram et al. 2013). The proteasomal subunit S5a/Rpn10 was identified as an essential component for proteasomal activity specifically at the centrosome in neurons to promote dendrite arbor elaboration. A deeper understanding of the molecular relationship between the centrosome and the proteasome could elucidate potential therapeutic targets in Parkinson’s disease progression and, thus, expand our understanding of neuronal development.
3. MT-ORGANIZING CENTER AND SPINDLE POLES
The centrosome is most commonly known as a microtubule-organizing center (MTOC). MTs appear to be organized at the centrosome in three different ways. They can be nucleated at this site from/by γ-tubulin ring complexes (γ-TuRCs) located in the PCM (Fig. 1). γ-TuRCs consist of γ-tubulin small complexes (γ-TuSCs) and accessory proteins (Doxsey 2001). Unlike MT growth in vitro, in which variable numbers of protofilaments are formed, centrosome-nucleated MTs typically comprise 13 protofilaments (reviewed in Doxsey 2001). γ-TuRCs form oligomers that have a slight helical geometry similar to the helical turns of the MT itself and are consistent with a role for the γ-TuRCs templating the assembly of 13-fold microtubule protofilaments to form an MT (Kollman et al. 2010). MTs (apparently centrosome-nucleated) can also be anchored at subdistal appendages of the centrosome in interphase cells, but little is known about how these MTs arise and anchor, other than them requiring ninein for their anchoring (Delgehyr et al. 2005). MTs can also be elongated from the older centriole during ciliogenesis.
The centrosome function expands when the interphase centrosome matures into a mitotic spindle pole. Centrosome maturation occurs every cell cycle and involves recruitment of signaling molecules to initiate centrosome separation and maturation, followed by the addition of PCM components to increase MT nucleation. The role of centrioles in mitotic spindle assembly appears to be dispensable during Drosophila development (Basto et al. 2006)—in this case, the cells rely on acentriolar MTOCs. However, in chicken DT40 cells, centrioles are required for timely spindle assembly and chromosome stability (Sir et al. 2013). In developing mouse embryos, acentriolar mitosis causes early embryonic lethality, arguing for an essential role for centrioles in mammalian development (Bazzi and Anderson 2014). To initiate spindle maturation in either Drosophila or mammalian cells, two well-studied kinases—Aurora A and Plk1—must be recruited to the spindle poles (reviewed in Barr and Gergely 2007), where they play crucial roles in spindle assembly and mitotic progression. Interestingly, Aurora A modifies several important cell cycle–related proteins that include CPEB (cytoplasmic polyadenylation element binding) to regulate translation of cyclin-B–Cdk1 for mitotic entry (Mendez and Richter 2001; Sasayama et al. 2005); Eg5, LATS2, and NDEL1, which are required for centrosome separation and maturation (Toji et al. 2004); and TACC (transforming acid coiled coil) for astral MT stability (Conte et al. 2003; Barros et al. 2005). Therefore, it is not surprising that defects in Aurora A signaling ultimately delay mitotic entry, induce monopolar spindle formation, and cause misaligned chromosomes (Hochegger et al. 2013). Plk1 seems to be more specific and directly phosphorylates substrates required for centrosome maturation and spindle assembly proteins (reviewed by Lens et al. 2010).
4. BASAL BODY
When a cell exits the cell cycle, during differentiation or because of a lack of nutrients (Goto et al. 2013), the centrosome relinquishes its main role as an MTOC and relocates from its perinuclear site to the apical plasma membrane where one of the two centrioles, the mother centriole, serves as a basal body for ciliogenesis (Fig. 2) (reviewed by Kobayashi and Dynlacht 2011; Hehnly and Doxsey 2012). The basal body can revert back to an MTOC and reenter the cell cycle by reabsorbing the cilia and moving away from the apical membrane.
As the centrosome is at the base of the primary cilium and is required for anchoring the cilium at the plasma membrane, it is not surprising that many centrosome proteins were found to be crucial for ciliogenesis (Graser et al. 2007; Mikule et al. 2007; Nigg and Raff 2009). The mother centriole appendages comprise a handful of centrosome proteins that have been implicated in the formation of cilia (e.g., Cep164, ODF2/Cenexin, Cep83, and centriolin; Gromley et al. 2003; Ishikawa et al. 2005; Graser et al. 2007; Chang et al. 2013; Tanos et al. 2013). The distal appendage molecular components, Cep164 and Cep83, are thought to be important for docking of the mother centriole at the plasma membrane during ciliogenesis. The role of the subdistal appendages is less defined. Interestingly, ODF2/cenexin localizes to both subdistal and distal appendages and contributes to the formation of both (Ishikawa et al. 2005; Chang et al. 2013). One interesting role for subdistal appendages is that they might act as a site to assemble a ciliary vesicle (Sorokin 1962), which is a membranous capsule proposed to be required for fusion of the cilium with the plasma membrane (Hehnly et al. 2013). Another recent study showed that ciliary components, including membrane vesicles, remain associated with the mother centriole during mitosis (Paridaen et al. 2013). Furthermore, it seems that the daughter cell that inherits the oldest centriole will be the first to form a primary cilium (Paridaen et al. 2013).
Besides the formation of the ciliary vesicle, the centrosome undergoes additional changes during ciliogenesis that include the establishment of a ciliary rootlet (Yang et al. 2002). During assembly of cilia, the centrosome (basal body) facilitates the formation of the ciliary rootlet. Rootletin—a structural component of the ciliary rootlet (Yang et al. 2002)—is also a component of the centrosome during G1–S cell cycle stages and is required for centrosome cohesion (Bahe et al. 2005). Interestingly, a lack of rootletin does not affect initial cilia development, but is implicated in long-term cilia stability in photoreceptors and, when lost, leads to retinal degeneration (Yang et al. 2005).
PCM components of the centrosome, such as pericentrin, are also implicated in ciliogenesis (for further details, see Delaval and Doxsey 2010). One possible explanation for the perturbation of ciliogenesis that accompanies the loss of pericentrin is that pericentrin interacts with components of the intraflagellar transport (IFT) system, IFT20 and IFT88 (Jurczyk et al. 2004). IFT is a cargo-trafficking pathway that functions inside cilia and contributes to their assembly and the delivery of signaling proteins (e.g., sonic hedgehog; reviewed by Sung and Leroux 2013). The IFT system efficiently delivers cargo to the cilium in a timely manner, which is important as the cilium itself lacks translational machinery. The cilium consists of two protein subcomplexes, IFT-A and IFT-B, which use the motor proteins kinesin 2 and dynein to move bidirectionally along MTs in the cilium.
An unexpected recent finding showed that formation of the cilium involves autophagy-regulated degradation of centrosome satellite proteins. Centriolar satellites (Fig. 1) are cytoplasmic granules that have been proposed to function in centrosome protein targeting and exchange, as well as communication between centrosomes and the cytoplasm. More specifically, the centriolar satellite protein oral-facial-digital syndrome 1 protein (OFD1) is specifically degraded by autophagy, and this degradation is important for ciliogenesis (Tang et al. 2013). The interaction of OFD1 with the autophagic protein LC3 is enhanced by another satellite protein, PCM1. It has also been shown that there is a mutual interdependence between ciliogenesis and autophagy, favoring a model in which autophagy influences ciliogenesis (Pampliega et al. 2013). Other support for this comes from the observation that cigarette smoke causes the loss or shortening of cilia, and this is correlated with an overall increase in autophagy (Lam et al. 2013). Inhibition of autophagy prevents cilia shortening in response to cigarette smoke, thus supporting the notion regarding an inhibitory effect of autophagy on ciliogenesis.
Most cell types contain primary cilia, implicating their importance in cellular function. One particularly interesting observation is the presence of signaling receptors at the primary cilium, suggesting a role as a specialized signaling “antenna.” Proteome analyses of nonmotile cilia have identified signaling components such as sonic hedgehog, smoothened and Wnt (Liu et al. 2007; Mayer et al. 2009; Ishikawa et al. 2012). These cilia-localized signaling cascades have been modeled to influence neuronal migration and cerebral cortical development (reviewed by Guemez-Gamboa et al. 2014). One proposed mechanism for how cilia might regulate neuronal migration is that they could act as an antenna sensing hormonal changes in the extracellular environment. Strikingly, a recent study revealed a branched cilium in neurons of the nematode Caenorhabditis elegans, suggesting that there could be some diversity in the range of morphology of primary cilia (Doroquez et al. 2014). Another example of the diversity of cilia comes from comparative proteomic studies (Liu et al. 2007). When compared with proteome analysis between photoreceptor sensory cilia and other cilia proteomes, a higher number of transport and light-perception components, such as photoreceptors and photoadaptor proteins, were detected in photoreceptor cilia. This specialized organization of distinct signaling components suggests that the cilium is an organelle that is “tunable” to specific cell types.
5. LOSS OF CENTROSOME OR CILIA FUNCTION LEADS TO CILIOPATHIES
Dysfunction of primary (sensory) cilia is associated with several human disorders, collectively termed ciliopathies. The primary cilium was thought to be the reason behind the syndromic phenotypes associated with their disruption, including obesity, cystic kidneys, polydactyly, situs inversus, and encephalocele, to name just a few. One recent example linking primary cilia with obesity is a study on the obesity-linked hormone leptin. Leptin is produced by adipocytes and was shown to stimulate elongation of cilia in the hypothalamus neurons in adulthood (Han et al. 2014). These findings suggested that leptin governs cilia length, possibly to increase the sensitivity of hypothalamic neurons to metabolic signals.
One specific ciliopathy, Bardet–Biedl syndrome (BBS), has at least nine associated “ciliopathy” phenotypes that include retinopathy, polydactyly, situs inversus, and developmental delays such as a hypoplastic (underdeveloped) cerebellum and obesity. The multiple phenotypes of BBS make it difficult to determine whether loss of cilia is the sole defect in this disorder. BBS can result from mutations in at least 14 different genes, some of which are involved in a protein complex called the BBSome that contributes to cilia structure, formation, and function. However, some BBS proteins have been implicated in cellular structures outside the cilium (reviewed by Vertii et al. 2015a). For instance, BBS4 and BBS6 both localize to the centrosome/basal body during interphase, when a cilium is emanating from the basal body, and during mitotic progression, when the cilium is not present (Kim et al. 2004, 2005). Based on these known BBS protein localizations to the centrosome, we propose that defects in centrosome function, leading to mitotic or cilia defects, might cocontribute to the etiology of ciliopathies.
6. THE ENDOCYTIC RECYCLING MACHINERY INTERACTS DIRECTLY WITH THE CENTROSOME AND PLAYS A ROLE IN CILIA FORMATION AND THE CELL CYCLE
Intercellular trafficking is dependent on centrosome structure (Fig. 2). One example of this is the recycling endosome and its interaction with mother centriole appendages (Hehnly et al. 2012). The appendages are required for the assembly of the ciliary vesicle, which forms at the mother centriole before the centriole docks to the plasma membrane from which the primary cilium emerges (Sorokin 1962). The recycling endosome contains two GTPases—Rab11 and Rab8—that regulate cargo recycling in and out of this organelle. Recently, these GTPases were shown to interact directly with a mother centriole appendage protein, cenexin (Hehnly et al. 2012, 2013; Chang et al. 2013). In addition, a Rab11–Rab8 GTPase cascade was shown to be required for formation of cilia, in which active Rab11 recruits the Rab8 GDP–GTP exchange factor (GEF) Rabin8 to activate Rab8 and, thus, induce cilia formation (Knödler et al. 2010; Westlake et al. 2011). Rab8 and Rab11 also interact with several well-defined proteins required for ciliogenesis such as the BBSome, the vesicle tethering complex known as the exocyst, and the mother centriole appendage protein cenexin (Fielding et al. 2005; Wu et al. 2005; Nachury et al. 2007; Hehnly et al. 2012, 2013; Chang et al. 2013). However, the molecular mechanism underlying the interplay between these molecular components during ciliogenesis is unknown. Early steps in ciliogenesis require the Rab8–Rab11 cascade and the membrane-shaping proteins EHD1 and EHD3 (Lu et al. 2015). We speculate that the interaction between the recycling endosome and its machinery with mother centriole appendages facilitates the organization of the Rab11–Rab8 GTPase cascade, ensuring the initiation of the formation of cilia at the appropriate time during the cell cycle.
Strikingly, a recent study implicated Rab11 and its role in dynein-based endosome motility in maturation of spindle poles and mitotic progression. Time-lapse imaging showed Rab11-associated endosomes organized around interphase centrosomes, mitotic spindles, and mitotic spindle poles (Hehnly and Doxsey 2014). The Rab11-decorated endosomes contained γ-tubulin and dynein, suggesting that endosomes might act as carriers for localizing MT-nucleating and spindle pole proteins to the mitotic spindle poles. Taken together, the results show that Rab11-associated endosomes are important in centrosome maturation (Hehnly and Doxsey 2014) and ciliogenesis (Knödler et al. 2010; Westlake et al. 2011), thus showing their role in centrosome function.
7. THE CENTROSOME AS A STRESS SENSOR
Signaling by centrosomes is crucial for cell cycle progression (Doxsey et al. 2005). Cyclin-dependent kinases (CDKs) are well-known regulators of cell cycle progression, and so it is not surprising that two cyclins—cyclin E and cyclin A—physically interact with and activate CDKs, including Cdk2 and Cdk1, and activate these kinases. Interestingly, cyclins E and A both display centrosome localization and are required not only for centriole duplication but also for DNA replication. Each of these cyclins, cyclin E and A, contain their own unique centrosome-localization signals, which are thought to spatially activate CDKs (Matsumoto and Maller 2004; Pascreau et al. 2010). Cyclin B also localizes to the centrosome. However, in this case, the sequestration at the centrosome serves to prevent cyclin B from conducting its nuclear function (Krämer et al. 2004) and does not directly affect the centrosome.
On induction of DNA damage, the pathway involving the proteins ATM, ATR, and Chk1 regulates the localization of cyclin B at centrosomes. Normal entry into mitosis occurs after activation of Cdk1, resulting in chromosome condensation in the nucleus and centrosome separation, as well as increased MT-nucleation activity in the cytoplasm. The centrosome-associated serine/threonine protein kinase Chk1 is proposed to colocalize with, and thus shield, centrosome-localized Cdk1 from unscheduled activation by the cytoplasmic tyrosine-protein phosphatase Cdc25B, thereby contributing to the accurate timing of the initial steps of cell division, including mitotic spindle formation (Krämer et al. 2004). However, on DNA damage, a Chk1-mediated checkpoint induces excessive formation of centriolar satellites that constitute assembly platforms for centrosomal proteins, which subsequently leads to centrosome amplification (Löffler et al. 2013). This proposed centrosome-inactivation checkpoint comprising centrosome amplification might lead to the elimination of cells by mitotic catastrophe in response to DNA damage. Furthermore, following UV-induced DNA damage, Chk1 first accumulates, and then becomes phosphorylated, at the centrosome (Löffler et al. 2007). The specific phosphorylation of Chk1 is a prerequisite for its degradation by the proteasome (Zhang et al. 2005). One interesting possibility is that, on DNA damage, the nuclear fraction of Chk1 accumulates at the centrosome, where it is then spatially, and in a timely manner, modified for degradation. This would be one of several examples showing that signaling activities involving centrosomes and nuclear events are linked in the response to cellular stress. For example, it appears that the disruption of centrosome structure results in a G1 arrest through a p38–p53 stress pathway (Mikule et al. 2007). Similarly, increasing p38 or p53 expression modulates PLK4 activity, thereby regulating centriole duplication (Nakamura et al. 2013).
8. CAN THE CENTROSOME DISCRIMINATE BETWEEN DIFFERENT TYPES OF STRESS?
Heat stress, like DNA damage, can have a deleterious effect on centrosome function. For instance, cells either undergoing heat stress, such as fever, or incurring DNA damage have a higher propensity to form a primary cilium (Villumsen et al. 2013), suggesting that multiple cellular stresses can cause cells to exit the cell cycle and remain in G0. The centrosome seems to be sensitive to stress. When comparing the DNA damage response in Drosophila embryos (Sibon et al. 2000) with the heat-stress response of mammalian cells (Vidair et al. 1993), both undergo dramatic but somewhat opposite changes in centrosome structure. For example, although UV stress causes centriolar satellite amplification and centrosome overduplication, heat stress in some cases causes disassembly of the PCM (Vidair et al. 1993; Brown et al. 1996b; Löffler et al. 2013). There have been several discrepancies between studies on changes in centrosome function that might have resulted from differences in heat-stress intensity and/or duration (Vidair et al. 1993; Brown et al. 1996b; Villumsen et al. 2013). Based on these differences, it is important to define the physiological relevance of heat stress in vivo on centrosome structure/function and correlate these phenotypes to what has been reported in the literature. Analysis of leukocytes from patients with febrile condition revealed decreased centrosome integrity compared with that of healthy controls (Vertii et al. 2015b). These data are supported by experiments in vitro with fever-mimicking temperature regimens, indicating that the centrosome is a heat-stress-sensitive organelle. Moreover, this sensitivity is unique to the centrosome, as other nonmembranous organelles, the midbody, and kinetochore appear to be heat-stress-tolerant (Vertii et al. 2015b). The heat-stress-related defects in centrosome integrity resulted in functional consequences for the centrosome, such as its ability to nucleate MTs and polarize during formation of the immunological synapse (IS; see below).
9. CENTROSOMES IN THE IMMUNE RESPONSE
Recent studies have shown that centrosome reorientation occurs during formation of the IS. IS formation is a necessary step within the immune response that includes membrane rearrangement at the sites where a T cell contacts an antigen-presenting cell (APC), resulting in activation of the T cell (Kloc et al. 2013). Cytotoxic T lymphocyte (CTL) or natural killer (NK) cells form one type of IS that can eliminate infected or tumorigenic cells by releasing cytolytic granules. Another type of IS occurs when T helper cells recognize an antigen presented by the APC through its T-cell receptor (TCR). The IS then assembles into a highly organized structure that contains a peripheral and central supramolecular activation complex (pSMAC and cSMAC, respectively) (Dustin et al. 2010; Thauland and Parker 2010). Once formation of the IS is initiated, the centrosome reorients toward the cSMAC within the IS and proceeds to dock at the plasma membrane where it can control secretion of lytic granules (Fig. 2) (Stinchcombe et al. 2006). At the same time that the centrosome docks at the plasma membrane, secretory granules move in a dynein-dependent manner toward the centrosome (Mentlik et al. 2010). Centrosome reorientation and movement toward the IS depends on several molecular factors that include MTs; MT motors such as dynein and kinesin; membrane components such as diacylglycerol; and signaling components that include protein kinase C (PKC), casein kinase 1 delta, Lck, Fyn, and Zap-70 (Knox et al. 1993; Kuhné et al. 2003; Combs et al. 2006; Martín-Cófreces et al. 2006; Quann et al. 2009; Bertrand et al. 2010; Tsun et al. 2011; Zyss et al. 2011; Liu et al. 2013; Jenkins et al. 2014). The molecular interplay between all these components has yet to be deciphered. However, recent evidence illustrates that MTs are required for centrosomes to orient themselves at the IS, and that this process is biphasic with centrosome polarization toward, and docking at, the IS (Yi et al. 2013). This work suggests an interesting model whereby centrosome reorientation might influence polarity formation in the IS. Future studies are required to investigate whether reorientation of the centrosome sets up T-cell polarity or whether T-cell polarity initiates centrosome reorientation.
Recently, similarities have been discerned between IS formation and cilia formation. For instance, in both cases, the centrosome is required to reorient toward a polarized membrane, either the apical membrane in the context of cilia formation or the IS in T cells. Both these processes involve hedgehog signaling either to function (Rohatgi et al. 2007; Tasouri and Tucker 2011) or to dock the centrosome at the plasma membrane (Stinchcombe et al. 2006; de la Roche et al. 2013). The similarities between IS formation and cilia formation provide a rationale to test whether proteins involved in cilia formation are also required in IS formation (e.g., IFT proteins, mother centriole appendage proteins). Along these lines, a recent finding has implicated an IFT protein—IFT20—in assembly of the IS in T cells (Finetti et al. 2009). Specifically, IFT20 is implicated as a central regulator of TCR recycling to the IS, possibly through an interplay between IFT20 and the Rab GTPase network that controls endosomal recycling (Finetti et al. 2014). Recycling endosomes, specifically ones containing either the Rab8 or Rab11 GTPases, have also been implicated in formation of cilia (Knödler et al. 2010). Therefore, we propose that the centrosome might be the control center for organizing the endosomal pathway containing IFT20 (Hehnly et al. 2012; Finetti et al. 2014) to initiate either cilia formation or IS formation at the right time and place.
10. MAINTAINING CENTROSOME INTEGRITY REQUIRES CHAPERONE MACHINERY
A high percentage of centrosomal proteins are >150 kDa in size and contain large hydrophobic coiled-coil motifs that are important in centrosome assembly. However, these characteristics imply a risk of rapid aggregation under cellular stress, suggesting that the centrosome is a highly unstable macromolecular complex. Maintaining centrosome integrity is essential for cell cycle progression and cilia formation. One proposed mechanism for maintaining centrosome integrity, and thus function, is through chaperone assistance. Chaperones are important components that aid folding of newly synthesized proteins, prevent aggregation of misfolded proteins, and target misfolded proteins for proteasome-mediated degradation (Horwich 2014). In mammalian cells, there are multiple chaperones, including heat-shock protein (Hsp) families: Hsp100, Hsp90, Hsp70, the chaperonin complex TRiC/CCT, and small chaperones such as crystallins (Kampinga et al. 2009). Some of these, such as Hsp70 and Hsp90, use ATP to prevent protein aggregation and facilitate proper folding of target proteins, whereas others, such as the small chaperones Hsp27 and αB crystallin, are ATP-independent but still efficient in their chaperone function.
The molecular chaperonin system differs dramatically from other chaperones (Kim et al. 2013). Chaperonins have been implicated in facilitating molecular components that assist in centrosome function. The chaperonin TRiC/CCT is responsible for folding tubulin and actin intermediates, as well as the PLK1 kinase (Llorca et al. 2000; Liu et al. 2005; Muñoz et al. 2011), and localizes to the centrosome (Brown et al. 1996a). Interestingly, CCT chaperonins have been identified as molecular components contributing to the onset of BBS (see Sec. 5 above). Two protein complexes implicated in the etiology of BBS consist of an octameric complex, the BBSome, and a handful of chaperonin-TRiC/CCT-like proteins (BBS6, BBS10, BBS12) (Nachury et al. 2007; Loktev et al. 2008; Jin et al. 2010; Wei et al. 2012). Any defect in protein complex assembly can result in BBS (Katsanis 2004; Yang et al. 2008; Billingsley et al. 2010). Many of the components that assist in formation of the BBSome localize to the centrosome and include BBS6 and the CCT proteins CCT1, CCT4, CCT5, and CCT8 (Kim et al. 2005; Seo et al. 2010; Zhang et al. 2012, 2013). Whether chaperonins require centrosome localization to interact with BBSome components is unknown. We speculate that the centrosome, which already contains many chaperone components, acts as a checkpoint to investigate the integrity of the BBSome complex.
Like the chaperonin system, the Hsp70–Hsp90 chaperone machinery localizes to the centrosome (Wigley et al. 1999). This localization suggests that Hsp70–Hsp90 might be involved in maintaining centrosome structure. In fact, when centrosome structure is compromised (by depletion of 13 specific centrosome proteins; Mikule et al. 2007), the cells arrest in G1 phase. Strikingly, this same cell cycle arrest occurs when Hsp/Hsc70 is depleted (Powers et al. 2008). This finding suggests that the Hsp/Hsc70 depletion, like depletion of centrosome proteins, compromises centrosome structure and, thus, leads to cell cycle arrest. Such an intriguing correlation led to the hypothesis that chaperones have a bona fide role in centrosome structure/function, in addition to their role in being recruited to the centrosome under stress. For example, under heat stress, the PCM of the centrosome is disrupted (Vidair et al. 1993; Hut et al. 2005). This disruption occurs concomitant with Hsp70 recruitment to the centrosome, suggesting a role for this chaperone in centrosome protection. Furthermore, ectopic expression of Hsp70 protects the PCM when under heat stress (Brown et al. 1996a,b). However, as increased cellular levels of chaperones will protect multiple cellular organelles from stress-related defects, from these studies it is difficult to determine whether Hsp70 expression is directly protecting the centrosome or whether this effect is indirect. Specific targeting of Hsp70 to the centrosome rescues centrosomal defects during stress, most likely through its ability to nucleate MTs and to serve as a basal body for polarizing toward the IS (Vertii et al. 2015b).
11. CONCLUSION
The emerging roles of the centrosome in underpinning a myriad of cellular functions might be related to the possession by the centrosome of many unique structural components. For example, the centrosome nucleates most of its MTs from the PCM and anchors MTs at the subdistal appendages, which are also implicated in organizing recycling endosomes. Furthermore, the distal appendages are linked to a separate centrosome function that involves centriole docking to the plasma membrane for both cilia formation and the formation of the IS. Although these different centrosome substructures can be linked to different cellular processes, we still do not know whether there is cross talk between the substructures themselves. For instance, could the subdistal appendages regulate PCM function? Support for this idea comes from the observation that several subdistal appendage proteins become more PCM-like (e.g., ninein) during maturation of the spindle pole, suggesting that these molecular players “toggle” between their roles at the appendages and their roles at the PCM. However, the role of appendage proteins at the PCM is unclear, although we do know that the appendage protein ninein is required for symmetric cell division (Wang et al. 2009; Dauber et al. 2012). We propose that chaperone components guard these different centrosome structures and assist in the assembly and disassembly of the many substructures of centrosomes.
Centrosome multitasking involves dynamic molecular rearrangements and, sometimes, movement of the entire centrosome during cell migration or ciliogenesis. These changes are probably due to MT-based transport to and from the centrosome. However, an alternative method to reorganize centrosome structure could involve local degradation of protein pools through centrosome-localized proteasome activity. One can imagine that maintaining the centrosome requires dynamic transport of material to and from the centrosome, the initiation of newly synthesized material at the centrosome, and the degradation of material no longer needed. If any of these pathways are improperly regulated, the fate of the cell will ultimately be decided by the centrosome. Therefore, it will be important to gain a greater understanding of the molecular pathways that feed into the centrosome and how the centrosome ultimately influences them.
Footnotes
Editors: Thomas D. Pollard and Robert D. Goldman
Additional Perspectives on The Cytoskeleton available at www.cshperspectives.org
REFERENCES
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. 2003. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426: 570–574. [DOI] [PubMed] [Google Scholar]
- Badano JL, Teslovich TM, Katsanis N. 2005. The centrosome in human genetic disease. Nat Rev Genet 6: 194–205. [DOI] [PubMed] [Google Scholar]
- Bahe S, Stierhof Y-D, Wilkinson CJ, Leiss F, Nigg EA. 2005. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J Cell Biol 171: 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr AR, Gergely F. 2007. Aurora-A: The maker and breaker of spindle poles. J Cell Sci 120: 2987–2996. [DOI] [PubMed] [Google Scholar]
- Barros TP, Kinoshita K, Hyman AA, Raff JW. 2005. Aurora A activates D-TACC-Msps complexes exclusively at centrosomes to stabilize centrosomal microtubules. J Cell Biol 170: 1039–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. 2006. Flies without centrioles. Cell 125: 1375–1386. [DOI] [PubMed] [Google Scholar]
- Bazzi H, Anderson KV. 2014. Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proc Natl Acad Sci 111: E1491–E1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand F, Esquerré M, Petit A-E, Rodrigues M, Duchez S, Delon J, Valitutti S. 2010. Activation of the ancestral polarity regulator protein kinase Cζ at the immunological synapse drives polarization of Th cell secretory machinery toward APCs. J Immunol 185: 2887–2894. [DOI] [PubMed] [Google Scholar]
- Billingsley G, Bin J, Fieggen KJ, Duncan JL, Gerth C, Ogata K, Wodak SS, Traboulsi EI, Fishman GA, Paterson A, et al. 2010. Mutations in chaperonin-like BBS genes are a major contributor to disease development in a multiethnic Bardet–Biedl syndrome patient population. J Med Genet 47: 453–463. [DOI] [PubMed] [Google Scholar]
- Brito DA, Gouveia SM, Bettencourt-Dias M. 2012. Deconstructing the centriole: Structure and number control. Curr Opin Cell Biol 24: 4–13. [DOI] [PubMed] [Google Scholar]
- Brown CR, Hong-Brown LQ, Doxsey SJ, Welch WJ. 1996a. Molecular chaperones and the centrosome. A role for HSP 73 in centrosomal repair following heat shock treatment. J Biol Chem 271: 833–840. [DOI] [PubMed] [Google Scholar]
- Brown CR, Doxsey SJ, Hong-Brown LQ, Martin RL, Welch WJ. 1996b. Molecular chaperones and the centrosome. A role for TCP-1 in microtubule nucleation. J Biol Chem 271: 824–832. [DOI] [PubMed] [Google Scholar]
- Chang J, Seo SG, Lee KH, Nagashima K, Bang JK, Kim BY, Erikson RL, Lee K-W, Lee HJ, Park J-E, et al. 2013. Essential role of Cenexin1, but not Odf2, in ciliogenesis. Cell Cycle 12: 655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CT, Hehnly H, Yu Q, Farkas D, Zheng G, Redick S, Hung H, Samtani R, Jurczyk A, Akbarian S, et al. 2014. A unique set of centrosome proteins requires pericentrin for spindle-pole localization and spindle orientation. Curr Biol 24: 2327–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs J, Kim SJ, Tan S, Ligon LA, Holzbaur ELF, Kuhn J, Poenie M. 2006. Recruitment of dynein to the Jurkat immunological synapse. Proc. Natl Acad Sci 103: 14883–14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte N, Delaval B, Ginestier C, Ferrand A, Isnardon D, Larroque C, Prigent C, Séraphin B, Jacquemier J, Birnbaum D. 2003. TACC1-chTOG-Aurora A protein complex in breast cancer. Oncogene 22: 8102–8116. [DOI] [PubMed] [Google Scholar]
- Dauber A, Lafranchi SH, Maliga Z, Lui JC, Moon JE, McDeed C, Henke K, Zonana J, Kingman GA, Pers TH, et al. 2012. Novel microcephalic primordial dwarfism disorder associated with variants in the centrosomal protein ninein. J Clin Endocrinol Metab 97: E2140–E2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Roche M, Ritter AT, Angus KL, Dinsmore C, Earnshaw CH, Reiter JF, Griffiths GM. 2013. Hedgehog signaling controls T cell killing at the immunological synapse. Science 80: 1247–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaval B, Doxsey SJ. 2010. Pericentrin in cellular function and disease. J Cell Biol 188: 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgehyr N, Sillibourne J, Bornens M. 2005. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J Cell Sci 118: 1565–1575. [DOI] [PubMed] [Google Scholar]
- Dictenberg JB, Zimmerman W, Sparks CA, Young A, Vidair C, Zheng Y, Carrington W, Fay FS, Doxsey SJ. 1998. Pericentrin and γ-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J Cell Biol 141: 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroquez DB, Berciu C, Anderson JR, Sengupta P, Nicastro D. 2014. A high-resolution morphological and ultrastructural map of anterior sensory cilia and glia in Caenorhabditis elegans. Elife 3: e01948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey S. 2001. Re-evaluating centrosome function. Nat Rev Mol Cell Biol 2: 688–698. [DOI] [PubMed] [Google Scholar]
- Doxsey S, McCollum D, Theurkauf W. 2005. Centrosomes in cellular regulation. Annu Rev Cell Dev Biol 21: 411–434. [DOI] [PubMed] [Google Scholar]
- Dustin ML, Chakraborty AK, Shaw AS. 2010. Understanding the structure and function of the immunological synapse. Cold Spring Harb Perspect Biol 2: a002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding AB, Schonteich E, Matheson J, Wilson G, Yu X, Hickson GRX, Srivastava S, Baldwin SA, Prekeris R, Gould GW. 2005. Rab11-FIP3 and FIP4 interact with Arf6 and the exocyst to control membrane traffic in cytokinesis. EMBO J 24: 3389–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finetti F, Paccani SR, Riparbelli MG, Giacomello E, Perinetti G, Pazour GJ, Rosenbaum JL, Baldari CT. 2009. Intraflagellar transport is required for polarized recycling of the TCR/CD3 complex to the immune synapse. Nat Cell Biol 11: 1332–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finetti F, Patrussi L, Masi G, Onnis A, Galgano D, Lucherini OM, Pazour GJ, Baldari CT. 2014. Specific recycling receptors are targeted to the immune synapse by the intraflagellar transport system. J Cell Sci 127: 1924–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Glover DM. 2012. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol 2: 120104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H, Inoko A, Inagaki M. 2013. Cell cycle progression by the repression of primary cilia formation in proliferating cells. Cell Mol Life Sci 70: 3893–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S, Stierhof Y-D, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. 2007. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol 179: 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley A, Jurczyk A, Sillibourne J, Halilovic E, Mogensen M, Groisman I, Blomberg M, Doxsey S. 2003. A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J Cell Biol 161: 535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudi R, Haycraft CJ, Bell PD, Li Z, Vasu C. 2015. Centrobin-mediated regulation of CPAP level limits centriole length during elongation stage. J Biol Chem 290: 6890–6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guemez-Gamboa A, Coufal NG, Gleeson JG. 2014. Primary cilia in the developing and mature brain. Neuron 82: 511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard P, Hachet V, Majubu N, Neves A, Demurtas D, Olieric N, Fluckiger I, Yamada A, Kihara K, Nishida Y, et al. 2013. Native architecture of the centriole proximal region reveals features underlying its 9-fold radial symmetry. Curr Biol 23: 1620–1628. [DOI] [PubMed] [Google Scholar]
- Han YM, Kang GM, Byun K, Ko HW, Kim J, Shin M, Kim H, Gil SY, Yu JH, Lee B, et al. 2014. Leptin-promoted cilia assembly is critical for normal energy balance. J Clin Invest 124: 2193–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehnly H, Doxsey S. 2012. Polarity sets the stage for cytokinesis. Mol Biol Cell 23: 7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehnly H, Doxsey S. 2014. Rab11 endosomes contribute to mitotic spindle organization and orientation. Dev Cell 28: 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehnly H, Chen C-T, Powers CM, Liu H-L, Doxsey S. 2012. The centrosome regulates the Rab11-dependent recycling endosome pathway at appendages of the mother centriole. Curr Biol 22: 1944–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehnly H, Hung H-F, Doxsey S. 2013. One among many: ODF2 isoform 9, a.k.a. Cenexin-1, is required for ciliogenesis. Cell Cycle 12: 1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochegger H, Hégarat N, Pereira-Leal JB. 2013. Aurora at the pole and equator: Overlapping functions of Aurora kinases in the mitotic spindle. Open Biol 3: 120185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich AL. 2014. Molecular chaperones in cellular protein folding: The birth of a field. Cell 157: 285–288. [DOI] [PubMed] [Google Scholar]
- Hung H-F, Hehnly H, Doxsey S. 2016. The mother centriole appendage protein cenexin modulates lumen formation through spindle orientation. Curr Biol 26: 793–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hut HMJ, Kampinga HH, Sibon OCM. 2005. Hsp70 protects mitotic cells against heat-induced centrosome damage and division abnormalities. Mol Biol Cell 16: 3776–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Kubo A, Tsukita S, Tsukita S. 2005. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat Cell Biol 7: 517–524. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Thompson J, Yates JR, Marshall WF. 2012. Proteomic analysis of mammalian primary cilia. Curr Biol 22: 414–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen L, Vanselow K, Skogs M, Toyoda Y, Lundberg E, Poser I, Falkenby LG, Bennetzen M, Westendorf J, Nigg EA, et al. 2011. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J 30: 1520–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins MR, Stinchcombe JC, Au-Yeung BB, Asano Y, Ritter AT, Weiss A, Griffiths GM. 2014. Distinct structural and catalytic roles for Zap70 in formation of the immunological synapse in CTL. Elife 3: e01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, White SR, Shida T, Schulz S, Aguiar M, Gygi SP, Bazan JF, Nachury MV. 2010. The conserved Bardet–Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell 141: 1208–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurczyk A, Gromley A, Redick S, San Agustin J, Witman G, Pazour GJ, Peters DJM, Doxsey S. 2004. Pericentrin forms a complex with intraflagellar transport proteins and polycystin-2 and is required for primary cilia assembly. J Cell Biol 166: 637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanguay RM, Bruford EA, Cheetham ME, Chen B, Hightower LE. 2009. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones 14: 105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsanis N. 2004. The oligogenic properties of Bardet–Biedl syndrome. Hum Mol Genet 13Spec No 1: R65–R71. [DOI] [PubMed] [Google Scholar]
- Kim JC, Badano JL, Sibold S, Esmail MA, Hill J, Hoskins BE, Leitch CC, Venner K, Ansley SJ, Ross AJ, et al. 2004. The Bardet–Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat Genet 36: 462–470. [DOI] [PubMed] [Google Scholar]
- Kim JC, Ou YY, Badano JL, Esmail MA, Leitch CC, Fiedrich E, Beales PL, Archibald JM, Katsanis N, Rattner JB, et al. 2005. MKKS/BBS6, a divergent chaperonin-like protein linked to the obesity disorder Bardet–Biedl syndrome, is a novel centrosomal component required for cytokinesis. J Cell Sci 118: 1007–1020. [DOI] [PubMed] [Google Scholar]
- Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. 2013. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem 82: 323–355. [DOI] [PubMed] [Google Scholar]
- Kitagawa D, Vakonakis I, Olieric N, Hilbert M, Keller D, Olieric V, Bortfeld M, Erat MC, Flückiger I, Gönczy P, et al. 2011. Structural basis of the 9-fold symmetry of centrioles. Cell 144: 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M, Kubiak JZ, Li XC, Ghobrial RM. 2013. The newly found functions of MTOC in immunological response. J Leukoc Biol 95: 1–14. [DOI] [PubMed] [Google Scholar]
- Knödler A, Feng S, Zhang J, Zhang X, Das A, Peränen J, Guo W. 2010. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc Natl Acad Sci 107: 6346–6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox JD, Mitchel REJ, Brown DL. 1993. Effects of taxol and taxol/hyperthermia treatments on the functional polarization of cytotoxic T lymphocytes. Cell Motil Cytoskeleton 24: 129–138. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Dynlacht BD. 2011. Regulating the transition from centriole to basal body. J Cell Biol 193: 435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Polka JK, Zelter A, Davis TN, Agard DA. 2010. Microtubule nucleating γ-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature 466: 879–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A, Mailand N, Lukas C, Syljuåsen R.G, Wilkinson CJ, Nigg EA, Bartek J, Lukas J. 2004. Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat Cell Biol 6: 884–891. [DOI] [PubMed] [Google Scholar]
- Kuhné MR, Lin J, Yablonski D, Mollenauer MN, Ehrlich LIR, Huppa J, Davis MM, Weiss A. 2003. Linker for activation of T cells, ζ-associated protein-70, and Src homology 2 domain-containing leukocyte protein-76 are required for TCR-induced microtubule-organizing center polarization. J Immunol 171: 860–866. [DOI] [PubMed] [Google Scholar]
- Lam HC, Cloonan SM, Bhashyam AR, Haspel JA, Singh A, Sathirapongsasuti JF, Cervo M, Yao H, Chung AL, Mizumura K, et al. 2013. Histone deacetylase 6-mediated selective autophagy regulates COPD-associated cilia dysfunction. J Clin Invest 123: 5212–5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo S, Hasegan M, Gupta GD, Pelletier L. 2012. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat Cell Biol 14: 1148–1158. [DOI] [PubMed] [Google Scholar]
- Lens SM, Voest EE, Medema RH. 2010. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer 10: 825–841. [DOI] [PubMed] [Google Scholar]
- Liu X, Lin C-Y, Lei M, Yan S, Zhou T, Erikson RL. 2005. CCT chaperonin complex is required for the biogenesis of functional Plk1. Mol Cell Biol 25: 4993–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Tan G, Levenkova N, Li T, Pugh EN, Rux JJ, Speicher DW, Pierce EA. 2007. The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics 6: 1299–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Kapoor TM, Chen JK, Huse M. 2013. Diacylglycerol promotes centrosome polarization in T cells via reciprocal localization of dynein and myosin II. Proc Natl Acad Sci 110: 11976–11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca O, Martín-Benito J, Ritco-Vonsovici M, Grantham J, Hynes GM, Willison KR, Carrascosa JL, Valpuesta JM. 2000. Eukaryotic chaperonin CCT stabilizes actin and tubulin folding intermediates in open quasi-native conformations. EMBO J 19: 5971–5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler H, Bochtler T, Fritz B, Tews B, Ho AD, Lukas J, Bartek J, Krämer A. 2007. DNA damage-induced accumulation of centrosomal Chk1 contributes to its checkpoint function. Cell Cycle 6: 2541–2548. [DOI] [PubMed] [Google Scholar]
- Löffler H, Fechter A, Liu FY, Poppelreuther S, Krämer A. 2013. DNA damage–induced centrosome amplification occurs via excessive formation of centriolar satellites. Oncogene 32: 2963–2972. [DOI] [PubMed] [Google Scholar]
- Loktev AV, Zhang Q, Beck JS, Searby CC, Scheetz TE, Bazan JF, Slusarski DC, Sheffield VC, Jackson PK, Nachury MV. 2008. A BBSome subunit links ciliogenesis, microtubule stability, and acetylation. Dev Cell 15: 854–865. [DOI] [PubMed] [Google Scholar]
- Lu Q, Insinna C, Ott C, Stauffer J, Pintado PA, Rahajeng J, Baxa U, Walia V, Cuenca A, Hwang YS, et al. 2015. Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation. Nat Cell Biol 17: 228–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Cófreces NB, Sancho D, Fernández E, Vicente-Manzanares M, Gordón-Alonso M, Montoya MC, Michel F, Acuto O, Alarcón B, Sánchez-Madrid F. 2006. Role of Fyn in the rearrangement of tubulin cytoskeleton induced through TCR. J Immunol 176: 4201–4207. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Maller JL. 2004. A centrosomal localization signal in cyclin E required for Cdk2-independent S phase entry. Science 306: 885–888. [DOI] [PubMed] [Google Scholar]
- Mayer U, Küller A, Daiber PC, Neudorf I, Warnken U, Schnölzer M, Frings S, Möhrlen F. 2009. The proteome of rat olfactory sensory cilia. Proteomics 9: 322–334. [DOI] [PubMed] [Google Scholar]
- Mendez R, Richter JD. 2001. Translational control by CPEB: A means to the end. Nat. Rev Mol Cell Biol 2: 521–529. [DOI] [PubMed] [Google Scholar]
- Mennella V, Keszthelyi B, McDonald KL, Chhun B, Kan F, Rogers GC, Huang B, Agard DA. 2012. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat Cell Biol 14: 1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella V, Agard DA, Huang B, Pelletier L. 2014. Amorphous no more: Subdiffraction view of the pericentriolar material architecture. Trends Cell Biol 24: 188–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentlik AN, Sanborn KB, Holzbaur EL, Orange JS. 2010. Rapid lytic granule convergence to the MTOC in natural killer cells is dependent on dynein but not cytolytic commitment. Mol Biol Cell 21: 2241–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikule K, Delaval B, Kaldis P, Jurcyzk A, Hergert P, Doxsey S. 2007. Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nat Cell Biol 9: 160–170. [DOI] [PubMed] [Google Scholar]
- Muñoz IG, Yébenes H, Zhou M, Mesa P, Serna M, Park AY, Bragado-Nilsson E, Beloso A, de Cárcer G, Malumbres M, et al. 2011. Crystal structure of the open conformation of the mammalian chaperonin CCT in complex with tubulin. Nat Struct Mol Biol 18: 14–19. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peränen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, et al. 2007. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129: 1201–1213. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Saito H, Takekawa M. 2013. SAPK pathways and p53 cooperatively regulate PLK4 activity and centrosome integrity under stress. Nat Commun 4: 1775. [DOI] [PubMed] [Google Scholar]
- Nigg E, Raff JW. 2009. Centrioles, centrosomes, and cilia in health and disease. Cell 139: 663–678. [DOI] [PubMed] [Google Scholar]
- Pagan JK, Marzio A, Jones MJK, Saraf A, Jallepalli PV, Florens L, Washburn MP, Pagano M. 2015. Degradation of Cep68 and PCNT cleavage mediate Cep215 removal from the PCM to allow centriole separation, disengagement and licensing. Nat Cell Biol 17: 31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampliega O, Orhon I, Patel B, Sridhar S, Díaz-Carretero A, Beau I, Codogno P, Satir BH, Satir P, Cuervo AM. 2013. Functional interaction between autophagy and ciliogenesis. Nature 502: 194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paridaen JTML, Wilsch-Bräuninger M, Huttner WB. 2013. Asymmetric inheritance of centrosome-associated primary cilium membrane directs ciliogenesis after cell division. Cell 155: 333–344. [DOI] [PubMed] [Google Scholar]
- Pascreau G, Eckerdt F, Churchill MEA, Maller JL. 2010. Discovery of a distinct domain in cyclin A sufficient for centrosomal localization independently of Cdk binding. Proc Natl Acad Sci 107: 2932–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers MV, Clarke PA, Workman P. 2008. Dual targeting of HSC70 and HSP72 inhibits HSP90 function and induces tumor-specific apoptosis. Cancer Cell 14: 250–262. [DOI] [PubMed] [Google Scholar]
- Puram SV, Kim AH, Park HY, Anckar J, Bonni A. 2013. The ubiquitin receptor S5a/Rpn10 links centrosomal proteasomes with dendrite development in the mammalian brain. Cell Rep 4: 19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quann EJ, Merino E, Furuta T, Huse M. 2009. Localized diacylglycerol drives the polarization of the microtubule-organizing center in T cells. Nat Immunol 10: 627–635. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. 2007. Patched1 regulates Hedgehog signaling at the primary cilium. Science 317: 372–376. [DOI] [PubMed] [Google Scholar]
- Sasayama T, Marumoto T, Kunitoku N, Zhang D, Tamaki N, Kohmura E, Saya H, Hirota T. 2005. Over-expression of Aurora-A targets cytoplasmic polyadenylation element binding protein and promotes mRNA polyadenylation of Cdk1 and cyclin B1. Genes Cells 10: 627–638. [DOI] [PubMed] [Google Scholar]
- Scheer U. 2014. Historical roots of centrosome research: Discovery of Boveri’s microscope slides in Würzburg. Philos Trans R Soc Lond B Biol Sci 369: 20130469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Baye LM, Schulz NP, Beck JS, Zhang Q, Slusarski DC, Sheffield VC. 2010. BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome assembly. Proc Natl Acad Sci 107: 1488–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibon OC, Kelkar A, Lemstra W, Theurkauf WE. 2000. DNA-replication/DNA-damage-dependent centrosome inactivation in Drosophila embryos. Nat Cell Biol 2: 90–95. [DOI] [PubMed] [Google Scholar]
- Sir J-H, Pütz M, Daly O, Morrison CG, Dunning M, Kilmartin JV, Gergely F. 2013. Loss of centrioles causes chromosomal instability in vertebrate somatic cells. J Cell Biol 203: 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 2012. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open 1: 965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin S. 1962. Centrioles rudimentary and smooth and the formation of cilia muscle by fibroblasts. J Cell Biol 15: 363–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe JC, Majorovits E, Bossi G, Fuller S, Griffiths GM. 2006. Centrosome polarization delivers secretory granules to the immunological synapse. Nature 443: 462–465. [DOI] [PubMed] [Google Scholar]
- Sung C-H, Leroux MR. 2013. The roles of evolutionarily conserved functional modules in cilia-related trafficking. Nat Cell Biol 15: 1387–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Lin MG, Stowe TR, Chen S, Zhu M, Stearns T, Franco B, Zhong Q. 2013. Autophagy promotes primary ciliogenesis by removing OFD1 from centriolar satellites. Nature 502: 254–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos BE, Yang H-J, Soni R, Wang W-J, Macaluso FP, Asara JM, Tsou M-FB. 2013. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev 27: 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasouri E, Tucker KL. 2011. Primary cilia and organogenesis: Is Hedgehog the only sculptor? Cell Tissue Res 345: 21–40. [DOI] [PubMed] [Google Scholar]
- Thauland TJ, Parker DC. 2010. Diversity in immunological synapse structure. Immunology 131: 466–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toji S, Yabuta N, Hosomi T, Nishihara S, Kobayashi T, Suzuki S, Tamai K, Nojima H. 2004. The centrosomal protein Lats2 is a phosphorylation target of Aurora-A kinase. Genes Cells 9: 383–397. [DOI] [PubMed] [Google Scholar]
- Tsun A, Qureshi I, Stinchcombe JC, Jenkins MR, de la Roche M, Kleczkowska J, Zamoyska R, Griffiths GM. 2011. Centrosome docking at the immunological synapse is controlled by Lck signaling. J Cell Biol 192: 663–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertii A, Bright A, Delaval B, Hehnly H, Doxsey S. 2015a. New frontiers: Discovering cilia-independent functions of cilia proteins. EMBO Rep 16: e201540632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertii A, Zimmerman W, Ivshina M, Doxsey S. 2015b. Centrosome-intrinsic mechanisms modulate centrosome integrity during fever. Mol Biol Cell 26: 3451–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidair CA, Doxsey SJ, Dewey WC. 1993. Heat shock alters centrosome organization leading to mitotic dysfunction and cell death. J Cell Physiol 154: 443–455. [DOI] [PubMed] [Google Scholar]
- Villumsen BH, Danielsen JR, Povlsen L, Sylvestersen KB, Merdes A, Beli P, Yang Y-G, Choudhary C, Nielsen ML, Mailand N, et al. 2013. A new cellular stress response that triggers centriolar satellite reorganization and ciliogenesis. EMBO J 32: 3029–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tsai J-W, Imai JH, Lian W-N, Vallee RB, Shi S-H. 2009. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature 461: 947–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Zhang Y, Li Y, Zhang Q, Ling K, Hu J. 2012. The BBSome controls IFT assembly and turnaround in cilia. Nat Cell Biol 14: 950–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlake CJ, Baye LM, Nachury MV, Wright KJ, Ervin KE, Phu L, Chalouni C, Beck JS, Kirkpatrick DS, Slusarski DC, et al. 2011. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc Natl Acad Sci 108: 2759–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigley WC, Fabunmi RP, Lee MG, Marino CR, Muallem S, DeMartino GN, Thomas PJ. 1999. Dynamic association of proteasomal machinery with the centrosome. J Cell Biol 145: 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Mehta SQ, Pichaud F, Bellen HJ, Quiocho FA. 2005. Sec15 interacts with Rab11 via a novel domain and affects Rab11 localization in vivo. Nat Struct Mol Biol 12: 879–885. [DOI] [PubMed] [Google Scholar]
- Yang J, Liu X, Yue G, Adamian M, Bulgakov O, Li T. 2002. Rootletin, a novel coiled-coil protein, is a structural component of the ciliary rootlet. J Cell Biol 159: 431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Gao J, Adamian M, Wen X-H, Pawlyk B, Zhang L, Sanderson MJ, Zuo J, Makino CL, Li T. 2005. The ciliary rootlet maintains long-term stability of sensory cilia. Mol Cell Biol 25: 4129–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yang Y, Zhao P, Chen K, Chen B, Lin Y, Guo F, Chen Y, Liu X, Lu F, et al. 2008. A novel mutation in BBS7 gene causes Bardet–Biedl syndrome in a Chinese family. Mol Vis 14: 2304–2308. [PMC free article] [PubMed] [Google Scholar]
- Yi J, Wu X, Chung AH, Chen JK, Kapoor TM, Hammer J. 2013. Centrosome repositioning in T cells is biphasic and driven by microtubule end-on capture-shrinkage. J Cell Biol 202: 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, Otterness DM, Chiang GG, Xie W, Liu YC, Mercurio F, Abraham RT. 2005. Genotoxic stress targets human Chk1 for degradation by the ubiquitin-proteasome pathway. Mol Cell 19: 607–618. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Yu D, Seo S, Stone EM, Sheffield VC. 2012. Intrinsic protein-protein interaction-mediated and chaperonin-assisted sequential assembly of stable Bardet–Biedl syndrome protein complex, the BBSome. J Biol Chem 287: 20625–20635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Nishimura D, Vogel T, Shao J, Swiderski R, Yin T, Searby C, Carter CS, Kim G, Bugge K, et al. 2013. BBS7 is required for BBSome formation and its absence in mice results in Bardet–Biedl syndrome phenotypes and selective abnormalities in membrane protein trafficking. J Cell Sci 126: 2372–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ren Y, Jiang Q, Feng J. 2003. Parkin is recruited to the centrosome in response to inhibition of proteasomes. J Cell Sci 116: 4011–4019. [DOI] [PubMed] [Google Scholar]
- Zimmerman WC, Sillibourne J, Rosa J, Doxsey SJ. 2004. Mitosis-specific anchoring of γ tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol Biol Cell 15: 3642–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C, Li J, Bai Y, Gunning WT, Band V, Gao Q. 2005. Centrobin: A novel daughter centriole-associated protein that is required for centriole duplication. J Cell Biol 171: 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyss D, Ebrahimi H, Gergely F. 2011. Casein kinase I δ controls centrosome positioning during T cell activation. J Cell Biol 195: 781–797. [DOI] [PMC free article] [PubMed] [Google Scholar]