Abstract
Targeted nucleases have provided researchers with the ability to manipulate virtually any genomic sequence, enabling the facile creation of isogenic cell lines and animal models for the study of human disease, and promoting exciting new possibilities for human gene therapy. Here we review three foundational technologies—clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein 9 (Cas9), transcription activator-like effector nucleases (TALENs), and zinc-finger nucleases (ZFNs). We discuss the engineering advances that facilitated their development and highlight several achievements in genome engineering that were made possible by these tools. We also consider artificial transcription factors, illustrating how this technology can complement targeted nucleases for synthetic biology and gene therapy.
Three technologies—CRISPR-Cas9, TALE nucleases, and zinc-finger nucleases—have facilitated a genome-editing revolution. But several challenges (e.g., effectively treating human diseases) remain.
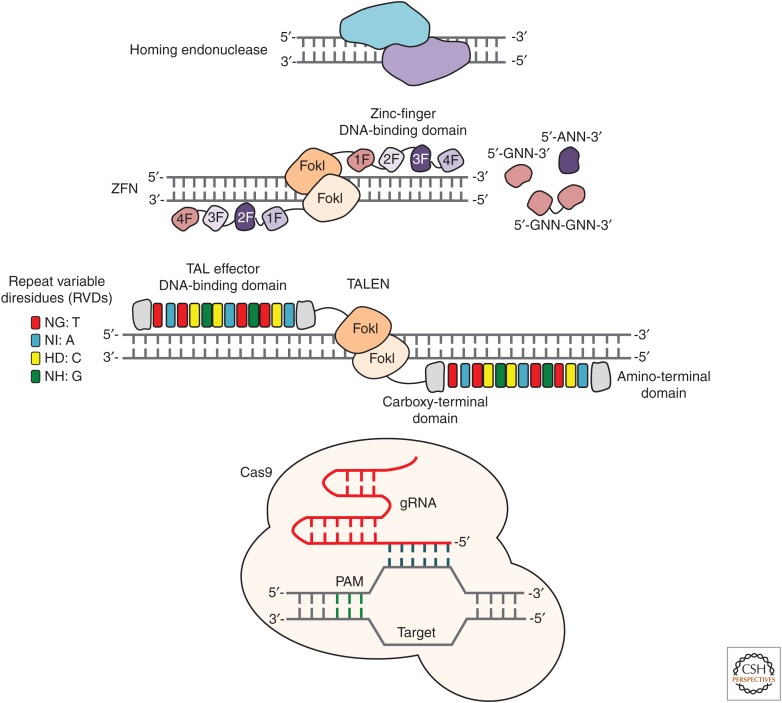
In recent years, the emergence of highly versatile genome-editing technologies has provided investigators with the ability to rapidly and economically introduce sequence-specific modifications into the genomes of a broad spectrum of cell types and organisms. The core technologies now most commonly used to facilitate genome editing, shown in Figure 1, are (1) clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein 9 (Cas9), (2) transcription activator-like effector nucleases (TALENs), (3) zinc-finger nucleases (ZFNs), and (4) homing endonucleases or meganucleases.
Figure 1.
Genome-editing technologies. Cartoons illustrating the mechanisms of targeted nucleases. From top to bottom: homing endonucleases, zinc-finger nucleases (ZFNs), transcription activator-like effector (TALE) nucleases (TALENs), and clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein 9 (Cas9). Homing endonucleases generally cleave their DNA substrates as dimers, and do not have distinct binding and cleavage domains. ZFNs recognize target sites that consist of two zinc-finger binding sites that flank a 5- to 7-base pair (bp) spacer sequence recognized by the FokI cleavage domain. TALENs recognize target sites that consist of two TALE DNA-binding sites that flank a 12- to 20-bp spacer sequence recognized by the FokI cleavage domain. The Cas9 nuclease is targeted to DNA sequences complementary to the targeting sequence within the single guide RNA (gRNA) located immediately upstream of a compatible protospacer adjacent motif (PAM). DNA and protein are not drawn to scale.
In particular, the ease with which CRISPR-Cas9 and TALENs can be configured to recognize new genomic sequences has driven a revolution in genome editing that has accelerated scientific breakthroughs and discoveries in disciplines as diverse as synthetic biology, human gene therapy, disease modeling, drug discovery, neuroscience, and the agricultural sciences.
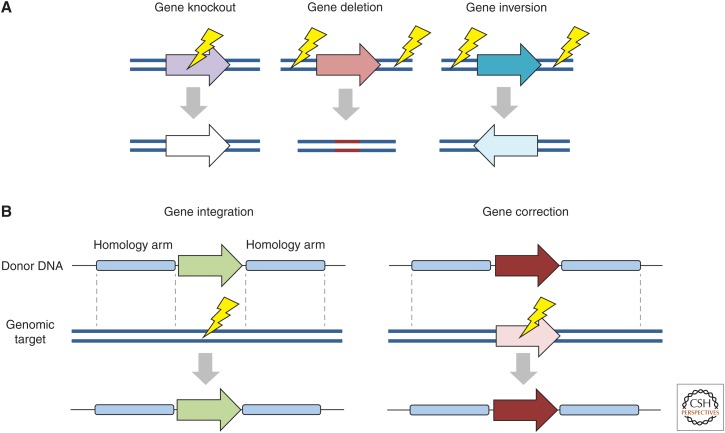
The diverse array of genetic outcomes made possible by these technologies is the result, in large part, of their ability to efficiently induce targeted DNA double-strand breaks (DSBs). These DNA breaks then drive activation of cellular DNA repair pathways and facilitate the introduction of site-specific genomic modifications (Rouet et al. 1994; Choulika et al. 1995). This process is most often used to achieve gene knockout via random base insertions and/or deletions that can be introduced by nonhomologous end joining (NHEJ) (Fig. 2A) (Bibikova et al. 2002). Alternatively, in the presence of a donor template with homology to the targeted chromosomal site, gene integration, or base correction via homology-directed repair (HDR) can occur (HDR) (Fig. 2B) (see Fig. 2 for an overview of other possible genome-editing outcomes) (Bibikova et al. 2001, 2003; Porteus and Baltimore 2003; Urnov et al. 2005). Indeed, the broad versatility of these genome-modifying enzymes is evidenced by the fact that they also serve as the foundation for artificial transcription factors, a class of tools capable of modulating the expression of nearly any gene within a genome.
Figure 2.
Genome-editing outcomes. Targeted nucleases induce DNA double-strand breaks (DSBs) that are repaired by nonhomologous end joining (NHEJ) or, in the presence of donor template, homology-directed repair (HDR). (A) In the absence of a donor template, NHEJ introduces small base insertions or deletions that can result in gene disruption. When two DSBs are induced simultaneously, the intervening genomic sequence can be deleted or inverted. (B) In the presence of donor DNA (plasmid or single-stranded oligonucleotide), recombination between homologous DNA sequences present on the donor template and a specific chromosomal site can facilitate targeted integration. Lightning bolts indicate DSBs.
Here we review key principles of genome editing, emphasizing many of the engineering advances that have laid the groundwork for the creation, refinement, and implementation of the current suite of genome-modifying tools. We also provide an overview of the achievements made possible by genome editing, illustrating how this technology can enable advances throughout the life sciences.
TARGETED NUCLEASES
Zinc-Finger Nucleases
ZFNs, which are fusions between a custom-designed Cys2-His2 zinc-finger protein and the cleavage domain of the FokI restriction endonuclease (Kim et al. 1996), were the first targeted nuclease to achieve widespread use (Porteus and Carroll 2005; Urnov et al. 2010). ZFNs function as dimers, with each monomer recognizing a specific “half site” sequence—typically nine to 18 base pairs (bps) of DNA—via the zinc-finger DNA-binding domain (Fig. 1). Dimerization of the ZFN proteins is mediated by the FokI cleavage domain, which cuts DNA within a five- to seven-bp spacer sequence that separates two flanking zinc-finger binding sites (Smith et al. 2000). Each ZFN is typically composed of three or four zinc-finger domains, with each individual domain composed of ∼30 amino acid residues that are organized in a ββα motif (Pavletich and Pabo 1991). The residues that facilitate DNA recognition are located within the α-helical domain and typically interact with three bps of DNA, with occasional overlap from an adjacent domain (Wolfe et al. 2000). Using methods such as phage display (Choo and Klug 1994; Jamieson et al. 1994; Wu et al. 1995), a large number of zinc-finger domains recognizing distinct DNA triplets have been identified (Segal et al. 1999; Dreier et al. 2001, 2005; Bae et al. 2003). These domains can be fused together in tandem using a canonical linker peptide (Liu et al. 1997) to generate polydactyl zinc-finger proteins that can target a wide range of possible DNA sequences (Beerli et al. 1998, 2000a; Kim et al. 2009). In addition to this “modular assembly” approach to zinc-finger construction, selection-based methods for constructing zinc-finger proteins have also been reported (Greisman and Pabo 1997; Isalan et al. 2001; Hurt et al. 2003; Magnenat et al. 2004), including those that consider context-dependent interactions between adjacent zinc-finger domains, such as oligomerized pool engineering (OPEN) (Maeder et al. 2008). In addition, specialized sets of validated two-finger, zinc-finger modules have been used to assemble zinc-finger arrays (Kim et al. 2009; Bhakta et al. 2013), including those that take context-dependent effects into account (Sander et al. 2011b; Gupta et al. 2012).
One major concern associated with the use of ZFNs for genome editing (in addition to all targeted nucleases) is off-target mutations (Gabriel et al. 2011; Pattanayak et al. 2011). As a result, several approaches have been undertaken to enhance their specificity. Among the most successful of these has been the creation of obligate heterodimeric ZFN architectures that rely on charge–charge repulsion to prevent unwanted homodimerization of the FokI cleavage domain (Miller et al. 2007; Doyon et al. 2011), thereby minimizing the potential for ZFNs to dimerize at off-target sites. Additionally, protein-engineering methods have been used to enhance the cleavage efficiency of the FokI cleavage domain (Guo et al. 2010). One particularly promising approach for improving ZFN specificity is to deliver them into cells as protein. Because of the intrinsic cell-penetrating activity of zinc-finger domains (Gaj et al. 2014a), ZFN proteins themselves are inherently cell-permeable and can facilitate gene editing with fewer off-target effects when applied directly onto cells as purified protein compared to when expressed within cells from nucleic acids (Gaj et al. 2012). Modified ZFN proteins endowed with improved cell-penetrating activity have since been described (Liu et al. 2015a). ZFNickases can also facilitate gene correction in the absence of a DSB (Kim et al. 2012; Ramirez et al. 2012; Wang et al. 2012). These enzymes, which consist of one catalytically inactivated ZFN monomer in combination with a second native ZFN monomer, can stimulate HDR by nicking or cleaving one strand of DNA and are derived from a concept first illustrated by Stoddard and colleagues using homing endonucleases (McConnell Smith et al. 2009).
Unlike TALENs and CRISPR-Cas9, the difficulty associated with constructing zinc-finger arrays has hindered their widespread adoption in unspecialized laboratories. In particular, it remains challenging to create zinc-finger domains that can effectively recognize all DNA triplets, especially those of the 5′-CNN-3′ and 5′-TNN-3′ variety. As a result, ZFNs lack the target flexibility inherent to more recent genome-editing platforms. Nevertheless, the potential for ZFNs to mediate specific and efficient genome editing is evidenced by ongoing clinical trials based on ZFN-mediated knockout of the human immunodeficiency virus (HIV)-1 coreceptor CCR5 for treatment of HIV/acquired immune deficiency syndrome (AIDS) (Tebas et al. 2014) and a planned clinical trial based on site-specific integration of the factor IX gene into the albumin locus to treat hemophilia B (Clinical Trial ID: NCT02695160) (Sharma et al. 2015).
TALE Nucleases
TALE proteins are bacterial effectors. In 2009, the code used by TALE proteins to recognize DNA was uncovered (Boch et al. 2009; Moscou and Bogdanove 2009). In a matter of months, this discovery enabled the creation of custom TALENs capable of modifying nearly any gene. Like ZFNs, TALENs are modular in form and function, comprised of an amino-terminal TALE DNA-binding domain fused to a carboxy-terminal FokI cleavage domain (Christian et al. 2010; Miller et al. 2011). Also like ZFNs, dimerization of TALEN proteins is mediated by the FokI cleavage domain, which cuts within a 12- to 19-bp spacer sequence that separates each TALE binding site (Fig. 1) (Miller et al. 2011). TALEs are typically assembled to recognize between 12- to 20-bps of DNA, with more bases typically leading to higher genome-editing specificity (Guilinger et al. 2014a). The TALE-binding domain consists of a series of repeat domains, each ∼34 residues in length. Each repeat contacts DNA via the amino acid residues at positions 12 and 13, known as the repeat variable diresidues (RVDs) (Boch et al. 2009; Moscou and Bogdanove 2009). Unlike zinc fingers, which recognize DNA triplets, each TALE repeat recognizes only a single bp, with little to no target site overlap from adjacent domains (Deng et al. 2012; Mak et al. 2012). The most commonly used RVDs for assembling synthetic TALE arrays are: NI for adenine, HD for cytosine, NG for thymine, and NN or HN for guanine or adenine (Boch et al. 2009; Moscou and Bogdanove 2009; Cong et al. 2012; Streubel et al. 2012). TALE DNA-binding domains can be constructed using a variety of methods, with the most straightforward approach being Golden Gate assembly (Cermak et al. 2011). However, high-throughput TALE assembly methods have also been developed, including FLASH assembly (Reyon et al. 2012), iterative capped assembly (Briggs et al. 2012), and ligation independent cloning (Schmid-Burgk et al. 2013), among others. More recent advances in TALEN assembly, though, have focused on the development of methods that can enhance their performance, including specificity profiling to uncover nonconventional RVDs that improve TALEN activity (Guilinger et al. 2014a; Yang et al. 2014; Juillerat et al. 2015; Miller et al. 2015), directed evolution as means to refine TALE specificity (Hubbard et al. 2015), and even fusing TALE domains to homing endonuclease variants to generate chimeric nucleases with extended targeting specificity (discussed in more detail below) (Boissel et al. 2014).
Compared to ZFNs, TALENs offer two distinct advantages for genome editing. First, no selection or directed evolution is necessary to engineer TALE arrays, dramatically reducing the amount of time and experience needed to assemble a functional nuclease. Second, TALENs have been reported to show improved specificity and reduced toxicity compared to some ZFNs (Mussolino et al. 2014), potentially because of their increased affinity for target DNA (Meckler et al. 2013) or perhaps a greater energetic penalty for associating with base mismatches. However, TALENs are substantially larger than ZFNs, and have a highly repetitive structure, making their efficient delivery into cells through the use of lentivirus (Holkers et al. 2013) or a single adeno-associated virus (AAV) particle challenging. Methods for overcoming these limitations have emerged as TALENs can be readily delivered into cells as mRNA (Mahiny et al. 2015; Mock et al. 2015) and even protein (Cai et al. 2014; Liu et al. 2014a), although alternative codon usage and amino acid degeneracy can also be leveraged to express RVD arrays that might be less susceptible to recombination (Kim et al. 2013a). In addition, adenoviral vectors have also proven particularly useful for mediating TALEN delivery to hard-to-transfect cell types (Holkers et al. 2014; Maggio et al. 2016).
CRISPR-Cas9
The CRISPR-Cas9 system, which has a role in adaptive immunity in bacteria (Horvath and Barrangou 2010; Marraffini and Sontheimer 2010), is the most recent addition to the genome-editing toolbox. In bacteria, the type-II CRISPR system provides protection against DNA from invading viruses and plasmids via RNA-guided DNA cleavage by Cas proteins (Wiedenheft et al. 2012; Sorek et al. 2013). Short segments of foreign DNA are integrated within the CRISPR locus and transcribed into CRISPR RNA (crRNA), which then anneal to trans-activating crRNA (tracrRNA) to direct sequence-specific degradation of pathogenic DNA by the Cas9 protein (Jinek et al. 2012). In 2012, Charpentier, Doudna, and co-workers reported that target recognition by the Cas9 protein only requires a seed sequence within the crRNA and a conserved protospacer-adjacent motif (PAM) upstream of the crRNA binding site (Jinek et al. 2012). This system has since been simplified for genome engineering (Cho et al. 2013; Cong et al. 2013; Jinek et al. 2013; Mali et al. 2013b) and now consists of only the Cas9 nuclease and a single guide RNA (gRNA) containing the essential crRNA and tracrRNA elements (Fig. 1). Because target site recognition is mediated entirely by the gRNA, CRISPR-Cas9 has emerged as the most flexible and user-friendly platform for genome editing, eliminating the need for engineering new proteins to recognize each new target site. The only major restriction for Cas9 target site recognition is that the PAM motif—which is recognized by the Cas9 nuclease and is essential for DNA cleavage—be located immediately downstream of the gRNA target site. The PAM sequence for the Streptococcus pyogenes Cas9, for example, is 5′-NGG-3′ (although in some cases 5′-NAG-3′ can be tolerated) (Hsu et al. 2013; Jiang et al. 2013; Mali et al. 2013a). Several studies have now shed light on the structural basis of DNA recognition by Cas9, revealing that the heteroduplex formed by the gRNA and its complementary strand of DNA is housed in a positively charged groove between the two nuclease domains (RuvC and HNH) within the Cas9 protein (Nishimasu et al. 2014), and that PAM recognition is mediated by an arginine-rich motif present in Cas9 (Anders et al. 2014). Doudna and colleagues have since proposed that DNA strand displacement induces a structural rearrangement within the Cas9 protein that directs the nontarget DNA strand into the RuvC active site, which then positions the HNH domain near target DNA (Jiang et al. 2016), enabling Cas9-mediated cleavage of both DNA strands.
The Cas9 nuclease and its gRNA can be delivered into cells for genome editing on the same or separate plasmids, and numerous resources have been developed to facilitate target site selection and gRNA construction, including E-CRISP (Heigwer et al. 2014), among others. Although Cas9 boasts the highest ease of use among the targeted nuclease platforms, several reports have indicated that it could be prone to inducing off-target mutations (Cradick et al. 2013; Fu et al. 2013). To this end, considerable effort has been devoted to improving the specificity of this system, including using paired Cas9 nickases (Mali et al. 2013a; Ran et al. 2013), which increase gene-editing specificity by requiring the induction of two sequential and adjacent nicking events for DSB formation, or truncated gRNA that are more sensitive to mismatches at the genomic target site than a full-length gRNA (Fu et al. 2014). Off-target cleavage has also been reduced by controlling the dosage of either the Cas9 protein or gRNA within the cell (Hsu et al. 2013), or even by using Cas9 variants configured to enable conditional genome editing, such as a rapamycin-inducible split-Cas9 architecture (Zetsche et al. 2015b) or a Cas9 variant that contains a strategically placed small-molecule-responsive intein domain (Davis et al. 2015). Nucleofection (Kim et al. 2014) or transient transfection (Zuris et al. 2015) of a preformed Cas9 ribonucleoprotein complex has also been shown to reduce off-target effects, enabling DNA-free gene editing in primary human T cells (Schumann et al. 2015), embryonic stem cells (Liu et al. 2015b), Caenorhabditis elegans gonads (Paix et al. 2015), mouse (Menoret et al. 2015; Wang et al. 2015a) and zebrafish embryos (Sung et al. 2014), and even plant protoplasts (Woo et al. 2015). The incorporation of specific chemical modifications known to protect RNA from nuclease degradation and stabilize secondary structure can further enhance Cas9 ribonucleoprotein activity (Hendel et al. 2015; Rahdar et al. 2015). In a clever marriage of genome-editing platforms, the FokI cleavage domain has even been fused to an inactivated Cas9 variant to generate hybrid nucleases that require protein dimerization for DNA cleavage (Guilinger et al. 2014b; Tsai et al. 2014), theoretically increasing CRISPR-Cas9 specificity. Similarly, fusing Cas9 to DNA-binding domains has also proven effective at improving its specificity (Bolukbasi et al. 2015). Finally, several studies have recently showed that protein engineering can broadly enhance Cas9 specificity (Kleinstiver et al. 2016; Slaymaker et al. 2016) and even alter its PAM requirements (Kleinstiver et al. 2015), the latter having the potential to enable creation of customized variants of Cas9 for allele-specific gene editing, although Cas9 orthologs (Cong et al. 2013; Esvelt et al. 2013; Hou et al. 2013; Ran et al. 2015) or alternative CRISPR systems (Zetsche et al. 2015a) with unique PAM specificities have been uncovered in nature.
Homing Endonucleases
Homing endonucleases, also known as meganucleases, represent the final member of the targeted nuclease family. These enzymes have been reviewed at length elsewhere (Silva et al. 2011; Stoddard 2014) but, briefly, members of the LAGLIDADG family of endonucleases—so named for the conserved amino acid motif present within these enzymes that interacts with DNA—are a collection of naturally occurring enzymes that recognize and cleave long DNA sequences (14–40 bps) (Fig. 1). These enzymes make extensive sequence-specific contacts with their DNA substrate (Stoddard 2011), and thus typically show exquisite specificity. However, unlike ZFNs and TALENs, the binding and cleavage domains in homing endonucleases are not modular. This overlap in form and function make their repurposing challenging, and limits their utility for more routine applications of genome editing. More recently megaTALs—fusions of a rare-cleaving homing endonuclease to a TALE-binding domain—have been reported to induce highly specific gene modifications (Boissel et al. 2014; Lin et al. 2015a). These enzymes have enabled integration of antitumor and anti-HIV factors into the human CCR5 gene in both primary T cells and hematopoietic stem/progenitor cells (Sather et al. 2015), as well as disruption of endogenous T-cell receptor elements in T cells (Osborn et al. 2016), indicating their potential for enabling and enhancing immunotherapies.
GENOME-EDITING APPLICATIONS
Engineering Cell Lines and Organisms
Before the emergence of engineered nucleases, genetically modifying mammalian cell lines was labor intensive, costly, and often times limited to laboratories with specialized expertise. However, with the advent of cost-effective and user-friendly gene-editing technologies, custom cell lines carrying nearly any genomic modification can now be generated in simply a matter of weeks. Examples of some of the outcomes that have become routine because of the emergence of targeted nucleases include gene knockout (Santiago et al. 2008; Mali et al. 2013b), gene deletion (Lee et al. 2010), gene inversion (Xiao et al. 2013), gene correction (Urnov et al. 2005; Ran et al. 2013), gene addition (Moehle et al. 2007; Hockemeyer et al. 2011; Hou et al. 2013), and even chromosomal translocation (Fig. 2) (Torres et al. 2014). In addition to cell line engineering, targeted nucleases have also expedited the generation of genetically modified organisms, facilitating the rapid creation of transgenic zebrafish (Doyon et al. 2008; Sander et al. 2011a; Hwang et al. 2013), mice (Cui et al. 2011; Wang et al. 2013; Wu et al. 2013), rats (Geurts et al. 2009; Tesson et al. 2011; Li et al. 2013), monkeys (Liu et al. 2014c), and livestock (Hauschild et al. 2011; Carlson et al. 2012), which together have the capacity to accelerate human disease modeling and the discovery of new therapeutics.
Targeted nucleases have also emerged as powerful tools for plant engineering (Baltes and Voytas 2015). Both TALENs and CRISPR-Cas9 have been used to modify multiple alleles within hexaploid bread wheat to confer heritable resistance to powdery mildew (Wang et al. 2014b). In another study, TALENs were used to knock out nonessential genes in the fatty acid metabolic pathway in soybean to generate a simplified plant cell with reduced metabolic components (Haun et al. 2014). Of special note, two recent reports showed that purified nuclease proteins can be introduced directly into plant protoplasts, enabling the introduction of germline-transmissible modifications that are virtually indistinguishable from naturally occurring (Luo et al. 2015; Woo et al. 2015). This technical advance could help to overcome certain regulatory hurdles associated with the use of transgenic crops. Finally, targeted nucleases have also been used to inactivate pathogenic genes to prevent viral (Lin et al. 2014) or parasitic (Ghorbal et al. 2014) infection, as well as to introduce knockin-specific factors capable of imparting pathogen resistance (Wu et al. 2015).
Intriguingly, targeted nucleases could also serve as conduits for curbing mosquito- or insect-borne diseases through a technique known as gene drive (Burt 2003; Sinkins and Gould 2006), which harnesses genome editing to facilitate the introduction of a specific gene or mutation that can then confer a particular phenotype into a host and also be transmitted to its progeny (Windbichler et al. 2011). Gene drives have now been tested in the malaria vector mosquitos Anopheles stephensi (Gantz et al. 2015) and Anopheles gambiae (Hammond et al. 2016) as a means for achieving population control and to prevent disease transmission, respectively. However, owing to the ease with which CRISPR-Cas9 can be programmed (Gantz and Bier 2015), debate has ignited on the potential societal and environmental impact of this technology (Esvelt et al. 2014; Akbari et al. 2015), spurring the development of safeguard elements that could help to minimize the risk of gene-edited organisms escaping from the laboratory (DiCarlo et al. 2015).
Synthetic Biology and Genome-Scale Engineering
Targeted nucleases also offer a facile means for generating modified bacterial and yeast strains for synthetic biology, including metabolic pathway engineering. Bacterial species of the order Actinomycetales, for instance, are one of the most important sources of industrially relevant secondary metabolites. However, many Actinomycetales species are recalcitrant to genetic manipulation, a fact that has severely hampered their use for metabolic engineering. CRISPR-Cas9 has been used to inactivate multiple genes in actinomycetes (Tong et al. 2015), indicating its ability to enable the creation of designer bacterial strains with enhanced metabolite production capabilities. CRISPR has also facilitated multiplexed metabolic pathway engineering in yeast at high efficiencies (Jakociunas et al. 2015a,b), as well as random mutagenesis of yeast chromosomal DNA for phenotypic screening of desired mutants (Ryan et al. 2014). Indeed, genome-wide CRISPR-based knockout screens hold tremendous potential for functional genomics (Hilton and Gersbach 2015), having facilitated the discovery of genomic loci that confer drug resistance to cells (Koike-Yusa et al. 2014; Shalem et al. 2014; Wang et al. 2014a; Zhou et al. 2014), uncovered how cells can control induction of the host immune response (Parnas et al. 2015), provided new insights into the genetic basis of cellular fitness (Hart et al. 2015; Wang et al. 2015b), and even shed light on how certain viruses induce cell death (Ma et al. 2015). Genome-wide CRISPR screens can also facilitate the discovery of functional noncoding elements (Kim et al. 2013b; Korkmaz et al. 2016), and provide a means for studying the structure and evolution of the human genome. In a remarkable example of the latter, Shendure and colleagues used Cas9 to mediate integration of short randomized DNA sequences into the BRCA1 and DBR1 genes (Findlay et al. 2014). They then measured the functional consequences of these mutations on fitness, achieving an unprecedented look at some of the factors driving genome and disease evolution. Finally, CRISPR screens have even proven effective in vivo, enabling the identification of factors involved in zebrafish development (Shah et al. 2015) and disease progression in mice (Chen et al. 2015).
Therapeutic Genome Editing
Genome editing itself also holds tremendous potential for treating the underlying genetic causes of certain diseases (Cox et al. 2015; Porteus 2015; Maeder and Gersbach 2016). In one of the most successful examples of this to date, ZFN-mediated disruption of the HIV coreceptor CCR5 was used to engineer HIV resistance into both CD4+ T cells (Perez et al. 2008) and CD34+ hematopoietic stem/progenitor cells (HSPCs) (Holt et al. 2010), proving safe and well-tolerated in a phase I clinical trial that infused these gene-modified T cells into individuals with HIV/AIDS (Tebas et al. 2014). In addition to enabling the introduction of gene modification that can enhance autologous cell therapies, targeted nucleases can also be combined with viral vectors—including AAV—to mediate genome editing in situ (Gaj et al. 2016). For instance, delivery of an AAV vector encoding a ZFN pair designed to target a defective copy of the factor IX gene, along with its repair template, led to efficient gene correction in mouse liver, increasing factor IX protein production in both neonatal (Li et al. 2011) and adult (Anguela et al. 2013) models of the disease. In vivo genome editing also recently enabled the restoration of dystrophin gene expression and the rescue of muscle function in mouse models of Duchenne muscular dystrophy (Long et al. 2015; Nelson et al. 2015; Tabebordbar et al. 2015). Therapeutic gene editing in a mouse model of human hereditary tyrosinemia has also been reported using both hydrodynamic injection of plasmid DNA encoding CRISPR-Cas9 (Yin et al. 2014) and by combining nanoparticle-mediated delivery of Cas9-encoding mRNA with AAV-mediated delivery of the DNA template for gene correction (Yin et al. 2016). More recently, a dual particle AAV system, wherein one AAV vector carried the Cas9 nuclease and a second harbored the gRNA and donor repair template, was able to mediate correction of a disease-causing mutation in the ornithine transcarbamylase gene in the liver of a neonatal model of the disease (Yang et al. 2016). This work, in particular, showed that therapeutic levels of gene correction could be achieved in a regenerating tissue even when using multiple AAV particles. Although highly promising, numerous hurdles still need to be overcome for in vivo applications of genome editing to reach its full potential. Chief among these are methods that can facilitate nuclease delivery or expression to only diseased cells or tissues, and the development of new strategies that can enhance HDR in disease-associated postmitotic cells in vivo.
TARGETED TRANSCRIPTION FACTORS
Tools for Modulating Gene Expression
The modular qualities of zinc-finger and TALE proteins, in addition to the highly flexible DNA recognition ability of CRISPR-Cas9, also provide investigators with the ability to modulate the expression of nearly any gene from its promoter or enhancer sequences via their fusion to transcriptional activator and repressor protein domains. Among the first fully synthetic transcriptional effector proteins to be generated (Beerli et al. 1998) were those based on the fusion of engineered zinc-finger proteins with either the Herpes simplex–derived transactivation domain (Sadowski et al. 1988) or the Krüppel-associated box (KRAB) repression protein (Margolin et al. 1994). Over the course of the next 15 years, zinc-finger-based transcriptional modulators were expanded and featured several other types of effector domains (Beerli and Barbas 2002), including, for example, the Dnmt3a methyltransferase domain (Rivenbark et al. 2012; Siddique et al. 2013) and the ten-eleven translocation methylcytosine dioxygenase 1 (TET1) (Chen et al. 2014), which can modulate transcription via targeted methylation or demethylation, respectively. As a natural extension of zinc-finger transcription factors, and further drawing on the parallels with zinc-finger proteins, TALE transcription factors have also emerged as an especially effective platform for achieving targeted transcriptional modulation (Miller et al. 2011; Zhang et al. 2011). Effector domains are generally fused to the carboxyl terminus of the synthetic TALE array and, contrary to the longer sequence typically required for efficient modulation by zinc-finger transcription factors, TALEs have been reported to regulate gene expression with as few as 10.5 repeats (Boch et al. 2009). Like zinc fingers, TALEs are also compatible with numerous epigenetic modifiers, including the TET1 hydroxylase catalytic domain (Maeder et al. 2013b) and the lysine-specific histone demethylase 1 (LSD1) (Mendenhall et al. 2013) domains, which have been used for targeted CpG demethylation and histone demethylation, respectively. In particular, the ease with which a large number of TALEs can be constructed has enabled the discovery that tiling a promoter sequence with combinations of synthetic transcription factors can lead to a synergistic increase in gene expression (Maeder et al. 2013b; Perez-Pinera et al. 2013). And, like zinc fingers (Beerli et al. 2000b; Pollock et al. 2002; Magnenat et al. 2008; Polstein and Gersbach 2012), TALE activators have also been successfully engineered to regulate gene expression in response to external (Mercer et al. 2014) or endogenous (Li et al. 2012) chemical stimuli, optical signals (Konermann et al. 2013), and even proteolytic cues (Copeland et al. 2016; Lonzaric et al. 2016).
Because of the exquisite ease with which it can be programmed, the CRISPR-Cas9 system has also been adapted for transcriptional modulation through fusion of specific effector domains to a catalytically inactivated variant of the Cas9 protein (Qi et al. 2013). Deactivation is achieved by introducing two amino acid substitutions, D10A and H840A, into the RuvC and NHN endonuclease domains of Cas9, respectively. Although unable to cleave DNA, this mutant, referred to as dCas9, retains its ability to bind DNA in an RNA-directed manner. Effector domains are fused to the carboxyl terminus of the dCas9 protein and can modulate gene expression from either strand of the targeted DNA sequence (Farzadfard et al. 2013; Maeder et al. 2013a; Perez-Pinera et al. 2013). Genome-scale activation studies have indicated that the most robust levels of activation are generally observed when dCas9 activators are targeted to -400 to -50 bp upstream from the transcriptional start site (Gilbert et al. 2014; Hu et al. 2014). Additionally, dCas9 can inhibit gene expression by simply blocking transcriptional initiation or elongation through a process known as CRISPR interference (Qi et al. 2013), although fusing dCas9 to transcriptional repressor domains can also lead to efficient silencing from the promoter (Gilbert et al. 2013; Zalatan et al. 2015). Much like zinc fingers and TALEs, methods for achieving conditional gene modulation using dCas9 have also been reported, including the fusion of a dihydrofolate reductase destabilization domain to dCas9, which can provide chemical control over activation, enabling cellular reprogramming or differentiation (Balboa et al. 2015). Light-inducible dCas9-based systems capable of providing optical control of gene expression provide another means for achieving conditional control of gene expression (Nihongaki et al. 2015; Polstein and Gersbach 2015).
Although flexible, first-generation dCas9 activators were routinely found to display suboptimal levels of activation. As a result, the development of second-generation CRISPR activators quickly emerged as a highly active area of research. One particularly elegant approach for overcoming the low activation thresholds inherent within first-generation systems was by strategically inserting an RNA aptamer within a functionally inert region of the gRNA. This aptamer recruits specific activation helper proteins that work in concert with a dCas9 activator to enhance transcription (Konermann et al. 2015; Zalatan et al. 2015). Other strategies based on directly fusing additional helper activation domains to dCas9 have also been shown to enhance transcription (Chavez et al. 2015). Targeted acetylation of histone proteins within a promoter or enhancer sequence via epigenome editing using the catalytic core of the human acetyltransferase p300 fused to dCas9 can also lead to robust levels of gene activation (Hilton et al. 2015). Similarly, dCas9 repressor proteins targeted to distal regulatory elements have been found to facilitate chromatin remodeling and gene repression via epigenomic modification (Thakore et al. 2015). Finally, by simply reducing the length of the gRNA, catalytically active variants of Cas9 can stimulate transcription without inducing DNA breaks (Dahlman et al. 2015; Kiani et al. 2015), enabling orthogonal gene knockout and activation with the same Cas9 variant in a single cell.
Applications of Targeted Transcriptional Regulation
Early work on the use of engineered zinc-finger transcription factors revealed that synthetic transcriptional modulators are effective tools for a broad range of applications, enabling such tasks as inhibiting viral replication (Papworth et al. 2003; Reynolds et al. 2003; Segal et al. 2004; Eberhardy et al. 2006), modulating the expression of disease-associated loci (Graslund et al. 2005; Wilber et al. 2010), inducing angiogenesis for accelerated wound healing (Rebar et al. 2002), and genomic screening of cellular targets for cancer progression and drug resistance (Park et al. 2003; Blancafort et al. 2005, 2008). Facilitated by many of the insights gained from zinc-finger transcription factor technology, both TALEs and CRISPR-Cas9 have now further expanded the possibilities of engineered transcriptional activators and repressors. For example, TALEs and CRISPR-Cas9 have enabled rapid construction of custom genetic circuits and logic gates (Gaber et al. 2014; Lebar et al. 2014; Liu et al. 2014b), complex gene regulation networks (Nielsen and Voigt 2014; Nissim et al. 2014), and even facilitated cellular reprogramming (Gao et al. 2013) and the differentiation of mouse embryonic fibroblasts to skeletal myocytes (Chakraborty et al. 2014). dCas9 transcriptional effectors have even been used to efficiently mediate repression and activation of endogenous genes in Drosophila (Lin et al. 2015b) and in plant cells (Piatek et al. 2015). Both TALE and Cas9 activators have also been configured to stimulate transcription of latent HIV (Zhang et al. 2015; Ji et al. 2016; Limsirichai et al. 2016; Perdigao et al. 2016; Saayman et al. 2016), indicating their potential to work in concert with antiretroviral therapy for eradicating HIV infection. Importantly, because of the ease with which the CRISPR-Cas9 system can be used, genome-wide screens using Cas9 transcriptional activators (Gilbert et al. 2014; Konermann et al. 2015) and repressors (Gilbert et al. 2014) can be easily implemented to discover genes involved in a number of diverse processes, including drug resistance and cancer metastasis. In particular, CRISPR-based genome-scale screening methods have the potential to overcome many of the technical hurdles associated with other contemporary screening technologies, such as cDNA libraries and RNAi, indicating its potential for facilitating drug discovery and basic biological research.
CONCLUSIONS
Despite the successes already achieved, many challenges remain before the full potential of genome editing can be realized. First and foremost are the development of new tools capable of introducing genomic modifications in the absence of DNA breaks. Targeted recombinases (Akopian et al. 2003; Mercer et al. 2012), which can be programmed to recognize specific DNA sequences (Gaj et al. 2013; Sirk et al. 2014; Wallen et al. 2015) and even integrate therapeutic factors into the human genome (Gaj et al. 2014b), are one such option. More recent work has indicated that single-base editing without DNA breaks can be achieved using an engineered Cas9 nickase complex (Komor et al. 2016), although it remains unknown how effective this technology is in therapeutically relevant settings. By linking genomic modifications induced by targeted nucleases to their own self-degradation, self-inactivating vectors are also poised to improve the specificity of genome editing, especially because the frequency of off-target modifications can be directly proportional to the duration of cellular exposure to a nuclease (Pruett-Miller et al. 2009). In addition, much of the knowledge behind genome engineering has been obtained in immortalized cell lines. However, in the case of regenerative medicine, it is highly desirable to genetically manipulate progenitor or stem-cell populations, both of which can differ markedly from transformed cell lines with respect to their epigenome or three-dimensional organization of their genomic DNA. These differences can have profound effects on the ability of genome-editing tools to either modify a specific sequence or regulate gene expression. Although the union between genome engineering and regenerative medicine is still in its infancy, realizing the full potential of these technologies in stem/progenitor cells requires that their functional landscape be fully explored in these genetic backgrounds. Only then will genome editing technologies truly be able to reprogram cell fate and behavior for the next generation of advances in synthetic biology and gene therapy.
ACKNOWLEDGMENTS
We gratefully acknowledge the support and mentorship of the late Carlos F. Barbas, III (1964–2014). This work is supported by the National Institutes of Health (F32GM113446 to T.G.) and ShanghaiTech University (to J.L.). We apologize to those whose important contributions were not cited because of space constraints.
Footnotes
Editors: Daniel G. Gibson, Clyde A. Hutchison III, Hamilton O. Smith, and J. Craig Venter
Additional Perspectives on Synthetic Biology available at www.cshperspectives.org
REFERENCES
- Akbari OS, Bellen HJ, Bier E, Bullock SL, Burt A, Church GM, Cook KR, Duchek P, Edwards OR, Esvelt KM, et al. 2015. Safeguarding gene drive experiments in the laboratory. Science 349: 927–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akopian A, He J, Boocock MR, Stark WM. 2003. Chimeric recombinases with designed DNA sequence recognition. Proc Natl Acad Sci 100: 8688–8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders C, Niewoehner O, Duerst A, Jinek M. 2014. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513: 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anguela XM, Sharma R, Doyon Y, Miller JC, Li H, Haurigot V, Rohde ME, Wong SY, Davidson RJ, Zhou S, et al. 2013. Robust ZFN-mediated genome editing in adult hemophilic mice. Blood 122: 3283–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae KH, Kwon YD, Shin HC, Hwang MS, Ryu EH, Park KS, Yang HY, Lee DK, Lee Y, Park J, et al. 2003. Human zinc fingers as building blocks in the construction of artificial transcription factors. Nat Biotechnol 21: 275–280. [DOI] [PubMed] [Google Scholar]
- Balboa D, Weltner J, Eurola S, Trokovic R, Wartiovaara K, Otonkoski T. 2015. Conditionally stabilized dCas9 activator for controlling gene expression in human cell reprogramming and differentiation. Stem Cell Rep 5: 448–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltes NJ, Voytas DF. 2015. Enabling plant synthetic biology through genome engineering. Trends Biotechnol 33: 120–131. [DOI] [PubMed] [Google Scholar]
- Beerli RR, Barbas CF III. 2002. Engineering polydactyl zinc-finger transcription factors. Nat Biotechnol 20: 135–141. [DOI] [PubMed] [Google Scholar]
- Beerli RR, Segal DJ, Dreier B, Barbas CF III. 1998. Toward controlling gene expression at will: Specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc Natl Acad Sci 95: 14628–14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli RR, Dreier B, Barbas CF III. 2000a. Positive and negative regulation of endogenous genes by designed transcription factors. Proc Natl Acad Sci 97: 1495–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli RR, Schopfer U, Dreier B, Barbas CF III. 2000b. Chemically regulated zinc finger transcription factors. J Biol Chem 275: 32617–32627. [DOI] [PubMed] [Google Scholar]
- Bhakta MS, Henry IM, Ousterout DG, Das KT, Lockwood SH, Meckler JF, Wallen MC, Zykovich A, Yu Y, Leo H, et al. 2013. Highly active zinc-finger nucleases by extended modular assembly. Genome Res 23: 530–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, Chandrasegaran S. 2001. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol 21: 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Golic M, Golic KG, Carroll D. 2002. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161: 1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Beumer K, Trautman JK, Carroll D. 2003. Enhancing gene targeting with designed zinc finger nucleases. Science 300: 764. [DOI] [PubMed] [Google Scholar]
- Blancafort P, Chen EI, Gonzalez B, Bergquist S, Zijlstra A, Guthy D, Brachat A, Brakenhoff RH, Quigley JP, Erdmann D, et al. 2005. Genetic reprogramming of tumor cells by zinc finger transcription factors. Proc Natl Acad Sci 102: 11716–11721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blancafort P, Tschan MP, Bergquist S, Guthy D, Brachat A, Sheeter DA, Torbett BE, Erdmann D, Barbas CF III. 2008. Modulation of drug resistance by artificial transcription factors. Mol Cancer Ther 7: 688–697. [DOI] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. 2009. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326: 1509–1512. [DOI] [PubMed] [Google Scholar]
- Boissel S, Jarjour J, Astrakhan A, Adey A, Gouble A, Duchateau P, Shendure J, Stoddard BL, Certo MT, Baker D, et al. 2014. megaTALs: A rare-cleaving nuclease architecture for therapeutic genome engineering. Nucleic Acids Res 42: 2591–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolukbasi MF, Gupta A, Oikemus S, Derr AG, Garber M, Brodsky MH, Zhu LJ, Wolfe SA. 2015. DNA-binding-domain fusions enhance the targeting range and precision of Cas9. Nat Methods 12: 1150–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs AW, Rios X, Chari R, Yang L, Zhang F, Mali P, Church GM. 2012. Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res 40: e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A. 2003. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc Biol Sci 270: 921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Bak RO, Mikkelsen JG. 2014. Targeted genome editing by lentiviral protein transduction of zinc-finger and TAL-effector nucleases. eLife 3: e01911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, Voytas DF, Long CR, Whitelaw CB, Fahrenkrug SC. 2012. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci 109: 17382–17387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. 2011. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Ji H, Kabadi AM, Gersbach CA, Christoforou N, Leong KW. 2014. A CRISPR/Cas9-based system for reprogramming cell lineage specification. Stem Cell Rep 3: 940–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, Iyer EPR, Lin S, Kiani S, Guzman CD, Wiegand DJ, et al. 2015. Highly efficient Cas9-mediated transcriptional programming. Nat Methods 12: 326–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Kazemier HG, de Groote ML, Ruiters MH, Xu GL, Rots MG. 2014. Induced DNA demethylation by targeting Ten-Eleven Translocation 2 to the human ICAM-1 promoter. Nucleic Acids Res 42: 1563–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, Scott DA, Song J, Pan JQ, Weissleder R, et al. 2015. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160: 1246–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. 2013. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31: 230–232. [DOI] [PubMed] [Google Scholar]
- Choo Y, Klug A. 1994. Toward a code for the interactions of zinc fingers with DNA: Selection of randomized fingers displayed on phage. Proc Natl Acad Sci 91: 11163–11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choulika A, Perrin A, Dujon B, Nicolas JF. 1995. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol Cell Biol 15: 1968–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. 2010. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186: 757–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Zhou R, Kuo YC, Cunniff M, Zhang F. 2012. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat Commun 3: 968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland MF, Politz MC, Johnson CB, Markley AL, Pfleger BF. 2016. A transcription activator-like effector (TALE) induction system mediated by proteolysis. Nat Chem Biol 12: 254–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DB, Platt RJ, Zhang F. 2015. Therapeutic genome editing: Prospects and challenges. Nat Med 21: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cradick TJ, Fine EJ, Antico CJ, Bao G. 2013. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res 41: 9584–9592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Ji D, Fisher DA, Wu Y, Briner DM, Weinstein EJ. 2011. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat Biotechnol 29: 64–67. [DOI] [PubMed] [Google Scholar]
- Dahlman JE, Abudayyeh OO, Joung J, Gootenberg JS, Zhang F, Konermann S. 2015. Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease. Nat Biotechnol 33: 1159–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KM, Pattanayak V, Thompson DB, Zuris JA, Liu DR. 2015. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat Chem Biol 11: 316–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng D, Yan C, Pan X, Mahfouz M, Wang J, Zhu JK, Shi Y, Yan N. 2012. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science 335: 720–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo JE, Chavez A, Dietz SL, Esvelt KM, Church GM. 2015. Safeguarding CRISPR-Cas9 gene drives in yeast. Nat Biotechnol 33: 1250–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, et al. 2008. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol 26: 702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD, Holmes MC. 2011. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods 8: 74–79. [DOI] [PubMed] [Google Scholar]
- Dreier B, Beerli RR, Segal DJ, Flippin JD, Barbas CF III. 2001. Development of zinc finger domains for recognition of the 5′-ANN-3′ family of DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem 276: 29466–29478. [DOI] [PubMed] [Google Scholar]
- Dreier B, Fuller RP, Segal DJ, Lund CV, Blancafort P, Huber A, Koksch B, Barbas CF III. 2005. Development of zinc finger domains for recognition of the 5′-CNN-3′ family DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem 280: 35588–35597. [DOI] [PubMed] [Google Scholar]
- Eberhardy SR, Goncalves J, Coelho S, Segal DJ, Berkhout B, Barbas CF III. 2006. Inhibition of human immunodeficiency virus type 1 replication with artificial transcription factors targeting the highly conserved primer-binding site. J Virol 80: 2873–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. 2013. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods 10: 1116–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt KM, Smidler AL, Catteruccia F, Church GM. 2014. Concerning RNA-guided gene drives for the alteration of wild populations. eLife 10.7554/eLife.03401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzadfard F, Perli SD, Lu TK. 2013. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth Biol 2: 604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J. 2014. Saturation editing of genomic regions by multiplex homology-directed repair. Nature 513: 120–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. 2013. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31: 822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. 2014. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32: 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaber R, Lebar T, Majerle A, Ster B, Dobnikar A, Bencina M, Jerala R. 2014. Designable DNA-binding domains enable construction of logic circuits in mammalian cells. Nat Chem Biol 10: 203–208. [DOI] [PubMed] [Google Scholar]
- Gabriel R, Lombardo A, Arens A, Miller JC, Genovese P, Kaeppel C, Nowrouzi A, Bartholomae CC, Wang J, Friedman G, et al. 2011. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol 29: 816–823. [DOI] [PubMed] [Google Scholar]
- Gaj T, Guo J, Kato Y, Sirk SJ, Barbas CF III. 2012. Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat Methods 9: 805–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Mercer AC, Sirk SJ, Smith HL, Barbas CF III. 2013. A comprehensive approach to zinc-finger recombinase customization enables genomic targeting in human cells. Nucleic Acids Res 41: 3937–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Liu J, Anderson KE, Sirk SJ, Barbas CF III. 2014a. Protein delivery using Cys2-His2 zinc-finger domains. ACS Chem Biol 9: 1662–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Sirk SJ, Tingle RD, Mercer AC, Wallen MC, Barbas CF III. 2014b. Enhancing the specificity of recombinase-mediated genome engineering through dimer interface redesign. J Am Chem Soc 136: 5047–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Epstein BE, Schaffer DV. 2016. Genome engineering using adeno-associated virus: Basic and clinical research applications. Mol Ther 24: 458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantz VM, Bier E. 2015. The mutagenic chain reaction: A method for converting heterozygous to homozygous mutations. Science 348: 442–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantz VM, Jasinskiene N, Tatarenkova O, Fazekas A, Macias VM, Bier E, James AA. 2015. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc Natl Acad Sci 112: E6736–E6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Yang J, Tsang JC, Ooi J, Wu D, Liu P. 2013. Reprogramming to pluripotency using designer TALE transcription factors targeting enhancers. Stem Cell Rep 1: 183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, et al. 2009. Knockout rats via embryo microinjection of zinc-finger nucleases. Science 325: 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio JJ. 2014. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol 32: 819–821. [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154: 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. 2014. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159: 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graslund T, Li X, Magnenat L, Popkov M, Barbas CF III. 2005. Exploring strategies for the design of artificial transcription factors: Targeting sites proximal to known regulatory regions for the induction of γ-globin expression and the treatment of sickle cell disease. J Biol Chem 280: 3707–3714. [DOI] [PubMed] [Google Scholar]
- Greisman HA, Pabo CO. 1997. A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites. Science 275: 657–661. [DOI] [PubMed] [Google Scholar]
- Guilinger JP, Pattanayak V, Reyon D, Tsai SQ, Sander JD, Joung JK, Liu DR. 2014a. Broad specificity profiling of TALENs results in engineered nucleases with improved DNA-cleavage specificity. Nat Methods 11: 429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger JP, Thompson DB, Liu DR. 2014b. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol 32: 577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Gaj T, Barbas CF III. 2010. Directed evolution of an enhanced and highly efficient FokI cleavage domain for zinc finger nucleases. J Mol Biol 400: 96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Christensen RG, Rayla AL, Lakshmanan A, Stormo GD, Wolfe SA. 2012. An optimized two-finger archive for ZFN-mediated gene targeting. Nat Methods 9: 588–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond A, Galizi R, Kyrou K, Simoni A, Siniscalchi C, Katsanos D, Gribble M, Baker D, Marois E, Russell S, et al. 2016. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat Biotechnol 34: 78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, et al. 2015. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163: 1515–1526. [DOI] [PubMed] [Google Scholar]
- Haun W, Coffman A, Clasen BM, Demorest ZL, Lowy A, Ray E, Retterath A, Stoddard T, Juillerat A, Cedrone F, et al. 2014. Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol J 12: 934–940. [DOI] [PubMed] [Google Scholar]
- Hauschild J, Petersen B, Santiago Y, Queisser AL, Carnwath JW, Lucas-Hahn A, Zhang L, Meng X, Gregory PD, Schwinzer R, et al. 2011. Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proc Natl Acad Sci 108: 12013–12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heigwer F, Kerr G, Boutros M. 2014. E-CRISP: Fast CRISPR target site identification. Nat Methods 11: 122–123. [DOI] [PubMed] [Google Scholar]
- Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, Steinfeld I, Lunstad BD, Kaiser RJ, Wilkens AB, et al. 2015. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol 33: 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton IB, Gersbach CA. 2015. Enabling functional genomics with genome engineering. Genome Res 25: 1442–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE. 2015. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33: 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, et al. 2011. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 29: 731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holkers M, Maggio I, Liu J, Janssen JM, Miselli F, Mussolino C, Recchia A, Cathomen T, Goncalves MA. 2013. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res 41: e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holkers M, Maggio I, Henriques SF, Janssen JM, Cathomen T, Goncalves MA. 2014. Adenoviral vector DNA for accurate genome editing with engineered nucleases. Nat Methods 11: 1051–1057. [DOI] [PubMed] [Google Scholar]
- Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, Crooks GM, Kohn DB, Gregory PD, Holmes MC, et al. 2010. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol 28: 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327: 167–170. [DOI] [PubMed] [Google Scholar]
- Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA. 2013. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci 110: 15644–15649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31: 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Lei Y, Wong WK, Liu S, Lee KC, He X, You W, Zhou R, Guo JT, Chen X, et al. 2014. Direct activation of human and mouse Oct4 genes using engineered TALE and Cas9 transcription factors. Nucleic Acids Res 42: 4375–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard BP, Badran AH, Zuris JA, Guilinger JP, Davis KM, Chen L, Tsai SQ, Sander JD, Joung JK, Liu DR. 2015. Continuous directed evolution of DNA-binding proteins to improve TALEN specificity. Nat Methods 12: 939–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt JA, Thibodeau SA, Hirsh AS, Pabo CO, Joung JK. 2003. Highly specific zinc finger proteins obtained by directed domain shuffling and cell-based selection. Proc Natl Acad Sci 100: 12271–12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. 2013. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31: 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isalan M, Klug A, Choo Y. 2001. A rapid, generally applicable method to engineer zinc fingers illustrated by targeting the HIV-1 promoter. Nat Biotechnol 19: 656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakociunas T, Bonde I, Herrgard M, Harrison SJ, Kristensen M, Pedersen LE, Jensen MK, Keasling JD. 2015a. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab Eng 28: 213–222. [DOI] [PubMed] [Google Scholar]
- Jakociunas T, Rajkumar AS, Zhang J, Arsovska D, Rodriguez A, Jendresen CB, Skjodt ML, Nielsen AT, Borodina I, Jensen MK, et al. 2015b. CasEMBLR: Cas9-facilitated multiloci genomic integration of in vivo assembled DNA parts in Saccharomyces cerevisiae. ACS Synth Biol 4: 1226–1234. [DOI] [PubMed] [Google Scholar]
- Jamieson AC, Kim SH, Wells JA. 1994. In vitro selection of zinc fingers with altered DNA-binding specificity. Biochemistry 33: 5689–5695. [DOI] [PubMed] [Google Scholar]
- Ji H, Jiang Z, Lu P, Ma L, Li C, Pan H, Fu Z, Qu X, Wang P, Deng J, et al. 2016. Specific reactivation of latent HIV-1 by dCas9-SunTag-VP64-mediated guide RNA targeting the HIV-1 promoter. Mol Ther 24: 508–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. 2013. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31: 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Taylor DW, Chen JS, Kornfeld JE, Zhou K, Thompson AJ, Nogales E, Doudna JA. 2016. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 351: 867–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. 2013. RNA-programmed genome editing in human cells. eLife 2: e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juillerat A, Pessereau C, Dubois G, Guyot V, Marechal A, Valton J, Daboussi F, Poirot L, Duclert A, Duchateau P. 2015. Optimized tuning of TALEN specificity using non-conventional RVDs. Sci Rep 5: 8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiani S, Chavez A, Tuttle M, Hall RN, Chari R, Ter-Ovanesyan D, Qian J, Pruitt BW, Beal J, Vora S, et al. 2015. Cas9 gRNA engineering for genome editing, activation and repression. Nat Methods 12: 1051–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Cha J, Chandrasegaran S. 1996. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci 93: 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. 2009. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res 19: 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Kim S, Kim DH, Choi BS, Choi IY, Kim JS. 2012. Precision genome engineering with programmable DNA-nicking enzymes. Genome Res 22: 1327–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Kweon J, Kim A, Chon JK, Yoo JY, Kim HJ, Kim S, Lee C, Jeong E, Chung E, et al. 2013a. A library of TAL effector nucleases spanning the human genome. Nat Biotechnol 31: 251–258. [DOI] [PubMed] [Google Scholar]
- Kim YK, Wee G, Park J, Kim J, Baek D, Kim JS, Kim VN. 2013b. TALEN-based knockout library for human microRNAs. Nat Struct Mol Biol 20: 1458–1464. [DOI] [PubMed] [Google Scholar]
- Kim S, Kim D, Cho SW, Kim J, Kim JS. 2014. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 24: 1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales AP, Li Z, Peterson RT, Yeh JR, et al. 2015. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523: 481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. 2016. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529: 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C, Yusa K. 2014. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol 32: 267–273. [DOI] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. 2016. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. 2013. Optical control of mammalian endogenous transcription and epigenetic states. Nature 500: 472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. 2015. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517: 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkmaz G, Lopes R, Ugalde AP, Nevedomskaya E, Han R, Myacheva K, Zwart W, Elkon R, Agami R. 2016. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat Biotechnol 34: 192–198. [DOI] [PubMed] [Google Scholar]
- Lebar T, Bezeljak U, Golob A, Jerala M, Kadunc L, Pirs B, Strazar M, Vucko D, Zupancic U, Bencina M, et al. 2014. A bistable genetic switch based on designable DNA-binding domains. Nat Commun 5: 5007. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Kim E, Kim JS. 2010. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res 20: 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Haurigot V, Doyon Y, Li T, Wong SY, Bhagwat AS, Malani N, Anguela XM, Sharma R, Ivanciu L, et al. 2011. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature 475: 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Moore R, Guinn M, Bleris L. 2012. Transcription activator-like effector hybrids for conditional control and rewiring of chromosomal transgene expression. Sci Rep 2: 897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Teng F, Li T, Zhou Q. 2013. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat Biotechnol 31: 684–686. [DOI] [PubMed] [Google Scholar]
- Limsirichai P, Gaj T, Schaffer DV. 2016. CRISPR-mediated activation of latent HIV-1 expression. Mol Ther 24: 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SR, Yang HC, Kuo YT, Liu CJ, Yang TY, Sung KC, Lin YY, Wang HY, Wang CC, Shen YC, et al. 2014. The CRISPR/Cas9 system facilitates clearance of the intrahepatic HBV templates in vivo. Mol Ther Nucleic Acids 3: e186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Chen H, Luo L, Lai Y, Xie W, Kee K. 2015a. Creating a monomeric endonuclease TALE-I-SceI with high specificity and low genotoxicity in human cells. Nucleic Acids Res 43: 1112–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Ewen-Campen B, Ni X, Housden BE, Perrimon N. 2015b. In vivo transcriptional activation using CRISPR/Cas9 in Drosophila. Genetics 201: 433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Segal DJ, Ghiara JB, Barbas CF III. 1997. Design of polydactyl zinc-finger proteins for unique addressing within complex genomes. Proc Natl Acad Sci 94: 5525–5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Gaj T, Patterson JT, Sirk SJ, Barbas CF III. 2014a. Cell-penetrating peptide-mediated delivery of TALEN proteins via bioconjugation for genome engineering. PLoS ONE 9: e85755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zeng Y, Liu L, Zhuang C, Fu X, Huang W, Cai Z. 2014b. Synthesizing AND gate genetic circuits based on CRISPR-Cas9 for identification of bladder cancer cells. Nat Commun 5: 5393. [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhou X, Zhu Y, Chen ZF, Yu B, Wang Y, Zhang CC, Nie YH, Sang X, Cai YJ, et al. 2014c. Generation of a monkey with MECP2 mutations by TALEN-based gene targeting. Neurosci Bull 30: 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Gaj T, Wallen MC, Barbas CF III. 2015a. Improved cell-penetrating zinc-finger nuclease proteins for precision genome engineering. Mol Ther Nucleic Acids 4: e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Gaj T, Yang Y, Wang N, Shui S, Kim S, Kanchiswamy CN, Kim JS, Barbas CF III. 2015b. Efficient delivery of nuclease proteins for genome editing in human stem cells and primary cells. Nat Protoc 10: 1842–1859. [DOI] [PubMed] [Google Scholar]
- Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, Olson EN. 2015. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351: 400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonzaric J, Lebar T, Majerle A, Mancek-Keber M, Jerala R. 2016. Locked and proteolysis-based transcription activator-like effector (TALE) regulation. Nucleic Acids Res 44: 1471–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Li J, Stoddard TJ, Baltes NJ, Demorest ZL, Clasen BM, Coffman A, Retterath A, Mathis L, Voytas DF, et al. 2015. Non-transgenic plant genome editing using purified sequence-specific nucleases. Mol Plant 8: 1425–1427. [DOI] [PubMed] [Google Scholar]
- Ma H, Dang Y, Wu Y, Jia G, Anaya E, Zhang J, Abraham S, Choi JG, Shi G, Qi L, et al. 2015. A CRISPR-based screen identifies genes essential for West-Nile-virus-induced cell death. Cell Rep 12: 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Gersbach CA. 2016. Genome-editing technologies for gene and cell therapy. Mol Ther 24: 430–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. 2008. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell 31: 294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. 2013a. CRISPR RNA-guided activation of endogenous human genes. Nat Methods 10: 977–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Linder SJ, Reyon D, Angstman JF, Fu Y, Sander JD, Joung JK. 2013b. Robust, synergistic regulation of human gene expression using TALE activators. Nat Methods 10: 243–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio I, Stefanucci L, Janssen JM, Liu J, Chen X, Mouly V, Goncalves MA. 2016. Selection-free gene repair after adenoviral vector transduction of designer nucleases: rescue of dystrophin synthesis in DMD muscle cell populations. Nucleic Acids Res 44: 1449–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnenat L, Blancafort P, Barbas CF III. 2004. In vivo selection of combinatorial libraries and designed affinity maturation of polydactyl zinc finger transcription factors for ICAM-1 provides new insights into gene regulation. J Mol Biol 341: 635–649. [DOI] [PubMed] [Google Scholar]
- Magnenat L, Schwimmer LJ, Barbas CF III. 2008. Drug-inducible and simultaneous regulation of endogenous genes by single-chain nuclear receptor-based zinc-finger transcription factor gene switches. Gene Ther 15: 1223–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahiny AJ, Dewerth A, Mays LE, Alkhaled M, Mothes B, Malaeksefat E, Loretz B, Rottenberger J, Brosch DM, Reautschnig P, et al. 2015. In vivo genome editing using nuclease-encoding mRNA corrects SP-B deficiency. Nat Biotechnol 33: 584–586. [DOI] [PubMed] [Google Scholar]
- Mak AN, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL. 2012. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science 335: 716–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. 2013a. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 31: 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013b. RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin JF, Friedman JR, Meyer WK, Vissing H, Thiesen HJ, Rauscher FJ III. 1994. Kruppel-associated boxes are potent transcriptional repression domains. Proc Natl Acad Sci 91: 4509–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet 11: 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell Smith A, Takeuchi R, Pellenz S, Davis L, Maizels N, Monnat RJ Jr, Stoddard BL. 2009. Generation of a nicking enzyme that stimulates site-specific gene conversion from the I-AniI LAGLIDADG homing endonuclease. Proc Natl Acad Sci 106: 5099–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckler JF, Bhakta MS, Kim MS, Ovadia R, Habrian CH, Zykovich A, Yu A, Lockwood SH, Morbitzer R, Elsaesser J, et al. 2013. Quantitative analysis of TALE-DNA interactions suggests polarity effects. Nucleic Acids Res 41: 4118–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM, Williamson KE, Reyon D, Zou JY, Ram O, Joung JK, Bernstein BE. 2013. Locus-specific editing of histone modifications at endogenous enhancers. Nat Biotechnol 31: 1133–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menoret S, De Cian A, Tesson L, Remy S, Usal C, Boule JB, Boix C, Fontaniere S, Creneguy A, Nguyen TH, et al. 2015. Homology-directed repair in rodent zygotes using Cas9 and TALEN engineered proteins. Sci Rep 5: 14410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer AC, Gaj T, Fuller RP, Barbas CF 3rd. 2012. Chimeric TALE recombinases with programmable DNA sequence specificity. Nucleic Acids Res 40: 11163–11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer AC, Gaj T, Sirk SJ, Lamb BM, Barbas CF III. 2014. Regulation of endogenous human gene expression by ligand-inducible TALE transcription factors. ACS Synth Biol 3: 723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. 2007. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol 25: 778–785. [DOI] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. 2011. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29: 143–148. [DOI] [PubMed] [Google Scholar]
- Miller JC, Zhang L, Xia DF, Campo JJ, Ankoudinova IV, Guschin DY, Babiarz JE, Meng X, Hinkley SJ, Lam SC, et al. 2015. Improved specificity of TALE-based genome editing using an expanded RVD repertoire. Nat Methods 12: 465–471. [DOI] [PubMed] [Google Scholar]
- Mock U, Machowicz R, Hauber I, Horn S, Abramowski P, Berdien B, Hauber J, Fehse B. 2015. mRNA transfection of a novel TAL effector nuclease (TALEN) facilitates efficient knockout of HIV co-receptor CCR5. Nucleic Acids Res 43: 5560–5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle EA, Rock JM, Lee YL, Jouvenot Y, DeKelver RC, Gregory PD, Urnov FD, Holmes MC. 2007. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci 104: 3055–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. 2009. A simple cipher governs DNA recognition by TAL effectors. Science 326: 1501. [DOI] [PubMed] [Google Scholar]
- Mussolino C, Alzubi J, Fine EJ, Morbitzer R, Cradick TJ, Lahaye T, Bao G, Cathomen T. 2014. TALENs facilitate targeted genome editing in human cells with high specificity and low cytotoxicity. Nucleic Acids Res 42: 6762–6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Rivera RMC, Madhavan S, Pan X, Ran FA, Yan WX, et al. 2015. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351: 403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AA, Voigt CA. 2014. Multi-input CRISPR/Cas genetic circuits that interface host regulatory networks. Mol Syst Biol 10: 763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihongaki Y, Yamamoto S, Kawano F, Suzuki H, Sato M. 2015. CRISPR-Cas9-based photoactivatable transcription system. Chem Biol 22: 169–174. [DOI] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. 2014. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156: 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK. 2014. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol Cell 54: 698–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn MJ, Webber BR, Knipping F, Lonetree CL, Tennis N, DeFeo AP, McElroy AN, Starker CG, Lee C, Merkel S, et al. 2016. Evaluation of TCR gene editing achieved by TALENs, CRISPR/Cas9, and megaTAL nucleases. Mol Ther 24: 570–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Rasoloson D, Seydoux G. 2015. High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics 201: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papworth M, Moore M, Isalan M, Minczuk M, Choo Y, Klug A. 2003. Inhibition of herpes simplex virus 1 gene expression by designer zinc-finger transcription factors. Proc Natl Acad Sci 100: 1621–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KS, Lee DK, Lee H, Lee Y, Jang YS, Kim YH, Yang HY, Lee SI, Seol W, Kim JS. 2003. Phenotypic alteration of eukaryotic cells using randomized libraries of artificial transcription factors. Nat Biotechnol 21: 1208–1214. [DOI] [PubMed] [Google Scholar]
- Parnas O, Jovanovic M, Eisenhaure TM, Herbst RH, Dixit A, Ye CJ, Przybylski D, Platt RJ, Tirosh I, Sanjana NE, et al. 2015. A genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell 162: 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V, Ramirez CL, Joung JK, Liu DR. 2011. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods 8: 765–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich NP, Pabo CO. 1991. Zinc finger-DNA recognition: Crystal structure of a Zif268-DNA complex at 2.1 A. Science 252: 809–817. [DOI] [PubMed] [Google Scholar]
- Perdigao P, Gaj T, Santa-Marta M, Barbas CF III, Goncalves J. 2016. Reactivation of latent HIV-1 expression by engineered TALE transcription factors. PLoS ONE 11: e0150037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, et al. 2008. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol 26: 808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P, Ousterout DG, Brunger JM, Farin AM, Glass KA, Guilak F, Crawford GE, Hartemink AJ, Gersbach CA. 2013. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nat Methods 10: 239–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatek A, Ali Z, Baazim H, Li L, Abulfaraj A, Al-Shareef S, Aouida M, Mahfouz MM. 2015. RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol J 13: 578–589. [DOI] [PubMed] [Google Scholar]
- Pollock R, Giel M, Linher K, Clackson T. 2002. Regulation of endogenous gene expression with a small-molecule dimerizer. Nat Biotechnol 20: 729–733. [DOI] [PubMed] [Google Scholar]
- Polstein LR, Gersbach CA. 2012. Light-inducible spatiotemporal control of gene activation by customizable zinc finger transcription factors. J Am Chem Soc 134: 16480–16483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polstein LR, Gersbach CA. 2015. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol 11: 198–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus MH. 2015. Towards a new era in medicine: Therapeutic genome editing. Genome Biol 16: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus MH, Baltimore D. 2003. Chimeric nucleases stimulate gene targeting in human cells. Science 300: 763. [DOI] [PubMed] [Google Scholar]
- Porteus MH, Carroll D. 2005. Gene targeting using zinc finger nucleases. Nat Biotechnol 23: 967–973. [DOI] [PubMed] [Google Scholar]
- Pruett-Miller SM, Reading DW, Porter SN, Porteus MH. 2009. Attenuation of zinc finger nuclease toxicity by small-molecule regulation of protein levels. PLoS Genet 5: e1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahdar M, McMahon MA, Prakash TP, Swayze EE, Bennett CF, Cleveland DW. 2015. Synthetic CRISPR RNA-Cas9-guided genome editing in human cells. Proc Natl Acad Sci 112: E7110–E7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez CL, Certo MT, Mussolino C, Goodwin MJ, Cradick TJ, McCaffrey AP, Cathomen T, Scharenberg AM, Joung JK. 2012. Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Res 40: 5560–5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, et al. 2013. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154: 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et al. 2015. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520: 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebar EJ, Huang Y, Hickey R, Nath AK, Meoli D, Nath S, Chen B, Xu L, Liang Y, Jamieson AC, et al. 2002. Induction of angiogenesis in a mouse model using engineered transcription factors. Nat Med 8: 1427–1432. [DOI] [PubMed] [Google Scholar]
- Reynolds L, Ullman C, Moore M, Isalan M, West MJ, Clapham P, Klug A, Choo Y. 2003. Repression of the HIV-1 5′ LTR promoter and inhibition of HIV-1 replication by using engineered zinc-finger transcription factors. Proc Natl Acad Sci 100: 1615–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. 2012. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol 30: 460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivenbark AG, Stolzenburg S, Beltran AS, Yuan X, Rots MG, Strahl BD, Blancafort P. 2012. Epigenetic reprogramming of cancer cells via targeted DNA methylation. Epigenetics 7: 350–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet P, Smih F, Jasin M. 1994. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol 14: 8096–8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan OW, Skerker JM, Maurer MJ, Li X, Tsai JC, Poddar S, Lee ME, DeLoache W, Dueber JE, Arkin AP, et al. 2014. Selection of chromosomal DNA libraries using a multiplex CRISPR system. eLife 10.7554/eLife.03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saayman SM, Lazar DC, Scott TA, Hart JR, Takahashi M, Burnett JC, Planelles V, Morris KV, Weinberg MS. 2016. Potent and targeted activation of latent HIV-1 using the CRISPR/dCas9 activator complex. Mol Ther 24: 488–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M. 1988. GAL4-VP16 is an unusually potent transcriptional activator. Nature 335: 563–564. [DOI] [PubMed] [Google Scholar]
- Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR. 2011a. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol 29: 697–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, et al. 2011b. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat Methods 8: 67–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, Holmes MC, Guschin D, Waite A, Miller JC, et al. 2008. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci 105: 5809–5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sather BD, Romano Ibarra GS, Sommer K, Curinga G, Hale M, Khan IF, Singh S, Song Y, Gwiazda K, Sahni J, et al. 2015. Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Sci Transl Med 7: 307ra156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Burgk JL, Schmidt T, Kaiser V, Honing K, Hornung V. 2013. A ligation-independent cloning technique for high-throughput assembly of transcription activator-like effector genes. Nat Biotechnol 31: 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann K, Lin S, Boyer E, Simeonov DR, Subramaniam M, Gate RE, Haliburton GE, Ye CJ, Bluestone JA, Doudna JA, et al. 2015. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc Natl Acad Sci 112: 10437–10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal DJ, Dreier B, Beerli RR, Barbas CF III. 1999. Toward controlling gene expression at will: Selection and design of zinc finger domains recognizing each of the 5′-GNN-3′ DNA target sequences. Proc Natl Acad Sci 96: 2758–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal DJ, Goncalves J, Eberhardy S, Swan CH, Torbett BE, Li X, Barbas CF III. 2004. Attenuation of HIV-1 replication in primary human cells with a designed zinc finger transcription factor. J Biol Chem 279: 14509–14519. [DOI] [PubMed] [Google Scholar]
- Shah AN, Davey CF, Whitebirch AC, Miller AC, Moens CB. 2015. Rapid reverse genetic screening using CRISPR in zebrafish. Nat Methods 12: 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343: 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Anguela XM, Doyon Y, Wechsler T, DeKelver RC, Sproul S, Paschon DE, Miller JC, Davidson RJ, Shivak D, et al. 2015. In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood 126: 1777–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique AN, Nunna S, Rajavelu A, Zhang Y, Jurkowska RZ, Reinhardt R, Rots MG, Ragozin S, Jurkowski TP, Jeltsch A. 2013. Targeted methylation and gene silencing of VEGF-A in human cells by using a designed Dnmt3a-Dnmt3L single-chain fusion protein with increased DNA methylation activity. J Mol Biol 425: 479–491. [DOI] [PubMed] [Google Scholar]
- Silva G, Poirot L, Galetto R, Smith J, Montoya G, Duchateau P, Paques F. 2011. Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr Gene Ther 11: 11–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkins SP, Gould F. 2006. Gene drive systems for insect disease vectors. Nat Rev Genet 7: 427–435. [DOI] [PubMed] [Google Scholar]
- Sirk SJ, Gaj T, Jonsson A, Mercer AC, Barbas CF III. 2014. Expanding the zinc-finger recombinase repertoire: Directed evolution and mutational analysis of serine recombinase specificity determinants. Nucleic Acids Res 42: 4755–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. 2016. Rationally engineered Cas9 nucleases with improved specificity. Science 351: 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Bibikova M, Whitby FG, Reddy AR, Chandrasegaran S, Carroll D. 2000. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res 28: 3361–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Lawrence CM, Wiedenheft B. 2013. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem 82: 237–266. [DOI] [PubMed] [Google Scholar]
- Stoddard BL. 2011. Homing endonucleases: From microbial genetic invaders to reagents for targeted DNA modification. Structure 19: 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard BL. 2014. Homing endonucleases from mobile group I introns: Discovery to genome engineering. Mob DNA 5: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streubel J, Blucher C, Landgraf A, Boch J. 2012. TAL effector RVD specificities and efficiencies. Nat Biotechnol 30: 593–595. [DOI] [PubMed] [Google Scholar]
- Sung YH, Kim JM, Kim HT, Lee J, Jeon J, Jin Y, Choi JH, Ban YH, Ha SJ, Kim CH, et al. 2014. Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Genome Res 24: 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabebordbar M, Zhu K, Cheng JKW, Chew WL, Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, et al. 2015. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 351: 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, Spratt SK, Surosky RT, Giedlin MA, Nichol G, et al. 2014. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med 370: 901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesson L, Usal C, Menoret S, Leung E, Niles BJ, Remy S, Santiago Y, Vincent AI, Meng X, Zhang L, et al. 2011. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol 29: 695–696. [DOI] [PubMed] [Google Scholar]
- Thakore PI, D’Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, Reddy TE, Crawford GE, Gersbach CA. 2015. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods 12: 1143–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Charusanti P, Zhang L, Weber T, Lee SY. 2015. CRISPR-Cas9 based engineering of actinomycetal genomes. ACS Synth Biol 4: 1020–1029. [DOI] [PubMed] [Google Scholar]
- Torres R, Martin MC, Garcia A, Cigudosa JC, Ramirez JC, Rodriguez-Perales S. 2014. Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR-Cas9 system. Nat Commun 5: 3964. [DOI] [PubMed] [Google Scholar]
- Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. 2014. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol 32: 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. 2005. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435: 646–651. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. 2010. Genome editing with engineered zinc finger nucleases. Nat Rev Genet 11: 636–646. [DOI] [PubMed] [Google Scholar]
- Wallen MC, Gaj T, Barbas CF III. 2015. Redesigning recombinase specificity for safe harbor sites in the human genome. PLoS ONE 10: e0139123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Friedman G, Doyon Y, Wang NS, Li CJ, Miller JC, Hua KL, Yan JJ, Babiarz JE, Gregory PD, et al. 2012. Targeted gene addition to a predetermined site in the human genome using a ZFN-based nicking enzyme. Genome Res 22: 1316–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hu YC, Markoulaki S, Welstead GG, Cheng AW, Shivalila CS, Pyntikova T, Dadon DB, Voytas DF, Bogdanove AJ, et al. 2013. TALEN-mediated editing of the mouse Y chromosome. Nat Biotechnol 31: 530–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. 2014a. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343: 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL. 2014b. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32: 947–951. [DOI] [PubMed] [Google Scholar]
- Wang L, Shao Y, Guan Y, Li L, Wu L, Chen F, Liu M, Chen H, Ma Y, Ma X, et al. 2015a. Large genomic fragment deletion and functional gene cassette knock-in via Cas9 protein mediated genome editing in one-cell rodent embryos. Sci Rep 5: 17517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. 2015b. Identification and characterization of essential genes in the human genome. Science 350: 1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA. 2012. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482: 331–338. [DOI] [PubMed] [Google Scholar]
- Wilber A, Tschulena U, Hargrove PW, Kim YS, Persons DA, Barbas CF III, Nienhuis AW. 2010. A zinc-finger transcriptional activator designed to interact with the γ-globin gene promoters enhances fetal hemoglobin production in primary human adult erythroblasts. Blood 115: 3033–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windbichler N, Menichelli M, Papathanos PA, Thyme SB, Li H, Ulge UY, Hovde BT, Baker D, Monnat RJ Jr, Burt A, et al. 2011. A synthetic homing endonuclease-based gene drive system in the human malaria mosquito. Nature 473: 212–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe SA, Nekludova L, Pabo CO. 2000. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct 29: 183–212. [DOI] [PubMed] [Google Scholar]
- Woo JW, Kim J, Kwon SI, Corvalan C, Cho SW, Kim H, Kim SG, Kim ST, Choe S, Kim JS. 2015. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 33: 1162–1164. [DOI] [PubMed] [Google Scholar]
- Wu H, Yang WP, Barbas CF III. 1995. Building zinc fingers by selection: Toward a therapeutic application. Proc Natl Acad Sci 92: 344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Liang D, Wang Y, Bai M, Tang W, Bao S, Yan Z, Li D, Li J. 2013. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell 13: 659–662. [DOI] [PubMed] [Google Scholar]
- Wu H, Wang Y, Zhang Y, Yang M, Lv J, Liu J, Zhang Y. 2015. TALE nickase-mediated SP110 knockin endows cattle with increased resistance to tuberculosis. Proc Natl Acad Sci 112: E1530–E1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A, Wang Z, Hu Y, Wu Y, Luo Z, Yang Z, Zu Y, Li W, Huang P, Tong X, et al. 2013. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res 41: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang Y, Yuan P, Zhou Y, Cai C, Ren Q, Wen D, Chu C, Qi H, Wei W. 2014. Complete decoding of TAL effectors for DNA recognition. Cell Res 24: 628–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang L, Bell P, McMenamin D, He Z, White J, Yu H, Xu C, Morizono H, Musunuru K, et al. 2016. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol 34: 334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. 2014. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol 32: 551–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Song CQ, Dorkin JR, Zhu LJ, Li Y, Wu Q, Park A, Yang J, Suresh S, Bizhanova A, et al. 2016. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol 34: 328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, et al. 2015. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 160: 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, et al. 2015a. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163: 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Volz SE, Zhang F. 2015b. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat Biotechnol 33: 139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. 2011. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol 29: 149–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yin C, Zhang T, Li F, Yang W, Kaminski R, Fagan PR, Putatunda R, Young WB, Khalili K, et al. 2015. CRISPR/gRNA-directed synergistic activation mediator (SAM) induces specific, persistent and robust reactivation of the HIV-1 latent reservoirs. Sci Rep 5: 16277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, Wei W. 2014. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature 509: 487–491. [DOI] [PubMed] [Google Scholar]
- Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen ZY, Liu DR. 2015. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol 33: 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]