Abstract
Cell-free protein synthesis (CFPS) technologies have enabled inexpensive and rapid recombinant protein expression. Numerous highly active CFPS platforms are now available and have recently been used for synthetic biology applications. In this review, we focus on the ability of CFPS to expand our understanding of biological systems and its applications in the synthetic biology field. First, we outline a variety of CFPS platforms that provide alternative and complementary methods for expressing proteins from different organisms, compared with in vivo approaches. Next, we review the types of proteins, protein complexes, and protein modifications that have been achieved using CFPS systems. Finally, we introduce recent work on genetic networks in cell-free systems and the use of cell-free systems for rapid prototyping of in vivo networks. Given the flexibility of cell-free systems, CFPS holds promise to be a powerful tool for synthetic biology as well as a protein production technology in years to come.
Recombinant proteins can be produced inexpensively and rapidly using cell-free protein synthesis platforms. This technology will have many applications in the synthetic biology field (e.g., construction of genetic circuits).
Cell-free protein synthesis (CFPS) technology was first used more than 50 years ago by Nirenberg and Matthaei to decipher the genetic code (Nirenberg and Matthaei 1961). In the late 1960s and early 1970s, CFPS was used to help elucidate the regulatory mechanisms of the Escherichia coli lactose (Chambers and Zubay 1969) and tryptophan (Zalkin et al. 1974) operons. Now, in the last two decades, cell-free protein expression platforms have experienced a surge in development to meet the increasing demand for inexpensive and rapid recombinant protein expression technologies, which has resulted in the development of numerous highly active CFPS platforms (Carlson et al. 2012).
This renewed interest in CFPS technology was motivated by the advantages offered by this methodology for the production of recombinant proteins. In particular, the open reaction environment allows for the addition or removal of substrates for protein synthesis, as well as precise, online reaction monitoring. Furthermore, the CFPS reaction environment can be wholly directed toward and optimized for the production of the protein product of interest. In this way, CFPS platforms separate catalyst synthesis (cell growth) from catalyst usage (protein synthesis), representing a significant departure from cell-based processes that rely on microscopic cellular “reactors.” CFPS effectively decouples the cell’s objectives (growth and reproduction) from the engineer’s objectives (protein overexpression and simple product purification). Overall, the nature of CFPS technology allows for shortened protein synthesis timelines and increased flexibility for the addition or removal of natural or synthetic components compared with in vivo approaches. The versatility of CFPS makes it especially attractive for fundamental discovery and high-throughput screening applications.
The ability to prioritize the engineer’s objectives in CFPS has further motivated recent applications of CFPS technology to the exciting and ever-growing field of synthetic biology. For instance, cell-free synthetic biology approaches have enabled development of an in vitro prototyping environment for characterization of synthetic parts or genetic networks (Siegal-Gaskins et al. 2014; Takahashi et al. 2014; Chappell et al. 2015). The open environment and reduced complexity of cell-free systems has also made it possible to develop quantitative models describing cell-free genetic network performance and perform machine learning optimization of CFPS (Caschera et al. 2011; Siegal-Gaskins et al. 2014). Additionally, the absence of cell viability constraints has made CFPS an attractive technology for expanding the possible applications of synthetic biology. Recent advances in cell-free synthetic biology include the incorporation of nonnatural chemistries into biological polymers (Goerke and Swartz 2009; Bundy and Swartz 2010; Albayrak and Swartz 2013a; Hong et al. 2014a, 2015), in vitro assembly of complex biological machines and devices (Matthies et al. 2011), and the development of minimal cells (Shin and Noireaux 2012; Stano and Luisi 2013; Caschera and Noireaux 2014a). Excitingly, cell-free technology has also transitioned beyond the laboratory bench, both to the industrial scale for therapeutic production (Zawada et al. 2011; Yin et al. 2012) and to a low-cost, user-friendly format for diagnostic applications (Pardee et al. 2014).
In this review, we focus on the application of CFPS technology to synthetic biology. More detailed reviews on the development of CFPS technology and the types of proteins produced in cell-free systems have been published recently (Katzen et al. 2005; Carlson et al. 2012; Chong 2014; Harbers 2014; Hong et al. 2014a; Lian et al. 2014; Zemella et al. 2015). Here, we begin by introducing the various CFPS platforms and discuss their technological capabilities. We then outline the types of proteins, protein complexes, and protein modifications that have been achieved using CFPS technologies. Finally, we discuss cutting-edge cell-free synthetic biology applications.
MULTIPLE CELL-FREE PROTEIN SYNTHESIS TECHNOLOGIES ENABLE PRODUCTION OF DIVERSE PROTEINS
The recent technological renaissance has resulted in a variety of highly active CFPS platforms for expression of proteins from diverse organisms. Although E. coli and wheat germ extracts have been predominantly used in a high-throughput format, all CFPS platforms have the potential to be used for high-throughput screening of DNA libraries and gene products from diverse organisms for biological discovery and synthetic biology applications.
CFPS systems carry out protein synthesis by harnessing the biological catalysts for translation, protein folding, and energy generation from prokaryotic or eukaryotic cells. When combined with a DNA template, amino acids, an RNA polymerase, an adenosine triphosphate (ATP)-regeneration system, salts, and other buffers or environmental stabilizers (e.g., HEPES), these complex biological catalytic ensembles carry out sustained protein synthesis in vitro (Fig. 1) (Jewett et al. 2008).
Figure 1.
Cell-free protein synthesis (CFPS) platforms allow for increased flexibility and shortened process timelines to create a variety of high-value recombinant proteins. This technology provides platforms from numerous organisms with varied complexity to meet the needs of the specific target proteins. CFPS also decouples catalyst synthesis and catalyst usage, traditionally interconnected during in vivo recombinant protein production schemes. For CFPS, catalyst synthesis involves cell growth, cell lysis, and extract processing that removes genomic DNA to create crude extract. The crude extract may be frozen for future use or used directly for catalyst usage. This process requires NTPs, DNA, amino acids, and an energy source to produce various proteins with applications in the synthetic biology field.
There are two categories of CFPS platforms, which represent bottom-up and top-down approaches to in vitro protein synthesis. Protein synthesis using purified recombinant elements (PURE) systems build the protein synthesis ensemble from the bottom up. The PURE approach involves purifying the molecular components required for protein synthesis and subsequently adding them to CFPS reactions (Shimizu et al. 2014). Crude extract systems represent the alternative, top-down approach. Crude cell extract is generated by clarifying whole-cell lysate via centrifugation to remove genomic DNA, insoluble biological elements, and unlysed cells. In some platforms, additional measures are taken to degrade endogenous messenger RNAs (mRNAs) (Pratt 1984). Importantly, the crude extract contains all of the biological components required for translation, protein folding, and energy regeneration (e.g., ribosomes, tRNAs, chaperones, metabolic enzymes, elongation factor-Tu [EF-Tu], translation initiation factors, etc.). Crude lysate CFPS platforms have been developed using cells from a number of organisms. Each crude extract CFPS platform has advantages and disadvantages depending on the desired application (Table 1) (Zemella et al. 2015).
Table 1.
Advantages and disadvantages of existing CFPS technologies
| Platform | Advantages | Disadvantages | Representative yields (μg/mL) | Applications |
|---|---|---|---|---|
| PURE | 1. No nucleases or proteases remaining after component purification 2. Flexible and modular 3. Commercially available |
1. Expensive 2. Cannot activate endogenous metabolism 3. His purification |
GFP: 380a β-galactosidase: 4400b (Kazuta et al. 2014) |
Minimal cells (Matsubayashi and Ueda 2014) Complex proteins (Kuruma et al. 2012) nsAAs (Murakami et al. 2006) |
| Escherichia coli extract (ECE) | 1. High-batch yields 2. Low-cost extract preparation and reagents 3. Commercially available 4. Genetic tools available for strain modifications or synthetic biology 5. CFPS scales linearly >106 L (Zawada et al. 2011) |
1. Limited PTMs | GFP: 2300a (Caschera and Noireaux 2014b) GM-CSF: 700a (Zawada et al. 2011) VLP: 356a (Bundy et al. 2008) |
High-throughput format (Stafford et al. 2014) Antibodies (Groff et al. 2014) Vaccines (Ng et al. 2012) Diagnostics (Pardee et al. 2014) nsAAs (Goerke and Swartz 2009; Ozawa et al. 2012) Genetic circuits (Noireaux et al. 2003; Chappell et al. 2013) |
| Saccharomyces cerevisiae extract (SCE) | 1. Simple, low-cost extract preparation 2. Cotranslational folding mechanisms present 3. Genetic tools available for strain modifications or synthetic biology |
1. No PTMs shown 2. Low batch yields |
Luc: 8.9a (Choudhury et al. 2014) GFP: 17b (Schoborg et al. 2014) HPV L1 protein: 60a (Wang et al. 2008) |
Complex proteins (unpubl.) |
| Wheat germ extract (WGE) | 1. High yields 2. Proven for synthesis of many types of eukaryotic proteins, including membrane proteins 3. Continuous exchange reactions last ≤60 hours |
1. Extract preparation is long and labor-intensive 2. Difficult technology transfer |
GFP: 1600a (Harbers 2014), 9700b (Endo and Sawasaki 2006) | High-throughput format (Goshima et al. 2008) Vaccines (Tsuboi et al. 2008) Expressed proteomes (Nozawa and Tozawa 2014) Structural characterization (Bernhard and Tozawa 2013) |
| Tobacco BY-2 cell extract (BYE) | 1. Greatly simplified extract preparation compared with WGE 2. Continuous exchange reactions last ≤18 hours |
1. Lower yields than WGE 2. No PTMs shown |
Luc: 270a (Buntru et al. 2014b) YFP: 400b Human mAb: 150a Hb-EGF: 25a (Buntru et al. 2014a) |
Complex proteins (Buntru et al. 2014a) |
| Insect cell extract (ICE) | 1. PTMs possible, including glycosylation 2. Proven for synthesis of membrane proteins 3. Commercially available |
1. Low batch yields 2. Requires more extract (50% CFPS reaction volume vs. 25% for ECE) |
YFP: 10a (Katzen and Kudlicki 2006), 50b (Stech et al. 2014) | Complex proteins (Katzen and Kudlicki 2006; Suzuki et al. 2007; Orth et al. 2011; Stech et al. 2012; Ezure et al. 2014; Fenz et al. 2014) |
| CHO, K562, and HeLa extracts | 1. PTMs possible, including glycosylation 2. Proven for synthesis of membrane proteins 3. Mammalian cells are preferred expression platform for human proteins |
1. Low batch yields 2. Requires more extract (50% CFPS reaction volume vs. 25% for ECE) |
Luc: 50a (CHO), 20a (K562) (Brodel et al. 2015) Glutathione-S-transferase: 50b (HeLa) (Mikami et al. 2006) |
Complex proteins (Mikami et al. 2006; Stech et al. 2013; Brodel et al. 2015) |
| Leishmania tarentolae extract (LTE) | 1. Showed advantages for expressing parasitic proteins 2. Background translation mostly repressed |
1. No PTMs shown 2. Limited number and types of protein synthesized |
GFP: 300a (Mureev et al. 2009) | Complex proteins (Mureev et al. 2009) Expressed proteomes (Kovtun et al. 2010) |
Hb-EGF, Heparin-binding EGF-like growth factor; EPO, erythropoietin; GFP, green fluorescent protein; Luc, luciferase; mAb, monoclonal antibody; YFP, yellow fluorescent protein; PTM, posttranslational modification; nsAAs, nonstandard amino acids; CFPS, cell-free protein synthesis.
aBatch reaction.
bContinuous exchange reaction.
PURE Systems
PURE technology was pioneered by Ueda and colleagues (Shimizu et al. 2001). The PURE approach reconstitutes the transcriptional, translational, and energy-generation machinery from E. coli by purifying recombinantly expressed histidine (His)-tagged components and adding them to the CFPS reaction mixture. E. coli ribosomes are purified using sucrose-density gradient centrifugation (Shimizu et al. 2014).
PURE CFPS systems are commercially available and widely used for both fundamental discovery and synthetic biology applications. Protein synthesis yields from the PURE system have been optimized recently (Jackson et al. 2014a) and specialized PURE systems have been developed for continuous substrate replenishment (Jackson et al. 2014b, 2015), and to produce membrane proteins (Niwa et al. 2015), as well as disulfide bond- and nonstandard amino acid (nsAA)-containing proteins (Shimizu et al. 2005; Murakami et al. 2006). PURE technology has proven particularly useful for the isolated study of recombinant proteins and protein complexes (Kuruma et al. 2010, 2012; Matsubayashi et al. 2014) because of the simplicity of the system. Furthermore, PURE technology has played a pivotal role in the development of minimal cells (Nishimura et al. 2012, 2014; Stano and Luisi 2013; Caschera and Noireaux 2014a; Matsubayashi and Ueda 2014). However, the expense and cross-reactivity of the His-tag-based component purification process limits its utility in some cases.
Microbial Crude Extract Systems
E. coli and Saccharomyces cerevisiae are attractive hosts for CFPS and cell-free synthetic biology because (1) they are easily fermentable, and (2) they are model organisms. As model organisms, there exists a wealth of experimental tools and genetic information for both E. coli and S. cerevisiae to aid synthetic biology efforts.
The prokaryotic E. coli crude extract (ECE) system is one of the most widely adopted platforms for CFPS. This technology is commercially available and is used at the industrial scale (Zawada et al. 2011; Yin et al. 2012; Groff et al. 2014). ECE has been widely adopted for two main reasons: its high batch yields, with up to 2.3 g/L of green fluorescent protein (GFP) reported (Caschera and Noireaux 2014b), and the fast, scalable, and cost-effective extract preparation process (Swartz 2006). In addition, simple and rapid methods for extract preparation have been developed (Shrestha et al. 2012; Kwon and Jewett 2015). Furthermore, ECE reactions can use nucleoside monophosphates and inexpensive energy substrates, such as glucose or starch to regenerate ATP (Calhoun and Swartz 2005a,b; Lian et al. 2014). ECE has been used to synthesize recombinant bacterial and eukaryotic proteins containing posttranslational modifications (PTMs) (Guarino and DeLisa 2012) and nsAAs (Goerke and Swartz 2009; Bundy and Swartz 2010; Ozawa et al. 2012; Albayrak and Swartz 2013a,b; Hong et al. 2014b, 2015). Furthermore, ECE has been adapted to a high-throughput format for production of antibody variants for drug development and screening studies (Stafford et al. 2014), demonstrating its use for screening or functional genomic studies. Finally, ECE has been shown to scale linearly over a 106 L range in reaction volumes (Zawada et al. 2011), which helped motivate the adoption of this technology for large-scale protein synthesis (Yin et al. 2012; Groff et al. 2014; Zimmerman et al. 2014). Although bacterial CFPS systems are able to produce proteins that would be difficult or impossible in cells, further reductions in cost would enable wider adoption for industrial biomanufacturing.
A second microbial CFPS system uses extract from S. cerevisiae (baker’s yeast) to catalyze protein synthesis. S. cerevisiae cells contain eukaryotic folding machinery that can be beneficial for recombinant production of complex eukaryotic proteins. The first S. cerevisiae extract (SCE) system was pioneered by Iizuka et al. (1994) more than 20 years ago. Recently, the Jewett laboratory developed and optimized a scalable, low-cost SCE preparation method (Hodgman and Jewett 2013; Choudhury et al. 2014). Many efforts have been made to improve the efficiency and reduce the cost of SCE CFPS (Gan and Jewett 2014; Hodgman and Jewett 2014; Schoborg et al. 2014; Anderson et al. 2015). Together, these advances have resulted in a relatively low-cost eukaryotic CFPS technology with protein yields comparable to other eukaryotic platforms (Table 1). However, further improvements in protein synthesis yields will be necessary to encourage adoption of yeast CFPS for synthetic biology applications.
Plant, Animal, and Insect Crude Extract Systems
Crude extract CFPS technologies derived from higher eukaryotes offer advantages for complex protein production. Importantly, these extracts enable complex cotranslational folding mechanisms and the addition of PTMs not currently possible using microbial platforms. However, batch yields from these systems are typically at least an order of magnitude lower, whereas the system cost is much higher, compared with ECE (Table 1). Additionally, applications of these technologies to synthetic biology are currently limited. One challenge preventing high-yielding eukaryotic CFPS is the difficulty of enabling endogenous translation initiation mechanisms (e.g., 5′ capping) in vitro.
Two main plant-based crude extract CFPS systems have been developed to date are wheat germ extract (WGE) and tobacco BY-2 cell extract (BYE). The WGE platform, which is derived from wheat-seed embryos (Madin et al. 2000), has achieved both high yields (Endo and Sawasaki 2006) and high-throughput expression (Sawasaki et al. 2002) of recombinant proteins. Typically, this system is used with continuous substrate replenishment (continuous exchange) (Jackson et al. 2014b, 2015). For example, Goshima et al. (2008) used WGE to synthesize ∼13,000 human proteins in a single study. WGE was also used to synthesize bioactive proteins of diverse species origin, including Arabidopsis (Nozawa et al. 2009) and the malaria parasite genus Plasmodium (Tsuboi et al. 2008), without codon optimization in most cases. WGE is the highest-yielding eukaryotic CFPS platform, with yields of up to 1.6 mg/mL GFP in batch (Harbers 2014) and 9.7 mg/mL GFP with continuous exchange reported (Endo and Sawasaki 2006). Notably, the active lifetime of WGE is remarkably long—up to 60 h in continuous exchange reactions that replenish substrates (Endo and Sawasaki 2006)—because most nucleases and proteases are removed during extract preparation. However, the primary drawback of WGE is the extensive extract preparation process, which takes 4–5 days (Madin et al. 2000).
The BYE CFPS system was recently developed as an alternative plant-based CFPS platform. Preparation of BYE is greatly simplified compared with WGE and can be completed in just 4–5 h. Like WGE, BYE has also been used to synthesize proteins from diverse organisms, including a transmembrane growth factor, a glycosylated Apergillus niger enzyme, and a bioactive human antibody (Buntru et al. 2014b). However, unlike WGE, mg/mL yields from BYE have not yet been achieved (Table 1).
The insect cell extract (ICE) CFPS platform was developed using extract from Spodoptera frugiperda (fall army worm) cells and is commercially available. The first ICE system was developed by Ezure et al. (2006). A second ICE system, developed by Katzen and Kudlicki (2006), produced extracts containing endoplasmic reticulum (ER) vesicles that retained glycosylation and signal sequence processing activity. ICE has since been used to produce glycoproteins and membrane proteins (Katzen and Kudlicki 2006; Kubick et al. 2009; Sachse et al. 2014; Stech et al. 2014).
More recently, CFPS platforms were developed from mammalian cell lines. Mammalian cells are currently the preferred platform for large-scale in vivo production of human proteins bearing PTMs. The Kubick laboratory has described CFPS technologies using extracts from Chinese hamster ovary (CHO) cells and human K562 cells, a myelogenous leukemia cell line (Brodel et al. 2015). In addition, Mikami et al. (2006) developed a CFPS platform using lysate from HeLa cells, a cervical cancer cell line. Mammalian crude extract CPFS platforms provide a potentially attractive alternative for the synthesis and high-throughput screening of recombinant human proteins.
Parasitic Organism Crude Extract Systems
The Leishmania tarentolae extract (LTE) platform, developed by Mureev and colleagues, is the only platform that uses extracts from a parasitic organism. Parasites are attractive candidates for functional proteomic studies because, in some organisms, >50% of proteins have no identifiable homology or predictable function. However, in vivo expression of genes from parasitic or infectious organisms can be challenging because of differences in codon usage and protein synthesis machinery (Mureev et al. 2009). Parasitic organism crude extract systems are potentially advantageous for generating expressed parasite proteomes. In one instance, a parasitic crude extract CFPS system outperformed ECE in bioactive yields of proteins from Plasmodium falciparum, a malaria parasite (Kovtun et al. 2010).
L. tarentolae, a single-celled lizard parasite, was chosen for CFPS technology development for two main reasons: (1) it is easily fermentable, and (2) all endogenous mRNAs share the same leader sequence. The latter feature enables complete repression of background translation via inclusion of an antileader oligonucleotide (Mureev et al. 2009). LTE offers a rapid and scalable extract preparation process (Kovtun et al. 2011; Johnston and Alexandrov 2014), and is one of the highest yielding eukaryotic systems in batch; yields of up to 300 µg/mL GFP have been reported (Mureev et al. 2009). Importantly, Mureev and colleagues also developed species-independent translation sequences (SITS) to bypass translational regulation in LTE. The inclusion of SITS allowed cell-free synthesis of GFP in prokaryotic and eukaryotic extracts, including LTE, ECE, WGE, SCE, and ICE, using the same DNA template (Mureev et al. 2009; Kovtun et al. 2011; Johnston and Alexandrov 2014). LTE has been used to synthesize mammalian, L. tarentolae, and P. falciparum proteins.
Despite recent technological advances, there remain challenges facing CFPS technology that need to be addressed. Yields of bioactive human proteins from CFPS are sometimes lower than those achievable in vivo. In particular, no eukaryotic CFPS platform is currently capable of producing mg/mL quantities of protein in a batch. Increasing eukaryotic CFPS batch yields to 0.5 mg/mL or greater is an important challenge for two reasons: (1) These platforms have the potential to achieve higher soluble, bioactive yields of recombinant human proteins than bacterial platforms, and (2) eukaryotic CFPS systems could be useful as prototyping tools for genetic circuit characterization or production of recombinant proteins in eukaryotic cells. Furthermore, most crude extract CFPS platforms are derived from model organisms, and only the L. tarentolae platform is derived from a parasitic organism. Crude extract platforms from nonmodel organisms could have usage for functional genomics studies. Finally, development and optimization of species-independent translation initiation strategies, such as SITS, or novel cap-independent translation initiation sequences (Gan and Jewett 2016) could further increase protein yields and enable expression of proteins from diverse organisms. Addressing these challenges will be important to encourage broad adoption of CFPS technology for fundamental discovery and synthetic biology applications.
COMPLEX PROTEINS AND PROTEIN ASSEMBLIES
Cell-Free Production of Complex Proteins
Most CFPS platforms have been applied to the production of complex proteins. CFPS systems offer distinct advantages over in vivo protein production for applications that require more precise control over the protein synthesis reaction conditions. Such applications include bispecific antibodies, antibody–drug conjugates (ADCs), vaccines, and membrane proteins.
Therapeutics and Vaccines
ECE has been used extensively for production of bioactive recombinant therapeutic proteins, including granulocyte macrophage colony-stimulating factor (GM-CSF), onconase, antibodies, bispecific antibodies, and antibody–drug conjugates (Zawada et al. 2011; Yin et al. 2012; Groff et al. 2014; Stafford et al. 2014; Xu et al. 2014; Zimmerman et al. 2014; Cai et al. 2015; Salehi et al. 2016). Recently, a method for in vitro display of antibody fragments was developed in ECE, providing an exciting new technology for high-throughput production and screening of antibody candidates (Stafford et al. 2014; Cai et al. 2015). Excitingly, recent work has resulted in the development of microfluidic systems to enable CFPS reactions to be performed at the point-of-care (Sullivan et al. 2016; Timm et al. 2016). These systems would enable rapid production of medicines on demand and opens the door to bedside synthesis of personalized protein therapeutics.
The eukaryotic WGE, ICE, CHO extract, and K562 extract systems have also been used to produce therapeutic proteins. WGE and ICE have been used to synthesize bioactive antibody fragments, and ICE has been used to produce bioactive tissue-type plasminogen activator (Kawasaki et al. 2003; Stech et al. 2012, 2014). Furthermore, ICE, CHO extract, and K562 extract have been used to synthesize glycosylated erythropoietin (Stech et al. 2013; Brodel et al. 2015).
An especially promising application for CFPS systems is the high-throughput screening and in vitro production of vaccine antigens and virus-like particle (VLP) antigens. The WGE platform was applied to the synthesis of 124 P. falciparum genes of potential interest as vaccine antigens, and 75% were successfully synthesized without codon optimization (Tsuboi et al. 2008). ECE has also been used to synthesize vaccines and VLPs, including a B-cell lymphoma vaccine (Ng et al. 2012), anti-influenza VLPs (Lu et al. 2014), and anti-hepatitis B VLPs (Bundy et al. 2008), as well as a virus-like nanoparticle vaccine scaffold (Lu et al. 2015). Notably, the VLPs were produced at significantly higher yields than those possible in vivo, with yields improved up to 15-fold (Bundy et al. 2008). These studies show the potential of CFPS systems as novel vaccine discovery tools and platforms for large-scale vaccine production. Looking forward, because CFPS systems are abiotic, they are potentially well suited for portable and on-demand production of vaccines in resource-poor areas. This is an exciting future application area for CFPS technology.
Membrane Proteins
CFPS systems offer advantages for the production of membrane proteins. Newly synthesized membrane proteins can be stabilized in vitro by including membrane mimics (e.g., surfactants, liposomes, nanodiscs) to the cell-free reaction either posttranslationally, to solubilize proteins, or cotranslationally, to prevent aggregation (Junge et al. 2011; Sachse et al. 2014; Zieleniecki et al. 2016). A recent study even showed that supplying an oil–water interface by encapsulating CFPS reactions in emulsions was sufficient to synthesize functional single-span membrane proteins (Yunker et al. 2016). The ECE, WGE, ICE, CHO extract, and K562 extract systems as well as the PURE system have all been used to synthesize membrane proteins including ATP synthase (Matthies et al. 2011), G-protein-coupled receptors (Kaiser et al. 2008; Wang et al. 2011), membrane proteins from hepatitis C virus (Fogeron et al. 2015), and epidermal growth factor receptor (Stech et al. 2013). The synthesis of membrane proteins is an excellent application for CFPS technology, and has been useful for the structural biology community (Bernhard and Tozawa 2013). Overall, enhanced control over the protein synthesis reaction in vitro enables significantly higher yields of soluble, active protein compared with in vivo expression.
Taken together, these studies show the ability of CFPS technology to produce a variety of recombinant proteins with diverse complexity. ECE and WGE in particular show the potential of CFPS technologies for high-throughput production of many different kinds of proteins of interest for therapeutic development, fundamental discovery, or use as synthetic biological parts.
Macromolecule Production and Assembly in Cell-Free Systems
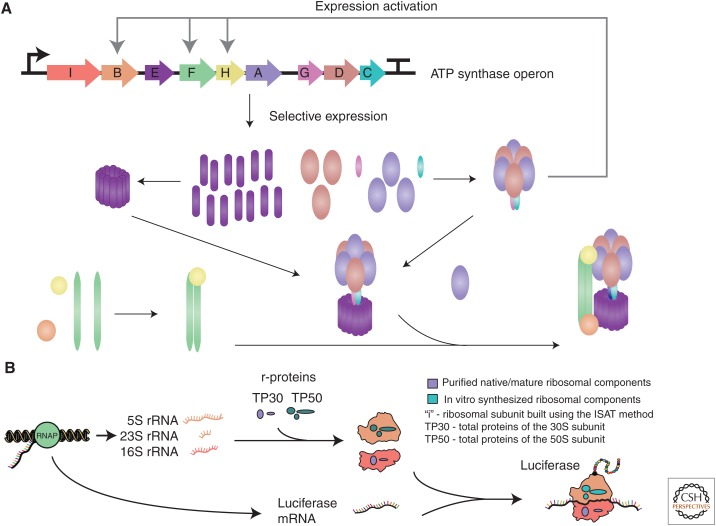
Much progress has been made toward in vivo synthesis of protein assemblies (Selkrig et al. 2014; Du et al. 2015); however, cellular viability constrains the complexity of the assemblies that can be overproduced. Moreover, cellular complexity may obscure fundamental properties of intricate assembly processes. Recently, CFPS systems have been extended for the production of complex protein assemblies. This advance offers interesting opportunities for studying macromolecule self-assembly and developing synthetic biological devices. CFPS platforms circumvent in vivo macromolecule production and assembly limitations in three key ways. First, the cell-free environment accelerates protein engineering efforts by enabling high-throughput experimentation and simplifying the purification of individual subunits. Second, CFPS platforms are flexible, ranging from crude extract to fully purified transcription–translation systems making it possible to study assembly processes in a controlled reaction environment. Third, CFPS platforms provide an additional level of complexity by coupling encoded protein expression to assembly of protein ensembles, which may elucidate the role of genetic regulation on subunit stoichiometries and assembly mechanisms (Fig. 2A).
Figure 2.
Assembly of macromolecules in cell-free protein synthesis (CFPS) reactions. (A) Complete in vitro assembly of adenosine triphosphate (ATP) ATP synthase with hypothesized genetic regulation. The ATP operon is added to a crude Escherichia coli extract and transcribed into a single messenger RNA (mRNA). Proteins are expressed at various levels determined by operon regulation. Matthies et al. (2011) suggest synthesis of correctly assembled ATP synthase complexes is dependent on specific expression levels of the subunits, correlated to the subunit stoichiometry in the complex. It is hypothesized that intermediate assemblies may also activate the expression of other subunits in the operon, allowing for sequential assembly processes (Kucharczyk et al. 2009). Thus, combined expression and assembly in CFPS systems allows for an additional level of assembly complexity for analysis. (B) In vitro integrated synthesis, assembly, and translation (iSAT) method of constructing ribosomes enables synthesis of active firefly luciferase in a one-pot reaction. iSAT enables one-step coactivation of ribosomal RNA (rRNA) transcription, assembly of ribosomal subunits, and synthesis of active protein by these ribosomes in same compartment. This process begins with T7 RNAP polymerase transcribing rRNA and luciferase mRNA. Ribosomal subunits are reconstituted from mature rRNA and ribosomal components previously purified or synthesized in vitro. Newly assembled ribosomes translate mRNA encoding the reporter protein luciferase to assess its activity.
Synthesis and Assembly of Naturally Occurring Macromolecules
Early demonstrations of protein assembly in CFPS systems produced macromolecules such as a five-subunit E. coli RNA polymerase (Asahara and Chong 2010) and a two subunit hepatitis B core antigen virus-like particle (Bundy et al. 2008). Even these simple assemblies have many applications. For instance, VLPs have been produced in cell-free systems at higher yields than those possible in vivo. This advance can be further applied to the discovery of novel vaccines, and provides straightforward production scale-up processes (Bundy et al. 2008).
As protein production capacity and reaction lifetime increased in CFPS systems, more proteins could be synthesized at once (Zemella et al. 2015). This allowed for the construction of complex structures, such as the pioneering work of Matthies et al. (2011) that performed the synthesis and assembly of Caldalkalibacillus thermarum ATP synthase, a 542-kDa membrane protein consisting of 25 individual proteins arranged into eight subunits. Cell-free production and assembly of ATP synthase was made possible by supplementing detergents to crude E. coli extract (Fig. 2A). This demonstration opens the way for the construction of novel energy generating biodevices that can be rapidly prototyped in vitro. Multistep coordination of protein ensembles has also been successfully shown in vitro. For instance, Fujiwara et al. (2013) simulated chromosomal DNA replication in a cell-free system by expressing 13 essential genes. In another study, Shin et al. (2012) showed complete in vitro DNA replication, synthesis, and assembly of bacteriophage T7 particles from expression of 60 genes. This is remarkable considering only 35 of the ∼60 proteins encoded by the T7 genome have known function (Chan et al. 2005). Full synthesis and assembly of other phages and VLPs have since been shown at higher yields (Garamella et al. 2016) and showcase the use of CFPS in screening efforts (Lu et al. 2015). Overall, this work reaffirms the value of CFPS platforms for interrogating native self-assembly processes and synthesizing self-replicating biological machines in vitro.
Beyond protein machines, the ability to assemble macromolecules with RNA and protein has also significantly advanced. For instance, the E. coli ribosome, a large protein/RNA assembly, has been assembled in vitro using a prokaryotic CFPS system (Fig. 2B) (Jewett et al. 2013). By adding DNA encoding the ribosomal RNA (rRNA) and purified ribosomal proteins to an E. coli extract lacking native ribosomes, in vitro assembled ribosomes are capable of translation under physiological conditions. Tuning transcription (Fritz and Jewett 2014) and using a continuous-exchange platform to replenish substrates and remove toxic byproducts (Liu et al. 2014) has resulted in superfolder GFP (sfGFP) yields of up to 7.5 µmol/L. This new ribosome construction platform, termed iSAT for integrated synthesis, assembly, and translation of ribosomes, makes possible new ways to probe, dissect, and understand ribosome biogenesis. In addition, it contributes meaningfully toward efforts to build minimal cells and construct engineered ribosomes with novel and useful properties.
Assembly and Evolution of Biological Devices (Biodevices)
To enable in vitro engineering of protein assemblies to create biodevices, cell-free systems can be combined with detection hardware. This is crucial because high molecular structure resolution is needed to follow assembly processes at the nanometer length scale. Interestingly, Heyman and colleagues developed a device that simultaneously synthesizes, assembles, and images nanotubes. This is achieved by patterning anti-hemagglutinin (HA) antibodies onto a TEM grid, trapping CFPS-synthesized nanotube precursor proteins tagged with HA (Heyman et al. 2012). This showed that coupling synthesis to assembly in the presence of an anchoring site could enable on-chip visualization of protein assembly. Microfluidic affinity assays have also been paired with CFPS platforms, enabling 14,792 simultaneous, on-chip experiments to exhaustively measure protein–protein interactions of 43 Streptococcus pneumoniae proteins in quadruplicate (Gerber et al. 2009). This study found several physical interactions between proteins that had been previously unreported. In sum, cell-free systems provide a flexible environment for expanding understanding of native assembly processes and the repertoire of synthetic protein assemblies.
SYNTHETIC PATHWAYS IN CFPS SYSTEMS ENABLE SITE-SPECIFIC MODIFICATIONS OF PROTEINS FOR NOVEL FUNCTIONALITIES
The development of highly active CFPS platforms has enabled the adaptation of CFPS technology toward synthesizing proteins with synthetic modifications and novel functionalities. Synthetic pathways have been added to confer the ability of cotranslational and posttranslational modification of recombinant proteins. The ability to site-specifically modify proteins could have many uses in fundamental and applied science. For example, the cotranslational incorporation of acetyllysine (AcK) in vivo has produced recombinant histone proteins that revealed new understanding of epigenetic regulation (Neumann et al. 2009). Site-specific modification will also have usage in the development of therapeutics bearing (a)biological modifications (i.e., phosphorylation, glycosylation, PEGylation, drug conjugates).
Cell-free production of modified proteins complements in vivo efforts and offers many advantages. In particular, modulation of substrate concentrations permits direct monitoring of their effects on protein modifications. This is especially important because synthetic protein modification pathways involve multiple proteins that can show cross talk with the native system. Additionally, an open and abiotic environment allows for the addition of substrates and production of byproducts that cannot be used in vivo because of cell-membrane permeability or cellular toxicity limitations. CFPS platforms offer greater control over the modification process and expand the types of chemistries that can be added to proteins.
Cotranslational Incorporation of Nonstandard Amino Acids
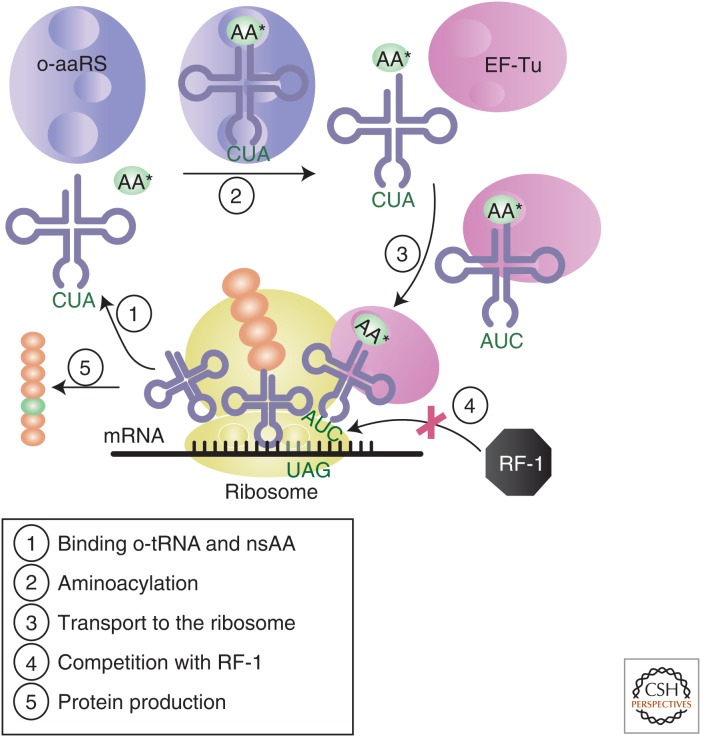
The cotranslational incorporation of nsAAs into proteins expands the chemistry of life. Such an expansion has enabled the incorporation of (a)biological groups into proteins such as biophysical probes (Cornish et al. 1994) (spin-labeled, fluorescent molecules, and photoactivatable cross linkers), redox active groups (Alfonta et al. 2003), and natural PTMs (Neumann et al. 2009; Nguyen et al. 2009; Virdee et al. 2011; Oza et al. 2015). To date, more than 100 nsAAs have been cotranslationally incorporated into proteins (O’Donoghue et al. 2013). This has been possible with the introduction of an orthogonal translation system (OTS), which is capable of genetically encoding nsAAs. Traditionally, OTSs consist of an engineered, orthogonal tRNA/aminoacyl tRNA synthetase (o-tRNA/o-aaRS) pair derived from a phylogenetically distant organism, often Methanocaldococcus jannaschii. The o-tRNA has a modified anticodon specific to the UAG stop codon (Fig. 3), whereas the o-aaRS is evolved to bind and aminoacylate the nsAA of interest to the o-tRNA. This process, which was initially pioneered by Schultz and colleagues (Wang et al. 2006), is called amber suppression as it allows the nsAA to be encoded at the UAG (amber) stop codon. An additional, engineered EF-Tu may be required when the nsAA of interest is too bulky to fit into the native EF-Tu amino acid binding pocket or its charge causes inefficient binding (Park et al. 2011). Although it is worth noting that both the WGE (Kiga et al. 2002) and the ICE (Ezure et al. 2014) systems have been used to incorporate nsAAs, for the purposes of this review, we will focus on the extensive work that has been performed in the ECE CFPS platform.
Figure 3.
Schematic representation of cotranslational incorporation of a nonstandard amino acid (nsAA) using an orthogonal translation system and amber suppression. The orthogonal aminoacyl-tRNA synthetase (o-aaRS) first binds its cognate nsAA and cognate o-tRNA. The o-aaRS then catalyzes the aminoacylation of the o-tRNA. The aminoacyl-tRNA (aa-tRNA) is then released from the o-aaRS and transported to the ribosome by the elongation factor-Tu (EF-Tu). The aa-tRNA associates with the A-site of the ribosome and its anticodon binds the complementary triplet codon of the messenger RNA (mRNA). The ribosome then ligates the nsAA to the growing peptide chain. When release factor 1 (RF-1) outcompetes the aa-tRNA for binding at the UAG amber stop codon, the protein is truncated, which results in a decrease of nsAA incorporation efficiency. This problem has been overcome by recoding all TAG codons to the synonymous TAA codon, permitting the deletion of RF-1 (Johnson et al. 2011; Loscha et al. 2012b; Ohtake et al. 2012; Lajoie et al. 2013).
The Swartz laboratory has made many contributions to the development of CFPS platforms for nsAA incorporation, especially in the area of substrate optimization (Goerke and Swartz 2009; Bundy and Swartz 2010). Albayrak and Swartz produced ∼1.5 mg/mL of sfGFP protein containing p-azido-phenylalnine (pAzF; azide, “click” chemistry, photocrosslinker) (Chin et al. 2002) and p-acetylphenylalanine (pAcF; keto, orthogonal reactivity) (Lemke 2011) with a 50%–88% amber suppression efficiency (Albayrak and Swartz 2013a). This success was possible by optimizing the concentration of o-tRNA, which was identified as a limiting factor of nsAA incorporation in CFPS reactions (Albayrak and Swartz 2013b). To develop a practical, cost-effective method for supplying more o-tRNA to the reaction, the o-tRNA was coexpressed in the CFPS reaction with the modified protein. The same method was validated by Hong et al. (2014b) and proved to reduce cost and setup time of nsAA CFPS reactions. It has also been found that increasing o-aaRS concentrations can improve the production of nsAA-containing proteins in CFPS reactions. This not surprising considering that engineered o-aaRS have 500- to 7000-fold lower catalytic efficiencies, as compared with their native counterparts (O’Donoghue et al. 2013). High concentrations of o-aaRS is only possible in vitro as o-aaRSs are known to be toxic in vivo at medium to high concentrations (Hong et al. 2014a). Despite this, Chemla and colleagues have shown that endogenously expressed pyrrolysyl OTS can allow for maximal suppression efficiency for CFPS nsAA incorporation (Chemla et al. 2015).
The efficiency of nsAA incorporation is greatly limited by nsAA aminoacylation efficiency (O’Donoghue et al. 2013). Further evolution of o-aaRS/o-tRNA pairs could increase catalytic efficiencies. Using in vitro compartmentalized directed evolution strategies (Nishikawa et al. 2012), CFPS systems can, in principle, assay larger libraries sizes compared with in vivo evolution methods, which are limited by transformation efficiencies. Additionally, o-aaRS evolved in vitro have the potential to reach lower Km values by limiting the concentration of nsAA. By comparison, intracellular concentrations of nsAAs are often high because of limited nsAA export and catabolism mechanisms, limiting the Km values attainable via in vivo evolution strategies.
Major advances have also removed a significant obstacle of amber suppression: nsAA-o-tRNA competition with release factor 1 (RF-1) (Johnson et al. 2011; Loscha et al. 2012b; Ohtake et al. 2012; Lajoie et al. 2013). Endogenous RF-1 recognizes amber codons and subsequently activates hydrolysis of peptidyl-tRNA to release the peptide chain. During nsAA incorporation using amber suppression technology, RF-1 competes with the nsAA-o-tRNA, resulting in a significant amount of truncated product and reduced nsAA incorporation efficiencies. By recoding TAG codons from seven essential genes to the synonymous TAA ochre codons, the RF-1 gene was deleted without dramatically effecting cellular growth. The RF-1 deletion strain increased O-phosphoserine incorporation by 120-fold in vivo (Heinemann et al. 2012). A strain with RF-1 knocked out and 13 essential gene TAG codons recoded to TAA was recently developed as a chassis for CFPS. As a result, a 250% improvement was observed compared with the parent stain with RF-1 present (Hong et al. 2014b). More recently, researchers developed an RF-1 deletion strain in which all 321 amber codons were recoded to TAA, completely freeing the amber codon to encode nsAAs (Lajoie et al. 2013). Although successful insertion of multiple nsAAs is possible without these modifications (Loscha et al. 2012a; Ozawa et al. 2012), a total recoded CFPS chassis has allowed for the incorporation of five pAcF residues into sfGFP (Hong et al. 2015) without additional extract processing. Further development of this chassis can potentially allow tens of nsAAs to be incorporated into a single protein.
Despite the many advances of nsAA incorporation technology, there is room for improvement and development for novel applications. At present, it is not possible to incorporate more than one kind of nsAA into a single protein in vitro without significant extract processing (Oh et al. 2014). Enabling incorporation of multiple types of nsAAs at multiple locations in recombinant proteins are important technological challenges for cell-free systems that need to be tackled in the near future. The development of further evolved OTSs for improved nsAA incorporation is another important research direction that could be performed in vitro, in which the design space is not constrained by cell viability. In addition, genome-engineering efforts to remove negative effectors of in vitro protein synthesis (e.g., nucleases, proteases) will increase yields of nsAA-containing proteins in recoded chassis strains (Hong et al. 2015). Beyond extract-based systems, it should be noted that purified approaches offer much more freedom of design and control of the translation apparatus for genetic reprogramming (Forster et al. 2003; Murakami et al. 2006; Goto et al. 2011; Passioura and Suga 2013).
Posttranslational Incorporation of Glycans and Metal Centers
PTMs can be performed in CFPS systems by coactivating enzyme pathways and protein synthesis in the cell lysate. The synthetic pathways can be synthesized in vivo in the chassis strain before extract preparation, in vitro via CFPS, or purified and added to the CFPS reaction. These efforts have enabled the in vitro production of proteins bearing glycans (sugars) and accessorized with metal centers.
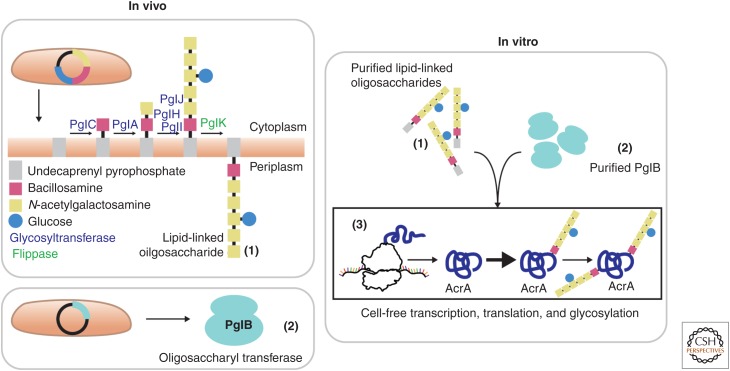
Glycosylation is particularly important for the production of recombinant protein therapeutics, as improper glycosylation can adversely affect the therapeutic activity or circulation half-life of a therapeutic (Li and d’Anjou 2009). Glycans are synthesized by enzymes called glycosyltransferases (GTs) and attached to proteins by oligosaccharyltransferases (OSTs), which together comprise the glycosylation machinery of the cell (Fig. 4).
Figure 4.
Glycoprotein production in cell-free systems. The Campylobacter jejuni N-glycan biosynthesis pathway, which contains glycosyltransferases and a flippase, is expressed in vivo in Escherichia coli. These enzymes assemble sugar monomers (bacillosamine, N-acetylglucosamine, glucose) onto a lipid anchor (undecaprenyl pyrophosphate) in the membrane to form lipid-linked oligosaccharides (LLOs) (top). Similarly, the C. jejuni oligosaccharyltransferase (OST), PglB, is expressed in vivo in E. coli (bottom). LLOs and PglB are purified and added to the in vitro glycoprotein synthesis reaction. Addition of purified LLOs and PglB to the cell-free protein synthesis (CFPS) reaction results in the synthesis of fully glycosylated AcrA, a C. jejuni glycoprotein (Guarino and DeLisa 2012).
Glycosylation is possible in some eukaryotic CFPS systems, including ICE, CHO extract, and K562 extract (Kubick et al. 2009; Stech et al. 2013, 2014; Brodel et al. 2015). However, these platforms harness the endogeneous machinery to carry out glycosylation. As a result, this approach restricts the possible glycan structures to those naturally synthesized by the host cells, which may not precisely resemble those found on the recombinant protein of interest. In addition, one would need to learn how to recapitulate protein trafficking to achieve some glycosylation patterns. Recent in vivo glycoengineering efforts have shown that cellular glycosylation machinery can be engineered to synthesize desired glycan structures (Hamilton et al. 2006; Fisher et al. 2011; Valderrama-Rincon et al. 2012; Meuris et al. 2014). More recent efforts are exploiting the open cell-free reaction environment for glycoengineering to enable synthetic glycosylation pathways. In a pioneering study, Guarino and DeLisa (2012) showed that glycosylated proteins can be synthesized in vitro by adding purified bacterial lipid-linked oligosaccharides and the PglB OST to a CPFS reaction. Yields of between 50–100 µg/mL of AcrA, a Campylobacter jejuni glycoprotein, were achieved. This result provides a potential path for the incorporation of synthetic, human glycosylation in cell-free systems through the transfer of synthetic enzyme pathways to bacterial cell-free systems. However, significant development of this technology is still needed.
Metalloproteins are important catalysts in biology (Kunamneni et al. 2008). Typically, in vivo production of metalloproteins involves apoprotein production and purification, followed by in vitro metal loading (Lawton and Rosenzweig 2011). CFPS systems potentially offer one-pot synthesis of metal-loaded metalloproteins by addition of free metal atoms and protein chaperones to in vitro reaction mixture. The first example of this approach was the in vitro synthesis of the FeFe hydrogenase metalloprotein (Boyer et al. 2006; Kuchenreuther et al. 2014). Three FeS cluster-containing proteins, HydE, HydF, and HydG, are required to assemble the FeFe hydrogenase. This enzyme pathway was reconstituted in vitro using E. coli lysates containing HydE, HydF, and HydG. When the purified apo-form of the FeFe hydrogenase was added to the reaction, the active hydrogenase was synthesized at yields of 100–200 µg/mL (Kuchenreuther et al. 2014). This study provided evidence that enzymatic pathways can be reconstituted in vitro, but the integration of in vitro protein synthesis into this system was not shown. More recently, Li and colleagues showed that two bacterial and two archaeal multicopper oxidase (MCO) enzymes could be synthesized in vitro, with three of the four enzymes produced at yields of greater than 1 mg/mL. They further showed that active MCO could be synthesized in a one-pot in vitro reaction containing 100 µM CuSO4, or by posttranslational addition of CuSO4 to the reaction without the need for purification. MCO synthesis was also scaled from 15 µL to 100 µL without impacting productivity or protein solubility (Li et al. 2016). This study shows the potential of CFPS as a novel method for production of metalloproteins at high titers.
Synthetic PTM mechanisms and enzymatic pathways in cell-free systems have great potential for expanding the types of controllable protein modifications that are possible. Further work in this area will expand opportunities for producing modified proteins with a variety of (a)biological modifications in vitro.
BUILDING UP TO GENETIC NETWORKS IN CELL-FREE SYSTEMS
In addition to using cell-free synthetic biology for the production of individual proteins or assembly of macromolecules, CFPS systems are now used for the construction of sophisticated genetic networks, often referred to as genetic circuits (Brophy and Voigt 2014). Initial efforts to design and implement synthetic genetic networks dominated early in vivo synthetic biology efforts (Gardner et al. 2000), serving as a forward engineering approach for studying natural gene regulation and controlling cellular behavior. Much progress has been made toward developing foundational genetic network modules, such as genetic switches (Bayer and Smolke 2005; Ham et al. 2006, 2008), logic gates (Rackham and Chin 2005; Anderson et al. 2007; Win and Smolke 2008), and memory modules (Friedland et al. 2009), as well as engineering cells with practical applications in the areas of bioremediation (Gilbert et al. 2003), biosensing (Rajendran and Ellington 2008), biofuel production (Inokuma et al. 2010), and therapeutic applications (Ro et al. 2006). Despite the aforementioned progress, translational in vivo synthetic biology is often limited by cellular viability for three key reasons. First, in vivo genetic networks are constrained to relatively small networks owing to restrictions on cellular metabolic load and a limited number of genetic parts. Second, limited high-throughput methods for optimizing regulatory networks slow in vivo genetic network design cycles. Third, interference between host regulation and synthetic networks (cross talk) results in poor computational modeling, reducing prediction capabilities as the network grows in size. Fortunately, many of these challenges can be addressed by implementing genetic networks in vitro using CFPS platforms.
For instance, synthetic gene networks can be studied in isolation as all of the endogenous DNA and mRNA are removed during lysate preparation. This eliminates the issue of cross talk between endogenous regulation and the synthetic network. In addition, the open nature of cell-free systems allows integration with hardware for high-throughput assays and on-board reaction monitoring. Consequently, automation will reduce current design–build–test cycle time, and will provide new biological modules for in vivo characterization. Furthermore, compartmentalizing CFPS reactions within synthetic vesicles provides an opportunity to study artificial cells (Caschera and Noireaux 2016) and the fundamental properties of gene regulation in confined environments.
Implementing Genetic Networks In Vitro
Simple regulatory elements, such as inducible promoters (Noireaux et al. 2003), transcriptional switches (Kim et al. 2006), and multistage cascades (Noireaux et al. 2003), were the first modules to be implemented in vitro. However, because current CFPS systems are optimized for mRNA stability and protein overproduction, in vitro genetic networks are inadequate when constructing oscillatory circuits (Elowitz and Leibler 2000) or larger networks. Complex genetic circuitry requires resources to be efficiently directed toward downstream networks, as well as component half-lives to mimic those found in cells. This is made clear when considering that in vivo global mRNA half-life is ∼6.8 min (Selinger et al. 2003), whereas, in vitro mRNA half-life is ∼13 min (Shin and Noireaux 2010). This discrepancy causes overall limitations of translational machinery for CFPS reactions as mRNA is accumulated in the reaction (Selinger et al. 2003). To overcome these issues, larger genetic circuits have been implemented in vitro by using purified MazF, a sequence-specific endoribonuclease, which has been shown to incrementally inactivate mRNA (Shin and Noireaux 2010). In particular, AND gates and negative feedback loops were developed in an E. coli cell-free expression system using this method (Shin and Noireaux 2012). Karig and colleagues also showed negative feedback loops but without mRNA activation mechanisms, and consequently saw unwanted accumulation of mRNA that ultimately affected the dynamics of the network (Karig et al. 2012). CFPS protein half-life has also been found to be much longer than in in vivo systems because of the reduced proteolytic activity of the CFPS system. This last aspect is important for constructing oscillatory circuits and needs to be further investigated.
Additionally, characterization and optimization of regulatory elements for cell-free systems have been explored (Shin and Noireaux 2012; Garamella et al. 2016). This provides more transcriptional handles to design higher-order genetic networks. Moreover, T7 promoter variants have also been developed in a cell-free context for expanding regulation options and improved switch-like activity (Karig et al. 2012; Iyer et al. 2013). To limit the number of synthesized proteins required for a genetic network, alternative transcriptional control mechanisms, such as ligand-sensitive transcriptional controls, have also been shown (Iyer and Doktycz 2014). Finally, RNA-based genetic circuits represent another solution to limit energy loss from protein production and provide the benefit of quicker response times (Takahashi et al. 2014; Chappell et al. 2015).
The open nature of CFPS also allows for in situ monitoring of genetic networks (Norred et al. 2015). In particular, binary FRET probes have been used to monitor real-time mRNA dynamics (Niederholtmeyer et al. 2013). Fluorescent RNA aptamers have also been used as an alternative simple strategy for monitoring RNA in real-time (Chizzolini et al. 2014). High-throughput analysis of biomolecular interactions can also be performed by encapsulating CFPS reactions into nanoliter droplets and examining interactions via an immunoassay (Rendl et al. 2013). Additionally, HPLC analyses for monitoring metabolite concentrations are straightforward to use because CFPS reactions can be run directly on an HLPC or MS machine.
Collectively, recent efforts hold promise for using more complex circuitry and real-world applications (Pardee et al. 2014). Because of the open environment of cell-free systems and short implementation times, CFPS systems can also be used as a complementary route to prototyping genetic networks in an in vitro environment before in vivo implementation.
Opportunities in Prototyping
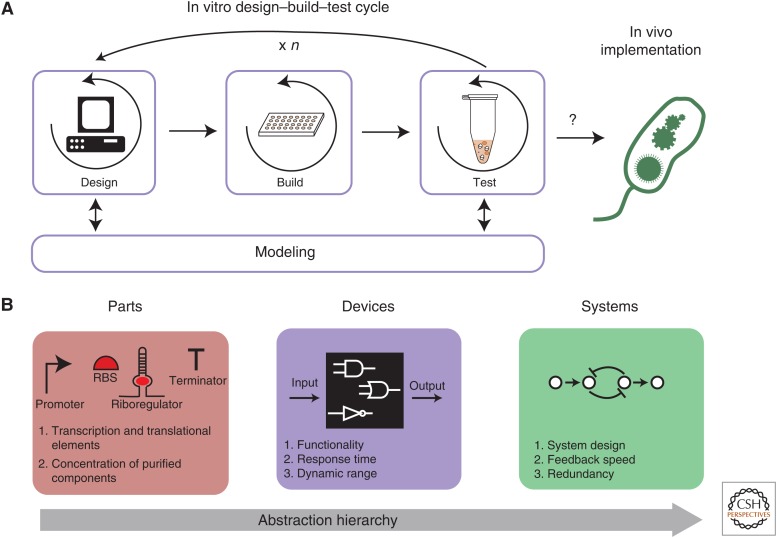
In vivo synthetic biology applies molecular biology tools to forward-engineer cellular behavior. This is performed first through in silico design of genetic networks, construction of those networks, followed by testing in vivo (Litcofsky et al. 2012; Sleight and Sauro 2013). However, in vivo engineering cycles require a significant amount of time and financial resources (Ro et al. 2006). In contrast, in vitro biomolecular prototyping promises to improve the overall efficiency of the design–build–test cycle (Fig. 5A) (Sun et al. 2014). There are several demonstrations of prototyping circuits in vitro for in vivo implementation, such as negative feedback loops (Karig et al. 2012), multi-input regulated T7 promoters (Iyer et al. 2013), riboswitches (Martini and Mansy 2014), and oscillators (Niederholtmeyer et al. 2015). However, it is difficult to predict correlations between parameters (e.g., promoters/RBS strengths) found in vitro and predicted values in vivo. For instance, it was found that characterizing promoter strength in vitro must be performed with circularized DNA for there is a correlation to in vivo promoter strengths (Chappell et al. 2013); however, this is limited to low to medium strength promoters. Additional work is needed to find unknown parameters involved with the transition from in vitro to in vivo implementation.
Figure 5.
In vitro prototyping of genetic networks. (A) Overview of in vitro prototyping for speeding up in vivo design–build–test cycles. In vitro prototyping allows for a genetic part or network to be quickly screened for specific characteristics in vitro before implementation in vivo. First, in the “Design” stage, the genetic parts or networks are designed, informed by computational models or literature. Next, in the “Build” stage, several designs are built. In the “Test” phase, built designs are assayed in vitro. If the Test stage does not yield the desired behavior, one reinitiates the cycle n number of times until the desired characteristic is achieved. The Test stage can also inform the modeling and allow for better models for the Design stage. Once the desired characteristics are found, top candidates that behave as expected can be implemented in vivo, with an increased likeliness of being functional. (B) Variables for characterization and optimization. In vitro prototyping can occur by characterizing or optimizing various levels on the genetic network “abstraction hierarchy.” At the most basic level, transcriptional and translational parts or purified components of a network can be analyzed for functionality. In vitro prototyping can also be applied at the device level to assess input–output relationships. Finally, an in vitro systems level analysis allows for an isolated study of how multiple genetic devices feed into each other and a preview of the overall network behavior.
Despite these issues, in vitro prototyping techniques are compelling and provide a potentially rapid alternative to in vivo systems for circuit design and testing (Niederholtmeyer et al. 2015; de Los Santos et al. 2016). For instance, in vitro prototyping can be accelerated through microscale reactions in micro-well plates and microfluidic systems. Indeed, these methodologies decrease prototyping time with real-time simultaneous or sequential screening of many system parameters (Lee et al. 2007). Additionally, complementary, predictive computational models are being developed to better understand CFPS’ limitations in prototyping (Karzbrun et al. 2011). This work has confirmed experimental results (Noireaux et al. 2003) that, in CFPS, potential design limitations arise from improperly balanced energy usage. Using a continuous exchange set-up can partially address this issue, and has been proven to be a reliable solution for prototyping (Noireaux et al. 2003). Prototyping efforts have even moved beyond a laboratory environment with development of a CFPS system that is both portable and stable because of its ability to react on paper (Pardee et al. 2014). This allows for a low-cost prototyping environment and provides opportunities in using CFPS for portable, inexpensive diagnostic applications.
Improvements in CFPS systems have opened new opportunities for cell-free synthetic biology, including studying genetic networks in isolation, characterizing new regulatory elements and speeding up design–build–test cycles for in vivo synthetic biology. The size of genetic networks achievable in CFPS systems will only increase as CFPS systems are further developed to meet the specific needs of in vitro genetic networks (e.g., downstream resourcing and matching in vivo component half-lives). Additionally, computational modeling of cell-free reactions will further improve in silico design of in vitro networks and assist in understanding the genetic regulation in isolation. In sum, the development of in vitro genetic networks holds much potential for the synthetic biology field as it complements many in vivo efforts.
CONCLUSIONS AND FUTURE OUTLOOK
Overall, the renewed scientific interest in CFPS in the past two decades has resulted in drastically increased batch yields, active reaction durations, and reaction volumes (Carlson et al. 2012). The variety of different CFPS technology platforms has further enabled the in vitro production of proteins with diverse complexity and species origin. CFPS technology will be invaluable in the near future as an “organism protein factory.” In this way, CFPS technology will aid efforts to determine the gene products for the many organisms whose genomes have been sequenced or will be sequenced in the near future, owing to the rapidly decreasing cost of DNA sequencing technology. For example, the Sargasso Seas expedition identified >1.2 million genes, many of which have unknown function and unknown potential as synthetic parts (Venter et al. 2004).
CFPS technology will also have an increasing role in complementing in vivo synthetic biology efforts. Because of its flexibility and high-throughput potential, cell-free systems are perfectly suited to synthesize and assay large libraries, not only for genetic networks as surveyed herein, but also for evolutionary or proteomics studies (Takemori et al. 2015). This will provide in vivo efforts with enriched mutant libraries and accelerated methods for preparing large-scale protein libraries. In sum, CFPS holds tremendous potential to transform our ability to rapidly synthesize and engineer recombinant proteins and its applications in the synthetic biology field will only grow in the future.
ACKNOWLEDGMENTS
The authors would like to acknowledge Jennifer A. Schoborg, Erik D. Carlson, and Filippo Caschera for assistance in editing this manuscript. Select figures in this text were adapted from Jennifer A. Schoborg and Erik D. Carlson. We gratefully acknowledge the National Science Foundation (NSF) (MCB-0943393), the Office of Naval Research (N00014-11-1-0363), the Defense Advanced Research Projects Agency Young Faculty Award (DARPA YFA) Program (N66001-11-1-4137), the Army Research Office (W911NF-11-1-0445), the NSF Materials Network Grant (DMR-1108350), the DARPA Living Foundries Program (N66001-12-C-4211), the David and Lucile Packard Foundation (2011-37152), Advanced Research Projects Agency-Energy (ARPA-E) (DE-AR0000435), and the Chicago Biomedical Consortium with support from the Searle Funds at the Chicago Community Trust for support. J.G.P. is funded by the NSF Graduate Research Fellowship (DGE-1144469). J.C.S. is funded, in part, by the Northwestern Biotechnology Training Program supported by the National Institutes of Health (NIH) (T32GM008449).
Footnotes
Editors: Daniel G. Gibson, Clyde A. Hutchison III, Hamilton O. Smith, and J. Craig Venter
Additional Perspectives on Synthetic Biology available at www.cshperspectives.org
REFERENCES
- Albayrak C, Swartz JR. 2013a. Cell-free co-production of an orthogonal transfer RNA activates efficient site-specific non-natural amino acid incorporation. Nucleic Acids Res 41: 5949–5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albayrak C, Swartz JR. 2013b. Using E. coli-based cell-free protein synthesis to evaluate the kinetic performance of an orthogonal tRNA and aminoacyl-tRNA synthetase pair. Biochem Biophys Res Commun 431: 291–295. [DOI] [PubMed] [Google Scholar]
- Alfonta L, Zhang Z, Uryu S, Loo JA, Schultz PG. 2003. Site-specific incorporation of a redox-active amino acid into proteins. J Am Chem Soc 125: 14662–14663. [DOI] [PubMed] [Google Scholar]
- Anderson JC, Voigt CA, Arkin AP. 2007. Environmental signal integration by a modular AND gate. Mol Syst Biol 3: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MJ, Stark JC, Eric Hodgman C, Jewett MC. 2015. Energizing eukaryotic cell-free protein synthesis with glucose metabolism. FEBS Lett 589: 1723–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahara H, Chong S. 2010. In vitro genetic reconstruction of bacterial transcription initiation by coupled synthesis and detection of RNA polymerase holoenzyme. Nucleic Acids Res 38: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer TS, Smolke CD. 2005. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat Biotechnol 23: 337–343. [DOI] [PubMed] [Google Scholar]
- Bernhard F, Tozawa Y. 2013. Cell-free expression—Making a mark. Curr Opin Struct Biol 23: 374–380. [DOI] [PubMed] [Google Scholar]
- Boyer ME, Wang CW, Swartz JR. 2006. Simultaneous expression and maturation of the iron-sulfur protein ferredoxin in a cell-free system. Biotechnol Bioeng 94: 128–138. [DOI] [PubMed] [Google Scholar]
- Brodel AK, Wustenhagen DA, Kubick S. 2015. Cell-free protein synthesis systems derived from cultured mammalian cells. Methods Mol Biol 1261: 129–140. [DOI] [PubMed] [Google Scholar]
- Brophy JAN, Voigt CA. 2014. Principles of genetic circuit design. Nat Meth 11: 508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundy BC, Swartz JR. 2010. Site-specific incorporation of p-propargyloxyphenylalanine in a cell-free environment for direct protein–protein click conjugation. Bioconjug Chem 21: 255–263. [DOI] [PubMed] [Google Scholar]
- Bundy BC, Franciszkowicz MJ, Swartz JR. 2008. Escherichia coli–based cell-free synthesis of virus-like particles. Biotechnol Bioeng 100: 28–37. [DOI] [PubMed] [Google Scholar]
- Buntru M, Vogel S, Spiegel H, Schillberg S. 2014a. Tobacco BY-2 cell-free lysate: An alternative and highly-productive plant-based in vitro translation system. BMC Biotechnol 14: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buntru M, Vogel S, Stoff K, Spiegel H, Schillberg S. 2014b. A versatile coupled cell-free transcription–translation system based on tobacco BY-2 cell lysates. Biotechnol Bioeng 112: 867–878. [DOI] [PubMed] [Google Scholar]
- Cai Q, Hanson JA, Steiner AR, Tran C, Masikat MR, Chen R, Zawada JF, Sato AK, Hallam TJ, Yin G. 2015. A simplified and robust protocol for immunoglobulin expression in Escherichia coli cell-free protein synthesis systems. Biotechnol Prog 31: 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun KA, Swartz JR. 2005a. An economical method for cell-free protein synthesis using glucose and nucleoside monophosphates. Biotechnol Prog 21: 1146–1153. [DOI] [PubMed] [Google Scholar]
- Calhoun KA, Swartz JR. 2005b. Energizing cell-free protein synthesis with glucose metabolism. Biotechnol Bioeng 90: 606–613. [DOI] [PubMed] [Google Scholar]
- Carlson ED, Gan R, Hodgman CE, Jewett MC. 2012. Cell-free protein synthesis: Applications come of age. Biotechnol Adv 30: 1185–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caschera F, Noireaux V. 2014a. Integration of biological parts toward the synthesis of a minimal cell. Curr Opin Chem Biol 22: 85–91. [DOI] [PubMed] [Google Scholar]
- Caschera F, Noireaux V. 2014b. Synthesis of 2.3 mg/ml of protein with an all Escherichia coli cell-free transcription-translation system. Biochimie 99: 162–168. [DOI] [PubMed] [Google Scholar]
- Caschera F, Noireaux V. 2016. Compartmentalization of an all–E. coli cell-free expression system for the construction of a minimal cell. Artif Life 2: 1–11. [DOI] [PubMed] [Google Scholar]
- Caschera F, Bedau MA, Buchanan A, Cawse J, de Lucrezia D, Gazzola G, Hanczyc MM, Packard NH. 2011. Coping with complexity: Machine learning optimization of cell-free protein synthesis. Biotechnol Bioeng 108: 2218–2228. [DOI] [PubMed] [Google Scholar]
- Chambers DA, Zubay G. 1969. The stimulatory effect of cyclic adenosine 3′5′-monophosphate on DNA-directed synthesis of β-galactosidase in a cell-free system. Proc Natl Acad Sci 63: 118–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan LY, Kosuri S, Endy D. 2005. Refactoring bacteriophage T7. Mol Syst Biol 1: 2005.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J, Jensen K, Freemont PS. 2013. Validation of an entirely in vitro approach for rapid prototyping of DNA regulatory elements for synthetic biology. Nucleic Acids Res 41: 3471–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J, Takahashi MK, Lucks JB. 2015. Creating small transcription activating RNAs. Nat Chem Biol 11: 214–220. [DOI] [PubMed] [Google Scholar]
- Chemla Y, Ozer E, Schlesinger O, Noireaux V, Alfonta L. 2015. Genetically expanded cell-free protein synthesis using endogenous pyrrolysyl orthogonal translation system. Biotechnol Bioeng 112: 1663–1672. [DOI] [PubMed] [Google Scholar]
- Chin JW, Santoro SW, Martin AB, King DS, Wang L, Schultz PG. 2002. Addition of p-azido-l-phenylalanine to the genetic code of Escherichia coli. J Am Chem Soc 124: 9026–9027. [DOI] [PubMed] [Google Scholar]
- Chizzolini F, Forlin M, Cecchi D, Mansy SS. 2014. Gene position more strongly influences cell-free protein expression from operons than T7 transcriptional promoter strength. ACS Synth Biol 3: 363–371. [DOI] [PubMed] [Google Scholar]
- Chong S. 2014. Overview of cell-free protein synthesis: Historic landmarks, commercial systems, and expanding applications. Curr Protoc Mol Biol 108: 16.30.1– 16.30.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury A, Hodgman CE, Anderson MJ, Jewett MC. 2014. Evaluating fermentation effects on cell growth and crude extract metabolic activity for improved yeast cell-free protein synthesis. Biochem Eng J 91: 140–148. [Google Scholar]
- Cornish VW, Benson DR, Altenbach CA, Hideg K, Hubbell WL, Schultz PG. 1994. Site-specific incorporation of biophysical probes into proteins. Proc Natl Acad Sci 91: 2910–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Los Santos EL, Meyerowitz JT, Mayo SL, Murray RM. 2016. Engineering transcriptional regulator effector specificity using computational design and in vitro rapid prototyping: Developing a vanillin sensor. ACS Synth Biol 5: 287–295. [DOI] [PubMed] [Google Scholar]
- Du D, van Veen HW, Luisi BF. 2015. Assembly and operation of bacterial tripartite multidrug efflux pumps. Trends Microbiol 23: 311–319. [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Leibler S. 2000. A synthetic oscillatory network of transcriptional regulators. Nature 403: 335–338. [DOI] [PubMed] [Google Scholar]
- Endo Y, Sawasaki T. 2006. Cell-free expression systems for eukaryotic protein production. Curr Opin Biotechnol 17: 373–380. [DOI] [PubMed] [Google Scholar]
- Ezure T, Suzuki T, Higashide S, Shintani E, Endo K, Kobayashi S, Shikata M, Ito M, Tanimizu K, Nishimura O. 2006. Cell-free protein synthesis system prepared from insect cells by freeze-thawing. Biotechnol Prog 22: 1570–1577. [DOI] [PubMed] [Google Scholar]
- Ezure T, Nanatani K, Sato Y, Suzuki S, Aizawa K, Souma S, Ito M, Hohsaka T, von Heijine G, Utsumi T, et al. 2014. A cell-free translocation system using extracts of cultured insect cells to yield functional membrane proteins. PLoS ONE 9: e112874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenz SF, Sachse R, Schmidt T, Kubick S. 2014. Cell-free synthesis of membrane proteins: Tailored cell models out of microsomes. Biochim Biophys Acta 1838: 1382–1388. [DOI] [PubMed] [Google Scholar]
- Fisher AC, Haitjema CH, Guarino C, Celik E, Endicott CE, Reading CA, Merritt JH, Ptak AC, Zhang S, DeLisa MP. 2011. Production of secretory and extracellular N-linked glycoproteins in Escherichia coli. Appl Environ Microbiol 77: 871–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogeron ML, Badillo A, Jirasko V, Gouttenoire J, Paul D, Lancien L, Moradpour D, Bartenschlager R, Meier BH, Penin F, et al. 2015. Wheat germ cell-free expression: Two detergents with a low critical micelle concentration allow for production of soluble HCV membrane proteins. Protein Expr Purif 105: 39–46. [DOI] [PubMed] [Google Scholar]
- Forster AC, Tan Z, Nalam MN, Lin H, Qu H, Cornish VW, Blacklow SC. 2003. Programming peptidomimetic syntheses by translating genetic codes designed de novo. Proc Natl Acad Sci 100: 6353–6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland AE, Lu TK, Wang X, Shi D, Church G, Collins JJ. 2009. Synthetic gene networks that count. Science 324: 1199–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz BR, Jewett MC. 2014. The impact of transcriptional tuning on in vitro integrated rRNA transcription and ribosome construction. Nucleic Acids Res 42: 6774–6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara K, Katayama T, Nomura SM. 2013. Cooperative working of bacterial chromosome replication proteins generated by a reconstituted protein expression system. Nucleic Acids Res 41: 7176–7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan R, Jewett MC. 2014. A combined cell-free transcription-translation system from Saccharomyces cerevisiae for rapid and robust protein synthe. Biotechnol J 9: 641–651. [DOI] [PubMed] [Google Scholar]
- Gan R, Jewett MC. 2016. Evolution of translation initiation sequences using in vitro yeast ribosome display. Biotechnol Bioeng 10.1002/bit.25933. [DOI] [PubMed] [Google Scholar]
- Garamella J, Marshall R, Rustad M, Noireaux V. 2016. The all E. coli TX-TL toolbox 2.0: A platform for cell-free synthetic biology. ACS Synth Biol 5: 344–355. [DOI] [PubMed] [Google Scholar]
- Gardner TS, Cantor CR, Collins JJ. 2000. Construction of a genetic toggle switch in Escherichia coli. Nature 403: 339–342. [DOI] [PubMed] [Google Scholar]
- Gerber D, Maerkl SJ, Quake SR. 2009. An in vitro microfluidic approach to generating protein-interaction networks. Nat Methods 6: 71–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert ES, Walker AW, Keasling JD. 2003. A constructed microbial consortium for biodegradation of the organophosphorus insecticide parathion. Appl Microbiol Biotechnol 61: 77–81. [DOI] [PubMed] [Google Scholar]
- Goerke AR, Swartz JR. 2009. High-level cell-free synthesis yields of proteins containing site-specific non-natural amino acids. Biotechnol Bioeng 102: 400–416. [DOI] [PubMed] [Google Scholar]
- Goshima N, Kawamura Y, Fukumoto A, Miura A, Honma R, Satoh R, Wakamatsu A, Yamamoto J, Kimura K, Nishikawa T, et al. 2008. Human protein factory for converting the transcriptome into an in vitro-expressed proteome. Nat Methods 5: 1011–1017. [DOI] [PubMed] [Google Scholar]
- Goto Y, Katoh T, Suga H. 2011. Flexizymes for genetic code reprogramming. Nat Protoc 6: 779–790. [DOI] [PubMed] [Google Scholar]
- Groff D, Armstrong S, Rivers PJ, Zhang J, Yang J, Green E, Rozzelle J, Liang S, Kittle JD Jr, Steiner AR, et al. 2014. Engineering toward a bacterial “endoplasmic reticulum” for the rapid expression of immunoglobulin proteins. mAbs 6: 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino C, DeLisa MP. 2012. A prokaryote-based cell-free translation system that efficiently synthesizes glycoproteins. Glycobiology 22: 596–601. [DOI] [PubMed] [Google Scholar]
- Ham TS, Lee SK, Keasling JD, Arkin AP. 2006. A tightly regulated inducible expression system utilizing the fim inversion recombination switch. Biotechnol Bioeng 94: 1–4. [DOI] [PubMed] [Google Scholar]
- Ham TS, Lee SK, Keasling JD, Arkin AP. 2008. Design and construction of a double inversion recombination switch for heritable sequential genetic memory. PLoS ONE 3: e2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton SR, Davidson RC, Sethuraman N, Nett JH, Jiang Y, Rios S, Bobrowicz P, Stadheim TA, Li H, Choi BK, et al. 2006. Humanization of yeast to produce complex terminally sialylated glycoproteins. Science 313: 1441–1443. [DOI] [PubMed] [Google Scholar]
- Harbers M. 2014. Wheat germ systems for cell-free protein expression. FEBS Lett 588: 2762–2773. [DOI] [PubMed] [Google Scholar]
- Heinemann IU, Rovner AJ, Aerni HR, Rogulina S, Cheng L, Olds W, Fischer JT, Soll D, Isaacs FJ, Rinehart J. 2012. Enhanced phosphoserine insertion during Escherichia coli protein synthesis via partial UAG codon reassignment and release factor 1 deletion. FEBS Lett 586: 3716–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyman Y, Buxboim A, Wolf SG, Daube SS, Bar-Ziv RH. 2012. Cell-free protein synthesis and assembly on a biochip. Nat Nanotechnol 7: 374–378. [DOI] [PubMed] [Google Scholar]
- Hodgman CE, Jewett MC. 2013. Optimized extract preparation methods and reaction conditions for improved yeast cell-free protein synthesis. Biotechnol Bioeng 110: 2643–2654. [DOI] [PubMed] [Google Scholar]
- Hodgman CE, Jewett MC. 2014. Characterizing IGR IRES-mediated translation initiation for use in yeast cell-free protein synthesis. N Biotechnol 31: 499–505. [DOI] [PubMed] [Google Scholar]
- Hong SH, Kwon YC, Jewett MC. 2014a. Non-standard amino acid incorporation into proteins using Escherichia coli cell-free protein synthesis. Front Chem 2: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SH, Ntai I, Haimovich AD, Kelleher NL, Isaacs FJ, Jewett MC. 2014b. Cell-free protein synthesis from a release factor 1 deficient Escherichia coli activates efficient and multiple site-specific nonstandard amino acid incorporation. ACS Synth Biol 3: 398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SH, Kwon YC, Martin RW, Des Soye BJ, de Paz AM, Swonger KN, Ntai I, Kelleher NL, Jewett MC. 2015. Improving cell-free protein synthesis through genome engineering of Escherichia coli lacking release factor 1. Chembiochem 16: 844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka N, Najita L, Franzusoff A, Sarnow P. 1994. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol Cell Biol 14: 7322–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuma K, Liao JC, Okamoto M, Hanai T. 2010. Improvement of isopropanol production by metabolically engineered Escherichia coli using gas stripping. J Biosci Bioeng 110: 696–701. [DOI] [PubMed] [Google Scholar]
- Iyer S, Doktycz MJ. 2014. Thrombin-mediated transcriptional regulation using DNA aptamers in DNA-based cell-free protein synthesis. ACS Synth Biol 3: 340–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer S, Karig DK, Norred SE, Simpson ML, Doktycz MJ. 2013. Multi-input regulation and logic with T7 promoters in cells and cell-free systems. PLoS ONE 8: e78442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson K, Kanamori T, Ueda T, Fan ZH. 2014a. Protein synthesis yield increased 72 times in the cell-free PURE system. Integr Biol (Camb) 6: 781–788. [DOI] [PubMed] [Google Scholar]
- Jackson K, Khnouf R, Fan ZH. 2014b. Cell-free protein synthesis in microfluidic 96-well plates. Methods Mol Biol 1118: 157–168. [DOI] [PubMed] [Google Scholar]
- Jackson K, Jin S, Fan ZH. 2015. Optimization of a miniaturized fluid array device for cell-free protein synthesis. Biotechnol Bioeng 112: 2459–2467. [DOI] [PubMed] [Google Scholar]
- Jewett MC, Calhoun KA, Voloshin A, Wuu JJ, Swartz JR. 2008. An integrated cell-free metabolic platform for protein production and synthetic biology. Mol Syst Biol 4: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett MC, Fritz BR, Timmerman LE, Church GM. 2013. In vitro integration of ribosomal RNA synthesis, ribosome assembly, and translation. Mol Syst Biol 9: 678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DB, Xu J, Shen Z, Takimoto JK, Schultz MD, Schmitz RJ, Xiang Z, Ecker JR, Briggs SP, Wang L. 2011. RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat Chem Biol 7: 779–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston WA, Alexandrov K. 2014. Production of eukaryotic cell-free lysate from Leishmania tarentolae. Methods Mol Biol 1118: 1–15. [DOI] [PubMed] [Google Scholar]
- Junge F, Haberstock S, Roos C, Stefer S, Proverbio D, Dotsch V, Bernhard F. 2011. Advances in cell-free protein synthesis for the functional and structural analysis of membrane proteins. N Biotechnol 28: 262–271. [DOI] [PubMed] [Google Scholar]
- Kaiser L, Graveland-Bikker J, Steuerwald D, Vanberghem M, Herlihy K, Zhang S. 2008. Efficient cell-free production of olfactory receptors: Detergent optimization, structure, and ligand binding analyses. Proc Natl Acad Sci 105: 15726–15731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karig DK, Iyer S, Simpson ML, Doktycz MJ. 2012. Expression optimization and synthetic gene networks in cell-free systems. Nucleic Acids Res 40: 3763–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzbrun E, Shin J, Bar-Ziv RH, Noireaux V. 2011. Coarse-grained dynamics of protein synthesis in a cell-free system. Phys Rev Lett 106: 048104. [DOI] [PubMed] [Google Scholar]
- Katzen F, Kudlicki W. 2006. Efficient generation of insect-based cell-free translation extracts active in glycosylation and signal sequence processing. J Biotechnol 125: 194–197. [DOI] [PubMed] [Google Scholar]
- Katzen F, Chang G, Kudlicki W. 2005. The past, present and future of cell-free protein synthesis. Trends Biotechnol 23: 150–156. [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Gouda MD, Sawasaki T, Takai K, Endo Y. 2003. Efficient synthesis of a disulfide-containing protein through a batch cell-free system from wheat germ. Eur J Biochem 270: 4780–4786. [DOI] [PubMed] [Google Scholar]
- Kazuta Y, Matsuura T, Ichihashi N, Yomo T. 2014. Synthesis of milligram quantities of proteins using a reconstituted in vitro protein synthesis system. J Biosci Bioeng 118: 554–557. [DOI] [PubMed] [Google Scholar]
- Kiga D, Sakamoto K, Kodama K, Kigawa T, Matsuda T, Yabuki T, Shirouzu M, Harada Y, Nakayama H, Takio K, et al. 2002. An engineered Escherichia coli tyrosyl-tRNA synthetase for site-specific incorporation of an unnatural amino acid into proteins in eukaryotic translation and its application in a wheat germ cell-free system. Proc Natl Acad Sci 99: 9715–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, White KS, Winfree E. 2006. Construction of an in vitro bistable circuit from synthetic transcriptional switches. Mol Syst Biol 2: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun O, Mureev S, Johnston W, Alexandrov K. 2010. Towards the construction of expressed proteomes using a Leishmania tarentolae based cell-free expression system. PloS ONE 5: e14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun O, Mureev S, Jung W, Kubala MH, Johnston W, Alexandrov K. 2011. Leishmania cell-free protein expression system. Methods 55: 58–64. [DOI] [PubMed] [Google Scholar]
- Kubick S, Gerrits M, Merk H, Stiege W, Erdmann VA, Larry D. 2009. In vitro synthesis of posttranslationally modified membrane proteins. In Current topics in membranes (ed. DeLucas L), Vol. 63, pp. 25–49. Elsevier, Amsterdam. [Google Scholar]
- Kucharczyk R, Zick M, Bietenhader M, Rak M, Couplan E, Blondel M, Caubet SD, di Rago JP. 2009. Mitochondrial ATP synthase disorders: Molecular mechanisms and the quest for curative therapeutic approaches. Biochim Biophys Acta 1793: 186–199. [DOI] [PubMed] [Google Scholar]
- Kuchenreuther JM, Shiigi SA, Swartz JR. 2014. Cell-free synthesis of the H-cluster: A model for the in vitro assembly of metalloprotein metal centers. Methods Mol Biol 1122: 49–72. [DOI] [PubMed] [Google Scholar]
- Kunamneni A, Plou FJ, Ballesteros A, Alcalde M. 2008. Laccases and their applications: A patent review. Recent Pat Biotechnol 2: 10–24. [DOI] [PubMed] [Google Scholar]
- Kuruma Y, Suzuki T, Ueda T. 2010. Production of multi-subunit complexes on liposome through an E. coli cell-free expression system. Methods Mol Biol 607: 161–171. [DOI] [PubMed] [Google Scholar]
- Kuruma Y, Suzuki T, Ono S, Yoshida M, Ueda T. 2012. Functional analysis of membranous Fo-a subunit of F1Fo-ATP synthase by in vitro protein synthesis. Biochem J 442: 631–638. [DOI] [PubMed] [Google Scholar]
- Kwon YC, Jewett MC. 2015. High-throughput preparation methods of crude extract for robust cell-free protein synthesis. Sci Rep 5: 8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie MJ, Rovner AJ, Goodman DB, Aerni HR, Haimovich AD, Kuznetsov G, Mercer JA, Wang HH, Carr PA, Mosberg JA, et al. 2013. Genomically recoded organisms expand biological functions. Science 342: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton TJ, Rosenzweig AC. 2011. Detection and characterization of a multicopper oxidase from Nitrosomonas europaea. Methods Enzymol 496: 423–433. [DOI] [PubMed] [Google Scholar]
- Lee KH, Joung HA, Ahn JH, Kim KO, Oh IS, Shin YB, Kim MG, Kim DM. 2007. Real-time monitoring of cell-free protein synthesis on a surface plasmon resonance chip. Anal Biochem 366: 170–174. [DOI] [PubMed] [Google Scholar]
- Lemke EA. 2011. Site-specific labeling of proteins for single-molecule FRET measurements using genetically encoded ketone functionalities. Methods Mol Biol 751: 3–15. [DOI] [PubMed] [Google Scholar]
- Li H, d’Anjou M. 2009. Pharmacological significance of glycosylation in therapeutic proteins. Curr Opin Biotechnol 20: 678–684. [DOI] [PubMed] [Google Scholar]
- Li J, Lawton TJ, Kostecki JS, Nisthal A, Fang J, Mayo SL, Rosenzweig AC, Jewett MC. 2016. Cell-free protein synthesis enables high yielding synthesis of an active multicopper oxidase. Biotechnol J 11: 212–218. [DOI] [PubMed] [Google Scholar]
- Lian Q, Cao H, Wang F. 2014. The cost-efficiency realization in the Escherichia coli-based cell-free protein synthesis systems. Appl Biochem Biotechnol 174: 2351–2367. [DOI] [PubMed] [Google Scholar]
- Litcofsky KD, Afeyan RB, Krom RJ, Khalil AS, Collins JJ. 2012. Iterative plug-and-play methodology for constructing and modifying synthetic gene networks. Nat Methods 9: 1077–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Fritz BR, Anderson MJ, Schoborg JA, Jewett MC. 2014. Characterizing and alleviating substrate limitations for improved in vitro ribosome construction. ACS Synth Biol 4: 454–462. [DOI] [PubMed] [Google Scholar]
- Loscha KV, Herlt AJ, Qi R, Huber T, Ozawa K, Otting G. 2012a. Multiple-site labeling of proteins with unnatural amino acids. Angew Chem Int Ed Engl 51: 2243–2246. [DOI] [PubMed] [Google Scholar]
- Loscha KV, Herlt AJ, Qi R, Huber T, Ozawa K, Otting G. 2012b. Multiple-site labeling of proteins with unnatural amino acids. Angew Chem Int Ed Engl 51: 2243–2246. [DOI] [PubMed] [Google Scholar]
- Lu Y, Welsh JP, Swartz JR. 2014. Production and stabilization of the trimeric influenza hemagglutinin stem domain for potentially broadly protective influenza vaccines. Proc Natl Acad Sci 111: 125–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Chan W, Ko BY, VanLang CC, Swartz JR. 2015. Assessing sequence plasticity of a virus-like nanoparticle by evolution toward a versatile scaffold for vaccines and drug delivery. Proc Natl Acad Sci 112: 12360–12365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madin K, Sawasaki T, Ogasawara T, Endo Y. 2000. A highly efficient and robust cell-free protein synthesis system prepared from wheat embryos: Plants apparently contain a suicide system directed at ribosomes. Proc Natl Acad Sci 97: 559–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini L, Mansy SS. 2014. Measuring riboswitch activity in vitro and in artificial cells with purified transcription-translation machinery. Methods Mol Biol 1111: 153–164. [DOI] [PubMed] [Google Scholar]
- Matsubayashi H, Ueda T. 2014. Purified cell-free systems as standard parts for synthetic biology. Curr Opin Chem Biol 22: 158–162. [DOI] [PubMed] [Google Scholar]
- Matsubayashi H, Kuruma Y, Ueda T. 2014. In vitro synthesis of the E. coli Sec translocon from DNA. Angew Chem Int Ed Engl 53: 7535–7538. [DOI] [PubMed] [Google Scholar]
- Matthies D, Haberstock S, Joos F, Dotsch V, Vonck J, Bernhard F, Meier T. 2011. Cell-free expression and assembly of ATP synthase. J Mol Biol 413: 593–603. [DOI] [PubMed] [Google Scholar]
- Meuris L, Santens F, Elson G, Festjens N, Boone M, Dos Santos A, Devos S, Rousseau F, Plets E, Houthuys E, et al. 2014. GlycoDelete engineering of mammalian cells simplifies N-glycosylation of recombinant proteins. Nat Biotechnol 32: 485–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami S, Masutani M, Sonenberg N, Yokoyama S, Imataka H. 2006. An efficient mammalian cell-free translation system supplemented with translation factors. Protein Expr Purif 46: 348–357. [DOI] [PubMed] [Google Scholar]
- Murakami H, Ohta A, Ashigai H, Suga H. 2006. A highly flexible tRNA acylation method for non-natural polypeptide synthesis. Nat Methods 3: 357–359. [DOI] [PubMed] [Google Scholar]
- Mureev S, Kovtun O, Nguyen UT, Alexandrov K. 2009. Species-independent translational leaders facilitate cell-free expression. Nat Biotechnol 27: 747–752. [DOI] [PubMed] [Google Scholar]
- Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. 2009. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell 36: 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PP, Jia M, Patel KG, Brody JD, Swartz JR, Levy S, Levy R. 2012. A vaccine directed to B cells and produced by cell-free protein synthesis generates potent antilymphoma immunity. Proc Natl Acad Sci 109: 14526–14531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DP, Garcia Alai MM, Kapadnis PB, Neumann H, Chin JW. 2009. Genetically encoding Nɛ-methyl-l-lysine in recombinant histones. J Am Chem Soc 131: 14194–14195. [DOI] [PubMed] [Google Scholar]
- Niederholtmeyer H, Xu L, Maerkl SJ. 2013. Real-time mRNA measurement during an in vitro transcription and translation reaction using binary probes. ACS Synth Biol 2: 411–417. [DOI] [PubMed] [Google Scholar]
- Niederholtmeyer H, Sun ZZ, Hori Y, Yeung E, Verpoorte A, Murray RM, Maerkl SJ. 2015. Rapid cell-free forward engineering of novel genetic ring oscillators. eLife 4: e09771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirenberg MW, Matthaei JH. 1961. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc Natl Acad Sci 47: 1588–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa T, Sunami T, Matsuura T, Yomo T. 2012. Directed evolution of proteins through in vitro protein synthesis in liposomes. J Nucleic Acids 2012: 923214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Matsuura T, Nishimura K, Sunami T, Suzuki H, Yomo T. 2012. Cell-free protein synthesis inside giant unilamellar vesicles analyzed by flow cytometry. Langmuir 28: 8426–8432. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Tsuru S, Suzuki H, Yomo T. 2014. Stochasticity in gene expression in a cell-sized compartment. ACS Synth Biol 4: 566–576. [DOI] [PubMed] [Google Scholar]
- Niwa T, Sasaki Y, Uemura E, Nakamura S, Akiyama M, Ando M, Sawada S, Mukai SA, Ueda T, Taguchi H, et al. 2015. Comprehensive study of liposome-assisted synthesis of membrane proteins using a reconstituted cell-free translation system. Sci Rep 5: 18025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noireaux V, Bar-Ziv R, Libchaber A. 2003. Principles of cell-free genetic circuit assembly. Proc Natl Acad Sci 100: 12672–12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norred SE, Caveney PM, Retterer ST, Boreyko JB, Fowlkes JD, Collier CP, Simpson ML. 2015. Sealable femtoliter chamber arrays for cell-free biology. J Vis Exp 10.3791/52616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa A, Tozawa Y. 2014. Modifications of wheat germ cell-free system for functional proteomics of plant membrane proteins. Methods Mol Biol 1072: 259–272. [DOI] [PubMed] [Google Scholar]
- Nozawa A, Matsubara Y, Tanaka Y, Takahashi H, Akagi T, Seki M, Shinozaki K, Endo Y, Sawasaki T. 2009. Construction of a protein library of Arabidopsis transcription factors using a wheat cell-free protein production system and its application for DNA binding analysis. Biosci Biotechnol Biochem 73: 1661–1664. [DOI] [PubMed] [Google Scholar]
- O’Donoghue P, Ling J, Wang YS, Soll D. 2013. Upgrading protein synthesis for synthetic biology. Nat Chem Biol 9: 594–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SJ, Lee KH, Kim HC, Catherine C, Yun H, Kim DM. 2014. Translational incorporation of multiple unnatural amino acids in a cell-free protein synthesis system. Biotechnol Bioproc Eng 19: 426–432. [Google Scholar]
- Ohtake K, Sato A, Mukai T, Hino N, Yokoyama S, Sakamoto K. 2012. Efficient decoding of the UAG triplet as a full-fledged sense codon enhances the growth of a prfA-deficient strain of Escherichia coli. J Bacteriol 194: 2606–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth JH, Schorch B, Boundy S, Ffrench-Constant R, Kubick S, Aktories K. 2011. Cell-free synthesis and characterization of a novel cytotoxic pierisin-like protein from the cabbage butterfly Pieris rapae. Toxicon 57: 199–207. [DOI] [PubMed] [Google Scholar]
- Oza JP, Aerni HR, Pirman NL, Barber KW, ter Haar CM, Rogulina S, Amrofell MB, Isaacs FJ, Rinehart J, Jewett MC. 2015. Robust production of recombinant phosphoproteins using cell-free protein synthesis. Nat Commun 6: 8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa K, Loscha KV, Kuppan KV, Loh CT, Dixon NE, Otting G. 2012. High-yield cell-free protein synthesis for site-specific incorporation of unnatural amino acids at two sites. Biochem Biophys Res Commun 418: 652–656. [DOI] [PubMed] [Google Scholar]
- Pardee K, Green AA, Ferrante T, Cameron DE, DaleyKeyser A, Yin P, Collins JJ. 2014. Paper-based synthetic gene networks. Cell 159: 940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HS, Hohn MJ, Umehara T, Guo LT, Osborne EM, Benner J, Noren CJ, Rinehart J, Soll D. 2011. Expanding the genetic code of Escherichia coli with phosphoserine. Science 333: 1151–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passioura T, Suga H. 2013. Flexizyme-mediated genetic reprogramming as a tool for noncanonical peptide synthesis and drug discovery. Chemistry 19: 6530–6536. [DOI] [PubMed] [Google Scholar]
- Pratt J. 1984. Coupled transcription-translation in prokaryotic cell-free systems. In Transcription and translation: A practical approach (ed. Hames BD, Higgins SJ), pp. 179–209. Oxford University Press, Cambridge. [Google Scholar]
- Rackham O, Chin JW. 2005. Cellular logic with orthogonal ribosomes. J Am Chem Soc 127: 17584–17585. [DOI] [PubMed] [Google Scholar]
- Rajendran M, Ellington AD. 2008. Selection of fluorescent aptamer beacons that light up in the presence of zinc. Anal Bioanal Chem 390: 1067–1075. [DOI] [PubMed] [Google Scholar]
- Rendl M, Brandstetter T, Ruhe J. 2013. Time-resolved analysis of biological reactions based on heterogeneous assays in liquid plugs of nanoliter volume. Anal Chem 85: 9469–9477. [DOI] [PubMed] [Google Scholar]
- Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, et al. 2006. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440: 940–943. [DOI] [PubMed] [Google Scholar]
- Sachse R, Dondapati SK, Fenz SF, Schmidt T, Kubick S. 2014. Membrane protein synthesis in cell-free systems: From bio-mimetic systems to bio-membranes. FEBS Lett 588: 2774–2781. [DOI] [PubMed] [Google Scholar]
- Salehi AS, Smith MT, Bennett AM, Williams JB, Pitt WG, Bundy BC. 2016. Cell-free protein synthesis of a cytotoxic cancer therapeutic: Onconase production and a just-add-water cell-free system. Biotechnol J 11: 274–281. [DOI] [PubMed] [Google Scholar]
- Sawasaki T, Ogasawara T, Morishita R, Endo Y. 2002. A cell-free protein synthesis system for high-throughput proteomics. Proc Natl Acad Sci 99: 14652–14657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoborg JA, Hodgman CE, Anderson MJ, Jewett MC. 2014. Substrate replenishment and byproduct removal improve yeast cell-free protein synthesis. Biotechnol J 9: 630–640. [DOI] [PubMed] [Google Scholar]
- Selinger DW, Saxena RM, Cheung KJ, Church GM, Rosenow C. 2003. Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res 13: 216–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkrig J, Leyton DL, Webb CT, Lithgow T. 2014. Assembly of β-barrel proteins into bacterial outer membranes. Biochim Biophys Acta 1843: 1542–1550. [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. 2001. Cell-free translation reconstituted with purified components. Nat Biotechnol 19: 751–755. [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Kanamori T, Ueda T. 2005. Protein synthesis by pure translation systems. Methods 36: 299–304. [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Kuruma Y, Kanamori T, Ueda T. 2014. The PURE system for protein production. Methods Mol Biol 1118: 275–284. [DOI] [PubMed] [Google Scholar]
- Shin J, Noireaux V. 2010. Study of messenger RNA inactivation and protein degradation in an Escherichia coli cell-free expression system. J Biol Eng 4: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Noireaux V. 2012. An E. coli cell-free expression toolbox: Application to synthetic gene circuits and artificial cells. ACS Synth Biol 1: 29–41. [DOI] [PubMed] [Google Scholar]
- Shin J, Jardine P, Noireaux V. 2012. Genome replication, synthesis, and assembly of the bacteriophage T7 in a single cell-free reaction. ACS Synth Biol 1: 408–413. [DOI] [PubMed] [Google Scholar]
- Shrestha P, Holland TM, Bundy BC. 2012. Streamlined extract preparation for Escherichia coli-based cell-free protein synthesis by sonication or bead vortex mixing. Biotechniques 53: 163–174. [DOI] [PubMed] [Google Scholar]
- Siegal-Gaskins D, Tuza ZA, Kim J, Noireaux V, Murray RM. 2014. Gene circuit performance characterization and resource usage in a cell-free “breadboard.” ACS Synth Biol 3: 416–425. [DOI] [PubMed] [Google Scholar]
- Sleight SC, Sauro HM. 2013. Randomized BioBrick assembly: A novel DNA assembly method for randomizing and optimizing genetic circuits and metabolic pathways. ACS Synth Biol 2: 506–518. [DOI] [PubMed] [Google Scholar]
- Stafford RL, Matsumoto ML, Yin G, Cai Q, Fung JJ, Stephenson H, Gill A, You M, Lin SH, Wang WD, et al. 2014. In vitro Fab display: A cell-free system for IgG discovery. Protein Eng Des Sel 27: 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stano P, Luisi PL. 2013. Semi-synthetic minimal cells: Origin and recent developments. Curr Opin Biotechnol 24: 633–638. [DOI] [PubMed] [Google Scholar]
- Stech M, Merk H, Schenk JA, Stocklein WF, Wustenhagen DA, Micheel B, Duschl C, Bier FF, Kubick S. 2012. Production of functional antibody fragments in a vesicle-based eukaryotic cell-free translation system. J Biotechnol 164: 220–231. [DOI] [PubMed] [Google Scholar]
- Stech M, Brodel AK, Quast RB, Sachse R, Kubick S. 2013. Cell-free systems: Functional modules for synthetic and chemical biology. Adv Biochem Eng Biotechnol 137: 67–102. [DOI] [PubMed] [Google Scholar]
- Stech M, Quast RB, Sachse R, Schulze C, Wustenhagen DA, Kubick S. 2014. A continuous-exchange cell-free protein synthesis system based on extracts from cultured insect cells. PLoS ONE 9: e96635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CJ, Pendleton ED, Sasmor HH, Hicks WL, Farnum JB, Muto M, Amendt EM, Schoborg JA, Martin RW, Clark LG, et al. 2016. A cell-free expression and purification process for rapid production of protein biologics. Biotechnol J 11: 238–248. [DOI] [PubMed] [Google Scholar]
- Sun ZZ, Yeung E, Hayes CA, Noireaux V, Murray RM. 2014. Linear DNA for rapid prototyping of synthetic biological circuits in an Escherichia coli based TX-TL cell-free system. ACS Synth Biol 3: 387–397. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Ito M, Ezure T, Shikata M, Ando E, Utsumi T, Tsunasawa S, Nishimura O. 2007. Preparation of N-acylated proteins modified with fatty acids having a specific chain length using an insect cell-free protein synthesis system. Biosci Biotechnol Biochem 71: 261–264. [DOI] [PubMed] [Google Scholar]
- Swartz J. 2006. Developing cell-free biology for industrial applications. J Ind Microbiol Biotechnol 33: 476–485. [DOI] [PubMed] [Google Scholar]
- Takahashi MK, Chappell J, Hayes CA, Sun ZZ, Kim J, Singhal V, Spring KJ, Al-Khabouri S, Fall CP, Noireaux V, et al. 2014. Rapidly characterizing the fast dynamics of RNA genetic circuitry with cell-free transcription-translation (TX-TL) systems. ACS Synth Biol 4: 503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemori N, Takemori A, Matsuoka K, Morishita R, Matsushita N, Aoshima M, Takeda H, Sawasaki T, Endo Y, Higashiyama S. 2015. High-throughput synthesis of stable isotope-labeled transmembrane proteins for targeted transmembrane proteomics using a wheat germ cell-free protein synthesis system. Mol Biosyst 11: 361–365. [DOI] [PubMed] [Google Scholar]
- Timm AC, Shankles PG, Foster CM, Doktycz MJ, Retterer ST. 2016. Toward microfluidic reactors for cell-free protein synthesis at the point-of-care. Small 12: 810–817. [DOI] [PubMed] [Google Scholar]
- Tsuboi T, Takeo S, Iriko H, Jin L, Tsuchimochi M, Matsuda S, Han ET, Otsuki H, Kaneko O, Sattabongkot J, et al. 2008. Wheat germ cell-free system-based production of malaria proteins for discovery of novel vaccine candidates. Infect Immun 76: 1702–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valderrama-Rincon JD, Fisher AC, Merritt JH, Fan YY, Reading CA, Chhiba K, Heiss C, Azadi P, Aebi M, DeLisa MP. 2012. An engineered eukaryotic protein glycosylation pathway in Escherichia coli. Nat Chem Biol 8: 434–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66–74. [DOI] [PubMed] [Google Scholar]
- Virdee S, Kapadnis PB, Elliott T, Lang K, Madrzak J, Nguyen DP, Riechmann L, Chin JW. 2011. Traceless and site-specific ubiquitination of recombinant proteins. J Am Chem Soc 133: 10708–10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xie J, Schultz PG. 2006. Expanding the genetic code. Annu Rev Biophys Biomol Struct 35: 225–249. [DOI] [PubMed] [Google Scholar]
- Wang X, Liu J, Zheng Y, Li J, Wang H, Zhou Y, Qi M, Yu H, Tang W, Zhao WM. 2008. An optimized yeast cell-free system: Sufficient for translation of human papillomavirus 58 L1 mRNA and assembly of virus-like particles. J Biosci Bioeng 106: 8–15. [DOI] [PubMed] [Google Scholar]
- Wang X, Corin K, Baaske P, Wienken CJ, Jerabek-Willemsen M, Duhr S, Braun D, Zhang S. 2011. Peptide surfactants for cell-free production of functional G protein–coupled receptors. Proc Natl Acad Sci 108: 9049–9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win MN, Smolke CD. 2008. Higher-order cellular information processing with synthetic RNA devices. Science 322: 456–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Lee J, Tran C, Heibeck TH, Wang WD, Yang J, Stafford RL, Steiner AR, Sato AK, Hallam TJ. 2014. Production of bispecific antibodies in “Knobs-into-Holes” using a cell-free expression system. mAbs 7: 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin G, Garces ED, Yang J, Zhang J, Tran C, Steiner AR, Roos C, Bajad S, Hudak S, Penta K, et al. 2012. Aglycosylated antibodies and antibody fragments produced in a scalable in vitro transcription-translation system. mAbs 4: 217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunker PJ, Asahara H, Hung KC, Landry C, Arriaga LR, Akartuna I, Heyman J, Chong S, Weitz DA. 2016. One-pot system for synthesis, assembly, and display of functional single-span membrane proteins on oil–water interfaces. Proc Natl Acad Sci 113: 608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalkin H, Yanofsky C, Squires CL. 1974. Regulated in vitro synthesis of Escherichia coli tryptophan operon messenger ribonucleic acid and enzymes. J Biol Chem 249: 465–475. [PubMed] [Google Scholar]
- Zawada JF, Yin G, Steiner AR, Yang J, Naresh A, Roy SM, Gold DS, Heinsohn HG, Murray CJ. 2011. Microscale to manufacturing scale-up of cell-free cytokine production—A new approach for shortening protein production development timelines. Biotechnol Bioeng 108: 1570–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemella A, Thoring L, Hoffmeister C, Kubick S. 2015. Cell-free protein synthesis: Pros and cons of prokaryotic and eukaryotic systems. Chembiochem 16: 2420–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieleniecki JL, Nagarajan Y, Waters S, Rongala J, Thompson V, Hrmova M, Koper I. 2016. Cell-free synthesis of a functional membrane transporter into a tethered bilayer lipid membrane. Langmuir 32: 2445–2449. [DOI] [PubMed] [Google Scholar]
- Zimmerman ES, Heibeck TH, Gill A, Li X, Murray CJ, Madlansacay MR, Tran C, Uter NT, Yin G, Rivers PJ, et al. 2014. Production of site-specific antibody-drug conjugates using optimized non-natural amino acids in a cell-free expression system. Bioconjug Chem 25: 351–361. [DOI] [PubMed] [Google Scholar]