Abstract
We review the evolution and structure of members of the transforming growth factor β (TGF-β) family, antagonistic or agonistic modulators, and receptors that regulate TGF-β signaling in extracellular environments. The growth factor (GF) domain common to all family members and many of their antagonists evolved from a common cystine knot growth factor (CKGF) domain. The CKGF superfamily comprises six distinct families in primitive metazoans, including the TGF-β and Dan families. Compared with Wnt/Frizzled and Notch/Delta families that also specify body axes, cell fate, tissues, and other families that contain CKGF domains that evolved in parallel, the TGF-β family was the most fruitful in evolution. Complexes between the prodomains and GFs of the TGF-β family suggest a new paradigm for regulating GF release by conversion from closed- to open-arm procomplex conformations. Ternary complexes of the final step in extracellular signaling show how TGF-β GF dimers bind type I and type II receptors on the cell surface, and enable understanding of much of the specificity and promiscuity in extracellular signaling. However, structures suggest that when GFs bind repulsive guidance molecule (RGM) family coreceptors, type I receptors do not bind until reaching an intracellular, membrane-enveloped compartment, blurring the line between extra- and intracellular signaling. Modulator protein structures show how structurally diverse antagonists including follistatins, noggin, and members of the chordin family bind GFs to regulate signaling; complexes with the Dan family remain elusive. Much work is needed to understand how these molecular components assemble to form signaling hubs in extracellular environments in vivo.
All TGF-β family members share a common growth factor (GF) domain. The distinct activities of TGF-β family members are not determined by GF-receptor interactions alone but by a network of interacting partners.
Multicellular organisms require an elaborate system of intercellular communication to coordinate cellular actions. In developing embryos, the ability of cells to sense and respond to such communication is vital for the establishment of the overall body plan and to pattern tissues, whereas, in adults, it is required for diverse processes including repair of damaged tissues and regulation of immune responses. In mammals, the 33 genes of the transforming growth factor β (TGF-β) family each encode a polypeptide comprising a secretion signal peptide, a ∼250-residue prodomain, and a ∼110-residue growth factor (GF) domain. Family members include bone morphogenetic proteins (BMPs), growth and differentiation factors (GDFs), activins, and TGF-βs. We discuss the emerging concept that the distinctive activities of TGF-β family GFs are determined not just by signaling type I and type II receptors, which show varying degrees of promiscuity for GFs, but also by multiple binding proteins and enzymes. These modulators greatly diversify signaling activity by adding another layer of signaling that occurs in extracellular environments. They control not only whether GFs reach their receptors on cells, but also whether additional components are present within GF-receptor complexes. Receptors include not only the classical type I and type II receptors, but also type III coreceptors and repulsive guidance molecule (RGM) coreceptors. Binding proteins comprise the cognate prodomains, antagonists such as noggin, follistatin, chordin, and Dan proteins, anchoring molecules such as latent TGF-β binding protein (LTBP), and activators such as integrins. Enzymes include proprotein convertases (PCs) that cleave the prodomain from the GF, either intracellularly during biosynthesis or extracellularly, and tolloid and matrix metalloproteases. Thus, molecular recognition in the TGF-β family is not singularly achieved by GF-receptor interactions, but by a network of interactions with multiple partners. Box 1 summarizes the proteins, interactions, relationships, and terms used in this review.
BOX 1. PRODOMAIN, GROWTH FACTOR (GF), AND RECEPTOR STRUCTURES AND EVOLUTIONARY TREES.
Structures are described in sections on Structures and Functions of TGF-β Family Procomplexes, and Specificity and Promiscuity in GF-Receptor Interactions. For evolutionary trees, sequences were aligned using MAFFT version 7 and the E-INS-i strategy with BLOSUM30 matrices and gap penalties of 1.53 to 2.5 (Katoh and Standley 2013). Trees were calculated as implemented on the same MAFFT server with the NJ method using all gap-free sites, the JTT model, estimation of α, and 1000 bootstrap samples. In each tree, branch lengths are to the scale shown in the bars. 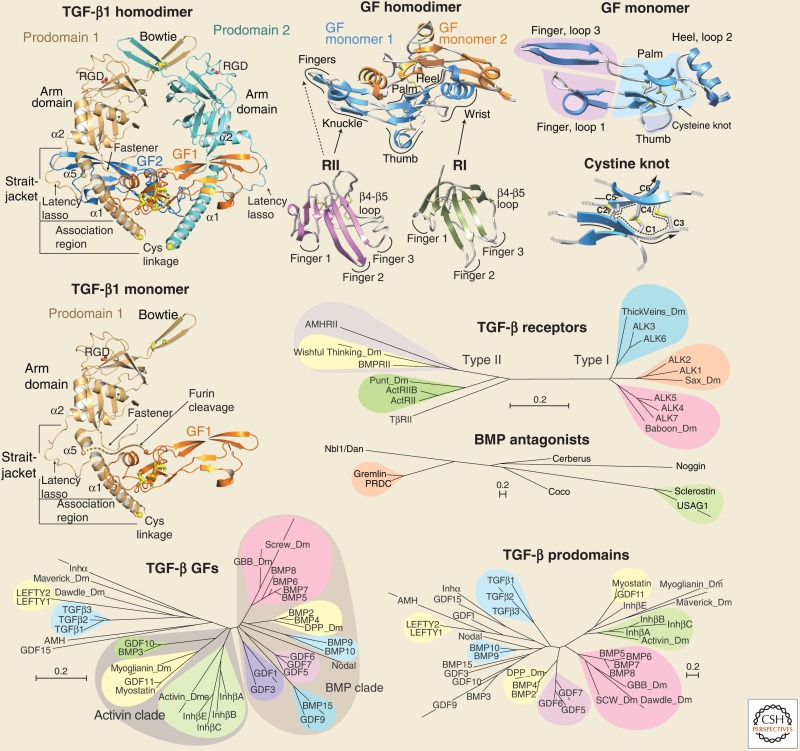
The complex network of interactions described here parallels that found through high-throughput mapping of interactomes (Li et al. 2004; Rual et al. 2005; Guruharsha et al. 2011), which has shown that regulation is maintained by a protein–protein interaction network in which some proteins serve as hubs and interact with a larger number of interaction partners to regulate activity (Jeong et al. 2000). These findings shift our understanding of “linear” signaling cascades toward complex signaling networks, which may better explain the functional diversity encoded by TGF-β family proteins in multicellular animals.
Our review has four sections following the introduction. The protein domain from which TGF-β GFs and many of their inhibitors are constructed evolved in early metazoans before the development of bilateral symmetry (see section on a Brief History of TGF-β Family Evolution). GFs are biosynthesized as dimeric prodomain–GF complexes, and the prodomain has an important role in storage and release (activation) of the GF (see section on Structures and Functions of TGF-β Family Procomplexes). Type I and type II receptors and coreceptors bind and discriminate between GFs and signal into cells (see section on Specificity and Promiscuity in GF-Receptor Interactions). Finally, a large number of inhibitors form complexes with the GFs to antagonize or modulate signaling (see section on Regulation of TGF-β/BMP Signaling by Modulator Proteins—BMPs as Signaling Hubs).
A BRIEF HISTORY OF TGF-β FAMILY EVOLUTION
Function, three-dimensional structure, and amino acid sequence are all related. Thus, tracing the evolution of the TGF-β family through its sequence relationships is an excellent introduction to function. The TGF-β family and its receptors and antagonists evolved in parallel with the Wnt/Frizzled and Delta/Notch pathways. Together, these extracellular ligands and receptors create the networks that establish multicellular animal body plans. They specify anteroposterior, dorsoventral (bilateral), and left–right axes, details of individual organs, and regulate development and homeostasis.
The CKGF Domain and Its Families
The cystine knot (CK) growth factor (CKGF) domain has a specific three-dimensional fold and sequence. Other proteins contain CKs and thus the CK alone does not define the CKGF domain; the definition of this domain also includes the topological relationship between β-strand order in β-sheets and amino acid sequence. The CKGF domain can thus be recognized both by sequence and by structure (Roch and Sherwood 2014), similarly to epidermal growth factor (EGF) or immunoglobulin (Ig) domains. And just as the latter families have diverse functions, many proteins containing CKGF domains lack growth factor activity. Most TGF-β family members act as differentiation factors rather than growth factors and some such as inhibins and Leftys antagonize activities of other members. However, just as TGF-β is eponymous for the family as a whole, its signaling moiety is also eponymous for what we term the GF domain. All family members contain a GF domain that can form a GF dimer; thus, we term this the GF domain whether or not it has signaling activity.
The CKGF domain contains two long β-ribbons (β-sheets with two antiparallel β-strands) and a CK (GF monomer, Box 1). Two closely spaced pairs of cysteines in adjacent, parallel β-strands disulfide-link to form a ring composed of two peptide backbone segments and two disulfides. Another disulfide passes through the ring, linking two additional polypeptide segments (cystine knot, Box 1). Thus, the knot ties together four polypeptide segments that are distal in sequence, and forms a highly stable CK core from which three long loops emanate (GF monomer, Box 1).
The CKGF domain superfamily is present in the earliest metazoans. Like the Wnt/Frizzled and Delta/Notch families, the CKGF family is not found in protists, and first appears in primitive metazoans that lack a bilateral axis. Representatives are present in four distinct prebilaterian phyla, that is, Porifera (sponges), Cnidaria (coral and hydra), Ctenophora (comb jellies), and Placozoa (Trichoplax) (Roch and Sherwood 2014). The CKGF superfamily is composed of six groups (families) with the CKGF domain as the primary structural feature, that is, with or without a prodomain and without any other folded domain (Adamska et al. 2007; Roch and Sherwood 2014). These are (1) the TGF-β family, (2) the Dan family of BMP antagonists, (3) the glycoprotein hormone family (GPH), for example, the pituitary hormones follicle-stimulating hormone, luteinizing hormone, and thyroid-stimulating hormone, (4) bursicon hormones, which are limited to invertebrates, (5) the platelet-derived growth factor (PDGF) family, including vascular endothelial growth factors (VEGFs), and (6) the nerve growth factors (NGFs) (Roch and Sherwood 2014). CKGF domains are also found in noggin, in the CCN family, and as carboxy-terminal CK domains.
From Prebilaterians to the Emergence of TGF-β Itself in Deuterostomes
Of these six families, the first five are represented in prebilaterian phyla. Moreover, sequences characteristic of specific Dan family members represented in Cnidaria, Porifera, Ctenophora, and Placazoa are detectable in humans. In other words, statistically significant relationships between sequences can be used to infer orthology so that prebilaterian Dan family members can be linked to specific proteins or subfamilies of related proteins in humans, including gremlin, sclerostin, and uterine sensitization-associated gene 1 (USAG1) (Roch and Sherwood 2014). TGF-β family members and their type I and type II receptors are also present in nonbilaterians, but orthology with particular family members in chordates is unclear. In contrast, Smad orthologues corresponding to Smad 1 or 5, Smad 2 or 3, the co-Smad Smad4, and inhibitory Smads (I-Smads) can be identified in nonbilaterians (Herpin et al. 2004; Huminiecki et al. 2009; Pang et al. 2011).
The next step in evolution is marked by the definition of a new body axis, the bilateral axis, which creates dorsoventral asymmetry and also enables development of left–right asymmetry. Bilaterian phyla are grouped by nucleotide and protein sequence into three branches that extend previous embryological classification (Blair and Hedges 2005; Dunn et al. 2008; Hejnol et al. 2009). The protostome branch is now subdivided into the Ecdysozoa (e.g., Caenorhabditis elegans and Drosophila melanogaster) and Lophotrochozoa (e.g., molluscs and annelids) branches. Deuterostomes represent a later evolving group of phyla. Molecular phylogeny confirms deuterostomes as a branch with three phyla, Echinodermata (e.g., sea urchins), Chordata (vertebrates, urochordates, and cephalochordates), and Hemichordata (acorn worms).
Key innovations in the evolution of deuterostomes from lower bilaterians include the emergence of pharyngeal gill slits and an inversion in the dorsoventral axis of the body plan (Holland et al. 2015; Lowe et al. 2015). Notably, the BMP and chordin gradients that establish dorsoventral polarity also invert in deuterostomes (Lowe et al. 2015).
Despite the presence of multiple TGF-β family members, TGF-β family receptors, and Smads, TGF-β is clearly absent from early bilaterians. TGF-β itself first appears in the genome-based evolutionary record with the emergence of deuterostomes. Lower deuterostomes, including sea urchins, tunicates, and hemichordates, have only a single TGF-β, and in all cases its prodomain has an RGD motif (Robertson and Rifkin 2013). Mammals have three TGF-βs. TGF-β1 and -β3 contain RGD motifs and bind and are activated by integrins. TGF-β2 diverged from TGF-β3 after the divergence of TGF-β1. Thus, the lack of the Arg of the RGD motif in TGF-β2 is an acquired, not ancestral, characteristic. Furthermore, the acidic Asp residue retained in TGF-β2 is the only key residue required for integrin binding, and thus it is possible that TGF-β2 is activated by a yet unidentified integrin.
These evolutionary events shed important light on the TGF-β family and answer one of the commonly asked questions in the field, “Why are there so many antagonists of BMPs and activins but not TGF-βs?” Currently sequenced nonbilaterian metazoans have as many or more members of the CKGF Dan family than of the CKGF TGF-β family itself. Therefore, in prebilaterians, BMP antagonist and TGF-β family members coevolved in the presence of one another (Roch and Sherwood 2014), and in the presence of other developmental pathways such as Wnt (Adamska et al. 2007). These signaling families matured and diversified much further in bilaterian invertebrates, to the point where members in Drosophila can be grouped with specific TGF-β subfamilies in humans (TGF-β GFs, Box 1). In contrast, TGF-β itself evolved much later, long after the diversification of bilaterian phyla, at the emergence of deuterostomes. Integrins, like the CKGF superfamily, are present in both prebilaterian and early bilaterian metazoans, in which they are already well diversified. Thus, TGF-β emerged in a context in which the key tasks of bilaterian axis and organ specification by complex signaling networks were well established. Within this context, TGF-β evolved to take advantage of a new regulatory mechanism, in which integrins activate signaling. Such a new mechanism may have been required because the regulatory hubs involving BMP antagonists were already well established in the specification of body axes and organs, and their use by a radical new CKGF member might have thrown a wrench into this machinery.
Family Trees
The family trees in Box 1 show the diversification of family members in humans and D. melanogaster for TGF-β family GFs and prodomains (separately), BMP antagonists, and TGF-β family type I and type II receptors. These trees were constructed using rigorous methods appropriate for defining phylogenetic relationships, as implemented in MAFFT (Katoh and Standley 2013). Scale bars show the extent of sequence divergence, which for any two members is given by adding the lengths of the branches that separate them. However, the trees show ontogenetic relationships, not phylogenetic relationships. Thus, the trees in Box 1 show sequence divergence as a consequence of functional and structural diversification, rather than as a consequence of separation of animal species in evolutionary time. Furthermore, the shapes of the trees in Box 1 provide insights into how function and structure have driven diversification of the subfamilies. Separate trees for the TGF-β family prodomains and GFs emphasize that the prodomains are more divergent; when the scale bars in Box 1 are taken into account, the prodomains are about fourfold more divergent than the GFs. Thus, the prodomains have been under greater functional or structural pressure for divergence than the GFs. Also, these pressures must differ, because branch lengths and clustering within subfamilies differ; however, because of higher sequence identity for the GFs, subfamilies can be defined with more statistical significance.
Clustering using neighbor joining with bootstrap calculations (here) or Bayesian analysis (Pang et al. 2011) support significant relationships among TGF-β family GF domains (color-coded in Box 1). A BMP clade includes six subfamilies. Human BMP-5, 6, 7, and 8 and Drosophila Glass bottom boat (Gbb) and Screw cluster in one group that is significantly related to the cluster with human BMP-2 and 4 and Drosophila Decapentaplegic. Four other clusters contain BMP-9 and 10; GDF-5, 6, and 7; GDF-9 and BMP-15; and GDF-1 and 3. Nodal falls in the same BMP clade. An activin clade includes three subgroups: Drosophila activin and human inhibin β-subunits, which dimerize to form activins, GDF-11, myostatin (GDF-8) and possibly Drosophila Myoglianin, and BMP-3 and GDF-10. Further family members in the left side of the GF family tree in Box 1 are difficult to place, but lie closer to the activin than BMP clade. All have unique features. TGF-βs and anti-Müllerian hormone (AMH) each have unique type II receptors. AMH and GDF-15 have atypical prodomains that are the longest and shortest in the family, with 434 and 170 residues, respectively. Lefty 1 and 2 are cleaved by furin after the α2-helix in the prodomain, rather than between the prodomain and GF domain (Fig. 1), and are thus predicted to have their type II receptor binding sites permanently blocked (Shi et al. 2011; Mi et al. 2015). Inhibin-α is inhibitory; when it heterodimerizes with inhibin β-subunits it forms inhibin.
Figure 1.
Sequence alignment of the transforming growth factor β (TGF-β) family. Sequences were aligned with MAFFT (Katoh and Standley 2013), as described (Shi et al. 2011), with slight structural realignment of TGF-β1 and bone morphogenetic protein (BMP)-9 procomplexes (Mi et al. 2015). Structure elements of pro-TGF-β1 (Shi et al. 2011) and pro-BMP-9 (Mi et al. 2015) are labeled and shown as colored lines (upper and lower lines, respectively). Potential proprotein convertase (PC) and tolloid cleavage sites are marked with green and red circles, respectively. Potentially N-glycosylated asparagines in all family members and known phosphorylated serines in mature BMP-15 and growth and differentiation factor 9 (GDF-9) (Tibaldi et al. 2010) are shown in bold red. All 33 human TGF-β family polypeptides are shown, except BMP-8B; the almost identical BMP-8A is shown as BMP-8. Three well-characterized Drosophila melanogaster BMPs are included: Decapentaplegic (Dpp), Screw (Scw), and Glass bottom boat (Gbb). Closely related family members appear in adjacent rows.
Type I and type II receptors for TGF-β family GFs have structurally homologous ligand-binding ectodomains and homologous dual specificity kinase cytoplasmic domains, and thus may be compared in the same tree (TGF-β receptors, Box 1). The longer branches of type II receptors show that they are more diverse than the type I receptors; the receptor trees reflect primarily the cytoplasmic domain because of its much greater number of aligned positions. The human type I receptors show three clades, each with a Drosophila relative. Activin receptor-like kinase (ALK) 1 and 2 are closely related to Saxophone (Sax), ALK3 and 6 are related to Thickveins, and ALK4, 5 and 7 are relatives to Baboon. The ALK1/2, ALK3/6, and ALK4–7 subfamilies couple to two distinct groups of receptor-regulated Smads (R-Smads), with the ALK1/2 and ALK3/6 subfamilies coupling to R-Smad1, 5, 8 and the ALK4/5/7 subfamily coupling to R-Smad2 and 3. Type II receptors also show three groups. BMPRII and AMHRII relate to Wishful thinking, and ActRII and ActRIIB ally with Punt. In contrast, among all type I and type II receptors, only TβRII lacks a Drosophila relative, consistent with the recent deuterostome origin of TGF-β. Moreover, TβRII has no close human relatives.
The Dan family of antagonists against BMPs and Wnts has seven members in humans and shares some similarity with noggin in its CKGF domain (BMP antagonists, Box 1). Although present in many prebilaterians, the Dan family was lost in insects.
Did the Prodomain Contribute to the Evolutionary Success of the TGF-β Family?
Although the origins of the prodomain of TGF-β remain unclear, it may have been an important contributor to the success of the TGF-β family branch of the CKGF superfamily. In evolutionary terms, success may be defined not only as lack of extinction, but also as diversification and radiation into a protein family with a large number of members in higher organisms. By the criterion of number of members alone, the TGF-β family has been more successful than any of its kindred hormone or GF families in the CKGF superfamily (Roch and Sherwood 2014). The TGF-β family also radiated more than other extracellular protein families of parallel importance in developmental specification. With 33 gene family members in mammals, the TGF-β family is more numerous than the 19 Wnts, which have 10 Frizzled receptors, or the total of five Deltas and Jaggeds, which have four Notch receptors.
The prodomain, including its arm domain and straitjacket appendages (TGF-β1 homodimer, Box 1) is clearly defined in the sequences of prebilaterian TGF-β family members (Adamska et al. 2007; Roch and Sherwood 2014). The CKGF domains of the TGF-β family dimerize in a specific manner and associate with two prodomain monomers; the prodomain arm domain and straitjacket extensions together wrap around the GF and block receptor and antagonist binding (TGF-β1 homodimer and GF homodimer, Box 1).
The common focus in the TGF-β family field on the structures and activities of the mature proteins might suggest that the prodomain functions primarily in proper folding and dimerization of GF domains during biosynthesis; however, evolution suggests otherwise. Among the six CKGF groups described above, the GPHs, bursicons, and BMP antagonists have no prodomains and many are disulfide-linked dimers. Thus, prodomains are not essential for folding or dimerization of proteins in the CKGF superfamily. Moreover, the glial-derived neurotrophic factors (GDNFs), including neurturin, artemin, and persephin, have GF domains that are very similar in sequence to those of the TGF-β family, dimerize through the equivalent cysteine, and have similar structures (Wang et al. 2006). Yet, GDNF prodomains are much shorter (41–75 residues) than TGF-β family prodomains (∼250 residues), and have no detectable sequence homology (Shi et al. 2011). Two other CKGF families, the nerve growth factor (NGF) and PDGF families, have prodomains of ∼100 residues that are functionally important and structurally characterized. The PDGF prodomain remains bound after cleavage and shields the receptor binding site (Shim et al. 2010). In contrast, the NGF prodomain is poorly ordered and dissociates from the GF after cleavage; nonetheless, the prodomain has an important regulatory role because it alters the stoichiometry and symmetry of NGF binding to its receptor p75NTR (Feng et al. 2010).
These evolutionary comparisons show that CKGF domains do not inherently require prodomains for biosynthesis or dimerization and that prodomains of even smaller size than those of TGF-β have important functional roles in the PDGF and NGF families. Among the six CKGF protein families (see section on CKGF Domain and Its Families), the TGF-β gene family has both the largest prodomain and the largest number of members in vertebrates (33 in humans), whereas bursicons lack prodomains and are extinct in deuterostomes. It is thus tempting to propose a prodomain-centric view that the evolutionary success of the TGF-β family is in part a result of its large and complex prodomain, which enables complex regulation of biological function during signaling in extracellular environments that can be layered onto cell-surface and intracellular signaling in control of agonism and antagonism. The following section highlights the important functional role of the prodomains in the TGF-β family.
STRUCTURES AND FUNCTIONS OF TGF-β FAMILY PROCOMPLEXES
Biosynthesis and Latency
TGF-β family members are synthesized with large, ∼250 residue amino-terminal prodomains that are required for proper folding and dimerization of the smaller, ∼110 residue carboxy-terminal GF domains (Fig. 1) (Gentry and Nash 1990; Gray and Mason 1990). During prodomain and GF domain folding and disulfide bond formation in the endoplasmic reticulum (ER), TGF-βs 1–3 also become disulfide-linked to LTBPs. Studies on TGF-β1, activin, and AMH suggest that the carboxy-terminal GF domain folds either concomitantly with, or subsequently to, the amino-terminal prodomain (Gray and Mason 1990; Belville et al. 2004; Walton et al. 2009).
Processing by PCs (Miyazono et al. 1991) occurs following transit out of the ER. All TGF-β family members have one or more PC cleavage sites (green dots, Fig. 1) (Constam 2014). PCs are a family of secreted, or more commonly membrane-bound, Golgi serine proteases. Seven PCs cleave after a basic residue (Arg or Lys) (Seidah and Prat 2012). These include PC1, PC2, furin, PC4, PC5, PACE4, and PC7 (Seidah and Prat 2012). Most PC family members reside in the Golgi and most TGF-β family members are cleaved between the prodomain and GF domain in the Golgi prior to secretion. However, some PC family members are secreted, and some TGF-β family members, including nodal and myostatin, are cleaved extracellularly (Anderson et al. 2008; Blanchet et al. 2008; Constam 2014).
PC cleavage is regulated by many factors, including association with other proteins (Blanchet et al. 2008), but most PCs cleave after the motif (R/K)-Xn-(R/K), in which Xn is a 0, 2, 4, or 6 amino acid spacer, and R (Arg) is highly favored over K (Lys) immediately before the cleavage site (Seidah and Prat 2012). Often, multiple PC cleavage motifs are present (Fig. 1) and are used (Israel et al. 1992; Akiyama et al. 2012; Tilak et al. 2014).
The tendency of the prodomains and GFs to dissociate after secretion varies greatly among the TGF-β family. TGF-β, GDF-8 (myostatin), and GDF-11 procomplexes are so stable that their GFs are kept latent by the associating prodomain, whereas the BMP-2 prodomain readily dissociates during GF purification (Hammonds et al. 1991; Israel et al. 1992). On the other hand, the prodomains and GFs of BMP-7 and BMP-9 largely remain associated during purification (Brown et al. 2005; Gregory et al. 2005), and many BMP prodomains bind their GFs with high affinity (Sengle et al. 2008).
Structures of Prodomain–GF Complexes
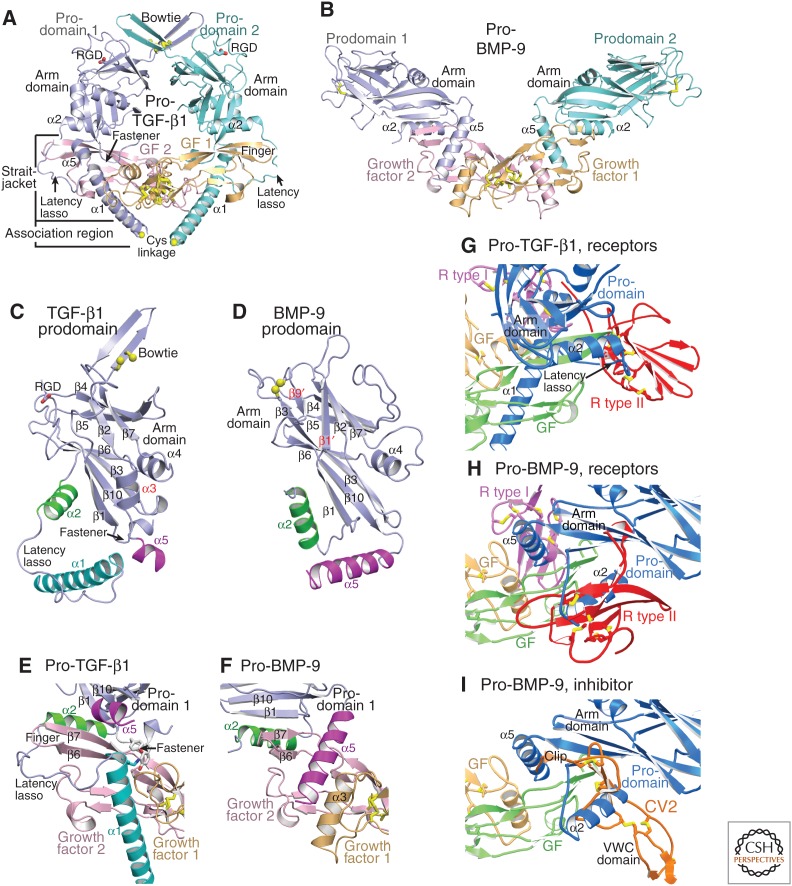
Structures of TGF-β1 (Shi et al. 2011) and BMP-9 (Mi et al. 2015) procomplexes, which share only 11% prodomain sequence identity, illuminate TGF-β family diversity (Fig. 2A,B). The prodomain contains an arm domain flanked by shorter, partially α-helical, amino- and carboxy-terminal straitjacket elements (Fig. 2C,D). TGF-β1 and BMP-9 procomplexes reveal overall crossed-arm and open-arm conformations, respectively, with markedly different orientations between arm domain monomers (Fig. 2A,B). In the open-arm conformation of pro-BMP-9, the two arm domains point away from one another and are not in contact (Fig. 2B). In the crossed-arm conformation of pro-TGF-β1, the arm domains point toward one another and dimerize at the bowtie (Fig. 2A). In contrast to the arm domain, the segments amino-terminal (α1-helix, latency lasso, and fastener) and carboxy-terminal (α5-helix) to the arm domain interact differently with the GF domain in the two conformations (Fig. 2A–F).
Figure 2.
Procomplexes of transforming growth factor (TGF)-β1 and bone morphogenetic protein 9 (BMP-9) have crossed-arm and open-arm conformations, respectively. Crystal structures are shown in cartoon representation. Some labels are shown in the color of the corresponding proteins or protein segment. Cysteine side chains are shown as yellow sticks and in some cases with sulfur atom spheres. (A,B) Overall structures of pro-TGF-β1 (Shi et al. 2011) (A), and pro-BMP-9 (Mi et al. 2015) (B) in identical orientations with superimposition on GF dimers. Yellow spheres in A show Cys residues that disulfide-bond to latent TGF-β binding protein (LTBP) or GARP. (C,D) The prodomains in identical orientations after superimposition on the arm domains. Arm domain secondary structural elements common or unique to TGF-β1 and BMP-9 are labeled in black and red, respectively. Amino-terminal and carboxy-terminal appendages to the arm domain are labeled and are, in order, the α1-helix, latency lasso, α2-helix, and fastener (all amino-terminal) and the α5-helix (carboxy-terminal). (E,F) Interacting regions of the prodomain and GF shown in identical orientations after superimposition on GF dimers. (G–I) Competition of the prodomain with receptor and inhibitor binding. Complexes are shown in identical orientations after superimposition on GF dimers. For clarity, GFs in receptor and inhibitor complexes are omitted. (G) TGF-β1 procomplex (Shi et al. 2011) superimposed on TGF-β1 complex with ALK5 (R type I) and TβRII (R type II) (Radaev et al. 2010). (H,I) BMP-9 procomplex (Mi et al. 2015) superimposed on (H) BMP-9 complex with ALK1 (R type I) and ActRIIB (R type II) (Townson et al. 2012) or superimposed on (I) BMP-2 complex with a crossveinless 2(CV2)VWC domain fragment (Zhang et al. 2008).
Despite differences in orientation, the arm domains of TGF-β1 and BMP-9 show overall similar folds. TGF-β1 and BMP-9 arm domains have two β-sheets that partially overlap in the hydrophobic core. Long meandering loops and a substantial α4-helix cover the remainder of the core (Fig. 2C,D). The β1-strand of the arm domain hydrogen bonds to the β7-strand in the GF finger to form a super-β-sheet (Fig. 2E,F). This super-β-sheet knits the prodomain and GF together. Substantial twisting of the arm β1-strand between the crossed-arm and open-arm conformations enables the arm domain to reorient by ∼90°, while nonetheless maintaining equivalent super-β-sheet hydrogen bonds in TGF-β and BMP-9 procomplexes (Fig. 2A,B,E,F).
A long loop at the opposite end of the arm domain special to pro-TGF-β1, β2, and β3 contains cysteines that disulfide link the two arm domains together to stabilize the crossed-arm procomplex conformation (Fig. 2A). The disulfide-linked region is termed the bowtie knot; the region that follows the knot and includes the RGD motif is termed the bowtie tail. The bowtie tail greatly changes in conformation when bound to integrins (Dong et al. 2014; X Dong, B Zhao, and TA Springer, unpubl.).
Elements amino terminal to the arm domain surround the GF dimer in the crossed-arm conformation of pro-TGF-β and form a straitjacket (Fig. 2A,E) (Shi et al. 2011). The α1-helix is buried between the two GF monomers in an extensive interface that includes the amphipathic, carboxy-terminal end of the α1-helix (Fig. 2A,E). Hydrophobic residues in three turns of the α1-helix interface with aromatic residues on one GF monomer. α1-helix burial helps stabilize the crossed-arm conformation with its unique arm domain orientation, and displaces the GF α3-helix from its usual position in the interface between the two monomers. The latency lasso loosely surrounds the GF finger (Fig. 2A,E). The fastener, a short segment just before the arm domain, binds to the α1-helix and completes GF encirclement (TGF-β1 homodimer, Box 1, Fig. 2A,C,E). These elements form a straitjacket that prevents receptor binding (Fig. 2G) and maintains TGF-β latency, as verified by mutation (Shi et al. 2011).
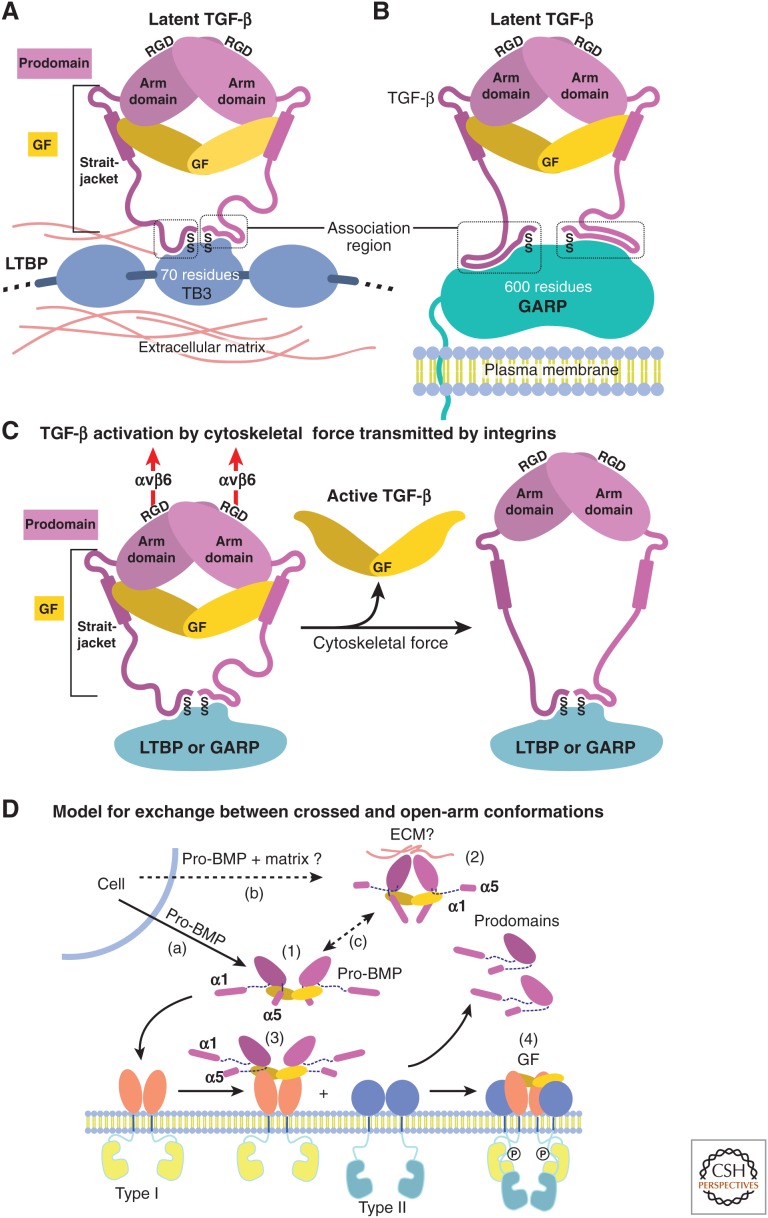
In native pro-TGF-β complexes with LTBP and the cell-surface leucine-rich repeat protein 32 (LRRC32 or GARP), cysteines in an “association region” that comprises the most amino-terminal portion of each prodomain (Figs.1 and 2A) link to two cysteines in a single LTBP or GARP molecule in a 2:1 complex (Shi et al. 2011; Wang et al. 2012). Thus, the prodomains are covalently linked together, directly through the bowtie knot at one end of the wreath-shaped procomplex, and indirectly through an intermediate protein on the opposite side of the procomplex (Fig. 2A). Unpublished pro-TGF-β crystal structures show that, in the absence of an LTBP or GARP partner, the association region of ∼10 residues is unstructured or varies in structure depending on the crystal lattice (X Dong, B Zhao, and TA Springer, unpubl.). Proper disulfide bond formation is guided by noncovalent interactions surrounding the cysteines. Because the TGF-β binding-like (TB3) domain in LTBP and the LRRC domain in GARP that disulfide-link to pro-TGF-β have no structural similarity, the association region must adopt different conformations when linked to these different partners. Furthermore, because the complex is 2:1, each prodomain monomer must associate with a different cysteine and surrounding region in the binding partner, resulting in a different structure in the association region of each monomer (shown in Fig. 3A,B).
Figure 3.
Models for growth factor (GF) latency, activation, and binding to receptors. (A,B) Schematic representation of how anchoring molecules present pro-transforming growth factor β1 (TGF-β1) in the extracellular matrix (ECM) or on cell surfaces, and how the association region at the amino terminus of the prodomain must take on different conformations to bind distinct anchoring molecules and two different sites within each anchor molecule. (C) Schematic representation of pro-TGF-β1 activation (Shi et al. 2011). Integrins, such as αvβ6, attached to the actin cytoskeleton exert traction force against RGDLXXL/I motifs in pro-TGF-β1 and -β3, which are covalently linked to anchor molecules in the ECM or on the cell surface. The most force-susceptible structural region is the straitjacket and its latency lasso, which will be elongated. Consequently, TGF-β1 is released, and shifts conformation to its free form. (D) Models for conformational change between crossed-arm, latent, and open-arm nonlatent conformations of pro-BMPs (Mi et al. 2015). (1) Open-arm, nonlatent conformation; (2) putative crossed-arm conformation of pro-BMPs stabilized by association with ECM components; (3,4) step-wise binding to type I (3) and type II (4) receptors. Putative pathways for secretion (a,b) and conformational interconversion (c) are marked.
In the open-arm conformation of pro-BMP-9, the prodomain α1-helix and latency lasso are not visualized (Fig. 2B) (Mi et al. 2015). Provocatively, the α1-helix sequence and especially its amphipathic signature are well conserved between BMP-9 and TGF-β procomplexes and among the majority of TGF-β family members (Fig. 1). This suggests that pro-BMP-9 may assume an alternative, crossed-arm conformation with an ordered α1-helix that resembles pro-TGF-β. Models show that such a conformation is plausible for pro-BMP-9 (schematized in Fig. 3D), and movies show a pathway for interconversion between open-arm and crossed-arm conformations of pro-BMP-9 (see Movie 1) (Mi et al. 2015).
Movie 1. A feasible pathway for conformational change of pro-BMP-9 between open-arm and crossed-arm conformations. A crossed-arm model of pro-BMP-9 and movie were constructed as described (Mi et al. 2015). The movie, when looped, displays a putative reversible conformational change from crossed-arm to open-arm and open-arm to crossed-arm. Structures are shown in ribbon cartoon with yellow disulfide bonds. BMP-9 is shown with magenta and orange monomers. Prodomain monomers are in blue and green. Different shades are used for different elements. For the prodomain monomer on the left, the arm domain and α2-helix are light blue, the α1-helix and latency loop are marine, and the α5-helix is dark blue. The crossed-arm pro-BMP-9 model accommodates the longer α5-helix of pro-BMP-9 in the same position as the shorter α5-helix of pro-TGF-β1. The open-arm pro-BMP-9 model adds the α1-helix and the straitjacket elements to the pro-BMP-9 crystal structure in positions in which they do not clash.
The sequence of the prodomain α2-helix is conserved in all 33 TGF-β family polypeptides (Fig. 1). The α2-helix has a unique role at the interface between the arm and GF domains. The α2-helix is amphipathic, and its hydrophobic face shields hydrophobic residues in the GF fingers from solvent. Between the crossed-arm and open-arm conformations, the α2-helix maintains its position relative to the GF fingers (Fig. 2A,B,E,F). In contrast, the α2-helix moves ∼90° with respect to the arm domain between crossed- and open-arm conformations (Fig. 2C,D). Reorientation of the α2-helix in pro-BMP-9 allows it to interact with the arm domain in a way not seen in pro-TGF-β1 (Mi et al. 2015).
The α5-helix in pro-BMP-9 is much longer than in pro-TGF-β1 (Fig. 1), orients differently relative to the arm domain, and binds to a similar region of the GF domain as the α1-helix in pro-TGF-β1 (Fig. 2A,B,E,F) (Mi et al. 2015). The BMP-9 prodomain α5 helix inserts into the hydrophobic groove formed by the fingers of one GF monomer and α3-helix of the other GF monomer (Fig. 2F). Thus, instead of displacing the GF α3-helix as the prodomain α1-helix does in pro-TGF-β1, the α5-helix in pro-BMP-9 binds adjacent to the GF α3-helix (Fig. 2F), enabling a relaxed, mature-like GF conformation in pro-BMP-9. In pro-TGF-β1 the α5-helix also binds the GF; it fills a small gap between the arm domain and α1-helix (Fig. 2E) (Shi et al. 2011).
A remarkable feature of the pro-TGF-β dimer is a swap in the GF monomer that each prodomain monomer embraces. The swap was suggested based on the structure of PC-cleaved pro-TGF-β1, in which prodomain residues preceding the PC cleavage site are disordered (Shi et al. 2011). In a structure with the PC site mutated (B Zhao, X Dong, and TA Springer, unpubl.), the linkage is also flexible, as expected for a polypeptide that is susceptible to PC cleavage. However, the disordered polypeptide is only long enough to span one of the two possible prodomain–GF connections (TGF-β1 monomer, Box 1). This establishes that the most intimate contacts are between pro- and GF domains of different polypeptide chain monomers in the precursor in the ER. Thus, the straitjacket, α2-helix, arm domain, and α5-helix from prodomain monomer 1 surround the GF from monomer 2 (Fig. 2A).
Prodomain–GF swapping has important biological implications, because it can provide a mechanism for preferential formation of heterodimers over homodimers when a cell synthesizes monomers for two different TGF-β family members. In zebrafish embryos, BMP-2/7 heterodimers pattern the dorsoventral axis, whereas the respective BMP-2 and BMP-7 homodimers are insufficient (Little and Mullins 2009). Overexpression of recombinant human BMPs also suggests greater activity of BMP-2/5, 2/6, 2/7, and 4/7 heterodimers than the corresponding BMP homodimers (Israel et al. 1996). Complementary interactions between swapped prodomains and GF domains must also be important among inhibin α- and β-subunits, which form both activating activin β homodimers and inhibitory inhibin αβ heterodimers (Walton et al. 2009).
Competition between Prodomains, Receptors, and Antagonists for GF Binding
Prodomains encoded by the 33 TGF-β family genes have important yet poorly understood roles, including extending the range of BMP signaling in vivo (Cui et al. 2001; Harrison et al. 2011; Akiyama et al. 2012; Constam 2014). Many prodomains form strong complexes with their GFs (Brown et al. 2005; Sengle et al. 2008), and some of these bind tightly enough to compete for binding of the GFs to their cognate receptors (Sengle et al. 2008; Walton et al. 2009; Mi et al. 2015). Interestingly, the prodomains seem to compete mainly with type II receptors but rarely with type I receptors (Sengle et al. 2008; Hauburger et al. 2009; Mi et al. 2015).
The strong effect of BMP prodomains on type II receptor binding can be explained by the pro-BMP-9 structure (Fig. 2H). The prodomain arm domain and α2-helix occupy the type II receptor-binding site. In contrast, only the prodomain α5-helix blocks the type I receptor binding site (Fig. 2H). The α5-helix binds weakly as suggested by the relatively small surface on the GF that it buries, and by its weak electron density in certain crystallization conditions (Mi et al. 2015). These structural results together with receptor competition experiments suggest that, among BMPs, type I receptors may displace the prodomain α5-helix from the GF, whereas the prodomain α2-helix and arm domain remain bound to the GF and inhibit type II receptor binding (Sengle et al. 2008; Mi et al. 2015).
Chordin and chordin-related BMP inhibitors contain one or several von Willebrand factor C (VWC) domains as BMP-inhibiting moieties. Most interestingly, the binding of such a domain of the chordin-member crossveinless 2 to BMP-2 (Zhang et al. 2008) is reminiscent of prodomain binding to BMP-9 (Fig. 2I). The VWC domain binds to the GF fingers similarly to the α2-helix and arm domain, whereas an amino-terminal appendage called clip binds to the same site as the prodomain carboxy-terminal appendage, the α5-helix (Fig. 2I, further described in section on Chordin Family Members). It is, therefore, theoretically possible that prodomains could help deliver GFs to receptors by shielding GFs from inhibitors; a similar mechanism has been shown for chordins (Ashe and Levine 1999; De Robertis et al. 2000; Harland 2001).
Integrin- and Force-Dependent Release of TGF-β1 from the Prodomain
TGF-βs evolved in deuterostomes with a unique integrin-dependent mechanism of activation. In the crossed-arm conformation of pro-TGF-β1, binding to both type I and type II receptors is completely blocked (Fig. 2G) (Shi et al. 2011). An integrin-binding RGDLXX(L/I) motif in the prodomains of TGF-β1 and 3 locates to the shoulder region of their arm domains, between the β7- and β10-strands of the arm domain (Fig. 1). Integrins αvβ6 and αvβ8 bind this motif with ∼10 nM affinity, which is extremely high for integrins (Dong et al. 2014). Integrin binding is not sufficient to release and thus to activate TGF-β (Shi et al. 2011). Anchoring pro-TGF-β covalently to LTBP in the extracellular matrix (ECM) or covalently to GARP on cell surfaces is required for activation (Munger et al. 1999; Annes et al. 2004; Yoshinaga et al. 2008; Wang et al. 2012). Furthermore, activation requires the cytoplasmic domain of the integrin β-subunit, which connects to the actin cytoskeleton and exerts traction (Munger et al. 1999). These and other experiments (Wipff et al. 2007; Wipff and Hinz 2008) suggest that traction force exerted by integrins on the RGDLXX(L/I) motif in the arm domain, and its resistance on the opposite side of the prodomain by association region cysteines that link to LTBP and GARP, is required for activation (Fig. 3C). The results suggest a model in which applied traction force distorts the straitjacket region and results in release of TGF-β (Shi et al. 2011).
Conformational Change in Procomplexes as a Mechanism for Regulating Activity in the TGF-β Family
Some members of the TGF-β family may be able to access both open-arm and crossed-arm procomplex conformations (Fig. 3D). BMP-9 may be one of these; its procomplex structure shows an open-arm conformation (Fig. 3D, structure 1), whereas strong conservation of its α1-helix sequence suggests the presence of a distinct, putative crossed-arm conformation (Fig. 3D, structure 2) (Mi et al. 2015). Modeling of a crossed-arm BMP-9 procomplex shows that the prodomain α1-helix and GF can associate in a crossed-arm conformation that recapitulates the hydrophobic interface in pro-TGF-β1 (see Movie 1) (Mi et al. 2015). BMP-9 regulates vascular development. Mutations in its prodomain can cause syndromes resembling hereditary hemorrhagic telangiectasia (HHT) caused by mutations in receptors for BMP-9 (David et al. 2009; Castonguay et al. 2011; Wooderchak-Donahue et al. 2013). The location of mutations associated with an HHT-like condition in the BMP-9 prodomain α1-helix and in the subsequent loop supports the hypothesis of a crossed-arm conformation. A similar embrace between the prodomain α1-helix and mature domains in inhibin α and βA is shown by mapping of disruptive mutations to the hydrophobic face of the amphipathic α1-helix (Walton et al. 2009). Thus, multiple family members besides pro-TGF-β1 may adopt an α1-helix–associated, putative crossed-arm conformation.
Conformational changes between latent, crossed-arm conformations and nonlatent, open-arm conformations may regulate whether procomplexes are stored in the ECM in a latent form or released in a nonlatent form for signaling (Fig. 3D) (Mi et al. 2015). Because the crossed-arm conformation is tensed with more surface area buried between the GF and prodomain, and with the GF surrounded by a straitjacket, it may correspond to a latent conformation. In contrast, the open-arm conformation may correspond to a nonlatent conformation. This idea is consistent with latency of crossed-arm pro-TGF-β1 (Shi et al. 2011) and nonlatency of open-arm pro-BMP-7 and -9 (Mi et al. 2015). Although most members of the TGF-β family are nonlatent when overexpressed as recombinant proteins (Fig. 3D, pathway a, structure 1) (Brown et al. 2005; Sengle et al. 2011; Mi et al. 2015), latency may differ in vivo, in which cells cosynthesize TGF-β family members with components of the ECM (Fig. 3D, pathway b). Prodomains can bind to ECM components, including heparin, proteoglycans, LTBP, and fibrillin (Gregory et al. 2005; Anderson et al. 2008; Sengle et al. 2008, 2011; Li et al. 2010); one or more of these may bind to procomplexes, and thereby stabilize the crossed-arm conformation (Fig. 3D, structure 2) and enable the GF domain, which is very short-lived in vivo, to reach storage concentrations as high as 100 ng/g in demineralized bone (Pietrzak et al. 2006). The short lifetimes of free BMPs, together with significant storage depots of BMPs in bone, and colocalization of GFs with prodomains (Sampath and Reddi 1981, 1984; Gregory et al. 2005; Pietrzak et al. 2006), suggest latency in vivo. Furthermore, purification of BMPs showed a 60-fold increase in total activity in the first two steps, consistent with possible purification away from an inhibitor such as the prodomain (Luyten et al. 1989). Release from storage depots in vivo may be associated with conformational change to the open-arm conformation, which is ready for receptor or inhibitor binding (Fig. 3D, pathway c).
Among different family members, differences in structural elements, including those corresponding to the straitjacket and fastener, may regulate whether crossed-arm or open-arm conformations predominate. The prodomain α1 and α5-helices are likely to compete for overlapping binding sites on the GF in pro-BMP-9 (Mi et al. 2015). Therefore, the relative strengths of α1- and α5-helix binding among different TGF-β family members may regulate which conformation is preferred (Fig. 3D, pathway c; see Movie 1). In a single family member, binding to ECM components, and proteolytic removal by PC or tolloid proteases of helical segments amino- or carboxy-terminal to the arm domain may regulate which state predominates. Many family members contain known or putative PC or tolloid/BMP protease cleavage sites in these segments (Fig. 1). The open-arm conformation of BMP-9 can readily bind to type I receptors, with displacement of the α5-helix (Fig. 3D, structure 3) (Mi et al. 2015). The final step in signaling could then be complete prodomain dissociation followed by binding to type II receptors (Fig. 3D, structure 4).
Implications of Prodomain Diversity for Structure and Function of the TGF-β Family
What can we deduce from sequence (Shi et al. 2011) and structural alignment (Mi et al. 2015) of all 33 human TGF-β family polypeptides (Fig. 1)? The minimum TGF-β family prodomain, exemplified by the short (169 residues) prodomain of GDF-15, contains an α2-helix and an arm domain core containing the β1, β2, β3, β6, β7, and β10-strands and the α4-helix (Figs. 1 and 2C,D). The β4 and β5-strands on the edge of the arm domain β-sandwich closest to the arm–arm interface in pro-TGF-β1 are dispensable, as they are absent in GDF-15 (Fig. 1). All members except GDF-6, GDF-15, and possibly AMH contain an α1-helix with an amphipathic signature (Fig. 1). A third of the family contains substantial sequence carboxy-terminal to the arm domain (Fig. 1) that might fulfill a function similar to the α5-helix in competing with the α1-helix for GF binding.
Prodomain sequences corresponding to the association region in TGF-β1 reveal interesting peculiarities (Fig. 1). An abundance of Arg residues in BMP-5, 6, 7, and 8 suggests possible binding to heparan sulfate proteoglycans. Many members have long, hydrophilic association regions that are likely unfolded, unless they associate with another macromolecule. Unusual features include continuous repeats of 7 to 11 Ala residues (GDF-11 and BMP-6), 7 Pro residues (inhibin βB) or 10 Gly residues (GDF-7). In similar positions to the LTBP or GARP-linked Cys of TGF-β1, one or two Cys are present in the association regions of myostatin, GDF-11, inhibin-βA, βB, βC, and βE, inhibin-α, BMP-6, and BMP-8. These unexplained features highlight how much remains to be learned about the prodomains of the TGF-β family, both as recombinant proteins, and in their native context in tissues in which our understanding of their binding partners remains incomplete. The GF domains in the family also show great variation in sequence and structure and can bind to ECM components as well as to receptors, as described in the next section.
SPECIFICITY AND PROMISCUITY IN GF-RECEPTOR INTERACTIONS
GF Structures
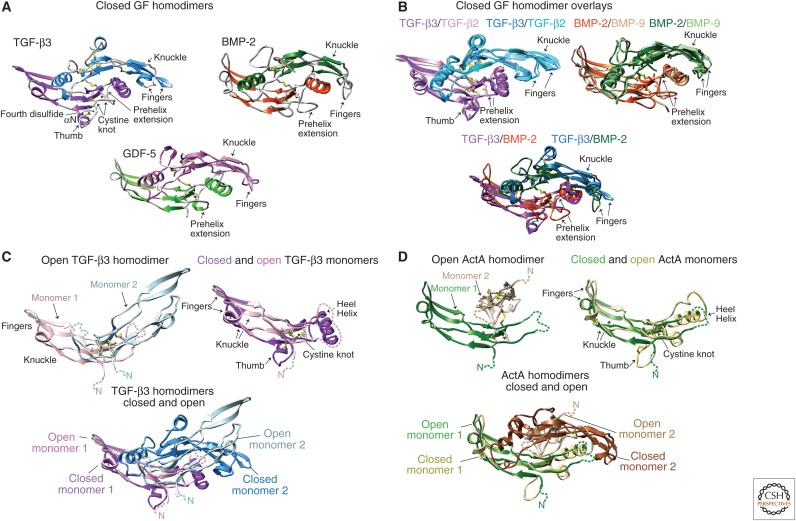
Three-dimensional structures have been reported for more than ten different TGF-β family GFs (Table 1, Fig. 4). The GFs consist of two extended monomers joined together in most, but not all cases, by a single interchain disulfide bond. The monomers adopt the shape of a curled left hand, with the heel of one hand packing into the palm of the other (GF homodimer, Box 1). The heel is formed by a 3-4 turn α-helix (GF monomer, Box 1). The fingers are formed by two loops that extend from the CK and adopt a β-strand conformation in most family members (GF monomer, Box 1). Finger nomenclature differs in the family. For TGF-βs each β-strand is counted as one finger, and thus TGF-βs are described as having four fingers; whereas for BMP, GDF, and activin GFs, and DAN family antagonists (see section on The Dan Family), each two-stranded β-ribbon is counted as one finger, and thus these CKGF domains are described as having two fingers. The thumb, when present, is formed by a short amino-terminal α-helix. TGF-βs, AMH and activins have a fourth disulfide that tethers the one-turn amino-terminal thumb α-helix to the amino-terminal end of β-strand 1. This disulfide causes the amino-terminal α-helix to extend away from the fingers to form a pronounced thumb (Fig. 4A, TGF-β3). The thumb region of BMP-2 has basic, heparin-binding residues and is disordered in crystal structures (Scheufler et al. 1999); GDF-5 lacks sequence for the thumb (Figs. 1 and 4A).
Table 1.
Structures that illuminate transforming growth factor β (TGF-β) family signaling
| Molecules | NMR or X-ray (resolution, Å) | PDB entry | References |
|---|---|---|---|
| Growth factors | |||
| TGF-β1 | NMR | 1KLC | Hinck et al. 1996 |
| TGF-β2 | X-ray (2.0, 1.95) | 2TGI, 1TFG | Daopin et al. 1992; Schlunegger and Grutter 1992; Daopin et al. 1993 |
| TGF-β3 | X-ray (3.3, 2.0) | 1TGK, 1TGJ | Mittl et al. 1996 |
| BMP-2 | X-ray (2.7, 2.65) | 3BMP, 1REU | Scheufler et al. 1999; Keller et al. 2004 |
| BMP-3 | X-ray (2.21) | 2QCQ | Allendorph et al. 2007 |
| BMP-6 | X-ray (2.49, 2.50, 2.1) | 2QCW, 2R52, 2R53 | Allendorph et al. 2007; Saremba et al. 2008 |
| BMP-7 | X-ray (2.8, 2.0) | 1BMP, 1LXI | Griffith et al. 1996; Greenwald et al. 2003 |
| BMP-9 | X-ray (2.33) | 1ZKZ, 4MPL | Brown et al. 2005; Wei et al. 2014 |
| Activin A | X-ray (2.0) | 2ARV | Harrington et al. 2006 |
| GDF-5 | X-ray (2.28, 2.40) | 1WAQ, 2BH5 | Nickel et al. 2005; Schreuder et al. 2005 |
| Nodal/BMP-2 chimera | X-ray (1.91) | 4N1D | Esquivies et al. 2014 |
| Activin A/BMP-2 chimera | X-ray (2.14) | 4MID | Yoon et al. 2014 |
| Procomplexes | |||
| Pro-TGF-β1 | X-ray (3.05 Å) | 3RJR | Shi et al. 2011 |
| Pro-BMP-9 | X-ray (3.3 Å, 3.25 Å) | 4YCG, 4YCI | Mi et al. 2015 |
| Receptors | |||
| ActRII | X-ray (1.5) | 1BTE | Greenwald et al. 1999 |
| TβRII | NMR, X-ray (1.05) | 1PLO, 1KS6, 1M9Z | Boesen et al. 2002; Deep et al. 2003; Marlow et al. 2003 |
| BMPRII | X-ray (1.2, 1.45) | 2HLR, 2HLQ | Mace et al. 2006 |
| ALK1 | NMR | 2LCR | Mahlawat et al. 2012 |
| ALK3 | NMR, X-ray (2.7) | 3K3G, 3NH7 | Klages et al. 2008; Harth et al. 2010 |
| ALK5 | NMR | 2L5S | Zuniga et al. 2011 |
| Antagonists | |||
| Sclerostin | NMR | 2K8P, 2KD3 | Veverka et al. 2009; Weidauer et al. 2009 |
| PRDC | X-ray (2.25) | 4JPH | Nolan et al. 2013 |
| Nbl1/Dan | X-ray (2.5, 1.8) | 4X1J, 4YU8a | Nolan et al. 2013 |
| CV2 fragment | NMR | 2MBK | Fiebig et al. 2013 |
| Coreceptors | |||
| Betaglycan fragment | X-ray (2.0, 2.7) | 3QW9, 4AJV | Lin et al. 2011; Diestel et al. 2013 |
| Cripto fragment | NMR | 2J5H | Calvanese et al. 2006 |
| Complexes | |||
| BMP-2:ALK3 | X-ray (2.90, 1.86) | 1ES7, 1REW | Kirsch et al. 2000; Keller et al. 2004 |
| BMP-2:ALK3/ALK6 chimera | X-ray (2.5, 2.4, 2.6) | 2QJB, 2QJ9, 2QJA | Kotzsch et al. 2008 |
| TGF-β3:TβRII | X-ray (2.15) | 1KTZ | Hart et al. 2002 |
| Activin A:ActRIIB | X-ray (3.1, 2.3) | 1NYU, 1S4Y | Thompson et al. 2003; Greenwald et al. 2004 |
| BMP-7:ActRII | X-ray (3.3) | 1LX5 | Greenwald et al. 2003 |
| GDF-5:ALK6 | X-ray (2.1) | 3EVS | Kotzsch et al. 2009 |
| GDF-5:ALK3 | X-ray (2.28) | 3QB4 | Klammert et al. 2015 |
| BMP-2:ActRII:ALK3 | X-ray (2.2) | 2GOO | Allendorph et al. 2006 |
| BMP-2:ActRIIB:ALK3 | X-ray (1.85, 1.92) | 2H62, 2H64 | Weber et al. 2007 |
| TGF-β3:TβRII:ALK5 | X-ray (3.0) | 2PJY | Groppe et al. 2008 |
| TGF-β3:Antibody Fab | X-ray (3.1) | 3EO1 | Grütter et al. 2008 |
| TGF-β1:TβRII:ALK5 | X-ray (3.0) | 3KFD | Radaev et al. 2010 |
| BMP-9:ActRIIB:ALK1 | X-ray (3.36) | 4FAO | Townson et al. 2012 |
| BMP-2:CV2 fragment | X-ray (2.7 Å) | 3BK3 | Zhang et al. 2008 |
| BMP-7:Noggin | X-ray (2.42) | 1M4U | Groppe et al. 2002 |
| GDF-8:Follistatin 288 | X-ray (2.15 Å) | 3HH2 | Cash et al. 2009 |
| Activin A:Follistatin 288 | X-ray (2.8 Å) | 2B0U | Thompson et al. 2005 |
| Activin A:Follistatin-like 3 | X-ray (2.48 Å) | 3B4V | Stamler et al. 2008 |
| Activin A:Follistatin Fs12 | X-ray (2.0 Å) | 2ARP | Harrington et al. 2006 |
| Activin A:Follistatin 315 | X-ray (3.40) | 2P6A | Lerch et al. 2007 |
| GDF-8:Follistatin-like 3 | X-ray (2.4 Å) | 3SEK | Cash et al. 2012 |
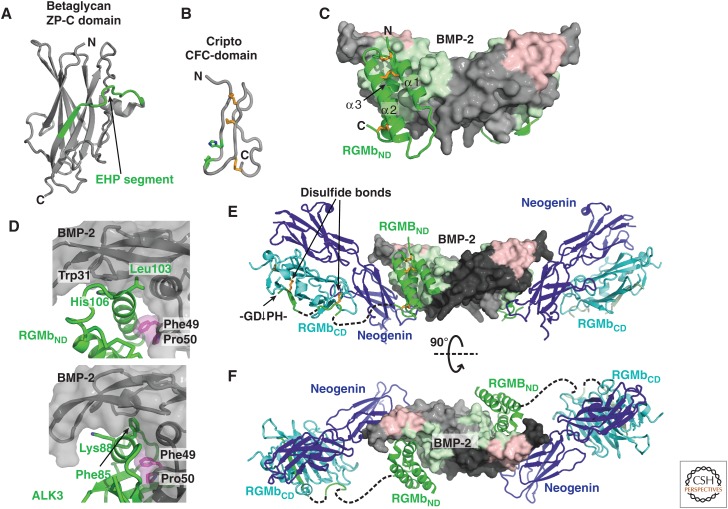
| RGMa:BMP-2 | X-ray (3.2) | 4UHY | Healey et al. 2015 |
| RGMb:BMP-2 | X-ray (2.85, 2.8) | 4UHZ, 4UIO | Healey et al. 2015 |
| RGMc:BMP-2 | X-ray (2.35) | 4UI1 | Healey et al. 2015 |
| RGMb:BMP-2:Neogenin | X-ray (3.15) | 4UI2 | Healey et al. 2015 |
| RGMb:Neogenin | X-ray (2.3, 6.6, 2.8) | 4BQ6, 4BQ7, 4BQ8 | Bell et al. 2013 |
| Norrin:Frizzled4 | X-ray (3.0) | 5BQC | Chang et al. 2015 |
NMR, Nuclear magnetic resonance; BMP, bone morhphogenetic factor; GDF, growth and differentiation factor.
aR.J. Owens, Oxford Protein Production Facility.
Figure 4.
Structures of transforming growth factor β (TGF-β) family growth factors (GFs). All structures are of disulfide-linked GF homodimers. Ribbon diagrams are color-coded by monomer (with names in black) or color-coded by monomer or GF (with corresponding color-coded names). Cysteine side chains are shown as yellow sticks. (A) Closed GF structures. (B) Superposition of closed GFs. (C) Comparison of open (1KTZ) and closed (1TGK) TGF-β3 dimers and monomers from homodimer crystal structures. (D) Comparison of open (1NYU) and closed (2B0U) activin A dimers and monomers from homodimer crystal structures. Structures are of TGF-β2 (Daopin et al. 1992), TGF-β3 (Mittl et al. 1996; Hart et al. 2002), growth and differentiation factor 5 (GDF-5) (Nickel et al. 2005), bone morphogenetic protein 2 (BMP-2) (Scheufler et al. 1999), BMP-9 (Brown et al. 2005), and activin A (Thompson et al. 2003).
TGF-β family proteins can be expressed as both homodimers and heterodimers. When a cell synthesizes two polypeptide subunits that then heterodimerize, prodomain-GF swapping (see section on Structures of Prodomain–GF Complexes) provides a mechanism for facilitating heterodimer formation (Israel et al. 1996). Inhibin subunits are interesting because dimerization of their β-subunits generates activins, whereas β-subunit heterodimerization with the inhibin α-subunit forms inhibins. Recombinant BMP heterodimers, including BMP-4/7 (Aono et al. 1995; Suzuki et al. 1997; Nishimatsu and Thomsen 1998) and BMP-2/7 (Buijs et al. 2012; Zheng et al. 2012), are more active than their homodimers both in cultured cells and in animals (Aono et al. 1995; Israel et al. 1996; Suzuki et al. 1997; Nishimatsu and Thomsen 1998; Zhu et al. 2006; Buijs et al. 2012; Zheng et al. 2012). However, we know little about the relative abundance of BMP homodimers and heterodimers in vivo, and structures for biologically relevant TGF-β family heterodimers are currently lacking.
The GF domains of GDF-3, GDF-9, and BMP-15 lack the cysteine residue that forms the interchain disulfide, but they can still form stable homodimers. BMP-15 and GDF-9 can form heterodimers either during biosynthesis or by exchange between preformed homodimers (Liao et al. 2003; McNatty et al. 2005). Their heterodimers have more pronounced biological activities than the homodimers (Mottershead et al. 2013, 2015; Peng et al. 2013). Curiously, some GFs including BMP-9 contain the cysteine for the interchain disulfide, yet their homodimers fail to be quantitatively disulfide-linked (Wei et al. 2014).
GF homodimers within the same subfamily have similar backbone conformations, as illustrated with TGF-β2 overlayed on TGF-β3, or BMP-2 overlayed on BMP-9 (Fig. 4B). GFs belonging to different subfamilies also share similarity, especially in the region of the CK and the fingers extending to the knuckle, but tend to have significant differences in the heel α3-helix, the “prehelix extension,” and the fingertips (Fig. 4B, overlay of TGF-β3 and BMP-2). Such differences correlate with sequence differences; for example, in the prehelix extension before the heel α3-helix, inhibin-βs are five residues longer, and BMPs and GDFs are three residues longer than TGF-βs (Fig. 1). Furthermore, the structural differences in the heel, prehelix extension, and fingertips are functionally important, because these regions comprise the primary binding sites, not only for the ectodomains of the signaling receptors, but also for prodomains and modulator proteins, which have distinct preferences for specific subclasses of GFs, or, in some cases, even for single GFs.
Structural studies have shown that not all family members are stable as “closed” dimers (Fig. 4A,B) in which the heel region of one monomer extensively packs onto the palm region of the opposing monomer. NMR shows that TGF-β3 is very dynamic (“open”), with little order in the regions including the heel α3-helix that are important in the dimer interface (Bocharov et al. 2000, 2002; Huang et al. 2014). Although extensive NMR data have been collected on both TGF-β1 and TGF-β3, an NMR model of the dimer could only be constructed for TGF-β1 (Hinck et al. 1996). In agreement, the TGF-β1 solution structure is closed, whereas TGF-β3 can crystallize in an open conformation, with little contact between monomers except near the interchain disulfide bond (Fig. 4C) (Hart et al. 2002). However, TGF-β3 also crystallizes in the closed conformation (Mittl et al. 1996). Similarly, activin A has been captured in alternative conformations that show large variation and asymmetry in monomer–monomer orientation (Fig. 4D) (Thompson et al. 2003; Greenwald et al. 2004). For both TGF-β3 and activin A, the less ordered, “open” conformation shows disorder at the dimerization-proximal end of the monomer. The other end of the monomer, with its β-sheets and CK is more stable, and makes the overall shapes of closed and open monomers similar (Fig. 4C,D). TGF-β3 in complex with type I and type II receptors shows a closed structure, in agreement with binding of type I receptors near the monomer–monomer interface, requiring ordering of this region (Groppe et al. 2008). Similarly, when activin A is surrounded by two follistatin monomers, it assumes a symmetric, closed conformation (see section on follistatin) (Thompson et al. 2005; Harrington et al. 2006).
Open forms may represent the preferred conformation for several family TGF-β family proteins, and might impart them with their distinctive activities, for example by altering the kinetics and/or affinities of binding to the signaling receptors or by exposing residues that allow recognition by a secreted antagonist. The latter was suggested by a study of TGF-β chimeras, in which cultured dermal fibroblasts migrate in response to TGF-β3 or a TGF-β3-β1-β3 chimera that adopted the open form, but not in response to TGF-β1 or a TGF-β1-β3-β1 chimera that adopted the closed form (Huang et al. 2014).
Structures of Type I and Type II Receptor Ectodomains
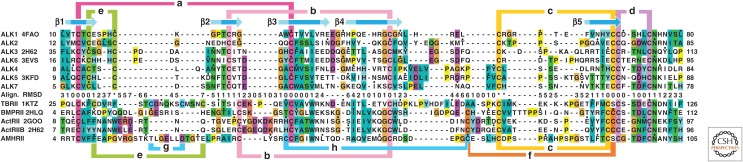
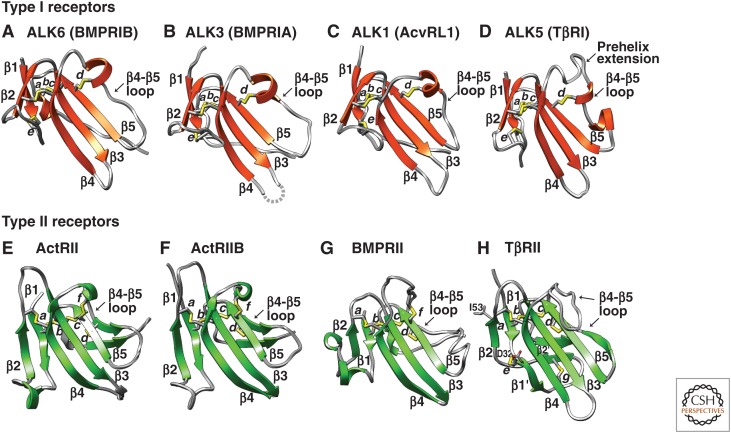
TGF-β family GF dimers initiate signaling by binding to type I and type II receptors. The receptor ectodomains of ∼100 residues are cysteine-rich. Homology in sequence and structure is detectable between type I and type II receptors in both their ectodomains and kinase domains, and thus they arose from a common ancestor (Box 1, Fig. 5). Four of the seven mammalian type I receptors and four of the five mammalian type II receptors are structurally characterized (Table 1, Fig. 6). Because the receptor ectodomain fold has three fingers (RI and RII, Box 1) and the superfamily includes neuro- and cardiotoxins, it is sometimes termed the three-finger toxin fold (Greenwald et al. 1999). The fold comprises a central three-stranded β4-β3-β5 antiparallel β-sheet flanked on its convex surface by a smaller two-stranded β1-β2 antiparallel β-sheet, and along the edge of β-strand 5 by an extended loop of variable structure that connects β-strands 4 and 5 (Fig. 6).
Figure 5.
Alignment of type I and type II receptor ectodomains by structure and sequence. Type I and type II receptors with structures (labeled with PDB codes) were aligned to one another structurally using DeepAlign (Wang et al. 2013); that is, only residues that are in equivalent positions in three-dimensional structures are aligned in the same column in the sequence alignment. Cα atom root mean square deviation (RMSD, Å) of each type I and type II receptor structure to their average position in the structural superposition is shown at each position in the alignment. Gaps were closed to minimize the length of the alignment; corresponding positions lack an RMSD value. Asterisks in the RMSD row show positions in which type I receptor cysteines are poorly aligned with one another structurally and were manually aligned. Human type I and type II receptors were also aligned separately from one another by sequence using MAFFT (Katoh and Standley 2013). Sequences not represented by structures were then aligned to those in the structural alignment. Color code is modified from ClustalX. Known disulfide bonds are color coded and lettered, except for the “h” disulfide in AMHRII, which is predicted by the structural alignment (see section on Structures of Type I and Type II Receptor Ectodomains). β-strand arrows above the alignment shown with dark or light shading show positions where all or some structures, respectively, have β-structure.
Figure 6.
Type I and type II receptors of the transforming growth factor β (TGF-β) family. Structures of type I (A–D) and II (E–H) receptor ectodomains are shown as ribbon diagrams, with disulfide bonds depicted as sticks with yellow sulfur atoms. Disulfide bond letters and β-strand numbers correspond to those in Fig. 5. A loop without reliable electron density in ALK3 is shown as a dashed line. Structures are for ALK6 (Kotzsch et al. 2009), ALK3 (Kirsch et al. 2000), ALK1 (Townson et al. 2012), ALK5 (Groppe et al. 2008), ActRII (Greenwald et al. 1999), ActRIIB (Thompson et al. 2003), BMPRII (Mace et al. 2006), and TβRII (Boesen et al. 2002).
Type I and type II receptors all share a common set of four disulfide bonds termed a, b, c, and d (Figs. 5 and 6). The e–g disulfides, which are present in only some receptors (Figs. 5 and 6), alter structures to enable distinctive GF binding modalities. Thus, the f disulfide, which is present in ActRII, ActRIIB, and BMPRII (Figs. 5 and 6E–G), braces the amino-terminal portion of the β4-β5 loop so that it points away from the convex surface of the β4-β3-β5 sheet. This is important for the function of ActRII (Greenwald et al. 2003; Allendorph et al. 2006) and ActRIIB (Thompson et al. 2003; Weber et al. 2007), and likely BMPRII as well (Yin et al. 2008), as these receptors use the exposed face of the β4-β3-β5 sheet, along with the β4-β5 loop, to bind their cognate GFs. The e disulfide is present in all type I receptors, as well as TβRII (Figs. 5 and 6A–D,H). The e disulfide in type I receptors fulfills an analogous role as the f disulfide in type II receptors, as it precludes the β1-β2 loop from occluding the convex surface of the β4-β3-β5 sheet, which along with the critical β4-β5 loop, binds cognate GFs (Kirsch et al. 2000).
The TGF-β type II receptor, TβRII, is unique among the type I and type II receptors of the family in that it binds its cognate GFs through a distinct interface, namely, an edge β-strand on the smaller β1-β2-sheet, the β2-strand, and the flanking β1 and β1′ strands (Figs. 6H and 7F) (Hart et al. 2002; Groppe et al. 2008; Radaev et al. 2010). Use of this alternative interface is promoted by two important structural differences in TβRII compared with ActRII, ActRIIB, and BMPRII. First, the β4-β5 loop is extended by seven to eight residues in TβRII relative to other type II receptors and lacks the f disulfide (Figs. 5 and 6H). The absence of this disulfide allows the extended β4-β5 loop to fold onto the concave surface of the β4-β3-β5 sheet, thus blocking GF binding through this interface. Second, the β1-β2 loop in TβRII is extended by more than 10 residues compared with all other type I and type II receptors of the family, except AMHRII (Fig. 5). The β1-β2 loop is further stabilized by both the e disulfide at its base, as in type I receptors, and the TβRII-specific g disulfide near its tip (Figs. 5 and 6H). The extended loop wraps onto the convex surface of the β4-β3-β5 sheet and packs tightly against it. This unusual structural feature is likely important for proper positioning and bracing of the short β2-strand, which forms the primary structural element used by TβRII to bind the three TGF-β isoforms.
Figure 7.
Representative growth factor (GF) complexes with type I and type II receptors. Structures are shown for representative binary (A–C) or ternary (D–F) complexes as ribbon diagrams with pink and light blue GF monomers, and type I and type II receptor ectodomains in orange-red and green, respectively. Orthogonal views are shown above and below one another in each panel. The ActRIIB:activin A structure model with a closed activin GF is made by superimposing open monomer complexes from the ActRIIB:activin A structure (Thompson et al. 2003) on the closed activin dimer from the activin A:Fst288 structure (Thompson et al. 2005). Panel F (inset) shows the interactions of Pro55, Asp57, Arg58, and Pro59 in the ALK5 pre-helix extension with Lys97 on TGF-β3 and Asp118 in TβRII, and of ALK5 with Phe24 in the amino-terminal tail of TβRII. Structures are for ALK6:GDF-5 (Kotzsch et al. 2009), ActRII:BMP-7 (Greenwald et al. 2003), ActRIIB:activin A (Thompson et al. 2003), ActRIIB:ALK3:BMP-2 (Weber et al. 2007), ActRIIB:ALK1:BMP-9 (Townson et al. 2012), and TβRII:ALK5:TGF-β3 (Groppe et al. 2008) complexes. N-term, amino terminal.
The AMHRII type II receptor is unusual in that it includes only one of the two cysteines that form the f disulfide found in other family type II receptors, including ActRII, ActRIIB, and BMPRII (Fig. 5). However, this cysteine is predicted to form a disulfide unique to AMHRII with another cysteine that immediately precedes the carboxy-terminal cysteine of the conserved a disulfide (Fig. 5). This putative h disulfide is predicted by the structure and sequence-based alignment in Figure 5, because aligned residues in homologous type II receptors have Cα atom separations and side chain orientations consistent with disulfide bond formation.
Structures of GF Complexes with Receptors
TGF-β family GF dimers greatly outnumber the type I and type II receptors, and thus, multiple GFs must share the same type I or II receptors, or in some cases, share both type I and type II receptors. Nonetheless, sequence and structure provide a basis for understanding this sharing; particular subfamilies of type I and type II receptors (TGF-β receptors, Box 1) preferentially bind and transduce signals for particular subfamilies of GFs (TGF-β GFs, Box 1). Thus, the closely related type I receptors ALK3 and ALK6 bind and transduce signals for the BMP clade comprising the BMP-5, 6, and 7 subfamily, the BMP-2 and 4 subfamily, or the GDF-5, 6, and 7 subfamily, yet not for the activin or the myostatin and GDF-11 subfamilies in the activin clade or for TGF-β (ten Dijke et al. 1994b). The type I receptors ALK4 and ALK5, which diverge from ALK3 and ALK6 (TGF-β receptors, Box 1), show the opposite GF binding properties; they bind and transduce signals for the activin subfamily and the myostatin and GDF-11 subfamily in the activin clade, and for TGF-β, but not for BMP-5, 6, and 7, BMP-2 and 4, or GDF-5, 6, and 7 in the BMP clade (ten Dijke et al. 1994a). The three type II receptors ActRII, ActRIIB, and BMPRII bind and transduce signals for BMP-5, 6, and 7, BMP-2 and 4, GDF-5, 6, and 7. Furthermore, the receptors ActRII and ActRIIB also interact with activin A and B (receptor binding data for activin C and E is unclear) as well as myostatin, and thus act across clades, yet do not bind or signal for the TGF-βs or AMH (Liu et al. 1995). In contrast, TβRII and AMHRII are highly specific or exclusive for binding and transducing signals for the TGF-βs and AMH, respectively (Baarends et al. 1994; Liu et al. 1995). TβRII does not ally with any other type II receptors in the family tree and lies closest to the type I receptors; however, AMHRII is the most divergent from the consensus type II receptor sequence (TGF-β receptors, Box 1).
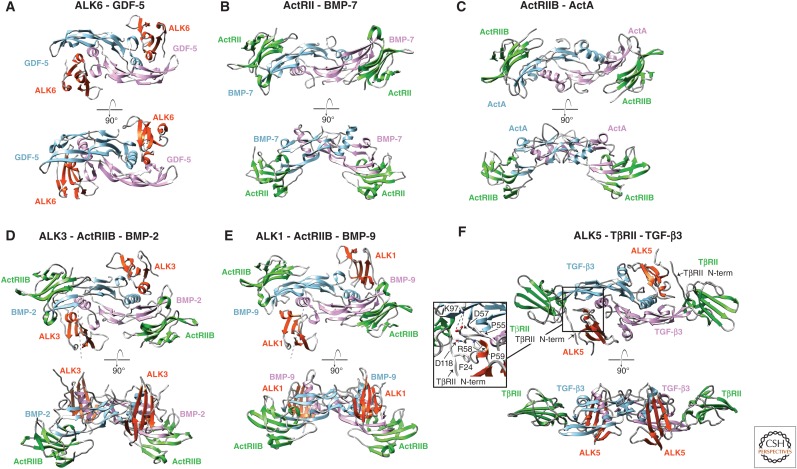
Among receptor ectodomain:GF complexes (Table 1), binary type I or II receptor:GF complexes and ternary type I receptor:type II receptor:GF complexes show nearly identical binding positions for type I and type II receptors (Fig. 7). Type II receptors bind the distal ends of the GF dimer, at the knuckle (ActRII and ActRIIB) or fingertips (TβRII) of the GF. Thus, residues within a single monomer determine type II receptor binding specificity. In contrast, type I receptors bind across the dimer interface at the wrist, and thus can show selectivity for open versus closed homodimers or homodimer versus heterodimer (Fig. 7). The GF:receptor structures provide insight into many patterns of receptor:GF binding that have been reported.
Promiscuous binding of many GFs from the BMP and activin clades by ActRII and ActRIIB, for example, is evident from their identical orientations for binding BMP-7, activin A, BMP-2, and BMP-9 dimers (Fig. 7B–E) (Greenwald et al. 2003, 2004; Thompson et al. 2003; Allendorph et al. 2006; Weber et al. 2007; Townson et al. 2012). Consistent with their similar β4-β3-β5 sheet concave surfaces and β4-β5 loop occluding f disulfide (Fig. 6E,F), ActRII and ActRIIB complement the convex knuckle of the GF in a manner that is nearly indistinguishable (Fig. 7B–E). Superpositions of ActRIIB binding to three different GFs and of ActRII and ActRIIB binding to two different GFs further show essentially identical orientations in all cases (Fig. 8E–G). Promiscuity of ActRII and ActRIIB for binding a large number of GFs agrees with the similarity in the receptor structures and knuckle regions of the GFs, which are among their most structurally conserved features. Promiscuity is further promoted by the predominantly hydrophobic composition of the interface (Fig. 9E–G). Among 23 residues in BMP-2 that directly contact ActRIIB in the structure of the BMP-2:ActRIIB:ALK3 complex, substitution with alanine of only six residues perturbed binding (Kirsch et al. 2000; Weber et al. 2007). Among these, only a Leu is conserved among the diverse GFs bound by these receptors (Leu residues 92, 90, and 125 in activin A, BMP-2, and BMP-7, respectively, in Fig. 9E–G). Thus, the similar overall shape and hydrophobicity of the knuckle in GFs that bind ActRII and ActRIIB enables the promiscuity that is possible for predominantly hydrophobic interfaces (Weber et al. 2007).
Figure 8.
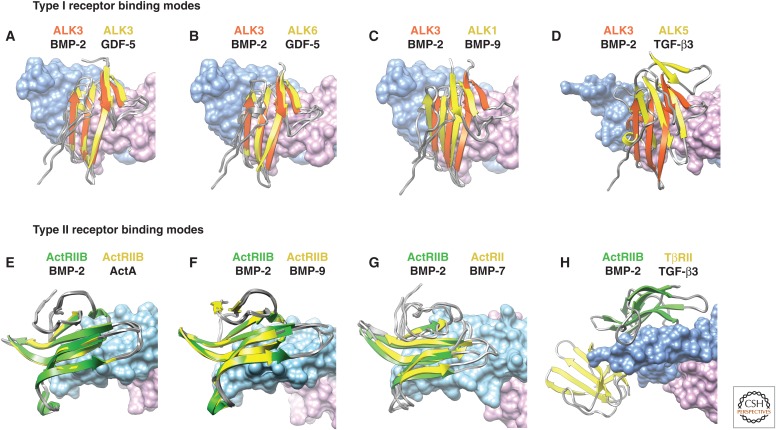
Relative positioning of type I and type II receptor ectodomains on growth factor (GF) dimers. Positions of type I receptors on GF dimers in complexes are compared by superimposition using GF dimers on the ALK3:BMP-2 complex (A–D); similarly, positions of type II receptors on GF dimers in complexes are compared by superimposition using GF dimers on the ActRIIB:BMP-2 complex (E–H). Receptor names are color-coded as in their ribbon diagrams. GFs used for superposition are shown with black names below names of their bound receptors. GF dimers are shown as surfaces, with monomers shaded pink and light blue. Overlaid type I and type II receptors are in yellow, whereas ALK3 and ActRIIB are in orange–red and green, respectively. Complex structures are for ALK3:GDF-5 (Klammert et al. 2015), ALK6:GDF-5 (Kotzsch et al. 2009), ALK1:ActRIIB:BMP-9 (Townson et al. 2012), ALK5:TβRII:TGF-β3 (Groppe et al. 2008), ALK3: BMP-2 (Kirsch et al. 2000), ActRIIB:ActA (Thompson et al. 2003), ActRII:BMP-7 (Greenwald et al. 2003), and ALK3:ActRIIB:BMP-2 (Weber et al. 2007).
Figure 9.
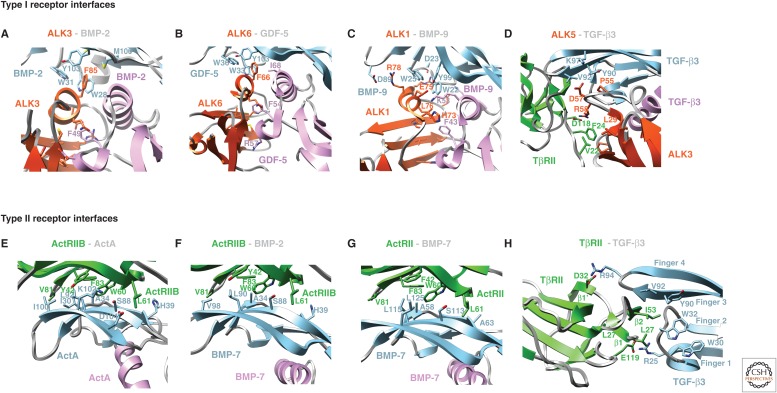
Type I and type II receptor–GF interfaces. Interfaces show growth factors (GFs) bound to type I receptors (A–D) or type II receptors (E–H). Cartoon diagrams show GFs as pink and light blue monomers, and type I and type II receptors in red–orange and green, respectively, with key side chains in stick with carbons in the same colors, red oxygens and blue nitrogens. Structures correspond to those in the Figure 8 legend.
How TβRII specifically binds TGF-β1–3 is evident from the structure of the TGF-β3:TβRII:ALK5 ternary complex (Figs. 6H, 7F, and 9H) (Groppe et al. 2008). TβRII binds in the cleft between the fingers of the GF, using primarily the β2, but also the flanking β1 and β1′ strands. Remarkably, the TβRII binding site on the GF does not overlap with those of ActRII and ActRIIB (Fig. 8H). The ability of TβRII to bind a distinct site on the GF is driven by differences in receptor structure that change the overall shape of the binding surface (see section on structures of type I and type II receptor ectodomains); sequence and loop shape differences in the finger region of TGF-βs relative to other GFs enable specificity (Fig. 4B, lower) (De Crescenzo et al. 2006; Baardsnes et al. 2009). The interface that stabilizes TβRII in the cleft between the TGF-β fingertips is unique relative to that used by ActRII and ActRIIB, and is characterized by an inner hydrophobic portion, with Ile53 of TβRII packing into a shallow pocket in TGF-β formed by Trp32, Tyr90, and Val92 (Fig. 9H) (Groppe et al. 2002; Radaev et al. 2010). The inner hydrophobic portion is flanked on either edge by hydrogen-bonded ion pairs between the side chain carboxylate groups of Asp32 and Glu119 on TβRII and the side chain guanidinium groups of Arg25 and Arg94 on the fingertip loops of TGF-β (Fig. 9H). The two arginine residues contribute >30% of the total binding energy and are invariant in TGF-β1 and 3, which bind TβRII with high affinity (De Crescenzo et al. 2006; Baardsnes et al. 2009). The two arginine residues are conservatively replaced by lysine in TGF-β2, which binds TβRII with low affinity, and substitution of these with arginine increases the affinity of TGF-β2 to that of TGF-β1 and 3 (De Crescenzo et al. 2006; Baardsnes et al. 2009). Relative to all other GFs, TGF-βs have a one-residue insertion in the loop bearing Arg25, and differ in the length and sequence of the loop bearing Arg94 (Fig. 1), correlating with the different shapes of fingertip loops (Fig. 4B, lower) and creating high specificity for TβRII.
Type I receptor ectodomain:GF complexes also provide insights into specificity, but the differences are more subtle. Thus, complexes containing ALK1, 3, 5, and 6 with GFs as diverse as GDF-5, BMP-2, BMP-9 and TGF-β3, show that the type I receptors are positioned similarly, although not identically, at the wrists of the GFs (Fig. 7A,D–F) (Kirsch et al. 2000; Groppe et al. 2008; Kotzsch et al. 2009; Townson et al. 2012). The closely related type I receptors ALK3 and ALK6 (TGF-β receptors, Box 1) bind similarly at the wrist and contact each GF monomer roughly equally (Kirsch et al. 2000; Keller et al. 2004; Kotzsch et al. 2009; Klammert et al. 2015). ALK1 and ALK5 belong to subfamilies distinct from that containing ALK3 and ALK6 (Box 1). The structural features that cause ALK1 and ALK5 binding positions to shift relative to the ALK3/6 subfamily (Fig. 8A–D) are discussed in the next two paragraphs.
The type I receptor ALK1 is divergent in sequence from ALK3 and ALK6 (TGF-β receptors, Box 1) and binds and transduces signals for BMP-9 and BMP-10, but not other BMPs and GDFs (David et al. 2007). ALK1 binds both monomers at the wrist, but the directionality of the strands that comprise its central β4-β3-β5 sheet is rotated relative to that of ALK3 and ALK6 (Mahlawat et al. 2012; Townson et al. 2012), as shown by the dashed lines in Figure 7D,E. Repositioning ALK1 relative to ALK3 and ALK6 is primarily driven by a two-residue shortening in the segment between the Cys of the c disulfide at the carboxy-terminal end of β-strand 4 and the Cys of the d disulfide in the 310 helix of the β4-β5 loop (Figs. 5 and 6). Shortening causes this segment of ALK1 to be more rigid and shifted closer to β-strand 5 than in ALK3 and ALK6 (Fig. 6A–C) (Mahlawat et al. 2012; Townson et al. 2012). Loop shortening in ALK1, together with conformational changes in the prehelix extension and other loops in the wrist region of BMP-9 (Fig. 4B), leads to steric clashes when ALK1 is positioned onto BMP-9 in the same manner as ALK3 on BMP-2 (Mahlawat et al. 2012; Townson et al. 2012). The structure of the BMP-9:ActRIIB:ALK1 complex shows that these clashes are alleviated when ALK1 binds BMP-9 by a rotation of ∼20° relative to ALK3 bound to BMP-2 (Fig. 7D,E) (Townson et al. 2012).
ALK5 (also known as TβRI) is in a type I receptor subfamily distinct from the ALK1 and ALK 3/6 subfamilies (TGF-β receptors, Box 1). Compared with these receptors, ALK5 binds more toward the TGF-β fingertips, which enables it to additionally contact TβRII (Fig. 7F, upper panel) (Groppe et al. 2008). TβRII has a longer amino-terminal segment than other type II receptors (Fig. 5), and a portion of this segment becomes ordered by this contact, with Val22 and Phe24 in TβRII binding to a hydrophobic pocket in ALK5 (Fig. 7F, inset). The repositioning of ALK5 relative to ALK3 and ALK6 is, like ALK1 repositioning, driven by a modification of the segment between the c disulfide cysteine in the carboxy-terminal end of β-strand 4 and the d disulfide cysteine in the 310 helix within the β4-β5 loop, but in this case with a five-residue extension rather than a two-residue shortening (Figs. 5 and 6). The PRDRP sequence motif in the prehelix extension, between the c and d disulfide in Alk5, is rigid and forms a tight turn at its amino-terminal cis Pro-55 (Figs. 5, 6D, and 7F, inset) (Zuniga et al. 2011). The extension precludes ALK5 from binding similarly to ALK3 due to clashes with hydrophobic residues in the wrist. These clashes cause ALK5 to reposition so that it shifts toward the fingertips in which it binds in the cleft between TGF-β and TβRII. Repositioning orchestrates a number of interactions with TGF-β and TβRII that are required for cooperative assembly of the TGF-β signaling complex (Fig. 7F, inset) (Groppe et al. 2008; Radaev et al. 2010; Zuniga et al. 2011). In summary, alternative positioning of ALK1 and ALK5 provide distinct interfaces and stabilizing interactions that enable segregation of GF specificities of the ALK1 and ALK5 subfamilies from the ALK3 and ALK6 subfamily.
Further details on amino acid sidechain and structural features that underlie the selectivities displayed by type I and type II receptors, despite promiscuity in binding multiple GFs, are summarized in Table 2. These details extend to differences among ActRII, ActRIIB, and BMPRII, and between ALK3 and ALK6 (Table 2).
Table 2.
Structural basis of GF binding preferences by ActRII, ActRIIB, ALK3, and ALK6
| Preference | Interactions that promote binding to preferred GF | References |
|---|---|---|
| ActRII and ActRIIB bind activins 100- to 1000-fold more avidly than BMPs and GDFs | Intramolecular salt bridge between Lys102 and Asp104 on finger 4 of activin A promotes hydrophobic and H-bond interactions with ActRIIB; ion pairs of Glu111 and Arg87 of activin A with Lys37 and Asp62 of ActRIIB | Thompson et al. 2003; Allendorph et al. 2007; Weber et al. 2007 |
| BMPRII binds BMPs and GDFs, but does not bind activins | BMPRII Tyr113 promotes binding to BMP-2, BMP-7, and GDF-5, but inhibits binding to activin A | Greenwald et al. 2003; Yin et al. 2008 |
| ALK6 binds GDF-5 10- to 20-fold more avidly than ALK3 | Rotation of ALK6 by about 9° brings its β1-β2 loop close to Arg57, which protrudes from the pre-helical loop of GDF-5; the equivalent loop of ALK3 has a different conformation that cannot accommodate Arg57 | Keller et al. 2004; Kotzsch et al. 2009 |
GDF, Growth differentiation factor; BMP, bone morphogenetic protein.
Coreceptors
Several families of coreceptors regulate GF signaling. One subfamily comprises betaglycan and endoglin, which show sequence homology with one another. They contain amino-terminal orphan and carboxy-terminal zona pellucida (ZP) domains (Gougos and Letarte 1990; Bork and Sander 1992; Esparza-Lopez et al. 2001), single-pass transmembrane domains, and short cytoplasmic domains of <50 amino acid residues. Both coreceptors can be shed from the cell surface as soluble fragments (Lamarre et al. 1994; Hawinkels et al. 2010).
Betaglycan interacts with TGF-βs, BMP-2 and 4, GDF-5, and inhibins (Lewis et al. 2000; Esparza-Lopez et al. 2001; Kirkbride et al. 2008). Most importantly, betaglycan binds the three TGF-βs and, hence, was identified as the TGF-β type III receptor. It was initially thought to predominantly function by presenting TGF-β2 to TβRII, and thereby compensate for TGF-β2’s low intrinsic affinity for TβRII (Lopez-Casillas et al. 1993). In agreement, betaglycan knockout and TGF-β2 knockout mice each show perinatal lethality and display many phenotypic characteristics in common (Stenvers et al. 2003). Because the cytoplasmic domain of betaglycan can interact with β-arrestin and mediate internalization of TGF-βs and their receptors, betaglycan can also directly modulate TGF-β signaling (Chen et al. 2003; McLean et al. 2013).
Betaglycan’s ZP domain comprises amino-terminal (ZP-N) and larger carboxy-terminal (ZP-C) immunoglobulin-like subdomains. Of these, only ZP-C interacts with TGF-β (Wiater et al. 2006). Structural and functional analyses of the betaglycan ZP-C domain suggest that a short peptide segment, named EHP, harbors the main binding determinants of the betaglycan ZP-C subdomain for TGF-β (Fig. 10A) (Lin et al. 2011; Diestel et al. 2013). Understanding how betaglycan interacts with TGF-β GFs to provide high affinity (Mendoza et al. 2009) will require structures of its orphan domain, and GF complexes with the orphan and ZP-C domains.
Figure 10.
Coreceptors for transforming growth factor β (TGF-β) family members. (A) The carboxy-terminal zona pellucida (ZP-C) domain of betaglycan (3QW9) (Lin et al. 2011). (B) The coreceptor cripto comprises small epidermal growth factor (EGF)-like and CFC domains. The CFC domain of cripto adopts an extended architecture lacking secondary structure and is stabilized by three disulfide bonds (yellow sticks) (2J5H, Calvanese et al. 2006). Two residues involved in binding of the CFC-domain to ALK4, His104, and Trp107, are shown with green side chains. (C) The amino-terminal domain (ND) of RGMb (RGMbND) in cartoon (green), bound to a BMP-2 dimer (surface, with type I and type II receptor binding sites in light green and pink, respectively) (PDB 4UI0 [Healey et al. 2015]). (D) Similar structural elements are used in the interaction of BMP-2 with RGMbND (upper panel) and ALK3 (lower panel). Leu103, conserved in all three repulsive guidance molecules (RGMs), engages in a hydrophobic knob-into-hole interaction resembling that of the phenylalanine residue conserved among BMP type I receptors (Phe85 in ALK3 and Phe66 in ALK6). Moreover, the Phe-Pro motif (surface and side chains in magenta) found in Smad1-, 5-, and/or 8-activating BMPs forms a hydrophobic patch between the 1st and 3rd helix of the RGM three-helix bundle to mimic interaction with ALK3. His106 in RGMb, conserved in all three RGMs, makes a π-stacking interaction with a conserved Trp in bone morphogenetic protein (BMP) and renders BMP-binding by RGMs pH-sensitive. (E) RGM binds via RGMND (green cartoon) to BMP (gray and black monomer surfaces with the type I and type II receptor sites in light green and pink, respectively) and via RGMBCD (cyan cartoon) to neogenin (dark blue cartoon) (PDB 4UI2 [Healey et al. 2015]). RGMbCD links to RGMND with a flexible linker (dashed lines) that is only partially observed in the crystal structure (green cartoon with disulfides to RGMbCD in gold stick). The acid-labile Asp-Pro bond (marked with GD↓PH) is at the carboxy-terminal end of β-strand 1 of the β-sandwich of RGMbCD. This β-strand and two disulfide bonds maintain covalent linkage between RGMbND and RGMbCD despite cleavage at the Asp-Pro bond. (F) As in E, but viewed along the dyad of the BMP GF dimer.
Endoglin has unique biological functions compared with betaglycan (Nassiri et al. 2011). Its importance in vascular development is shown by loss-of-function mutations in the vascular disorder HHT type 1 (HHT1) (Ardelean and Letarte 2015). BMP-9 and -10 are similarly important in vasculogenesis (Chen et al. 2013), and mutations in the BMP-9 prodomain and ALK1 cause similar HHT syndromes (Tillet and Bailly 2014). Within the TGF-β family, endoglin has high affinity and specificity for BMP-9 and -10 (Castonguay et al. 2011). It binds both BMPs through its orphan domain, does not compete or synergize with ALK1, but competes with type II receptors (Castonguay et al. 2011; Alt et al. 2012). BMP-9, BMP-10, endoglin, and ALK1 may form a regulatory circuit to control angiogenesis (Tillet and Bailly 2014).
Cripto is the eponymous member of a small protein family (Ciardiello et al. 1991) and serves as a coreceptor for several TGF-β family GF dimers (Yeo and Whitman 2001; Cheng et al. 2003, 2004; Gray et al. 2003; Chen et al. 2006). Cripto and its family members are attached to the cell membrane by a glycosylphosphatidylinositol (GPI)-moiety (Minchiotti et al. 2000). Like betaglycan and endoglin, cripto can be shed, but by GPI phospholipase D rather than by proteolysis (Watanabe et al. 2007). Cripto functions in switching between nodal and activin signaling, which use the same type I and type II receptors (Rosa 2002). As nodal requires cripto for binding to ALK4, whereas activins do not, nodal signaling is repressed in cells lacking cripto, whereas activins can signal via ALK4 and ActRII/IIB (Yeo and Whitman 2001). Structurally, cripto comprises two consecutive small cysteine-rich domains, an amino-terminal EGF-like domain and a CFC-domain. Both domains contain three disulfides required for folding. Although the EGF domain is involved in binding to nodal, the CFC domain interacts with the type I receptor ALK4 (Adkins et al. 2003; Calvanese et al. 2006). A structure for the CFC domain in its unbound form shows that two residues implicated in interaction with ALK4, His104 and Trp107, form a patch on the protein surface (Fig. 10B) (Calvanese et al. 2006). However, further insights into how cripto bridges nodal to ALK4 require a structure of the nodal:ALK4:cripto ternary complex.
RGMs, initially identified for their functions in guiding retinal axons and neural tube closure (Muller et al. 1996; Monnier et al. 2002) are BMP-specific coreceptors (Babitt et al. 2005; Samad et al. 2005). Mammals have three RGM family members, RGMa, RGMb/DRAGON and RGMc/hemojuvelin (HJV) (Camus and Lambert 2007). They are highly conserved, with 40%–50% amino acid sequence identity, and thus have identical protein architectures. RGMs both interact with the type I transmembrane protein neogenin, a netrin family receptor, to mediate neuronal functions (Matsunaga et al. 2004; Rajagopalan et al. 2004), and bind BMPs to modulate BMP signaling. The relationship between these two functions is poorly understood, but both neogenin and BMP binding are required for the proper function of HJV in regulating systemic iron stores (Kuns-Hashimoto et al. 2008; Zhang et al. 2009).
RGMs contain an amino-terminal domain (ND) with three disulfide-linked α-helices that binds BMPs, a flexible linker, a carboxy-terminal domain (CD) with two tightly packed β-sheets that contains an acid-labile Asp-Pro peptide bond and binds to neogenin, a PC cleavage site, and a carboxy-terminal GPI membrane anchor (Bell et al. 2013; Healey et al. 2015). Soluble forms of RGMa and RGMc result from processing by the PCs furin and/or SKI-1 (subtilisin kexin isozyme-1) (Tassew et al. 2012).
How do RGMs stimulate BMP signaling? Structural and microscopy studies suggest that endocytosis of BMPs by RGMs and neogenin promotes type I and type II receptor signaling in an intracellular compartment (Healey et al. 2015). The first 120 amino-terminal residues of RGMs, RGMND, fold into an antiparallel three-helical bundle that binds BMPs at the receptor type I site (wrist epitope) (Fig. 10C). When complexes of RGMs bound to BMP-2 are compared with ALK3 bound to BMP-2, the third helix of RGMND mimics the interaction of the short α-helix of ALK3 with BMP-2 (Fig. 10D). One key element for the RGM–BMP interaction is a histidine residue conserved in all RGM proteins, His106 in RGMb, which forms a π-stacking interaction with the second Trp of the W-X-X-W motif in BMPs (Fig. 10D). Binding of RGMs to BMPs is inhibited at pH <6.5, presumably because of histidine protonation. This feature points toward a mechanism by which RGMs might stimulate BMP signaling. Because RGMs block BMP binding to type I but not type II receptors, it is proposed that RGM–BMP–BMPRII complexes form at the cell surface, and that these are then endocytosed in a clathrin-dependent manner. As type I receptors, including ALK3, are constitutively endocytosed, endosomes might form that contain RGM–BMP–type RII complexes as well as free ALK3. Because of endosomal acidification, the RGM coreceptor would be released from its pH-dependent complex with BMP and the type II receptor, enabling the type I receptor to replace RGM in the complex, as type I receptor interaction with BMPs is not pH-sensitive (Heinecke et al. 2009; Healey et al. 2015). Signaling would thus occur in the cytoplasmic environment as for receptors on the cell surface, but on the cytoplasmic face of a distinct membrane compartment where other colocalizing molecules might differ (Healey et al. 2015). This might alter the shape of signaling as shown for some G protein-coupled receptors, in which a second wave of signaling occurs on endosomes that moves the site of second messenger production relative to effectors (Tsvetanova et al. 2015).
And how does neogenin influence the BMP-RGM interaction? The ternary complex structure of BMP-2:RGMb:neogenin not only shows that RGMs can simultaneously bind BMPs and neogenin (Fig. 10E,F), but also reveals that binding of type II receptors to this complex is again not blocked. Thus, formation of quaternary complexes consisting of RGMs, neogenin, BMP, and type II receptors is possible (Healey et al. 2015). Although RGMCD binding to neogenin brings neogenin into close proximity to BMP bound to RGMND, no direct interactions between BMP and neogenin are observed (Fig. 10E,F) (Healey et al. 2015). The modular architecture of RGMs with their two domains independently binding two different partners, BMPs and neogenin, of which BMP is a dimer and neogenin is dimerized on RGM binding (Bell et al. 2013), suggest that RGMs could bridge individual complexes and facilitate formation of supramolecular clusters. Indeed, high-resolution microscopy shows that BMP-2 mediates clustering of RGM–neogenin complexes into large assemblies at the cell surface (Healey et al. 2015). The physiological significance of supramolecular cluster formation is unknown, although this may enable BMPs to modulate neogenin signaling, and neogenin to modulate BMP signaling, adding more complexity to BMP receptor activation (Mueller 2015).
In summary, TGF-β and BMP coreceptors are functionally and structurally diverse, and the presence or absence of a coreceptor can have large qualitative (changing gene expression) and quantitative effects (sensitizing cells for a GF) on downstream signaling.
REGULATION OF TGF-β/BMP SIGNALING BY MODULATOR PROTEINS—BMPs AS SIGNALING HUBS
Structural Diversity of TGF-β/BMP Antagonists
A hallmark of the TGF-β/BMP family is the plethora of extracellular modulator proteins that bind GFs and regulate signaling. More than 25 modulator proteins are known. Attempts have been made to classify TGF-β/BMP modulator proteins structurally. A common hallmark is the large number of cysteine residues. Although the diversity of these modulators is stunning, they can be grouped, based on structural features and sequence homology, into six distinct families: noggin, members of the Dan family (Fig. 11), the follistatin family, the CCN family, the chordin family, and twisted gastrulation (Tsg). The Dan and CCN families as well as noggin have CKGF domains. Notably, the diversity of BMP and activin antagonists contrasts with the paucity or absence of modulators of TGF-β itself, perhaps correlating with the distinct integrin-dependent mechanism of TGF-β activation.
Figure 11.
Structural alignment of Dan family monomers to cystine knot (CK) growth factor (CKGF) superfamily representatives with similar monomer–monomer orientations. (A–G) CKGF monomers in cartoon with α-helices in cyan and β-strands in magenta. Disulfides are shown in stick and are color-coded with their cysteines numbered as in panel H. Cysteines in intermolecular disulfides are indicated by showing the partner Cys in the other monomer with its number preceded by a dash and followed by a hyphen. All monomers but sclerostin are from dimeric structures. A frizzled domain and glycan cocrystallized with norrin are shown in cartoon and stick, respectively. Monomers are in identical orientations, and aligned in two columns. (H) Structure-based sequence alignment. Cysteines disulfide-bonded to one another are in identical colors, except those that participate in interchain disulfides in some families and intrachain disulfides in others (positions 5, 6, and 11) are shown in red. Disulfides unique to particular families are shown in thinner silver stick and colored silver in panel H. Other residues are colored according to ClustalX. CK disulfides are labeled between the two rows of the alignment and link Cys residues distant in sequence; the three long loops between CK disulfides are also marked. For alignments to CKGF families with monomers dimerized in different orientations (neurotrophin in the nerve growth factor [NGF] family, and platelet-derived growth factor [PDGF] in the PDGF family) (see Zhou and Springer 2014). CKGF and CTCK monomers (PDB codes are shown in panel H) were aligned to one another structurally using DeepAlign (Wang et al. 2013). Structurally equivalent residues are shown aligned by sequence as described in Figure 5 legend. Cα atom root mean square deviation (RMSD, Å) of monomers in each position relative to the average at each position are shown below the alignment. Gaps were closed to minimize the length of the alignment; corresponding positions lack an RMSD value.
Noggin—The BMP Antagonist Paradigm
Noggin, a component of the Spemann organizer (Smith and Harland 1992), antagonizes the ventralizing activity of BMPs in dorsoventral patterning and regulates skeletogenesis (Zimmerman et al. 1996; Brunet et al. 1998). Noggin binds to the same sites on BMP-7 as the BMP type I and type II receptors, and thereby impedes BMP signaling (Fig. 12A–D) (Groppe et al. 2002). Noggin contains a CKGF domain like TGF-β GF domains, but has a 10- rather than 8-membered CK ring (Fig. 12A). Furthermore, its monomers dimerize through a Cys residue in a position (cysteine 11 in Fig. 11G,H) that differs from that in TGF-β family proteins, and through an additional α-helical domain (Fig. 12A). The helices lie on the convex side of noggin, opposite the concave BMP binding site, and support noggin’s C-clamp-like curvature (Fig. 12A,B). Thus, whereas the TGF-β CKGF dimer is linked by monomers that overlap in the palm and heel (GF homodimer, Box 1), and dimerize through cysteine 6, the noggin dimer links the two CKGF domain monomers more distally toward their carboxy-terminal end at cysteine 11 (Fig. 11F,G), leading to a longer and more curved dimer that straddles and clamps the BMP dimer (Fig. 12A,B).
Figure 12.
The bone morphogenetic protein (BMP) modulator noggin, members of the Dan family, and sclerostin. (A) Noggin binds BMPs (PDB 1M4U [Groppe et al. 2002]) in a clamp-like manner with the fingers of its cystine knot (CK) growth factor (CKGF) domain (red) blocking type II receptor binding, and a peptide segment termed clip (green) shielding the type I receptor epitopes. Hydrophobic Pro35 of noggin, engaging in the knob-into-hole interaction motif is shown as a gold sphere. The noggin dimer is formed by interactions between the α-helical dimerization domains and stabilized by an intermolecular disulfide bond (asterisk). Disulfides are shown with gold sticks and the CK motif (box) is shown in detail. (B) As in A but viewed down the dyad axis of the noggin and GF dimers. (C and D) A prototypical BMP ligand-receptor complex, with type I and type II receptors as green and red ribbons, respectively, and the GF dimer as gray surface (PDB 2H64 [Weber et al. 2007]) shown in similar orientations as in A and B. (E) Phylogenetic tree of the Dan family (upper panel) and schematic of domain organization (lower panel) with the CKGF domain boxed, its eight disulfides indicated by black lines, and the segments forming three loops colored as in panels F-H. The highly diverse segments amino- and carboxyl termini to the CKGF domain are shown as horizontal lines. (F) Sclerostin NMR structure (PDB 2KD3 [Weidauer et al. 2009]). The cartoon representation (upper) shows the NXI motif (red spheres) and the Cα atom trace (lower) shows the ensemble of 15 structures highlighting the flexibility of loop 2. Loops are colored as in E (lower). (G) PRDC (PDB 4JPH [Nolan et al. 2013]). The cartoon representation (upper, with one monomer colored as in E, lower) shows cysteines as gold sticks, with the unpaired Cys boxed. The lower panel shows PRDC as a surface, with the α-helical amino-terminal segments as blue Cα atom traces. Hydrophobic residues, which on exchange for alanine attenuate binding to BMP binding, are colored red. (H) Two Nbl1/Dan crystal structures (PDB 4X1J [Nolan et al. 2015], upper) and (PDB 4YU8, Table 1, lower). One monomer is colored as in E (lower). The other monomer(s) associates as in PRDC (gray, upper) or in a different manner, with two nearby monomers in the crystal lattice shown in two shades of gray (lower).
Distinct structural elements in noggin block the BMP type I (wrist) and type II (knuckle) receptor epitopes (Fig. 12A–D). A 25-residue segment within the amino-terminal region of noggin is spatially separated from the CKGF and α-helical domains in a complex with BMP, fastens to more than one BMP monomer in a U-shape, and is therefore termed the “clip” (Fig. 12A) (Groppe et al. 2002). In this segment, residues Pro35 to Ser38 snugly fold into the center of the wrist epitope, and block binding to type I receptors. The clip then turns over the fingers of the BMP and runs over the periphery of the knuckle epitope. In contrast, the noggin fingers at the end of its C-clamp extend over the fingertips of the BMP and shield a large part of the knuckle epitope, thereby blocking access and binding of type II receptors. Hydrophobic interactions dominate both the fingertip and clip interfaces, similarly to type I and type II receptor interfaces with BMPs. Moreover, Pro35 in the noggin clip, which is highly conserved among different species, extends into a hydrophobic pocket in the wrist epitope of BMP-7, mimicking the knob-into-hole interaction found in various BMP-type I receptor interactions (Fig. 12A–D) (Hatta et al. 2000; Kirsch et al. 2000; Groppe et al. 2002; Kotzsch et al. 2009). The importance of Pro35 for the BMP-neutralizing activity of noggin is highlighted by its mutation in skeletal malformation diseases (Gong et al. 1999; Mangino et al. 2002).
The mechanism of noggin binding to BMPs preserves much of the chemistry of receptor binding to BMPs, including shared hydrophobic interfaces and the knob-into-hole packing seen with most type I receptors; however, details of the distribution and arrangement of the structural elements differ. Remarkably, the noggin–BMP complex partially shares its building principle with the TGF-β1 proprotein complex; the folded noggin CKGF domain, like the prodomain arm domain, blocks the type II receptor site, and the single element clip in noggin, like the prodomain straitjacket, blocks type I receptor access to the GF (Fig. 12A; Box 1). In both noggin and the TGF-β1 prodomain, a central domain together with intermolecular disulfide bonds forms a covalent homodimer, which explains the extremely tight binding to the GF. Moreover, both interact with the ECM; the TGF-β1 prodomain is covalently linked to LTBP, and noggin has heparin binding sites to interact with the ECM (Groppe et al. 2002; Paine-Saunders et al. 2002) that can locally retain a bound GF to limit its radius of action.
Noggin binds in vitro to most BMPs with no apparent dissociation, which suggests that the concentration of active ligand would be effectively decreased for all BMPs that it targets (Zimmerman et al. 1996; Seemann et al. 2009; Song et al. 2010; Schwaerzer et al. 2012). However, in cell-based assays some BMPs, for example, BMP-6, have been described as noggin-resistant, although their affinity to noggin is in the same range as for nonresistant ligands (Song et al. 2010). Furthermore, mutations that render BMPs noggin-resistant only marginally decrease their affinity to noggin (Song et al. 2010; Schwaerzer et al. 2012), possibly indicating that the inhibition mechanism is more complex in vivo.
The Dan Family—A Heterogeneous Group of Antagonists for BMPs and/or Wnt Factors
With seven known members in mammals, Dan modulators are the largest family of TGF-β family antagonists (Nolan and Thompson 2014). They include neuroblastoma suppressor of tumorigenicity 1 (Nbl1, Dan, DAND1), gremlin/Drm (DAND2), protein related to Dan and cerberus (PRDC, DAND3, gremlin2), cerberus (DAND4), and coco/dante (DAND5) (Fig. 12E). These five members are the smallest proteins known to modulate BMP signaling. Sclerostin and USAG1 (also known as wise, ectodin, and SOSD) also belong to the Dan modulator family (BMP antagonists, Box 1, Figs. 11 and 12E), but they antagonize Wnt and not BMP signaling. Dan family members share a central CKGF domain of 90 residues that may be substantially elaborated with amino- and carboxy-terminal extensions that add another ∼100 residues (Fig. 11A–C, 12E, lower). Extensions include α-helices in PRDC (Nolan et al. 2013) and unstructured regions in sclerostin (Veverka et al. 2009; Weidauer et al. 2009). These might become structured when bound to GFs or ECM components and might resemble in this respect the long amino-terminal extensions in many TGF-β family prodomains (Fig. 1).
All Dan and sclerostin family proteins contain an 8-membered CK ring with the same sequence motif as in TGF-β family CKGF domains (C-X-G-X-C and C-X-C) (Figs. 11 and 12E). Besides the six CK cysteines, Dan proteins contain two additional cysteines (designated C2 and C8 in Fig. 11) that disulfide-link the two β-ribbon fingers at the fingertips (Figs. 11 and 12F,G). The same eight Cys residues are shared with carboxy-terminal CK (CTCK) domains, which are closely related to CKGF domains. CTCK domains have been structurally characterized in von Willebrand factor (Zhou and Springer 2014) and norrin (Ashe and Levine 1999; Chang et al. 2015). Three long loops in CKGF and CTCK domains link the CK residues (GF monomer, Box 1, Fig. 11). The Dan family (Nolan et al. 2013; Nolan et al. 2015), sclerostin (Veverka et al. 2009; Weidauer et al. 2009), and CTCK domains have similarly curved, long β-ribbon fingers (loops 1 and 3) stabilized by the fingertip C2-C8 disulfide (Figs. 11A–C,E and 12F–H). Human chorionic gonadotrophin (HCG) in the hormone CKGF family and noggin have similarly long, disulfide-stabilized fingers, but the disulfides are between nonequivalent cysteines (Fig. 11D,G,H). Loop 2 of TGF-β family GF domains forms the heel α-helix; in contrast, loop 2 of PRDC, Nbl1, and CTCK domains extends far from the CK, forms a β-ribbon in some members, and continues the curvature of loops 1 and 3 (Figs. 11 and 12G,H).
Different loop 2 structures correlate with the amount of overlap between monomers when they dimerize. TGF-β family CKGF domains dimerize through the cysteine in position C6 (C6-C6′ disulfide), whereas CTCK domains dimerize through reciprocal C5-C6′ and C6-C5′ disulfides and an additional C11-C11′ disulfide (Fig. 11). Because C5 and C6 are two residues apart in a C5-X-C6-C7 sequence that includes C7 of the CK, the dimer interface slides two residues in CTCK dimers so that their knots are two β-ribbon positions further apart than in TGF-β GF dimers. The dimer interface in the Dan family proteins PRDC and Nbl1 and in HCG slides similarly to that in CTCK domains, correlating with their similarly shaped loop 2 (Fig. 11). However, these Dan proteins and HCG form dimers that are not disulfide-linked. Furthermore, although PRDC and Nbl1 contain one or two Cys residues equivalent in sequence and structure to those in positions C5 and C11 in CTCK domains, they do not form equivalent disulfides. PRDC has a C5 cysteine that curiously has no disulfide bond partner, whereas in Nbl1 C5 and C11 form an intramolecular disulfide that contrasts with intermolecular disulfides in CTCK domains (Fig. 11A,B,E).
In contrast to Dan proteins, which are shown or considered to be dimers (Stanley et al. 1998; Pearce et al. 1999; Kattamuri et al. 2012), sclerostin and USAG1 are both monomeric (Kusu et al. 2003; Winkler et al. 2003; Lintern et al. 2009). In the absence of a dimer interface, loop 2 of sclerostin is not stabilized by van der Waals interactions and is highly flexible and disordered (Fig. 12F). Although sclerostin was initially described as a BMP antagonist (Kusu et al. 2003; Winkler et al. 2003), it functions as an antagonist of the canonical Wnt pathway and binds and blocks the Wnt coreceptor LRP5/6 (van Bezooijen et al. 2004, 2007; Krause et al. 2010; Boschert et al. 2013). Similarly, the close sclerostin relative USAG1 inhibits Wnt signaling (Itasaki et al. 2003). Structure–function studies show that the NXI motif, present in loop 2 of sclerostin and USAG1 but not the Dan clade (Figs. 11H and 12F), binds to the first β-propeller domain in the extracellular domain of LRP6 and thereby competes with binding of Wnt GFs (Bourhis et al. 2011; Holdsworth et al. 2012; Boschert et al. 2013).
In contrast to the monomeric Wnt antagonists sclerostin and USAG1, the CTCK family member norrin is a dimeric Wnt agonist. Norrin stimulates canonical Wnt signaling by binding frizzled4, LRP6, and glycosaminoglycans (GAGs). A synthetic heparin glycan mimetic binds to both Frizzled and the norrin fingertips, providing a model for how ECM components may contribute to the formation of signaling complexes (Fig. 11E) (Ke et al. 2013; Chang et al. 2015). As described in the section on the brief history of TGF-β family evolution, the TGF-β and Wnt signaling pathways coevolved in primitive metazoans. Remarkably, CKGF/CTCK domains evolved to serve as both agonists and antagonists in each of these pathways. The ECM has an important organizing role in both families. Binding sites for GAGs are at the amino termini of some GFs (Ruppert et al. 1996), in the prodomains of activin A and myostatin (Li et al. 2010; Sengle et al. 2011), in the dimerization domain in noggin (Groppe et al. 2002), and along one side of the extended sclerostin molecule harboring residues from all three loops (Veverka et al. 2009). Therefore, such sites must have evolved independently multiple times.
Although all dimeric Dan proteins are effective BMP antagonists, coco and cerberus also block nodal and Wnt signaling (Glinka et al. 1997; Piccolo et al. 1999; Bell et al. 2003). Cerberus binds and inhibits Wnt, nodal and BMPs at mutually noncompetitive sites. Furthermore, the amino-terminal portion of cerberus is required for BMP binding and Wnt antagonism whereas the carboxy-terminal portion including the CKGF domain is sufficient for nodal antagonism (Piccolo et al. 1999).
The prototypical Dan protein PRDC forms a homodimer that is not disulfide-linked, despite its odd number of Cys residues (Fig. 12G) (Nolan et al. 2013). Dimerization of PRDC is between extremely long β-strands in each monomer that begin in loop 1 and extend through the CK into loop 2 (Figs. 11 and 12G). These join with antiparallel β-strands in the other halves of loops 1 and 2 to form an extensive “intermolecular” four-stranded β-sheet, and build a large dimer interface in common with CTCK domains and HCG (Fig. 12G) (Ke et al. 2013; Zhou and Springer 2014; Chang et al. 2015). Additionally, fingers 1 and 2 form two β-ribbons that extend from the central four-stranded β-sheet, reminiscent of the wrist in TGF-β GFs (Nolan et al. 2013). However, instead of an overall butterfly-shaped architecture like TGF-β dimers, the PRDC monomers (Fig. 11A) assemble to form a longer arc-like structure (Fig. 12G).
The structure of the CKGF domain of Nbl1 is very similar to that of PRDC, despite only 35% sequence identity (Figs. 11A,B and 12G,H) (Nolan et al. 2015). Like PRDC, Nbl1 fails to form disulfide-linked dimers, with the C5 and C11 cysteines forming an intramolecular disulfide (Fig. 11B). This is remarkable because the C5 Cys in one Nbl1 monomer is positioned very close to the C11 Cys of the other monomer in the dimer, and thus only slight structural adjustment would be required to form intermolecular disulfides similar to those in the CTCK domains of VWF and norrin (Fig. 11E) (Ke et al. 2013; Zhou and Springer 2014; Chang et al. 2015). Dimerization in all of these structures is over essentially identical interfaces, and gives the dimers an arc-like shape, although the Nbl1 dimer is more curved and shows an S-like shape compared with PRDC (Fig. 12G,H, upper) (Nolan et al. 2015). Both PRDC and Nbl1 form noncovalent dimers in solution (Kattamuri et al. 2012). However, another structure of Nbl1 (PDB entry 4YU8, Table 1) fails to show the same dimerization interface in crystal lattices (Fig. 12H, lower).
Without structural data on a Dan protein bound to BMP or nodal, it is unclear whether Dan proteins inhibit GF signaling by impeding binding of both receptors like noggin, or of only one receptor type, as shown for Tsg (Zhang et al. 2007). Furthermore, the architecture of PRDC and Nbl1 differs from other structurally characterized BMP modulators and lacks C-clamp- or clip-like structures. However, a contiguous patch of hydrophobic residues on the convex surface of PRDC is required for binding and inhibition of BMP signaling in vitro (Fig. 12G, lower) (Nolan et al. 2013). Several of these residues are conserved in Nbl1, which binds BMP-2 and 7 and GDF-5 with different affinities compared with PRDC. The importance of these residues in encoding BMP selectivity was shown by exchange of patch residues from PRDC into Nbl1, which yielded an Nbl1 variant mimicking PRDC (Nolan et al. 2015). Interestingly, in PRDC the hydrophobic patch responsible for BMP binding seems to be covered by a dynamic α-helix in the amino-terminal segment (Fig. 12G, lower). A similar helix is not observed in the Nbl1 structure (Fig. 12H) (Nolan et al. 2015).
Follistatin Antagonizes Activins as Well as BMPs
Follistatin (Fst) was initially described as a protein that antagonizes activin signaling. However, follistatin also binds BMPs and GDFs, including BMP-2, 4, 6, and 7 as well as myostatin/GDF-11, albeit with lower affinities (Iemura et al. 1998; Amthor et al. 2002, 2004; Glister et al. 2004; Schneyer et al. 2008). Fst is a modular protein comprising a unique 65-residue amino-terminal domain (Fs0) followed by three follistatin domains (Fs1, Fs2, Fs3) of 70 to 85 residues each (Fig. 13A,B). Follistatin domains are in turn each composed of EGF-like and Kazal subdomains (Hohenester et al. 1997; Schneyer et al. 2004).
Figure 13.
Modulators of the follistatin and chordin families. (A,B) Two follistatin-288 molecules shown in ribbon cartoon wrap around myostatin shown as a surface (PDB 3HH2 [Cash et al. 2009]). Fs0 (green) blocks access to the type I receptor epitope (light green) with a knob-into-hole interaction indicated by a gold sphere; Fs1 (red) and Fs2 (magenta) block the type II receptor-binding site (pink). (C–E) Open and closed conformations of activin A. (C,D) Shows activin monomers as light and dark gray surfaces in different orientations (dashed lines) in an open complex with an Fs1-2 fragment of follistatin (C, PDB 2ARP [Harrington et al. 2006]) and in a closed conformation with FSTL3 (D, PDB 3B4V [Stamler et al. 2008]). (E) Compares monomer–monomer orientations in activin A and myostatin dimers by superimposing on one monomer (gray) and showing the orientation of the other monomer (colored and numbered). The colored and numbered monomers are from dimeric structures of myostatin bound to follistatin-288 (red, 1), activin A bound to FSTL3 (cyan, 2) and activin-A bound to the follistatin Fs1-2 fragment (blue, 3). (F) Chordin family members are mosaics of tandem domains. Although von Willebrand factor C (VWC) domain(s) mediate bone morphogenetic protein (BMP) binding, other domains present include chordin (CHRD), von Willebrand type D (VWD), trypsin inhibitor-like (TIL), insulin-like growth factor binding protein (IGFBP), thrombospondin (TSP) and cystine knot growth factor (CKGF). (G) VWC domain of collagen IIA (left) and VWC domain 1 of CV2 (right). Amino-terminal peptide segments and VWC subdomains 1 and 2 are green, red and blue, respectively. A structurally variable segment of VWC subdomain 1 is shown in yellow. (H) The VWC1 domain of CV2 shown in ribbon with knob-into-hole residue shown as gold sphere bound to BMP-2 shown as surface with type I and type II receptor sites colored green and pink, respectively (PDB 3BK3 [Zhang et al. 2008]). (I) Similarity in architecture of VWC1 domain of CV2 bound to BMP-2 in crystals (left) and free in solution (right panel, NMR ensemble of 10 structures, PDB 2MBK [Fiebig et al. 2013]).
Fs domains together with other domains are present in follistatin-like (FSTL) proteins. Although FSTL-1 and 3 are known to regulate activin and BMP signaling, FSTL-4, FSTL-5, SMOC1, and SMOC2 are not yet characterized for effects on activin and BMP signaling. Furthermore, SMOC1 and SMOC2 contain additional domains characteristic of the SPARC family (Schneyer et al. 2004; Bradshaw 2012). The follistatin family thus includes modulators with similar mechanisms of action (binding to GFs and blocking receptor binding) and similar specificities for activins and BMPs, yet its members are built from several types of domains that are assembled into distinctive architectures. Three isoforms of follistatin, Fst288, Fst303, and Fst315 have identical amino-terminal regions, but end at carboxy-terminal residues 288, 303, or 315, respectively, thus creating further complexity. They bind activin with different affinities, and longer isoforms partially lack the ability to bind to heparin and proteoglycans (Kumar 2005).
Structures of follistatin and FSTL-3 bound to activin A or myostatin provide insights into inhibitory mechanisms and specificity (Thompson et al. 2005; Harrington et al. 2006; Cash et al. 2009, 2012). Although follistatin and FSTL-3 block type I and type II receptor binding sites, they bind the GF domain vastly differently from noggin (Figs. 12A and 13A). Follistatin wraps around activin on the perimeter of the dyad (two-fold rotation axis) of the GF dimer (Fig. 13B). In contrast, noggin binds on one side of the dyad axis. Viewed normal to this axis (as in Fig. 12A), noggin resembles a C-shaped clamp with the ends of the clamp gripping the GF fingers. In the case of follistatin, two modulator molecules embrace the GF such that almost its entire surface seems covered (Fig. 13A,B). In contrast to noggin, follistatin is a monomer when not bound to the GF. However, on binding, two follistatin monomers (structures of follistatin 288 are shown) tightly interact in a head-to-tail fashion and form a stable, noncovalent dimer (Thompson et al. 2005). The type I receptor epitope of activin is blocked by the unique amino-terminal domain (Fs0) of follistatin (Fig. 13A). Here, the key element is Phe52 located in the second α-helix of the Fs0 domain, which makes a knob-into-hole interaction with the wrist epitope of activin A similarly to type I receptors that bind BMPs (Hatta et al. 2000; Keller et al. 2004). Fs1, and primarily Fs2, shield the type II receptor epitope by mainly using hydrophobic interactions as found in complexes of activins and BMPs bound to their type II receptors (Fig. 13A,B). The final domain, Fs3, wraps around the fingertips of activin A and engages in predominantly polar contacts with the amino-terminal Fs0 domain of the other follistatin monomer, thus stabilizing encirclement of the ligand (Fig. 13A,B).
The selectivity of follistatin for activins and BMPs is explained by a central core of GF amino acids in the binding sites for the Fs0 and Fs1/2 domains that are conserved in activins and BMPs but not in TGF-βs (Thompson et al. 2005). The contribution of individual follistatin domains to binding differs depending on the GF (Cash et al. 2009). For example, when bound to myostatin compared with activin A, the Fs0 domain shifts more into the GF wrist epitope to better interact. To avoid a clash with Fs0, Fs3 in the other monomer shifts away from myostatin. Additionally, the ability of activin A to show both closed and open conformations (see section on GF structures) is reflected in differences in monomer–monomer orientation in complexes with different follistatin fragments (Fig. 13C–E) (Thompson et al. 2003, 2005; Greenwald et al. 2004; Harrington et al. 2006).
The Fs0 domain in follistatin and FSTLs may play a role in discriminating between high-affinity ligands (i.e., activins, myostatin, and GDF-11), and low-affinity binders i.e., BMP-2, 4, 7, and others). The type I receptor ALK3 still interacts with BMP-2 when the latter is bound to follistatin, suggesting that the Fs0 domain does not bind to or cannot block the wrist epitope in BMPs (Iemura et al. 1998). Lacking this interaction might also explain the lower affinities of follistatin binding to BMPs and indicate that the inhibition of BMPs by follistatin is functionally different from that of activins (see also Stamler et al. 2008). A role for the Fs3 domain in BMP binding is suggested by the finding that FSTL-3, which lacks Fs3 and thus cannot fully encircle activin A and myostatin (Fig. 13D) (Stamler et al. 2008; Cash et al. 2012), binds activin A and myostatin with high affinity, but cannot bind BMPs (Sidis et al. 2006).
Chordin Family Members Exert a Dual Modulator Function with Both Pro- and Anti-BMP Activities
Members of the chordin family are modular proteins with VWC domains (Fig. 13F). VWC domains (Bork 1993), also confusingly known as cysteine-rich domains in chordin (Francois et al. 1994; Sasai et al. 1994), are 60 to 75 residues long and are found, often in multiple tandem repeats, in >70 human proteins, including the TGF-β family modulator proteins chordin, ventroptin, chordin-like 2, kielin, and crossveinless 2 (CV2, also known as BMP endothelial regulator or BMPER) (Fig. 13F) (Garcia Abreu et al. 2002). Chordin is a large multidomain protein that contains chordin (CHRD) domains in addition to VWC domains (Fig. 13F). Expression of chordin fragments in embryos showed that BMP’s dorsalizing activity and BMP binding reside in its VWC domains (Larrain et al. 2000). Structures of VWC domains in collagen IIA and CV2 show two subdomains (Fig. 13G,H) (O’Leary et al. 2004; Zhang et al. 2008). Subdomain 2 is highly dynamic (Fig. 13I, right) (O’Leary et al. 2004), which may be related to the finding that six tandem VWC domains in von Willebrand factor give rise to a region with an extended, highly flexible structure (Springer 2014).
An amino-terminal fragment of CV2 bound to BMP-2 exemplifies how a chordin family member binds and inhibits BMPs (Fig. 13G,H) (Zhang et al. 2008). The CV2 fragment comprises an amino-terminal segment (termed clip, as in the structurally distinct noggin) and VWC domain 1. In the 2:2 complex, the clip folds into the type I receptor epitope, whereas the amino-terminal subdomain of VWC1 binds the type II receptor site, showing how CV2 inhibits BMP binding to both types of receptors (Zhang et al. 2007, 2008). The interaction mode resembles attachment of a paperclip (CV2) to a sheet of paper (BMP) with the amino-terminal peptide and VWC subdomain 1 folding over opposite sides of the GF finger (Fig. 13H), and shares some similarities with the noggin–BMP interaction (Fig. 12A). In both cases, a linear peptide in an amino-terminal segment, clip, efficiently interferes with type I receptor binding, and a more globular domain occupies the type II receptor binding site. If the clip were highly flexible in the unbound state, it would be puzzling how this short motif could contribute significantly to BMP binding. However, NMR analysis of the VWC1 domain of CV2 in its unbound state reveals that the clip segment is pre-oriented to form a hook that facilitates cooperative binding to BMP-2 (Fig. 13I) (Fiebig et al. 2013).
In contrast to Dan family members, most VWC domains and most mosaic proteins with VWC domains do not act as BMP antagonists. Furthermore, the various VWC domains in chordin, chordin-like 2 and CV2 (Fig. 13F) bind BMPs with varying affinity, and some do not bind BMPs at all (Zhang et al. 2007). Among the five VWC domains of CV2, only VWC1 binds BMPs, and among the four VWC domains of chordin, only the first, third and the fourth interact with BMPs (Zhang et al. 2007). Furthermore, some VWC domains only bind to the type I receptor site, and some interact with only the type II receptor site, whereas others, like the first VWC domain of CV2, block both sites (Zhang et al. 2007, 2008). Moreover, binding architectures differ. For instance, CV2 and chordin-like 2 bind BMPs symmetrically in 2:2 complexes, whereas chordin interacts with BMPs in a 1:2 asymmetric complex (Zhang et al. 2007).
Additionally, chordin family members regulate BMP activity by a mechanism far more complex than for any other modulator family (Ashe and Levine 1999). Chordin and family members exert dual roles, displaying antagonistic (anti-BMP) (Francois et al. 1994) as well as agonistic (pro-BMP) (Ross et al. 2001) roles depending on context (Larrain et al. 2001). For instance, very strong expression of CV2 in Drosophila occurs during formation of the wing crossvein, which requires high BMP signaling activity (Conley et al. 2000). This dual activity might be explained for chordin by formation of ternary complexes with BMPs and Tsg, which by itself is a modulator protein that can block BMP signaling (Oelgeschlager et al. 2000; Chang et al. 2001; Ross et al. 2001; Scott et al. 2001). Ternary complexes might be transported to locations with low BMP expression, in which the pro-BMP effect is then mediated when the GF is released from the complex (Ashe and Levine 1999; De Robertis et al. 2000; Harland 2001). Localized release is achieved when chordin is proteolytically cleaved by zinc metalloproteases (Piccolo et al. 1997). In this context, Tsg binds and neutralizes the resulting chordin fragments, which otherwise would block BMP signaling (Larrain et al. 2001). Thus, in vivo Tsg can toggle chordin between a BMP-antagonistic and a BMP-agonistic function, and therefore functions as a switch rather than inhibitor as observed in vitro. CV2 can similarly interact with Tsg to yield a stronger BMP antagonist (Zakin et al. 2008). However, its pro-BMP role is due to its ability to directly interact with chordin and chordin-Tsg-BMP complexes (Ambrosio et al. 2008). By binding to the ECM via its heparin-binding site, CV2 does not diffuse far from its site of expression and can thus sequester and locally concentrate chordin–Tsg–BMP complexes, which when destroyed by tolloid, lead to a highly focused release of BMP ligand (Ambrosio et al. 2008; Zakin et al. 2010; for review, see O’Connor et al. 2006). Binding of Tsg and CV2 to chordin fragments blunts virtually all residual BMP-antagonistic activities of these three modulator proteins. Notably, CV2 binds to chordin’s second VWC domain, which itself does not bind BMPs. In CV2 a complex epitope comprising the first four VWC domains is responsible for binding to chordin (Zhang et al. 2010). These findings highlight the multifaceted binding properties of the VWC domains within the chordin family.
This section has shown the high structural diversity of BMP modulators. Binding to a shared interaction partner, in this case BMPs, does not require identical protein structures, can be achieved by different domains and architectures, and has evolved independently multiple times. The diversity of interaction partners also highlights the hub-like character of the TGF-β/BMP GFs, which they recognize. As the structures of many modulator proteins including Tsg and members of the multidomain CCN family have not yet been determined, even greater structural diversity among BMP binding partners is likely to be discovered in the future.
PERSPECTIVE
This review summarized our knowledge of the structures of proteins that regulate TGF-β family signaling in extracellular environments, and illustrated how the CKGF domain evolved to both agonize and antagonize TGF-β family and Wnt signaling. Structures of prodomain complexes with GF dimers suggest a new paradigm for regulation of release of GFs by conversion from crossed-arm to open-arm procomplex conformations. TGF-β family and other pathway components evolved in the earliest metazoans, and integrin- and force-dependent release of the TGF-β GF from the procomplex evolved in deuterostomes. However, how GF release from other procomplexes occurs in vivo remains unclear. The conformational change model requires extension beyond the two exemplary procomplexes reported, and hypotheses that binding to ECM components or prodomain cleavage by proteases regulate procomplex conformational changes and GF release need to be tested. The final extracellular step in signaling activation (i.e., binding of TGF-β family GFs to type I and type II receptors at the cell surface) is well characterized structurally with ternary complexes. These studies enable insight into the specificity and promiscuity in extracellular signaling, but await characterization of receptor complexes with biological GF heterodimers. Furthermore, structural understanding of how coreceptors bind GFs remains incomplete. One advance in this area shows how RGM family members bind GFs, yet lacks a bound type II receptor in the structure, and raises questions by suggesting that type I receptors do not bind until reaching an intracellular, membrane-enveloped compartment. This finding blurs the line between extracellular and intracellular signaling. Work on modulator proteins has revealed how structurally diverse antagonists including follistatins, noggin, and chordin fragments can bind GFs, although complexes with the Dan family remain elusive. We still lack ternary complexes with other ECM components, and structural understanding of how chordin members can both antagonize and agonize BMP signaling.
One of the greatest strengths in the field, the use of recombinant proteins, also led to one of its greatest weaknesses, the current paucity of work on physiological protein complexes from tissues. Many TGF-β family and modulator proteins were discovered by molecular cloning or genetics as cDNAs or genes, and the glycoproteins they encode have not been characterized in vivo. Weaknesses in structural characterization include a lack of complex structures showing how procomplexes, GFs, and modulator proteins bind ECM components to form signaling hubs and regulate extracellular signaling. Additionally, all structural studies to date on receptor complexes used homodimers, even though heterodimers are important, and structural biology shows that the type I receptor binds the interface between the two GF monomers. The two such type I sites on a heterodimer would be distinct from one another, as well as from those in homodimers. In vivo, the type I and type II receptors in the receptor–ligand complex need not be the same at the two sites on a GF dimer, even for homodimers, creating further complexity in signaling. Structures of such asymmetric complexes with homo- and heterodimeric ligands are particularly challenging. Furthermore, an understanding of signaling hubs in the extracellular environment will require new tools and techniques for isolating hub proteins from, and characterizing them in, their native environments.
ACKNOWLEDGMENTS
T.A.S. is deeply grateful to Dr. Li-Zhi Mi for help with Figures 1 to 3 and is supported by National Institutes of Health (NIH) Grants AR067288 and CA210920. A.P.H is supported by NIH grants GM58670 and CA054174. T.D.M. is supported by Deutsche Forschungsgemeinschaft (DFG) grants MU1095/3-2 and MU1095/5-1.
Footnotes
Editors: Rik Derynck and Kohei Miyazono
Additional Perspectives on The Biology of the TGF-β Family available at www.cshperspectives.org
REFERENCES
- Adamska M, Degnan SM, Green KM, Adamski M, Craigie A, Larroux C, Degnan BM. 2007. Wnt and TGF-↓ expression in the sponge Amphimedon queenslandica and the origin of metazoan embryonic patterning. PLoS ONE 2: e1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkins HB, Bianco C, Schiffer SG, Rayhorn P, Zafari M, Cheung AE, Orozco O, Olson D, De Luca A, Chen LL, et al. 2003. Antibody blockade of the Cripto CFC domain suppresses tumor cell growth in vivo. J Clin Invest 112: 575–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama T, Marques G, Wharton KA. 2012. A large bioactive BMP ligand with distinct signaling properties is produced by alternative proconvertase processing. Sci Signal 5: ra28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allendorph GP, Vale WW, Choe S. 2006. Structure of the ternary signaling complex of a TGF-β superfamily member. Proc Natl Acad Sci 103: 7643–7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allendorph GP, Isaacs MJ, Kawakami Y, Izpisua Belmonte JC, Choe S. 2007. BMP-3 and BMP-6 structures illuminate the nature of binding specificity with receptors. Biochemistry 46: 12238–12247. [DOI] [PubMed] [Google Scholar]
- Alt A, Miguel-Romero L, Donderis J, Aristorena M, Blanco FJ, Round A, Rubio V, Bernabeu C, Marina A. 2012. Structural and functional insights into endoglin ligand recognition and binding. PLoS ONE 7: e29948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosio AL, Taelman VF, Lee HX, Metzinger CA, Coffinier C, De Robertis EM. 2008. Crossveinless-2 Is a BMP feedback inhibitor that binds chordin/BMP to regulate Xenopus embryonic patterning. Dev Cell 15: 248–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amthor H, Christ B, Rashid-Doubell F, Kemp CF, Lang E, Patel K. 2002. Follistatin regulates bone morphogenetic protein-7 (BMP-7) activity to stimulate embryonic muscle growth. Dev Biol 243: 115–127. [DOI] [PubMed] [Google Scholar]
- Amthor H, Nicholas G, McKinnell I, Kemp CF, Sharma M, Kambadur R, Patel K. 2004. Follistatin complexes myostatin and antagonises myostatin-mediated inhibition of myogenesis. Dev Biol 270: 19–30. [DOI] [PubMed] [Google Scholar]
- Anderson SB, Goldberg AL, Whitman M. 2008. Identification of a novel pool of extracellular pro-myostatin in skeletal muscle. J Biol Chem 283: 7027–7035. [DOI] [PubMed] [Google Scholar]
- Annes JP, Chen Y, Munger JS, Rifkin DB. 2004. Integrin αVβ6-mediated activation of latent TGF-β requires the latent TGF-β binding protein-1. J Cell Biol 165: 723–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aono A, Hazama M, Notoya K, Taketomi S, Yamasaki H, Tsukuda R, Sasaki S, Fujisawa Y. 1995. Potent ectopic bone-inducing activity of bone morphogenetic protein-4/7 heterodimer. Biochem Biophys Res Commun 210: 670–677. [DOI] [PubMed] [Google Scholar]
- Ardelean DS, Letarte M. 2015. Anti-angiogenic therapeutic strategies in hereditary hemorrhagic telangiectasia. Front Genet 6: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe HL, Levine M. 1999. Local inhibition and long-range enhancement of Dpp signal transduction by Sog. Nature 398: 427–431. [DOI] [PubMed] [Google Scholar]
- Baardsnes J, Hinck CS, Hinck AP, O’Connor-McCourt MD. 2009. TβR-II discriminates the high- and low-affinity TGF-β isoforms via two hydrogen-bonded ion pairs. Biochemistry 48: 2146–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baarends WM, van Helmond MJ, Post M, van der Schoot PJ, Hoogerbrugge JW, de Winter JP, Uilenbroek JT, Karels B, Wilming LG, Meijers JH, et al. 1994. A novel member of the transmembrane serine/threonine kinase receptor family is specifically expressed in the gonads and in mesenchymal cells adjacent to the mullerian duct. Development 120: 189–197. [DOI] [PubMed] [Google Scholar]
- Babitt JL, Zhang Y, Samad TA, Xia Y, Tang J, Campagna JA, Schneyer AL, Woolf CJ, Lin HY. 2005. Repulsive guidance molecule (RGMa), a DRAGON homologue, is a bone morphogenetic protein co-receptor. J Biol Chem 280: 29820–29827. [DOI] [PubMed] [Google Scholar]
- Bell E, Munoz-Sanjuan I, Altmann CR, Vonica A, Brivanlou AH. 2003. Cell fate specification and competence by Coco, a maternal BMP, TGFβ and Wnt inhibitor. Development 130: 1381–1389. [DOI] [PubMed] [Google Scholar]
- Bell CH, Healey E, van Erp S, Bishop B, Tang C, Gilbert RJ, Aricescu AR, Pasterkamp RJ, Siebold C. 2013. Structure of the repulsive guidance molecule (RGM)-neogenin signaling hub. Science 341: 77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belville C, Van Vlijmen H, Ehrenfels C, Pepinsky B, Rezaie AR, Picard JY, Josso N, di Clemente N, Cate RL. 2004. Mutations of the anti-mullerian hormone gene in patients with persistent mullerian duct syndrome: Biosynthesis, secretion, and processing of the abnormal proteins and analysis using a three-dimensional model. Mol Endocrinol 18: 708–721. [DOI] [PubMed] [Google Scholar]
- Blair JE, Hedges SB. 2005. Molecular phylogeny and divergence times of deuterostome animals. Mol Biol Evol 22: 2275–2284. [DOI] [PubMed] [Google Scholar]
- Blanchet MH, Le Good JA, Mesnard D, Oorschot V, Baflast S, Minchiotti G, Klumperman J, Constam DB. 2008. Cripto recruits Furin and PACE4 and controls Nodal trafficking during proteolytic maturation. EMBO J 27: 2580–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocharov EV, Blommers MJ, Kuhla J, Arvinte T, Burgi R, Arseniev AS. 2000. Sequence-specific 1H and 15N assignment and secondary structure of transforming growth factor β3. J Biomol NMR 16: 179–180. [DOI] [PubMed] [Google Scholar]
- Bocharov EV, Korzhnev DM, Blommers MJ, Arvinte T, Orekhov VY, Billeter M, Arseniev AS. 2002. Dynamics-modulated biological activity of transforming growth factor β3. J Biol Chem 277: 46273–46279. [DOI] [PubMed] [Google Scholar]
- Boesen CC, Radaev S, Motyka SA, Patamawenu A, Sun PD. 2002. The 1.1 Å crystal structure of human TGF-β type II receptor ligand binding domain. Structure 10: 913–919. [DOI] [PubMed] [Google Scholar]
- Bork P. 1993. The modular architecture of a new family of growth regulators related to connective tissue growth factor. FEBS Lett 327: 125–130. [DOI] [PubMed] [Google Scholar]
- Bork P, Sander C. 1992. A large domain common to sperm receptors (Zp2 and Zp3) and TGF-β type III receptor. FEBS Lett 300: 237–240. [DOI] [PubMed] [Google Scholar]
- Boschert V, van Dinther M, Weidauer S, van Pee K, Muth EM, Ten Dijke P, Mueller TD. 2013. Mutational analysis of sclerostin shows importance of the flexible loop and the cystine-knot for Wnt-signaling inhibition. PLoS ONE 8: e81710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhis E, Wang W, Tam C, Hwang J, Zhang Y, Spittler D, Huang OW, Gong Y, Estevez A, Zilberleyb I, et al. 2011. Wnt antagonists bind through a short peptide to the first β-propeller domain of LRP5/6. Structure 19: 1433–1442. [DOI] [PubMed] [Google Scholar]
- Bradshaw AD. 2012. Diverse biological functions of the SPARC family of proteins. Int J Biochem Cell Biol 44: 480–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MA, Zhao Q, Baker KA, Naik C, Chen C, Pukac L, Singh M, Tsareva T, Parice Y, Mahoney A, et al. 2005. Crystal structure of BMP-9 and functional interactions with pro-region and receptors. J Biol Chem 280: 25111–25118. [DOI] [PubMed] [Google Scholar]
- Brunet LJ, McMahon JA, McMahon AP, Harland RM. 1998. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science 280: 1455–1457. [DOI] [PubMed] [Google Scholar]
- Buijs JT, van der Horst G, van den Hoogen C, Cheung H, de Rooij B, Kroon J, Petersen M, van Overveld PG, Pelger RC, van der Pluijm G. 2012. The BMP2/7 heterodimer inhibits the human breast cancer stem cell subpopulation and bone metastases formation. Oncogene 31: 2164–2174. [DOI] [PubMed] [Google Scholar]
- Calvanese L, Saporito A, Marasco D, D’Auria G, Minchiotti G, Pedone C, Paolillo L, Falcigno L, Ruvo M. 2006. Solution structure of mouse Cripto CFC domain and its inactive variant Trp107Ala. J Med Chem 49: 7054–7062. [DOI] [PubMed] [Google Scholar]
- Camus LM, Lambert LA. 2007. Molecular evolution of hemojuvelin and the repulsive guidance molecule family. J Mol Evol 65: 68–81. [DOI] [PubMed] [Google Scholar]
- Cash JN, Rejon CA, McPherron AC, Bernard DJ, Thompson TB. 2009. The structure of myostatin:follistatin 288: Insights into receptor utilization and heparin binding. EMBO J 28: 2662–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash JN, Angerman EB, Kattamuri C, Nolan K, Zhao H, Sidis Y, Keutmann HT, Thompson TB. 2012. Structure of myostatin follistatin-like 3: N-terminal domains of follistatin-type molecules exhibit alternate modes of binding. J Biol Chem 287: 1043–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castonguay R, Werner ED, Matthews RG, Presman E, Mulivor AW, Solban N, Sako D, Pearsall RS, Underwood KW, Seehra J, et al. 2011. Soluble endoglin specifically binds bone morphogenetic proteins 9 and 10 via its orphan domain, inhibits blood vessel formation, and suppresses tumor growth. J Biol Chem 286: 30034–30046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Holtzman DA, Chau S, Chickering T, Woolf EA, Holmgren LM, Bodorova J, Gearing DP, Holmes WE, Brivanlou AH. 2001. Twisted gastrulation can function as a BMP antagonist. Nature 410: 483–487. [DOI] [PubMed] [Google Scholar]
- Chang TH, Hsieh FL, Zebisch M, Harlos K, Elegheert J, Jones EY. 2015. Structure and functional properties of Norrin mimic Wnt for signalling with Frizzled4, Lrp5/6, and proteoglycan. eLife 4: 10.7554/eLife.06554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Kirkbride KC, How T, Nelson CD, Mo J, Frederick JP, Wang XF, Lefkowitz RJ, Blobe GC. 2003. β-arrestin 2 mediates endocytosis of type III TGF-β receptor and down-regulation of its signaling. Science 301: 1394–1397. [DOI] [PubMed] [Google Scholar]
- Chen C, Ware SM, Sato A, Houston-Hawkins DE, Habas R, Matzuk MM, Shen MM, Brown CW. 2006. The Vg1-related protein Gdf3 acts in a Nodal signaling pathway in the pre-gastrulation mouse embryo. Development 133: 319–329. [DOI] [PubMed] [Google Scholar]
- Chen H, Brady Ridgway J, Sai T, Lai J, Warming S, Chen H, Roose-Girma M, Zhang G, Shou W, Yan M. 2013. Context-dependent signaling defines roles of BMP9 and BMP10 in embryonic and postnatal development. Proc Natl Acad Sci 110: 11887–11892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SK, Olale F, Bennett JT, Brivanlou AH, Schier AF. 2003. EGF-CFC proteins are essential coreceptors for the TGF-β signals Vg1 and GDF1. Genes Dev 17: 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SK, Olale F, Brivanlou AH, Schier AF. 2004. Lefty blocks a subset of TGFβ signals by antagonizing EGF-CFC coreceptors. PLoS Biol 2: E30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciardiello F, Dono R, Kim N, Persico MG, Salomon DS. 1991. Expression of cripto, a novel gene of the epidermal growth factor gene family, leads to in vitro transformation of a normal mouse mammary epithelial cell line. Cancer Res 51: 1051–1054. [PubMed] [Google Scholar]
- Conley CA, Silburn R, Singer MA, Ralston A, Rohwer-Nutter D, Olson DJ, Gelbart W, Blair SS. 2000. Crossveinless 2 contains cysteine-rich domains and is required for high levels of BMP-like activity during the formation of the cross veins in Drosophila. Development 127: 3947–3959. [DOI] [PubMed] [Google Scholar]
- Constam DB. 2014. Regulation of TGFβ and related signals by precursor processing. Semin Cell Dev Biol 32: 85–97. [DOI] [PubMed] [Google Scholar]
- Cui Y, Hackenmiller R, Berg L, Jean F, Nakayama T, Thomas G, Christian JL. 2001. The activity and signaling range of mature BMP-4 is regulated by sequential cleavage at two sites within the prodomain of the precursor. Genes Dev 15: 2797–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daopin S, Piez KA, Ogawa Y, Davies DR. 1992. Crystal structure of transforming growth factor-β 2: An unusual fold for the superfamily. Science 257: 369–373. [DOI] [PubMed] [Google Scholar]
- Daopin S, Li M, Davies DR. 1993. Crystal structure of TGF-β 2 refined at 1.8 Å resolution. Proteins 17: 176–192. [DOI] [PubMed] [Google Scholar]
- David L, Mallet C, Mazerbourg S, Feige JJ, Bailly S. 2007. Identification of BMP9 and BMP10 as functional activators of the orphan activin receptor-like kinase 1 (ALK1) in endothelial cells. Blood 109: 1953–1961. [DOI] [PubMed] [Google Scholar]
- David L, Feige JJ, Bailly S. 2009. Emerging role of bone morphogenetic proteins in angiogenesis. Cytokine Growth Factor Rev 20: 203–212. [DOI] [PubMed] [Google Scholar]
- De Crescenzo G, Hinck CS, Shu Z, Zuniga J, Yang J, Tang Y, Baardsnes J, Mendoza V, Sun L, Lopez-Casillas F, et al. 2006. Three key residues underlie the differential affinity of the TGFβ isoforms for the TGFβ type II receptor. J Mol Biol 355: 47–62. [DOI] [PubMed] [Google Scholar]
- Deep S, Walker KP III, Shu Z, Hinck AP. 2003. Solution structure and backbone dynamics of the TGFβ type II receptor extracellular domain. Biochemistry 42: 10126–10139. [DOI] [PubMed] [Google Scholar]
- De Robertis EM, Larrain J, Oelgeschlager M, Wessely O. 2000. The establishment of Spemann’s organizer and patterning of the vertebrate embryo. Nat Rev Genet 1: 171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diestel U, Resch M, Meinhardt K, Weiler S, Hellmann TV, Mueller TD, Nickel J, Eichler J, Muller YA. 2013. Identification of a novel TGF-β-binding site in the zona pellucida C-terminal (ZP-C) domain of TGF-β-receptor-3 (TGFR-3). PLoS ONE 8: e67214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Hudson E, Lu C, Springer TA. 2014. Structural determinants of integrin β-subunit specificity for latent TGF-β. Nat Struct Mol Biol 21: 1091–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M, Edgecombe GD, et al. 2008. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452: 745–749. [DOI] [PubMed] [Google Scholar]
- Esparza-Lopez J, Montiel JL, Vilchis-Landeros MM, Okadome T, Miyazono K, Lopez-Casillas F. 2001. Ligand binding and functional properties of betaglycan, a co-receptor of the transforming growth factor-β superfamily. Specialized binding regions for transforming growth factor-β and inhibin A. J Biol Chem 276: 14588–14596. [DOI] [PubMed] [Google Scholar]
- Esquivies L, Blackler A, Peran M, Rodriguez-Esteban C, Izpisua Belmonte JC, Booker E, Gray PC, Ahn C, Kwiatkowski W, Choe S. 2014. Designer nodal/BMP2 chimeras mimic nodal signaling, promote chondrogenesis, and reveal a BMP2-like structure. J Biol Chem 289: 1788–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Kim T, Ozkan E, Light M, Torkin R, Teng KK, Hempstead BL, Garcia KC. 2010. Molecular and structural insight into proNGF engagement of p75NTR and sortilin. J Mol Biol 396: 967–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig JE, Weidauer SE, Qiu LY, Bauer M, Schmieder P, Beerbaum M, Zhang JL, Oschkinat H, Sebald W, Mueller TD. 2013. The clip-segment of the von Willebrand domain 1 of the BMP modulator protein Crossveinless 2 is preformed. Molecules 18: 11658–11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois V, Solloway M, O’Neill JW, Emery J, Bier E. 1994. Dorsal-ventral patterning of the Drosophila embryo depends on a putative negative growth factor encoded by the short gastrulation gene. Genes Dev 8: 2602–2616. [DOI] [PubMed] [Google Scholar]
- Garcia Abreu J, Coffinier C, Larrain J, Oelgeschlager M, De Robertis EM. 2002. Chordin-like CR domains and the regulation of evolutionarily conserved extracellular signaling systems. Gene 287: 39–47. [DOI] [PubMed] [Google Scholar]
- Gentry LE, Nash BW. 1990. The pro domain of pre-pro-transforming growth factor β1 when independently expressed is a functional binding protein for the mature growth factor. Biochemistry 29: 6851–6857. [DOI] [PubMed] [Google Scholar]
- Glinka A, Wu W, Onichtchouk D, Blumenstock C, Niehrs C. 1997. Head induction by simultaneous repression of Bmp and Wnt signalling in Xenopus. Nature 389: 517–519. [DOI] [PubMed] [Google Scholar]
- Glister C, Kemp CF, Knight PG. 2004. Bone morphogenetic protein (BMP) ligands and receptors in bovine ovarian follicle cells: Actions of BMP-4, -6 and -7 on granulosa cells and differential modulation of Smad-1 phosphorylation by follistatin. Reproduction 127: 239–254. [DOI] [PubMed] [Google Scholar]
- Gong Y, Krakow D, Marcelino J, Wilkin D, Chitayat D, Babul-Hirji R, Hudgins L, Cremers CW, Cremers FP, Brunner HG, et al. 1999. Heterozygous mutations in the gene encoding noggin affect human joint morphogenesis. Nat Genet 21: 302–304. [DOI] [PubMed] [Google Scholar]
- Gougos A, Letarte M. 1990. Primary structure of endoglin, an RGD-containing glycoprotein of human endothelial cells. J Biol Chem 265: 8361–8364. [PubMed] [Google Scholar]
- Gray AM, Mason AJ. 1990. Requirement for activin A and transforming growth factor-β1 pro-regions in homodimer assembly. Science 247: 1328–1330. [DOI] [PubMed] [Google Scholar]
- Gray PC, Harrison CA, Vale W. 2003. Cripto forms a complex with activin and type II activin receptors and can block activin signaling. Proc Natl Acad Sci 100: 5193–5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald J, Fischer WH, Vale WW, Choe S. 1999. Three-finger toxin fold for the extracellular ligand-binding domain of the type II activin receptor serine kinase. Nat Struct Biol 6: 18–22. [DOI] [PubMed] [Google Scholar]
- Greenwald J, Groppe J, Gray P, Wiater E, Kwiatkowski W, Vale W, Choe S. 2003. The BMP7/ActRII extracellular domain complex provides new insights into the cooperative nature of receptor assembly. Mol Cell 11: 605–617. [DOI] [PubMed] [Google Scholar]
- Greenwald J, Vega ME, Allendorph GP, Fischer WH, Vale W, Choe S. 2004. A flexible activin explains the membrane-dependent cooperative assembly of TGF-β family receptors. Mol Cell 15: 485–489. [DOI] [PubMed] [Google Scholar]
- Gregory KE, Ono RN, Charbonneau NL, Kuo CL, Keene DR, Bachinger HP, Sakai LY. 2005. The prodomain of BMP-7 targets the BMP-7 complex to the extracellular matrix. J Biol Chem 280: 27970–27980. [DOI] [PubMed] [Google Scholar]
- Griffith DL, Keck PC, Sampath TK, Rueger DC, Carlson WD. 1996. Three-dimensional structure of recombinant human osteogenic protein 1: Structural paradigm for the transforming growth factor β superfamily. Proc Natl Acad Sci 93: 878–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groppe J, Greenwald J, Wiater E, Rodriguez-Leon J, Economides AN, Kwiatkowski W, Affolter M, Vale WW, Izpisua Belmonte JC, Choe S. 2002. Structural basis of BMP signalling inhibition by the cystine knot protein Noggin. Nature 420: 636–642. [DOI] [PubMed] [Google Scholar]
- Groppe J, Hinck CS, Samavarchi-Tehrani P, Zubieta C, Schuermann JP, Taylor AB, Schwarz PM, Wrana JL, Hinck AP. 2008. Cooperative assembly of TGF-β superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol Cell 29: 157–168. [DOI] [PubMed] [Google Scholar]
- Grütter C, Wilkinson T, Turner R, Podichetty S, Finch D, McCourt M, Loning S, Jermutus L, Grutter MG. 2008. A cytokine-neutralizing antibody as a structural mimetic of 2 receptor interactions. Proc Natl Acad Sci 105: 20251–20256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruharsha KG, Rual JF, Zhai B, Mintseris J, Vaidya P, Vaidya N, Beekman C, Wong C, Rhee DY, Cenaj O, et al. 2011. A protein complex network of Drosophila melanogaster. Cell 147: 690–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammonds RG Jr, Schwall R, Dudley A, Berkemeier L, Lai C, Lee J, Cunningham N, Reddi AH, Wood WI, Mason AJ. 1991. Bone-inducing activity of mature BMP-2b produced from a hybrid BMP-2a/2b precursor. Mol Endocrinol 5: 149–155. [DOI] [PubMed] [Google Scholar]
- Harland RM. 2001. Developmental biology. A twist on embryonic signalling. Nature 410: 423–424. [DOI] [PubMed] [Google Scholar]
- Harrington AE, Morris-Triggs SA, Ruotolo BT, Robinson CV, Ohnuma S, Hyvonen M. 2006. Structural basis for the inhibition of activin signalling by follistatin. EMBO J 25: 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison CA, Al-Musawi SL, Walton KL. 2011. Prodomains regulate the synthesis, extracellular localisation and activity of TGF-β superfamily ligands. Growth Factors 29: 174–186. [DOI] [PubMed] [Google Scholar]
- Hart PJ, Deep S, Taylor AB, Shu Z, Hinck CS, Hinck AP. 2002. Crystal structure of the human TβR2 ectodomain–TGF-β3 complex. Nat Struct Biol 9: 203–208. [DOI] [PubMed] [Google Scholar]
- Harth S, Kotzsch A, Hu J, Sebald W, Mueller TD. 2010. A selection fit mechanism in BMP receptor IA as a possible source for BMP ligand-receptor promiscuity. PLoS ONE 5: e13049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta T, Konishi H, Katoh E, Natsume T, Ueno N, Kobayashi Y, Yamazaki T. 2000. Identification of the ligand-binding site of the BMP type IA receptor for BMP-4. Biopolymers 55: 399–406. [DOI] [PubMed] [Google Scholar]
- Hauburger A, von Einem S, Schwaerzer GK, Buttstedt A, Zebisch M, Schraml M, Hortschansky P, Knaus P, Schwarz E. 2009. The pro-form of BMP-2 interferes with BMP-2 signalling by competing with BMP-2 for IA receptor binding. FEBS J 276: 6386–6398. [DOI] [PubMed] [Google Scholar]
- Hawinkels LJ, Kuiper P, Wiercinska E, Verspaget HW, Liu Z, Pardali E, Sier CF, ten Dijke P. 2010. Matrix metalloproteinase-14 (MT1-MMP)-mediated endoglin shedding inhibits tumor angiogenesis. Cancer Res 70: 4141–4150. [DOI] [PubMed] [Google Scholar]
- Healey EG, Bishop B, Elegheert J, Bell CH, Padilla-Parra S, Siebold C. 2015. Repulsive guidance molecule is a structural bridge between neogenin and bone morphogenetic protein. Nat Struct Mol Biol 22: 458–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinecke K, Seher A, Schmitz W, Mueller TD, Sebald W, Nickel J. 2009. Receptor oligomerization and beyond: A case study in bone morphogenetic proteins. BMC Biol 7: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejnol A, Obst M, Stamatakis A, Ott M, Rouse GW, Edgecombe GD, Martinez P, Baguna J, Bailly X, Jondelius U, et al. 2009. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc Biol Sci 276: 4261–4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herpin A, Lelong C, Favrel P. 2004. Transforming growth factor-β-related proteins: An ancestral and widespread superfamily of cytokines in metazoans. Dev Comp Immunol 28: 461–485. [DOI] [PubMed] [Google Scholar]
- Hinck AP, Archer SJ, Qian SW, Roberts AB, Sporn MB, Weatherbee JA, Tsang ML, Lucas R, Zhang BL, Wenker J, et al. 1996. Transforming growth factor β1: Three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor β2. Biochemistry 35: 8517–8534. [DOI] [PubMed] [Google Scholar]
- Hohenester E, Maurer P, Timpl R. 1997. Crystal structure of a pair of follistatin-like and EF-hand calcium-binding domains in BM-40. EMBO J 16: 3778–3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth G, Slocombe P, Doyle C, Sweeney B, Veverka V, Le Riche K, Franklin RJ, Compson J, Brookings D, Turner J, et al. 2012. Characterization of the interaction of sclerostin with the low density lipoprotein receptor-related protein (LRP) family of Wnt co-receptors. J Biol Chem 287: 26464–26477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland ND, Holland LZ, Holland PW. 2015. Scenarios for the making of vertebrates. Nature 520: 450–455. [DOI] [PubMed] [Google Scholar]
- Huang T, Schor SL, Hinck AP. 2014. Biological activity differences between TGF-β1 and TGF-β3 correlate with differences in the rigidity and arrangement of their component monomers. Biochemistry 53: 5737–5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huminiecki L, Goldovsky L, Freilich S, Moustakas A, Ouzounis C, Heldin CH. 2009. Emergence, development and diversification of the TGF-β signalling pathway within the animal kingdom. BMC Evol Biol 9: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iemura S, Yamamoto TS, Takagi C, Uchiyama H, Natsume T, Shimasaki S, Sugino H, Ueno N. 1998. Direct binding of follistatin to a complex of bone-morphogenetic protein and its receptor inhibits ventral and epidermal cell fates in early Xenopus embryo. Proc Natl Acad Sci 95: 9337–9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel DI, Nove J, Kerns KM, Moutsatsos IK, Kaufman RJ. 1992. Expression and characterization of bone morphogenetic protein-2 in Chinese hamster ovary cells. Growth Factors 7: 139–150. [DOI] [PubMed] [Google Scholar]
- Israel DI, Nove J, Kerns KM, Kaufman RJ, Rosen V, Cox KA, Wozney JM. 1996. Heterodimeric bone morphogenetic proteins show enhanced activity in vitro and in vivo. Growth Factors 13: 291–300. [DOI] [PubMed] [Google Scholar]
- Itasaki N, Jones CM, Mercurio S, Rowe A, Domingos PM, Smith JC, Krumlauf R. 2003. Wise, a context-dependent activator and inhibitor of Wnt signalling. Development 130: 4295–4305. [DOI] [PubMed] [Google Scholar]
- Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL. 2000. The large-scale organization of metabolic networks. Nature 407: 651–654. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley D. 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattamuri C, Luedeke DM, Nolan K, Rankin SA, Greis KD, Zorn AM, Thompson TB. 2012. Members of the DAN family are BMP antagonists that form highly stable noncovalent dimers. J Mol Biol 424: 313–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke J, Harikumar KG, Erice C, Chen C, Gu X, Wang L, Parker N, Cheng Z, Xu W, Williams BO, et al. 2013. Structure and function of Norrin in assembly and activation of a Frizzled 4-Lrp5/6 complex. Genes Dev 27: 2305–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller S, Nickel J, Zhang JL, Sebald W, Mueller TD. 2004. Molecular recognition of BMP-2 and BMP receptor IA. Nat Struct Mol Biol 11: 481–488. [DOI] [PubMed] [Google Scholar]
- Kirkbride KC, Townsend TA, Bruinsma MW, Barnett JV, Blobe GC. 2008. Bone morphogenetic proteins signal through the transforming growth factor-β type III receptor. J Biol Chem 283: 7628–7637. [DOI] [PubMed] [Google Scholar]
- Kirsch T, Sebald W, Dreyer MK. 2000. Crystal structure of the BMP-2-BRIA ectodomain complex. Nat Struct Biol 7: 492–496. [DOI] [PubMed] [Google Scholar]
- Klages J, Kotzsch A, Coles M, Sebald W, Nickel J, Muller T, Kessler H. 2008. The solution structure of BMPR-IA reveals a local disorder-to-order transition upon BMP-2 binding. Biochemistry 47: 11930–11939. [DOI] [PubMed] [Google Scholar]
- Klammert U, Mueller TD, Hellmann TV, Wuerzler KK, Kotzsch A, Schliermann A, Schmitz W, Kuebler AC, Sebald W, Nickel J. 2015. GDF-5 can act as a context-dependent BMP-2 antagonist. BMC Biol 13: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzsch A, Nickel J, Seher A, Heinecke K, van Geersdaele L, Herrmann T, Sebald W, Mueller TD. 2008. Structure analysis of bone morphogenetic protein-2 type I receptor complexes reveals a mechanism of receptor inactivation in juvenile polyposis syndrome. J Biol Chem 283: 5876–5887. [DOI] [PubMed] [Google Scholar]
- Kotzsch A, Nickel J, Seher A, Sebald W, Muller TD. 2009. Crystal structure analysis reveals a spring-loaded latch as molecular mechanism for GDF-5-type I receptor specificity. EMBO J 28: 937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause C, Korchynskyi O, de Rooij K, Weidauer SE, de Gorter DJ, van Bezooijen RL, Hatsell S, Economides AN, Mueller TD, Lowik CW, et al. 2010. Distinct modes of inhibition by sclerostin on bone morphogenetic protein and Wnt signaling pathways. J Biol Chem 285: 41614–41626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar TR. 2005. Too many follistatins: Racing inside and getting out of the cell. Endocrinology 146: 5048–5051. [DOI] [PubMed] [Google Scholar]
- Kuns-Hashimoto R, Kuninger D, Nili M, Rotwein P. 2008. Selective binding of RGMc/hemojuvelin, a key protein in systemic iron metabolism, to BMP-2 and neogenin. Am J Physiol Cell Physiol 294: C994–C1003. [DOI] [PubMed] [Google Scholar]
- Kusu N, Laurikkala J, Imanishi M, Usui H, Konishi M, Miyake A, Thesleff I, Itoh N. 2003. Sclerostin is a novel secreted osteoclast-derived bone morphogenetic protein antagonist with unique ligand specificity. J Biol Chem 278: 24113–24117. [DOI] [PubMed] [Google Scholar]
- Lamarre J, Vasudevan J, Gonias SL. 1994. Plasmin cleaves betaglycan and releases a 60 kDa transforming growth factor-β complex from the cell surface. Biochem J 302: 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrain J, Bachiller D, Lu B, Agius E, Piccolo S, De Robertis EM. 2000. BMP-binding modules in chordin: A model for signalling regulation in the extracellular space. Development 127: 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrain J, Oelgeschlager M, Ketpura NI, Reversade B, Zakin L, De Robertis EM. 2001. Proteolytic cleavage of Chordin as a switch for the dual activities of Twisted gastrulation in BMP signaling. Development 128: 4439–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch TF, Shimasaki S, Woodruff TK, Jardetzky TS. 2007. Structural and biophysical coupling of heparin and activin binding to follistatin isoform functions. J Biol Chem 282: 15930–15939. [DOI] [PubMed] [Google Scholar]
- Lewis KA, Gray PC, Blount AL, MacConell LA, Wiater E, Bilezikjian LM, Vale W. 2000. Betaglycan binds inhibin and can mediate functional antagonism of activin signalling. Nature 404: 411–414. [DOI] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, et al. 2004. A map of the interactome network of the metazoan C. elegans. Science 303: 540–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Shimono C, Norioka N, Nakano I, Okubo T, Yagi Y, Hayashi M, Sato Y, Fujisaki H, Hattori S, et al. 2010. Activin A binds to perlecan through its pro-region that has heparin/heparan sulfate binding activity. J Biol Chem 285: 36645–36655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao WX, Moore RK, Otsuka F, Shimasaki S. 2003. Effect of intracellular interactions on the processing and secretion of bone morphogenetic protein-15 (BMP-15) and growth and differentiation factor-9. Implication of the aberrant ovarian phenotype of BMP-15 mutant sheep. J Biol Chem 278: 3713–3719. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Hu Y, Zhu J, Woodruff TK, Jardetzky TS. 2011. Structure of betaglycan zona pellucida (ZP)-C domain provides insights into ZP-mediated protein polymerization and TGF-β binding. Proc Natl Acad Sci 108: 5232–5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lintern KB, Guidato S, Rowe A, Saldanha JW, Itasaki N. 2009. Characterization of wise protein and its molecular mechanism to interact with both Wnt and BMP signals. J Biol Chem 284: 23159–23168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little SC, Mullins MC. 2009. Bone morphogenetic protein heterodimers assemble heteromeric type I receptor complexes to pattern the dorsoventral axis. Nat Cell Biol 11: 637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Ventura F, Doody J, Massague J. 1995. Human type II receptor for bone morphogenic proteins (BMPs): Extension of the two-kinase receptor model to the BMPs. Mol Cell Biol 15: 3479–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Casillas F, Wrana JL, Massague J. 1993. Betaglycan presents ligand to the TGF-β signaling receptor. Cell 73: 1435–1444. [DOI] [PubMed] [Google Scholar]
- Lowe CJ, Clarke DN, Medeiros DM, Rokhsar DS, Gerhart J. 2015. The deuterostome context of chordate origins. Nature 520: 456–465. [DOI] [PubMed] [Google Scholar]
- Luyten FP, Cunningham NS, Ma S, Muthukumaran N, Hammonds RG, Nevins WB, Woods WI, Reddi AH. 1989. Purification and partial amino acid sequence of osteogenin, a protein initiating bone differentiation. J Biol Chem 264: 13377–13380. [PubMed] [Google Scholar]
- Mace PD, Cutfield JF, Cutfield SM. 2006. High resolution structures of the bone morphogenetic protein type II receptor in two crystal forms: Implications for ligand binding. Biochem Biophys Res Commun 351: 831–838. [DOI] [PubMed] [Google Scholar]
- Mahlawat P, Ilangovan U, Biswas T, Sun LZ, Hinck AP. 2012. Structure of the Alk1 extracellular domain and characterization of its bone morphogenetic protein (BMP) binding properties. Biochemistry 51: 6328–6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangino M, Flex E, Digilio MC, Giannotti A, Dallapiccola B. 2002. Identification of a novel NOG gene mutation (P35S) in an Italian family with symphalangism. Hum Mutat 19: 308. [DOI] [PubMed] [Google Scholar]
- Marlow MS, Brown CB, Barnett JV, Krezel AM. 2003. Solution structure of the chick TGFβ type II receptor ligand-binding domain. J Mol Biol 326: 989–997. [DOI] [PubMed] [Google Scholar]
- Matsunaga E, Tauszig-Delamasure S, Monnier PP, Mueller BK, Strittmatter SM, Mehlen P, Chedotal A. 2004. RGM and its receptor neogenin regulate neuronal survival. Nat Cell Biol 6: 749–755. [DOI] [PubMed] [Google Scholar]
- McLean S, Bhattacharya M, Di Guglielmo GM. 2013. βarrestin2 interacts with TβRII to regulate Smad-dependent and Smad-independent signal transduction. Cell Signal 25: 319–331. [DOI] [PubMed] [Google Scholar]
- McNatty KP, Juengel JL, Reader KL, Lun S, Myllymaa S, Lawrence SB, Western A, Meerasahib MF, Mottershead DG, Groome NP, et al. 2005. Bone morphogenetic protein 15 and growth differentiation factor 9 co-operate to regulate granulosa cell function. Reproduction 129: 473–480. [DOI] [PubMed] [Google Scholar]
- Mendoza V, Vilchis-Landeros MM, Mendoza-Hernandez G, Huang T, Villarreal MM, Hinck AP, Lopez-Casillas F, Montiel JL. 2009. Betaglycan has two independent domains required for high affinity TGF-β binding: Proteolytic cleavage separates the domains and inactivates the neutralizing activity of the soluble receptor. Biochemistry 48: 11755–11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi LZ, Brown CT, Gao Y, Tian Y, Le VQ, Walz T, Springer TA. 2015. Structure of bone morphogenetic protein 9 procomplex. Proc Natl Acad Sci 24: 3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchiotti G, Parisi S, Liguori G, Signore M, Lania G, Adamson ED, Lago CT, Persico MG. 2000. Membrane-anchorage of Cripto protein by glycosylphosphatidylinositol and its distribution during early mouse development. Mech Dev 90: 133–142. [DOI] [PubMed] [Google Scholar]
- Mittl PR, Priestle JP, Cox DA, McMaster G, Cerletti N, Grutter MG. 1996. The crystal structure of TGF-β 3 and comparison to TGF-β 2: Implications for receptor binding. Protein Sci 5: 1261–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K, Olofsson A, Colosetti P, Heldin CH. 1991. A role of the latent TGF-β1-binding protein in the assembly and secretion of TGF-β1. EMBO J 10: 1091–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnier PP, Sierra A, Macchi P, Deitinghoff L, Andersen JS, Mann M, Flad M, Hornberger MR, Stahl B, Bonhoeffer F, et al. 2002. RGM is a repulsive guidance molecule for retinal axons. Nature 419: 392–395. [DOI] [PubMed] [Google Scholar]
- Mottershead DG, Harrison CA, Mueller TD, Stanton PG, Gilchrist RB, McNatty KP. 2013. Growth differentiation factor 9:bone morphogenetic protein 15 (GDF9:BMP15) synergism and protein heterodimerization. Proc Natl Acad Sci 110: E2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottershead DG, Sugimura S, Al-Musawi SL, Li JJ, Richani D, White MA, Martin GA, Trotta AP, Ritter LJ, Shi J, et al. 2015. Cumulin, an oocyte-secreted heterodimer of the transforming growth factor-β family, is a potent activator of granulosa cells and improves oocyte quality. J Biol Chem 290: 24007–24020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller TD. 2015. RGM co-receptors add complexity to BMP signaling. Nat Struct Mol Biol 22: 439–440. [DOI] [PubMed] [Google Scholar]
- Muller BK, Jay DG, Bonhoeffer F. 1996. Chromophore-assisted laser inactivation of a repulsive axonal guidance molecule. Curr Biol 6: 1497–1502. [DOI] [PubMed] [Google Scholar]
- Munger JS, Huang X, Kawakatsu H, Griffiths MJD, Dalton SL, Wu J, Pittet J-F, Kaminski N, Garat C, Matthay MA, et al. 1999. The integrin αvβ6 binds and activates latent TGFβ1: A mechanism for regulating pulmonary inflammation and fibrosis. Cell 96: 319–328. [DOI] [PubMed] [Google Scholar]
- Nassiri F, Cusimano MD, Scheithauer BW, Rotondo F, Fazio A, Yousef GM, Syro LV, Kovacs K, Lloyd RV. 2011. Endoglin (CD105): A review of its role in angiogenesis and tumor diagnosis, progression and therapy. Anticancer Res 31: 2283–2290. [PubMed] [Google Scholar]
- Nickel J, Kotzsch A, Sebald W, Mueller TD. 2005. A single residue of GDF-5 defines binding specificity to BMP receptor IB. J Mol Biol 349: 933–947. [DOI] [PubMed] [Google Scholar]
- Nishimatsu S, Thomsen GH. 1998. Ventral mesoderm induction and patterning by bone morphogenetic protein heterodimers in Xenopus embryos. Mech Dev 74: 75–88. [DOI] [PubMed] [Google Scholar]
- Nolan K, Thompson TB. 2014. The DAN family: Modulators of TGF-β signaling and beyond. Prot Sci 23: 999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan K, Kattamuri C, Luedeke DM, Deng X, Jagpal A, Zhang F, Linhardt RJ, Kenny AP, Zorn AM, Thompson TB. 2013. Structure of protein related to Dan and Cerberus: Insights into the mechanism of bone morphogenetic protein antagonism. Structure 21: 1417–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan K, Kattamuri C, Luedeke DM, Angerman EB, Rankin SA, Stevens ML, Zorn AM, Thompson TB. 2015. Structure of neuroblastoma suppressor of tumorigenicity 1 (NBL1): Insights for the functional variability across bone morphogenetic protein (BMP) antagonists. J Biol Chem 290: 4759–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor MB, Umulis D, Othmer HG, Blair SS. 2006. Shaping BMP morphogen gradients in the Drosophila embryo and pupal wing. Development 133: 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelgeschlager M, Larrain J, Geissert D, De Robertis EM. 2000. The evolutionarily conserved BMP-binding protein Twisted gastrulation promotes BMP signalling. Nature 405: 757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary JM, Hamilton JM, Deane CM, Valeyev NV, Sandell LJ, Downing AK. 2004. Solution structure and dynamics of a prototypical chordin-like cysteine-rich repeat (von Willebrand Factor type C module) from collagen IIA. J Biol Chem 279: 53857–53866. [DOI] [PubMed] [Google Scholar]
- Paine-Saunders S, Viviano BL, Economides AN, Saunders S. 2002. Heparan sulfate proteoglycans retain Noggin at the cell surface: A potential mechanism for shaping bone morphogenetic protein gradients. J Biol Chem 277: 2089–2096. [DOI] [PubMed] [Google Scholar]
- Pang K, Ryan JF, Baxevanis AD, Martindale MQ. 2011. Evolution of the TGF-β signaling pathway and its potential role in the ctenophore, Mnemiopsis leidyi. PLoS ONE 6: e24152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce JJ, Penny G, Rossant J. 1999. A mouse cerberus/Dan-related gene family. Dev Biol 209: 98–110. [DOI] [PubMed] [Google Scholar]
- Peng J, Li Q, Wigglesworth K, Rangarajan A, Kattamuri C, Peterson RT, Eppig JJ, Thompson TB, Matzuk MM. 2013. Growth differentiation factor 9:bone morphogenetic protein 15 heterodimers are potent regulators of ovarian functions. Proc Natl Acad Sci 110: E776–E785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo S, Agius E, Lu B, Goodman S, Dale L, De Robertis EM. 1997. Cleavage of chordin by xolloid metalloprotease suggests a role for proteolytic processing in the regulation of Spemann organizer activity. Cell 91: 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo S, Agius E, Leyns L, Bhattacharyya S, Grunz H, Bouwmeester T, De Robertis EM. 1999. The head inducer Cerberus is a multifunctional antagonist of Nodal, BMP and Wnt signals. Nature 397: 707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrzak WS, Woodell-May J, McDonald N. 2006. Assay of bone morphogenetic protein-2, -4, and -7 in human demineralized bone matrix. J Craniofac Surg 17: 84–90. [DOI] [PubMed] [Google Scholar]
- Radaev S, Zou Z, Huang T, Lafer EM, Hinck AP, Sun PD. 2010. Ternary complex of transforming growth factor-β1 reveals isoform-specific ligand recognition and receptor recruitment in the superfamily. J Biol Chem 285: 14806–14814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan S, Deitinghoff L, Davis D, Conrad S, Skutella T, Chedotal A, Mueller BK, Strittmatter SM. 2004. Neogenin mediates the action of repulsive guidance molecule. Nat Cell Biol 6: 756–762. [DOI] [PubMed] [Google Scholar]
- Robertson IB, Rifkin DB. 2013. Unchaining the beast; insights from structural and evolutionary studies on TGFβ secretion, sequestration, and activation. Cytokine Growth Factor Rev 24: 355–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roch GJ, Sherwood NM. 2014. Glycoprotein hormones and their receptors emerged at the origin of metazoans. Genome Biol Evol 5: 1466–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa FM. 2002. Cripto, a multifunctional partner in signaling: Molecular forms and activities. Sci STKE 2002: e47. [DOI] [PubMed] [Google Scholar]
- Ross JJ, Shimmi O, Vilmos P, Petryk A, Kim H, Gaudenz K, Hermanson S, Ekker SC, O’Connor MB, Marsh JL. 2001. Twisted gastrulation is a conserved extracellular BMP antagonist. Nature 410: 479–483. [DOI] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. 2005. Towards a proteome-scale map of the human protein–protein interaction network. Nature 437: 1173–1178. [DOI] [PubMed] [Google Scholar]
- Ruppert R, Hoffmann E, Sebald W. 1996. Human bone morphogenetic protein 2 contains a heparin-binding site which modifies its biological activity. Eur J Biochem 237: 295–302. [DOI] [PubMed] [Google Scholar]
- Samad TA, Rebbapragada A, Bell E, Zhang Y, Sidis Y, Jeong SJ, Campagna JA, Perusini S, Fabrizio DA, Schneyer AL, et al. 2005. DRAGON, a bone morphogenetic protein co-receptor. J Biol Chem 280: 14122–14129. [DOI] [PubMed] [Google Scholar]
- Sampath TK, Reddi AH. 1981. Dissociative extraction and reconstitution of extracellular matrix components involved in local bone differentiation. Proc Natl Acad Sci 78: 7599–7603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath TK, Reddi AH. 1984. Distribution of bone inductive proteins in mineralized and demineralized extracellular matrix. Biochem Biophys Res Commun 119: 949–954. [DOI] [PubMed] [Google Scholar]
- Saremba S, Nickel J, Seher A, Kotzsch A, Sebald W, Mueller TD. 2008. Type I receptor binding of bone morphogenetic protein 6 is dependent on N-glycosylation of the ligand. FEBS J 275: 172–183. [DOI] [PubMed] [Google Scholar]
- Sasai Y, Lu B, Steinbeisser H, Geissert D, Gont LK, De Robertis EM. 1994. Xenopus chordin: A novel dorsalizing factor activated by organizer-specific homeobox genes. Cell 79: 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheufler C, Sebald W, Hulsmeyer M. 1999. Crystal structure of human bone morphogenetic protein-2 at 2.7 Å resolution. J Mol Biol 287: 103–115. [DOI] [PubMed] [Google Scholar]
- Schlunegger MP, Grutter MG. 1992. An unusual feature revealed by the crystal structure at 2.2 Å resolution of human transforming growth factor-β2. Nature 358: 430–434. [DOI] [PubMed] [Google Scholar]
- Schneyer A, Sidis Y, Xia Y, Saito S, del Re E, Lin HY, Keutmann H. 2004. Differential actions of follistatin and follistatin-like 3. Mol Cell Endocrinol 225: 25–28. [DOI] [PubMed] [Google Scholar]
- Schneyer AL, Sidis Y, Gulati A, Sun JL, Keutmann H, Krasney PA. 2008. Differential antagonism of activin, myostatin and growth and differentiation factor 11 by wild-type and mutant follistatin. Endocrinology 149: 4589–4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreuder H, Liesum A, Pohl J, Kruse M, Koyama M. 2005. Crystal structure of recombinant human growth and differentiation factor 5: Evidence for interaction of the type I and type II receptor-binding sites. Biochem Biophys Res Commun 329: 1076–1086. [DOI] [PubMed] [Google Scholar]
- Schwaerzer GK, Hiepen C, Schrewe H, Nickel J, Ploeger F, Sebald W, Mueller T, Knaus P. 2012. New insights into the molecular mechanism of multiple synostoses syndrome (SYNS): Mutation within the GDF5 knuckle epitope causes noggin-resistance. J Bone Miner Res 27: 429–442. [DOI] [PubMed] [Google Scholar]
- Scott IC, Blitz IL, Pappano WN, Maas SA, Cho KW, Greenspan DS. 2001. Homologues of Twisted gastrulation are extracellular cofactors in antagonism of BMP signalling. Nature 410: 475–478. [DOI] [PubMed] [Google Scholar]
- Seemann P, Brehm A, Konig J, Reissner C, Stricker S, Kuss P, Haupt J, Renninger S, Nickel J, Sebald W, et al. 2009. Mutations in GDF5 reveal a key residue mediating BMP inhibition by NOGGIN. PLoS Genet 5: e1000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidah NG, Prat A. 2012. The biology and therapeutic targeting of the proprotein convertases. Nat Rev Drug Discov 11: 367–383. [DOI] [PubMed] [Google Scholar]
- Sengle G, Ono RN, Lyons KM, Bachinger HP, Sakai LY. 2008. A new model for growth factor activation: Type II receptors compete with the prodomain for BMP-7. J Mol Biol 381: 1025–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengle G, Ono RN, Sasaki T, Sakai LY. 2011. Prodomains of transforming growth factor β (TGFβ) superfamily members specify different functions: Extracellular matrix interactions and growth factor bioavailability. J Biol Chem 286: 5087–5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Zhu J, Wang R, Chen X, Mi L, Walz T, Springer TA. 2011. Latent TGF-β structure and activation. Nature 474: 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim A, Liu H, Focia P, Chen X, Lin P, He X. 2010. Structures of a platelet-derived growth factor/propeptide complex and a platelet-derived growth factor/receptor complex. Proc Natl Acad Sci 107: 11307–11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidis Y, Mukherjee A, Keutmann H, Delbaere A, Sadatsuki M, Schneyer A. 2006. Biological activity of follistatin isoforms and follistatin-like-3 is dependent on differential cell surface binding and specificity for activin, myostatin, and bone morphogenetic proteins. Endocrinology 147: 3586–3597. [DOI] [PubMed] [Google Scholar]
- Smith WC, Harland RM. 1992. Expression cloning of noggin, a new dorsalizing factor localized to the Spemann organizer in Xenopus embryos. Cell 70: 829–840. [DOI] [PubMed] [Google Scholar]
- Song K, Krause C, Shi S, Patterson M, Suto R, Grgurevic L, Vukicevic S, van Dinther M, Falb D, Ten Dijke P, et al. 2010. Identification of a key residue mediating bone morphogenetic protein (BMP)-6 resistance to noggin inhibition allows for engineered BMPs with superior agonist activity. J Biol Chem 285: 12169–12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer TA. 2014. von Willebrand factor, Jedi knight of the bloodstream. Blood 124: 1412–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamler R, Keutmann HT, Sidis Y, Kattamuri C, Schneyer A, Thompson TB. 2008. The structure of FSTL3.activin A complex. Differential binding of N-terminal domains influences follistatin-type antagonist specificity. J Biol Chem 283: 32831–32838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley E, Biben C, Kotecha S, Fabri L, Tajbakhsh S, Wang CC, Hatzistavrou T, Roberts B, Drinkwater C, Lah M, et al. 1998. DAN is a secreted glycoprotein related to Xenopus cerberus. Mech Dev 77: 173–184. [DOI] [PubMed] [Google Scholar]
- Stenvers KL, Tursky ML, Harder KW, Kountouri N, Amatayakul-Chantler S, Grail D, Small C, Weinberg RA, Sizeland AM, Zhu HJ. 2003. Heart and liver defects and reduced transforming growth factor β2 sensitivity in transforming growth factor β type III receptor-deficient embryos. Mol Cell Biol 23: 4371–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Kaneko E, Maeda J, Ueno N. 1997. Mesoderm induction by BMP-4 and -7 heterodimers. Biochem Biophys Res Commun 232: 153–156. [DOI] [PubMed] [Google Scholar]
- Tassew NG, Charish J, Seidah NG, Monnier PP. 2012. SKI-1 and Furin generate multiple RGMa fragments that regulate axonal growth. Dev Cell 22: 391–402. [DOI] [PubMed] [Google Scholar]
- ten Dijke P, Yamashita H, Ichijo H, Franzen P, Laiho M, Miyazono K, Heldin CH. 1994a. Characterization of type I receptors for transforming growth factor-β and activin. Science 264: 101–104. [DOI] [PubMed] [Google Scholar]
- ten Dijke P, Yamashita H, Sampath TK, Reddi AH, Estevez M, Riddle DL, Ichijo H, Heldin CH, Miyazono K. 1994b. Identification of type I receptors for osteogenic protein-1 and bone morphogenetic protein-4. J Biol Chem 269: 16985–16988. [PubMed] [Google Scholar]
- Thompson TB, Woodruff TK, Jardetzky TS. 2003. Structures of an ActRIIB:activin A complex reveal a novel binding mode for TGF-β ligand:receptor interactions. EMBO J 22: 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson TB, Lerch TF, Cook RW, Woodruff TK, Jardetzky TS. 2005. The structure of the follistatin:activin complex reveals antagonism of both type I and type II receptor binding. Dev Cell 9: 535–543. [DOI] [PubMed] [Google Scholar]
- Tibaldi E, Arrigoni G, Martinez HM, Inagaki K, Shimasaki S, Pinna LA. 2010. Golgi apparatus casein kinase phosphorylates bioactive Ser-6 of bone morphogenetic protein 15 and growth and differentiation factor 9. FEBS Lett 584: 801–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilak A, Nelsen SM, Kim HS, Donley N, McKnite A, Lee H, Christian JL. 2014. Simultaneous rather than ordered cleavage of two sites within the BMP4 prodomain leads to loss of ligand in mice. Development 141: 3062–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillet E, Bailly S. 2014. Emerging roles of BMP9 and BMP10 in hereditary hemorrhagic telangiectasia. Front Genet 5: 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townson SA, Martinez-Hackert E, Greppi C, Lowden P, Sako D, Liu J, Ucran JA, Liharska K, Underwood KW, Seehra J, et al. 2012. Specificity and structure of a high affinity activin receptor-like kinase 1 (ALK1) signaling complex. J Biol Chem 287: 27313–27325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetanova NG, Irannejad R, von Zastrow M. 2015. G protein-coupled receptor (GPCR) signaling via heterotrimeric G proteins from endosomes. J Biol Chem 290: 6689–6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bezooijen RL, Roelen BA, Visser A, van der Wee-Pals L, de Wilt E, Karperien M, Hamersma H, Papapoulos SE, ten Dijke P, Lowik CW. 2004. Sclerostin is an osteocyte-expressed negative regulator of bone formation, but not a classical BMP antagonist. J Exp Med 199: 805–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bezooijen RL, Svensson JP, Eefting D, Visser A, van der Horst G, Karperien M, Quax PH, Vrieling H, Papapoulos SE, ten Dijke P, et al. 2007. Wnt but not BMP signaling is involved in the inhibitory action of sclerostin on BMP-stimulated bone formation. J Bone Miner Res 22: 19–28. [DOI] [PubMed] [Google Scholar]
- Veverka V, Henry AJ, Slocombe PM, Ventom A, Mulloy B, Muskett FW, Muzylak M, Greenslade K, Moore A, Zhang L, et al. 2009. Characterization of the structural features and interactions of sclerostin: Molecular insight into a key regulator of Wnt-mediated bone formation. J Biol Chem 284: 10890–10900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton KL, Makanji Y, Wilce MC, Chan KL, Robertson DM, Harrison CA. 2009. A common biosynthetic pathway governs the dimerization and secretion of inhibin and related transforming growth factor β (TGFβ) ligands. J Biol Chem 284: 9311–9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Baloh RH, Milbrandt J, Garcia KC. 2006. Structure of artemin complexed with its receptor GFRα3: Convergent recognition of glial cell line-derived neurotrophic factors. Structure 14: 1083–1092. [DOI] [PubMed] [Google Scholar]
- Wang R, Zhu J, Dong X, Shi M, Lu C, Springer TA. 2012. GARP regulates the bioavailability and activation of TGF-β. Mol Biol Cell 23: 1129–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Ma J, Peng J, Xu J. 2013. Protein structure alignment beyond spatial proximity. Sci Rep 3: 1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Bianco C, Strizzi L, Hamada S, Mancino M, Bailly V, Mo W, Wen D, Miatkowski K, Gonzales M, et al. 2007. Growth factor induction of Cripto-1 shedding by glycosylphosphatidylinositol-phospholipase D and enhancement of endothelial cell migration. J Biol Chem 282: 31643–31655. [DOI] [PubMed] [Google Scholar]
- Weber D, Kotzsch A, Nickel J, Harth S, Seher A, Mueller U, Sebald W, Mueller TD. 2007. A silent H-bond can be mutationally activated for high-affinity interaction of BMP-2 and activin type IIB receptor. BMC Struct Biol 7: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Salmon RM, Upton PD, Morrell NW, Li W. 2014. Regulation of bone morphogenetic protein 9 (BMP9) by redox-dependent proteolysis. J Biol Chem 289: 31150–31159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidauer SE, Schmieder P, Beerbaum M, Schmitz W, Oschkinat H, Mueller TD. 2009. NMR structure of the Wnt modulator protein Sclerostin. Biochem Biophys Res Commun 380: 160–165. [DOI] [PubMed] [Google Scholar]
- Wiater E, Harrison CA, Lewis KA, Gray PC, Vale WW. 2006. Identification of distinct inhibin and transforming growth factor β-binding sites on betaglycan: Functional separation of betaglycan co-receptor actions. J Biol Chem 281: 17011–17022. [DOI] [PubMed] [Google Scholar]
- Winkler DG, Sutherland MK, Geoghegan JC, Yu C, Hayes T, Skonier JE, Shpektor D, Jonas M, Kovacevich BR, Staehling-Hampton K, et al. 2003. Osteocyte control of bone formation via sclerostin, a novel BMP antagonist. EMBO J 22: 6267–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wipff PJ, Hinz B. 2008. Integrins and the activation of latent transforming growth factor β1—An intimate relationship. Eur J Cell Biol 87: 601–615. [DOI] [PubMed] [Google Scholar]
- Wipff PJ, Rifkin DB, Meister JJ, Hinz B. 2007. Myofibroblast contraction activates latent TGF-β1 from the extracellular matrix. J Cell Biol 179: 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooderchak-Donahue WL, McDonald J, O’Fallon B, Upton PD, Li W, Roman BL, Young S, Plant P, Fulop GT, Langa C, et al. 2013. BMP9 mutations cause a vascular-anomaly syndrome with phenotypic overlap with hereditary hemorrhagic telangiectasia. Am J Hum Genet 93: 530–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo C, Whitman M. 2001. Nodal signals to Smads through Cripto-dependent and Cripto-independent mechanisms. Mol Cell 7: 949–957. [DOI] [PubMed] [Google Scholar]
- Yin H, Yeh LC, Hinck AP, Lee JC. 2008. Characterization of ligand-binding properties of the human BMP type II receptor extracellular domain. J Mol Biol 378: 191–203. [DOI] [PubMed] [Google Scholar]
- Yoon BH, Esquivies L, Ahn C, Gray PC, Ye SK, Kwiatkowski W, Choe S. 2014. An activin A/BMP2 chimera, AB204, displays bone-healing properties superior to those of BMP2. J Bone Miner Res 29: 1950–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga K, Obata H, Jurukovski V, Mazzieri R, Chen Y, Zilberberg L, Huso D, Melamed J, Prijatelj P, Todorovic V, et al. 2008. Perturbation of transforming growth factor (TGF)-β1 association with latent TGF-β binding protein yields inflammation and tumors. Proc Natl Acad Sci 105: 18758–18763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakin L, Metzinger CA, Chang EY, Coffinier C, De Robertis EM. 2008. Development of the vertebral morphogenetic field in the mouse: Interactions between crossveinless-2 and twisted gastrulation. Dev Biol 323: 6–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakin L, Chang EY, Plouhinec JL, De Robertis EM. 2010. Crossveinless-2 is required for the relocalization of Chordin protein within the vertebral field in mouse embryos. Dev Biol 347: 204–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JL, Huang Y, Qiu LY, Nickel J, Sebald W. 2007. von Willebrand factor type C domain-containing proteins regulate bone morphogenetic protein signaling through different recognition mechanisms. J Biol Chem 282: 20002–20014. [DOI] [PubMed] [Google Scholar]
- Zhang JL, Qiu LY, Kotzsch A, Weidauer S, Patterson L, Hammerschmidt M, Sebald W, Mueller TD. 2008. Crystal structure analysis reveals how the chordin family member crossveinless 2 blocks BMP-2 receptor binding. Dev Cell 14: 739–750. [DOI] [PubMed] [Google Scholar]
- Zhang AS, Yang F, Wang J, Tsukamoto H, Enns CA. 2009. Hemojuvelin-neogenin interaction is required for bone morphogenic protein-4-induced hepcidin expression. J Biol Chem 284: 22580–22589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JL, Patterson LJ, Qiu LY, Graziussi D, Sebald W, Hammerschmidt M. 2010. Binding between Crossveinless-2 and Chordin von Willebrand factor type C domains promotes BMP signaling by blocking Chordin activity. PLoS ONE 5: e12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wang L, Zhang X, Zhang X, Gu Z, Wu G. 2012. BMP2/7 heterodimer can modulate all cellular events of the in vitro RANKL-mediated osteoclastogenesis, respectively, in different dose patterns. Tissue Eng Part A 18: 621–630. [DOI] [PubMed] [Google Scholar]
- Zhou YF, Springer TA. 2014. Highly reinforced structure of a C-terminal dimerization domain in von Willebrand factor. Blood 123: 1785–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Kim J, Cheng C, Rawlins BA, Boachie-Adjei O, Crystal RG, Hidaka C. 2006. Noggin regulation of bone morphogenetic protein (BMP) 2/7 heterodimer activity in vitro. Bone 39: 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman LB, De Jesus-Escobar JM, Harland RM. 1996. The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell 86: 599–606. [DOI] [PubMed] [Google Scholar]
- Zuniga JE, Ilangovan U, Mahlawat P, Hinck CS, Huang T, Groppe JC, McEwen DG, Hinck AP. 2011. The TβR-I pre-helix extension is structurally ordered in the unbound form and its flanking prolines are essential for binding. J Mol Biol 412: 601–618. [DOI] [PMC free article] [PubMed] [Google Scholar]