In this Perspective, Wahl and Fischer discuss three studies that have now provided insights into how an assembly factor specifically recognizes substrate RNA molecules and enables their usage for assembly of Sm-class uridine-rich small nuclear RNA–protein complexes.
Keywords: Gemin5, snRNA, spliceosome, structure, SMN complex, WD40 domain, X-ray crystal structure
Abstract
Macromolecular complexes, rather than individual biopolymers, perform many cellular activities. Faithful assembly of these complexes in vivo is therefore a vital challenge of all cells, and its failure can have fatal consequences. To form functional complexes, cells use elaborate measures to select the “right” components and combine them into working entities. How assembly is achieved at the molecular level is unclear in many cases. Three groups (Jin and colleagues, pp. 2391–2403; Xu and colleagues, pp. 2376–2390; and Tang and colleagues in Cell Research) have now provided insights into how an assembly factor specifically recognizes substrate RNA molecules and enables their usage for assembly of Sm-class uridine-rich small nuclear RNA–protein complexes.
Uridine-rich small nuclear RNA–protein complexes (UsnRNPs) are major building blocks of the spliceosome, a dynamic macromolecular machine that catalyzes the removal of introns from eukaryotic precursor messenger RNA (pre-mRNA) molecules (Wahl et al. 2009). To cope with the task of splicing, there are high steady-state levels of cellular UsnRNPs in higher eukaryotes, reaching up to an estimated concentration of 10 µM per nucleus of a somatic cell. Therefore, efficient production of UsnRNPs and maintenance of their steady-state levels are crucial for eukaryotic cells.
Sm-class UsnRNPs contain a particle-specific snRNA, a set of seven common Sm proteins, and a variable number of additional proteins unique to each complex (Will and Lührmann 2001). Upon synthesis by RNA polymerase II, snRNAs receive an m7G cap. For assembly of UsnRNPs, the m7G-capped UsnRNAs are transiently exported to the cytoplasm. The Sm proteins then bind to a single-stranded uridine-rich sequence of the snRNAs, the Sm site, resulting in the formation of a toroidal Sm core (Pomeranz Krummel et al. 2009; Weber et al. 2010; Leung et al. 2011). This core is a common structural denominator of all UsnRNPs and enables several subsequent steps in the biogenesis pathway, including the hypermethylation of the m7G cap of the snRNAs to an m2,2,7G cap and the nuclear import of the assembled UsnRNP core particles. After further maturation, UsnRNPs are eventually incorporated into spliceosomes.
The cytosolic assembly of UsnRNP cores requires an amazingly large number of trans-acting factors. These are organized in two multisubunit machineries: the protein arginine methyltransferase 5 (PRMT5) complex and the survival motor neuron (SMN) complex. The PRMT5 complex acts early in the assembly pathway, facilitating the formation of higher-order Sm protein complexes (Chari et al. 2008). These building blocks are handed over to the SMN complex, whose major function is to combine the Sm proteins with the cognate snRNAs to form the Sm cores (Chari et al. 2009; Matera and Wang 2014). The SMN complex comprises the name-giving protein SMN along with eight other proteins, termed Gemin2–8 and unrip.
While SMN and Gemin2 have been implicated in Sm protein binding (Zhang et al. 2011; Grimm et al. 2013), only little was so far known about the function of the other SMN complex subunits during UsnRNP formation. Two studies in this issue of Genes & Development (Jin et al. 2016; Xu et al. 2016) and a report published in Cell Research (Tang et al. 2016) now provide insights into a possible role of Gemin5 during Sm core RNP assembly. This WD40 repeat protein has been shown to specifically bind UsnRNAs (Battle et al. 2006) and the m7G cap (Bradrick and Gromeier 2009) and hence was implicated in the selection of RNA substrates for the assembly reaction. Detailed interaction studies based on U4 snRNA in the present studies further revealed that the N-terminal tandem WD40 domains of Gemin5 can bind Sm site-containing RNA molecules and m7G cap moieties independently in vitro and in vivo (Jin et al. 2016; Tang et al. 2016; Xu et al. 2016). However, a 3′ stem–loop of U4 snRNA, previously also implicated in Gemin5 binding (Lau et al. 2009), was dispensable for stable complex formation. Based on these findings, the investigators of all three studies went on to determine crystal structures of the WD40 region of Gemin5 alone or in complex with various RNA molecules bearing complete or truncated Sm sites as well as with m7G cap mimics (Fig. 1A–D). The structures show that the two WD40 domains are tightly connected, with the N terminus of the protein completing the WD40 propeller of the second WD40 domain. The m7G cap is accommodated in a specific pocket on the second WD40 domain, while Sm site regions of the RNAs bind predominantly to the first WD40 domain by a combination of stacking interactions, hydrogen bonding, and electrostatic contacts with the RNA backbone. Extensive mutational studies in combination with binding assays were conducted by all three groups to elucidate the contributions of specific protein and RNA residues to the observed interactions.
Figure 1.
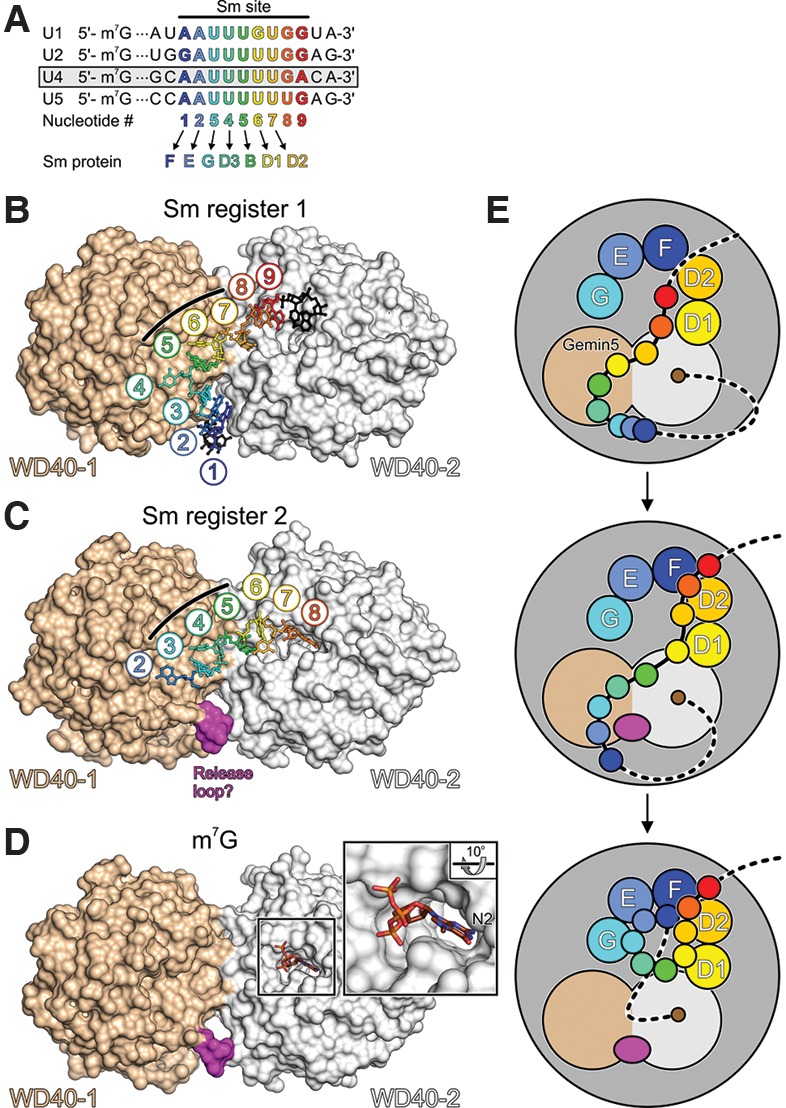
Possible role of Gemin5 in UsnRNP assembly. (A) Alignment of the Sm sites and neighboring nucleotides of the major spliceosomal U1, U2, U4, and U5 snRNAs. U4 snRNA, used in the present studies, is boxed. Sm site nucleotides are numbered 1–9 and colored blue to red in a 5′-to-3′ direction. Sm proteins F, E, G, D3, B, D1, and D2 are assembled on nucleotides 1–7 of the Sm sites to form Sm cores. (B,C) Structures of the Gemin5 N-terminal tandem WD40 domains in complex with a 13-nucleotide (nt) (B) and a 7-nt (C) oligomer containing a complete or partial Sm site, respectively (based on Jin et al. 2016). The two RNA molecules bind with different registers across a positively charged concave surface formed by both WD40 domains. Three consecutive uridines (U5, U6, and U7 or U3, U4, and U5 of the Sm site) bind in an analogous manner and at identical pockets to the center of the protein (black lines). Nucleotides 3′ of U7 in B lack extensive contacts to the protein. U8 in C occupies a pocket on the second WD40 domain that also accommodates the m7G cap (see D). (D) Binding of an m7GpppG mimic of a 5′-capped snRNA to a pocket on the second WD40 domain (based on Xu et al. 2016). The second guanine base is disordered in the structure. (Close-up) N2 of m7G is buried in the pocket. An m2,2,7G cap bearing two additional methyl groups on N2 could not be accommodated due to the disruption of hydrogen bonds and steric hindrance. (E) Model for the role of Gemin5 in Sm core assembly. Gemin5 could bind snRNAs concomitantly via the m7G cap and the Sm site. Nucleotides 3′ of a Sm site would be free to engage in initial contacts to the partially assembled Sm proteins on the SMN complex. A shift in the register of Sm site binding on Gemin5 could feed Sm site nucleotides 6 and 7 into their binding pockets on D1 and D2, respectively. The shift in register could involve a loop of the first WD40 domain occupying a position where Sm site nucleotides 1–3 are initially bound on Gemin5. After subsequent handover of Sm site nucleotides 1, 2, and 3 to Sm proteins F, E, and G, respectively, Gemin5 might still hold onto the cap, protecting it from hypermethylation until the D3–B heterodimer has been assembled on Sm site nucleotides 4 and 5. After assembly of the Sm core has been completed, another handover of the cap from Gemin5 to the cap methyltransferase TGS1 would have to occur in this scenario.
Surprisingly, two different registers were observed for RNAs bound to the Gemin5 WD40 domains. The two binding modes agree with two shifted bands observed in electrophoretic gel mobility shift assays (Jin et al. 2016). The most extensive contacts to Gemin5 were seen with a 13-nucleotide (nt) RNA bearing a central Sm site (Fig. 1B; Jin et al. 2016). In that structure, the three 5′-terminal residues of the Sm site (A1–U3) form an extended stack between the two WD40 domains. The following U4–U6 residues circle across the first WD40 domain, and nucleotide U7 binds to a pocket on the second WD40 domain, while further downstream residues lack extensive protein contacts. One group (Tang et al. 2016) observed a shorter RNA oligonucleotide comprising a complete Sm site plus an additional downstream residue bound in the same manner. However, in the hands of the other two groups (Jin et al. 2016; Xu et al. 2016), short oligonucleotides comprising Sm site residues 1–7, 1–8, or 1–9 occupied identical pockets on the first WD40 domain but were shifted 2 nt in the 3′ direction relative to the first binding mode (Fig. 1C). In these latter structures, the G8 nucleotide came to lie in the pocket that also accommodates the m7G cap on the second WD40 domain (Fig. 1C). However, this latter interaction is presumably not realized by full-length snRNAs, as mutations designed to interfere with accommodation of residue 8 in the cap-binding pocket had essentially no effect on the RNA affinity (Jin et al. 2016; Xu et al. 2016). Furthermore, G8 did not occupy this pocket when the cocrystallized oligonucleotide contained more than one additional residue downstream, as shown by cocrystallization with an RNA bearing an extra 2 nt 3′ of the Sm site (Xu et al. 2016).
Apart from snRNP assembly, Gemin5 has been implicated in a number of other RNA-related processes (Pineiro et al. 2015). In vivo interaction and stability studies by Xu et al. (2016) add to this spectrum of Gemin5 functions by providing preliminary evidence for a putative additional role of Gemin5 in P-body-related snRNA surveillance. Thus, in principle, the two different RNA-binding modes seen in the present analyses might relate to different functional scenarios. However, as the central Sm site-binding region of Gemin5 is apparently designed to recognize three consecutive uridines (Fig. 1B,C, black lines), a condition that can be met in different ways by most Sm sites (Fig. 1A), it is tempting to speculate that they might represent different snapshots of snRNA handover from Gemin5 to the Sm proteins (Fig. 1E). Thus, Gemin5 might initially recognize snRNAs via two of their key features: an m7G cap and a uridine-rich Sm site. Although not directly shown by the structural analyses, concomitant recognition of the cap and Sm site could avoid binding of many other RNAs that harbor stretches of uridines. Holding on to the cap, Gemin5 might then allow sliding of the RNA into the partly assembled Sm protein ring on the SMN complex (Fig. 1E). Once contacts of RNA nucleotides downstream from the Sm site to the perimeter of the Sm proteins have been established, strong binding of the Sm proteins to the Sm site (Raker et al. 1999) might provide the driving force that pulls the RNA away from Gemin5. The handover might additionally be supported by a loop in the first WD40 domain of Gemin5, which can interfere with the binding of the 5′ end of the Sm site in the first register (residues A1–U3) and thus might help to “push” the RNA into the second register or away from Gemin5 (Fig. 1B,C).
At the end of the transaction, Gemin5 might still hold on to the m7G cap (Fig. 1E). Thus, after completion of Sm core assembly, another transfer of the m7G cap from Gemin5 to the methyltransferase TGS1, which catalyzes cap hypermethylation (Mouaikel et al. 2002) and interacts with the SMN protein (Mouaikel et al. 2003), might be required. Notably, binding affinities of Gemin5 for Sm site oligonucleotides as well as for the m7G cap were found in the low micromolar range (Jin et al. 2016; Xu et al. 2016). Thus, Gemin5 interacts with both key recognition elements on the snRNAs with only moderate strength, as would be suitable for transient binding and thus consistent with its putative snRNA-channeling functions.
Gemin5 is lacking in a number of organisms, which nevertheless assemble snRNPs efficiently, and snRNPs can be effectively assembled in vitro without Gemin5 (Raker et al. 1996; Raker et al. 1999; Kroiss et al. 2008), calling the importance of Gemin5-mediated snRNA selection or channeling for snRNP assembly into question. Importantly, work presented in the three reports now also provides interesting new tools to further study these and possibly other putative Gemin5 activities. For instance, a number of Gemin5 variants have been identified that selectively interfere with either cap or Sm site binding. By replacing wild-type Gemin5 with such variants, it may be possible to test to what extent Sm proteins are assembled on noncognate RNAs in vivo upon such manipulation. Furthermore, in light of the multiple putative transfer processes of snRNA elements initially bound by Gemin5, it would be interesting to study the effects of the DEAD-box RNA helicase Gemin3, which is also associated with the SMN complex, on these transitions. Finally, future studies should address the question of to what extent and how UsnRNP assembly is influenced by additional RNA-binding activity, as seen previously for the C-terminal coiled-coil region of Gemin5 (Fernandez-Chamorro et al. 2014), and by Gemin5 oligomerization via its C-terminal region (Xu et al. 2016).
Acknowledgments
We apologize to colleagues whose work could not be cited due to length restrictions. Work in the authors’ laboratories is supported by the Deutsche Forschungsgemeinschaft (grants SFB740, TRR 186, and WA1126/7-1 to M.C.W., and grant Fi573/7-2 to U.F.), the Bundesministerium für Bildung und Forschung (grant 05K10KEC to M.C.W.), and the Einstein Foundation Berlin (grant A-2012-140 to M.C.W.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.293084.116.
References
- Battle DJ, Lau CK, Wan L, Deng H, Lotti F, Dreyfuss G. 2006. The Gemin5 protein of the SMN complex identifies snRNAs. Mol Cell 23: 273–279. [DOI] [PubMed] [Google Scholar]
- Bradrick SS, Gromeier M. 2009. Identification of Gemin5 as a novel 7-methylguanosine cap-binding protein. PLoS One 4: e7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari A, Golas MM, Klingenhager M, Neuenkirchen N, Sander B, Englbrecht C, Sickmann A, Stark H, Fischer U. 2008. An assembly chaperone collaborates with the SMN complex to generate spliceosomal snRNPs. Cell 135: 497–509. [DOI] [PubMed] [Google Scholar]
- Chari A, Paknia E, Fischer U. 2009. The role of RNP biogenesis in spinal muscular atrophy. Curr Opin Cell Biol 21: 387–393. [DOI] [PubMed] [Google Scholar]
- Fernandez-Chamorro J, Pineiro D, Gordon JM, Ramajo J, Francisco-Velilla R, Macias MJ, Martinez-Salas E. 2014. Identification of novel non-canonical RNA-binding sites in Gemin5 involved in internal initiation of translation. Nucleic Acids Res 42: 5742–5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Chari A, Pelz JP, Kuper J, Kisker C, Diederichs K, Stark H, Schindelin H, Fischer U. 2013. Structural basis of assembly chaperone-mediated snRNP formation. Mol Cell 49: 692–703. [DOI] [PubMed] [Google Scholar]
- Jin W, Wang Y, Liu C-P, Yang N, Jin M, Cong Y, Wang M, Xu R-M. 2016. Structural basis for snRNA recognition by the double WD40 repeat domain of Gemin5. Genes Dev 10.1101/gad.291377.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroiss M, Schultz J, Wiesner J, Chari A, Sickmann A, Fischer U. 2008. Evolution of an RNP assembly system: a minimal SMN complex facilitates formation of UsnRNPs in Drosophila melanogaster. Proc Natl Acad Sci 105: 10045–10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CK, Bachorik JL, Dreyfuss G. 2009. Gemin5–snRNA interaction reveals an RNA binding function for WD repeat domains. Nat Struct Mol Biol 16: 486–491. [DOI] [PubMed] [Google Scholar]
- Leung AK, Nagai K, Li J. 2011. Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis. Nature 473: 536–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Wang ZF. 2014. A day in the life of the spliceosome. Nat Rev Mol Cell Biol 15: 108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineiro D, Fernandez-Chamorro J, Francisco-Velilla R, Martinez-Salas E. 2015. Gemin5: a multitasking RNA-binding protein involved in translation control. Biomolecules 5: 528–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouaikel J, Verheggen C, Bertrand E, Tazi J, Bordonne R. 2002. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol Cell 9: 891–901. [DOI] [PubMed] [Google Scholar]
- Mouaikel J, Narayanan U, Verheggen C, Matera AG, Bertrand E, Tazi J, Bordonne R. 2003. Interaction between the small-nuclear-RNA cap hypermethylase and the spinal muscular atrophy protein, survival of motor neuron. EMBO Rep 4: 616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K. 2009. Crystal structure of human spliceosomal U1 snRNP at 5.5 Å resolution. Nature 458: 475–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raker VA, Plessel G, Lührmann R. 1996. The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J 15: 2256–2269. [PMC free article] [PubMed] [Google Scholar]
- Raker VA, Hartmuth K, Kastner B, Lührmann R. 1999. Spliceosomal U snRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol Cell Biol 19: 6554– 6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Bharath SR, Piao S, Tan VQ, Bowler MW, Song H. 2016. Structural basis for specific recognition of pre-snRNA by Gemin5. Cell Res 10.1038/cr.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718. [DOI] [PubMed] [Google Scholar]
- Weber G, Trowitzsch S, Kastner B, Luhrmann R, Wahl MC. 2010. Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J 29: 4172–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Lührmann R. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol 13: 290–301. [DOI] [PubMed] [Google Scholar]
- Xu C, Ishikawa H, Izumikawa K, Li L, He H, Nobe Y, Yamauchi Y, Shahjee HM, Wu XH, Yu YT, et al. 2016. Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly. Genes Dev 10.1101/gad.288340.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, So BR, Li P, Yong J, Glisovic T, Wan L, Dreyfuss G. 2011. Structure of a key intermediate of the SMN complex reveals Gemin2′s crucial function in snRNP assembly. Cell 146: 384–395. [DOI] [PMC free article] [PubMed] [Google Scholar]


