In this study, Vuolo et al. investigate the mechanisms underlying the genetic basis for morphological diversity in leaf shape. They show that evolution of an enhancer element in the homeobox gene REDUCED COMPLEXITY (RCO) altered leaf shape by changing gene expression from the distal leaf blade to its base.
Keywords: compound leaf, leaf development, plant homeobox gene, regulatory evolution
Abstract
Here we investigate mechanisms underlying the diversification of biological forms using crucifer leaf shape as an example. We show that evolution of an enhancer element in the homeobox gene REDUCED COMPLEXITY (RCO) altered leaf shape by changing gene expression from the distal leaf blade to its base. A single amino acid substitution evolved together with this regulatory change, which reduced RCO protein stability, preventing pleiotropic effects caused by its altered gene expression. We detected hallmarks of positive selection in these evolved regulatory and coding sequence variants and showed that modulating RCO activity can improve plant physiological performance. Therefore, interplay between enhancer and coding sequence evolution created a potentially adaptive path for morphological evolution.
Understanding the genetic basis for evolutionary change is a fundamental problem in biology. Morphological diversity is often underpinned by cis-regulatory divergence of developmental genes and consequent spatiotemporal modification of their expression (Gompel et al. 2005; Hay and Tsiantis 2006; Prud'homme et al. 2006; Carroll 2008; Chan et al. 2010; Frankel et al. 2011; Studer et al. 2011; Arnoult et al. 2013; Rast-Somssich et al. 2015; Indjeian et al. 2016). However, the origin of specific cis-regulatory elements underlying morphological diversity is still poorly understood (Rebeiz et al. 2015). For example, it is unclear whether such cis elements tend to arise de novo from rapidly evolving sequences or through the co-option of existing conserved regulatory sequences (Rebeiz et al. 2011; Boyd et al. 2015; Villar et al. 2015). Furthermore, it has not been investigated whether and how coding sequences evolve in concert with regulatory changes to optimize gene function in a new expression domain. Finally, links between regulatory changes underlying morphological change and organismal physiology and fitness remain scarce.
Plant leaves present a useful genetic model to tackle these questions because they show substantial morphological variation (Shleizer-Burko et al. 2011; Bar and Ori 2014) and have considerable eco–physiological importance as the major site of photosynthetic carbon fixation in terrestrial ecosystems (Givnish 1978). The REDUCED COMPLEXITY (RCO) gene played a key role in leaf shape diversification in the crucifer family (Sicard et al. 2014; Vlad et al. 2014), to which the reference plant Arabidopsis thaliana belongs. RCO arose through gene duplication and encodes a class I homeobox leucine zipper protein. Its function was discovered in Cardamine hirsuta, where it acts to divide the leaf into distinct leaflets by locally repressing growth at the leaf margin, creating a complex shape. This species-specific activity of RCO arose by neofunctionalization following gene duplication of its ancestral paralog, LMI1, which is conserved in seed plants. Specifically, RCO acquired a novel expression domain within the growth zone at the base of the leaf, where growth repression—a conserved function of the RCO/LMI protein—exerts a greater effect on leaf shape (Fig. 1A). RCO was secondarily lost in A. thaliana, leading to leaf simplification, and its reintroduction in the A. thaliana genome was sufficient to increase leaf complexity (Vlad et al. 2014). Thus, RCO is a large effect gene underlying morphological diversity and offers an excellent system to explore the causes and consequences of morphological evolution. Here, we identify the specific molecular events underpinning the evolution of RCO function and provide evidence that modulating RCO activity can improve plant physiological performance.
Figure 1.
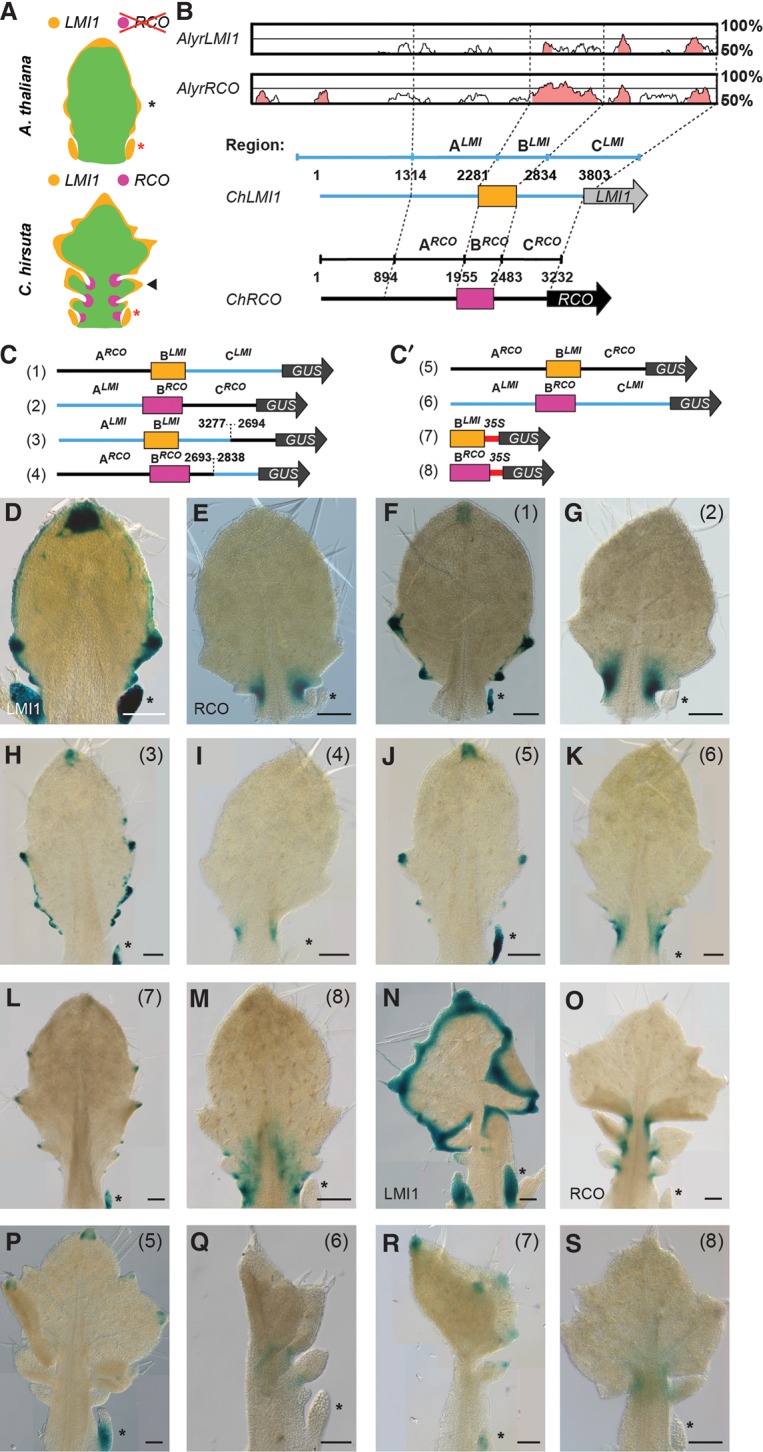
ChRCOenh500 is sufficient to drive proximal expression in the leaf lamina and evolved via modification of an enhancer driving distal expression in the leaf lamina. (A) Cartoon depicting leaf primordia of A. thaliana (top; black star indicates serration) and C. hirsuta (bottom; black arrowhead indicates leaflet). Red stars indicate stipules located at the leaf base. LMI1 (orange) and RCO (magenta) expression is shown. (B, top) mVISTA plot of Arabidopsis lyrata LMI1 and RCO upstream sequence aligned to C. hirsuta RCO, indicating sequence conservation (50%–100%) (mVISTA alignments with additional species are shown in Supplemental Fig. 1). Pink indicates conserved nucleotide sequences (CNSs; identity >70% within 100 base pairs [bp]). (Bottom) Subdivision of LMI1 and RCO upstream sequences into regions A, B, and C based on CNSs. ChLMI1 and ChRCO gene models (blue and black lines, respectively) with the CNS enhancer element of ∼500 bp indicated. (Orange) BLM1; (magenta) BRCO. (C,C′) Chimeric GUS reporter constructs (1–8) used to identify critical cis-regulatory elements in ChLMI1 and ChRCO upstream regions. (D–S) Representative A. thaliana (D–M) and C. hirsuta (N–S) GUS-stained leaves carrying reporter constructs depicted in C and C′ or full-length 3.8-kb ChLMI1 (D,N) and 3.2-kb ChRCO (E,O) upstream regions (see also Vlad et al. 2014). In each image, the construct used is indicated (1–8). The A. thaliana leaf depicted in L is more mature, hence the higher number of serrations. Each image contains at least one stipule (black star) to visualize the presence (LMI1) or absence (RCO) of expression. In each case, at least two independent T2 lines were analyzed with n > 5. Bar, 100 μm.
Results and Discussion
To understand whether discrete enhancer sequences explain the difference in expression between RCO and its ancestral paralog, LMI1, we analyzed the upstream sequences of LMI1 and RCO using transgenic assays. We first investigated whether discrete enhancer sequences are sufficient to explain the evolutionary shift in RCO expression with respect to its paralog, LMI1, and what their origin might be. We reasoned that if such enhancer elements exist in RCO, their introduction in LMI1 via chimeric constructs should recapitulate evolution and convert the ancestral distal expression pattern into the proximal one of RCO. We first defined upstream noncoding DNA fragments of RCO and LMI1 that were sufficient to drive reporter gene expression in the proximal and distal domains of the leaf lamina that characterize each gene (Supplemental Fig. 2A–E; Supplemental Table 3). Subsequently, we assayed the expression pattern of chimeric reporter genes where three individual segments (regions A, B, and C) of RCO and LMI1 upstream sequences were swapped between the two genes. (Fig. 1B–C′; Supplemental Table 3). We conducted these reporter gene assays in A. thaliana, as the upstream regulatory regions of C. hirsuta LMI1 and RCO recapitulate their respective distal and proximal expression patterns in the leaf lamina of A. thaliana (Fig. 1D,E; Supplemental Fig. 2B,D; Vlad et al. 2014).
These chimeric reporters had a binary readout: Each reporter yielded either the LMI1-type or the RCO-type expression pattern (Fig. 1F–K). The LMI1-type pattern was defined by expression in stipules and hydathodes, with weaker expression in the leaf margin. In comparison with this, the RCO-type pattern was expressed only at the base of the leaf blade. These observations indicated that specific sequences contributing to LMI1 expression might have been modified through evolution to produce the RCO expression pattern. In support of this idea, exchanging region BRCO for the corresponding LMI1 sequence converted the LMI1 expression pattern into the RCO pattern in both A. thaliana and the endogenous C. hirsuta context (Fig. 1K,Q). Conversely, introducing region BLMI into the RCO sequence resulted in an LMI1 expression pattern in both the A. thaliana and C. hirsuta contexts (Fig. 1J,P). Moreover, a reporter containing only region BLMI or BRCO coupled to a 50-base-pair (bp) CaMV 35S minimal promoter was sufficient to drive specific LMI1-type or RCO-type expression in A. thaliana (Fig. 1C′,L,M) and, in the case of BRCO, also in C. hirsuta (Fig. 1R,S). Notably, the corresponding region BLMI from Aethionema arabicum, an early divergent crucifer, is necessary for correct LMI1 gene expression (Supplemental Fig. 2G–H), also indicating that it was already functional before the divergence of Aethionema from other crucifers (Vlad et al. 2014). Thus, the 500-bp region BLMI or BRCO has a key function in determining the expression pattern of its respective downstream gene. Consequently, we call this region the RCO or LMI1 500-bp enhancer (ChRCOenh500 or ChLMI1enh500).
To test to what degree gene expression conferred by ChRCOenh500 is phenotypically relevant, we used it to express the RCO-coding sequence both in the LMI1 regulatory sequence context and using a 35S minimal promoter (Supplemental Table 3). Strikingly, both constructs increased leaf complexity in A. thaliana and rescued the rco mutant leaf phenotype in C. hirsuta (Fig. 2B–F). These findings demonstrate that ChRCOenh500 is necessary and sufficient to drive RCO function and that this sequence imparts morphologically relevant transcriptional information even in the context of a heterologous promoter. ChRCOenh500 must interact with additional sequences to ensure the correct level of RCO transcription, as the 2.3-kb fragment drives higher expression (Figs. 1M, 2C–F; Supplemental Fig. 2E). In summary, ChRCOenh500 recapitulates the RCO expression pattern, and its activity is sufficient to increase leaf complexity when transferred between two reproductively isolated species that diverged ∼30 million years ago (Vlad et al. 2014). Our findings demonstrate that a specific enhancer element in LMI1, which directs distal gene expression, neofunctionalized in the RCO duplicate gene to yield a novel expression pattern at the leaf base, resulting in a novel leaf form.
Figure 2.
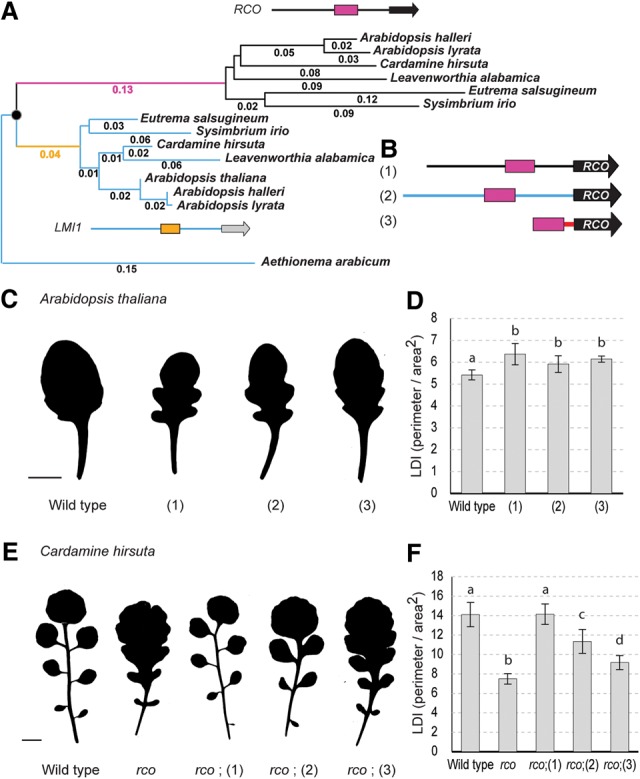
ChRCOenh500 evolved nonneutrally and is sufficient to increase leaf complexity when driving RCO in A. thaliana and C. hirsuta. (A) The RCO enhancer (magenta; depicted on a black gene model) experienced an increased base substitution rate (likelihood ratio test: ΔlnL = 9.64; P < 0.005) compared with the LMI1 enhancer (yellow; depicted on a blue gene model), consistent with positive selection (see the Supplemental Material for detailed analysis). Base substitution rates are indicated next to individual branches. (B) Constructs (1–3) used to express RCO in the RCO domain contain the upstream sequence of ChLMI1 (blue line), ChRCO (black line), or the CaMV 35S minimal promoter (red line) with enhancer BRCO (magenta). (C,E) Representative leaf 8 silhouettes from A. thaliana (C) and C. hirsuta (E) wild-type and transgenic plants. Bar, 1 cm. (D,F) Leaf dissection index ([LDI] perimeter/area2) calculated from A. thaliana (D) and C. hirsuta (F) leaf 8 silhouettes. Graphs indicate average LDI and standard deviation. Letters indicate significant differences between groups as indicated by ANOVA and post-hoc Tukey's test. P < 0.01. For constructs 2 and 3, at least three independent T2 lines were analyzed with n > 12.
Next, we investigated the evolutionary forces that led to the diversification of RCOenh500 from its LMI1 counterpart by comparing their sequence divergence patterns in a phylogenetic framework. We observed a significantly higher base substitution rate for this enhancer within the RCO clade than within the LMI1 clade (Fig. 2A). Using a modified branch site likelihood model adapted for noncoding regions (Wong and Nielsen 2004), we demonstrated that this accelerated evolution of RCOenh500 likely reflects the action of positive selection. These analyses, coupled with our functional data (Fig. 1B–S), are consistent with the idea that positive selection helped shape the RCO expression domain via acting on ChRCOenh500.
RCO/LMI1 proteins are potent growth repressors, and their broad expression results in miniature plants (Vlad et al. 2014). This raises the question of whether RCO enhancer evolution involved concomitant coding sequence diversification to alleviate potentially pleiotropic effects resulting from altered RCO expression. To address this question, we analyzed RCO-coding sequence diversification patterns from seven species. A phylogeny-based maximum likelihood ratio test (Yang 2007) identified signals of positive selection centered on the alanine and tyrosine residues at positions 48 (A48) and 56 (Y56), N-terminal to the homeodomain (Fig. 3A; Supplemental Table 1). To test the functional importance of these D48A and S56Y changes in the RCO protein, we generated A. thaliana plants expressing modified RCO genes (RCOgA48D/RCOgY56S/RCOgA48D-Y56S) where the native promoter drives RCO with the A48D and Y56S mutations individually or in combination. The leaf phenotype of plants expressing RCOY56S was indistinguishable from plants expressing RCO (Supplemental Fig. 3). However, plants expressing RCOA48D or RCOA48D-Y56S had more dissected leaves (Supplemental Fig. 3) and resembled plants expressing LMI1 in the RCO domain (Fig. 3B,C), indicating that the A48D mutation has a major effect on leaf form. The stronger effect of the A48D versus the Y56S change in RCO is consistent with a greater contrast between the properties of the derived and ancestral amino acids: Alanine (A) is nonpolar, neutral, and hydrophobic, and aspartic acid (D) is polar, acidic, and hydrophilic, whereas tyrosine (Y) and serine (S) are very similar. However, the possibility that the Y56S mutation might cause phenotypic effects under different growth conditions cannot be excluded.
Figure 3.
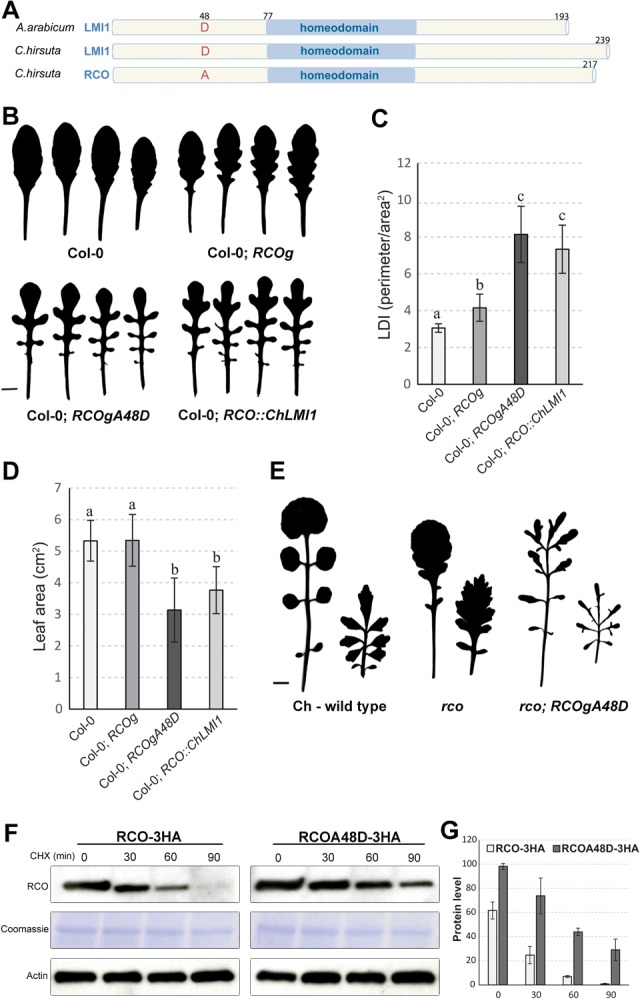
Positive selection dampened RCO protein function via a D > A transition that decreased protein stability. (A) Structure of LMI1/RCO protein in A. arabicum and C. hirsuta. (B–D) Silhouettes (B), leaf dissection index (C), and leaf area (D) of leaves 7 to 10 of A. thaliana wild-type, RCOg, RCOgA48D, and RCO::ChLMI1 plants. Error bars represent standard deviation based on at least 15 independent T1 lines. Letters indicate significant differences between groups as indicated by ANOVA and post-hoc Tukey's test. P < 0.01. (E) Silhouettes of rosette and cauline leaves of C. hirsuta wild type, rco, and rco; RCOgA48D. (F,G) Quantitative analysis of RCO-3HA and RCOA48D-3HA protein levels after cyclohexamide (CHX) treatment at the indicated time points. (F) Coomassie blue staining (SDS-PAGE slice containing a 57-kDa band) and anti-Actin immunoblotting indicate equal loading. (G) Protein level quantification of samples shown in F. Error bars represent standard deviation of the mean protein level using three biological replicates.
The increased leaf complexity in transgenic RCOgA48D and RCO::ChLMI1 plants was accompanied by a significant decrease in leaf area (Fig. 3B–D) compared with RCOg plants. Thus, the RCOA48D and LMI1 proteins are more potent than RCO, resulting in not only altered leaf shape but also compromised organ growth when expressed in the RCO domain. In contrast, the native RCO protein changes A. thaliana leaf shape without incurring an organ growth penalty (Fig. 3B–D). RCOA48D showed consistently higher potency than RCO in the endogenous C. hirsuta context: It rescued the rco mutant phenotype more effectively, increased wild-type leaf complexity, and reduced leaf size (Fig. 3E; Supplemental Fig. 4A–D). Reduction in cell size contributes to the reduced leaf surface of RCOgA48D (Supplemental Fig. 5), indicating that RCO/LMI1 proteins may repress growth at the whole-organ level by repressing cell growth. Taken together, these observations indicate that diversification of gene expression after duplication of the ancestral LMI1 gene entailed a risk of pleiotropic effects, detrimental to growth. We propose that these effects were counteracted by the D48A amino acid change, which dampened RCO protein potency. Two lines of evidence indicate that this dampening involved reduced protein stability. First, HA-tagged RCOA48D (RCOA48D-3HA) accumulated to higher levels in transgenic plants than HA-tagged RCO (RCO-3HA) (Supplemental Fig. 6A–C). Second, the degradation rate of RCO-3HA is higher than RCOA48D-3HA following de novo protein synthesis inhibition by cycloheximide (Fig. 3F,G). These findings highlight the importance of coordinated coding and regulatory sequence evolution for morphological variation. They also indicate that coupling protein and cis-regulatory evolution (Prud'homme et al. 2006; Stern and Orgogozo 2008; Chan et al. 2010; Frankel et al. 2011; Indjeian et al. 2016) can effectively minimize the pleiotropic effects of mutations in developmental genes. Notably, regulatory sequence variation in humans may minimize the detrimental effects of deleterious coding sequence mutations in highly expressed haplotypes (Lappalainen et al. 2011). Thus, coevolution of enhancers with their cognate coding sequences may be of broad significance across complex eukaryotes and at different evolutionary scales.
The hallmarks of positive selection in RCO indicate that it may have evolved adaptively. To investigate this hypothesis, we tested whether changes in RCO activity affected plant physiological performance. rco mutants showed reduced CO2 fixation (Fig. 4A), and introducing RCO into A. thaliana (RCOg genotype) was sufficient to increase CO2 fixation by 20%–25% (Fig. 4A). Furthermore, RCO positively influenced seed yield in both C. hirsuta and A. thaliana (Fig. 4B,C). RCO has a restricted expression pattern during plant development (Vlad et al. 2014) and is not expressed in the nutritive endosperm tissue of the seed but influences its size (Supplemental Fig. 7A–I). Therefore, the stimulatory effects of RCO on photosynthesis may ultimately influence resource allocation to seeds. Taken together, these findings strengthen the hypothesis that RCO evolved adaptively. These findings do not imply that complex leaves are superior to simple ones, as both forms occur readily in nature. Rather, they highlight the potential for complex leaves to perform better under certain conditions that may have been relevant during the evolutionary history of the species that we studied here (Piazza et al. 2010). Complex leaves are more prevalent under lower mean annual temperatures (Royer and Wilf 2006). Therefore, leaf margin geometry may influence the interplay between temperature and photosynthesis. The effects of RCO and leaf complexity on photosynthesis are unlikely to involve stomatal density (Supplemental Fig. 7J) but might arise from conditional improvement in some or all of the following processes: gas exchange, due to reduced air boundary layer thickness, as in other complex leaves (Royer and Wilf 2006); light capture, owing to reduced shading by older leaves (Niklas 1988); and hydraulics, owing to vasculature properties in a complex blade (Dengler and Kang 2001). Notably, okra-leaf cotton shows increased photosynthesis and leaf complexity (Wells et al. 1986) together with altered LMI1 expression (Chang et al. 2016). Therefore, our findings highlight the potential to improve photosynthesis via modulating RCO/LMI1 activity.
Figure 4.
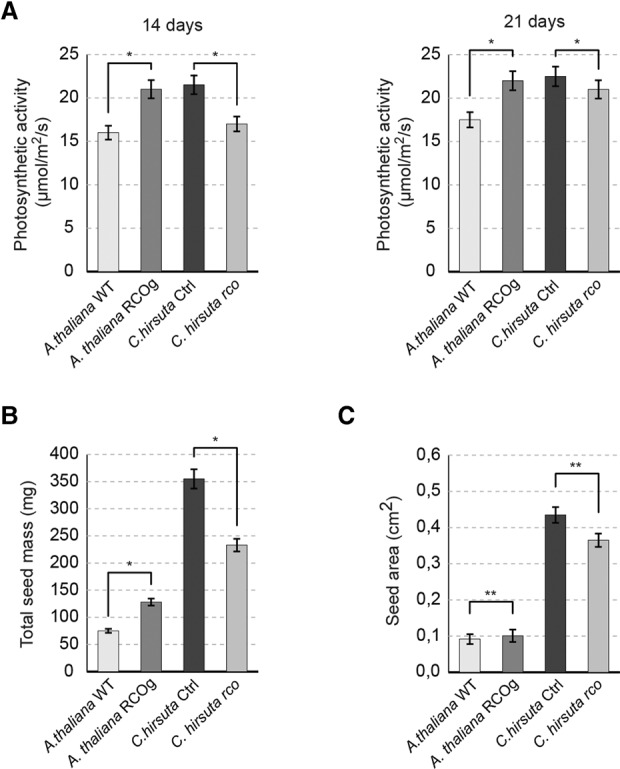
Alterations in RCO activity influence plant physiological performance. (A–C) Level of photosynthesis (CO2 absorbed per second, normalized to rosette area) after 14 and 21 d of growth (A), total seed mass per plant (B), and seed area (C) for A. thaliana wild type and RCOg and C. hirsuta control (Ctrl) and rco. Five replicates were measured per genotype at each time point to analyze photosynthetic activity. Total seed mass was estimated from five plants per genotype. The seed area was obtained from 50 seeds per genotype derived from five plants. Error bars represent standard deviation. A t-test was used to calculate significance. (N.S.) Not significant; (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
In conclusion, we show that neofunctionalization of an enhancer element coupled with targeted coding sequence diversification was instrumental in generating an altered leaf form with potential physiological benefits while at the same time minimizing pleiotropic effects. This type of trade-off—where molecular level functions are dampened to facilitate development of tissue- or organism-level traits—may be a pervasive feature of morphological evolution. For example, the activity of a key developmental enhancer in Ciona was found recently to be constrained by trade-offs between the specificity of gene activation and the level of transcriptional activity (Farley et al. 2015).
Materials and methods
Plants were cultivated in growth chambers under long-day (16-h d/8-h night) or short-day (8-h d/16-h night) conditions. A. thaliana and C. hirsuta were transformed using Agrobacterium tumefaciens floral dip transformations as in Hay et al. (2014). Histochemical detection of β-glucoronidase activity and subsequent visualization of samples were essentially according to Bilsborough et al. (2011). Selection tests on the promoter and coding sequences were conducted using a modified likelihood ratio test and phylogenetic analysis by maximum likelihood (PAML), respectively. Protein stability was determined after treatment with cycloheximide to inhibit protein synthesis. Gas exchange assays were conducted according to the LICOR 6400 xt manufacturer's protocol. A detailed description of the Materials and Methods is in the Supplemental Material.
Supplementary Material
Acknowledgments
We thank A. Hay, A. Hudson, C. Galinha, S. McCormick, and A. Runions for comments; N. Gompel for helpful discussions on pleiotropy; B. Walker and A. Weber for helpful discussion on photosynthesis; M. Alipour Kermani, L. Baumgarten, M. Cartolano, R. Pabst, B. Pieper, and U. Tartler for assistance; and P. Huijser for help with microscopy. This work was supported by Deutsche Forschungsgemeinschaft “Adaptomics” grants TS 229/1-1 (to M.T.) and Sonderforschungbereit (SFB) 680 (to M.T.), an EMBO Long-Term Fellowship (ALTF 502-2015 to R.A.M.), a core grant from the Max Planck Society (to M.T.), and a US National Science Foundation Plant Genome Research award (1238731 to C.D.B.). M.T. also acknowledges support of the Cluster of Excellence on Plant Sciences. F.V., R.A.M., and M.H. contributed equally to this work, and the order was determined by drawing lots. F.V., R.A.M., and M.H. designed and performed experiments and collected data. D.A.F. conducted evolutionary sequence analyses with input from C.D.B. and M.H. All authors analyzed the data. F.V., R.A.M., M.H., and M.T. wrote the manuscript with input from the other authors. M.T. designed and directed the study.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.290684.116.
Freely available online through the Genes & Development Open Access option.
References
- Arnoult L, Su KF, Manoel D, Minervino C, Magrina J, Gompel N, Prud'homme B. 2013. Emergence and diversification of fly pigmentation through evolution of a gene regulatory module. Science 339: 1423–1426. [DOI] [PubMed] [Google Scholar]
- Bar M, Ori N. 2014. Leaf development and morphogenesis. Development 141: 4219–4230. [DOI] [PubMed] [Google Scholar]
- Bilsborough GD, Runions A, Barkoulas M, Jenkins HW, Hasson A, Galinha C, Laufs P, Hay A, Prusinkiewicz P, Tsiantis M. 2011. Model for the regulation of Arabidopsis thaliana leaf margin development. Proc Natl Acad Sci 108: 3424–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd JL, Skove SL, Rouanet JP, Pilaz LJ, Bepler T, Gordan R, Wray GA, Silver DL. 2015. Human-chimpanzee differences in a FZD8 enhancer alter cell-cycle dynamics in the developing neocortex. Curr Biol 25: 772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36. [DOI] [PubMed] [Google Scholar]
- Chan YF, Marks ME, Jones FC, Villarreal G Jr, Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, et al. 2010. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327: 302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Fang L, Zhu Y, Wu H, Zhang Z, Liu C, Li X, Zhang T. 2016. Insights into interspecific hybridization events in allotetraploid cotton formation from characterization of a gene regulating leaf shape. Genetics 204: 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengler N, Kang J. 2001. Vascular patterning and leaf shape. Curr Opin Plant Biol 4: 50–56. [DOI] [PubMed] [Google Scholar]
- Farley EK, Olson KM, Zhang W, Brandt AJ, Rokhsar DS, Levine MS. 2015. Suboptimization of developmental enhancers. Science 350: 325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N, Erezyilmaz DF, McGregor AP, Wang S, Payre F, Stern DL. 2011. Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature 474: 598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givnish TJ. 1978. Ecological aspects of plant morphology: leaf form in relation to environment. Acta Biotheoretica 27: 83–142. [Google Scholar]
- Gompel N, Prud'homme B, Wittkopp PJ, Kassner VA, Carroll SB. 2005. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433: 481–487. [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. 2006. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat Genet 38: 942–947. [DOI] [PubMed] [Google Scholar]
- Hay AS, Pieper B, Cooke E, Mandakova T, Cartolano M, Tattersall AD, Ioio RD, McGowan SJ, Barkoulas M, Galinha C, et al. 2014. Cardamine hirsuta: a versatile genetic system for comparative studies. Plant J 78: 1–15. [DOI] [PubMed] [Google Scholar]
- Indjeian VB, Kingman GA, Jones FC, Guenther CA, Grimwood J, Schmutz J, Myers RM, Kingsley DM. 2016. Evolving new skeletal traits by cis-regulatory changes in bone morphogenetic proteins. Cell 164: 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T, Montgomery SB, Nica AC, Dermitzakis ET. 2011. Epistatic selection between coding and regulatory variation in human evolution and disease. Am J Hum Genet 89: 459–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklas KJ. 1988. The role of phyllotactic pattern as a developmental constraint on the interception of light by leaf surfaces. Evolution 42: 1–16. [DOI] [PubMed] [Google Scholar]
- Piazza P, Bailey CD, Cartolano M, Krieger J, Cao J, Ossowski S, Schneeberger K, He F, de Meaux J, Hall N, et al. 2010. Arabidopsis thaliana leaf form evolved via loss of KNOX expression in leaves in association with a selective sweep. Curr Biol 20: 2223–2228. [DOI] [PubMed] [Google Scholar]
- Prud'homme B, Gompel N, Rokas A, Kassner VA, Williams TM, Yeh SD, True JR, Carroll SB. 2006. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature 440: 1050–1053. [DOI] [PubMed] [Google Scholar]
- Rast-Somssich MI, Broholm S, Jenkins H, Canales C, Vlad D, Kwantes M, Bilsborough G, Dello Ioio R, Ewing RM, Laufs P, et al. 2015. Alternate wiring of a KNOXI genetic network underlies differences in leaf development of A. thaliana and C. hirsuta. Gene Dev 29: 2391–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Jikomes N, Kassner VA, Carroll SB. 2011. Evolutionary origin of a novel gene expression pattern through co-option of the latent activities of existing regulatory sequences. Proc Natl Acad Sci 108: 10036–10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Patel NH, Hinman VF. 2015. Unraveling the tangled skein: the evolution of transcriptional regulatory networks in development. Annu Rev Genomics Hum Genet 16: 103–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer D, Wilf P. 2006. Why do toothed leaves correlate with cold climates? Gas exchange at leaf margins provides new insights into a classic paleotemperature proxy. Int J Plant Sci 167: 11–18. [Google Scholar]
- Shleizer-Burko S, Burko Y, Ben-Herzel O, Ori N. 2011. Dynamic growth program regulated by LANCEOLATE enables flexible leaf patterning. Development 138: 695–704. [DOI] [PubMed] [Google Scholar]
- Sicard A, Thamm A, Marona C, Lee YW, Wahl V, Stinchcombe JR, Wright SI, Kappel C, Lenhard M. 2014. Repeated evolutionary changes of leaf morphology caused by mutations to a homeobox gene. Curr Biol 24: 1880–1886. [DOI] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. 2008. The loci of evolution: how predictable is genetic evolution? Evolution 62: 2155–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer A, Zhao Q, Ross-Ibarra J, Doebley J. 2011. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet 43: 1160–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar D, Berthelot C, Aldridge S, Rayner TF, Lukk M, Pignatelli M, Park TJ, Deaville R, Erichsen JT, Jasinska AJ, et al. 2015. Enhancer evolution across 20 mammalian species. Cell 160: 554–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad D, Kierzkowski D, Rast MI, Vuolo F, Dello Ioio R, Galinha C, Gan X, Hajheidari M, Hay A, Smith RS, et al. 2014. Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science 343: 780–783. [DOI] [PubMed] [Google Scholar]
- Wells R, Meredith WR, Williford JR. 1986. Canopy photosynthesis and its relationship to plant productivity in near-isogenic cotton lines differing in leaf morphology. Plant Physiol 82: 635–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WS, Nielsen R. 2004. Detecting selection in noncoding regions of nucleotide sequences. Genetics 167: 949–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


