Xu et al. show that the WD40 domain of Gemin5 is both necessary and sufficient for binding the Sm site of pre-snRNAs. They also determined the crystal structures of the WD40 domain of Gemin5 in complex with the Sm site or m7G cap of pre-snRNA.
Keywords: SMN complex, WD40 domain, X-ray crystal structure
Abstract
In cytoplasm, the survival of motor neuron (SMN) complex delivers pre-small nuclear RNAs (pre-snRNAs) to the heptameric Sm ring for the assembly of the ring complex on pre-snRNAs at the conserved Sm site [A(U)4–6G]. Gemin5, a WD40 protein component of the SMN complex, is responsible for recognizing pre-snRNAs. In addition, Gemin5 has been reported to specifically bind to the m7G cap. In this study, we show that the WD40 domain of Gemin5 is both necessary and sufficient for binding the Sm site of pre-snRNAs by isothermal titration calorimetry (ITC) and mutagenesis assays. We further determined the crystal structures of the WD40 domain of Gemin5 in complex with the Sm site or m7G cap of pre-snRNA, which reveal that the WD40 domain of Gemin5 recognizes the Sm site and m7G cap of pre-snRNAs via two distinct binding sites by respective base-specific interactions. In addition, we also uncovered a novel role of Gemin5 in escorting the truncated forms of U1 pre-snRNAs for proper disposal. Overall, the elucidated Gemin5 structures will contribute to a better understanding of Gemin5 in small nuclear ribonucleic protein (snRNP) biogenesis as well as, potentially, other cellular activities.
The spliceosome orchestrates the removal of introns from pre-mRNAs and processes them into distinct mRNA isoforms, contributing to an enormous repertoire of diverse mature mRNAs poised for translation into eukaryotic proteins (Chen and Moore 2014; Matera and Wang 2014). The core of this spliceosomal macromolecular machine is built from small nuclear ribonucleic proteins (snRNPs), which consist of small nuclear RNAs (snRNAs) and their respective associated proteins (Scotti and Swanson 2016). The biogenesis of major snRNPs requires the export of their pre-snRNA subcomponents from the nucleus for processing in the cytoplasm followed by the import of the mature snRNAs back into the nucleus (Will and Luhrmann 2001; Matera and Wang 2014). The maturation of the snRNAs in the cytoplasm consists of a series of tightly regulated, strictly ordered molecular events highlighted by the assembly of the Sm ring and hypermethylation of the m7G cap (Matera and Wang 2014). It has been reported that the survival of motor neuron (SMN) complex facilitates the assembly of the Sm ring complex on the Sm site [A(U)4–6G] of pre-snRNAs and safeguards against aberrant snRNP biogenesis and splicing-associated human syndromes, such as spinal muscular atrophy and cancers (Pellizzoni et al. 2002; Golembe et al. 2005; Matera and Wang 2014; Chabot and Shkreta 2016).
Gemin5, a core component of the SMN complex, is thought to confer the specificity of SMN-mediated pre-snRNA recognition. It has been proposed to be the pre-snRNA-binding and delivery component of the SMN complex (Battle et al. 2006b) and exhibits strict sequence specificity for the Sm site of pre-snRNAs (Yong et al. 2010). By means of deletion mapping, it has been suggested that the WD40 domain of Gemin5 mediates the specific binding to the Sm site and the adjacent variable 3′-terminal stem–loop structure of the pre-snRNAs (Lau et al. 2009). Surprisingly, the SMN complex is also capable of associating with a truncated form of U1 pre-snRNAs (U1-tfs), which lacks the Sm site but retains the m7G cap structure and shunts the U1-tfs into processing bodies (P bodies) for degradation (Ishikawa et al. 2014). Moreover, another study suggests that the WD40 domain of Gemin5 associates with the m7G cap structure (Bradrick and Gromeier 2009), a signature 5′ end cap cotranscriptionally added to the 5′ end of all transcripts from RNA polymerase II. Hence, although the significance of the unique binding event between the WD40 domain of Gemin5 and the Sm site of pre-snRNAs is well established for snRNA maturation (Matera et al. 2007; Muller-McNicoll and Neugebauer 2013), the biological significance of Gemin5's binding to pre-snRNAs via the m7G cap is still unclear.
WD40 domain-containing proteins participate in a broad spectrum of cellular processes, including signal transduction, cytoskeletal assembly, cell cycle control, chromatin dynamics, and transcription regulation. Malfunctions of these proteins contribute to the pathogenesis of various human diseases, such as cancer (Li and Roberts 2001). The WD40 domain serves as a rigid scaffold for protein–protein/DNA interactions and the coordination of downstream events, such as ubiquitinylation and histone methylation (Xu and Min 2011). To understand how the WD40 domain of Gemin5 recognizes the Sm site of pre-snRNAs and the m7G cap, we determined the crystal structures of the WD40 domain of Gemin5 in complex with either the Sm site RNA or the m7GpppG cap. We also found that Gemin5 binds U1-tfs via their m7G caps in vivo and shunts these defective pre-U1 snRNAs into P bodies for controlled RNA degradation. Overall, we propose a mechanistic model to explain the roles of cytoplasmic Gemin5 in delivering intact pre-snRNAs into the snRNP biogenesis pathway and defective pre-snRNAs for proper disposal.
Results
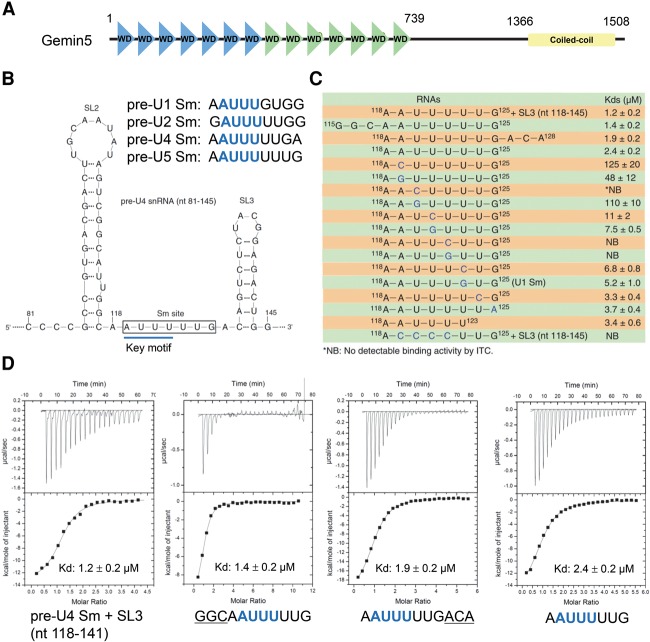
The WD40 domain of Gemin5 binds to the Sm site of pre-snRNAs
The SMN complex is indispensable for the cytoplasmic processing and maturation of snRNPs, the core components of the eukaryotic spliceosome, while Gemin5 is the only known component of the SMN complex that directly recognizes pre-snRNAs (Gubitz et al. 2002; Yong et al. 2010). Gemin5 is highly conserved in metazoans and consists of a WD40 domain at its N terminus and a coiled-coil domain at its C terminus (Fig. 1A; Supplemental Fig. S1). Deletion mapping analysis has suggested that the WD40 domain of Gemin5 is both necessary and sufficient for specific binding to pre-snRNAs via the conserved Sm site and the adjacent variable 3′-terminal stem–loop structure of pre-snRNAs (Yong et al. 2004, 2010), which is found only in snRNAs (Fig. 1B). To further investigate Gemin5-mediated recognition of pre-snRNAs, we measured by isothermal titration calorimetry (ITC) and compared the binding affinities and thermodynamic parameters of the human Gemin5 WD40 domain with different RNAs derived from U4 pre-snRNA (Fig. 1C; Supplemental Table S1). Our binding results displayed that a fragment of U4 pre-snRNA (nucleotides 118–125: 118AAUUUUUG125) binds to the WD40 domain of Gemin5 with a dissociation constant (Kd) of 2.4 µM (Fig. 1C, row 4), and the addition of extra nucleotides at the 5′ or 3′ end, including adding the 3′ stem–loop structure, enhanced their binding affinities only mildly, ∼1.3-fold to 2.0-fold (Fig. 1C [rows 1–4], D). Therefore, our binding data revealed that the Sm site of U4 pre-snRNA is sufficient for binding to the WD40 domain of Gemin5.
Figure 1.
The AUUU motif within the Sm site of pre-snRNAs is the major determinant for its binding interaction with Gemin5. (A) Domain architecture of full-length human Gemin5 protein. (B) The secondary structure of the human U4 pre-snRNA fragment (nucleotides 81–145). The Sm site of U4 pre-snRNA (119–125) is boxed. Sequence alignments of the Sm sites from different pre-snRNAs are presented at the top. The key 119AUUU122 motif within the different pre-snRNAs is colored in blue. (SL) Stem–loop of U4 pre-snRNA. (C) The binding affinities of the Gemin5 WD40 domain to different fragments and mutants of U4 pre-snRNA. (D) Representative ITC binding curves for the binding of the Gemin5 WD40 domain (amino acids 1–739) to different RNAs derived from U4 pre-snRNA. Derived Kds and respective standard deviations are also indicated. Additional nucleotides of U4 pre-snRNA preceding and following the Sm site are underlined.
Crystal structure of the WD40 domain of Gemin5 in complex with the Sm site RNA (118AAUUUUUG125)
To gain insights into the molecular mechanism of the Sm site recognition by the WD40 domain of Gemin5, we determined the crystal structures of the human Gemin5 WD40 domain alone and in complex with the Sm site RNA (118AAUUUUUG125) at 1.65 Å and 1.80 Å resolutions, respectively (Fig. 2A,B; Table 1).
Figure 2.
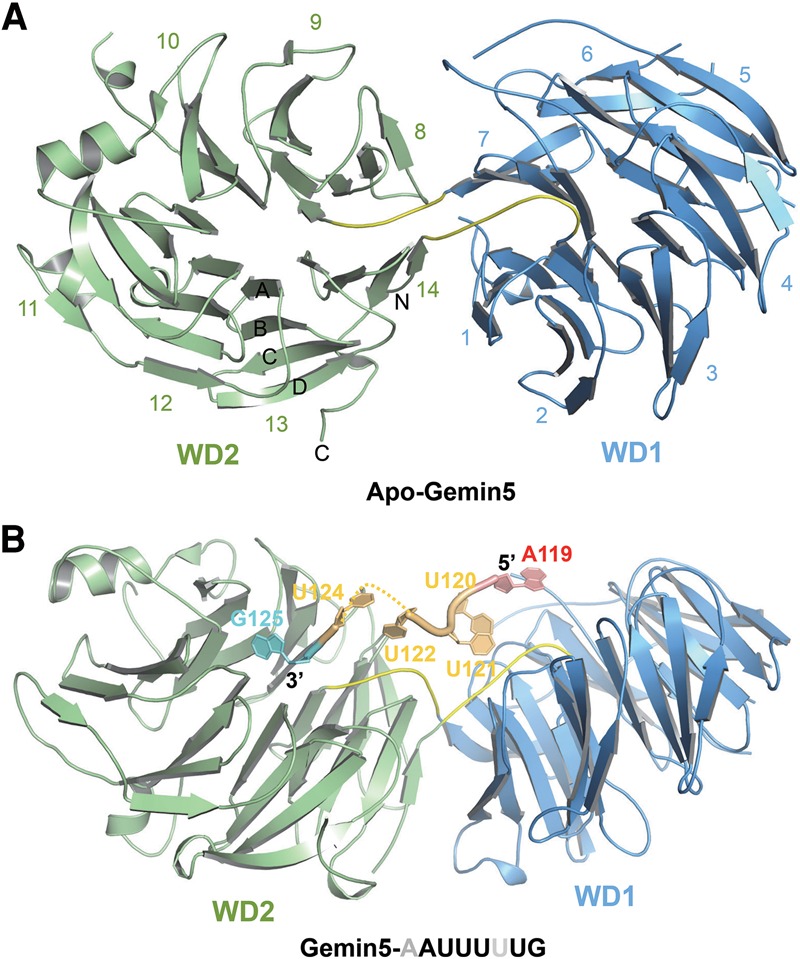
Crystal structures of human Gemin5 in apo form or in complex with the Sm site RNA. (A) The Gemin5 WD40 domain (amino acids 1–739), which contains two seven-bladed β propellers (colored in blue and green, respectively). Loops linking the two propellers are colored in yellow. (B) Gemin5 in complex with a Sm site RNA of U4 pre-snRNA (118AAUUUUUG125). Gemin5 is colored the same as in A. (Red) Adenosine; (orange) uridine; (cyan) guanosine. The missing U123 is denoted by dashes.
Table 1.
Data collection and refinement statistics
The structure of the WD40 domain of Gemin5 includes 14 WD40 repeats, which form two seven-bladed β propellers (named WD1 and WD2, respectively), and both WD40 β propellers of Gemin5 adopt the canonical WD40 fold (Fig. 2A; Smith et al. 1999; Stirnimann et al. 2010; Xu and Min 2011). The two β propellers are connected by two interpropeller loops, with the first repeat of WD1 packing against the last repeat of WD2, and the last repeat of WD1 packing against the first repeat of WD2 (Fig. 2A). The top surfaces of these two β propellers, tilted slightly against each other, face in the same general direction and form a positively charged concave. The cocrystal structure of Gemin5 and the Sm site RNA of the U4 (118AAUUUUUG125) complex revealed that this positively charged concave accommodates the Sm site RNA (Fig. 2B; Supplemental Fig. S2). The overall crystal structure of the WD40 domain of Gemin5 did not change significantly upon binding to the Sm site RNA, as the root mean square deviation (RMSD) between the backbones of the two Gemin5 structures is just ∼0.2 Å (Supplemental Fig. 3). In the Gemin5–118AAUUUUUG125 complex structure, the A119–U122 and U124–G125 nucleotides were modeled into electron density. A118 and most of U123 were not visible in the electron density map (Supplemental Fig. S4A).
A119–U122 is accommodated in the positively charged concave of the WD40 domain of Gemin5, while U124 points to the solvent, and G125 resides in a pocket of the top surface of WD2, respectively (Fig. 3A). The purine ring of the adenosine (A119 in U4 pre-snRNA; the first nucleotide in the Sm site) stacks with Tyr15 of Gemin5, and the amino group of its purine ring forms two water-mediated hydrogen bonds with the side chain of Glu197 of Gemin5 (Fig. 3B). Both Tyr15 and Glu197 are evolutionarily conserved in Gemin5 (Supplemental Fig. S1), and mutating Tyr15 or Glu197 to alanine reduced the binding affinity of Gemin5 to 118AAUUUUUG125 RNA by ∼7.5-fold and 16.8-fold, respectively (Fig. 4A). On the other hand, substitution of the A119 for C presumably would introduce electrostatic repulsion with Tyr15 of Gemin5 between their carbonyl groups, while substitution of the adenosine 119 for G (guanosine) or U (uridine) would introduce electrostatic repulsion with Glu197 of Gemin5 (Supplemental Fig. S5). The substitution of A119 with G or C dramatically reduced the binding of 118AAUUUUUG125 to Gemin5, although we could not exclude the possibility that the mutant RNA fragment may adopt a different conformation to bind Gemin5 (Fig. 1C). Hence, an adenosine nucleotide is strongly preferred in this binding pocket for the recognition of 118AAUUUUUG125 by Gemin5.
Figure 3.
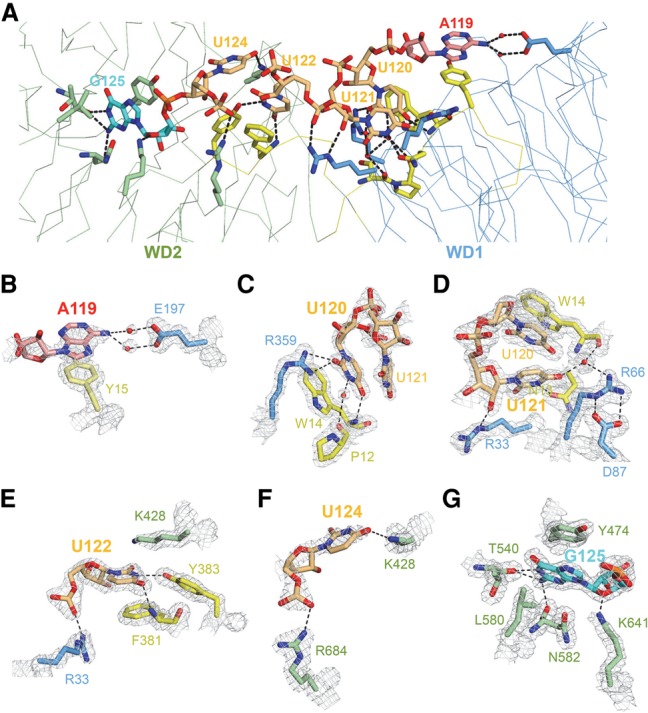
A relatively surface-exposed binding site characterized by respective base-specific interactions dictates Gemin5-mediated recognition of the Sm site of pre-snRNA. (A) Detailed depiction of intermolecular interactions between key residues of Gemin5 and the 119AUUUUUG125 Sm site RNA, with the two seven-bladed WD40 β propellers and the loops linking them colored in blue, green, and yellow, respectively. (Red) Adenosine; (orange) uridine. Gemin5 is shown as a ribbon. The RNA nucleotides and key residues of Gemin5 are shown as sticks. Key intermolecular interactions of Gemin5 residues with A119 (B), U120 (C), U121 (D), U122 (E), U124 (F), and G125 (G) of the Sm site RNA. In B–G, the models are overlaid on a 2Fo–Fc σ-weighted map contoured at 1.5 σ (gray cage).
Figure 4.
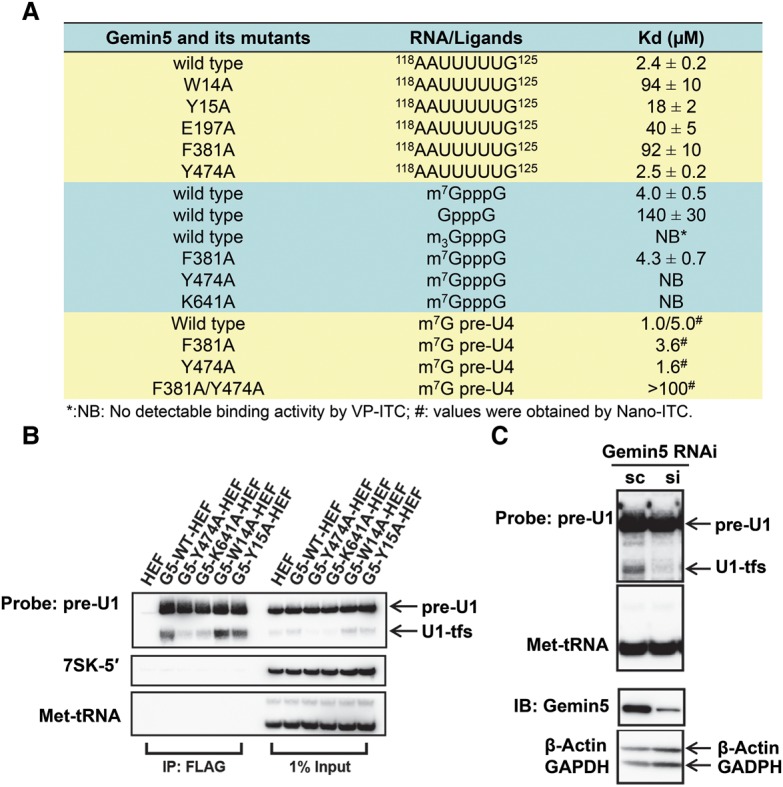
Gemin5-mediated recognition of the m7G cap of pre-snRNAs represents a novel independent binding event with unique cellular functions. (A) The binding affinities of the Gemin5 WD40 domain and its mutants toward the Sm site RNA, m7GpppG, GpppG, m3GpppG, and m7G-capped full-length U4 pre-snRNA, respectively. The Kd values were measured by ITC analysis, and the standard deviations of the Kd values are also indicated. (B) In vivo binding of Gemin5 wild type (G5-WT), Gemin5 m7G cap mutants (G5-Y474A and G5-K641A), and Gemin5 Sm site mutants (G5-W14A and G5-Y15A) to pre-U1 and U1-tfs, respectively. G5-WT-HEF (HA-TEV-Flag)-associated, G5-W14A-HEF-associated, G5-Y15A-HEF-associated, G5-Y474A-HEF-associated, and G5-K641A-HEF-associated RNA–protein complexes were prepared by immunoprecipitation and separated by SDS-PAGE. RNAs were detected by denaturing urea-PAGE–Northern blot analysis with the probes as indicated. (IP: Flag) The isolated RNA–protein complex; (1% input) the cell extract used for the immunoprecipitation. 7SK-5′ and Met-tRNA were employed as loading controls. (C) Evaluation of the impact of Gemin5 knockdown on U1-tfs in cells. Stealth siRNA against Gemin5 (si) or scRNA (sc; negative control) was transfected into 293T cells. The levels of pre-U1 and U1-tfs were examined by Northern blot analysis with Met-tRNA as the loading control. The knockdown of Gemin5 was confirmed by immunoblot (IB) with β-actin and GADPH as the loading controls.
The first uridine of the Sm site RNA (U120 in U4 pre-snRNA) stacks with the indole ring of Trp14, and one carbonyl group from its pyrimidine ring also forms a hydrogen bond with the main chain NH group of Trp14 (Fig. 3C). U120 also forms several hydrogen bonds with Ser11, Pro12, Trp14, and Arg359 of Gemin5 (Fig. 3C). Substitution of U120 for C, which is of similar size, would disrupt its hydrogen bond with Trp14 of Gemin5 and introduce steric clashes (Supplemental Fig. S6A). Similarly, substitution of U120 for a purine base (G or A) would disrupt the hydrogen bond network and introduce steric clashes with surrounding residues. Consistent with these findings, mutating U120 to either C or G or mutating Trp14 to alanine significantly diminished the binding ability of Gemin5 to the Sm site RNA (Figs. 1C, 4A; Supplemental Table S1). Taken together, U120, the first uridine of the Sm site RNA, is snugly accommodated in a uridine-specific binding pocket of Gemin5.
The second uridine (U121) of the Sm site RNA does not show stacking interactions with Gemin5 but still forms hydrogen bonds with Gemin5 through its pyrimidine and ribosyl moieties (Fig. 3D). The pyrimidine ring of U121 forms a direct hydrogen bond and a water-mediated hydrogen bond with Asn13 and Tyr14/Arg66 of Gemin5, respectively, while the ribosyl ring of U121 forms another hydrogen bond with the side chain of Arg33 of Gemin5 (Fig. 3D). Uridine appears to be favored at this position compared with other bases: Substitution of U121 by C would cause a steric clash with the side chain of Arg66 and disrupt the water-mediated hydrogen bond between Arg66 and the pyrimidine ring of U121 (Supplemental Fig. S6B), as would substitution of U121 by a purine nucleotide. Accordingly, mutation of U121 of the Sm site RNA to either C or G decreased its binding affinity to Gemin5 by about threefold and fivefold, respectively (Fig. 1C; Supplemental Table S1).
The third uridine (U122) of the Sm site RNA stacks with Phe381 of Gemin5 on one side and is flanked by the side chain of Lys428 on the other side (Fig. 3E). Furthermore, the base of U122 also forms two hydrogen bonds with the main chain NH group of Phe381 and the side chain hydroxylate group of Tyr383, respectively, while the phosphate moiety of U122 forms a hydrogen bond with the side chain of Arg33 of Gemin5 (Fig. 3E). Substitution of U122 for other nucleotides would disrupt those base-mediated hydrogen bonds with Phe381 and Tyr383 of Gemin5 and introduce electrostatic repulsion (Supplemental Fig. S6C). Therefore, the two base-mediated hydrogen bonds with Phe381 and Tyr383 are the structural determinants for sequence selectivity at this position (Fig. 3E). In accordance with this notion, our binding data indicate that mutation of U122 to either C or G abrogated the binding of the Sm site RNA to Gemin5 (Fig. 1C; Supplemental Table S1), while mutating Phe381 to alanine severely diminished the binding affinity between Gemin5 and Sm site RNA (Fig. 4A), underlining the importance of the third uridine of the Sm site RNA in Gemin5-mediated binding interaction with pre-snRNAs.
In contrast to the A119–U122 residues of the Sm site RNA, U123–G125 only make minor contributions to the Sm RNA binding. The majority of U123 is not visible in the electron density map, except its phosphate group, which does not directly interact with Gemin5. U124 points to the solvent and forms two hydrogen bonds with Lys428 and Arg684 via its base and phosphate groups, respectively (Fig. 3F). Although G125 of the Sm site RNA (118AAUUUUUG125) resides in a binding pocket at the top surface of WD2 (Fig. 3G), it seems that this binding pocket could accommodate only a terminal nucleotide, while G125 is an internal nucleotide in U4 pre-snRNA, implying that this G125 binding event is probably artificial. To investigate whether G125 of the Sm site RNA could also bind to Gemin5 as an internal nucleotide, we cocrystallized Gemin5 with a longer U4 pre-snRNA fragment, 118AAUUUUUGAG127, in which G125 becomes an internal nucleotide. Consistently, the RNA binding mode in this new complex is conserved with that of the Gemin5–118AAUUUUUG125 complex structure, except that G125 does not contact Gemin5 anymore, as expected (Supplemental Fig. S4B). In the new complex, the extra nucleotides at the 3′ end of G125 of the U4 pre-snRNA prevented the G125 residue from being positioned into that WD2 pocket of Gemin5 anymore. Since the accommodation of G125 by WD2 affects the orientation of 118AAUUUUUG125, it is possible that the longer U4 fragment may adopt an alternative conformation to contact Gemin5.
To evaluate the relative significance of the U123–G125 residues of U4 pre-snRNA in the interaction with Gemin5, we replaced U123, U124, or G125 of the Sm site RNA (118AAUUUUUG125) with other nucleotides and found that these mutations decreased the Sm site RNA's binding affinity to Gemin5 only slightly, by ∼1.4-fold to 3.3-fold (Fig. 1C), suggesting that U123–G125 make only minor contributions to the Gemin5 binding. We also tested the binding of the shorter Sm site RNA 118AAUUUU123 to Gemin5 and found that its binding affinity to Gemin5 was slightly weaker than 118AAUUUUUG125 (3.4 vs. 2.4 µM). In contrast, a Sm site RNA mutant (nucleotides 118–145) with the 119AUUU122 motif replaced with 119CCCC122 did not bind to Gemin5 at all (Fig. 1C). Taken together, our structural and mutagenesis data demonstrate that the oligouridine sequence within the Sm site of pre-snRNAs is a strong determinant of recognition by the Gemin5 WD40 domain.
In addition to the Gemin5–RNA complexes, only one WD40–DNA complex (DDB2) and two WD40–RNA complexes (Cwf17 and its human ortholog, U5-40K) have been reported (Supplemental Fig. S7; Scrima et al. 2008; Yan et al. 2015; Agafonov et al. 2016). Unlike Gemin5, DDB2 uses its top surface, and Cwf17 uses its side surface to contact the backbones of the respective nucleic acid ligands (Scrima et al. 2008; Yan et al. 2015). In contrast to the WD40 domain of Gemin5, neither DDB2 nor Cwf17 binds to DNA or RNA in a sequence-dependent manner.
Lau et al. (2009) reported that Trp286 of Gemin5 is key in recognizing the Sm site of pre-snRNAs. In our Gemin5–RNA complex crystal structures, Trp286 is buried inside the WD40 domain rather than solvent-exposed, so it does not directly contact any nucleotide but contributes to the formation of the WD1 β propeller. In our hands, the W286A mutant of Gemin5 (1–739) was insoluble, in contrast to the other mutants that we purified in this study. We speculate that the previously reported W286A mutant may have been similarly misfolded and therefore incapable of binding.
Crystal structure of the WD40 domain of Gemin5 in complex with m7GpppG
A previous study suggested that the immunoprecipitated Gemin5 protein from 293T cells associates either directly or indirectly with m7GpppG, a m7G cap mimic, but not GpppG (Bradrick and Gromeier 2009). Our ITC binding results show that the WD40 domain of Gemin5 exhibited a binding affinity to m7GpppG 35-fold stronger than to GpppG (4.0 vs. 140 µM) (Fig. 4A). Additionally, Gemin5's binding activity toward m3GpppG was undetectable by ITC (Fig. 4A). Collectively, our ITC binding data indicate that the WD40 domain of Gemin5 binds directly to m7G-containing RNAs with high selectivity.
We next determined the crystal structures of the WD40 domain of Gemin5 in complex with m7GpppG or GpppG at 1.85 Å and 1.80 Å resolutions, respectively (Table 1). The two complex structures could be readily superimposed with the apo form of Gemin5 with an RMSD of ∼0.3 Å, indicating that the binding of m7GpppG or GpppG to Gemin5 does not cause significant overall changes of Gemin5 (Supplemental Fig. S3). In both crystal structures, only one nucleotide (m7G or G) was resolved, and guanine was accommodated in a deep pocket at the WD2 top surface (Fig. 5A; Supplemental Figs. S4C, S8).
Figure 5.
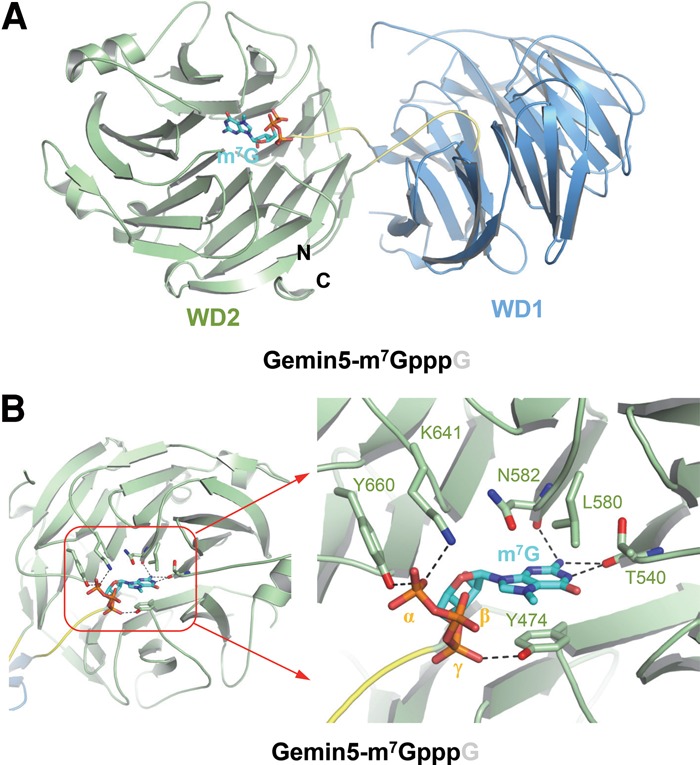
Crystal structure of the Gemin5 WD40 domain in complex with the m7G cap of pre-snRNA. (A) Gemin5 in complex with m7GpppG (cyan). The second guanosine was not resolved. (B) A detailed depiction of intermolecular interactions between key residues of Gemin5 and m7GpppG. m7G (cyan sticks) is accommodated into the top surface of the WD2 of Gemin5 (green). The three phosphate moieties (orange) following the m7G are denoted as γ, β, and α, respectively. Gemin5 is colored the same as in Figure 2A. The m7G-binding residues of Gemin5 (green) and the m7G (cyan) are shown as sticks. Hydrogen bonds are shown as black dashes.
The purine ring of m7G was sandwiched between the aromatic ring of Tyr474 and the side chains of Leu580 and Asn582 (Fig. 5B; Supplemental Fig. S9A), similar to the guanine of G125 in the Gemin5–118AAUUUUUG125 complex structure (Figs. 3G, 5B). Similar stacking has also been observed in other m7G or m3G RNA–protein complex structures, including CBC20–m7GpppG (Mazza et al. 2002), EIF4E–m7GpppG (Marcotrigiano et al. 1997), PARN–m7GpppG (Wu et al. 2009), Dcps–m7GpppG (Gu et al. 2004), and snurportin–m3GpppG (Supplemental Fig. S9B–F; Strasser et al. 2005). In addition to the stacking interaction, the m7G cap also forms several guanine-specific hydrogen bonds with Gemin5, including those between its purine 2 amino group and the backbone carbonyl groups of Asn582 and Thr540 and the hydrogen bond between its purine N1 group and the carbonyl group of Thr540 (Fig. 5B). Interestingly, the methyl moiety attached to the purine N7 of the m7G cap is in proximity to the acidic hydroxyl group of Tyr474, which would introduce an extra electrostatic interaction between the pronounced positive charge at the methylated N7 of m7GpppG and the side chain of Tyr474 (Fig. 5B; Supplemental Fig. S9A). This kind of methylation-mediated electrostatic interaction has also been observed in the CBC20–m7GpppG structure, in which the methyl moiety of the m7G cap is proximal to the hydroxyl group of Tyr43 in CBC20 (Supplemental Fig. S9B; Mazza et al. 2002). This additional electrostatic interaction would explain why Gemin5 preferentially binds to m7GpppG over GpppG (Figs. 4A, 5B; Supplemental Fig. S9A). In contrast, the 5′ m3G cap, doubly methylated at the 2 amino position (N2) in addition to single methylation at the N7position, would not only suffer the loss of the two hydrogen bonds with Thr540 and Asn582 of Gemin5 but also experience steric clashes with the backbone carbonyl groups of these amino acid residues. Accordingly, we did not detect any binding between Gemin5 and m3GpppG (Fig. 4A).
In addition to the m7G cap, the three phosphate groups of m7G also interact with Gemin5 (Fig. 5B). Specifically, the first (γ) and the third (α) phosphate groups form hydrogen bonds with the side chain hydroxyl groups of Tyr474 and Tyr660 of Gemin5, respectively. The α phosphate group forms a salt bridge with the side chain of Lys641 (Fig. 5B). Furthermore, the phosphate recognition mode in the Gemin5–m7GpppG complex structure is conserved in the Gemin5–GpppG complex structure (Supplemental Fig. S10). By evaluating the individual roles of the above key residues of Gemin5 in recognizing the m7GpppG cap, we found that neither the Y474A nor the K641A Gemin5 single mutant bound to the m7GpppG cap based on ITC analysis (Fig. 4A), underlining the essentiality of both the guanine- and phosphate-binding interactions in Gemin5-mediated m7GpppG cap recognition.
The 5′ m7GpppG cap and the Sm site of pre-snRNAs bind independently to Gemin5
Our structural and binding studies revealed that the m7GpppG cap and the Sm site RNA bound to two different sites of the WD40 domain of Gemin5. Given the well-established role of Gemin5-mediated pre-snRNA binding in the snRNP biogenesis pathway, we next aimed to investigate how these two binding events affect each other. Our ITC analysis shows that, even though the Y474A mutant of Gemin5 lost the m7GpppG cap-binding ability, it retained affinity to the Sm site RNA comparable with that of wild-type Gemin5 (Fig. 4A). On the other hand, the F381A mutant of Gemin5 displayed a significantly diminished affinity toward Sm site RNA compared with wild-type Gemin5 but bound to the m7GpppG cap almost as strongly as wild-type Gemin5 (Fig. 4A), implicating that these two binding events are largely independent.
To corroborate these findings, we synthesized full-length U4 pre-snRNA and added a 5′ m7G cap to mimic native m7G-capped U4 pre-snRNA. Our ITC binding results can be fit in a dual binding site model between the WD40 domain of Gemin5 and the full-length m7G-capped U4 pre-snRNA, with Kds of 1.0 and 5.0 µM (Supplemental Fig. S11A). Hence, Gemin5's interaction with the full-length U4 pre-snRNA exhibited similar binding affinities compared with respective affinities of Gemin5's binding to the Sm site RNA or the m7GpppG cap individually (Figs. 1C, 4A). Furthermore, the Sm site-binding mutant F381A of Gemin5 bound to m7G-capped U4 pre-snRNA with a Kd of 3.6 µM, similar to that of wild-type Gemin5's binding to m7GpppG (Fig. 4A; Supplemental Fig. S11B). On the other hand, the 5′ m7G-binding site mutant Y474A of Gemin5 bound to 5′ m7G-capped U4 pre-snRNA with a Kd of 1.6 µM, comparable with that of wild-type Gemin5's binding to Sm site RNA (Fig. 1C; Supplemental Fig. S11C). Moreover, the double mutant Y474A/F381A of Gemin5 displayed very weak affinity toward U4 pre-snRNA (Kd > 100 µM) (Supplemental Fig. S11D). Taken together, these results suggest that the binding of the 5′ m7GpppG cap and the binding of the Sm site RNA to wild-type Gemin5 are two virtually independent binding events. In addition, the Sm site and the m7G cap are the only two important sites within the full-length pre-snRNAs that are directly recognized by the WD40 domain of Gemin5.
In addition to the WD40 domain, Gemin5 also contains a C-terminal domain predicted to contain a coiled-coil region. To investigate whether the C-terminal domain binds to pre-snRNAs, we purified the C-terminal domain of Gemin5 (amino acids 843–1508) and found that it behaved as a polymer (∼600 kDa) in solution (Supplemental Fig. S12), suggesting that the coiled-coil domain could mediate homopolymerization of Gemin5. We then performed an EMSA gel shift assay to examine its binding to full-length U4 pre-snRNA. In contrast to its N-terminal WD40 domain, the C-terminal domain of Gemin5 did not display any detectable binding toward full-length U4 pre-snRNA (Supplemental Fig. S12).
Gemin5 recognizes m7G-capped pre-snRNAs in vivo
In order to validate the snRNA-binding partners of Gemin5 in vivo, we constructed a doxycycline-inducible cell line expressing HEF (HA-TEV-Flag)-tagged Gemin5 and purified the Gemin5-associated complex. The Gemin5-bound U1, U2, U4, and U5 pre-snRNAs and U1-tfs were then separated by urea-PAGE (Supplemental Fig. S13A) and subjected to in-gel RNase digestion followed by nano-scale liquid chromatography-mass spectrometry (LC-MS) analysis (Supplemental Fig. S13B–F). U1-tfs are ∼120 nucleotides in length, are truncated forms of U1 pre-snRNAs lacking the Sm site and the following stem–loop 4 (SL4) but retaining the m7G cap structure, and coexist with wild-type U1 pre-snRNAs in vivo (Ishikawa et al. 2014). Our LC-MS results show that Gemin5 bound to both m7G-capped and 5′ m3G-capped U1, U2, U4, and U5 snRNAs in cells (Supplemental Fig. S13B–F). The m7G-capped pre-snRNAs are almost undetectable in total snRNA analysis (<1% for U1/U2/U4 and ∼1.2% for U5), but Gemin5 enriched m7G-capped pre-snRNAs >100-fold, estimated from the detection limits of the LC-MS assay (Supplemental Fig. S14). Given the inability of Gemin5 to recognize the 5′ m3G cap structure as demonstrated above (Fig. 4A), Gemin5 might bind to the 5′ m3G-capped pre-snRNAs via their intact Sm site inside cells. Gemin5 could also bind and enrich the m7G-capped U1-tfs (Supplemental Fig. S12F), supporting the m7G cap-mediated recognition of U1-tfs by Gemin5 in vivo. As a control, SmB could recognize all regular snRNAs but not the U1-tfs (Supplemental Fig. S13B–F). Therefore, Gemin5 could bind independently to both the m7G cap and the Sm site of pre-snRNAs in vivo.
The m7G cap–Gemin5 interaction governs the fate of U1-tfs in cells
Our previous SMN pull-down coupled with nano-scale LC-tandem MS (LC-MS/MS) analysis had shown that the SMN complex could associate with novel U1-tfs (Ishikawa et al. 2014), and we show here that it is Gemin5 that is responsible for binding U1-tfs (Supplemental Fig. 13F). Furthermore, our RNA pull-down analysis revealed that the m7GpppG-binding site mutants Y474A and K641A of Gemin5 displayed significantly reduced in vivo binding to U1-tfs compared with wild-type Gemin5 as well as the Sm site-binding mutants W14A or Y15A of Gemin5 (Fig. 4A,B, left). On the other hand, the Sm or m7GpppG site mutants of Gemin5 did not show drastic changes in their in vivo affinities to full-length U1 pre-snRNAs (Fig. 4B, left), suggesting that Gemin5-mediated binding to either the m7GpppG cap or the Sm site of U1 pre-snRNAs alone provides sufficient affinity to pull down comparable amounts of U1 pre-snRNAs from cells.
Exogenous expression of the m7GpppG-binding site mutants Y474A and K641A of Gemin5 in cells significantly and selectively decreased U1-tfs levels, implying that these cap site mutants of Gemin5 had a dominant-negative effect on endogenous wild-type Gemin5, and the loss of the Gemin5-mediated m7G cap-binding events resulted in destabilization of U1-tfs in cells (Fig. 4B, right). None of these point mutations of Gemin5 affected in vivo binding of Gemin5 to other components of the SMN complex (including SMN, Gemin3, and Gemin6) or levels of other m7G cap-associated proteins (including CBP80, PHAX, and TGS1) (Supplemental Fig. S15A,B left panel). In addition, exogenous expression of these point mutants of Gemin5 in cells did not significantly affect the expression levels of all of these proteins (Supplemental Fig. S15B, right panel).
To examine the cellular impact of the loss of specific interaction between Gemin5 and the m7G cap on U1-tfs in cells, we depleted endogenous Gemin5 in cells. Interestingly, Gemin5 knockdown resulted in a drastic reduction of the endogenous level of U1-tfs in cells (Fig. 4C), further suggesting that Gemin5 stabilizes cellular U1-tfs and that Gemin5's interaction with U1-tfs represses the uncontrolled degradation of U1-tfs outside P bodies (Ishikawa et al. 2014). To confirm the effect of Gemin5–m7G cap interaction alone on U1-tfs, we cotransfected a truncated form of U1 pre-snRNA that lacks the Sm site and SL4 (y18Sn-ΔSmSL4; a U1-tfs mimic) with the wild type and the m7G cap mutants of Gemin5 (Y474A and K641A) (Supplemental Fig. S15C). Exogenous expression of the m7G cap-binding mutants of Gemin5 (Y474A and K641A) resulted in a significant reduction of cellular levels of the U1-tfs mimic (y18Sn-ΔSmSL4) as compared with the wild-type Gemin5, providing another layer of evidence that the recognition of the m7G cap of U1-tfs by Gemin5 stabilizes U1-tfs (Supplemental Fig. S15C). As a control, the cellular levels of 7SK-5′ and Met-tRNA controls did not change (Supplemental Fig. S15C). Collectively, these data support that Gemin5 binds directly to U1-tfs via their m7G caps and stabilizes these pre-snRNA mutants.
Discussion
Gemin5 of the SMN complex is responsible for recognizing pre-snRNAs and transferring them to the Sm ring complex for assembly. Our crystal structures, complemented with mutagenesis experiments, reveal that the WD40 domain of Gemin5 selectively recognizes the Sm site and the m7G cap mainly via the first WD40 β propeller and the second WD40 β propeller, respectively. Gemin5-mediated bimodal recognition of pre-snRNAs provides a stringent double-check mechanism for snRNP biogenesis. Remarkably, our data also revealed that the binding of Gemin5 and the m7G cap of defective pre-snRNAs, which lacks the Sm site, shunts these truncated pre-snRNAs away from the normal snRNP biogenesis pathway, thereby contributing indirectly to the selective generation of bona fide spliceosomes.
Gemin5 safeguards snRNP integrity by providing genuine pre-snRNAs to the Sm complex
Raker et al. (1996) provided evidence that the Sm ring complex can be assembled onto the Sm site RNA in vitro. During the assembly, the SmE–F–G trimer and the SmD1–D2 dimer are recruited to the Sm site RNA before enlisting the SmD3–B dimer, which completes the construction of the Sm ring complex. SmD2, SmG, and SmB display strictly uridine-specific pockets (Weber et al. 2010). Since the addition of the SmD3–B dimer is the final step in the formation of the heptameric ring (Raker et al. 1996), the proper assembly of snRNPs is initially thought to be likely dependent on SmD2 and SmG for recognizing the correct RNAs in vivo. However, it was later reported that substitutions at any uridine of U121–U124 within the Sm site RNA 118AAUUUUUGA126 surprisingly does not significantly affect the assembly process of the Sm ring complex on the Sm site RNA in vitro (Raker et al. 1999), thereby raising the risk of improper assembly. Given the very low frequency of RNA missplicing events in normal cells and thus the highly effective mechanism for generating bona fide cellular spliceosomes, we hypothesized the existence of an error-proof mechanism for capturing the correct RNAs in cells prior to the engagement of the components of the Sm ring complex with pre-snRNAs.
In this study, we provided mechanistic evidence that mostly the first WD40 β propeller of Gemin5 recognizes the Sm site RNA in a highly sequence-dependent binding fashion, and the precise docking of the Sm site of pre-snRNAs [A(U)4–6G, especially the 119AUUU122 key motif] to the exposed shallow groove of Gemin5 through base-specific interactions would accurately position the pre-snRNA for subsequent transfer from the SMN complex to the heptameric Sm ring complex during the Sm core assembly. Specifically, the recognition of oligo uridine at the Sm site of pre-snRNAs by Gemin5 probably prevents the misincorporation of other incorrect RNAs into snRNPs by compensating for the low sequence selectivity of the Sm ring complex. Therefore, our study provides mechanistic insights into how the SMN complex uses Gemin5 as a component to stringently scrutinize potential RNA substrates, leading to the tightly regulated delivery of pre-snRNAs into the Sm core assembly.
A possible model of pre-snRNA transfer from Gemin5 to the Sm ring complex
Gemin5 is well known for its association with pre-snRNAs prior to their incorporation into the Sm ring complex, leading to the biogenesis of snRNPs (Battle et al. 2006a,b). Our data presented above collectively indicate that the WD40 domain of Gemin5 binds to the Sm site of pre-snRNA in a sequence-specific manner. Notably, the published crystal structure of the U4 snRNP shows that the Sm site heptad (AUUUUUG) binds deep inside the central hole of the Sm heptameric ring, with basically each Sm protein recognizing one base (Weber et al. 2010; Leung et al. 2011). In contrast, our Gemin5–Sm site RNA complex structures reveal that, when loaded onto Gemin5, the Sm site RNA is mostly solvent-exposed (Fig. 2B). As a result, the pre-snRNA itself would likely endure necessary conformational changes during the transfer. Furthermore, Raker et al. (1999) found that regions outside the Sm site also facilitate the snRNP assembly, which is consistent with the observation that the 3′ sequence flanking the Sm site also makes contact with the Sm complex in the known snRNP structures (Weber et al. 2010; Leung et al. 2011). Therefore, it is plausible that the initial interaction of the Sm proteins with Gemin5-captured pre-snRNAs may occur at the free 3′ flanking sequence of the Sm site of these pre-snRNAs.
A working model of Gemin5 in quality control and maturation of pre-snRNAs
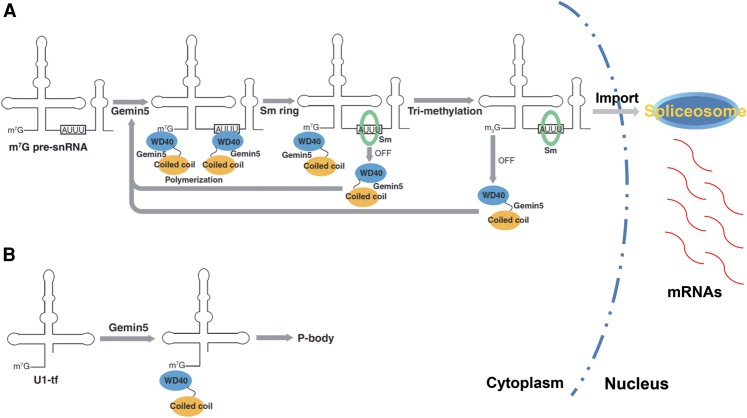
Given that Gemin5 can recognize pre-snRNAs at two independent sites (namely, the Sm site and the m7G cap), we cannot exclude the possibility that Gemin5 molecules could homopolymerize to cooperatively recognize a single pre-snRNA molecule in cells, suggesting the existence of Gemin5-mediated bimodal recognition signals for ensuring the correct RNAs are prevented from entering into the snRNP biogenesis pathway. In agreement with this notion, we also found that Gemin5 could recognize the defective form of pre-U1 snRNAs, U1-tfs, via binding to their m7G caps and shunts these defective Sm site-lacking pre-snRNAs away from the snRNP biogenesis pathway. As a P-body-associated protein, Gemin5 might direct U1-tfs to P bodies for organized degradation, although we could not exclude other possibilities (Fierro-Monti et al. 2006). Collectively, we propose a working model for Gemin5-guided quality control and maturation of pre-snRNAs inside the cytoplasm. On the one hand, through their N-terminal WD40 domains, two Gemin5 molecules can bind the same pre-snRNA molecule at the m7G cap and the Sm site, respectively. On the other hand, their C-terminal coiled-coil domains can homodimerize to stabilize their interactions with pre-snRNAs in the cytoplasmic microenvironment.
As the association of the SMN complex with the Sm proteins precedes its interaction with pre-snRNAs (Pellizzoni et al. 2002; Yong et al. 2002), this Gemin5-guided capture of cytoplasmic pre-snRNAs into the SMN complex brings the correct RNA molecules into close proximity to the Sm proteins, which have low intrinsic selectivity for pre-snRNAs. The Sm site-bound Gemin5 molecule is then displaced by the Sm proteins during Sm core assembly. The subsequent conversion of m7G to m3G at the 5′ cap of the pre-snRNA molecule abolishes the direct binding of the other Gemin5 to the matured snRNA, which is now ready to be imported into the nucleus for snRNP assembly (Fig. 6A). These Gemin5-guided mechanisms ensure the integrity of the maturation process for pre-snRNAs in the cytoplasm prior to their nuclear import for generation of a bona fide spliceosome. Overall, our findings elucidate the molecular mechanism behind Gemin5-mediated recognition of the Sm site of pre-snRNAs during snRNA maturation. More importantly, we provide evidence for a novel interaction event between the m7G cap of pre-snRNAs and the WD40 domain of Gemin5.
Figure 6.
A proposed model depicting Gemin5-driven molecular mechanisms responsible for the quality control and maturation of snRNAs inside the cytoplasm. (A) Through their N-terminal WD40 domains, two Gemin5 proteins recognize one pre-snRNA molecule via both the m7G cap and the Sm site, respectively, while the polymerization of these two Gemin5 molecules via their C-terminal coiled-coil domains stabilizes their binding interactions with the pre-snRNA molecule. As the association of the SMN complex with the Sm proteins precedes its interaction with pre-snRNAs (Pellizzoni et al. 2002; Yong et al. 2002), this Gemin5-guided capture of cytoplasmic pre-snRNAs into the SMN complex brings the correct RNA molecules into close proximity to the Sm proteins, which have an intrinsic low specificity for recognizing pre-snRNAs. The Sm site-bound Gemin5 molecule is then displaced by the Sm proteins during the Sm core assembly. The subsequent conversion of m7G to m3G at the 5′ cap of the pre-snRNAs leads to the dissociation of the other Gemin5 molecule. These Gemin5-guided mechanisms ensure the integrity of the maturation process for pre-snRNAs in the cytoplasm prior to their nuclear import for generation of bona fide spliceosomes. (B) Gemin5 recognizes U1-tfs via the m7G cap and escorts them to the P bodies for controlled RNA turnover, thereby shunting these U1 pre-snRNA mutants away from the normal U1 snRNP biogenesis pathway.
Interestingly, the maturation of snRNAs specifically inside the cytoplasm probably provides an advantage of compartmentation to avoid undesired assembly of random RNAs into snRNPs inside the nucleic acid-rich environment of the nucleus. This nuclear membrane shield, while effective, is likely not sufficient to prevent the production of spliceosomes containing defective RNA subcomponents. The novel functional role of Gemin5-mediated recognition of the m7G cap of U1-tfs in shunting these defective U1 pre-snRNA mutants into P bodies provides an additional surveillance mechanism inside the cytoplasm to ensure the integrity of U1 snRNA maturation (Fig. 6B). It is natural to speculate about the existence of other similar Gemin5-guided surveillance mechanisms for controlling the quality of other species of pre-snRNAs, and this deserves future investigation. Meanwhile, the functional significance of the Gemin5-mediated recognition of the m7G cap of intact pre-snRNAs in the normal snRNP biogenesis pathway will likely be of compelling interest to the field.
Other potential cellular functions of Gemin5
Given the low sequence selectivity of the Sm ring complex for uridine-rich RNAs in vitro (Wahl et al. 2009), our findings highlight the surveillance capacity of Gemin5 of the SMN complex in ensuring that Sm cores could be assembled only on pre-snRNAs, thereby safeguarding the cells from defective spliceosomes. However, besides pre-snRNAs, few other RNAs inside cells also contain the Sm site-like sequence motif. For example, Gemin5 in the nucleus can regulate the translation of the SMN proteins via its association with the 3′ Sm site-like sequence of SMN mRNAs (Workman et al. 2015). In addition, the telomerase RNA subunit TER1 also harbors the Sm site at its 3′ terminus (Seto et al. 1999; Leonardi et al. 2008; Gunisova et al. 2009), upon which the Sm and Lsm complexes assemble during telomerase biogenesis (Tang et al. 2012), implying a potential role of nuclear Gemin5 in recognition of TER1 at the Sm site. Thus, analogous to Gemin5's role in snRNA maturation, Gemin5's potential recognition of other cellular RNAs containing these Sm site-like sequence motifs in the nucleus may have important functional significances and deserves future investigations. Additionally, Gemin5 does not exist only as a component of the SMN complex but has also been found in the Gemin3–Gemin4–Gemin5 complex (Battle et al. 2007; Otter et al. 2007) and an IRES-specific ribonucleoprotein complex (Pacheco et al. 2009). Collectively, our findings in this study provide the structural foundation to facilitate the elucidation of the potentially pleiotropic cellular functions of Gemin5.
Materials and methods
Protein expression and purification of human Gemin5
The WD40 domain of Gemin5 (residues 1–739) was cloned into the vector pFBOH-LIC and expressed in Sf9 insect cells. Sf9 cells (0.8 L; 4.5 × 106 cells per milliliter) were infected with 8 mL of P3 recombinant baculovirus and grown in a 2.8-L shake flask at 100 rpm and 27°C. HyQ SFX insect serum medium containing 10 µg/mL gentamicin was used as the culture medium. Infected cells were harvested when viability dropped to 80%–85%, normally after 48 h. Harvested cells were washed with ice-cold PBS buffer, flash-frozen in liquid nitrogen, and stored at −80°C for purification. The recombinant protein, which contains an N-terminal 6xHis tag, was purified on a Ni-NTA resin (Qiagen) and further purified by Superdex 200 gel filtration column (GE Healthcare) in 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 1 mM DTT (dithiothreitol). Purified Gemin5 protein was concentrated to 15–20 mg/mL and stored at −80°C before further use.
Crystallization of apo Gemin5 and its complex with a Sm site RNA
The apo form of Gemin5 was crystallized in a condition of 0.1 M sodium citrate (pH 5.5–6.5) 0.2 M ammonium acetate, and 15% PEG 4000 using the sitting drop vapor diffusion method at 18°C by mixing 1 µL of the protein solution with 1 µL of the reservoir solution. For the Gemin5–118AAUUUUUG125 complex, the Gemin5 protein and RNA were mixed in a ratio of 1:1.5. The crystals, which appeared in 2–3 d in the same conditions as those of apo Gemin5, were then soaked with 0.5 mM 118AAUUUUUG125 RNA for another day in the same reservoir solution. The crystals of the Gemin5–118AAUUUUUGAG127 complex were obtained the same way.
For the Gemin5–m7GpppG or Gemin5–GpppG complex, m7GpppG and GpppG were purchased from Sigma-Aldrich. The crystals of either complex were obtained by soaking the crystal of apo Gemin5 with 1.0 mM m7GpppG or GpppG for 24 h using sitting drop in a condition of 0.1 M sodium citrate (pH 6.5), 0.2 M ammonium acetate, and 18% PEG 4000. Before flash-freezing crystals in liquid nitrogen, all crystals were soaked in a cryo-protectant consisting of 90% reservoir solution plus 10% glycerol.
Structure determination
Diffraction data were collected under cooling at 100 K as shown in Supplemental Table 1. Reflection intensities were integrated with the Denzo-ScalePack (Otwinowski and Minor 1997) or XDS (Kabsch 2010)–Pointless–Scala/Aimless (Evans and Murshudov 2013) pipeline. For each Gemin5–RNA complex, diffraction intensities from two similar crystals were merged. The Gemin5 apo structure was solved by molecular replacement using the programs Phaser (McCoy et al. 2007) and Molrep (Vagin and Teplyakov 2010) as well as SCWRL (Canutescu et al. 2003) models derived from Protein Data Bank (PDB) (Berman et al. 2000) entries 3OW8 (Xu and Min 2011), 2HES (Srinivasan et al. 2007), and 3DM0 (Ullah et al. 2008) on the FFAS03 server (Jaroszewski et al. 2005). ARP/wARP was used for phase improvement (Perrakis et al. 1997) and automated protein chain tracing (Perrakis et al. 1999). Phaser molecular replacement with apo Gemin5 coordinates solved the m7GpppG complex structure, whereas the RNA and GpppG complexes did not require a renewed molecular replacement search.
Restraints for the m7GpppG ligand were prepared with the program Grade server (http://grade.globalphasing.org/cgi-bin/grade/server.cgi) and coordinates from PDB entry 3HXI. Models were iteratively validated, rebuilt, and refined on the MolProbity (Chen et al. 2010) server in Coot (Emsley et al. 2010) and Refmac (Murshudov et al. 2011), respectively. Data collection and refinement statistics are summarized in Table 1 (Yang et al. 2004; Gildea et al. 2011).
Synthesis of the m7G-capped full-length U4 pre-snRNA
Full-length U4 pre-snRNA was synthesized from the DNA template by using the Megashortscript kit (Invitrogen) following the product instructions. RNeasy minicolumns (Qiagen) were used for further purification of synthesized snRNA. The capping kit (New England Biolabs) using the vaccinia virus capping enzyme was used to add m7G at the 5′ position of the synthesized U4 pre-snRNA. Next, the m7G-capped U4 pre-snRNAs were annealed and used for further experiments.
ITC binding assay
Measurements were performed at 25°C with a VP-ITC microcalorimeter (MicroCal, Inc.) or nano-ITC (TA, Inc.). All of the RNA fragments used in this study, except the m7G-capped U4 pre-snRNA, were purchased from Integrated DNA Technologies, Inc. RNAs were dissolved and dialyzed in buffer containing 20 mM Tris-HCl (pH 7.5) and 150 mM NaCl in diethylpyrocarbonate-treated RNase-free water. The concentrations of proteins and RNA fragments were estimated with absorbance spectroscopy using their extinction coefficients at 280 and 260 nm, respectively. VP-ITC was performed by injecting 10 µL of 0.2–0.44 mM RNA solution into a sample cell containing 7–15 µM protein. Fifteen to 25 injections were performed with a spacing of 180 sec and a reference power of 15 µcal/sec. Binding isotherms were plotted and analyzed with Origin software (MicroCal, Inc.) after subtraction of their respective RNA-only controls. The thermodynamic parameters, including the Kd, enthalpy (ΔH), entropy (ΔS), and free energy (ΔG), were calculated by fitting in a “one-site” model from the titration curves (Supplemental Table S1; Supplemental Fig. S16). The error bars for those parameters were obtained from the nonlinear least-squares curve fittings.
Nano-ITC was used to examine the binding of Gemin5 wild type or mutants to the m7G-capped U4 pre-snRNA. Two microliters of protein solution (120–150 µM) was injected into a sample cell containing 8 µM m7G-capped U4 pre-snRNA. Data were processed with NanoAnalyze (TA, Inc.) software and fitted in a “one-site” or “two-site” model after subtraction of protein-only controls (Supplemental Fig. S11).
Construction of plasmids expressing Gemin5 with a point mutation
Site-directed mutagenesis was performed using the expression plasmid encoding Gemin5 wild type fused with a C-terminal HEF tag (pcDNA5-FRT/TO-G5-WT-HEF) (Ishikawa et al. 2014). The plasmid DNA (50 ng) was amplified by PCR with the following set of mutagenesis primers: 5′-TCATAAGAAGACTGTAGCTACTTTAGCCTG-3′ and 5′-CAGGCTAAAGTAGCTACAGTCTTCTTATGA-3′ for G5-Y474A-HEF (corresponding to Gemin5 mutant Y474A), 5′-AGGGCATACGGCCGCGATTACCAGTGT-3′ and 5′-ACACTGGTAATCGCGGCCGTATGCCCT-3′ for G5-Y641A-HEF (Y641A), 5′-CTCCCCCAACGCGTACTGCGCCCGCTGCAG-3′ and 5′-CTGCAGCGGGCGCAGTACGCGTTGGGGGAG-3′ for G5-W14A-HEF (W14A), and 5′-CTCCCCCAACTGGGCCTGCGCCCGCTGCAG-3′ and 5′-CTGCAGCGGGCGCAGGCCCAGTTGGGGGAG-3′ for G5-Y15A- HEF (Y15A). After digestion with methylation-specific endonuclease Dpn I, the PCR product with point mutations was prepared by using Escherichia coli (DH5α). Point mutations were confirmed by DNA sequencing.
Cell culture and transfection
Flp-In T-REx 293 and 293T cells were cultured by standard methods in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum, 100 U of penicillin G (Sigma-Aldrich), and 100 µg/mL streptomycin (Wako) at 37°C in a humidified atmosphere of 95% air and 5% CO2. Expression plasmid was transfected into the cells using Lipofectamine 2000 or Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific).
Preparation of doxycycline-inducible cell lines expressing G5-WT-HEF or its point mutants
Flp-In T-REx-293 cells were cultured in DMEM at 37°C, seeded in a 24-well plate, and transfected with 2 µL of Lipofectamine 2000 containing 0.25 µg of the respective expression plasmid and 0.25 µg of pOG44 vector (Thermo Fisher Scientific). Once the cultured cells reached >90% confluence (∼24–48 h after transfection), hygromycin B (Thermo Fisher Scientific) was added to their DMEM (containing 10% fetal bovine serum) at a final concentration of 100 µg/mL. Cells were then cultured for another 2 wk in the presence of 100 µg/mL hygromycin B by changing the culture medium every 2 d. Culture of selected cell colonies was scaled up for further experiments.
Isolation of the RNA–protein complex associated with G5-WT-HEF or its point mutants
The Flp-In T-REx 293 cells containing G5-WT-HEF or its point mutants were induced for expression with 1 µg/mL doxycycline for 48 h in 150-mm dishes. The cells were then harvested, washed once with PBS, lysed by vigorous mixing for 30 sec in 1 mL of lysis buffer (50 mM Tris-HCl at pH 8.0 containing 150 mM NaCl, 0.5% [w/v] IGEPAL-CA630, 1 mM PMSF), and incubated for 20 min on ice. Soluble whole-cell lysates were prepared by centrifugation at 20,000g for 20 min at 4°C and incubated with 10–15 µL of anti-Flag M2 agarose beads (Sigma-Aldrich) for 3 h at 4°C with rotation. The beads were washed five times with 1 mL of lysis buffer and once with 50 mM Tris-HCl (pH 8.0) containing 150 mM NaCl, and then RNA–protein complexes (RNPs) were eluted from the beads with 500 µg/mL Flag peptide in the same buffer.
Extraction of proteins and RNAs from RNPs or soluble whole-cell lysates
RNPs or soluble whole-cell lysates were mixed vigorously with an equal volume of 2× RNA extraction buffer (10 mM Tris-HCl at pH 8.0 containing 7 M urea, 350 mM NaCl, 1% SDS, 10 mM EDTA) and buffered phenol/chloroform solution (pH 8.0). The aqueous and organic phases were separated by centrifugation at 20,000g for 30 min at 4°C. 2-propanol equal to the volume of the aqueous phase was added for precipitation of RNAs, whereas 4 vol of 2-propanol was added to organic phase for precipitation of proteins. Next, the RNAs or proteins were collected by centrifugation at 20,000g for 30 min at 4°C. Each precipitate was rinsed with 75% ethanol and air-dried.
Denaturing urea-PAGE and Northern blotting
RNAs were separated by urea-PAGE (7.5 M urea, 9% polyacrylamide) in 0.5× TBE running buffer at 12.5 V/cm for 2–3 h and transferred electrophoretically to a Hybond-N+ membrane (GE Healthcare) in 0.5× TBE using a semidry blotting apparatus. The membrane was dried, exposed to UV irradiation, and blocked with 40 µg/mL salmon sperm DNA in prehybridization buffer containing 5× SSC (saline–sodium citrate buffer), 20 mM NaH2PO4 (pH 7.5), 7% SDS, and 2× Denhardt's solution for at least 1 h. After blocking, membranes containing cross-linked RNAs were hybridized with their respective oligonucleotides labeled with biotin overnight at 50°C. The hybridized membrane was washed twice with wash buffer A (3× SSC, 25 mM NaH2PO4 at pH 7.5, 5% SDS) for 15 min and twice with wash buffer B (1× SSC, 1% SDS) for 15 min at 50°C. RNAs hybridized with biotin-labeled DNA were stained with a chemiluminescent nucleic acid detection module (Thermo Fisher Scientific) and detected with an LAS4000 luminescent image analyzer (GE Healthcare).
SDS-PAGE and Western blot analysis
Proteins were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Ishikawa et al. 2014). The membranes were blocked with 5% nonfat dried milk in TBST (Tris-buffered saline containing 0.1% [w/v] Tween 20) for 1 h and then incubated with their respective antibodies for 1 h at room temperature. The membranes were then washed three times with TBST for 10 min, incubated with a secondary antibody conjugated with horseradish peroxidase for 1 h at room temperature, and washed three times with TBST for 10 min. Clarity Western ECL substrate (Bio-Rad) was used for detection with an LAS4000.
Coexpression analysis of y18Sn-tagged pre-U1 constructs and Gemin5 mutants
Exogenously expressed y18Sn-tagged U1 and its derivative constructs were prepared as reported previously (Ishikawa et al. 2014). They were mixed with G5-WT-HEF or point mutant constructs, respectively, and transfected into 293T cells with Lipofectamine 2000 reagent. At 24 h after transfection, cells were harvested and collected by centrifugation at 1200g for 5 min at 4°C. Four-fifths of the collected cells were used for RNA extraction, while the remaining cells were used for protein extraction. Total RNAs were prepared with TRIzol reagent (Thermo Fisher Scientific), and total proteins were obtained with RIPA buffer (20 mM Tris-HCl at pH 8.0, 150 mM NaCl, 2mM EDTA, 1% sodium deoxycholate, 0.1% SDS, 1% Triton X-100). RNAs and proteins extracted from the cells were analyzed as described previously (Ishikawa et al. 2014).
RNAi
293T cells (5 × 105 cells) were cultured in 35-mm dishes until they reached 80% confluency and were then transfected with 5 µL of Lipofectamine RNAiMAX and 100 pmol of the following stealth siRNAs: 5′-CAAACCAUUCGAAUCUGGAGCUGUU-3′ and 5′-AACAGCUCCAGAUUCGAAUGGUUUG-3′ for Gemin5 (si) and 5′-CAAUUACAAGCGUCUCGAGUCAGUU-3′ and 5′- AACUGACUCGAGACGCUUGUAAUUG-3′ for Gemin5-negative control (sc). The cells were transferred to 100-mm dishes 24 h after transfection. The transfected cells were cultured in DMEM supplemented with 10% fetal bovine serum and harvested at 72 h after transfection. Proteins and RNAs were analyzed as described previously (Ishikawa et al. 2014).
MS-based RNA analysis
Gemin5 (G5-WT-HEF)-associated snRNAs were separated by 8 M urea/9% (w/v) PAGE followed by detection with SYBR Gold staining (Invitrogen) and in-gel digestion with RNase T1 (Taoka et al. 2010). SmB-associated RNAs were separated on a PLRP-S 300 Å column (2-mm I.D. × 100 mm in length, 3-mm particle size) (Polymer Laboratories) at 60°C by a 60-min linear gradient from 12% to 16% acetonitrile in 100 mM triethylammonium acetate (pH 7.0) containing 0.1 mM ammonium phosphate dibasic (Yamauchi et al. 2013). RNAs were detected by monitoring absorbance at A260 and were then digested with RNase T1 in 100 mM triethylammonium acetate (pH 7.0) for 60 min at 37°C. All of the resulting oligonucleotide mixtures were analyzed by the nano-flow LC-MS/MS system (Taoka et al. 2009; Nakayama et al. 2015) equipped with a genome-oriented database search engine, Ariadne (Nakayama et al. 2009). The Q-Exactive mass spectrometer (Thermo Scientific) was operated in a data-dependent mode so that it was automatically switched between MS and MS/MS acquisitions. Survey full-scan mass spectra (m/z 500–2000) were acquired with a mass resolution of 35,000 at m/z 200. For the five most intense mass peaks in a survey scan with an intensity that was >25,000 counts per second, each peak was isolated within a 3-m/z window for fragmentation. The mass peaks were fragmented by higher-energy collisional dissociation with a normalized collision energy of 20%. Ariadne-based RNA identification was performed using the genome database of Homo sapiens (reference assembly version GRCh37 obtained from ftp://ftp.ncbi.nlm.nih.gov/genomes/H_sapiens under the following parameters: maximum number of missed cleavages, 1; variable modification parameter, one methylation per RNA fragment for any residue; RNA mass tolerance, ±20 ppm; and MS/MS tolerance, ±50 ppm.
Atomic coordinates and structure factors for structures of Gemin5 in apo form and in complex with GpppG, m7GpppG, 118AAUUUUUG125, and 118AAUUUUUGAG127 have been deposited into the PDB with accession codes 5TEE, 5THA, 5TEF, 5GXH, and 5GXI, respectively.
Supplementary Material
Acknowledgments
We thank Dr. Tanja Cerovina, Dr. Wolfram Tempel, Dr. Aiping Dong, and Dr. Qi Zhang for technical assistance, and Peter J. Brown for stimulating discussion. We also thank Shelton S. Bradrick for kindly providing the DNA templates and plasmids and for helpful discussions. We also thank Dr. Rui-ming Xu for communication of his data and helpful discussion prior to publication. Results shown in this report were derived from work performed at Structural Biology Center at the Advanced Photon Source (APS). The General Medical Sciences and Cancer Institutes Structural Biology Facility at APS (GM/CA@APS) has been funded in whole or in part by federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). The APS is a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract number DE-AC02-06CH11357. The Structural Genomics Consortium is a registered charity (no. 1097737) that receives funds from AbbVie; Bayer Pharma AG; BoehringerIngelheim; Canada Foundation for Innovation; Eshelman Institute for Innovation; Genome Canada through the Ontario Genomics Institute; the Innovative Medicines Initiative (European Union/European Federation of Pharmaceutical Industries and Associations; Unrestricted Leveraging of Targets for Research Advancement and Drug Discovery [ULTRA-DD] grant no. 115766); Janssen, Merck, and Company; Novartis Pharma AG; Ontario Ministry of Economic Development and Innovation; Pfizer; São Paulo Research Foundation (FAPESP); Takeda; and the Wellcome Trust. This project was also funded by the National Nature Science Foundation of China (31570737) and Core Research for Evolutional Science and Technology (CREST) from the Japan Science and Technology Agency (JST). C.X. is supported by the Chinese Government “1000 Youth Talent Program.”
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.288340.116.
References
- Agafonov DE, Kastner B, Dybkov O, Hofele RV, Liu WT, Urlaub H, Luhrmann R, Stark H. 2016. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science 351: 1416–1420. [DOI] [PubMed] [Google Scholar]
- Battle DJ, Kasim M, Yong J, Lotti F, Lau CK, Mouaikel J, Zhang Z, Han K, Wan L, Dreyfuss G. 2006a. The SMN complex: an assembly machine for RNPs. Cold Spring Harb Symp Quant Biol 71: 313–320. [DOI] [PubMed] [Google Scholar]
- Battle DJ, Lau CK, Wan L, Deng H, Lotti F, Dreyfuss G. 2006b. The Gemin5 protein of the SMN complex identifies snRNAs. Mol Cell 23: 273–279. [DOI] [PubMed] [Google Scholar]
- Battle DJ, Kasim M, Wang J, Dreyfuss G. 2007. SMN-independent subunits of the SMN complex. Identification of a small nuclear ribonucleoprotein assembly intermediate. J Biol Chem 282: 27953–27959. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The Protein Data Bank. Nucleic Acids Res 28: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradrick SS, Gromeier M. 2009. Identification of gemin5 as a novel 7-methylguanosine cap-binding protein. PLoS One 4: e7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canutescu AA, Shelenkov AA, Dunbrack RL Jr. 2003. A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci 12: 2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot B, Shkreta L. 2016. Defective control of pre-messenger RNA splicing in human disease. J Cell Biol 212: 13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Moore MJ. 2014. The spliceosome: disorder and dynamics defined. Curr Opin Struct Biol 24: 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66: 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PR, Murshudov GN. 2013. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr 69: 1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierro-Monti I, Mohammed S, Matthiesen R, Santoro R, Burns JS, Williams DJ, Proud CG, Kassem M, Jensen ON, Roepstorff P. 2006. Quantitative proteomics identifies Gemin5, a scaffolding protein involved in ribonucleoprotein assembly, as a novel partner for eukaryotic initiation factor 4E. J Proteome Res 5: 1367–1378. [DOI] [PubMed] [Google Scholar]
- Gildea RJ, Bourhis LJ, Dolomanov OV, Grosse-Kunstleve RW, Puschmann H, Adams PD, Howard JA. 2011. iotbx.cif: a comprehensive CIF toolbox. J Appl Crystallogr 44: 1259–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golembe TJ, Yong J, Dreyfuss G. 2005. Specific sequence features, recognized by the SMN complex, identify snRNAs and determine their fate as snRNPs. Mol Cell Biol 25: 10989–11004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Fabrega C, Liu SW, Liu H, Kiledjian M, Lima CD. 2004. Insights into the structure, mechanism, and regulation of scavenger mRNA decapping activity. Mol Cell 14: 67–80. [DOI] [PubMed] [Google Scholar]
- Gubitz AK, Mourelatos Z, Abel L, Rappsilber J, Mann M, Dreyfuss G. 2002. Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J Biol Chem 277: 5631–5636. [DOI] [PubMed] [Google Scholar]
- Gunisova S, Elboher E, Nosek J, Gorkovoy V, Brown Y, Lucier JF, Laterreur N, Wellinger RJ, Tzfati Y, Tomaska L. 2009. Identification and comparative analysis of telomerase RNAs from Candida species reveal conservation of functional elements. RNA 15: 546–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Nobe Y, Izumikawa K, Yoshikawa H, Miyazawa N, Terukina G, Kurokawa N, Taoka M, Yamauchi Y, Nakayama H, et al. 2014. Identification of truncated forms of U1 snRNA reveals a novel RNA degradation pathway during snRNP biogenesis. Nucleic Acids Res 42: 2708–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaroszewski L, Rychlewski L, Li Z, Li W, Godzik A. 2005. FFAS03: a server for profile–profile sequence alignments. Nucleic Acids Res 33: W284–W288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. 2010. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr 66: 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CK, Bachorik JL, Dreyfuss G. 2009. Gemin5–snRNA interaction reveals an RNA binding function for WD repeat domains. Nat Struct Mol Biol 16: 486–491. [DOI] [PubMed] [Google Scholar]
- Leonardi J, Box JA, Bunch JT, Baumann P. 2008. TER1, the RNA subunit of fission yeast telomerase. Nat Struct Mol Biol 15: 26–33. [DOI] [PubMed] [Google Scholar]
- Leung AK, Nagai K, Li J. 2011. Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis. Nature 473: 536–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Roberts R. 2001. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell Mol Life Sci 58: 2085–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. 1997. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89: 951–961. [DOI] [PubMed] [Google Scholar]
- Matera AG, Wang Z. 2014. A day in the life of the spliceosome. Nat Rev Mol Cell Biol 15: 108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Terns RM, Terns MP. 2007. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol 8: 209–220. [DOI] [PubMed] [Google Scholar]
- Mazza C, Segref A, Mattaj IW, Cusack S. 2002. Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J 21: 5548–5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-McNicoll M, Neugebauer KM. 2013. How cells get the message: dynamic assembly and function of mRNA–protein complexes. Nat Rev Genet 14: 275–287. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67: 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama H, Akiyama M, Taoka M, Yamauchi Y, Nobe Y, Ishikawa H, Takahashi N, Isobe T. 2009. Ariadne: a database search engine for identification and chemical analysis of RNA using tandem mass spectrometry data. Nucleic Acids Res 37: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama H, Yamauchi Y, Taoka M, Isobe T. 2015. Direct identification of human cellular microRNAs by nanoflow liquid chromatography-high-resolution tandem mass spectrometry and database searching. Anal Chem 87: 2884–2891. [DOI] [PubMed] [Google Scholar]
- Otter S, Grimmler M, Neuenkirchen N, Chari A, Sickmann A, Fischer U. 2007. A comprehensive interaction map of the human survival of motor neuron (SMN) complex. J Biol Chem 282: 5825–5833. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Pacheco A, Lopez de Quinto S, Ramajo J, Fernandez N, Martinez-Salas E. 2009. A novel role for Gemin5 in mRNA translation. Nucleic Acids Res 37: 582–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L, Yong J, Dreyfuss G. 2002. Essential role for the SMN complex in the specificity of snRNP assembly. Science 298: 1775–1779. [DOI] [PubMed] [Google Scholar]
- Perrakis A, Sixma TK, Wilson KS, Lamzin VS. 1997. wARP: improvement and extension of crystallographic phases by weighted averaging of multiple-refined dummy atomic models. Acta Crystallogr D Biol Crystallogr 53: 448–455. [DOI] [PubMed] [Google Scholar]
- Perrakis A, Morris R, Lamzin VS. 1999. Automated protein model building combined with iterative structure refinement. Nat Struct Biol 6: 458–463. [DOI] [PubMed] [Google Scholar]
- Raker VA, Plessel G, Luhrmann R. 1996. The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J 15: 2256–2269. [PMC free article] [PubMed] [Google Scholar]
- Raker VA, Hartmuth K, Kastner B, Luhrmann R. 1999. Spliceosomal U snRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol Cell Biol 19: 6554–6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti MM, Swanson MS. 2016. RNA mis-splicing in disease. Nat Rev Genet 17: 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrima A, Konickova R, Czyzewski BK, Kawasaki Y, Jeffrey PD, Groisman R, Nakatani Y, Iwai S, Pavletich NP, Thoma NH. 2008. Structural basis of UV DNA-damage recognition by the DDB1–DDB2 complex. Cell 135: 1213–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. 1999. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401: 177–180. [DOI] [PubMed] [Google Scholar]
- Smith TF, Gaitatzes C, Saxena K, Neer EJ. 1999. The WD repeat: a common architecture for diverse functions. Trends Biochem Sci 24: 181–185. [DOI] [PubMed] [Google Scholar]
- Srinivasan V, Netz DJ, Webert H, Mascarenhas J, Pierik AJ, Michel H, Lill R. 2007. Structure of the yeast WD40 domain protein Cia1, a component acting late in iron-sulfur protein biogenesis. Structure 15: 1246–1257. [DOI] [PubMed] [Google Scholar]
- Stirnimann CU, Petsalaki E, Russell RB, Muller CW. 2010. WD40 proteins propel cellular networks. Trends Biochem Sci 35: 565–574. [DOI] [PubMed] [Google Scholar]
- Strasser A, Dickmanns A, Luhrmann R, Ficner R. 2005. Structural basis for m3G-cap-mediated nuclear import of spliceosomal UsnRNPs by snurportin1. EMBO J 24: 2235–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kannan R, Blanchette M, Baumann P. 2012. Telomerase RNA biogenesis involves sequential binding by Sm and Lsm complexes. Nature 484: 260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka M, Yamauchi Y, Nobe Y, Masaki S, Nakayama H, Ishikawa H, Takahashi N, Isobe T. 2009. An analytical platform for mass spectrometry-based identification and chemical analysis of RNA in ribonucleoprotein complexes. Nucleic Acids Res 37: e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka M, Ikumi M, Nakayama H, Masaki S, Matsuda R, Nobe Y, Yamauchi Y, Takeda J, Takahashi N, Isobe T. 2010. In-gel digestion for mass spectrometric characterization of RNA from fluorescently stained polyacrylamide gels. Anal Chem 82: 7795–7803. [DOI] [PubMed] [Google Scholar]
- Ullah H, Scappini EL, Moon AF, Williams LV, Armstrong DL, Pedersen LC. 2008. Structure of a signal transduction regulator, RACK1, from Arabidopsis thaliana. Protein Sci 17: 1771–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. 2010. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr 66: 22–25. [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718. [DOI] [PubMed] [Google Scholar]
- Weber G, Trowitzsch S, Kastner B, Luhrmann R, Wahl MC. 2010. Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J 29: 4172–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol 13: 290–301. [DOI] [PubMed] [Google Scholar]
- Workman E, Kalda C, Patel A, Battle DJ. 2015. Gemin5 binds to the survival motor neuron mRNA to regulate SMN expression. J Biol Chem 290: 15662–15669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Nilsson P, Henriksson N, Niedzwiecka A, Lim MK, Cheng Z, Kokkoris K, Virtanen A, Song H. 2009. Structural basis of m(7)GpppG binding to poly(A)-specific ribonuclease. Structure 17: 276–286. [DOI] [PubMed] [Google Scholar]
- Xu C, Min J. 2011. Structure and function of WD40 domain proteins. Protein Cell 2: 202–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Taoka M, Nobe Y, Izumikawa K, Takahashi N, Nakayama H, Isobe T. 2013. Denaturing reversed phase liquid chromatographic separation of non-coding ribonucleic acids on macro-porous polystyrene-divinylbenzene resins. J Chromatogr A 1312: 87–92. [DOI] [PubMed] [Google Scholar]
- Yan C, Hang J, Wan R, Huang M, Wong CC, Shi Y. 2015. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science 349: 1182–1191. [DOI] [PubMed] [Google Scholar]
- Yang H, Guranovic V, Dutta S, Feng Z, Berman HM, Westbrook JD. 2004. Automated and accurate deposition of structures solved by X-ray diffraction to the Protein Data Bank. Acta Crystallogr D Biol Crystallogr 60: 1833–1839. [DOI] [PubMed] [Google Scholar]
- Yong J, Pellizzoni L, Dreyfuss G. 2002. Sequence-specific interaction of U1 snRNA with the SMN complex. EMBO J 21: 1188–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong J, Golembe TJ, Battle DJ, Pellizzoni L, Dreyfuss G. 2004. snRNAs contain specific SMN-binding domains that are essential for snRNP assembly. Mol Cell Biol 24: 2747–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong J, Kasim M, Bachorik JL, Wan L, Dreyfuss G. 2010. Gemin5 delivers snRNA precursors to the SMN complex for snRNP biogenesis. Mol Cell 38: 551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.