Abstract
ChIA-PET is a high-throughput mapping technology that reveals long-range chromatin interactions and provides insights into the basic principles of spatial genome organization and gene regulation mediated by specific protein factors. Recently, we showed that a single ChIA-PET experiment provides information at all genomic scales of interest, from the high-resolution locations of binding sites and enriched chromatin interactions mediated by specific protein factors, to the low resolution of nonenriched interactions that reflect topological neighborhoods of higher-order chromosome folding. This multilevel nature of ChIA-PET data offers an opportunity to use multiscale 3D models to study structural-functional relationships at multiple length scales, but doing so requires a structural modeling platform. Here, we report the development of 3D-GNOME (3-Dimensional Genome Modeling Engine), a complete computational pipeline for 3D simulation using ChIA-PET data. 3D-GNOME consists of three integrated components: a graph-distance-based heat map normalization tool, a 3D modeling platform, and an interactive 3D visualization tool. Using ChIA-PET and Hi-C data derived from human B-lymphocytes, we demonstrate the effectiveness of 3D-GNOME in building 3D genome models at multiple levels, including the entire genome, individual chromosomes, and specific segments at megabase (Mb) and kilobase (kb) resolutions of single average and ensemble structures. Further incorporation of CTCF-motif orientation and high-resolution looping patterns in 3D simulation provided additional reliability of potential biologically plausible topological structures.
It is now well established that the organization of the eukaryotic genome is not random (Belmont 2014), and both the spatial placement and structural arrangement of chromosomes could contribute to transcriptional regulation (Wendt and Grosveld 2014). However, the detailed topology and 3D organization are still not well understood. ChIA-PET (Fullwood et al. 2009) and Hi-C (Lieberman-Aiden et al. 2009) are high-throughput, genome-wide mapping technologies which reveal long-range chromatin interactions and provide data which can be used to reconstruct the 3D structure of the genome. The outputs of these technologies are pairwise interaction frequencies (IFs) between genomic loci, and these must be converted to relative physical distances. This process is known as inferential structure determination (ISD), and it is a fundamental tool in various disciplines, including NMR-based protein structure studies (Rieping et al. 2005). However, applying well-established ISD techniques to 3D genome mapping data is not straightforward, as the precise relationship between IFs and physical distances is unknown and may differ according to specific chromosomal segments and locations in the nuclear space. Nevertheless, recent results suggest that chromosome structure prediction is robust with respect to experimental noise but more sensitive to different structural-functional relationships (Trussart et al. 2015).
There are a number of published methods for 3D chromatin modeling using Hi-C data (Serra et al. 2015). Most of the approaches share the same strategy to convert the matrix of IFs into a matrix of preferred pairwise distances, and then use these distances in an optimization algorithm to build a chromosome structure. Variations of this approach are implemented in several programs, including TADbit (Baù et al. 2011; Baù and Marti-Renom 2012), MOGEN (Trieu and Cheng 2014, 2016), MCMC5C (Rousseau et al. 2011), AutoChrom3D (Peng et al. 2013), BACH (Hu et al. 2013), PASTIS (Varoquaux et al. 2014), InfMod3DGen (Wang et al. 2015), and HSA (Zou et al. 2016). Other approaches include ChromSDE (Zhang et al. 2013), that used semidefinite programming, and ShRec3D (Lesne et al. 2014), that used a graph theory to normalize the input heat map. An alternative strategy, described in Meluzzi and Arya (2013), avoids the conversion between IFs and preferred distances and instead uses the IFs to define a Boolean matrix indicating pairwise contact status between loci, and then generates structures most consistent with this matrix. All the efforts cited above suggest that we are still in an early and very active stage of searching for the most suitable methodologies for predicting the 3D genome organization from experimental mapping results.
We recently showed that ChIA-PET data has several advantages over Hi-C data in mapping and modeling the human genome (Tang et al. 2015). Hi-C data is genome-wide but generally lacks in specificity and is typically in lower resolution. Although recent Hi-C experiments have achieved kilobase (kb) resolution (Rao et al. 2014), obtaining such resolution requires extremely deep sequencing of billions of reads, which is impractical and prohibitive to many applications. On the other hand, ChIA-PET achieves mapping resolutions at the element level (protein factor binding sites) and requires just moderate sequencing depth (Li et al. 2010). Previously, ChIA-PET data was only analyzed for high-frequency chromatin interactions mediated by specific protein factors (Fullwood et al. 2009; Li et al. 2012; Kieffer-Kwon et al. 2013). Recently, we showed that the nonenriched portion of the ChIA-PET data can also be used to generate whole-genome contact maps in very good agreement with Hi-C data (Tang et al. 2015). Thus, ChIA-PET offers a multiscale view of chromatin organization, from multi-megabase contacts at low resolution down to sub-megabase topologically associating domains (TADs) (Dixon et al. 2012; Nora et al. 2012) and further to the element level of individual protein binding sites (at 50–100 bp). In light of this observation, there is now a need for a computational tool which takes advantage of this wealth of ChIA-PET data in a coordinated fashion to generate both high-resolution structures from specific protein-mediated chromatin interactions and lower resolution of chromosomal folding from the Hi-C-like data component.
Here, we introduce a 3-Dimensional Genome Modeling Engine (3D-GNOME), an open source suite of software tools that executes the 3D simulation pipeline for ChIA-PET data. 3D-GNOME is composed of multiple components, including data normalization, 3D modeling, visualization, and annotation. We demonstrate the utility of this pipeline by analyzing the ChIA-PET data derived from GM12878 cells, a well-characterized lymphoblastoid cell line with a relatively normal karyotype and completely phased genome sequences from the 1000 Genomes Project (Pedregosa et al. 2011; Tang et al. 2015). We generate models for whole-genome and individual chromosome structures, either as a single best average, or as an ensemble of 3D conformations at various resolution scales, and show how including CTCF-binding motif orientations enhances structural modeling. In addition, we test our computational approach on Hi-C data sets for low-resolution models, which generated similar results as the ChIA-PET data, validating that 3D-GNOME is suitable for modeling both types of 3D mapping data sets.
Results
Overview of 3D-GNOME strategy and modules
Paired-end-tags (PETs) generated from ChIA-PET library sequencing are typically divided into one of the three categories (Fig. 1A; Supplemental Fig. S1). PET singletons were originally defined as interactions which appear only once in a given library when mapped to reference genome, but here, we include in this category all PET clusters with an interaction count below four. Singletons reflect the higher-order topological organization of the genome, and when binned, they generate long distance contact maps (i.e., heat maps) similar to Hi-C data. PET clusters are long-range, statistically significant interactions which represent specific interactions mediated by the protein of interest. Self-ligation PETs are short-range PETs derived from the same chromatin DNA fragment (Supplemental Fig. S2) and indicate the locations of protein binding sites, similar to ChIP-PET (Wei et al. 2006) and ChIP-seq (Johnson et al. 2007) data. Additional details are provided in the Supplemental Material.
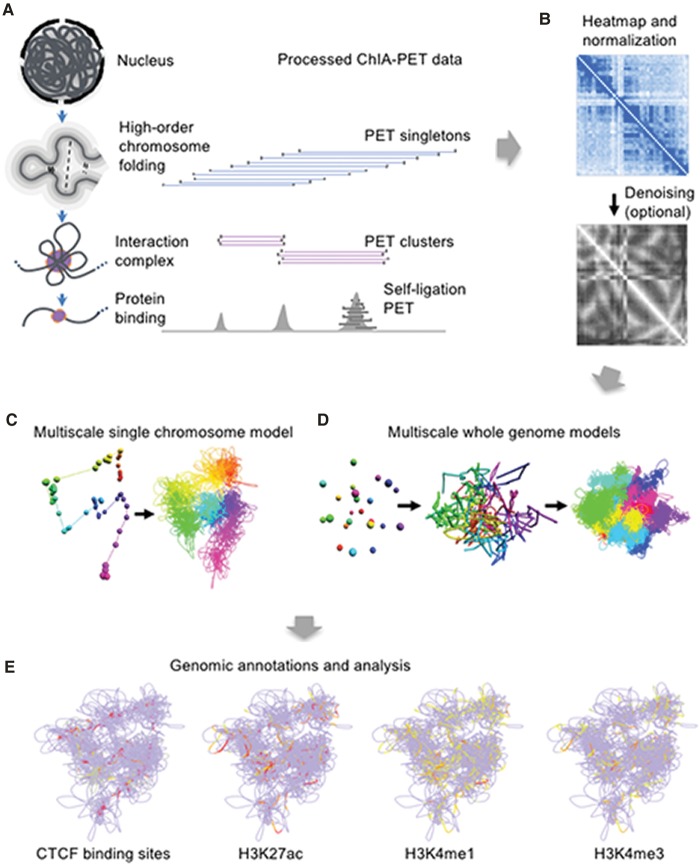
Figure 1.
Schematic overview of the 3D-GNOME pipeline. (A) Processed ChIA-PET library sequencing reads including the enriched PET clusters and the nonenriched PET singletons are the data sets to be used for 3D modeling. (B) PET reads are aggregated to construct 2D contact heat maps for each chromosome. Heat maps can be optionally de-noised using a graph distance (GD) normalization approach. (C) A model of a single chromosome (Chromosome 10) at low (left) and high (right) resolutions. (D) Whole-genome model at three different scales for GM12878 cells. (E) Structures are analyzed using a fully interactive 3D viewer. Genomic annotations may be overlaid on the structure to facilitate analysis. The marks depicted here were obtained from the ENCODE Project.
3D-GNOME is a pipeline for the complete 3D simulation analysis of ChIA-PET data: It takes as input a pair of bedpe-like files listing PET clusters and singletons and generates 2D contact heat maps and 3D structures. The platform consists of three independent but closely related modules (Fig. 1B–E). The first component (Fig. 1B) de-noises the data and generates low-resolution, HiC-like 2D heat maps, with options to normalize the IF matrix using a graph distance-based normalization. The second component (Fig. 1C,D) provides options for constructing 3D models of either individual chromosomes or the whole genome. The first option uses multidimensional scaling (MDS) to provide physical distance maps for rapid inference of the low-resolution 3D structure of individual chromosomes and the whole genome. The second option uses simulated annealing (SA) to construct 3D models at multiple scales, including high-resolution modeling using enriched chromatin interaction data, either independently from the first component (i.e., MDS analysis), or by using the low-resolution MDS chromosomal structures as initial conditions for high-resolution modeling (see Supplemental Fig. S3C–E). Lastly, the third component (Fig. 1E) is a set of web-based visualization and annotation tools for structural analysis. Although it is developed for ChIA-PET data, the pipeline is also suitable for modeling Hi-C data, with the caveat that models constructed using Hi-C data are necessarily limited to low-resolution structures. Although the modeling approach described here in the second component of the 3D-GNOME system is superficially analogous to some methods developed for Hi-C data, our approach is uniquely designed to take the advantage of the multiscale nature of ChIA-PET data and generates a group of related structures at varying resolutions, thus representing a noticeable advancement in this direction.
In the following sections, we describe the novel strategies that we employ in the first and second modules in 3D-GNOME, omitting most technical details, which can be found in the Methods section. The details of the visualization module can also be found in the Methods section.
Data-driven 2D contact heat maps
We have recently shown (Tang et al. 2015) that ChIA-PET data, including singletons that were previously discarded as background noise, in fact correlate well with the genome-wide signal from Hi-C experiments (Fig. 2; Supplemental Fig. S4). On this basis, we formulate a modeling strategy whereby singleton data is used to determine the higher-order, low-resolution structure of the genome, and the enriched chromatin interactions identified by PET clusters are used to determine high-resolution structures by refining the low-resolution structures. At each resolution scale, we must decide on the genomic span to use as the basic modeling units (individual beads in a typical “beads-and-springs” model of a polymer). We previously identified dense groups of overlapping CTCF-mediated loops, called chromatin contact domains (CCDs) (Tang et al. 2015), and we treat these experimentally defined domains as the basis for partitioning the genome. We then group these CCDs into “segments” (Supplemental Fig. S3A), where the size of the segment is tuned to match the desired resolution (around 2 Mb for the human genome) for modeling, and we treat these segments as the fundamental low-resolution unit. This clustering procedure is experimentally inspired by the recent CTCF biology (Rao et al. 2014; Sanborn et al. 2015; Tang et al. 2015) because it partitions the genome into highly interactive domains, thereby avoiding domain splitting that would occur with arbitrary binning.
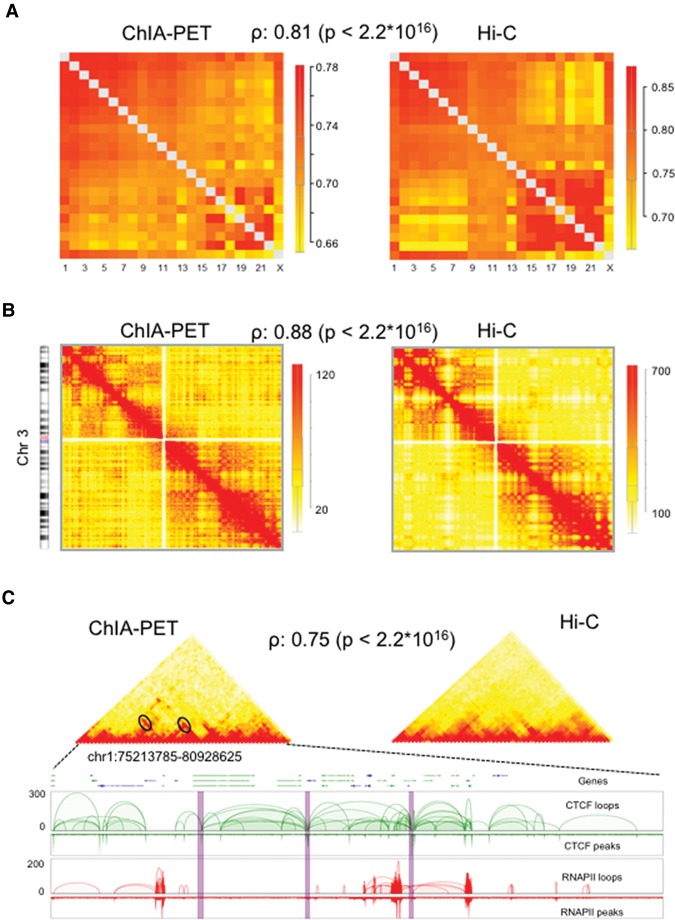
Figure 2.
Comparison of contact maps between ChIA-PET and Hi-C at multiscale. (A) Normalized inter-chromosomal contact heat maps between all pairs of chromosomes. The ChIA-PET and Hi-C show similar heat maps with a correlation coefficient of 0.73. The boxplot shows the normalized contact frequencies distribution. The color bar indicates the enrichment of contact frequency, where red indicates enriched contacts and yellow indicates depleted contacts. (B) The intra-chromosomal contact heat maps for Chr 3. The ChIA-PET and Hi-C showed similar contact heat maps with a correlation value of 0.67. The chromosome was binned at 1-Mb resolution. The boxplot shows the contact frequencies distribution. The color bar indicates the enrichment of contact frequency. Colors are as in A. (C) The contact heat maps of topological associated domains (TADs). The heat maps of two TADs display similar patterns between ChIA-PET and Hi-C. The bottom panel shows the interaction loop views for CTCF and Pol II PET clusters, respectively. Interaction hotspots are marked with ovals in contact heat maps and are highlighted with purple bars in 2D track views. TADs were binned at 50-kb resolution. CTCF and Pol II ChIA-PET data were combined to generate the heat maps.
We evaluate the utility of our binning scheme by comparing it with a conventional partition using bins of uniform size. Using Chromosome 6 as an example (Fig. 3A), although the higher-order heat map patterns are visually similar, careful inspection showed that the CCD-based heat map revealed more detailed contact structures with larger differences in the values of neighboring bins than the uniform-size heat map, which is suggestive of a better signal-to-noise ratio due to the use of the natural domain structure. To quantify this observation, we calculated the ratio of the value of neighboring bins for each heat map (see Methods) and plotted the resulting distributions (Fig. 3B). These distributions are significantly different (P < 2.2 × 10−16, Kolmogorov–Smirnov test), with the uniform-sized binning scheme exhibiting a much narrower range of values concentrated near 1, suggesting that uniform bins frequently divide the contact patterns from individual interactions, while CCD-based bins better describe the domains underlying these interactions (Fig. 3B).
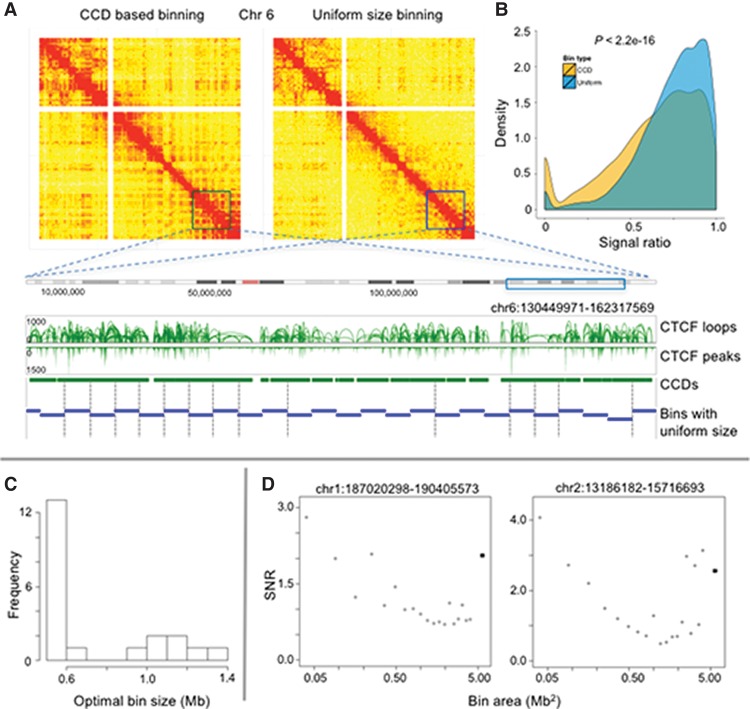
Figure 3.
Comparison of data-driven vs. uniform size genomic binning procedure. (A) Contact maps were generated using CCD-based binning (left) and uniform size binning (right) for Chromosome 6, respectively. The sizes of tiles in the contact map correspond to their genomic lengths. The same color scale was used for both contact maps. The squares in the contact maps were enlarged as a zoomed-in region, shown below in a genomic browser track view, in which the CCD-based bins are in green and the uniform-sized bins are in blue. The vertical dashed lines indicate the split of functional CCDs into pieces by uniform-sized bins. (B) Distribution of interaction frequency ratio for neighboring bins for all chromosomes by CCD (yellow) and uniform (blue) bins. (C) Histogram of the optimal uniform bin size for 21 long-range interactions identified in the ChIA-PET library. (D) Plots of the SNR as a function of uniform bin area for two of the long-range interactions. The values for the CCD-based bins are indicated by dark squares. The chromosome coordinates of each interaction are listed above the plots.
One complication with using a uniform binning scheme is choosing an ideal resolution that is high enough to avoid washing out interaction peaks but also low enough to avoid introducing spurious peaks due to noise in the data. Moreover, the ideal uniform bin size is likely to differ at each interaction locus. To illustrate this effect, we used 21 long-range (>2 Mb) and strong (IF > 8) interactions and constructed contact matrices at these loci using both CCD-based bins and uniform bins of various sizes (0.1–2 Mb). For each contact matrix, we calculated the signal-to-noise ratio (SNR) for the interaction peak, which we defined as the ratio between the matrix entry with the largest value and the background calculated as an average of its eight neighbors. The bin size that optimized the SNR varied across the 21 loci; for about half of the loci, the optimal bin size was 0.5–0.6 Mb, but the range was from 0.5 to 1.4 Mb (Fig. 3C). Intuitively, a high SNR is obtained for bins matching the size of interacting loci. As the size of interacting loci varies across the genome, a partition using uniform bin sizes would miss or underestimate the strength of interactions within regions smaller or larger than the bin size. In many cases, the CCD-based bins realized a near maximal SNR despite using much lower resolution bins (Fig. 3D). This analysis suggests that, at least at these loci, the CCD-based binning procedure better captures the underlying interaction patterns than the uniform binning scheme.
Although our analysis suggests that data-driven binning is an ideal scheme for 3D modeling at the resolution of chromatin domains, where the objective is to identify a small set of low-resolution bins with large SNRs, we note that CCD-based binning may not be ideal for all analyses, and a more extensive investigation of this issue is beyond the scope of our current study.
Simulation method
Low-resolution models are constructed by one of two methods, either simulated annealing or multidimensional scaling. In the SA approach, we first convert the interaction frequencies in the singleton heat maps into average distances between chromatin segments, following the conventional assumption that the two are related through , where i and j are the node indices, c is a scaling factor, and α is the scaling exponent. Given the preferred distances, we minimize a harmonic energy functional using SA to arrive at a best low-resolution structure. In the MDS approach, we first normalize the IF heat map to a graph distance (GD) heat map, use the GDs to infer physical distances, and use these physical distances to generate a structure using MDS. Our GD method is similar to previous works of Lesne et al. (2014), Fraser et al. (2009) and finally our own work (Pietal et al. 2015). The earlier work by Fraser et al. uses inverse IFs as the input to MDS, which yields low-quality structures with poor reproducibility. Lesne et al. improves over this formulation by computing the Floyd-Warshall shortest path over inverse IFs and then performing 3D reconstruction by using MDS. Our GD method generalizes the method of Lesne et al. by treating interaction frequencies directly as expected contact probabilities. Instead of using IFs to construct distances on a binary graph, we interpret IFs directly as the probability of connecting two nodes in the graph. All technical details are presented in the Methods. Supplemental Figure S5 shows examples of low-resolution structures for all 23 chromosomes.
Although motivated by ChIA-PET data, our low-resolution pipeline works equally well with Hi-C data. Supplemental Figures S6 and S7 show low-resolution models for all 23 chromosomes generated using MDS and based on either ChIA-PET (Supplemental Fig. S6) or Hi-C (Supplemental Fig. S7) data. There is general agreement between the ChIA-PET- and Hi-C-derived structures, which is yet another indication that ChIA-PET data recapitulates the salient features of Hi-C data.
High-resolution models are constructed in two steps, first by decomposing segments into individual PET interaction anchors and loops (the intervening space between two interacting anchors), converting PET IFs into preferred relative distances and minimizing a different energy functional, and then by filling in the regions between anchors with “subloop” nodes, and modeling the interaction between these nodes using standard polymer physics (stretching and bending energies).
The high-resolution models are then optionally refined as illustrated in Figure 4. It was recently reported that most CTCF interactions are aligned with their motifs in a “convergent” orientation, with a smaller portion aligned in a “tandem” conformation (Rao et al. 2014; Tang et al. 2015). Based on the structure of the CTCF dimer, we hypothesized that the chromatin interaction occurs with the CTCF binding motifs aligned, creating hairpin loop structures for convergent motifs and coiled loops for tandem motifs, as illustrated in Figure 4A. Thus, we included an interaction term that tends to align CTCF motifs, a consequence of which is more strictly organized CTCF clusters (Fig. 4B,C). As a second level of refinement, the loops between anchor regions are perturbed using PET singleton data to describe interactions in the loop region. Contact matrices (Fig. 4D) are constructed using a combination of the singleton data and polymer physics models (see Methods for details), and the IFs are converted to preferred distances. When incorporated in the model, these interactions modify the loops in various ways (Fig. 4E).
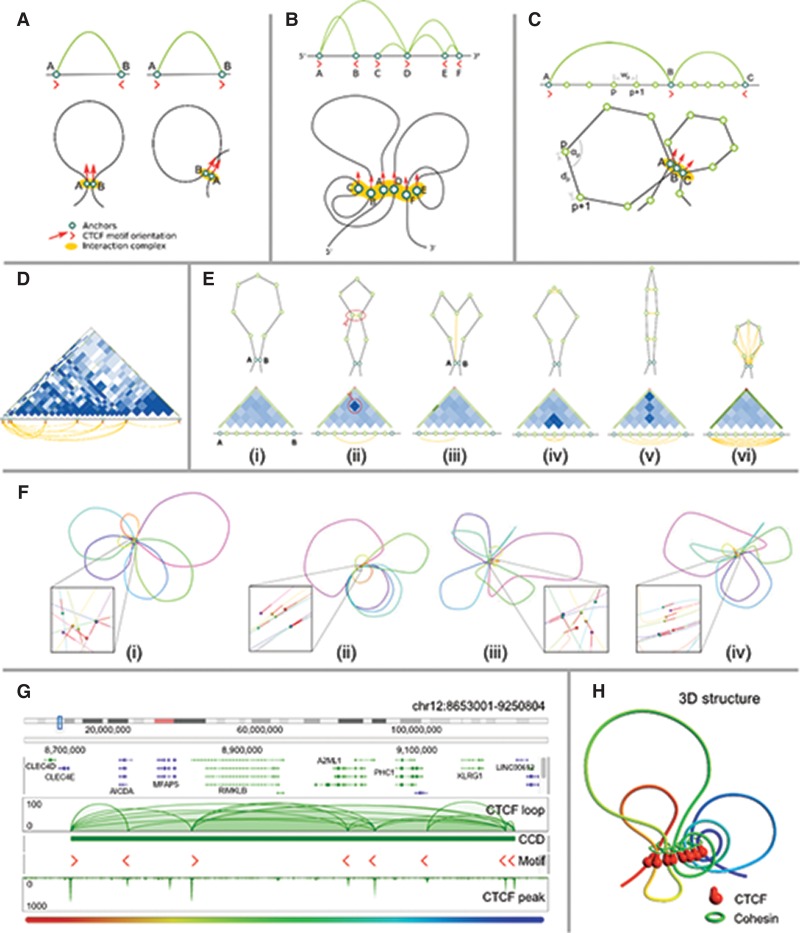
Figure 4.
Modeling of loop structures. (A) Two possible chromatin loop structures associated with CTCF motif orientation: hairpin loop structure resulting from the convergent CTCF motifs (left), and coiled loop structure resulting from tandem CTCF motifs (right). (B) Schematic representation of a CCD unit and possible chromatin loops stretch out of an interconnected interaction complex containing all the anchors bound by CTCF. (C) Simulation model on the subloop level. Genomic distance wp between two anchors of a loop (subloop) is translated to physical distance dp between the corresponding beads. During the simulation, the angles αi between two flanking segments (lines) are also considered. CTCF motif orientation could force the chromatin fiber to form different shapes of loops for convergent (A,B) and tandem (B,C) orientations. (D) An example of a subloop structure indicated by a heat map constructed by using all intra-loop PET singletons, with the heat maps generated assuming simple polymer physics. (E) Potential impacts of the intra-loop PET singleton data onto the likely shapes (i–vi) of chromatin loop. (F) Effect of including different subloop level energy terms onto the shapes of chromatin loops presented on a selected interaction block (Chr 17: 63226521–64565170). A zoomed-in view on the interaction complex is presented for each structure (spheres denote the anchors, red lines denote the CTCF motif orientations). (F[i]) In the base model, only loop segment lengths and angles between them are considered, resulting in regularly shaped and freely distributed loops. (F[ii]) Including the CTCF motif orientation in the model introduces more constraints and thus results in limited mobility of loops, which become aligned to each other. (F[iii]) Considering the subanchor heat map introduces more constraints on the loops’ shape and results in loops being more irregular. (F[iv]) Finally, including both CTCF motif orientation and subanchor heat maps yields irregular but aligned loops. For clarity, the simulation was restricted to two dimensions. (G) An example CCD identified at Chr 12: 8665991–9229876, with several anchors with well-defined CTCF motif orientations. (H) Predicted 3D structure for the CCD shown in G. A highly organized CTCF core is surrounded by chromatin loops of various sizes.
Figure 4F shows the effects of these refinements on the orientation and loop structure of a chromatin model (a 2D model was used for clarity). In a model without any additional interactions (Fig. 4F[i]), the PET anchors are unaligned and the loops are nearly circular. Including the motif information without the subloop heat map (Fig. 4F[ii]) strongly aligns the anchors without changing the character of the loops. Conversely, including the subloop heat map without the motif interaction (Fig. 4F[iii]) perturbs the loops without affecting alignment. When both interactions are turned on (Fig. 4F[iv]), the model recapitulates motif alignment with more realistic, irregular loops.
The inclusion of CTCF motif orientation and subloop refinement allows us to generate highly detailed high-resolution models. We modeled a CCD on Chromosome 12 of GM12878 cells (Fig. 4G), which generated a complex chromatin looping structure (Fig. 4H). The predicted structure shows the organized CTCF cluster resulting from motif alignment and an irregular looping structure most consistent with the data.
We validated our modeling approach using heat maps from simulated structures. A complete discussion of our validation experiments, the robustness of our approach, and parameter values can be found in the Supplemental Material.
Whole-genome modeling
The structure of the entire genome content—at the chromosomal, segmental, and subanchor levels—is presented in Figure 1D. Models at chromosomal and segmental levels provide a structural overview, highlighting the general shape and relative placement of chromosomes. The subanchor and loop level models provide a more detailed view of individual domains and chromatin loops. Despite not enforcing excluded volume in our simulation, chromosomes tended to segregate into discrete chromosomal territories. However, detailed examination revealed some physically impossible overlaps between chromatin loops, especially for large loops. This was a consequence of our decision to ignore long-range and inter-chromosomal interactions at the anchor level, as well as indiscriminate effects of excluded volume or constraints from the nuclear membrane. Future improved versions of 3D-GNOME will include features for these parameters, which can be turned on for more realistic modeling or turned off to increase computation speed.
Ensemble analysis
Given a large contact heat map of IFs, there could be several 3D structures which are consistent with the data. As ChIA-PET data represent an average over a large population of cells, generating an ensemble of consistent structures may be more representative of the diversity of chromatin conformations in the population. Using our hierarchical modeling approach allowed us to perform an ensemble analysis at multiple resolutions.
At each resolution, we generate 100 structures using different initial conditions, and then compare all pairs within the ensemble. To compare two structures, a and b, we use a distance measure given by
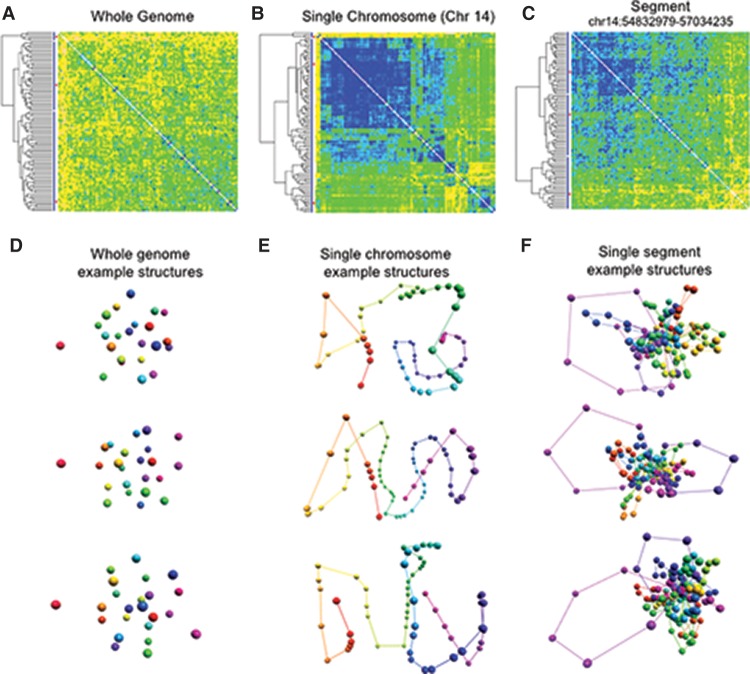
where DA (i, j) is the distance between beads i and j in structure A, and E(D(i, j)) is the expected distance between these beads given the heat map. This measure is insensitive to mirror symmetries and is thus more appropriate for chromosome data than the commonly used RMSD. We compute the distance between all pairs of structures in the ensemble and use these to build a pairwise distance matrix (Fig. 5). Clusters of similar structures are identified by performing a hierarchical clustering analysis on the pairwise distance matrix.
Figure 5.
Structural ensemble analysis for GM12878 cells at different resolutions. For each resolution, we generated 100 structures and constructed a matrix of pairwise distances between structures, which were hierarchically clustered. (A) The matrix of pairwise distances for the whole-genome models at the chromosome level. The differences between clusters are small, suggesting that the simulation on this level yields very similar results across different runs. (B) The matrix of pairwise distances between structures of Chromosome 14. Three large clusters dominate the set, and their extent is indicated by the red bars on the left. (C) The matrix of pairwise distances between three-dimensional structures for a subloop level model of a single TAD (Chr 14: 54832979–57034235). While there are visible differences between some of the structures, the clusters are not as well-defined, suggesting that several conformations of this region are present in the cell population. (D) Selected whole-genome structures from the three main subpopulation clusters identified in A. (E) Selected single chromosome structures from the main subpopulation clusters identified in B. (F) Selected single segment structures at the subloop level from the main subpopulation clusters identified in C.
At the whole-genome level, the pairwise distance matrix (Fig. 5A) shows only small differences between the models. The matrix does not decompose into any distinct clusters, suggesting that the simulation on this level yields very similar results across different runs. A possible explanation for this result is that, with only 23 beads, the number of restraints is on the same order as the degrees of freedom, and thus the system is characterized by a single, well-defined structure.
At the chromosome level, the clustering analysis consistently identified prominent subsets of structures. For example, the clustering analysis of Chromosome 14 indicated three large clusters categorized the majority of structures (Fig. 5B). From each of these main clusters, we selected representative structures for further study. The striking differences between the structures indicated that our clustering analysis indeed identified distinct subpopulations of structures.
To investigate high-resolution ensembles, we modeled the genomic region Chr 14: 54832979–57034235. We created 100 structures on the subloop level and performed the clustering as described above. There are easily discernable, visible differences between the structures, and the hierarchical clustering analysis did not identify any large, prominent clusters (Fig. 5C). This suggests that the local structure does not consist of a number of well-defined populations but instead dynamically samples a wide range of conformations. This behavior is to be expected in our simulation, because the scarcity of singleton data at the subloop level means the structure is dominated by the physics underlying a random polymer chain. Structures representing the three largest clusters for each level are presented in Figure 5, D–F.
Combining the previous results, the modeling suggests that the organization of the genome is most well-defined at the lowest resolution and becomes progressively more variable and dynamic at higher resolutions. This conclusion, which is obtained directly from the experimental data and static modeling, is in good agreement with our physical expectation that global structures undergo slower and less dramatic changes than the changes observed at high resolutions. We note, however, that each structure in an ensemble is still a representation of the population data, and at this point it is a hypothesis that these structures represent instances of actual structures in the population. To create an ensemble representing the true subpopulations (assuming that they exist), one would need to disentangle the contact heat map into a number of subpopulations heat maps, and methods for such a deconvolution are currently under investigation (Kalhor et al. 2012; Junier et al. 2015).
Discussion
We have demonstrated that the 3D-GNOME software suite performs well for 3D chromatin structure modeling using elements based on experimentally determined biological objects—CCDs and chromatin loops—instead of a uniform partitioning of the genome. Although the simulation algorithm was developed for ChIA-PET data, it can be adopted for any contact frequency data generated by other methods, as long as the location of chromatin contact domains and individual loops are provided for the high-resolution modeling. This can be inferred from the contact data itself or determined using a separate data set. In particular, there are now several software packages designed to identify topologically associating domains and loop regions from Hi-C data sets (for review, see Trussart et al. 2015), and these structures can be used in 3D-GNOME in the same manner as CCDs.
The three modules of 3D-GNOME are tightly integrated to provide a complete data analysis pipeline. The first software component consists of tools for 2D contact heat map generation and normalization using graph distance calculations. The graph distance calculations are required as input for the MDS algorithm but are otherwise completely optional. The second 3D-GNOME software component consists of structural modeling tools. If only low-resolution structures are of interest, then MDS may be preferred because of its computational speed. On the other hand, the Monte Carlo SA approach generates structures at multiple resolutions. This allows for a multiscale analysis of chromatin structure, whereby (1) the lowest resolution structure provides the relative positions of individual chromosomes in the nucleus, and (2) the segment level provides a low-resolution structure, while (3) the PET anchor and subanchor levels provide a high-resolution structure.
Finally, the third software component consists of a web-based, interactive 3D-viewer, used for visualizing and analyzing structures produced by the simulation platform. The viewer runs in any modern web browser and does not require any locally installed software. Furthermore, chromosomes can be displayed at different resolutions via a dropdown menu, and there are tools for highlighting regions of interest and overlaying genomic annotations. Examples of overlaid genome annotations are shown in Figure 1E, and a screenshot of the viewer is shown in Supplemental Figure S8. In addition to user-supplied annotations, we intend to integrate a wide range of genomic modification data from the ENCODE Project for direct use in the web-based viewer. Overlaying such annotation data on 3D structures can reveal patterns and clustering which are not apparent in the 2D heat maps (Hu et al. 2013). The general availability of such data in a web-based structural viewer is expected to greatly accelerate and enhance functional analysis.
The structures generated by our 3D-GNOME platform are robust to noise and to variations in the underlying parameters. It is important to note, however, that the true shapes of in vivo chromatin loops are not known, due to limitations in current experimental techniques and data interpretation. Therefore, the theoretical foundations of 3D-GNOME are based on current knowledge and physically reasonable assumptions from polymer theory. As new data becomes available, 3D-GNOME algorithms may need to be further improved. Nevertheless, our current platform contains useful and powerful analytical tools that will promote further research in this field.
Methods
ChIA-PET data
ChIA-PET data for GM12878 cells was obtained from Tang et al. (2015). PET interactions were identified and classified following the approach in Li et al. (2010), with modifications described by Tang et al. (2015), using a PET cutoff of 4—PET clusters with counts <4 are considered not significant, and these weak interactions are grouped with PET singletons. Although we prefer this analysis method, several methods are available for calling PET interactions (Paulsen et al. 2014; He et al. 2015; Phanstiel et al. 2015), and the output of any of these methods is suitable as input to 3D-GNOME.
The data processing pipeline identified 93,409 PET clusters as well as 35 million singletons from the CTCF ChIA-PET library (Supplemental Table S1). Similarly, we obtained 113,591 PET clusters and 53 million singletons from the Pol II ChIA-PET library (Supplemental Table S1).
2D contact heat map generation for comparison between ChIA-PET and Hi-C data
Hi-C data for the GM12878 cell line were obtained from Selvaraj et al. (2013). The expected number of inter-chromosomal interactions for each chromosome pair i, j was calculated by multiplying the number of inter-chromosomal PETs connected to chromosome i with the number of inter-chromosomal PETs connected to chromosome j and then dividing by the total number of inter-chromosomal PETs. Normalized contact frequencies were calculated by taking the actual number of inter-chromosomal PETs between chromosome i and j and then dividing by the expected value. For the intra-chromosomal contacts, each individual chromosome was divided into 1-Mb loci, and PET clusters and singletons were binned according to the location of both ends to produce a contact frequency matrix. The contact frequency matrix for topologically associated domains was generated in a similar manner.
Heat map generation using CCD-based binning and uniform size binning
CCDs were defined based on connectivity of CTCF loops for GM12878 cells, as described in Tang et al. (2015). Each chromosome has a variable number of CCDs (Supplemental Fig. S9), and the average size of CCDs is 980 kb. For the CCD binning scheme, each chromosome was partitioned according to its CCDs, and the CCDs were extended to encompass the gaps between them except for CCDs bordering the centromere. Heat map values were calculated using singleton data. Raw counts were first normalized to bin area to account for varying CCD sizes and then normalized according to Trieu and Cheng (2014). The normalized interaction count between region i and region j, Nij, is given by
where Fij = Cij/(Li Lj), Cij is the raw interaction count between regions i and j, and Li is the length of region i. The inverse of the second term in the product is the expected interaction frequency between regions i and j, assuming the expected interaction frequency is given by the number of interactions of each region divided by the total number of interactions (Trieu and Cheng 2014).
Graph distance map and multidimensional scaling
MDS is a general-purpose method for converting pairwise similarity matrices between objects to distances between those objects (Pietal et al. 2015). For our purposes, the objects are individual genomic loci and the similarity matrix is the heat map derived from singleton interactions. Our MDS component starts with the inter-chromosomal singleton heat maps normalization for the chromosome length. This is achieved by scaling all entries with a factor based on the respective chromosome sizes (see Supplemental Material for details).
Noise detection and removal
ChIA-PET contact maps are dominated by small values, which represent intrinsic background noise. MDS relies on the calculation of a graph distance between any two loci (described below), and for this calculation, background noise is particularly problematic because it generates spurious contacts between loci, which tend to collapse the overall structure. The histogram of heat map values, as shown in Supplemental Figure S10, shows that the noise is Gaussian distributed and generally much smaller than the values of true interactions. To remove this noise, we first use the Kolmogorov-Smirnov test to fit the small values in the histogram to a Gaussian (see Supplemental Material for details). The mean, µ, and standard deviation, σ, of this Gaussian are used to define a threshold, µ + nσ, with n a small positive integer. All heat map values smaller than this threshold are set to zero. The remaining values are scaled by the largest value in the heat map, which produces a frequency map with entries between 0 and 1. Finally, all diagonal entries are set equal to 1. All off-diagonal entries are also set equal to 1 if at least one of these entries is less than our contact threshold of 0.5. This step is required to ensure that the frequency map can be interpreted as a fully connected graph, as described below. The normalization procedure just described is applied to inter-chromosomal data and then separately to each intra-chromosomal heat map.
Graph distance calculation
The input to MDS is a matrix of graph distances between nodes. Given a graph of nodes connected by edges, the graph distance between any two nodes is simply an integer specifying the number of edges in the shortest path separating the nodes. If the frequency map generated above was truly binary (all values either 0 or 1), it would be trivial to construct a connected graph where each node represents a genomic loci and each edge an interaction. As our frequency map is not binary, we use well-established concepts from the field of fuzzy logic to define and calculate the graph distances.
Given a frequency map F, we construct the matrices of powers F2, F3, etc. The graph distance between nodes i and j, gij, is the smallest integer k, such that (Fk)ij is nonzero. One complication of this procedure is allowing for the use of modified forms of addition and multiplication, which are defined for fuzzy algebras. These modifications ensure that all operations produce values between 0 and 1 and can therefore be interpreted as frequencies. Multiplication is replaced by a T-norm, and addition by an S-norm. There are various T- and S-norms in common use, among which the simplest are the “probability norms,” namely,
We tried several T- and S-norms, including the probability norm, the Lukasiewicz norm, and the Hamacher norm, and found that they gave slightly different results. For graph distance calculations, matrix multiplication is performed as usual, but all instances of multiplication and addition are performed using the T- and S-norm, respectively. Supplemental Figure S11 shows the distribution of graph distances obtained using different T-norms.
Discrete to continuous graph distances
After calculating graph distances for all inter- and intra-chromosomal frequency maps, we use the scaling equation from Wang et al. (2015) to associate a physical diameter with nodes. This diameter should be directly proportional to the largest entry in the graph distance matrix. Thus, as a final step before MDS, we convert the discrete graph distance matrix into a continuous graph distance matrix by multiplying by the diameter and dividing by the maximum graph distance entry. This transformation generates a 2D graph distance matrix, which is a crude approximation of the real distance map and is thus fully suitable for further processing and refinement by the MDS. Supplemental Figure S12 shows a heat map of the graph distance matrix for the entire nucleus.
Multidimensional scaling
We run MDS with the fully processed graph distance map by using the MDS implementation in the Orange Data Mining package (Demšar et al. 2013). MDS uses the graph distances to construct a stress function and generates a 3D structure, which minimizes the stress function. The minimization procedure is very similar to Monte Carlo-based simulated annealing. Additional details are provided in the Supplemental Material. The output of MDS is a list of the spatial coordinates of beads representing all genomic loci defined in the graph distance map. Our script generates output files in the PDB format (Berman et al. 2000), which contains ALA/CA pseudoatoms for visualization in popular programs such as PyMOL (DeLano 2002) and Chimera (Pettersen et al. 2004). Additionally, a final distance map and graph distance map are generated in a PROTMAP2D format file, which is a slightly modified CASP RR format file (Pietal et al. 2007).
Supplemental Figure S13 shows heat map representations of the raw reads, the graph distances, and the result of MDS for Chromosomes 1 through 5. Supplemental Figure S14 shows the complete results for Chromosome 1, including the low-resolution 3D structure.
Multiscale Monte Carlo simulated annealing
Nucleome representation and partitioning
In our multiscale Monte Carlo approach, the human genome is represented as a hierarchical tree structure to take advantage of the multiscale nature of ChIA-PET data. A root node represents the entire nucleome. At all other levels, individual chromosomes are represented using a conventional beads-and-spring polymer model, where each bead represents a specific genomic location with well-defined initial and final genomic coordinates. The level below the root node consists of one bead for each chromosome. Thus, this level consists of 46 beads for diploid modeling, 23 beads for haploid modeling (the usual case in the absence of phased information), or a single bead for individual chromosome simulations.
Under each chromosome node are three additional levels, each representing the chromosome at increasing resolution. We use the following terminology to describe the beads at each level. At the lowest resolution level are “segments” which span ∼2 Mb. Next is the “anchor” level, where each bead represents an experimentally observed PET anchor. At the highest resolution are “subanchors,” which are beads representing the regions between observed anchors. The beads at each level have a parent-child relationship, where each low-resolution bead is associated with child beads, which span the same genomic region, as illustrated schematically in Supplemental Figure S15.
To construct the hierarchical tree, we begin with the anchor level. At this level beads are defined by the PET interaction anchors. Extremely long-range PET interactions (>2 Mb) are filtered at this level in order to facilitate the clustering described next.
We observed that PET interactions tend to cluster together such that there are genomic regions dense with numerous interactions which are separated by genomic regions devoid of interactions or with only weak interactions, as shown in Supplemental Figure S3A. We call the former regions chromatin contact domains. We hypothesize that CCDs possess a particular structure in which all the anchors belonging to the block are tightly packed together in an interaction complex with the chromatin loops extending on the sides.
The CCDs are not uniformly distributed across the genome but rather tend to cluster together. We aggregate CCDs that are separated by a relatively short genomic distance to define the segments, which are the fundamental objects of the lower resolution level above the anchor level. There are two reasons we use segments instead of CCDs as the nodes at the lower resolution level. First, CCDs that are very close to each other may in fact have interactions, which would be revealed with either deeper sequencing or by using a different protein factor to construct the ChIA-PET library. Secondly, the distribution of CCD sizes is very broad, ranging from 10 kb to 7 Mb (Supplemental Fig. S16). Clustering CCDs produces segments with a narrower size distribution, which both reduces size-related biases in the heat maps and allows for a more direct interpretation of predicted chromosome structures.
We note that, while the identification of CCDs is unique, their clustering into segments is arbitrary. The program will automatically cluster CCDs into segments based on the user's desired resolution, the size and density of the CCDs, and the size of intervening gaps. Automatic partitioning generally produces acceptable segments for modeling, but as an alternative, the program will also accept user-defined partitions based on a visual inspection of the CCDs. The results presented below were generated with a manual partition of the genome. See the Supplemental Material for more details.
To model the loops between PET anchors, we introduce a subanchor level, which consists of beads that fill in the genomic regions between anchors. We do this in the simplest possible manner, namely, the gap between anchors within a CCD is divided into a predefined number of regions. Here, we used either five or seven subanchors per loop. Note that the regions between CCDs are not modeled in this way, primarily because we do not have enough data to model these regions, but also because we do not expect substantial looping in these gaps.
Simulated annealing for structure reconstruction
The general procedure for generating a structure is the same for each level. Energy is defined, which is a function of the node positions, and Monte Carlo simulated annealing is used to find a structure, which minimizes the energy. In general, the energy can be written as
where the first term includes standard polymer interactions such as stretching and bending energies, and the second term includes all additional interactions imposed by the experimental data. The experimental data are used to define a preferred distance, dij, between each interacting pair of nodes i and j, and the energy is a function of these preferred distances and the actual distances, rij, between each pair of nodes. The exact energy function and the method of calculating preferred distances are different for each level, as described below. We work in a top-down approach, where lower resolution structures are constructed first and are used to inform the structures at higher resolutions.
Chromosome and segment levels
The methods at the chromosome and segment level are similar and we present them together. At the chromosome level, we use both inter-chromosomal PET interactions and singletons to construct inter-chromosomal heat maps. The value of the heat map entry between two chromosomes is the sum of the individual IFs for all PET interactions and singletons between those chromosomes. At the segment level, we use intra-chromosomal singletons and long-range PETs (those that were filtered out at the anchor level) to construct a heat map.
After binning, the heat maps are first normalized to account for bin size, the idea being that larger regions will naturally experience more interactions just by chance. Given two segments with lengths si and sj (in Mb), we define a normalized IF , where fij is the raw IF. Next, we rescale the heat map so that each row contains the same total count, based on the assumption that each genomic region should have equal “visibility.” Optionally, the heat map can be de-noised using the procedure described in the MDS section. The normalized IFs are then converted into preferred distances using an inverse power law , where c is a constant of proportionality. The best power for structural modeling has been extensively discussed in previous studies (Rousseau et al. 2011). Different polymer models predict different powers, but Rousseau et al. (2011) showed that structural predictions are robust over a range of parameters. We performed a ranking-based analysis to identify the parameter value α = 0.6 that yields the structures for which distances between beads best correspond to the contact frequencies (see Supplemental Material; Supplemental Fig. S17). We highlight the fact that the frequency-distance scaling is performed only on the chromosome and segment level, i.e., at the resolution of a few Mb. At the higher resolutions, we use genomic distance to define a physical distance, as described below. Some of the predicted distances are unrealistically large because of small IFs, which are a consequence of low binding affinity, poor mappability, or some other technical artifact. We therefore cut off large distance values using , where is the average heat map distance and ξ > 1 (typically, we use 2 < ξ < 3). Optionally, the pipeline can use the MDS procedure described above to generate preferred distances for low-resolution levels.
The energy function is , which is the standard harmonic potential where each interaction is represented by a spring connecting two beads. At the chromosome level, the initial configuration is determined by randomly placing each bead in the nuclear volume. Simulated annealing is run for several random initial configurations, and the configuration with the lowest energy is chosen as the best structure. At the segment level, the initial configuration is constructed by randomly positioning the segment beads within a sphere centered at the parent chromosome position of radius Rc. This initial configuration ensures segment beads begin near their preferred position and thus speeds convergence of the simulated annealing algorithm. Optionally, the low-resolution structure from MDS can be used as the initial configuration.
We note that inter-chromosomal interactions may or may not be used during the segment level simulation. If they are used then the model will accurately capture the relationship between genomic fragments on different chromosomes. However, it can be argued that such relations aren't very confident, partly because of the difficulty with obtaining phased diploid contact maps, and thus it may be preferable to use only the more confident intra-chromosomal interactions at this level. In this case, the chromosome conformations are constructed independent of each other but with a proper relative position in a nucleome. This does not modify our model, and either approach can be used depending on the quality of the data, depth of sequencing, or other biological and/or computational factors.
Anchor level
Only PET interactions are used when calculating the IF between two anchors, and the intervals between two anchors are the inferred loops. Anchor level heat maps are constructed for each segment individually because there are no PET interactions between anchors in different segments. This fact allows us to model all anchors within a segment independent of the other anchors and results in a significant increase in computational speed. IFs are converted into preferred distances using , where α, β, γ, and δ are constants. The energy function is identical in form to the one used at the chromosome and segment levels. The initial positions of anchors are determined in two steps. First, a Catmull-Rom interpolating spline between segment level beads is used to determine positions of CCDs, and then the anchors associated with a CCD are positioned randomly in a sphere centered at the CCD's position.
Subloop level
At the subloop level, we consider several contributions to the energy in order to properly generate the loops between anchors. First, in order to ensure that the physical size of a loop scales with its genomic span, we include a term which imposes a larger distance between sequential subanchor beads separated by a larger genomic span. All polymer models predict a power law relationship between arc length and physical size, and thus we use , where Ni,i+1 is the number of base pairs between subanchors i and i + 1. These preferred distances contribute a term to the total energy. Next, we include a bending energy, which prevents excessive curvature. This energy is , where is the unit vector pointing from subanchor i to subanchor i + 1.
We introduced the orientation of CTCF binding motifs into our computational algorithm in the following way. The genomic orientation of a motif (determined by whether the motif is directed upstream or downstream) is reflected in the structural, 3D motif orientation, which is defined as a unit vector tangent to the chromatin curve at the location of the motif. The vector points either “along” the fiber (from the 5′ to 3′ direction) or in the opposite direction, depending on the genomic motif orientation. These directions correspond to rightward and leftward motifs, respectively. We assume a pair of interacting anchors with CTCF motifs will preferentially align with their tangent vectors pointing in the same spatial direction (Fig. 4A,B). To account for this interaction, we include a third energy term based on the orientation of interacting anchors, , where is the orientation of the anchor i and P is a set of pairs of interactions in the current CCD. A schematic representation of the model containing these three terms is depicted in Figure 4C.
These energy terms can be used to model smooth, circular loops passing through the fixed anchor beads, but they do not account for interactions between subanchors in different loops. To determine the effect of these interactions, we build two heat maps, as illustrated in Supplemental Figure S28. First we use the intra-CCD singletons to construct a subanchor heat map (Supplemental Fig. S28B). This heat map is not directly used to compute preferred distances because it contains many null entries, which are simply consequences of the sparseness of interaction data at extremely high resolutions. To impute these missing values, we construct several structures using just the distance and bending energies (Supplemental Fig. S28C). For each structure, we construct a heat map using the distance between each pair of loci, and then these heat maps are averaged to produce a consensus distance heat map. Each entry in the distance map is then decreased in proportion to the corresponding entry in the singleton heat map to generate a refined distance heat map (Supplemental Fig. S28D), and these reduced distances are used to define the fourth energy term, . This term is similar to Edist but the sum is over all pairs of beads instead of just over neighbors. An example subanchor structure is shown in Supplemental Figure S28E.
Combining these terms, we arrive at Esubanchor = wdist Edist + wbend Ebend + worn Eorn + wheat Eheat, where wdist, wbend, worn, and wheat are weights assigned to particular energy terms.
Optimization algorithm
Monte Carlo simulated annealing proceeds in the conventional fashion, namely, at each step a random bead is chosen and shifted by a vector drawn at random from a sphere of a specified radius. The new energy is calculated, and the move is accepted if Enew ≤ Eold. If Enew > Eold, then the move is accepted with probability
where T is analogous to the temperature. This form differs from the Boltzmann form typically used in Metropolis Monte Carlo simulations, but any form is acceptable for simulated annealing, and this form is convenient because it is insensitive to the magnitude of the energies and thus provides more flexibility with parameter choices. The “temperature” is initialized to Tinit > 0, and is reduced after each step, Tnew = κTold, for some κ < 1. The simulation is checked every Nmilestone steps, and the simulation is stopped when the energy decrease since the last milestone is below a user-defined threshold.
Visualization tools
An essential component of any spatial modeling pipeline is the ability to visualize the resulting structures. We first developed a desktop application written in C++ to visualize the structures, using the Qt framework to create the user interface and OpenGL for 3D rendering. To improve portability, we then developed an HTML5 web application supporting three-dimensional interactive visualization. A screenshot of the application is shown in Supplemental Figure S8. We use the open source JavaScript library Three.js to enable WebGL based, GPU-accelerated three-dimensional graphics. The user interface is developed with the lightweight dat.gui library. Simulation output files in HDF5 format are served using the JSON RESTful Web Services API to dynamically deliver data to the client side browser. By choosing an HTML5 implementation for the user interface, we can ensure that any user with a modern browser can simply visit our applications web site in order to start using our application with interactive 3D graphics. This is in contrast to most other 3D modeling applications, which may have specific operating system requirements (usually requires desktop installer).
The viewer supports the full range of mouse-controlled 3D transformations, including translations, rotations, and zooms. The viewer was built to support the multiscale modeling philosophy of 3D-GNOME and allows the user to seamlessly transition between structures at different resolutions. Whole-genome visualization is provided up to the 2-Mb resolution, while individual chromosomes can be rendered at the 10- to 100-kb resolution. Structural analysis is aided through a variety of visualization options. At the highest resolution, the viewer can display the location of DNA binding proteins, which were used to generate the ChIA-PET library, and indicate the local interactions between genomic regions that are mediated by these factors. Specific regions of the chromosome may be highlighted by defining the genomic coordinates of interest.
Functional analysis is supported by allowing the user to upload a genomic annotation file in the standard bed format. The genome is then colored to reflect the annotation values in different regions, as shown in Figure 1E and Supplemental Figure S18. Such analysis can reveal patterns and clustering of histone marks or DNA methylation, which were not apparent in the 1D looping maps or the 2D heat map. We are currently working to provide the full data set of ENCODE-tabulated variations for various cell lines as default annotation options.
It is often of interest to compare genomic structural changes induced by an experimental protocol to a control genome, or to compare genomic structure at different time points after drug treatment or other perturbation. To facilitate such comparisons, the web site provides the option to simultaneously display multiple independent 3D viewers. Automatically identifying the optimal alignment between two large structures is a nontrivial operation and is currently not supported. However, it is often trivial for a user to visually align two chromosomes or regions of interest, for example, by highlighting specific regions to aid in alignment. Once a desired alignment is obtained, identical transformations can be enacted by manually entering translation vectors or rotation angles.
Software availability
The software that was used for this study is available for download (see Supplemental Material). The most recent version of the software can be found on the project web page (http://nucleus3d.cent.uw.edu.pl/3dgnome/) and the BitBucket repository (https://bitbucket.org/3dome/3dgnome) as a stand-alone software package. We also provide an open access to the developer versions of the components. The experimental data used in the study are available at the project home page.
Supplementary Material
Acknowledgments
D.P., P.S., M.S., and M.J.P. are supported by grants from the Polish National Science Centre (2014/15/B/ST6/05082 and 2013/09/B/NZ2/00121), and the European Cooperation in Science and Technology action (COST BM1405 and BM1408). P.S. and D.P. are supported by funds from National Leading Research Centre in Bialystok and the European Union under the European Social Fund. Y.R. is supported by the Director Innovation Fund from the Jackson Laboratory and National Cancer Institute (NCI) grant R01 CA186714. Y.R. is also supported by the Roux family as a Florine Roux Endowed Chair and Professor in Genomics and Computational Biology. All authors were supported by National Institutes of Health grant 1U54DK107967-01, “Nucleome Positioning System for Spatiotemporal Genome Organization and Regulation,” within the 4DNucleome NIH program. We thank Keith Sheppard and Mei Xiao for their contributions to the web-based viewer, particularly to the server side, and Gosia Popiel for help in preparing Figure 4.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.205062.116.
References
- Baù D, Marti-Renom MA. 2012. Genome structure determination via 3C-based data integration by the integrative modeling platform. Methods 58: 300–306. [DOI] [PubMed] [Google Scholar]
- Baù D, Sanyal A, Lajoie BR, Capriotti E, Byron M, Lawrence JB, Dekker J, Marti-Renom MA. 2011. The three-dimensional folding of the α-globin gene domain reveals formation of chromatin globules. Nat Struct Mol Biol 18: 107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont AS. 2014. Large-scale chromatin organization: the good, the surprising, and the still perplexing. Curr Opin Cell Biol 26: 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat T, Weissig H, Shindyalov IN, Bourne PE. 2000. The protein data bank. Nucleic Acids Res 28: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA: http://www.pymol.org/. [Google Scholar]
- Demšar J, Curk T, Erjavec A, Gorup Č, Hočevar T, Milutinovič M, Možina M, Polajnar M, Toplak M, Starič A, et al. 2013. Orange: data mining toolbox in Python. J Mach Learn Res 14: 2349–2353. [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser J, Rousseau M, Shenker S, Ferraiuolo MA, Hayashizaki Y, Blanchette M, Dostie J. 2009. Chromatin conformation signatures of cellular differentiation. Genome Biol 10: R37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. 2009. An oestrogen-receptor-α-bound human chromatin interactome. Nature 462: 58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Zhang MQ, Wang X. 2015. MICC: an R package for identifying chromatin interactions from ChIA-PET data. Bioinformatics 31: 3832–3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Deng K, Qin Z, Dixon J, Selvaraj S, Fang J, Ren B, Liu JS. 2013. Bayesian inference of spatial organizations of chromosomes. PLoS Comput Biol 9: e1002893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. 2007. Genome-wide mapping of in vivo protein-DNA interactions. Science 316: 1497–1502. [DOI] [PubMed] [Google Scholar]
- Junier I, Spill YG, Marti-Renom MA, Beato M, le Dily F. 2015. On the demultiplexing of chromosome capture conformation data. FEBS Lett 589: 3005–3013. [DOI] [PubMed] [Google Scholar]
- Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. 2012. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol 30: 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer-Kwon KR, Tang Z, Mathe E, Qian J, Sung MH, Li G, Resch W, Baek S, Pruett N, Grontved L, et al. 2013. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell 155: 1507–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesne A, Riposo J, Roger P, Cournac A, Mozziconacci J. 2014. 3D genome reconstruction from chromosomal contacts. Nat Methods 11: 1141–1143. [DOI] [PubMed] [Google Scholar]
- Li G, Fullwood MJ, Xu H, Mulawadi FH, Velkov S, Vega V, Ariyaratne PN, Mohamed YB, Ooi HS, Tennakoon C, et al. 2010. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol 11: R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. 2012. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148: 84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluzzi D, Arya G. 2013. Recovering ensembles of chromatin conformations from contact probabilities. Nucleic Acids Res 41: 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen J, Rødland EA, Holden L, Holden M, Hovig E. 2014. A statistical model of ChIA-PET data for accurate detection of chromatin 3D interactions. Nucleic Acids Res 42: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. 2011. Scikit-learn: machine learning in Python. J Mach Learn Res 12: 2825–2830. [Google Scholar]
- Peng C, Fu LY, Dong PF, Deng ZL, Li JX, Wang XT, Zhang HY. 2013. The sequencing bias relaxed characteristics of Hi-C derived data and implications for chromatin 3D modeling. Nucleic Acids Res 41: e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- Phanstiel DH, Boyle AP, Heidari N, Snyder MP. 2015. Mango: a bias-correcting ChIA-PET analysis pipeline. Bioinformatics 31: 3092–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietal MJ, Tuszynska I, Bujnicki JM. 2007. PROTMAP2D: visualization, comparison and analysis of 2D maps of protein structure. Bioinformatics 23: 1429–1430. [DOI] [PubMed] [Google Scholar]
- Pietal MJ, Bujnicki JM, Kozlowski LP. 2015. GDFuzz3D: a method for protein 3D structure reconstruction from contact maps, based on a non-Euclidean distance function. Bioinformatics 31: 3499–3505. [DOI] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieping W, Habeck M, Nilges M. 2005. Inferential structure determination. Science 309: 303–306. [DOI] [PubMed] [Google Scholar]
- Rousseau M, Fraser J, Ferraiuolo MA, Dostie J, Blanchette M. 2011. Three-dimensional modeling of chromatin structure from interaction frequency data using Markov chain Monte Carlo sampling. BMC Bioinformatics 12: 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn AL, Rao SS, Huang S-C, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. 2015. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci 112: E6456–E6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj S, Dixon JR, Bansal V, Ren B. 2013. Whole-genome haplotype reconstruction using proximity-ligation and shotgun sequencing. Nat Biotechnol 31: 1111–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra F, Di Stefano M, Spill YG, Cuartero Y, Goodstadt M, Bau D, Marti-Renom MA. 2015. Restraint-based three-dimensional modeling of genomes and genomic domains. FEBS Lett 589: 2987–2995. [DOI] [PubMed] [Google Scholar]
- Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, Szalaj P, Trzaskoma P, Magalska A, Wlodarczyk J, Ruszczycki B, et al. 2015. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell 163: 1611–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu T, Cheng J. 2014. Large-scale reconstruction of 3D structures of human chromosomes from chromosomal contact data. Nucleic Acids Res 42: e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu T, Cheng J. 2016. MOGEN: a tool for reconstructing 3D models of genomes from chromosomal conformation capturing data. Bioinformatics 32: 1286–1292. [DOI] [PubMed] [Google Scholar]
- Trussart M, Serra F, Baù D, Junier I, Serrano L, Marti-Renom MA. 2015. Assessing the limits of restraint-based 3D modeling of genomes and genomic domains. Nucleic Acids Res 43: 3465–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoquaux N, Ay F, Noble WS, Vert J-P. 2014. A statistical approach for inferring the 3D structure of the genome. Bioinformatics 30: i26–i33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Xu J, Zeng J. 2015. Inferential modeling of 3D chromatin structure. Nucleic Acids Res 43: e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C-L, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. 2006. A global map of p53 transcription-factor binding sites in the human genome. Cell 124: 207–219. [DOI] [PubMed] [Google Scholar]
- Wendt KS, Grosveld FG. 2014. Transcription in the context of the 3D nucleus. Curr Opin Genet Dev 25: 62–67. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li G, Toh KC, Sung WK. 2013. 3D chromosome modeling with semi-definite programming and Hi-C data. J Comput Biol 20: 831–846. [DOI] [PubMed] [Google Scholar]
- Zou C, Zhang Y, Ouyang Z. 2016. HSA: integrating multi-track Hi-C data for genome-scale reconstruction of 3D chromatin structure. Genome Biol 17: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.