Abstract
DNA methylation at the 5-position of cytosine (5mC) is an epigenetic modification that regulates gene expression and cellular plasticity in development and disease. The ten-eleven translocation (TET) gene family oxidizes 5mC to 5-hydroxymethylcytosine (5hmC), providing an active mechanism for DNA demethylation, and it may also provide its own regulatory function. Here we applied oxidative bisulfite sequencing to generate whole-genome DNA methylation and hydroxymethylation maps at single-base resolution in human normal liver and lung as well as paired tumor tissues. We found that 5hmC is significantly enriched in CpG island (CGI) shores while depleted in CGIs themselves, especially in active genes, which exhibit a bimodal distribution of 5hmC around CGI that corresponds to H3K4me1 modifications. Hydroxymethylation on promoters, gene bodies, and transcription termination regions (TTRs) showed strong positive correlation with gene expression within and across tissues, suggesting that 5hmC is a marker of active genes and could play a role in gene expression mediated by DNA demethylation. Comparative analysis of methylomes and hydroxymethylomes revealed that 5hmC is significantly enriched in both tissue-specific DMRs (t-DMRs) and cancer-specific DMRs (c-DMRs), and 5hmC is negatively correlated with methylation changes, especially in non-CGI-associated DMRs. These findings revealed novel reciprocity between epigenetic markers at CGI shores corresponding to differential gene expression in normal tissues and matching tumors. Overall, our study provided a comprehensive analysis of the interplay between the methylome, hydroxymethylome, and histone modifications during tumorigenesis.
DNA methylation is an important epigenetic modification that plays a role in diverse biological processes, including maintenance of genomic stability, gene silencing, embryonic development, and tumorigenesis (Jones 2012; Smith and Meissner 2013; Timp and Feinberg 2013). Recently, a family of ten-eleven translocation methylcytosine dioxygenase (TET) proteins was shown to oxidize 5mC (5-methylcytosine) to 5hmC (5-hydroxymethylcytosine) (Laird et al. 2013; Wu and Zhang 2014). TET-mediated 5hmC is abundant in a variety of mammalian tissues, such as brain and stem cells (Pastor et al. 2013), and plays a role in epigenetic reprogramming, cell differentiation, and tumorigenesis.
In contrast to a relatively constant 5mC level across tissues, 5hmC is highly tissue-specific (Nestor et al. 2012). 5hmC has been shown to be enriched in promoters, gene bodies, and distal cis-regulatory elements, such as enhancers, and thus potentially involved in regulation of tissue-specific gene expression (Pastor et al. 2013; Wu and Zhang 2014). However, how 5hmC regulates and shapes tissue-specific epigenomes through DNA demethylation remains largely unknown.
Significant global loss of hydroxymethylation has been observed in cancer, and disruption of TET-mediated DNA demethylation was proposed to contribute to tumorigenesis (Kudo et al. 2012; Pfeifer et al. 2013). Loss-of-function mutations in TET2 are found in myelodysplastic syndrome (Delhommeau et al. 2009; Langemeijer et al. 2009), and mutations in the isocitrate dehydrogenase IDH1/IDH2, co-factors of TET enzymes, are found in glioma, acute myeloid leukemia, and melanoma (Dang et al. 2010; Shibata et al. 2011). Down-regulation of TETs and IDHs is also found in several cancer types (Lian et al. 2012; Yang et al. 2013). By depletion of TET enzymes in a pluripotent embryonic carcinoma cell (ECC) model, it was proposed that disruption of TET-mediated 5hmC, which associated with a transcriptionally active chromatin environment, represented a crucial step toward aberrant gene silencing through DNA methylation in cancer cells (Putiri et al. 2014). Limited by the lack of high-resolution hydroxymethylomes in normal and matched-tumor tissues, dynamic changes in 5hmC and underlying mechanisms during carcinogenesis have not yet been elucidated.
Distinguishing 5hmC from 5mC is virtually impossible by traditional bisulfite conversion-based methods. Several strategies have been developed to label and enrich 5hmC, followed by sequencing with limited resolution (Laird et al. 2013; Putiri et al. 2014). Recently, two methods were implemented allowing for quantification of 5hmC at a single-base resolution. TET-assisted bisulfite sequencing (TAB-seq) (Yu et al. 2012) makes use of enzymatic reactions: β-glucosyltransferase to glucosylate 5hmC and TET1 for subsequent oxidation to produce 5-carboxylcytosine (5caC). In the final step of bisulfite treatment, both 5caC and unmodified cytosine are converted to uracil, allowing the identification of 5hmC, which is read as cytosine during sequencing. Oxidative bisulfite sequencing (oxBS-seq) (Booth et al. 2012) rather makes use of the highly selective chemical oxidant potassium perruthenate (KRuO4) to convert 5hmC to 5-formylcytosine (5fC), followed by bisulfite conversion and sequencing. This library is then compared to a traditional bisulfite sequencing library (BS-seq) constructed on the same sample to identify differences in 5mC that account for 5hmC positions. oxBS-seq has a relatively simple and fast experimental workflow and can obtain both the methylome and hydroxymethylome simultaneously.
Here, we utilized the oxBS-seq method to study, at single-base resolution, the methylomes and hydroxymethylomes of a normal and matching tumor set from human lung and liver tissues, providing a valuable resource for studying 5hmC landscapes in different tissues and understanding the roles of 5hmC during tumorigenesis. We performed a novel integrated analysis with RNA-seq, ChIP-seq data, genome-wide DNA methylation and hydroxymethylation maps in normal and tumor tissues. Analysis of differentially methylated regions (DMRs) in normal and tumor tissues revealed a negative correlation between changes of 5mC and 5hmC/H3K4me1. Together our findings demonstrated intricate gene expression regulation through the interplay of methylome, hydroxymethylome, and histone modifications during tissue differentiation and tumorigenesis.
Results
Generation of single-base resolution hydroxymethylation and methylation maps in matched human normal and tumor samples from lung and liver
We applied oxBS-seq and BS-seq to genomic DNA extracted from four human liver normal-tumor pairs and three human lung normal-tumor pairs (14 samples total). All libraries were sequenced to an average depth of 15.4 × per CpG cytosine (Supplemental Table S1). In order to evaluate bisulfite (BS) and oxidative bisulfite (oxBS) conversion rates, nonmethylated E. coli and CpG hydroxymethylated lambda phage DNA were spiked in as controls during library preparation. Based on spike-in controls, both high bisulfite (unmethylated cytosine to uracil, 99.66%) and high oxidative bisulfite conversion (5-hydroxymethycytosine to uracil, 96.57%) were observed (Supplemental Table S1).
According to the oxBS-seq method principle (Booth et al. 2013), the hydroxymethylation level of cytosines was calculated for each sample based on the differential methylation between oxBS-seq and the corresponding BS-seq libraries. Since human tissues, except for brain, exhibit a relatively low abundance of 5hmC (Nestor et al. 2012), high sequencing coverage was suggested in order to achieve an accurate 5hmC measurement at single CpG sites (Booth et al. 2013). Here, in order to confidently identify 5hmC-enriched regions and sites, we took advantage of the information from biological replicates and adjacent CpG sites, using an algorithm based on local likelihood smoothing (BSmooth) to identify consensus 5hmC regions and sites for each normal and tumor group. This method was successfully applied to DMR identification between groups with low sequencing coverage in our previous studies (Hansen et al. 2011, 2014).
In total, 89,437–297,724 5hmC regions containing 1,013,839–3,178,223 CpG sites were identified in normal tissues, while only 2255–37,159 5hmC regions containing 15,567–365,443 CpG sites were found in matching tumor tissues (Supplemental Tables S2–S6). The global hydroxymethylation levels of normal tissues are 2.27%–5.68% in liver and 1.94%–3.04% in lung, while significantly lower in the matched tumors (liver: 0.70%–2.07%; lung: 0.65%–1.07%). These results are similar to those achieved by mass spectrometry or antibody-based assays (Jin et al. 2011a; Lian et al. 2012; Yang et al. 2013). Considering the incomplete conversion of 5hmC to 5fC in oxBS-seq that could result in false negatives in 5hmC detection, the reported global 5hmC level here is a conservative underestimation. In contrast to the small variation in the global 5mC level among patients in normal tissues, the global 5hmC level showed considerable variation in both normal and tumor tissues (Supplemental Fig. S1).
To further verify our BSmooth method, we did deep sequencing (55 × coverage) for one liver normal sample and compared the estimated 5mC and 5hmC profiles based on 55 × data to that based on the original 15 × data. High correlations (5mC: 0.96; and 5hmC: 0.82) were observed between the two sets to data when smoothing both high and low coverage data (Supplemental Fig. S2). Additionally, we estimated 5mC and 5hmC levels based on high coverage data and using only CpGs at least 20 × coverage over 2-kb intervals across the whole genome and compared them to smoothed results based on low coverage data. We also found close agreement between them for both 5mC (0.90) and 5hmC (0.79). These results validated our BSmooth approach that estimates 5mC and 5hmC levels with high accuracy using relatively low coverage data. Furthermore, 15 identified liver cancer DMRs (c-DMRs) and 5hmC regions were picked out, and all 15 c-DMRs and 13 out of 15 5hmC regions were replicated using another cohort including six liver normal-cancer pairs (Supplemental Figs. S3, S4), again confirming the high accuracy of our approach for identifying both DMRs and 5hmCs.
Hydroxymethylation landscapes of human normal and malignant liver and lung
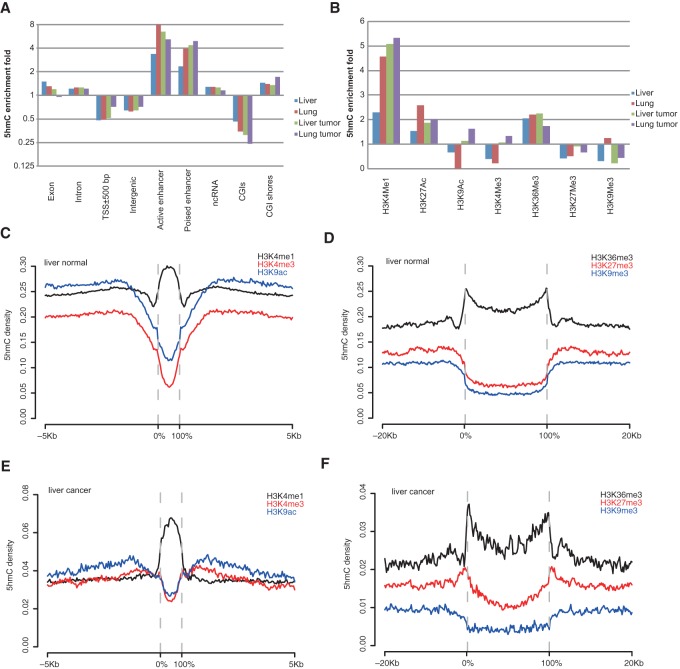
To explore the hydroxymethylation landscape, we first examined the genomic distribution of 5hmC in each tissue according to annotated genomic features. Compared to the whole-genome 5hmC average level, 5hmC was depleted in regions around transcriptional start sites (TSSs) and in intergenic regions. In contrast, 5hmC was highly enriched at enhancers, especially active enhancers, in all tissues (Fig. 1A), which is consistent with previous studies in stem cells and brain tissue (Yu et al. 2012; Wen et al. 2014). Notably, 5hmC was highly enriched at CGI shores while depleted at CGIs, which are largely overlapped with TSSs (Fig. 1A). Our and others’ previous studies showed that both c-DMRs and tissue DMRs (t-DMRs) are mostly located at CGI shores rather than at CGIs and that differential DNA methylation at CGI shores strongly correlates with differential gene expression when comparing different tissues as well as normal and tumor tissues (Irizarry et al. 2009; Hansen et al. 2011; Pathiraja et al. 2014). Together, those findings suggest that 5hmC could play a particular part in regulating DNA methylation at CGI shores.
Figure 1.
The hydroxymethylation landscapes of human normal and tumor tissues. (A,B) 5hmC fold enrichment in different genomic features (A) and histone modifications (B). 5hmC enrichment fold in each genomic feature was calculated as the ratio of 5hmC density of that feature to the genome-wide average 5hmC density. H3K9ac ChIP-seq data for lung normal is not available. (C–F) 5hmC density distribution across narrow and broad histone modifications and their flanking regions in liver normal (C,D) and liver tumor samples (E,F). Zero percent and 100% on the x-axis indicate the start and end of all called histone peaks/domains. For each liver/lung normal/cancer group, 5hmC density was averaged among biological replicates.
Next, we examined the relationship between 5hmC and chromatin organization, using chromatin immunoprecipitation sequencing (ChIP-seq) data from the Roadmap Epigenomics Project (Roadmap Epigenomics Consortium et al. 2015). In general, 5hmC positively correlated with active chromatin features, including active histone modification (H3K4me1/2, H3K27ac, and H3K36me3) and DNase hypersensitive sites, with the exception of promoter-associated marks (H3K4me3 and H2A.Z) (Fig. 1B–F; Supplemental Fig. S5). On the other hand, 5hmC showed negative correlation with repressive chromatin marks, such as H3K27me3 and H3K9me3 (Fig. 1B–F; Supplemental Fig. S5). Our analysis of 5hmC and chromatin organization further supports other studies indicating that 5hmC is an active epigenetic mark that corresponds to open chromatin (Wen et al. 2014).
Large hypomethylated blocks and small DMRs associated with loss of CGI methylation boundary stability is a common feature of the cancer epigenome (Timp and Feinberg 2013). To explore the role of 5hmC in DNA methylation change, we first examined the distribution of 5hmC among different cancer DMR categories. It is noteworthy that previous studies using the traditional bisulfite sequencing method cannot distinguish 5mC and 5hmC. Therefore, the previously identified DMRs are a mixture of both methylation and hydroxymethylation differences. Using data from oxBS-seq, we were able to identify DMRs by only taking methylation changes into account between normal and matched tumor samples. Identified DMRs were grouped into small-scale DMRs and large-scale blocks according to their length (see Methods). We found that in both liver and lung c-DMRs, 5hmC is enriched in hypermethylated blocks and small DMRs, but not in hypomethylated blocks (Supplemental Fig. S6A,B). We also compared the distribution of 5hmC among different t-DMR groups between normal liver and lung tissues and observed a similar pattern (Supplemental Fig. S6C). This result suggests that 5hmC and the responsible DNA demethylation pathway might be involved in methylation regulation on small-scale DMRs, while large-scale genomic hypomethylation may have different underlying mechanisms not involving active DNA demethylation.
5hmC distribution around genic regions
By examining 5mC and 5hmC distribution across gene regions in normal and malignant liver, we found that both 5mC and 5hmC formed a deep dip around TSSs (Supplemental Fig. S7A,B), which is consistent with the previous result of 5hmC depletion in CGIs and regions around TSSs (Fig. 1A; Wu and Zhang 2014). However, we further found that, in contrast to 5mC, 5hmC exhibited several significant peaks around gene regulatory regions. First, 5hmC formed a bimodal distribution around TSSs of genes. Second, 5hmC levels increased along the gene body toward the 3′ terminus of the gene and formed another peak at TTR right after transcriptional termination sites (TTSs) (Supplemental Fig. S7A,B). Although tumors experience global 5hmC loss compared to their normal counterparts, 5hmC enrichment around TSSs and TTSs still endured in liver tumors, perhaps suggesting a gene regulatory function (Supplemental Fig. S7B).
In line with these results, the distribution of histone modifications around genes strongly associated with the distribution of 5hmC. For example, H3K4me1 associated with 5hmC and was significantly enriched at the flanking regions of TSSs (Supplemental Fig. S7C), while H3K4me3 was enriched at TSSs where 5hmC was depleted (Supplemental Fig. S7D). H3K36me3, which is positively correlated with 5hmC, also showed an increasing modification level along the gene body toward the 3′ end of genes (Supplemental Fig. S7E). Given histone modifications were usually coupled with the status of gene expression and were not as stable as 5mC (Cedar and Bergman 2009), the strong associations between 5hmC and active histone modifications suggest that 5hmC may be a mark for active genes and can be more dynamically regulated by cells compared to 5mC.
Since CGIs and 5hmC overlap with gene regulatory regions such as the upstream region of TSSs, we further asked whether 5hmC enrichment at gene promoters is dependent on the existence of CGIs and their shores. To address this question, we compared 5hmC enrichment at active genes (FPKM > 1) with and without promoter CGIs. Compared to genes with promoter CGIs, those genes without promoter CGIs showed almost no 5hmC enrichment around TSSs (Supplemental Fig. S7F). This suggests that CGIs and their shores, rather than the location of transcriptional start sites, shape the 5hmC distribution pattern around TSSs.
Promoter, TTR, and gene-body hydroxymethylation positively correlated with gene expression
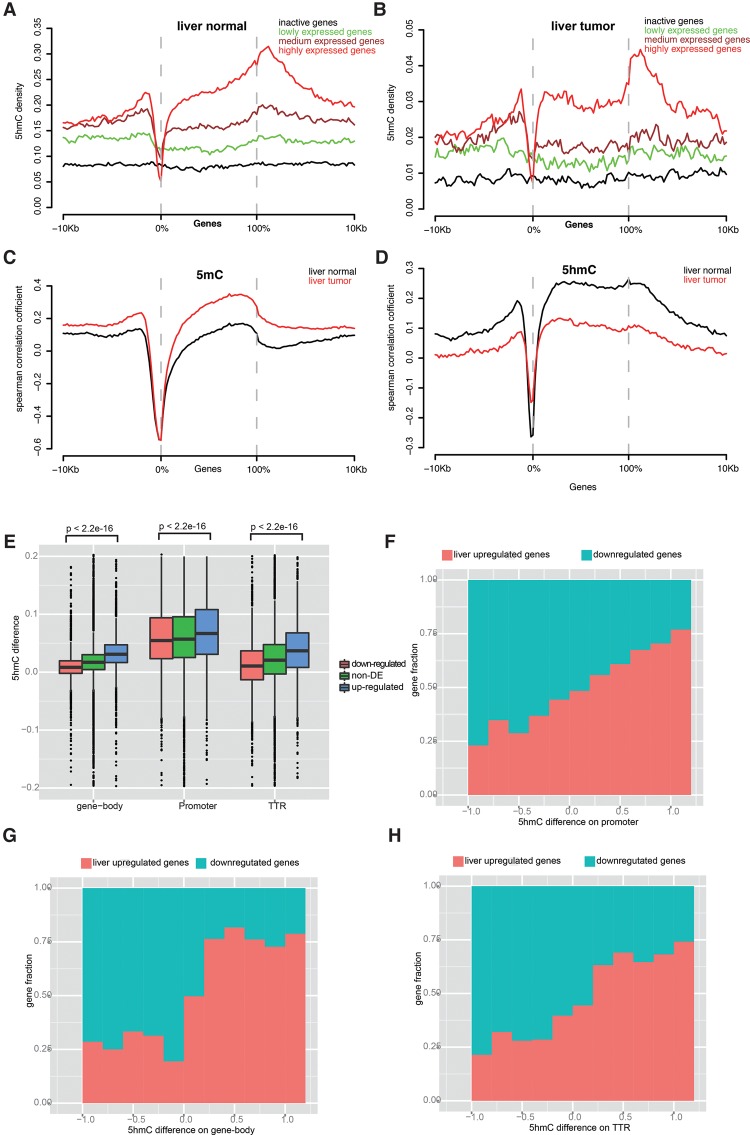
To determine the relationship between 5hmC and gene expression, we generated transcriptome profiles for samples used in this study by RNA-seq. Based on the 5hmC distribution around the genic region, we examined the gene regulatory role of hydroxymethylation at the promoter, TTR, and gene body. We classified all genes within tissues into four groups according to their expression levels and found that the presence of 5hmC on the promoter, gene body, and downstream TTS all show a significant positive correlation with gene expression. The Spearman correlation coefficients (r) between 5hmC and gene expression were 0.21, 0.30, and 0.47 for the promoter (2 kb upstream of the TSS), TTR (2 kb downstream from the TTS) and gene body, respectively. Furthermore, compared to inactive genes which have a relatively uniform 5hmC distribution throughout genic regions, active genes rather presented two 5hmC peaks in both the promoter and TTR, which became more obvious with increased gene expression (Fig. 2A,B). Positive correlation between transcriptional activity and gene-body 5hmC was reported by several previous studies using different assays (Jin et al. 2011b; Song et al. 2011; Mellen et al. 2012; Wen et al. 2014). However, among them, two studies with limited resolution of 5hmC profiling generated by enrichment-based methods failed to detect a positive correlation between promoter 5hmC and gene expression (Jin et al. 2011b; Mellen et al. 2012). Besides the gene body, our results based on single-base 5hmC maps showed a clear positive correlation between promoter/TTR 5hmC and gene expression, which is also supported by another study with high-resolution 5hmC maps (Wen et al. 2014). This result suggested that conclusions from previous studies using enrichment-based traditional methods may need to be reassessed via high-resolution maps.
Figure 2.
5hmC distribution around genic regions and 5hmC positively correlated with gene expression. (A,B) 5hmC distribution across active and inactive genes and their 10-kb flanking regions in liver normal (A) and liver tumor (B). All genes were classified into active and inactive genes. Active genes were further divided into three groups with equal numbers of genes by quintile based on gene expression (FPKM value): lowly, medium, and highly expressed genes. (C,D) Spearman correlation coefficient around genic region between 5mC (C), 5hmC (D), and gene expression. (E) 5hmC difference between normal liver and lung at three genomic features in liver: up-regulated, down-regulated, and non-DE genes. (F–H) Comparison of 5hmC difference between normal liver and lung on promoter (F), gene body (G), and TTR (H) and gene expression changes between tissues.
We then plotted Spearman correlation coefficients around genic regions between 5mC/5hmC and gene expression (Fig. 2C,D) and found a very different pattern between normal and tumor tissues, and between 5mC and 5hmC. For 5mC, except for regions around TSSs at which 5mC showed a strong negative correlation with gene expression in both normal and tumor tissues, tumors showed positive and much higher correlation coefficients than that in matched normal around the genic region. In contrast, for 5hmC, due to extensive loss of 5hmC, tumors showed positive but much lower correlation coefficients across genic regions compared to normal.
We next studied the relationship between the level of 5hmC and the difference of gene expression between the normal liver and lung by first identifying differentially expressed (DE) genes. We found 3771 up-regulated genes and 3952 down-regulated genes in liver compared to lung, and then calculated the difference in hydroxymethylation level between lung and liver (5hmCliver − 5hmClung) at the promoter, TTR, and gene body of those DE genes. Due to a much higher global hydroxymethylation level in liver than in lung, the average 5hmC level of promoter, TRR, and gene body in liver is higher than that of lung, regardless of whether the genes are up-regulated or down-regulated. However, we found that 5hmC differences at promoter, TRR, and gene body between liver and lung in up-regulated genes are significantly higher than that of down-regulated genes (Wilcoxon test, P < 2.2 × 10−16) (Fig. 2E). Furthermore, if we divided those DE genes into several groups according to the 5hmC difference between tissues, we found that the 5hmC difference between liver and lung tissues at promoter, TRR, and gene body showed positive correlation with the fraction of liver up-regulated genes (Fig. 2F–H). Genes in the liver with higher 5hmC level at any of the three gene regulatory region categories were more likely to be up-regulated in liver. In summary, these results reveal a unique 5hmC distribution pattern at genic regions and particular enrichment in active genes and suggest an important role for 5hmC in gene regulation both within tissues and across tissue types.
5hmC defines CGI shores and associates with H3K4me1 mark
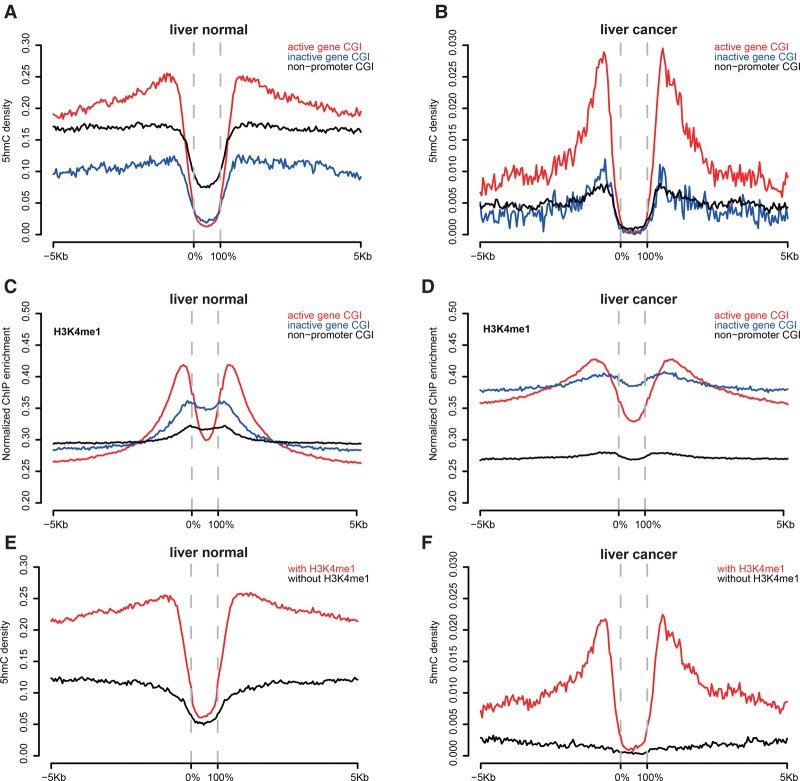
Our previous studies provided evidence that most t-DMRs and c-DMRs surprisingly overlap with CGI shores but not CGIs, which usually disrupt methylation boundaries of CGI boundaries, and that DNA methylation changes in CGI shores strongly correlate with changes in gene expression (Irizarry et al. 2009). The results in this study also confirmed the previous finding (Supplemental Fig. S8). Here, we examined the 5hmC distribution on CGIs and their shores according to their location and to the expression status of their associated genes. Interestingly, we found that 5hmC is only enriched in CGI shores of active genes, but not in CGI shores of inactive genes or nonpromoter CGI shores. 5hmC formed a unique bimodal distribution pattern around CGI shores of active genes in both normal and malignant liver (Fig. 3A,B). We obtained similar results in human stem cells and brain tissue by analyzing published whole-genome TAB-seq 5hmC data sets (Supplemental Fig. S9A,B), supporting a universal 5hmC bimodal distribution pattern around CGI shores of expressed genes regardless of tissue type and disease status.
Figure 3.
5hmC showed a bimodal distribution around CGIs and associated with H3K4me1. (A,B) 5hmC distribution around different CGI categories in liver normal (A) and liver tumor (B). (C,D) H3K4me1 enrichment around different CGI categories in liver normal (C) and liver tumor (D). (E,F) H3K4me1 associated with 5hmC enrichment in CGI shores in liver normal (E) and liver tumor (F).
To gain insight into the potential functional role of 5hmC in CGI shores, we analyzed the association of 5hmC distribution with a variety of active histone modification marks using publicly available ChIP-seq data (Roadmap Epigenomics Consortium et al. 2015). While H3K4me3, H3K9ac, and H3K27ac showed peaks at CGIs of active genes (Supplemental Fig. S9C–H), H3K4me1 showed bimodal peaks around CGI shores similar to that of the 5hmC (Fig. 3C,D), suggesting potential crosstalk between H3K4me1 and 5hmC at CGI shores. To thoroughly examine the association between 5hmC and H3K4me1, we divided all CGI shores into two groups—with or without H3K4me1 modification—and found that only CGI shores with H3K4me1 modification showed clear 5hmC bimodal peaks (Fig. 3E,F). CGIs with H3K4me1 on shores also showed H3K4me3 modification that tightly associated with active genes, again indicating positive correlation between 5hmC/H3K4me1 and gene expression (Supplemental Fig. S10). Taken together, these results suggest that 5hmC and the association with the H3K4me1 mark contribute to the function of CGI shores in regulating gene expression.
5hmC correlates positively with H3K4m1 and negatively with 5mC in both t-DMRs and c-DMRs
Consistent with the observation that liver showed a lower global 5mC methylation level than lung (62.08% vs. 65.32%, t-test, P-value = 0.01354), we identified more hypomethylated than hypermethylated t-DMRs in normal liver (Supplemental Tables S7–S8) for 5mC. We divided all t-DMRs into three groups: CGI-associated, CGI shore-associated, and other DMRs. Consistent with our previous study (Irizarry et al. 2009), very few t-DMRs were CGI-associated (2.9%), while most t-DMRs were CGI shore-associated (21.0%) or distant from CGIs (76.1%), the latter mostly within gene bodies (66.5%).
Our previous study showed that c-DMRs highly overlapped with t-DMRs (Irizarry et al. 2009). Following this point, we divided t-DMRs into hypermethylated and hypomethylated groups and overlapped the c-DMRs within each tissue. We further found that liver hypermethylated c-DMRs significantly overlapped with normal liver hypermethylated t-DMRs (liver vs. lung), compared to normal liver hypomethylated t-DMRs (13.4% vs. 2.1%, χ2 test, P < 2.2 × 10−16) (Supplemental Table S9). Similarly, lung hypermethylated c-DMRs significantly overlapped with normal lung hypermethylated t-DMRs (lung vs. liver), compared to normal lung hypomethylated t-DMRs (14.8% vs. 1.8%, χ2 test, P < 2.2 × 10−16) (Supplemental Table S9). In line with the above result, we also found that liver cancer down-regulated genes were significantly enriched in those genes comparatively up-regulated in normal liver compared to normal lung (42% vs. 6.1%, χ2 test, P < 2.2 × 10−16). A similar result was also observed in lung cancer (54.2% vs. 8.8%, χ2 test, P < 2.2 × 10−16). Taken together, these results suggested that the aberrant DNA hypermethylation might disrupt the normal tissue-specific methylation landscape during tumorigenesis and result in transcriptional silencing of genes that are usually actively expressed in normal tissues.
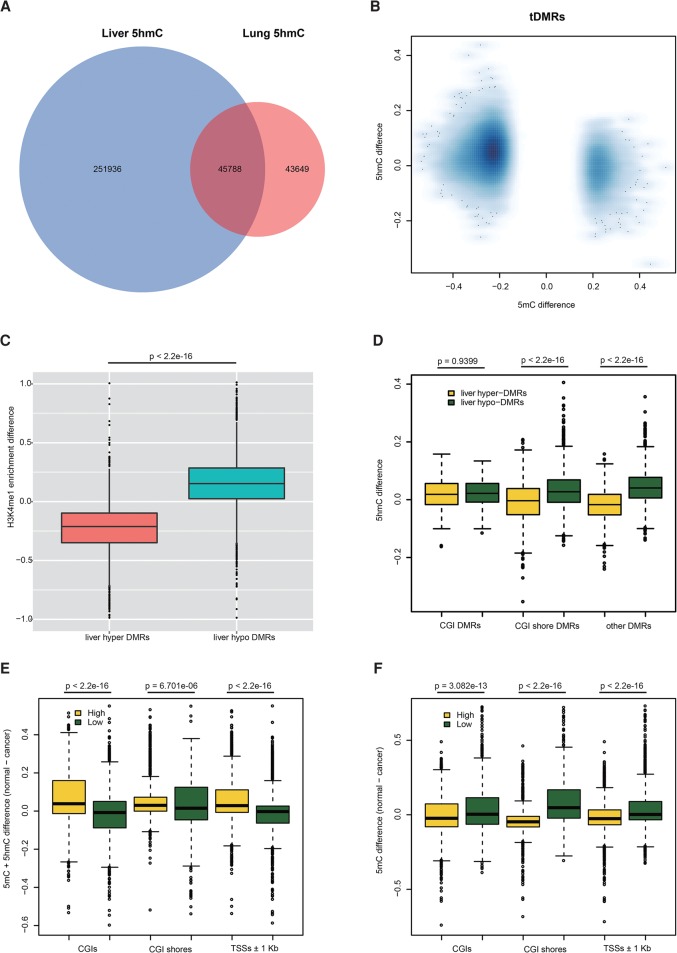
To determine the association between t-DMR and 5hmC, we overlapped t-DMRs with identified liver- and lung-specific 5hmC regions (Fig. 4A). We found that, in general, liver-hypermethylated t-DMRs were hypo-hydroxymethylated, while liver-hypomethylated t-DMRs were hyper-hydroxymethylated between normal liver and lung (5hmCliver − 5hmClung, −0.014 vs. 0.048, Wilcoxon test, P-value < 2.2 × 10−16), indicating a negative correlation between the 5mC difference and 5hmC difference on t-DMRs (Fig. 4B). Furthermore, we found that DNA methylation changes at t-DMRs also negatively correlated with the H3K4me1 difference (Fig. 4C). Consistently, t-DMRs where liver was comparatively hypermethylated overlapped with lung-specific H3K4me1 peaks, while t-DMRs where liver was comparatively hypomethylated overlapped with liver-specific H3K4me1 peaks (Supplemental Tables S7–S8). These results suggested strong associations among dynamic changes of 5mC, 5hmC, and H3K4me1, and more importantly, these H3K4me1-associated 5mC and 5hmC changes corresponded to changes in gene expression between tissue types as shown above.
Figure 4.
5mC change of t-DMRs and c-DMRs anticorrelated with 5hmC and H3K4me1 changes. (A) Venn diagram of identified liver and lung 5hmC regions. (B) Negative correlation between 5mC difference and 5hmC difference on t-DMRs. (C) Negative correlation between 5mC difference and H4K4me1 enrichment difference on t-DMRs. (D) 5hmC difference on different t-DMR categories. 5hmC difference on non-CGI t-DMRs showed a larger difference than that of CGI t-DMRs. (E) Cytosine modification difference (5mC + 5hmC) between normal and tumor at promoters with high and low 5hmC level in liver. (F) Only DNA methylation difference (5mC) between normal and tumor at promoters with high and low 5hmC level in liver. Promoters were ranked according to the 5hmC level, and the highest and lowest 10th percentiles of the promoter were used for our analysis.
Next, we explored c-DMRs in two types of cancer (Supplemental Tables S9–S11). We examined the association between changes in 5hmC and 5mC at c-DMRs. Similar to t-DMRs, 5hmC changes in c-DMRs also showed a strong negative correlation with 5mC changes (Fig. 5A,B; Supplemental Fig. S11A,B). In other words, a loss of 5hmC corresponded to a gain of 5mC and vice versa. For both t-DMRs and c-DMRs, we found that the negative correlation between 5hmC and 5mC changes is stronger in CGI shore-associated DMRs and other-DMR categories than CGI-associated DMRs (Fig. 4D; Supplemental Fig. S11C,D), which is consistent with depletion of 5hmC in CGIs. Again, similar to t-DMRs, the negative correlation between 5mC and H3K4me1 was also observed in c-DMRs (Supplemental Fig. S11E,F). Taken together, our results suggested that the gene expression pattern in different tissues could be regulated and achieved through intricate interplays among DNA methylation, hydroxymethylation, and histone modification such as H3K4me1.
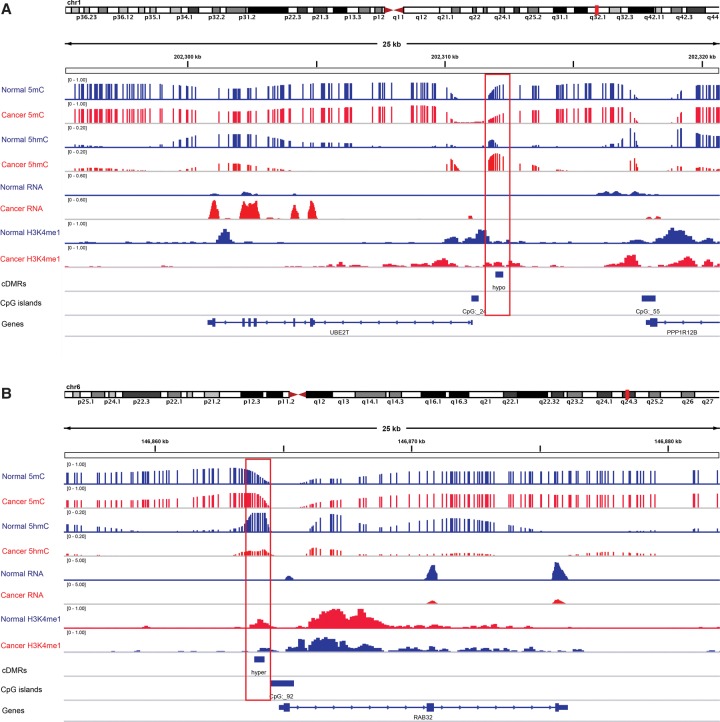
Figure 5.
Two examples of liver c-DMRs that showed 5mC changes were regulated through 5hmC and associated with gene expression changes. 5mC, 5hmC, and gene expression level of both liver normal (blue tracks) and cancer (red tracks) were displayed. Red boxes indicate the location of c-DMRs. (A) A hypomethylated liver c-DMR located in CpG island shores near the promoter of the UBE2T gene. (B) A hypermethylated c-DMR located in CpG island shores near the promoter of the RAB32 gene.
The negative correlation between 5mC and 5hmC observed here is not consistent with the finding, in a previous study using colon tissue, that high 5hmC promoters in normal tissue are prone to loss of DNA methylation in tumors thus resistant to DNA hypermethylation in cancer (Uribe-Lewis et al. 2015). However, that study used an Infinium27K array to obtain methylation profiles, which cannot distinguish 5mC and 5hmC. We performed a similar analysis using our data and found that promoters with high 5hmC levels in normal tissue indeed tend to lose DNA methylation in cancer, compared to that of low 5hmC promoters when considering 5mC and 5hmC together as DNA methylation differences (Fig. 4E; Supplemental Fig. S11G). However, when removing the effect of 5hmC and only using 5mC, we found the opposite pattern, that promoters with a high 5hmC level in normal tissue in fact tend to be hypermethylated in cancer, whereas low 5hmC promoters tend to be hypomethylated (Fig. 4F; Supplemental Fig. S11H). This result suggests that high 5hmC levels maintain promoter hypomethylation in normal tissue and that global loss of 5hmC could result in hypermethylation at promoters and dysregulation of gene expression in cancer. This result is consistent with the observation that there are more small hypermethylated c-DMRs than hypomethylated c-DMRs and again confirmed the anticorrelation between 5hmC and 5mC. This result also demonstrated the importance of distinguishing 5mC and 5hmC in DNA methylation analysis, especially for those tissues with relatively high 5hmC levels, and the conclusions from previous studies using traditional methods may need to be reassessed. That said, it is quite possible that the colon differs from other tissues, particularly given its low level of 5hmC in normal tissue, although one would need to use a method distinguishing 5hmC from 5mC in the 5mC assays, as done here.
Discussion
In this study, we have performed base-level resolution analysis of 5mC and 5hmC in normal and matching malignant tissues from human liver and lung. Our results revealed that 5hmC is an important epigenetic mark of active genes that is strongly associated with active histone modifications and could play a role in gene expression mediated by DNA demethylation. This integrated analysis showed that differential gene expression between cancer and normal tissues is strongly associated with a unique interplay between 5mC, 5hmC, and H3K4me1 that was strongest at CGI shores.
Our results showed 5hmC is significantly enriched in the CGI shore relative to the CGI itself and revealed an interesting 5hmC bimodal distribution pattern around the CGI. Supporting this finding, recent studies using hmeDIP-seq showed that 5hmC is enriched within the shores of promoter CpG islands in human normal colon tissues and an embryonic carcinoma cell line (Putiri et al. 2014; Uribe-Lewis et al. 2015). By overlaying these data with RNA-seq of the same tissues, we found that 5hmC is only enriched in CGI shores which are associated with active genes, and not with CGI shores which are associated with inactive genes or with non-promoter-associated CGI shores. In addition, this unique 5hmC distribution around CGIs also coupled with H3K4me1. Although our previous studies have reported that DNA methylation in CGI shores significantly regulates gene expression across tissues (Irizarry et al. 2009; Hansen et al. 2011), the underlying molecular mechanism remained unclear. Our present results demonstrate that CGIs and their shores function as important regulatory regions that are enriched with a variety of histone modifications, and these histone modifications around CGIs and their shores could subject them to regulation by TET-mediated DNA methylation.
Besides 5hmC bimodal peaks around TSSs, the 5hmC level increased along gene bodies and formed another peak right after transcriptional termination sites. Strikingly, the 5hmC level is even higher at TTSs than at promoter regions and is also positively correlated with gene expression, suggesting a potential novel regulatory mechanism that 5hmC around TTSs could affect gene expression by interacting with transcription termination factors and regulating RNA polymerase II (Pol II) processivity. Supporting this idea, an in vitro study showed DNA templates incorporated with 5fC and 5caC, oxidation variants of 5hmC, can dramatically reduce the transcription rate of Pol II and stall Pol II (Kellinger et al. 2012). Whether 5hmC enrichment at TTS contributes to oxidation variants that give rise to transcription termination in vivo requires further detailed investigation.
Our results further showed that many t-DMRs and c-DMRs were located in gene bodies and also significantly affected gene expression between tissues. Furthermore, those DMRs usually overlap with tissue-specific 5hmC and H3K4me1 peaks, which usually associate with distal regulatory elements and are used for defining enhancers. In addition, previous studies showed that intragenic enhancers within the gene body can regulate gene expression by acting as alternative promoters (Kowalczyk et al. 2012). Recent work also showed that extensive loss of 5hmC at enhancers mediated by Tet2 deletion was coupled with enhancer hypermethylation and affected gene expression during early stages of mouse stem cell differentiation (Hon et al. 2014). Taken together, these results suggest that a dynamic change in 5mC/5hmC at intragenic enhancer regions could be an important epigenetic mechanism in regulation of gene expression during tissue differentiation and tumorigenesis.
In cancer compared to normal tissue, 5hmC was quantitatively diminished (∼70%) and partially retained at regulatory regions. Moreover, the difference in 5hmC between euchromatic and heterochromatic regions in normal tissue is essentially lost in cancer. Similarly, the specific relationship between 5hmC and chromatin marks in normal tissue is largely erased in tumors. These results suggested that a 5hmC landscape change in cancer could associate with chromatin structure change and regulation of gene expression during tumorigenesis.
These results also clearly showed a strong anticorrelation between 5hmC and 5mC changes in both t-DMRs and c-DMRs, with this effect stronger in CGI shores. Global loss of 5hmC is considered to be a hallmark of cancer cells, and impairment in the TET-mediated DNA demethylation machinery is described in several tumor types (Delhommeau et al. 2009; Langemeijer et al. 2009; Dang et al. 2010; Shibata et al. 2011). Considering that TET enzymes preferentially bind CpG enriched regions, such as CGIs, dysfunction of TET-mediated DNA demethylation in cancer could explain why more hypermethylated c-DMRs are located in CGIs, which is also supported by previous results that loss of 5hmC in a TET-depleted human ECC line coincides with genes susceptible to aberrant hypermethylation.
The antagonistic role of 5mC and 5hmC in normal tissue, and the bimodal distribution of 5hmC at TSSs and at H3K4me1 sites associated with enhancers, suggests that DNA methylation is mediated by the balance and topological organization of 5mC and 5hmC, creating a mechanism for both stable gene expression but also substantial and abrupt changes during normal development. Finally, the DNA methylation maintenance machinery is robust and self-sustained. In agreement with this, DNMT1 mutations are rarely observed in cancer. Therefore, we postulate that disruption of 5hmC could lead to instability of methylation marks that are then selected for and maintained by the DNMT1 machinery to increase the proliferative advantage of cancer cells over normal.
Based on our results, we suggest that the majority of DNA loss in tumors could be due to passive demethylation, especially in large hypomethylated blocks where 5hmC is depleted. However, we also found both hyper- and hypomethylated c-DMRs at CGI shores where 5hmC is significantly enriched and associates with bimodal H3K4me1. Therefore, it is possible that active demethylation plays an important role in those particular regulatory elements. Future studies would be necessary in order to determine whether DNA demethylation is passive or active in tumorigenesis. For example, dynamic 5hmC profiling during different stages of tumorigenesis or during induced tumorigenesis in the absence of TET enzymes.
Methods
Preparation of hydroxymethylated lambda phage genome
One microgram of unmethylated lambda DNA (Promega) was treated with SssI methyltransferase (Zymo) overnight and cleaned up using Genomic DNA Clean & Concentrator (Zymo). To make sure all CpG sites of the lambda genome were fully methylated, SssI treatment and clean-up were repeated. Then, 500 ng CpG methylated lambda DNA was bisulfite-converted using the EZ DNA Methylation-Lightning kit following the manufacturer's manual (Zymo). Ten nanograms bisulfite-converted lambda DNA from the above step was used to perform whole-genome amplification with the GenoMatrix Whole Genome Amplification kit (Active Motif) using 5-hydroxymethylcytosine dNTP mix (Zymo) instead of the dNTP mix in the kit. To achieve as high a hydroxymethylation level as possible, the PCR reaction of whole-genome amplification was repeated for another three times using 5-hydroxymethylcytosine dNTP and the PCR product from the previous time as template every time.
oxBS-seq and BS-seq library preparation
Genomic DNA was extracted using the DNeasy Blood and Tissue kit (Qiagen). Four micrograms of genomic DNA plus spike-in 20 ng hydroxymethylated lambda phage and nonmethylated E. coli DNA control (Zymo) in 150 µL water was sheared into ∼10-kb fragments using g-Tube (Covaris) following the manufacturer's instructions. The sheared DNA was cleaned up using a GeneJET PCR Purification kit (Thermo Fisher Scientific) with a modified protocol (Protocol 03, TrueMethyl Preparation of High Molecular Weight gDNA, http://www.cambridge-epigenetix.com/en_US/resources/application-notes). Then, oxidative bisulfite- and only bisulfite-converted DNA templates were generated using the TrueMethyl 24 kit (Cambridge Epigenetix) for each sample according to the manufacturer's instructions. Last, oxBS- and BS-converted DNA were used to construct oxBS-seq and corresponding BS-seq libraries using the EpiGenome Methyl-seq kit (Epicentre) following the manufacturer's instructions.
oxBS-seq and BS-seq data processing
Paired-end HiSeq 2000 sequencing reads from oxBS-seq and BS-seq were aligned by the BSmooth bisulfite alignment pipeline (version 0.7.1) (Hansen et al. 2012) as previously described in detail (Hansen et al. 2011). Briefly, reads were aligned by Bowtie 2 (version 2.0.1) (Langmead and Salzberg 2012) against the human genome (hg19) that is used for Roadmap Epigenomics Project, as well as the lambda phage and E. coli genomes. After alignment, methylation measurements for each CpG were extracted from aligned reads. We filtered out measurements with mapping quality < 20 or nucleotide base quality on cytosine position < 10, and we also removed measurements from the 5′-most 7 nt of both mates. The methylation levels of lambda phage and E. coli genomes were used to access oxidation and bisulfite conversion rates, respectively.
t-DMR and c-DMR identification
To identify t-DMRs and c-DMRs that were only contributed by 5mC and do not involve 5hmC changes, oxBS-seq data were used for DMR identification by the bsseq package, which can borrow statistical power from neighboring CpG sites and biological replicates and was successfully applied to DMR identification even with low sequencing coverage in our previous studies (Hansen et al. 2011, 2014). We reasoned that oxBS data, measuring only 5mC, should behave similarly to standard WGBS data measuring the sum of 5mC and 5hmC, and we therefore applied BSmooth with standard parameters. Specifically, for small DMRs we used a smooth window containing either 70 CpGs or a width of 1 kb, whichever is larger, and for large DMRs (blocks) we used a smoothing window containing either 500 CpGs or a width of 20 kb, whichever is larger. Following smoothing, putative DMRs were identified using a t-statistic cutoff (see below). For this computation, we only considered CpGs with coverage of at least 5 in at least two samples in each group, and we only considered putative DMRs with a methylation difference of at least 20% (small DMRs) or 10% (large DMRs) and a length >5 kb. These arbitrarily chosen cutoffs are more stringent that what we have employed in earlier work (Hansen et al. 2011, 2012, 2014). Final DMRs depends on both the t-statistic cutoff and the procedure for assigning significance. Previously, we employed a very stringent permutation procedure, which controls the family-wise error rate; this is a much more stringent error rate than the widely used false discovery rate (FDR) (Hansen et al. 2014). In this work, we approached the problem differently. We systematically tested a range of t-statistic cutoffs (1.65, 2, 2.5, 3, 3.5, 4, 4.6) and for each cutoff we found putative DMRs as consecutive sets of CpGs with a t-statistics exceeding the cutoff. We compute P-values associated with each DMR using the following procedure. First we compute a CpG level P-value based on a standard t-test for the smoothed methylation data using an asymptotic reference distribution. Next, these CpG level P-values were combined into a region level P-value by using a modified Stouffer-Liptak method as implemented in the Comb-p software (Pedersen et al. 2012). Next, these regional level P-values were corrected for multiple testing and we kept regions with an FDR < 5%. This gives us a set of DMRs for each t-statistic cutoff. We then choose the cutoff that yielded the most differentially methylated regions, effectively optimizing empirical power (Supplemental Fig. S12). To assess the error rate, we permuted the sample labels and repeated the procedure (Supplemental Fig. S12), yielding almost no DMRs with an FDR < 5%, showing our low error rate.
5hmC region and site identification
According to the principle of oxidation bisulfite sequencing, hydroxymethylation can be ascertained by the methylation difference of oxBS-seq and corresponding BS-seq data for each sample. Since hydroxymethylation level is usually very low in human tissues, except for brain tissues, and it needs high sequencing depth to get accurate measurement, we used a similar smooth-based algorithm like the above DMR identification in this study to identify hydroxymethylation regions (5hmC regions). Effectively, this is identifying differences between BS-seq and oxBS-seq libraries. We smoothed both data types using default parameters from BSmooth (as above, a window size encompassing at least 1 kb or 70 CpGs). Assessment of significance was done as described above (Supplemental Fig. S12C,D). After obtaining 5hmC regions, only CpGs passing the t-statistic threshold within 5hmC regions were considered as hydroxymethycytosines (5hmC sites) and used for analysis in this study. In this study, 5hmC density is estimated by the proportion of 5hmC sites out of all CpGs within a certain genomic feature/region. 5hmC level for CpG sites or regions is estimated by the subtraction between BS-seq and oxBS-seq libraries based on smoothed values.
c-DMRs and 5hmC regions replications
Another liver cohort including six normal-cancer pairs was kindly provided by Professor Robert Albert Anders. Genomic DNA was purified using the DNeasy Blood and Tissue kit (Qiagen). One and one-half micrograms of DNA for each sample was used to generate oxidative bisulfite- and only bisulfite-converted DNA templates with the TrueMethyl 24 kit (Cambridge Epigenetix) following the manufacturer's instructions. Primers for bisulfite sequencing PCR were designed using MethPrimer (http://www.urogene.org/methprimer/), and sequences of all primers are listed in Supplemental Table S12. Locus-specific PCRs were performed by nested PCR using both oxBS- and BS-converted DNA as templates. Then, all amplicons from the same sample were pooled, and barcoded libraries were prepared with the TruSeq DNA PCR-Free Library Preparation kit (Illumina) following the manufacturer's instruction. Amplicon sequencing was performed on a MiSeq instrument (Illumina).
RNA-seq library preparation and data processing
Total RNA was extracted using the RNeasy Mini kit (Qiagen). RNA-seq libraries were constructed using the TruSeq Stranded mRNA LT Sample Prep kit (Illumina) according to the manufacturer's manual.
Paired-end HiSeq 2000 sequencing reads were aligned against the human genome (hg19) by OSA (version 2.0.1) (Hu et al. 2012) with default parameters. After alignment, only uniquely aligned reads were kept for further analysis. Gene annotation information was downloaded from GENCODE (http://www.gencodegenes.org/releases/19.html, release 19). Read counts for each gene of all samples were estimated using HTSeq (http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html) and then were used to identify differentially expressed genes using the DESeq2 package (Love et al. 2014). Genes with FDR < 0.01 and fold-change > 2 between groups were considered as DE genes.
ChIP-seq data processing
Uniformly processed ChIP-seq data used in this study including normal and tumor tissues of human liver and lung were downloaded from the Roadmap Epigenomics Project (https:// personal.broadinstitute.org/anshul/projects/roadmap/alignments/consolidated/).
For narrow histone modification peaks including H3K4me1, H3K4me3, H3K9ac, and H3K27ac, MACS2 was used for peak calling with default parameters (Zhang et al. 2008). For broad histone modifications peaks including H3K27me3, H3K36me3, and H3K9me3, large domains were identified using RSEG, which is based on the hidden Markov model (HMM) and specifically designed for identifying broad histone peaks (Song and Smith 2011).
Based on ChIP-seq, enhancers were defined as H3K4me1 peaks that are at least 2 kb away from any transcriptional start site of annotated genes. Enhancers overlapped with H3K27ac peaks were defined as active enhancers, while others were poised enhancers.
To plot each histone modification on CpG islands and their flanking regions, we divided flanking sequences into bins with fixed length (in bp) and CGIs themselves into bins with a fixed percentage of each length. ChIP enrichment was measured and normalized using a previous published method (Hawkins et al. 2010). In brief, the number of reads per kilobase of bin per million mapped reads (RPKM) was calculated for each ChIP and its input control (denoted as RPKMChIP and RPKMinput). ChIP enrichment is measured as ΔRPKM = RPKMChIP – RPKMinput, and ChIP enrichment regions should have ΔRPKM > 0. Then, all ΔRPKM were normalized to a scale between 0 and 1, and the average normalized ChIP enrichment signals across all bins were plotted for each histone mark.
Data access
The raw and processed oxBS-seq, BS-seq, and RNA-seq data sets generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE70091.
Supplementary Material
Acknowledgments
We thank Rebecca Curley, Rakel Tryggvadottir, Birna Berndsen, and Adrian Idrizi for guidance in library construction and sequencing assistance. This work was supported by National Institutes of Health (NIH) grant CA54358 to A.F.
Author contributions: X.L. and A.F. designed the research and wrote the manuscript; X.L., Y.L., and T.S. performed the experiments. X.L. performed the analysis, K.D.H. provided substantial statistical oversight and writing regarding our improved BSmooth method, and Y.L. reviewed the accuracy of the statistical code and analysis.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.211854.116.
References
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. 2012. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336: 934–937. [DOI] [PubMed] [Google Scholar]
- Booth MJ, Ost TWB, Beraldi D, Bell NM, Branco MR, Reik W, Balasubramanian S. 2013. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat Protoc 8: 1841–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. 2009. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet 10: 295–304. [DOI] [PubMed] [Google Scholar]
- Dang L, Jin S, Su SM. 2010. IDH mutations in glioma and acute myeloid leukemia. Trends Mol Med 16: 387–397. [DOI] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Valle VD, James C, Trannoy S, Masse A, Kosmider O, Le Couedic J-P, Robert F, Alberdi A. 2009. Mutation in TET2 in myeloid cancers. N Engl J Med 360: 2289–2301. [DOI] [PubMed] [Google Scholar]
- Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D. 2011. Increased methylation variation in epigenetic domains across cancer types. Nat Genet 43: 768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Langmead B, Irizarry RA. 2012. BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol 13: R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Sabunciyan S, Langmead B, Nagy N, Curley R, Klein G, Klein E, Salamon D, Feinberg AP. 2014. Large-scale hypomethylated blocks associated with Epstein-Barr virus–induced B-cell immortalization. Genome Res 24: 177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S. 2010. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 6: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon GC, Song C-X, Du T, Jin F, Selvaraj S, Lee AY, Yen CA, Ye Z, Mao S-Q, Wang B-A. 2014. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol Cell 56: 286–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Ge H, Newman M, Liu K. 2012. OSA: a fast and accurate alignment tool for RNA-Seq. Bioinformatics 28: 1933–1934. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M. 2009. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 41: 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Jiang Y, Qiu R, Rauch TA, Wang Y, Schackert G, Krex D, Lu Q, Pfeifer GP. 2011a. 5-hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res 71: 7360–7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Wu X, Li AX, Pfeifer GP. 2011b. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res 39: 5015–5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13: 484–492. [DOI] [PubMed] [Google Scholar]
- Kellinger MW, Song C-X, Chong J, Lu X-Y, He C, Wang D. 2012. 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat Struct Mol Biol 19: 831–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk MS, Hughes JR, Garrick D, Lynch MD, Sharpe JA, Sloane-Stanley JA, McGowan SJ, De Gobbi M, Hosseini M, Vernimmen D. 2012. Intragenic enhancers act as alternative promoters. Mol Cell 45: 447–458. [DOI] [PubMed] [Google Scholar]
- Kudo Y, Tateishi K, Yamamoto K, Yamamoto S, Asaoka Y, Ijichi H, Nagae G, Yoshida H, Aburatani H, Koike K. 2012. Loss of 5-hydroxymethylcytosine is accompanied with malignant cellular transformation. Cancer Sci 103: 670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird A, Thomson JP, Harrison DJ, Meehan RR. 2013. 5-hydroxymethylcytosine profiling as an indicator of cellular state. Epigenomics 5: 655–669. [DOI] [PubMed] [Google Scholar]
- Langemeijer SMC, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RAP, et al. 2009. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet 41: 838–842. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, Xu W, Tan L, Hu Y, Zhan Q. 2012. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 150: 1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. 2012. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151: 1417–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestor CE, Ottaviano R, Reddington J, Sproul D, Reinhardt D, Dunican D, Katz E, Dixon JM, Harrison DJ, Meehan R. 2012. Tissue-type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res 22: 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Aravind L, Rao A. 2013. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol 14: 341–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathiraja TN, Nayak SR, Xi Y, Jiang S, Garee JP, Edwards DP, Lee AV, Chen J, Shea MJ, Santen RJ, et al. 2014. Epigenetic reprogramming of HOXC10 in endocrine-resistant breast cancer. Sci Transl Med 6: 229ra241–229ra241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen BS, Schwartz DA, Yang IV, Kechris KJ. 2012. Comb-p: software for combining, analyzing, grouping and correcting spatially correlated P-values. Bioinformatics 28: 2986–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer GP, Kadam S, Jin S-G. 2013. 5-hydroxymethylcytosine and its potential roles in development and cancer. Epigenetics Chromatin 6: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putiri EL, Tiedemann RL, Thompson JJ, Liu C, Ho T, Choi JH, Robertson KD. 2014. Distinct and overlapping control of 5-methylcytosine and 5-hydroxymethylcytosine by the TET proteins in human cancer cells. Genome Biol 15: R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Kokubu A, Miyamoto M, Sasajima Y, Yamazaki N. 2011. Mutant IDH1 confers an in vivo growth in a melanoma cell line with BRAF mutation. Am J Pathol 178: 1395–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. 2013. DNA methylation: roles in mammalian development. Nat Rev Genet 14: 204–220. [DOI] [PubMed] [Google Scholar]
- Song Q, Smith AD. 2011. Identifying dispersed epigenomic domains from ChIP-Seq data. Bioinformatics 27: 870–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen C-H, Zhang W, Jian X, et al. 2011. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotech 29: 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timp W, Feinberg AP. 2013. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat Rev Cancer 13: 497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe-Lewis S, Stark R, Carroll T, Dunning M, Bachman M, Ito Y, Stojic L, Halim S, Vowler S, Lynch A, et al. 2015. 5-hydroxymethylcytosine marks promoters in colon that resist DNA hypermethylation in cancer. Genome Biol 16: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Li X, Yan L, Tan Y, Li R, Zhao Y, Wang Y, Xie J, Zhang Y, Song C, et al. 2014. Whole-genome analysis of 5-hydroxymethylcytosine and 5-methylcytosine at base resolution in the human brain. Genome Biol 15: R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. 2014. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156: 45–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J, Xu ZD, Zhu HG, Ling ZQ, Ye D, et al. 2013. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene 32: 663–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song C-X, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B. 2012. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149: 1368–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.