Abstract
Clinical exome sequencing routinely identifies missense variants in disease-related genes, but functional characterization is rarely undertaken, leading to diagnostic uncertainty1,2. For example, mutations in PPARG cause Mendelian lipodystrophy3,4 and increase risk of type 2 diabetes (T2D)5. While approximately one in 500 people harbor missense variants in PPARG, most are of unknown consequence. To prospectively characterize PPARγ variants we used highly parallel oligonucleotide synthesis to construct a library encoding all 9,595 possible single amino acid substitutions. We developed a pooled functional assay in human macrophages, experimentally evaluated all protein variants, and used the experimental data to train a variant classifier by supervised machine learning (http://miter.broadinstitute.org). When applied to 55 novel missense variants identified in population-based and clinical sequencing, the classifier annotated six as pathogenic; these were subsequently validated by single-variant assays. Saturation mutagenesis and prospective experimental characterization can support immediate diagnostic interpretation of newly discovered missense variants in disease-related genes.
A major challenge in clinical exome sequencing is determining pathogenicity of missense variants incidentally found in genes previously implicated in a severe genetic disease 1,2,6. Every exome contains ~200 missense variants that have never before been seen7. Few of these are in fact pathogenic, but functional testing is too slow and resource intensive for clinical use, leading to many Variants of Uncertain Significance (VUS)8. The lack of functional data and failure to explicitly incorporate information about ascertainment and prior probability can lead both to misdiagnosis6,9 (if a benign variant is presumed pathogenic) and overestimation of penetrance (if modestly functional variants are systematically excluded from disease databases).
The peroxisome proliferator-activated receptor γ (PPARγ) exemplifies the challenge of classifying newly identified variants even in a well-studied disease gene. Rare mutations in PPARG cause familial partial lipodystrophy 3 (FPLD3)3,4 and a common missense variant p.P12A, along with linked non-coding variants, associates with risk of T2D10,11. Molecular functions of PPARγ are well characterized12,13 including its role as the target of anti-diabetic thiazolidinedione medications. Approximately 0.2% of the general population carries a rare missense variant in PPARG, but only 20% of these variants are functionally significant and associated with metabolic disease5.
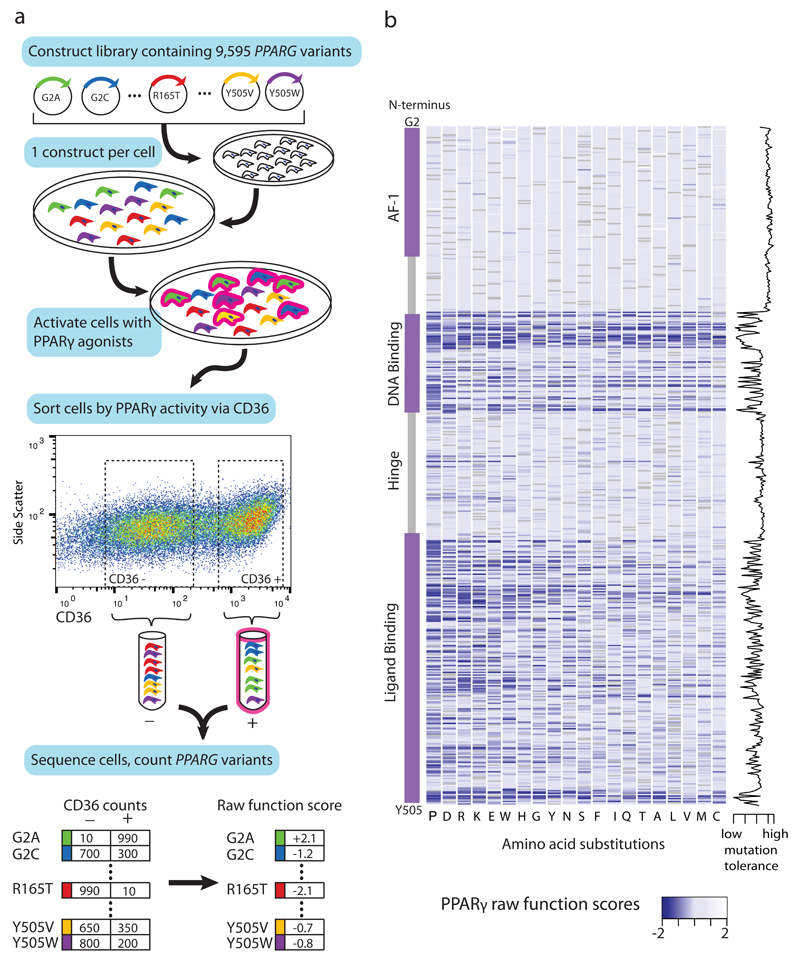
In order to enable functional interpretation of PPARγ variants identified in exome sequencing we constructed a cDNA library consisting of all possible amino acid substitutions in the protein (Figure 1A and Supplementary Figure 1). Based on the observation that primary human blood monocytes from patients with FPLD3 exhibit blunted PPARG response when stimulated with agonists ex vivo13, the construct library was introduced into human macrophages edited to lack the endogenous PPARG gene (Supplementary Figure 2). After stimulation with PPARγ agonists, cells were FACS sorted according to the level of expression of CD36, a canonical target of PPARγ in multiple tissues14,15 (Figure 1A). The sorted CD36+ and CD36- cell populations were sequenced to determine the distribution of each PPARG variant in relation to CD36 activity.
Figure 1. Comprehensive functional testing of 9,595 PPARγ amino acid variants.
a) A library of 9,595 PPARG constructs was synthesized, each construct containing one amino acid substitution. The construct library was introduced into THP-1 monocytes (edited to lack the endogenous PPARG gene) such that each cell received a single construct. This polyclonal population of THP-1 monocytes was differentiated to macrophages and stimulated with PPARγ agonists (rosiglitazone, PGJ2); the stimulated macrophages were separated via fluoresence activated cell sorting according to expression of the PPARγ response gene CD36 into low (-) and high (+) activity bins. Each bin of cells was subject to next-generation sequencing at the transgenic PPARG locus to identify and tabulate introduced variants. PPARγ variant counts in the CD36 low and CD36 high bins were used to calculate a functional score for all 9,595 variants. b) Raw PPARγ function scores for each of the 9,595 variants plotted according to amino acid position along the PPARγ sequence. “Blue” denotes that any amino acid change away from reference results in low CD36 function score, whereas ”white” denotes that amino acid changes do not alter function; “grey” denotes the reference amino acid. Function scores summed by amino acid position are plotted to the right, denoting tolerance for any amino acid substitution away from reference.
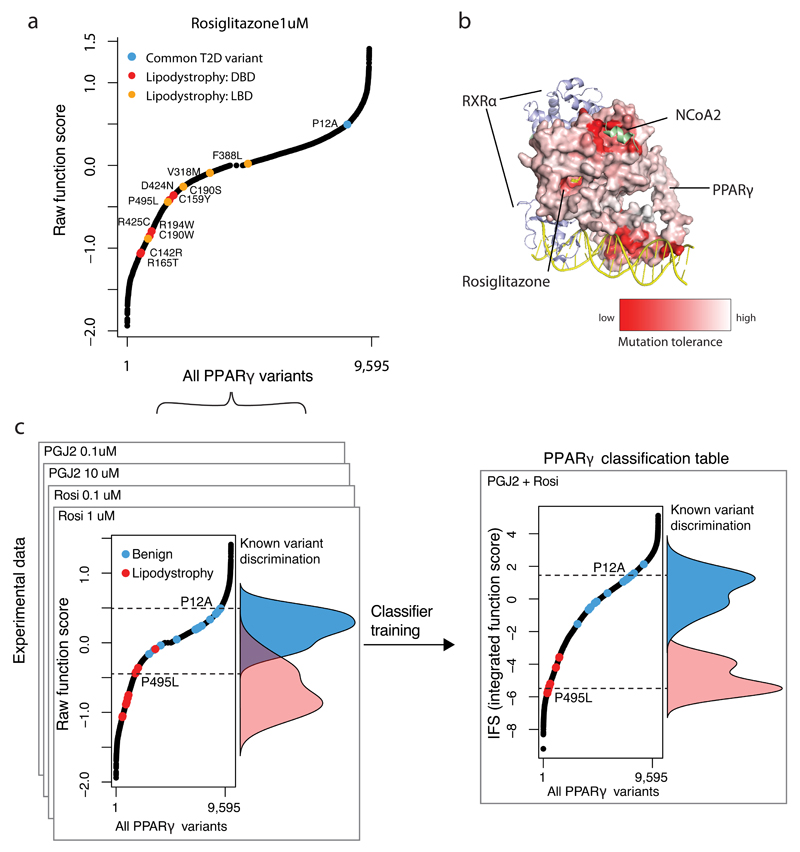
“Function scores” were generated for each amino acid substitution at each site in PPARγ (see Methods, Figure 1B, Figure 2A) based on the partitioning of variants into CD36+/- FACS populations. Over 99% of all possible amino acid substitutions in the protein were covered. Of the twenty possible amino acid substitutions at each site, change to proline was most likely to reduce function, and to cysteine was best tolerated, consistent with the known conformational effects of amino acid side chains on protein structure16. Each of the 505 amino positions in PPARγ was assigned a “tolerance score” by combining function scores of the 19 alternative amino acids at that position (Figure 1B). Tolerance scores were overlaid on the known crystal structure of PPARγ (Figure 2B)17,18 demonstrating that amino acid positions that are intolerant of substitution cluster at residues that contact DNA, co-activating proteins, and ligands (rosiglitazone) (Figure 1B, 2B).
Figure 2. Integrating experimental function to construct a PPARγ classification table.
a) Raw PPARγ function scores ranked for all 9,595 PPARγ variants tested. Highlighted in red are raw function scores of known lipodystrophy causing mutations if they reside in the DNA-binding domain (DBD) or in orange if they reside in the Ligand-binding domain (LBD). The common P12A variant is shown in blue. b) Mutation tolerance scores as described in Figure1 are shown color-coded and mapped onto the known crystal structure of PPARγ with RXRα, NCoA and Rosiglitazone. “Red” denotes that amino acid changes away from reference results in low CD36 function score, whereas ”white” denotes that amino acid changes do not alter function. c) Raw PPARγ function scores were obtained for 9,595 variants under four experimental conditions: 1) 1 μM Rosiglitazone, 2) 0.1 μM Rosiglitazone, 3) 10 μM Prostaglandin J2, and 4) 0.1 μM Prostaglandin J2. The function of known benign (n=13) and lipodystrophy-causing (n=11) variants are highlighted in blue and red respectively with their overall distributions overlaid. The raw function scores were combined into an integrated function score (IFS) after classifier training using linear discriminant analysis (LDA).
We next examined the function scores derived from the CD36/macrophage assay for those mutations previously reported in patients with lipodystrophy/insulin resistance and known to diminish PPARγ activity (Figure 2A). These pathogenic variants (Figure 2A, 2C), clustered in the PPARγ ligand-binding and DNA-binding domains19,4 and had function scores demonstrating enrichment in the CD36-“low” activity bin. In contrast, higher frequency variants including the common P12A variant had function scores demonstrating enrichment in the CD36-“high” activity bin (Figure 2C, Supplementary Table 1). The distribution of function scores for the pathogenic and common variants were significantly different (p < 6x10-7, KS test).
Linear discriminant analysis was used to combine function scores for each of the 9,595 variants across multiple agonist conditions (Figure 2C) into a classifier that maximized discrimination between the set of lipodystrophy-associated variants and the set of high frequency variants described above. The classifier emits the likelihood of each variant being drawn from either of the two classes (pathogenic or benign) and can be expressed as a continuous integrated function score (IFS) (Figure 2C-D).
As above and described in the Methods, the classifier was trained on pathogenic variants obtained from the published literature and benign variants from population-based sequencing20. In order to evaluate the performance of the model on independent data, we turned to novel variants obtained in population-based exome sequencing and sequencing of PPARG in patients referred to specialty clinics for possible lipodystrophy and early-onset diabetes. Specifically, we tested the predictions of functionality emitted by the classifier using standard assays and correlation to clinical phenotypes.
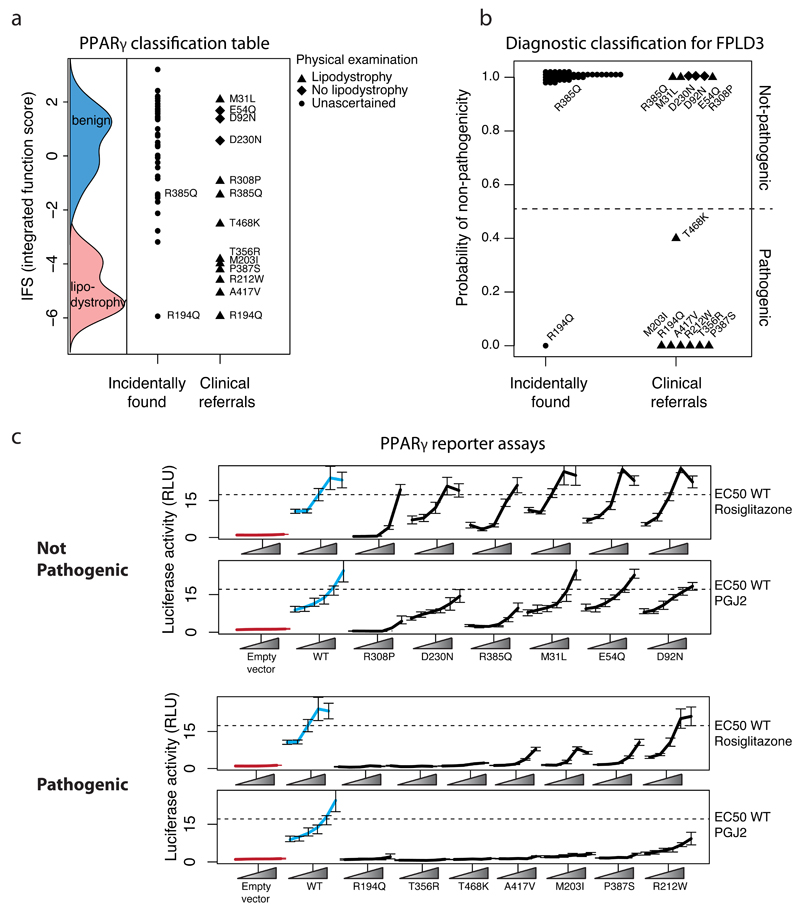
The classifier was applied to data from exome sequencing of 22,106 case/controls selected for study of early-onset myocardial infarction (MIGEN21). In total, 57 missense variants in PPARG were observed with minor allele frequency < 0.1%. Of these, 74% (n=42/57) were novel and thus had not previously been functionally characterized (Supplementary Table 1). In order to calculate a posterior probability of pathogenicity relevant to the clinical context in which the carriers were identified we combined the IFS of these variants with the estimated prevalence of FPLD3 in the general population (1:100,000-1:1,000,00019). One variant, p.R194Q, was estimated pathogenic with high posterior odds (benign:pathogenic) of 1:10,000. The individual who was heterozygous for p.R194Q carried a diagnosis of T2D and had fasting triglyceride levels in the 99th percentile (Supplementary Table 2). As described below, p.R194Q was independently identified in a separate individual referred for clinical features of lipodystrophy (Figure 3, and Supplementary Table 3) who similarly manifested T2D and severe hypertriglyceridemia. Moreover, the p.R194Q variant abolished PPARγ transactivation activity in standard assays (Figure 3C). The combination of clinical and functional data indicate that p.R194Q is likely pathogenic, and that the individual from MIGEN may have undiagnosed FPLD3.
Figure 3. Experimental and clinical classification of novel missense PPARG variants identified in sequenced individuals.
a) Variants identified in patients plotted according to their integrated function score (IFS) alongside the IFS distributions of known benign, and lipodystrophy associated variants. b) Diagnostic classification for Familial Partial Lipodystrophy 3 (FPLD3) expressed as posterior probability of non-pathogenicity of PPARG variants shown in (a). Posterior probability was calculated by combining IFS with prevalence of lipodystrophy in the general population (1:100,000) or from patients referred for lipodystrophy/familial diabetes (1:7). c) The variants identified in patients were individually recreated and tested for their ability to activate luciferase reporter constructs containing three, tandemly-repeated, copies of the PPRE from the Acyl-CoA oxidase gene linked to the thymidine kinase promoter under varying doses of pharmacologic (rosiglitazone) or endogenous (prostaglandin J2; PGJ2) ligands (mean +/- S.E.M n =5). Variants are grouped according to not-pathogenic/pathogenic designation in (b).
We next applied the classifier to variants ascertained from 335 patients referred to UK centers specializing in monogenic forms of diabetes and/or insulin resistance. Thirteen individuals were identified as carrying novel missense variants in PPARG (Supplementary Table 2 and 3), of whom 77% (10/13) had clinical features suggestive of lipodystrophy and associated metabolic derangement including severe insulin resistance, non-alcoholic fatty liver, dyslipidaemia and low serum adiponectin (Supplementary Table 3). The IFS for these thirteen variants were lower than those found in the population-based cohort (above and Figure 3A) (P<0.005 Student’s t-test). For each variant, the posterior probability of pathogenicity was calculated by combining the IFS for that variant and the prevalence of FPLD3 in patients ascertained in these specialty clinics (~1:7 as estimated from the Cambridge national lipodystrophy clinic records).
Three variants (p.E54Q, p.D92N, p.D230N) were found in patients without clinical features of lipodystrophy who had been referred for sequencing based on suspected monogenic diabetes. Despite a higher prior probability based on ascertainment in specialty clinics, these three variants were classified as benign with high confidence (posterior odds benign:pathogenic = 200:1) (Supplementary Table 2). Moreover, when tested individually in standard PPARγ reporter assays these variants showed function indistinguishable from wild-type PPARγ (Figure 3C). Thus, the rate of benign variant identification in individuals ascertained in specialty clinics (~1:110, n=335) was similar to the rate of benign variants identified in the MIGEN cohort (~1:200, n=22,106).
Three variants (p.M31L, p.R308P, p.R385Q) classified as benign with high confidence were found in individuals with clinical features of partial lipodystrophy. The p.M31L variant was found in a female proband with features of lipodystrophy and metabolic derangement (Supplementary Table 3); critically, her daughter had a very similar fat distribution and metabolic phenotype but did not carry the p.M31L variant. Thus, in this case, the phenotype did not segregate with genotype at PPARG. An individual with partial lipodystrophy carried p.R385Q, which was independently identified in a woman from the population-based cohort who had not developed T2D at age 61 (Supplementary Table 2). When tested in PPARγ reporter assays, these variants retained reporter activity, albeit subtly diminished under some conditions (Figure 3). The combination of functional testing, clinical data, and segregation / epidemiology suggests that p.M31L, p.R308P, and p.R385Q are likely incidental findings, although it is not possible to rule out that they act as partial risk-factors for metabolic phenotypes.
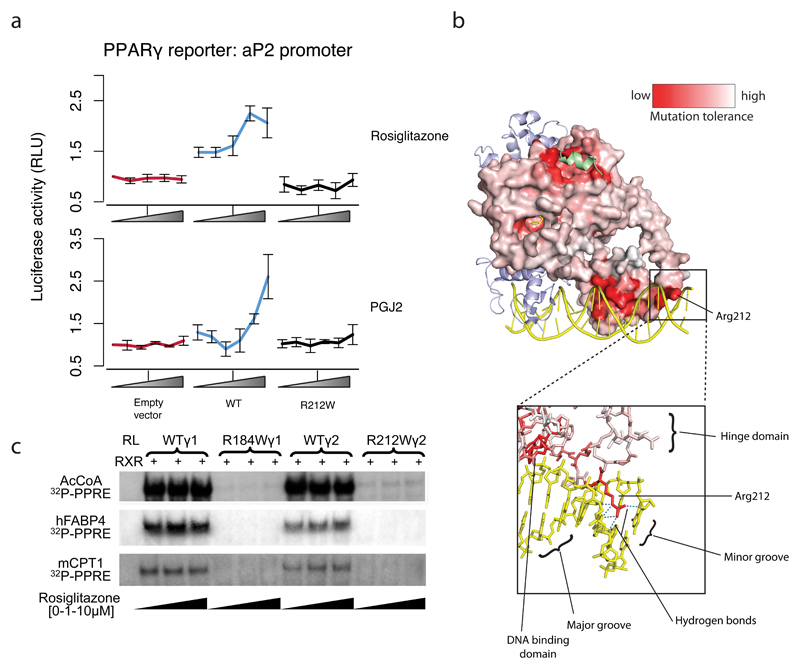
Six variants (p.R194Q, p.A417V, p.R212W, p.P387S, p.M203I, p.T356R) were found in patients with lipodystrophy and classified as pathogenic with high probability (posterior-odds benign:pathogenic = 1:>25,000). Five of the six were confirmed as defective in classical transactivation assays. The exception was p.R212W, where transactivation function when tested using a synthetic PPARγ response element (PPRE) was normal. However, R212W showed less activity in a reporter assay with an endogenous promoter (Figure 4A), and reduced in vitro binding to three PPREs (Figure 4B). The R212 side-chain forms multiple hydrogen-bond contacts in the minor-groove-bound DNA (Figure 4C), outside the main PPRE binding motif.These data indicate that R212W is likely a pathogenic variant despite not showing decreased activity in the traditional functional assay using a synthetic promoter.
Figure 4. Ability of PPARγ p.R212W to transactivate gene expression and bind DNA at endogenous enhancers.
a) Ability of PPARγ2 WT or R212W mutant to activate luciferase reporter constructs containing FABP4 promoter under varying doses of pharmacologic (rosiglitazone 0-1μM) or endogenous (prostaglandin J2; PGJ2 0-10μM) ligands (mean +/- S.E.M n = 5). b) Comparison of the DNA binding properties of in vitro translated wild type or mutant PPARγ proteins, tested in electrophoretic mobility shift assays using either γ1 (R184W) or γ2 (R212W) mutants and radiolabelled PPREs from the acyl coenzyme A oxidase (AcCoA: 5’ ggaccAGGACAaAGGTCAcgtt 3’ ), fatty acid binding protein 4 (FABP4: 5’aaacaCAGGCAaAGGTCAgagg 3’) or muscle carnitine palmitoyl transferase 1 (CPT1: 5’ atcggTGACCTtTTCCCTaca 3’) promoters with retinoid X receptor (RXR) and increasing concentrations of ligand (Rosiglitazone 0 to 10uM). RL, reticulocyte lysate. c) PPARγ colored by mutation tolerance scores obtained under stimulation with 1μM Rosiglitazone in THP-1 cells. As in Figure 2b, red represents sites that exhibited low CD36 response when mutated away from WT. Arginine 212 is highlighted which occurs in the ‘hinge’ region of PPARγ connecting the DNA binding and ligand binding domains. The positively charged arginine side chain extends into the minor groove of DNA forming multiple hydrogen bonds with bases.
Finally, p.T468K, found in a single patient with partial lipodystrophy, was classified by IFS as pathogenic with low confidence (posterior-odds benign:pathogenic = 2:3): its score fell in the overlapping tails of the benign and lipodystrophy-associated variant distributions. In PPARγ reporter assays, this variant demonstrated severely decreased function (Figure 3), supporting that p.T468K is likely a pathogenic variant.
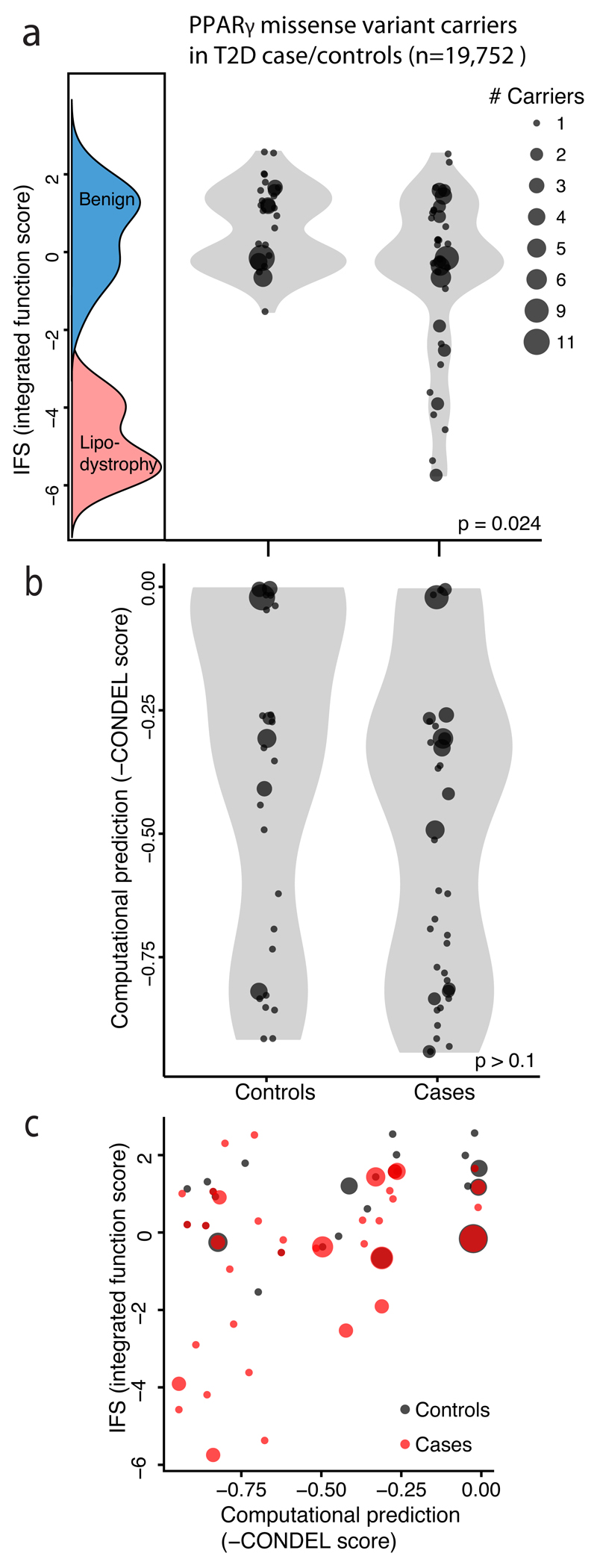
We previously reported that rare missense variants in PPARG that impair function in a single-variant adipocyte differentiation assay confer increased risk of T2D in the general population 5. We re-examined this relationship using functional annotation emitted by the classifier (i.e. IFS) for the original sample of 118 PPARG variant carriers ascertained from 19,752 T2D case/controls (Figure 5A). We observe a long tail of variants with low IFS in T2D cases but not controls (P =0.024, two-sample Kolmogorov-Smirnov test). We quantified this inverse relationship between IFS and T2D case status (logistic regression beta = -0.49 +/- SE 0.15, P=0.002). The odds ratio for T2D in carriers of variants with the lowest tertile of IFS (as compared to carriers of variants in the highest tertile) was 6.5 (95%CI 1.9 – 41) consistent with our previously published estimate5. The odds ratio for the middle vs highest tertile of IFS was 2.0 (95%CI 1.3 – 3.1) suggesting that PPARG variants with even moderately reduced IFS confer a modest increase in T2D risk. By contrast, a conventional predictor of mutation deleteriousness (CONDEL score22) failed to distinguish between likely pathogenic and benign variants (Figure 5b; P > 0.1 two-sample Kolmogorov-Smirnov test) by misclassifying many likely benign variants as pathogenic (Figure 5C).
Figure 5. Relationship of PPARγ function to T2D risk in the general population.
a) Missense PPARγ variants identified from 19,752 sequenced type 2 diabetes (T2D) case/controls plotted according to IFS (integrated functional score) from the PPARγ classification table alongside the IFS distributions of known benign, and lipodystrophy associated variants. Each point represents a missense variant; point size denote the number of individuals carrying that variant. Among the 118 individuals carrying missense PPARγ variants T2D cases contained a long tail of low-functioning missense variants, which was notably absent from the distribution of variants observed in T2D controls (p = 0.024 two-sample Kolmogorov-Smirnov test). b) When the same 118 individuals were plotted according to computational prediction of deleteriousness no difference is distributions of functional variants is seen among T2D cases vs controls (p > 0.1 two-sample Kolmogorov-Smirnov test). c) Scatterplot of IFS vs computational prediction scores for PPARγ missense variants from T2D case/controls as described above.
These data show that it is possible to experimentally characterize all possible missense variants in a mammalian gene and use the information to guide interpretation of VUS, a concept that has been previously applied to single protein domains23,24. Testing variants prospectively (that is, prior to their discovery in patients) overcomes barriers of time and scalability that have thus far made it impractical to incorporate experimental data into routine clinical variant interpretation. Furthermore, by simultaneously and consistently evaluating all variants in a single experiment, more valid comparisons can be made across variants as compared to data on different variants generated in different labs at different times.
The PPARG classifier annotated as benign nearly all variants (56/57) incidentally identified in a study of myocardial infarction. The one variant classified as pathogenic with high confidence (and confirmed by single variant laboratory experiments) was observed in an individual with hypertriglyceridemia and T2D, and independently observed in a patient with lipodystrophy, likely indicating FPLD325. In 12/13 cases referred for suspected lipodystrophy or monogenic diabetes and carrying a PPARG variant, the classifier provided immediate, high confidence information regarding the likelihood of a functional defect and a molecular diagnosis of FPLD3. In only a single case (p.T468K) did the classifier not provide a high confidence estimate and low-throughput laboratory assays fail to corroborate the pooled assay data13.
Systematic variant construction, pooled experimental characterization in relevant assays, and statistical integration with epidemiological data offer a generalizable approach to enable genome interpretation at clinically important genes, reducing overdiagnosis6,9 and diagnostic uncertainty8. Fully realizing such comprehensive approaches will require a complementary array of methods26. The PPARG construct library is easily shared so that others can generate and contribute function scores in other assays27, but as a transgene library it is not ideally suited for detecting functional effects of coding variation on splicing efficiency. Given the limitations on the library and because CD36 expression is unlikely to report on all the functions of PPARγ we have made the PPARγ classifier available as a web application (http://miter.broadinstitute.org) that can be updated as new genetic and functional data become available. Broadening this approach to other genes and diseases will require cellular assays that read out disease relevant characteristics, are robust and scalable, and the availability of training sets of pathogenic and benign variants. Such assays and variants exist for a number of Mendelian disease genes, making it possible to apply a similar approach to help interpret VUS for many other clinical situations.
Methods
Synthesis and assembly of 9,595 PPARG variant constructs
A library of all 9,595 possible single amino acid variants in PPARG was synthesized using a site-directed, multiplexed method (Mutagenesis by Integrated TilEs (MITE)28) adapted to render it suitable for saturation mutagenesis in mammalian cells. Detail is provided below where methodologic advancements were made permitting saturation mutagenesis of PPARG. First, the PPARG cDNA sequence (CCDS2609.1) was recoded (see Supplementary Table 4) to eliminate susceptibility to restriction enzymes and CRISPR/CAS9 targeting sgRNAs (see below) to enable a “delete and replace” strategy. As described previously, DNA oligonucleotides were synthesized on a programmable microarray, each oligonucleotide encoding a desired amino acid change but otherwise homologous to the template un-mutated PPARG in all other respects. Oligonucleotides were organized into ‘tiles’, where those within each tile differ in a central variable region but share identical 5’ and 3’ ends (see Supplementary Table 4). Tiles were staggered such that their variable regions collectively span the entire template. To ensure uniform amplification and reduce chimera formation for the longer PPARG template, the protocol was modified to amplify each tile by emulsion PCR (MICELLULA DNA Emulsion & Purification Kit; EURx). The resulting products were inserted into linearized plasmids (Phusion® High-Fidelity DNA Polymerase NEB M0530) that carry the remaining template sequence using multiplexed Gibson assembly (NEBuilder® HiFi DNA Assembly Master Mix, NEB, cat E2621L) according to the manufacturer’s protocol. A “frameshift cleaning” procedure was introduced given that the most common error mode during library construction (25-30% of constructs; data not shown) resulted from oligo synthesis errors causing 1-2 bp indels. The PPARG template vector was designed such that all PPARG constructs terminated with amber stop codons (i.e. TAG) and bore an in-frame zeocin resistance cassette (pUC57-PPARG-zeo; GenScript). Constructs bearing frame-shifting indels were depleted by transforming into an amber suppressor cloning host (TG1, Lucigen) and selecting the construct library under zeocin and kanamycin dual selection. Library plasmids were purified from >106 colonies to preserve complexity and the frameshift depleted PPARG transgenes excised from the zeocin resistance cassette. To enable mammalian cell transduction, the transgene library was transferred into a lenti-viral expression vector by simple restriction cloning and transfected into a packaging cell line to produce pooled lenti-virus according to standard protocols (pLXI_TRC401; http://www.broadinstitute.org/rnai/public/resources/protocols)5.
Deletion of endogenous PPARG in THP-1 monocytes using CRISPR/CAS9
The endonuclease Cas9 and sgRNAs targeting exon 6 of PPARG and exon 8 of a control gene, PHACTR1, were introduced into THP1 cells by lenti-viral transduction (see Supplementary Table 4). To quantify modification of the endogenous gene, genomic DNA was extracted at multiple time points, amplified by PCR around the PPARG sgRNA target site and Sanger sequenced (see Supplementary Table 4). Cutting efficiency was determined using the TIDE web tool for decomposition analysis of the sequencing traces29.
Twenty-one days after transduction of CRISPR/Cas9 with PPARG or control sgRNAs, cells were tested for PPARG response by gene (FABP4) and protein (CD36) expression to validate lack of functional endogenous PPARG. PPARG targeting sgRNA and control sgRNA treated THP1 cells were stimulated with 1 μMRosiglitazone in THP1 growth media (RPMI 1640 + 10% heat-inactivated FBS + 1% PenStrep + 0.1% BME) for 72 hours. mRNA was then extracted and quantified for FABP4 gene expression(nanoString Technologies). For CD36 protein expression, THP1 cells were stimulated with 50 ng/mL PMA and 1 μM of Rosiglitazone in growth media for 72 hours. Cells were then detached from the plate, washed and stained with a monoclonal antibody to CD36 according to the manufacturer’s protocol (Miltenyi 130-100-149) and subjected to flow cytometry.
Simultaneous testing of 9,595 PPARG variants in experimental assays
The PPARG construct library was introduced into a human monocytic cell line (THP-1: obtained from http://www.broadinstitute.org/achilles and tested mycoplasma negative) engineered through CRISPR/CAS9 to lack endogenous PPARG (Supplementary Figure 2) by pooled infection. While isoform 1 of PPARG is dominantly expressed in monocyte/macrophages, we expressed isoform 2, which is identical in sequence but encodes a protein with an additional 28 N-terminal amino acids. Both isoforms demonstrated identical ligand dependent activity. The pooled virus was diluted such that the multiplicity of infection (number of viral particles per cell) was 0.3 so that each monocyte would receive zero or a single PPARG variant. Uninfected cells were eliminated by selection with puromycin 2 μg/mL. Expression of the PPARG transgene was controlled by a doxycycline inducible promoter5. At least 107 cells were infected to ensure that each PPARG variant was independently represented in 1000 monocytes. The resulting polyclonal population of THP-1 monocytes containing the PPARG variant library was stimulated for 72 hours with 1) 50 μM phorbol ester (PMA) to induce differentiation into macrophages, 2) doxycycline 1 μg/mL to induce expression of PPARG constructs, and 3) low/high doses (based on ranges used in prior studies 13) of thiazolidinedione (Roziglitazone 0.1 μM/1 μM) or proposed natural ligand30 (Prostaglandin J2 (PGJ2) 0.1 μM/10 μM) to stimulate PPARG activity. The population of stimulated THP-1 macrophages was immuno-stained for CD36 (Miltenyi: 130-095-472), a cell surface protein that is a direct transcriptional target of PPARG 15. Using fluorescence activated cell sorting, stained cells were grouped into two activity bins separated by at least 5-10 fold expression of CD36 and selected to encompass equal numbers of cells (Supplementary Figure 3). For each stimulation condition, at least three replicates were generated, each with at least 5×106 cells sorted. To re-identify and quantitate the PPARG variants in the CD36 ‘high’ and ‘low’ bins, genomic DNA was extracted from the cells in each bin and the integrated proviral PPARG transgenes amplified by PCR and shotgun sequenced (Nextera, Illumina). Raw sequencing reads were aligned to the reference PPARG cDNA sequence (see Supplementary Table 4) and the number of occurrences of each amino acid at each position along the coding region counted and tabulated with a custom aligner. To minimize erroneous mutation calls, only codons that matched designed mutations and consisted of high quality base calls (Phred score > 30) were tabulated. Over 99 percent of the designed amino acid substitutions were observed at least 50 times for a given experimental condition (see Supplementary Figure 1). A raw function score was calculated based on the ratio of observed frequencies of each mutant amino acid in the two CD36 activity bins (see Figure 1).
Calculation of raw function score
Control experiments showed that variants deleterious to PPARG function were enriched in the CD36 low fraction and benign variants enriched in the CD36 high fraction. We constructed a likelihood function based on the log-odds of an amino acid variant in the CD36 high and low fractions. The log-odds for each amino acid variant was estimated by maximizing a likelihood function based on the observed counts of each amino acid variant in the CD36 high and low fractions as well as the total read depth at that amino acid position. Data were combined across experimental replicates after determining replicate variability (see Supplementary Figure 4). To avoid spuriously high or low log-odds estimates for any given variant, we constrained the log-odds estimate with a Gaussian prior whose parameters were estimated from data combined across all variants. See “Supplemental Note: Supplementary Analytic Methods” for detailed specification.
Construction of a PPARG classifier by supervised machine learning
To predict the likelihood of novel variants being benign and pathogenic, we developed a classifier based on raw function scores obtained across various experimental conditions. The synthesis of multiple experimental conditions was intended to span a greater range of possible activities of PPARγ than would be queried using a single condition. Specifically, we used linear discriminant analysis (MASS package in R 3.0) to train the classifier, adopting a two-class model. The model incorporates as parameters (a) raw function scores for each PPARγ variant as measured across the four experimental conditions (i.e. rosiglitazone (Rosi) and Prostaglandin J2 (PGJ2) at high and low doses) and (b) mutation tolerance scores calculated for each position in PPARG as measured across the four experimental conditions (see Figure 1B). Potential classifiers were systematically constructed on linear combinations of four of these eight parameters, with a requirement that one parameter be included from each experimental condition. Classifier models were built for each the 16 possible combinations of four parameters using a training set of pathogenic and benign PPARγ variants (see Supplementary Table 1). Pathogenic variants used to train the classifier were selected based on (a) segregation with FPLD3 and (b) prior demonstration of loss-of-function in cellular assays. Benign variants used to train the classifier were selected from among variants identified in 60,706 aggregated exome sequences20 at an allele frequency rendering them very unlikely to be causal for FPLD3 under a dominant model of inheritance and prevalence estimate ranging from 1:100,000 to 1:1,000,000 (P<0.05 1-tailed binomial probability n=121,412 chromosomes, p=10-5) (see Supplementary table 1). The performance of these 16 models was compared using a leave-one-out cross-validation (LOOCV) protocol with each model scored by its aggregate ability to correctly classify the “left-out” variant over all the cycles of LOOCV. The highest scoring model consisted of raw function scores for each possible variant obtained from three conditions (Rosi 1μM, Rosi 0.1μM, PGJ2 10μM) and mutation tolerance score for each position in PPARG obtained from PGJ2 0.1μM. This model was fit to the full training dataset for prospective evaluation of novel PPARG variants. The weighted sum of the four parameters in the final model, as fit by the LDA algorithm, is denoted as the integrated function score (IFS) (see Figure 2C and Supplementary Figure 5) and represents an aggregate measure of variant function over the four experimental conditions. For clinical prediction, the IFS was expressed as an odds (benign:pathogenic), which when multiplied by the estimated prior odds of FPLD3 based on the clinical situation (i.e. prevalence) yielded an estimated probability of pathogenicity. Because the final model was trained on the full set of available pathogenic and benign variants, its performance next required prospective evaluation on a completely independent set of variants. These variants were obtained from the population and clinic data described below, and evaluated as described in Figure 3.
Missense PPARG variants identified in population based exomes and clinically referred individuals
The study was conducted in accordance with the Declaration of Helsinki, and approved by research ethics committees; written informed consent was obtained from all participants.
Missense PPARG variants were extracted from 22,106 exomes (8,400 with early-onset coronary artery disease and 12,804 controls) sequenced by the Myocardial Genetics Consortium (MIGEN) as described elsewhere21. Study participants were ascertained from the following studies: ATVB, DHM, DUKE, JHS, ESP-EOMI, MedStar, OHS, PennCath, PROCARDIS, PROMIS, and REGICOR. Participants were of European ancestry (n=12,849; 58%), Asian ancestry (n=6,823; 31%), African ancestry (n=2,399; 11%), and “other or unknown” self-reported ethnicity (n=34; 0.2%). Twenty-two percent (n=4,258) reported a diagnosis of T2D.
Patients were referred to one of two UK centers (Cambridge: www.cuh.org.uk/national-severe-insulin-resistance-service or Exeter: www.diabetesgenes.org) which specialize in syndromes of severe insulin resistance and/or monogenic forms of diabetes. In clinically suspected FPLD3 cases, mutations in PPARG were identified in genomic DNA extracted from peripheral-blood leukocytes using PPARG amplification and sequencing. In patients for whom FPLD3 was not the primary clinical diagnosis, PPARG was sequenced as part of a targeted next-generation panel of 29 genes31 selected to improve diagnostic yield for suspected monogenic diabetes. Mutations were confirmed in index patients and, where possible, from family members. In all instances, the nomenclature used for missense variants is for isoform 2 of PPARG (transcript accession: NM_015869.4; protein accession: NP_056953.2).
Individual testing of PPARG variant function by transcriptional activity
The novel variants identified in patients with suspected familial lipodystrophy or diabetes were characterized using a well-established PPARG reporter containing three, tandemly-repeated, copies of the PPRE from the Acyl-CoA oxidase (AcCoA: 5’ ggaccAGGACAaAGGTCAcgtt 3’) gene upstream of the thymidine kinase (TK) promoter and luciferase. In brief, 293EBNA cells, cultured in DMEM/10%FCS were transfected with Lipofectamine2000 in 24-well plates and assayed for luciferase and β-galactosidase activity as described previously13 following a 36-hour incubation with or without ligand.
Supplementary Material
Acknowledgements
Supported by grants from the National Institute of Diabetes, Digestive, and Kidney Diseases (1K08DK102877-01, to Dr. Majithia; 1R01DK097768-01, to Dr. Altshuler), NIH/Harvard Catalyst (1KL2 TR001100-01, to Dr. Majithia), Broad Institute (SPARC award, to Dr. Majithia and Dr. Mikkelsen), and Wellcome Trust (#095564, to Dr. Chatterjee; #107064, to Dr. Savage).
We thank John Doench, Cong Zhu, Daniel O’Connell, Glenn Cowley, Meagan Sullender, Daniel MacArthur, Eric Minkel, Brendan Bulik-Sullivan and Joseph Avruch for helpful discussions, laboratory assistance and manuscript review.
Dedicated to the memory of Promila Nandi April 30, 1933 – December 27, 2013
Footnotes
URLs
http://miter.broadinstitute.org; PPARG missense variant lookup table
http://www.cuh.org.uk/national-severe-insulin-resistance-service
www.cuh.org.uk/national-severe-insulin-resistance-service
http://www.broadinstitute.org/rnai/public/resources/protocols; lentivirus
http://www.broadinstitute.org/achilles; cell lines
Author Contributions
A.R.M, T.M. and D.A. designed the study. A.R.M., B.T., M.A., and K.G. performed experiments with help from R.R., X.Z., M.F.B. and E.K. A.R.M and N.P. analyzed the data with help from B.T., T.S., G.P., K.A.P., M.D., and T.M. I.B., S.E., S.K., S.O.R., K.C. and D.B.S. contributed clinical data and genotypes. A.R.M and D.A. wrote the manuscript. D.B.S., S.O.R., K.C., E.D.R., and J.C.F revised the manuscript.
Competing financial interests
No competing financial interests
References
- 1.Majewski J, Schwartzentruber J, Lalonde E, Montpetit A, Jabado N. What can exome sequencing do for you? J Med Genet. 2011;48:580–9. doi: 10.1136/jmedgenet-2011-100223. [DOI] [PubMed] [Google Scholar]
- 2.Gahl WA, et al. The National Institutes of Health Undiagnosed Diseases Program: insights into rare diseases. Genet Med. 2012;14:51–9. doi: 10.1038/gim.0b013e318232a005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barroso I, et al. Dominant negative mutations in human PPARgamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature. 1999;402:880–3. doi: 10.1038/47254. [DOI] [PubMed] [Google Scholar]
- 4.Jeninga E, Gurnell M. Functional implications of genetic variation in human PPAR [gamma] Trends in Endocrinology & …. 2009 doi: 10.1016/j.tem.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Majithia AR, et al. Rare variants in PPARG with decreased activity in adipocyte differentiation are associated with increased risk of type 2 diabetes. Proc Natl Acad Sci U S A. 2014;111:13127–32. doi: 10.1073/pnas.1410428111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flannick J, et al. Assessing the phenotypic effects in the general population of rare variants in genes for a dominant Mendelian form of diabetes. Nature genetics. 2013;45:1380–1385. doi: 10.1038/ng.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tennessen JA, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–9. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McLaughlin HM, et al. A systematic approach to the reporting of medically relevant findings from whole genome sequencing. BMC Med Genet. 2014;15:134. doi: 10.1186/s12881-014-0134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manrai AK, et al. Genetic Misdiagnoses and the Potential for Health Disparities. N Engl J Med. 2016;375:655–65. doi: 10.1056/NEJMsa1507092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altshuler D, et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 11.Claussnitzer M, et al. Leveraging cross-species transcription factor binding site patterns: from diabetes risk loci to disease mechanisms. Cell. 2014;156:343–58. doi: 10.1016/j.cell.2013.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- 13.Agostini M, et al. Non-DNA binding, dominant-negative, human PPAR [gamma] mutations cause lipodystrophic insulin resistance. Cell metabolism. 2006;4:303–311. doi: 10.1016/j.cmet.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu S, et al. Adipocyte-specific gene expression and adipogenic steatosis in the mouse liver due to peroxisome proliferator-activated receptor gamma1 (PPARgamma1) overexpression. J Biol Chem. 2003;278:498–505. doi: 10.1074/jbc.M210062200. [DOI] [PubMed] [Google Scholar]
- 15.Tontonoz P, Nagy L, Alvarez JG, Thomazy VA, Evans RM. PPARgamma promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell. 1998;93:241–52. doi: 10.1016/s0092-8674(00)81575-5. [DOI] [PubMed] [Google Scholar]
- 16.Barnes MR, Gray IC, editors. Bioinformatics for geneticists. Wiley; Chichester, West Sussex, England; Hoboken, N.J: 2003. [Google Scholar]
- 17.Chandra V, et al. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature. 2008;456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.8. 2015 [Google Scholar]
- 19.Garg A. Acquired and inherited lipodystrophies. New England Journal of Medicine. 2004 doi: 10.1056/NEJMra025261. [DOI] [PubMed] [Google Scholar]
- 20.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016 doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myocardial Infarction Genetics Consortium, I et al. Inactivating mutations in NPC1L1 and protection from coronary heart disease. N Engl J Med. 2014;371:2072–82. doi: 10.1056/NEJMoa1405386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez-Perez A, Lopez-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet. 2011;88:440–9. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fowler DM, et al. High-resolution mapping of protein sequence-function relationships. Nat Methods. 2010;7:741–6. doi: 10.1038/nmeth.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Starita LM, et al. Massively Parallel Functional Analysis of BRCA1 RING Domain Variants. Genetics. 2015;200:413–22. doi: 10.1534/genetics.115.175802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Demir T, et al. Familial partial lipodystrophy linked to a novel peroxisome proliferator activator receptor -gamma (PPARG) mutation, H449L: a comparison of people with this mutation and those with classic codon 482 Lamin A/C (LMNA) mutations. Diabet Med. 2016 doi: 10.1111/dme.13061. [DOI] [PubMed] [Google Scholar]
- 26.Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014;513:120–3. doi: 10.1038/nature13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fowler DM, Fields S. Deep mutational scanning: a new style of protein science. Nat Methods. 2014;11:801–7. doi: 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melnikov A, Rogov P, Wang L, Gnirke A, Mikkelsen TS. Comprehensive mutational scanning of a kinase in vivo reveals substrate-dependent fitness landscapes. Nucleic Acids Res. 2014;42:e112. doi: 10.1093/nar/gku511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forman BM, et al. 15-Deoxy-delta 12, 14-prostaglandin J2 is a ligand for the adipocyte determination factor PPAR gamma. Cell. 1995;83:803–12. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 31.Ellard S, et al. Improved genetic testing for monogenic diabetes using targeted next-generation sequencing. Diabetologia. 2013;56:1958–63. doi: 10.1007/s00125-013-2962-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.