Abstract
Objective
Reproducibility is the cornerstone of the scientific method. However, currently, many systems biology models cannot easily be reproduced. This paper presents methods that address this problem.
Methods
We analyzed the recent Mycoplasma genitalium whole-cell (WC) model to determine the requirements for reproducible modeling.
Results
We determined that reproducible modeling requires both repeatable model building and repeatable simulation.
Conclusion
New standards and simulation software tools are needed to enhance and verify the reproducibility of modeling. New standards are needed to explicitly document every data source and assumption, and new deterministic parallel simulation tools are needed to quickly simulate large, complex models.
Significance
We anticipate that these new standards and software will enable researchers to reproducibly build and simulate more complex models, including WC models.
Index Terms: Systems biology, whole-cell, computational modeling, reproducibility, repeatability, standards, provenance
I. Introduction
Reproducibility is one of the central tenets of the scientific method. We define reproducibility as the ability to confirm a result via a completely independent test, including different investigators, experimental methods, and experimental machinery. In the context of systems biology, a simulation result is reproducible if the model which generated the result can be recreated from our collective scientific knowledge, including manuscripts, databases, code repositories, and supplementary materials, and the simulation result can be regenerated from descriptions of the model and the simulation experiment. This rigorous standard eliminates conclusions that are based on incorrect methods, machinery, or experiments, and ensures that scientific results are only accepted as facts once multiple scientists have thoroughly dismissed other potential explanations.
We define repeatability as the ability to regenerate a result given the same experimental machinery and conditions. In the context of systems biology, a simulation result is repeatable if the numerical result can be regenerated from descriptions of the model and the simulation experiment. Repeatability is a more lenient standard than reproducibility because it does not require regeneration of the model itself. Consequently, testing repeatability only eliminates scientific conclusions that are based on erroneous experiments, and cannot eliminate conclusions that are based on faulty models.
To illustrate the distinction between reproducibility and repeatability, consider the following example: A modeler Alice wants to investigate a discrepant simulation result published by Bob. Bob’s model predicts that knocking out regulator Y causes cancer, whereas Alice’s model indicates that cancer requires at least two knockouts. Fortunately, Alice can investigate Bob’s results because Bob published his model and simulation experiments in standard formats. Alice uses Bob’s model and simulation experiment files to repeat Bob’s simulation results, compares the two models, and finds that Bob’s model generates different predictions because it uses a different rate law to describe the effect of regulator Y. However, because Bob’s model file does not describe all of the experimental data and assumptions underlying his model, Alice cannot reproduce Bob’s rate laws and thus cannot fully resolve why their models generate different predictions.
The systems biology community, spearheaded by the Computational Modeling in Biology Network [1], has developed several standard formats to exchange models and repeat simulations [2]. Examples of these standards include CellML [3], the COMBINE archive [4], the Systems Biology Markup Language (SBML) [5], the Simulation Experiment Description Markup Language (SED-ML) [6], and the Systems Biology Graphical Notation (SBGN) [7]. These standards support several types of models including ordinary differential equation (ODE), flux balance analysis (FBA) [8], and logical models. Numerous software programs support these standards [9]. Furthermore, several model repositories, including BioModels [10] and the CellML Model Repository [11], share models that are encoded in these standards. In addition, the bioinformatics and computer science communities have developed several methods and tools, including Galaxy [12], Taverna [13], and VisTrails [14], to track the provenance of computational analyses, including recording all of the data sources and assumptions used to conduct analyses.
These standards and modeling software tools help researchers repeat most systems biology simulation experiments and reuse, modify, expand, and combine most systems biology models. However, these standards and software provide limited support for regenerating models because they do not record all design choices, such as experimental data sources and assumptions, that are used to build models,
Furthermore, researchers have begun to develop more complex models which cannot be represented by the existing standards or simulated by the existing standards-compliant software. For example, several aspects of the recent whole-cell (WC) model of Mycoplasma genitalium [15] cannot currently be represented by SBML [16]. In particular, SBML cannot represent the multi-algorithmic nature of the model. In addition, no SBML-compatible simulation software program supports all of the SBML packages needed to simulate WC models, including the Arrays [17], Distributions [18], Flux Balance Constraints [19], Hierarchical Model Composition [20], and Multistate and Multicomponent Species [21] packages.
Here, we present a path toward fully reproducible systems biology modeling, including WC modeling. First, we propose several requirements for reproducible modeling. Second, we outline several gaps in the existing standards and software for reproducible modeling and describe several new standards and software tools that are needed to achieve reproducible modeling. Third, we describe several methods for verifying repeatability. Achieving this vision will require significant research on standards and software development.
II. The Requirements for Reproducible Modeling
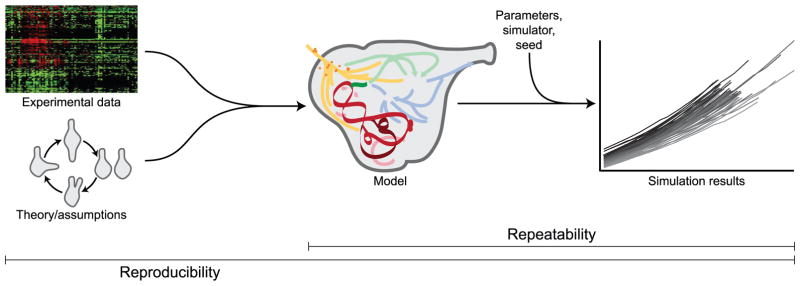
There are three requirements for fully reproducible systems biology modeling (Fig. 1). (1) Researchers should be able to regenerate models, including every species and reaction from the literature. Consequently, researchers should record the provenance of every data source and assumption used to build models, as well as save a copy of each data source to guarantee future access to every source. (2) Researchers should be able to regenerate statistically identical simulation results. Consequently, researchers should record every parameter value, algorithm, and simulation software option used to simulate models. (3) Researchers should ensure that multiple simulation software tools generate statistically identical results. This is particularly helpful for identifying errors in complex simulation software programs. This requires standard model description formats that support all systems biology models, including WC models. This would enable researchers to use different software programs to generate the same results.
Fig. 1.
Three requirements for reproducible systems biology modeling. (1) The provenance of every data source and assumption should be recorded so that models can be regenerated, including every species, reaction, and rate law. (2) Every simulation parameter value should be recorded so that numerical simulation results can be regenerated. (3) Multiple simulation software tools should generate statistically identical numerical results. Reproducibility is a stricter standard than repeatability. Models are considered repeatable, but not reproducible, if they satisfy just the second and third criteria.
III. Towards a Platform for Reproducible Modeling
Currently, each individual modeler is responsible for reproducibly conducting their own research. This places a substantial burden on individual researchers to make every step of their research reproducible. To ease this burden, we recommend that the systems biology community develops several software tools, databases, and standards to enable researchers to conveniently conduct reproducible research.
(1) More comprehensive experimental databases should be developed to provide the data needed to build systems models. For example, Karr et al. developed the WholeCellKB database [22] to organize the over 1,400 quantitative measurements used to build the M. genitalium WC model. These databases should be machine-readable so that they can be automatically queried and used to build models. These databases should also use data models similar to that of systems biology models to clarify the connection between models and their underlying data. (2) New model design tools and Systems Biology Ontology (SBO) [23] terms are needed to record how models are built, including every design choice, assumption, and data source. Currently, researchers can use SBML annotations and SBO terms to record several common assumptions, such as the rapid equilibrium between free enzymes and enzyme-substrate complexes in Michaelis-Menten kinetics. However, many researchers do not utilize these annotations and the SBO does not represent all possible assumptions. Thus, researchers should develop model design tools which automatically record assumptions and data sources by adopting provenance tracking techniques [12–14]. The SBO should also be expanded to represent more assumptions. (3) Where possible, researchers should use standard formats such as SBML and SED-ML rather than proprietary codes to describe models and simulation experiments. This should include annotation of the units of every parameter. (4) The existing standard model description formats should be expanded to represent all types of systems biology models, including WC models. For example, to describe WC models in a standard format, SBML could be expanded to support genomic sequence data and sequence-based reaction patterns or SBML could be linked with the Synthetic Biology Open Language (SBOL) [24], which is already capable of representing sequence data. (5) The existing standards-compliant simulation software programs should be expanded to support a wider variety of models. In particular, simulation software programs should be expanded to support the Arrays, Distributions, Flux Balance Constraints, Hierarchical Model Composition, and Multistate and Multicomponent Species SBML packages. Several simulation software programs, including BioUML [25] and iBioSim [26], have already begun to support these packages. Unfortunately, most simulation software developers have insufficient funding to implement every package. (6) Modeling workflows should be expanded to systematically verify the statistical repeatability of simulation results. (7) New model verification tools should be developed to automatically identify errors in models. We developed a suite of tests to systematically error-check our M. genitalium WC model1. These tests checked for simple problems such as undefined species, reaction mass and charge imbalance, and inconsistent reaction rates among submodels. Nevertheless, we found this test suite invaluable for debugging our model. These tests should be generalized and new software tools should be developed to help researchers systematically evaluate such tests. (8) Every model, simulation experiment, simulation result, and simulation software program should be reusable, extensible, documented, and published open-source [27]. (9) Journals should require and help authors permanently archive every reported data source and model.
A. Special Considerations for Stochastic Simulation
Stochastic simulations typically use pseudo-random number generators (PRNGs) which deterministically produce the same numbers when seeded with the same initial state. Therefore, we recommend that stochastic simulation software developers provide ways to set and record PRNG seeds so that modelers can repeat stochastic simulation results. This would enable researchers to repeat not only statistical distributions, but exact trajectories, which is invaluable for identifying and debugging errors in complex models and simulation algorithms.
Furthermore, different implementations of the same PRNG often differ in subtle ways. For example, the seed methods of the Boost C++ Library [28] and Python Standard Library implementations of the Mersenne-Twister algorithm differentially initialize the PRNGs’ internal states. Thus, we recommend the creation of standard PRNGs, including documentation of their algorithms, parameterizations, and initial states, and a test suite to help software developers verify their implementations.
B. Special Considerations for Multi-algorithm WC Models
Multi-algorithm WC models strive to represent every gene and cell function by combining multiple submodels of individual cellular pathways, each represented using different mathematics and each trained using different experimental data [29–31]. For example, the M. genitalium WC was composed of 28 submodels, including an FBA submodel which describes its metabolism, stochastic submodels which describe its transcription and translation, and an ODE submodel which describes its division. This multi-algorithm methodology is motivated by the desire to model biological systems as completely as possible, given our current knowledge, by simultaneously using fine-grained representations and coarse-grained representations to represent well- and poorly-characterized pathways, respectively.
Multi-algorithm modeling is a new methodology which still lacks a rigorous theoretical foundation. Consequently, significant work is needed to develop tools for building, simulating, and reproducing WC models. (1) The Hierarchical Model Composition SBML package should be extended to use the KiSAO ontology [32] to represent each submodel’s simulation algorithm. (2) Researchers should determine how to concurrently integrate submodels that share state. Researchers are currently exploring several potential methods to concurrently integrate submodels, including shared memory and parallel discrete event simulation [33]. (3) Researchers should develop a high-performance, reusable multi-algorithm simulator. (4) Additional tools should be developed to help researchers build and analyze WC models. (5) These programs should be implemented as separate tools and integrated into a comprehensive WC modeling platform so that software developers can contribute to individual tools and so that modelers can easily use alternative components. We anticipate that this platform will enable more researchers to engage in WC modeling and accelerate the WC modeling field.
C. Special Considerations for Parallel Simulation Software Programs
Multi-threaded and distributed computing are two important methods for accelerating the simulation of computationally expensive models such as WC models. Parallel programs use multiple threads and/or cores to simultaneously execute different parts of a simulation. The order in which these threads and/or cores complete computations depends on the operating environment, and the order in which computations complete can affect subsequent computations and, in turn, simulation results. Other sources of non-determinism in parallel computing include memory layout, delays caused by devices and processes, and user input. Thus, parallel programs can generate non-deterministic simulation results due to variable operating environments.
For example, if two WC submodels which are executed in parallel attempt to bind proteins to the same site on the chromosome, the submodel which completes its computations first will bind the protein to the chromosome. However, when the simulation is re-run under a different load on the computer, the other submodel may complete its computations first, resulting in the binding of a different protein the site. This small difference can lead to further differences between later time steps, resulting in different simulation results solely due to different operating environments.
Consequently, special care must be taken to create deterministic parallel simulation programs. Therefore, we briefly examine methods from computer science which have been developed to create deterministic parallel programs.
Several libraries have been developed to eliminate nondeterminism from multi-threaded programs. These include COREDET [35] and DTHREADS [36]. COREDET is a library for compiling C/C++ programs such that they behave deterministically. DTHREADS is a replacement for the standard UNIX Pthreads library. Both systems create deterministic programs by preventing information sharing between threads during parallel phases, and deterministically allowing sharing during serial phases. These systems also eliminate nondeterminism due to variable memory layout. DTHREADS achieves this by implementing a special deterministic memory allocator. COREDET deterministically allocates memory by compiling memory allocators with the COREDET library to generate deterministic memory allocators. COREDET and DTHREADS have both been shown to impose minimal overhead [36]. We recommend that simulation software developers implement options for deterministic, repeatable simulations, and we offer COREDET and DTHREADS as examples of how this can be achieved for multi-threaded simulation programs.
Computer scientists have also developed several practices to create deterministic distributed programs. (1) Inter-process communication messages should be deterministically executed to avoid race conditions [37]. Messages can be deterministically scheduled by using first-in-first-out message queues between each pair of communicating processes and blocking message receive operations [38]. Time-driven simulations can also deterministically schedule messages by ordering messages based on their simulation times [39]. Parallel shared-memory time step simulations can be made deterministic by synchronizing the end of time steps with barriers [40], such as implemented in OpenMP [41], and employing deterministic approaches to share time step state updates among threads. (2) Each individual process in a distributed simulation program should be deterministic. This can be achieved by following the recommendations in this manuscript. We recommend that simulation software developers adopt these practices to create deterministic distributed simulation programs.
Lastly, we recognize that it is challenging and time-consuming to implement deterministic parallel software. Thus, we recommend that each software developer evaluate the costs and benefits of implementing deterministic programs. However, we hope that more developers will choose to implement deterministic software, particularly as complex hybrid models, which need deterministic simulations for debugging and verification, become more popular.
IV. Methods for Verifying Repeatability
After adopting the methods outlined above to repeatably simulate models, we recommend that systems biology modelers and software developers verify repeatability by testing the equivalence of simulation results generated by multiple simulation software programs for the same model, and then correcting any discrepancies identified by this analysis. Bergmann and Sauro [42] and Evans et al. [43] have used this approach to assess the repeatability of models across several simulators, identifying errors in several simulation programs.
Model checking and unit testing are two formalisms that can be used to verify repeatability. Simulation-based model checkers, such as SPIN [44], are extensively used in electrical engineering to verify the behavior of models. Simulation-based checkers execute multiple simulations and then verify that the results are consistent with the user-supplied specification of the model’s behavior [45]. Model verification systems should be adopted to verify the repeatability of systems biology models. Unit testing is a powerful strategy for verifying the behavior of software. Unit testing could also be used to verify the repeatability of systems biology models and simulation programs by (1) using multiple simulators to execute multiple simulations and (2) verifying that their predictions are statistically identical.
A. Special Considerations for Stochastic Models
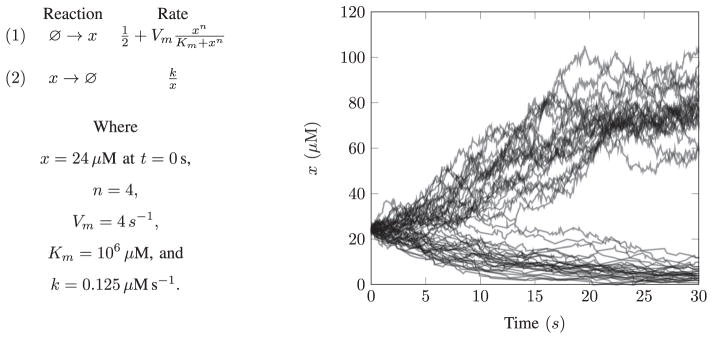
The repeatability of stochastic models can be assessed by verifying the statistical equivalence of simulation results across multiple simulators. Statistical equivalence should be tested using multivariate Kolmogorov-Smirnov tests [46]. Tests which only check the predicted mean and variance are insufficient because most model predictions cannot be fully specified by their mean and variance. For example, the bistable system illustrated in Fig. 2 cannot be specified by its mean and variance.
Fig. 2.
Mean and variance testing should not be used to verify the repeatability of stochastic models because most simulation results cannot be uniquely specified by these statistical metrics. For example, the bistable stochastic model described in (a) and illustrated in (b) cannot be specified by its mean and variance. Statistical equivalence should be tested using multivariate Kolmogorov-Smirnov tests. The bistable stochastic model described in (a) includes two reactions. Reaction (1) represents the production of species x and reaction (2) represents the degradation of x. Supplementary Material S1 contains Python and Antimony [34] code for this example.
However, it is often challenging to statistically verify the repeatability of complex algorithms and models across multiple simulators because large numbers of simulations are needed to statistically verify repeatability with high confidence, especially for models with rare events [47]. Instead, we recommend that researchers who are developing novel simulation algorithms and large scale models (1) utilize the methods described in the previous section to simulate models deterministically and (2) verify that each simulation software program can exactly regenerate its own numerical results. Although this approach does not ensure repeatability across simulators, it does facilitate the debugging of complex simulation algorithms.
B. Special Considerations for Hierarchical Models
The repeatability of hierarchical models, such as WC models, can be efficiently verified by taking advantage of their hierarchical structure. (1) Modelers should verify the repeatability of each submodel. This also facilitates debugging by beginning with small, easily testable units. (2) Modelers should verify the repeatability of the combined model.
C. Special Considerations for Multi-algorithm WC Models
As introduced above, multi-algorithm modeling is a new methodology which still requires significant fundamental research. Thus, multi-algorithm simulation algorithms will need to be extensively tested to verify their repeatability, as well as to understand their fundamental properties.
D. Special Considerations for Chaotic Models
Chaotic systems pose special challenges for repeatability and reproducibility. One criterion for judging whether a model is chaotic is the Lyapunov exponent, which measures the divergence between two trajectories starting at infinitesimally close initial conditions. A positive value indicates that the distance between the trajectories increases exponentially with time and implies that the model is chaotic. An extensive analysis of the reproducibility of chaotic systems is beyond the scope of this article. However, we suggest three guidelines for repeating simulations of chaotic systems. First, Lyapunov exponents should be used to gauge whether a model is chaotic. Second, statistical tests of the type discussed in Section IV should not be used to evaluate the reproducibility of chaotic models. Third, simulation software developers should use high precision to encode initial conditions and parameters so that chaotic model simulation results can be repeated.
V. Conclusion
Substantial work is needed to address the numerous challenges to enhancing the reproducibility of systems biology modeling, including developing new standards and simulation software tools. These standards and software tools should enable researchers to regenerate models from our scientific knowledge by recording the provenance of every model design decision, assumption, data source, parameter, and software option. These software tools should also enable researchers to regenerate simulation results by using pseudo-random number generators and deterministic multi-threading libraries. This requires expanded standards and software tools which support all systems biology models, including WC models. These simulation software tools should also be extensively tested to verify that they produce consistent simulation results. In turn, this requires a strong commitment among the scientific community to high-quality, open-source software development, including more emphasis on the publication of software programs in academic journals.
Supplementary Material
Acknowledgments
J. K. Medley was supported by National Institutes of Health grant R01 GM081070 and National Science Foundation grant 1355909. A. P. Goldberg was supported by the Icahn Institute for Genomics and Multiscale Biology. J. R. Karr was supported by National Science Foundation grant 1548123 and a James S. McDonnell Foundation Postdoctoral Fellowship Award in Studying Complex Systems. The content is solely the responsibility of the authors and does not necessarily represent the views of the National Institutes of Health, the National Science Foundation, or the James S. McDonnell Foundation.
We thank Graeme Gossel, Pedro Mendes, and Herbert Sauro for thoughtful discussions on reproducibility.
Biographies

J. Kyle Medley obtained his BSc in Mechanical Engineering from the University of Missouri-Kansas City and is currently pursuing a Ph.D. in Bioengineering from the University of Washington, USA. His research interests include modeling dynamical biophysical processes such as metabolism and gene expression regulation. He currently develops libRoadRunner, a software package for simulating dynamical models encoded in SBML.

Arthur P. Goldberg received his Ph.D. in Computer Science from the University of California, Los Angeles, USA in 1991 and his A.B. in Astronomy and Astrophysics from Harvard College, USA in 1977. Currently, Dr. Goldberg is an Associate Professor at the Icahn School of Medicine at Mount Sinai, USA. His research focuses on the development of high-performance simulation technologies for WC modeling.

Jonathan R. Karr received his Ph.D. in Biophysics and M.S. in Medicine from Stanford University, USA in 2014 and his B.S.s in Physics and Brain & Cognitive Sciences from the Massachusetts Institute of Technology, USA in 2006. Currently, Dr. Karr is a Fellow at the Icahn School of Medicine at Mount Sinai, USA. His research focuses on the development of comprehensive WC computational models and their application to bioengineering and medicine.
Contributor Information
J. Kyle Medley, Email: medleyj@uw.edu, Department of Bioengineering, University of Washington, Seattle, WA 98195, USA.
Arthur P. Goldberg, Email: arthur.goldberg@mssm.edu, Department of Genetics & Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA
Jonathan R. Karr, Email: karr@mssm.edu, Department of Genetics & Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA
References
- 1.Hucka M, Nickerson DP, Bader GD, Bergmann FT, Cooper J, Demir E, Garny A, Golebiewski M, Myers CJ, Schreiber F, et al. Promoting coordinated development of community-based information standards for modeling in biology: the COMBINE initiative. Front Bioeng Biotechnol. 2015;3 doi: 10.3389/fbioe.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dräger A, Palsson BØ. Improving collaboration by standardization efforts in systems biology. Front Bioeng Biotechnol. 2014;2 doi: 10.3389/fbioe.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cuellar AA, Lloyd CM, Nielsen PF, Bullivant DP, Nickerson DP, Hunter PJ. An overview of CellML 1.1, a biological model description language. Simulation. 2003;79(12):740–747. [Google Scholar]
- 4.COMBINE. COMBINE. 2012 [Online]. Available: http://co.mbine.org/events/COMBINE2012.
- 5.Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, Cuellar AA, Dronov S, Gilles E-D, Ginkel M, Gor V, Goryanin II, Hedley WJ, Hodgman TC, Hofmeyr J-H, Hunter PJ, Juty NS, Kasberger JL, Kremling A, Kummer U, Novère NL, Loew LM, Lucio D, Mendes P, Mjolsness ED, Nakayama Y, Nelson MR, Nielsen PF, Sakurada T, Schaff JC, Shapiro BE, Shimizu TS, Spence HD, Stelling J, Takahashi K, Tomita M, Wagner J, Wang J. The Systems Biology Markup Language (SBML): A medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19(4):524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 6.Waltemath D, Adams R, Bergmann F, Hucka M, Kolpakov F, Miller A, Moraru I, Nickerson D, Sahle S, Snoep J, Le Novère N. Reproducible computational biology experiments with SED-ML—the Simulation Experiment Description Markup Language. BMC Syst Biol. 2011;5(1):198. doi: 10.1186/1752-0509-5-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Novère N, Hucka M, Mi H, Moodie S, Schreiber F, Sorokin A, Demir E, Wegner K, Aladjem M, Wimalaratne SM, Bergman FT, Gauges R, Ghazal P, Kawaji H, Li L, Matsuoka Y, Villéger A, Boyd SE, Calzone L, Courtot M, Dogrusoz U, Freeman T, Funahashi A, Ghosh S, Jouraku A, Kim S, Kolpakov F, Luna A, Sahle S, Schmidt E, Watterson S, Wu G, Goryanin I, Kell DB, Sander C, Sauro H, Snoep JL, Kohn K, Kitano H. The Systems Biology Graphical Notation. Nat Biotechnol. 2009;27:735–741. doi: 10.1038/nbt.1558. [DOI] [PubMed] [Google Scholar]
- 8.Orth JD, Thiele I, Palsson BØ. What is flux balance analysis? Nat Biotechnol. 2010;28(3):245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hucka M, Bergmann FT, Keating SM, Smith LP. A profile of today’s SBML-compatible software. e-Science Workshops (eScienceW), 2011 IEEE Seventh International Conference on; IEEE; 2011. pp. 143–150. [Google Scholar]
- 10.Chelliah V, Juty N, Ajmera I, Ali R, Dumousseau M, Glont M, Hucka M, Jalowicki G, Keating S, Knight-Schrijver V, et al. BioModels: ten-year anniversary. Nucleic Acids Res. 2015;43(D1):D542–D548. doi: 10.1093/nar/gku1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lloyd CM, Lawson JR, Hunter PJ, Nielsen PF. The CellML model repository. Bioinformatics. 2008;24(18):2122–2123. doi: 10.1093/bioinformatics/btn390. [DOI] [PubMed] [Google Scholar]
- 12.Hillman-Jackson J, Clements D, Blankenberg D, Taylor J, Nekrutenko A, Team G. Using galaxy to perform large-scale interactive data analyses. Current protocols in bioinformatics. 2012:10–5. doi: 10.1002/0471250953.bi1005s38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oinn T, Addis M, Ferris J, Marvin D, Senger M, Greenwood M, Carver T, Glover K, Pocock MR, Wipat A, et al. Taverna: a tool for the composition and enactment of bioinformatics workflows. Bioinformatics. 2004;20(17):3045–3054. doi: 10.1093/bioinformatics/bth361. [DOI] [PubMed] [Google Scholar]
- 14.Callahan SP, Freire J, Santos E, Scheidegger CE, Silva CT, Vo HT. Vistrails: visualization meets data management. Proceedings of the 2006 ACM SIGMOD international conference on Management of data; ACM; 2006. pp. 745–747. [Google Scholar]
- 15.Karr JR, Sanghvi JC, Macklin DN, Gutschow MV, Jacobs JM, Bolival B, Jr, Assad-Garcia N, Glass JI, Covert MW. A whole-cell computational model predicts phenotype from genotype. Cell. 2012;150(2):389–401. doi: 10.1016/j.cell.2012.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waltemath D, Karr JR, Bergmann FT, Chelliah V, Hucka M, Krantz M, Liebermeister W, et al. Toward community standards and software for whole-cell modeling. doi: 10.1109/TBME.2016.2560762. In submission. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe L, Myers CJ. Efficient analysis of SBML models of cellular populations using arrays. ACS Synth Biol. 2016 doi: 10.1021/acssynbio.5b00242. [DOI] [PubMed] [Google Scholar]
- 18.Moodie SL, Smith LP, Le Novère N, Wilkinson D, Swat M, Keating S, Gillespie C. [accessed: 2016-02-26];The distributions package for SBML level 3. 2015 [Online]. Available: http://sourceforge.net/p/sbml/code/HEAD/tree/trunk/specifications/sbml-level-3/version-1/distrib/sbml-level-3-distrib-package-proposal.pdf?format=raw.
- 19.Olivier BG, Bergmann FT. The Systems Biology Markup Language (SBML) level 3 package: Flux balance constraints. J Integr Bioinform. 2015;12(2):269. doi: 10.2390/biecoll-jib-2015-269. [DOI] [PubMed] [Google Scholar]
- 20.Smith LP, Hucka M, Hoops S, Finney A, Ginkel M, Myers CJ, Moraru I, Liebermeister W. SBML Level 3 package: hierarchical model composition, version 1 release 3. J Integr Bioinform. 2015;12(2):268. doi: 10.2390/biecoll-jib-2015-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang F, Meier-Schellersheim M. SBML Level 3 Package Specification: Multistate/Multicomponent Species (Version 1, Release 0.1 Draft 369) [accessed: 2015-05-25];2015 [Online]. Available: http://sbml.org/Documents/Specifications/SBMLLevel3/Packages/multi.
- 22.Karr JR, Sanghvi JC, Macklin DN, Arora A, Covert MW. WholeCellKB: Model organism databases for comprehensive whole-cell models. Nucleic Acids Res. 2013;41(D1):D787–D792. doi: 10.1093/nar/gks1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juty N, le Novère N. Systems Biology Ontology. Enc Syst Biol. 2013:2063–2063. [Google Scholar]
- 24.Galdzicki M, Clancy KP, Oberortner E, Pocock M, Quinn JY, Rodriguez CA, Roehner N, Wilson ML, Adam L, Anderson JC, et al. The Synthetic Biology Open Language (SBOL) provides a community standard for communicating designs in synthetic biology. Nat Biotechnol. 2014;32(6):545–550. doi: 10.1038/nbt.2891. [DOI] [PubMed] [Google Scholar]
- 25.Kolpakov F. BioUML: visual modeling, automated code generation and simulation of biological systems. Proc BGRS. 2006;3:281–285. [Google Scholar]
- 26.Stevens JT, Myers CJ. Dynamic modeling of cellular populations within iBioSim. ACS Synth Biol. 2013;2(5):223–229. doi: 10.1021/sb300082b. [DOI] [PubMed] [Google Scholar]
- 27.Easterbrook SM. Open code for open science? Nature Geosci. 2014;7(11):779–781. [Google Scholar]
- 28.Schäling B. The boost C++ libraries. Boris Schäling; 2011. [Google Scholar]
- 29.Karr JR, Takahashi K, Funahashi A. The principles of whole-cell modeling. Curr Opin Microbiol. 2015;27:18–24. doi: 10.1016/j.mib.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Macklin DN, Ruggero NA, Covert MW. The future of whole-cell modeling. Curr Opin Biotechnol. 2014;28:111–115. doi: 10.1016/j.copbio.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carrera J, Covert MW. Why build whole-cell models? Trends Cell Biol. 2015;25(12):719–722. doi: 10.1016/j.tcb.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Courtot M, Juty N, Knüpfer C, Waltemath D, Zhukova A, Dräger A, Dumontier M, Finney A, Golebiewski M, Hastings J, et al. Controlled vocabularies and semantics in systems biology. Mol Syst Biol. 2011;7(1):543. doi: 10.1038/msb.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldberg AP, Chew YH, Karr JR. Prin Adv Discret Simul. ACM; 2016. Toward scalable whole-cell modeling of human cells. [Google Scholar]
- 34.Smith LP, Bergmann FT, Chandran D, Sauro HM. Antimony: a modular model definition language. Bioinformatics. 2009;25(18):2452–2454. doi: 10.1093/bioinformatics/btp401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bergan T, Anderson O, Devietti J, Ceze L, Grossman D. CoreDet: A compiler and runtime system for deterministic multithreaded execution. ACM SIGARCH Comput Archit News. 2010;38(1):53–64. [Google Scholar]
- 36.Liu T, Curtsinger C, Berger ED. Proc 23rd ACM Symposium Oper Syst Prin. ACM; 2011. Dthreads: Efficient deterministic multithreading; pp. 327–336. [Google Scholar]
- 37.Netzer RH, Miller BP. What are race conditions?: Some issues and formalizations. ACM Letters on Programming Languages and Systems (LOPLAS) 1992;1(1):74–88. [Google Scholar]
- 38.Chandy KM, Misra J. Asynchronous distributed simulation via a sequence of parallel computations. Communications of the ACM. 1981;24(4):198–206. [Google Scholar]
- 39.Jefferson DR. Virtual time. ACM Transactions on Programming Languages and Systems. 1985;7(3):404–425. [Google Scholar]
- 40.Brooks ED., III The butterfly barrier. International Journal of Parallel Programming. 1986;15(4):295–307. [Google Scholar]
- 41.Dagum L, Enon R. Openmp: an industry standard api for shared-memory programming. Computational Science & Engineering, IEEE. 1998;5(1):46–55. [Google Scholar]
- 42.Bergmann FT, Sauro HM. Comparing simulation results of SBML capable simulators. Bioinformatics. 2008;24(17):1963–1965. doi: 10.1093/bioinformatics/btn319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evans TW, Gillespie CS, Wilkinson DJ. The SBML discrete stochastic models test suite. Bioinformatics. 2008;24(2):285–286. doi: 10.1093/bioinformatics/btm566. [DOI] [PubMed] [Google Scholar]
- 44.Holzmann GJ. The model checker SPIN. IEEE Trans Softw Eng. 1997;23(5):279. [Google Scholar]
- 45.Kwiatkowska M, Norman G, Parker D. Prism 4.0: Verification of probabilistic real-time systems. Computer Aided Verification. 2011:585–591. [Google Scholar]
- 46.Justel A, Peña D, Zamar R. A multivariate kolmogorovsmirnov test of goodness of fit. Statit Probab Lett. 1997;35(3):251–259. [Google Scholar]
- 47.Kim KH, Qian H, Sauro HM. Nonlinear biochemical signal processing via noise propagation. J Chem Phys. 2013;139(14):144108. doi: 10.1063/1.4822103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.