Abstract
NEDD8 is a ubiquitin-like protein that activates the largest ubiquitin E3 ligase family, the cullin RING ligases. Many non-cullin neddylation targets have been proposed in recent years. However, overexpression of exogenous NEDD8 can trigger NEDD8 conjugation through the ubiquitylation machinery, which makes validating potential NEDD8 targets challenging. Here we re-evaluate these studies in light of the current understanding of the neddylation pathway, and suggest criteria for the identification of genuine neddylation substrates under homeostatic conditions. We describe the biological processes that might be regulated by non-cullin neddylation, and the utility of neddylation inhibitors for research and as potential therapies. Understanding the biological significance of non-cullin neddylation is an exciting research prospect primed to reveal fundamental insights.
Introduction
Reversible post-translational protein modifications are major regulators of eukaryotic cell functions. Examples include modifications by small chemical moieties such as phosphates, acetyl and methyl groups, conjugation of lipids and sugars, and the attachment of poly-peptides like the small protein ubiquitin and the 16 human ubiquitin like proteins (UBLs)1. Ubiquitin is the best characterized of these modifiers and is thought to be involved in the regulation of nearly every aspect of cellular homeostasis mainly through its functions in the ubiquitin-proteasome system2. Of the other UBLs, SUMO family members regulate the subcellular localization of proteins, transcription and DNA repair3; the Atg8 family members and ATG12 control autophagy4; and FAT10, ISG15 and FUB1 are regulators of immune and inflammatory responses5. URM1 can function as a regulator of oxidative stress and promotes protein translation through tRNA thiolation1.
The neural precursor cell expressed developmentally down-regulated protein 8 (NEDD8) is another UBL that has been principally characterized in the context of its major target, the ubiquitin E3 ligase family of the cullin RING ligases [G] (CRLs). Reports of non-cullin neddylation targets in recent years indicate that NEDD8 might have additional biological functions. However, as NEDD8 overexpression, which is routinely used experimentally, causes aberrant effects in cells, such targets are controversial. Thus, determining experimental criteria for assigning bona fide non-cullin NEDD8 targets is an important research challenge. To establish such criteria, we review the current mechanistic understanding of neddylation and deneddylation, as well as the knowledge of NEDD8-recognition domains. We then discuss in detail the emerging picture of biological processes that might be regulated by non-CRL neddylation.
Structure of the ubiquitin-like protein NEDD8
NEDD8 is a conserved, predominantly nuclear protein with a broad expression pattern in adult tissues6,7,8. Its expression is increased during mouse embryogenesis9,10 and its levels in the developing brain are higher than in adults6,11–14. Moreover, NEDD8 and the neddylation enzymes are over-expressed in human cancers8,15,16. Neddylation is essential in mammals17, plants18, fruit flies19, nematodes20, and fission yeast21. Budding yeast is the notable exception with a conserved but non-essential neddylation pathway22,23.
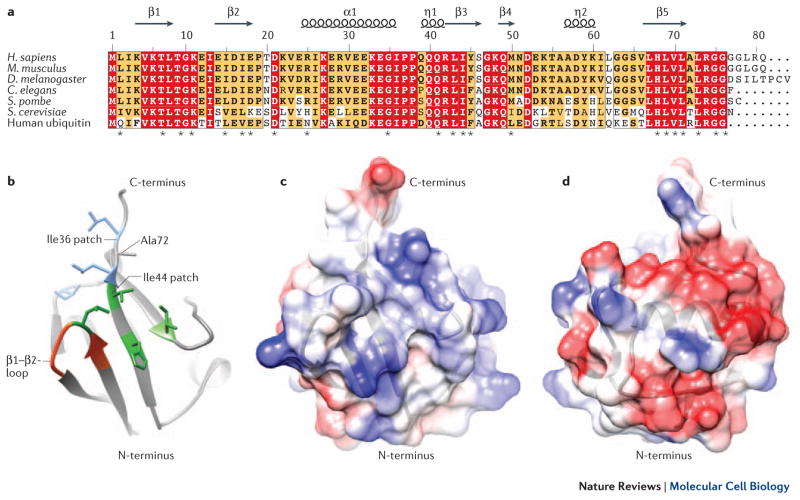
Ubiquitin and NEDD8 have the greatest similarity among all of the UBLs (the human orthologues are 59% identical)24,25 (Figure 1A). Despite this, ubiquitin and NEDD8 have non-interchangeable functions in cells as a result of small differences in their structures that mediate distinct interactions (discussed below). The overall structure of NEDD8 can be subdivided into a C-terminal tail and a globular ubiquitin-fold domain (Figure 1B). The tail is flexible in solution26, adopts different extended structures upon interaction with neddylation and deneddylation enzymes, and, similarly to ubiquitin, ends with a Gly-Gly sequence that becomes covalently bonded to targets27–29. The globular domain is characterized by four β-sheets interspersed by one α- and two 310-helices (helix 1, 2, and 3 respectively).
Figure 1. NEDD8 Structure.
A Structure-based multiple sequence alignment of NEDD8 from indicated species and human ubiquitin, highlighting identical residues (red) and similar residues (yellow). Asterisks signify residues found to be essential for NEDD8 function in S. pombe63. B Structural representation of the NEDD8 hydrophobic patches, contributing most of the known interfaces for interaction with binding partners (Table 1). The side chains contributing to the Ile36 patch are shown in blue, the residues of the Ile44 patch in green and the β1/β2-loop in red. Those interfaces are conserved in ubiquitin. Ala72, which is responsible for discrimination between NEDD8 and ubiquitin by the respective E1 enzymes, is indicated. C, D Structural representations of the NEDD8-specific charged surface patches. Acidic patches are depicted in red and basic surfaces in blue. These surfaces might be responsible for interactions that discriminate between NEDD8 and ubiquitin.
Two surface-exposed hydrophobic patches on ubiquitin, the Ile44 and Ile36 patches, have been shown to be essential for mediating protein–protein interactions30. Both of them are conserved in the NEDD8 structure (Figure 1B). The NEDD8 Ile44 patch comprises Leu8-Ile44-His68-Val70 and the Ile36 patch contains Leu8-Ile36-Leu71-Leu76. Another functionally important region in mammalian ubiquitin includes residues of and around the flexible loop between β1 and β2 and Lys6 and Lys1131. Here, we refer to this largely conserved region in NEDD8 as the β1/β2-loop (Figure 1B, Table 1). The intervening polar and charged surfaces are less conserved between ubiquitin and NEDD8, and probably influence their specific functions (Figure 1C, D).
Table 1.
NEDD8-interacting proteins*
| NEDD8-pathway enzymes | ||||
|---|---|---|---|---|
| NEDD8 binding partner | NEDD8 interaction interface | Binding partner interaction interface | Function | Reference(s) |
| NAE | Acidic face Ile44 patch C-terminal tail |
APP BP1 catalytic cysteine domain hydrophobic portion of UBA3 adenylation domain UBA3 adenylation domain and cross-over loop |
Chemical activation of the C-terminus of NEDD8 | 27 |
| DCNLs | Unknown | Unknown (UBA domain is not required in S. cerevisiae) | Unknown | 66 |
| DEN1 | Ile44 patch C-terminal tail Residues 45–56 |
channel formed by the N-termini of helices 2 and 7 N-terminus |
Deneddylation Processing of the C-terminus of NEDD8 | 28,29 |
| UCH-L3 | Unknown | Unknown | Processing of the C-terminus of NEDD8 | 173 |
| Downstream effectors | ||||
| Neddylated cullins | β1/β2-loop, Ile44 patch and C-terminal tail | winged-helix B domain | Stabilization of a freed RBX1 conformation and activation of CRL ubiquitin transfer activity | 85 |
| NUB/NUBL1 | Unknown | Uba domain; sequence A(X4)L(X10)L(X3)L in both NUB1 and NUBL1 | Targeting NEDD8 and neddylated proteins to the 26S proteasome for degradation |
89 96 127 |
| UBXD7 (Ubx5 in S. cerevisiae) | Neddylated cullins, through the Ile44 patch | UIM domain | Interaction with P97 |
91 92 |
| RNF168 | NEDD8 chains | MIU domain | DNA damage response | 64 |
| De Novo DNA methyltransferase (DNMT3B) | Stronger binding to neddylated proteins, including CUL4A | residues 532–583 of mouse DNMT3B responsible for NEDD8 binding | Increased DNMT3b-dependent DNA methylation | 93 |
| HGS | Unknown | UIM domain | Probable function in targeting neddylated proteins to clathrin-mediated endocytosis146 | 90 |
| Ariadne domain Ring-in-between-Ring E3s | Neddylated cullins | potentially the UBA domain | Activation of Ariadne RBRs | 95 |
| Non-proteasomal ubiquitin receptors and shuttles (such as RAD23, UBQLN1 and Ddi1) | Ile44 patch | UBA2 domain of human RAD23; UBA domain of human UBQLN1; UBA domain of S. cerevisiae Ddi1 | Unknown | 65 |
| BRAP2 | Stronger binding to a linear di-NEDD8 | RING domain might be involved | Unknown | 141 |
| Ubiquitin E2 enzymes (such as UBCH5, UBC7 and RAD6) | β1/β2-loop, Ile44 patch and C-terminal tail | portions of α1, α2 and backside (β1,2,3174) | Unknown | 26,175 |
| aryl hydrocarbon receptor (AhR) | Unknown | Unknown | Enhanced transcriptional activity | 94 |
Abbreviations: UBA - ubiquitin-associated domain; UIM - ubiquitin-interacting motif; MIU - motif interacting with ubiquitin
Mechanism of neddylation and deneddylation
The translation product of NEDD8, similar to other UBLs, is a precursor, which requires proteolytic processing to expose a C-terminal glycine residue. It is subsequently chemically activated by specific E1 and E2 enzymes, and eventually conjugated by an E3 enzyme to a lysine residue in the target protein through an isopeptide bond.
Proteolytic Processing
The budding yeast cysteine protease Yuh1 removes the C-terminal extensions of both ubiquitin and NEDD8 precursors, and is essential for cullin neddylation32. Its orthologue in mammals, ubiquitin C-terminal hydrolase 3 (UCH-L3) can also process both ubiquitin and NEDD833. A second cysteine protease, known as deneddylase 1 (DEN1), has high specificity for NEDD8 but not ubiquitin, and can process the NEDD8 precursor form, as well as deconjugate NEDD8 from some protein substrates, but not from the best-characterized NEDD8 targets, cullins32–37 (see below). The NEDD8 C-terminal maturation functions of DEN1 and UCH-L3 are probably redundant in vivo, as knockout of either enzyme does not lead to neddylation defects38,39.
NEDD8 E1 enzyme
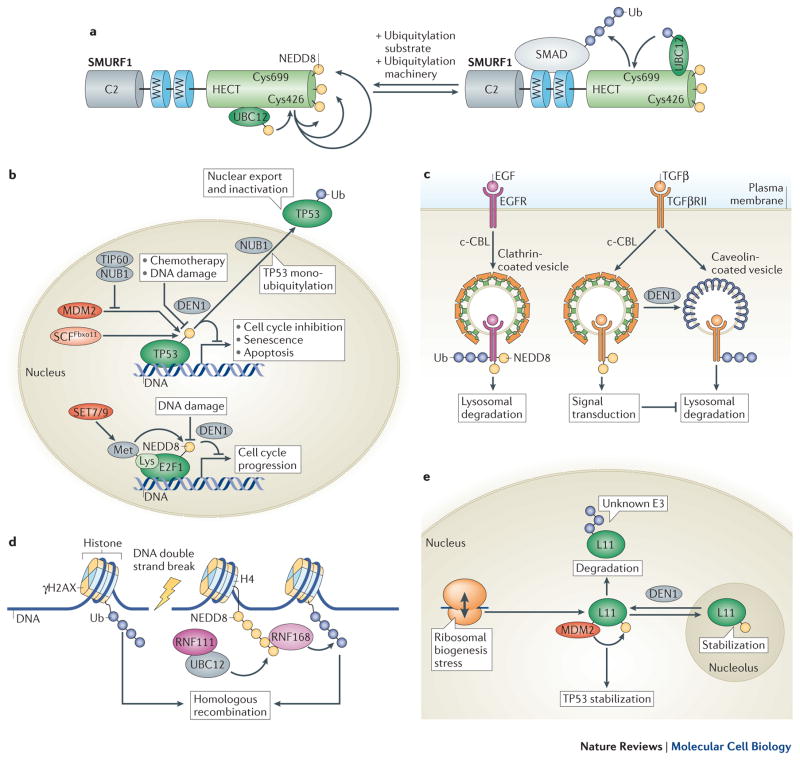
The NEDD8 E1 activating enzyme (NAE) initiates the NEDD8 transfer cascade (Figure 2A) by chemically activating the di-Gly-bearing C-terminus of NEDD8. NAE is a heterodimer comprising the amyloid-beta precursor protein binding protein 1 (APPBP1) and UBA3. NAE contains two distinct active sites: an adenylation site located in UBA3 and a cysteine transthiolation domain, composed of residues from both subunits. The two active site domains are separated by a large groove in the holo-enzyme27, referred to as an ‘open’ conformation. A third functionally important domain is a C-terminal ubiquitin fold domain (UFD) in UBA3, which binds the NEDD8 E2 enzymes (see below). The reactions catalyzed by NAE are ordered. Binding of ATP, Mg2+ and NEDD8 at the adenylation site of UBA3 is followed by AMP attachment to the C-terminus of NEDD8 (formation of NEDD8-adenylate), and the release of inorganic pyrophosphate. The C-terminus of NEDD8 is thus chemically activated and can be attacked by the catalytic cysteine of NAE. However, there is a ~ 35 Å gap between the active site cysteine of NAE and the activated C-terminus of NEDD8 in the open conformation27. Extrapolating from structural data of the SUMO E1 activating enzyme40, structural remodeling of NAE is probably required to result in a ‘closed’ state, characterized by a 130-degree rotation of the remodeled cysteine domain, thus positioning the catalytic cysteine of NAE sufficiently close to the NEDD8-adenylate.
Figure 2. The Neddylation and Deneddylation Pathways.
A NEDD8 activation by the heterodimeric NEDD8 E1 activating enzyme (NAE; a heterodimer of APPBP1 and UBA3) requires sequential binding of ATP and NEDD8 to the adenylation site of UBA3, where conjugation of AMP activates the C-terminus of NEDD8. NAE rearrangement into a closed conformation allows thiolation of the NEDD8 C-terminus to the NAE active site cysteine with AMP release. NAE then returns to its open conformation and adenylates a second NEDD8, resulting in conformational rearrangement of its ubiquitin-fold domain (UFD), binding of a NEDD8 E2 enzyme such as UBC12 and transfer of NEDD8 onto the E2 active site cysteine. B Neddylation of cullin (left) or other substrates (right). Cullin neddylation by the E3 ligase RBX1 is aided by DCNLs such as DCNL1 and occurs on a single conserved lysine residue. Neddylation of other substrates occurs on multiple lysines and is mediated mostly by RING E3 ligases. The details of the interaction between the NEDD8 E2 enzyme and a non-RBX RING E3 ligase are unclear. C Molecular models of UBCH5~Ub activated by CBL-B50 (left) and UBC12~NEDD8 activated by RBX154 (right). A linchpin arginine residue in CBL-B, which is conserved in many RING ubiquitin E3 ligases, would clash with a lysine in UBC12 such that CBL-B does not interact with and ubiquitylate UBC12. D Sequence alignment of RING domain E3 ligases that are reported to function as NEDD8 ligases (left) and of NEDD8 E2 enzymes and the ubiquitin E2 UBCH5 (right). The arrow indicates the E2 linchpin residue position and corresponding neighboring residue (red and blue denote side chain electrostatic potential). E Deneddylation of cullin by the eight-subunit COP9 signalosome (CSN) complex (left) and of non-cullin substrates by DEN1 (right). Besides cullin deneddylation, CSN binding to the C-terminal domain of cullins, mainly mediated through CSN2, keeps CRLs such as RBX1 inactive82.
The C-terminus of NEDD8 is consequently linked to the active site cysteine of NAE, while a second NEDD8 binds at the adenylation domain, and NAE re-adopts the open conformation41. Although it is not fully understood, the switch between the closed and open conformations of NAE might be triggered by remodeling and consequent loss of the ATP biding pocket of UBA3 in the closed state, which probably triggers release of AMP after NEDD8 attachment to the catalytic cysteine.
NEDD8 E2 enzymes
In metazoans, two NEDD8-specific E2 conjugating enzymes have been described – UBC12 (also known as UBE2M) and UBE2F. They both bind to the doubly NEDD8-loaded NAE, catalyzing a transthiolation reaction involving the transfer of NEDD8 from the active site cysteine of NAE onto the active site cysteine of the E2 enzyme (Figure 2A). The major interaction interface with the E2 enzyme is through the UFD of NAE42. Importantly, when NAE is doubly NEDD8-loaded, its UFD reorients and together with a portion of the adenylation domain forms a cryptic UBC12-binding groove41. In addition, UBC12 directly binds the NEDD8 moiety attached to the catalytic cysteine of NAE41. Unique to the NEDD8 cascade is a third E1–E2 interface, formed by the N-terminus of either E2 enzyme (UBC12 or UBE2F) binding in a groove of the adenylation domain of UBA343. Together, these binding interfaces position the catalytic cysteines of the NEDD8 E1 and E2 enzymes in close proximity for catalysis. As two of the binding sites for the E2 enzyme are only present in the doubly NEDD8-loaded NAE, they are lost after transthiolation and thus catalysis contributes to weakening E1–E2 binding and the release of the E2 enzyme from the E1.
NEDD8 E3 enzymes
NEDD8 E3 ligases catalyze the transfer of NEDD8 from the E2 enzyme onto the target protein. All currently reported NEDD8 E3 enzymes can also function as ubiquitin E3 enzymes (Supp. Table 1). All but one of the reported NEDD8 E3 enzymes are of the RING subclass44. The RING domain is found in more than 600 human E3 ligases and is structurally characterized by two or three interleaved zinc-coordination sites, which form a protein–protein interaction interface for various E2 enzymes. The best studied NEDD8 E3 ligases are the RING domain subunits RBX1, a component of CRL1, CRL2, CRL3 and CRL4 complexes, and its close homologue RBX2, a CRL5 component27,40,41,45,46. RBX1 can, in principle, work with both UBC12 and UBE2F, whereas RBX2 has specificity for UBE2F45,47. The same surface on the NEDD8 E2 enzymes is recognized by both the E3 RING domains and the UFD of NAE; the toggling of relative affinities ensures the uni-directionality of the process42,48.
The interaction between the E3 RING domain and E2-bound UBL catalyzes the transfer of the UBL from the E2 active site cysteine onto a lysine residue or the N-terminus of the target protein (Figure 2B). In the particular case of CRL-mediated neddylation, this step is further aided by DCN1-like proteins (see below). Recent structural studies have uncovered the catalytic mechanism for ubiquitin transfer, and its general conservation in the NEDD8 system. In the absence of an E3 ligase, ubiquitin-charged E2 enzymes adopt multiple conformations, sampling various configurations of ubiquitin relative to the E249. Conformational “wobbling” of ubiquitin relative to the E2 reduces reactivity of the thioester bond, and may also reduce accessibility to the active site of the E2 enzyme. Upon binding to an E3 RING domain, the ubiquitin-loaded E2 adopts an activated conformation in which the RING domain binds both the E2 and ubiquitin and the thiolated C-terminus of ubiquitin is immobilized to increase reactivity through an attack by the substrate’s lysine residue41,50–53. A key structural feature of ubiquitylation intermediates is a RING “linchpin arginine” that stabilizes the activated E2~ubiquitin intermediate (Figure 2C, left). The RBX1 RING domain binds UBC12~NEDD8 with a similar overall architecture to the ubiquitylation reaction, but has several unique features that establish exquisite specificity for both neddylation and the cullin acceptor lysine (Figure 2C, right)54. Among the differences from structurally characterized ubiquitylation intermediates is that a canonical RING linchpin arginine would electrostatically repel human UBC12 (Figure 2C). Notably, the majority of reported non-RBX-family NEDD8 E3 ligases, including c-CBL, RNF111, MDM2 and DmIAP1, do contain a linchpin arginine (Figure 2D, Supp Table 1), and this raises the question of whether they bind and activate UBC12~NEDD8 through an unknown mechanism, or whether they promote NEDD8 transfer from alternative E2s, potentially UBE2F.
NEDD8 chains
Ubiquitin, SUMO and other UBLs can in principle form chains on their substrates by first being attached to a target protein and then themselves functioning as an acceptor for a further UBL. Many proposed NEDD8 substrates, including cullins, are thought to be mainly mono-neddylated on a single or several conserved lysines. Cullins can, however, be hyper-neddylated in vitro55–58 and a proteomic study revealed that this is a mixture of neddylation on multiple C-terminal lysines and NEDD8 chain formation59. Moreover, it was found that NEDD8 can form chains through Lys11, Lys22, Lys27, Lys48, Lys54 and Lys6059–62. NEDD8 chains could pre-form on the active site of UBC1257.
The physiological significance of NEDD8 chains is unclear. On the one hand, a mutagenesis study in fission yeast found that mutating all lysines in NEDD8 to arginines (to prevent chain formation) did not compromise cell viability under normal and stress conditions, including DNA damage, and microtubule and proteasome stress63. On the other hand, a recent study in cultured human cells reported that Lys22- and Lys48-linked NEDD8 chains form on histone H4 following DNA damage and regulate downstream events64. Intriguingly, NEDD8 can form mixed chains with ubiquitin in budding yeast and human cells and can function as a chain-terminator65. The probable explanation for this is that Lys60 is conserved in NEDD8 but not ubiquitin and prevents NEDD8 from being a good ubiquitin acceptor26. Thus, the exact mechanism, potential substrates and physiological significance of NEDD8 chain formation remain important future research directions.
Neddylation regulators
Neddylation is regulated in vivo by several mechanisms. The five defective in cullin neddylation 1 (DCN1)-like proteins (DCNLs) are probably the main regulators of neddylation in humans66. They bind both cullins and NEDD8 E2 enzymes, thereby increasing the kinetic efficiency of the neddylation reaction47,67,68 (Figure 2B). They do so by specifically recognizing a site in the cullin substrate close to its neddylation site as well as the acetylated N-terminus of the E2 enzyme UBC12, and thus DCNLs probably structurally constrain the RBX1–UBC12~NEDD8 complex to orientations that are optimally suited for NEDD8 transfer68,69. However, we know little about the roles of individual DCNLs. It is conceivable that different DCNLs regulate different cullins in vivo. For example the only known DCNL orthologue in budding yeast, Dcn1, is important for the neddylation of Cul1 and Cul3, but not the CUL4 orthologue Rtt10170. The human DCNL paralogues differ in subcellular localization. In order to function as a co-E3 DCNL1 has to co-localize with UBC12, which is mainly a nuclear protein71,72. DCNL1 and DCNL2 are themselves mono-ubiquitylated in a reaction requiring their UBA domain, which directs their nuclear export72. DCNL3, by contrast, has a lipid-modified domain and contributes to the neddylation of CRL3 complexes at membranes73.
At present it remains unknown whether N-terminal acetylation of E2 enzymes and/or DCNLs are involved in the neddylation of non-cullin proteins. This is, however, a possibility, as the overexpression of DCNL1 in human cells results in a marked increase in levels of neddylated proteins72.
Budding yeast Tfb3 (MAT1 in mammals) is a component of the transcription initiation factor TFIIH, which functions both in transcription and nucleotide excision repair74. Tfb3 comprises a RING domain, which regulates the neddylation of CUL3 and CUL4 but not CUL1 orthologues70. Similarly to RBX1, Tfb3 interacts with Ubc12 and lacks the lynchpin arginine (Figure 2D), which indicates that it may directly activate Ubc12. The mechanism of action of Tfb3, and whether it also has a role in neddylation in higher eukaryotes, remain to be established.
Neddylation can be inhibited during bacterial infection. Cif from enteropathogenic Escherichia coli and its Burkholderia pseudomallei homolog CHBP are deamidases, which convert the conserved Gln40 residue of NEDD8 to a Glu75. Deamidated NEDD8 is incapable of activating CRL activity and can induce macrophage-specific apoptosis76.
Deneddylation
CSN5 is the major cullin deneddylase (Figure 2E)77. It comprises a metalloprotease active site within a JAB1/MPN/MOV34 metalloenzyme (JAMM) motif [G]78. However, its active site is auto-inhibited when CSN5 is in isolation and its assembly into the eight-subunit COP9 signalosome (CSN) complex is required for catalytic activity79,80. Curiously, CSN5 is also autoinhibited in the fully assembled CSN complex81. What releases this auto-inhibition is not fully understood, but since the major interaction interface for deneddylation occurs between the CSN subunits CSN2 and CSN4 and the C-terminus of the cullin, rather than between NEDD8 and CSN5, active site remodeling of CSN5 most probably occurs allosterically upon binding of the neddylated cullin substrate to the CSN complex81,82. Neither CSN5 in isolation nor the CSN complex has appreciable affinity for free NEDD8 and they are very inefficient in processing its precursor form or chemically synthesized C-terminal derivatives37,78,80,82. Furthermore, the CSN complex stably binds deneddylated CRLs and sterically inhibits the ability of RBX1 to activate E2 enzymes. CRL substrates can compete away CSN and thus trigger their own degradation82–84.
DEN1 is a cysteine protease that selectively binds NEDD834,37 and has complementary deneddylation activities to the CSN complex in vivo38 (Figure 2E). As discussed above, DEN1 can process the NEDD8 precursor form, but it has a negligible activity to deconjugate a single NEDD8 from cullin substrates. However, DEN1 is more active in deneddylating hyper-neddylated cullins to leave mono-neddylation behind37, which indicates that it might prevent aberrant polyneddylation. The most probable explanation for the inability of DEN1 to remove a single NEDD8 from cullins is that, among other interfaces, it recognizes NEDD8 through its Ile44 patch28, which is probably inaccessible in a mono-neddylated substrate as this patch also interacts with the cullin85. Furthermore, DEN1 can remove NEDD8 from many postulated non-cullin substrates both in vitro and in vivo (Supp Table 1).
Finally, several putative deneddylases have been reported but further characterization is lacking at present86,87. USP21 was initially reported as capable of deconjugating both ubiquitin and NEDD8, but a structural and biochemical study suggested that steric clashes and repulsive forces would prevent USP21 from binding NEDD888. It is unknown whether UCH-L3 can deneddylate proteins also or only functions to process NEDD8 precursors.
NEDD8-interacting proteins
It is reasonable to assume that as for ubiquitin, specific NEDD8-binding proteins recognize neddylated substrates and signal downstream effects. Indeed, several NEDD8-interacting proteins have been reported (Table 1). However, in most cases we are still lacking knowledge of the molecular determinants of the interface, its specificity over other UBLs and how NEDD8 binding translates into a biological function. Nevertheless, we can begin to address two key questions. Which surfaces/residues in NEDD8 and NEDD8 chains function as interaction interfaces, and what domains in NEDD8-binding proteins recognize them? As summarized in Table 1, two classes of NEDD8-interacting proteins can be distinguished. The first comprises NEDD8-pathway enzymes (discussed in the previous section) and the second is composed of downstream effectors (discussed below).
This second class of NEDD8-interacting proteins comprises scaffold proteins as well as enzymes that exert downstream signalling effects by binding free NEDD8 and/or neddylated proteins. The exact functions are not well studied but there are examples of substrate recruitment via NEDD8 to the 26S proteasome65,89, endocytic vesicles90 and p97 [G]91,92. Moreover, NEDD8 binds to effectors of DNA methylation93, transcription94 and DNA damage repair64 as well as activators of Ariadne domain RING-between-RING E3 ligases95.
In most studied cases, such interactions occur through discrete small domains of downstream effectors, which bind to the hydrophobic patch around Ile44 of NEDD8. Examples of such domains include ubiquitin-associated (UBA) domains [G] of yeast and human proteasomal ubiquitin-binding proteins65, NUB1/NUBL196, ubiquitin interaction motif (UIM) domains [G] of UBXD791,92 and HGS90, as well the motif interacting with ubiquitin (MIU) [G], which recognizes NEDD8 chains64. The reported affinities of these domains for NEDD8 are in the μM range, similar to the interactions of such domains with ubiquitin. Thus it remains to be investigated whether and how such interfaces can discriminate between NEDD8 and ubiquitin. Detailed future studies of NEDD8-specific binding domains are warranted not only to learn more about their cellular functions and biochemical properties, but also with the hope that they may function as affinity tags for enriching endogenously neddylated proteins for proteomic or biochemical studies. A further important question is whether free NEDD8 or neddylated proteins are primarily recognized. As there are several reports of the recognition of neddylated cullins91,92,95, structural studies are required to resolve the issue of whether the Ile44 patch of NEDD8 is released from the WHB domain in cullins to be recognized by other proteins.
Neddylation through the ubiquitin pathway
Both the ubiquitylation and neddylation pathways exhibit great specificity in cells under homeostatic conditions. Such specificity is largely conferred by a single amino acid difference in the C-termini of the two UBLs — Ala72 in NEDD8, and Arg72 in ubiquitin (Figure 1A, B) — which is recognized by their respective E1 enzymes, thus ensuring that the correct UBLs are passed on to the respective E2 and E3 enzymes and the correct substrates24,97–99. However, the ubiquitin E1 enzyme UBA1 can activate NEDD8, although NEDD8 is a significantly worse substrate for UBA1 than ubiquitin24,100. Thus, detectable activation of NEDD8 by UBA1 occurs if NEDD8 is in excess. Importantly, once activated by UBA1, NEDD8 can be trans-thiolated to most ubiquitin-specific E2 enzymes24,100 and is thus conjugated to multiple ubiquitylation substrates either by itself or as a part of mixed ubiquitin–NEDD8 chains62,101 (Box 1).
Box 1. Identification of genuine in vivo neddylation substrates: experimental approaches and challenges.
An increase of free NEDD8 over ubiquitin can occur in an overexpression experiment or as a consequence of cellular stress or pathology. Such ratio perturbation results in the activation of NEDD8 through the ubiquitin E1 enzyme UBA1, and to conjugation of NEDD8 through the ubiquitylation machinery62,100,101 (see figure). This can lead to erroneous assignments of neddylation substrates.
A genuine neddylation target should comply with the following criteria (see also158):
Covalent attachment of NEDD8 through its C-terminal glycine to a Lys residue in the target protein
Detectable neddylation under homeostatic conditions and at endogenous levels of NEDD8 and substrate expression
Neddylation of the proposed target is sensitive to MLN4924 treatment under conditions that block cullin neddylation but not ubiquitylation, and NEDD8 is thus activated by NAE.
Ideally, studies of a neddylation target should further define:
The specific NEDD8 E2 and E3 enzyme(s)
The regulation and biological consequences of neddylation.
Immunoprecipitations with specific antibodies are the preferred approach to study modifications of endogenous proteins, but genome editing159 can be used to introduce affinity-tagged constructs under the endogenous promoter. NAE-mediated neddylation is most reliably demonstrated with the small molecule inhibitor MLN4924102 (Box 2) and should include proof that the treatment has no effect on other cellular functions. NAE depletion by RNAi is generally much less specific and, if used, should be controlled by a rescue experiment with an RNAi-resistant construct. In both cases, however, indirect effects through inhibition of CRL activity should be excluded. Identification of a NEDD8-modification should best proceed by mass spectrometry, using the LysC protease, which, as opposed to trypsin, enables discrimination between ubiquitin, NEDD8 and ISG1560. This approach identifies the neddylated lysines and indicates whether NEDD8 forms chains. Further caution is needed if the neddylation target is also targeted by ubiquitin on the same lysine(s). In such cases, the relative abundance of the two modifications would be a crucial indicator of their physiological significance. Important follow-up experiments include identifying the E2 and E3 enzyme(s) that are involved in neddylation of the substrate. To this end, the structural restraints shown in Figure 2 C, D54 should be considered. Finally, neddylation of the target protein may be regulated by physiological stimuli, and specific cellular phenotype(s) should be observed upon generation of non-neddylatable mutants.
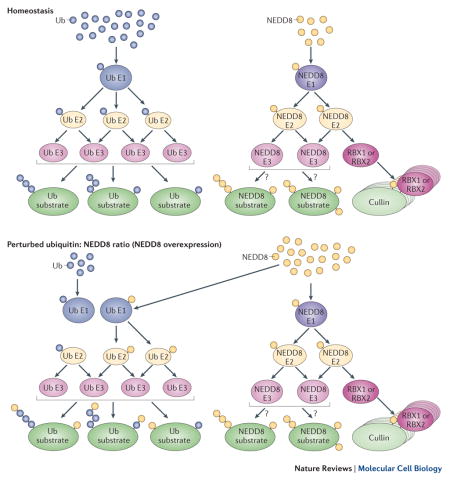
Current thinking is that NEDD8 conjugation through ubiquitin cascades has no role under physiological conditions, as the concentrations of free ubiquitin and NEDD8 in cells are almost equal100. However, an excess of NEDD8 over ubiquitin occurs in many cancers or cancer-derived cell lines or can be artificially generated upon ectopic overexpression of NEDD8 — a method that is frequently used in the search for novel neddylation targets. Therefore, conclusions about neddylation substrates and/or NEDD8 chain formation under physiological conditions should not be drawn from experiments involving NEDD8 overexpression.
In the hope to aid future experimental design, we propose a set of criteria for concluding that a cellular protein is a physiological neddylation target (Box 1). We have re-evaluated the published studies with regards to these criteria (Supp Table 1). The inevitable conclusion is that the only substrates that currently fulfill all criteria as neddylation substrates are the members of the cullin family. Nevertheless, there may be other genuinely neddylated substrates, and we have classified these proposed substrates according to their biological roles below. Several reported substrates pre-date the development of the NAE-specific inhibitor MLN4924102 and we would thus encourage additional validation experiments using this powerful tool (Box 2).
Box 2. Pharmacological Inhibition of Neddylation.
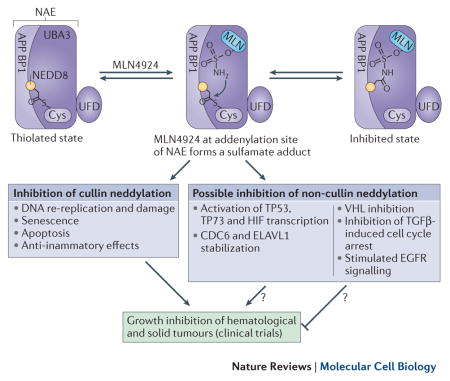
Overexpression of NEDD8 and the consequent aberrant activation of the neddylation pathway and CRL activity can drive the progression of numerous cancers8,15,16,160–163, as well as inflammatory and autoimmune diseases164,165. Moreover, the CRL system can be ‘hijacked’ by HIV or hepatitis virus proteins, which facilitate infection by targeting host proteins for degradation7,11. Pharmacological inhibition of CRLs and/or neddylation and deneddylation are thus active research areas and a small molecule NAE inhibitor, MLN4924102, is currently undergoing several clinical trials166.
MLN4924 resembles AMP and preferentially binds the adenylation site of a NAE molecule that already has a NEDD8 bound at its active site cysteine. MLN4924 attacks this NEDD8, forming a sulfamate adduct167 and inhibiting neddylation and CRL activity (Figure). The major cellular phenotype of MLN4924 treatment is DNA re-replication102,168, due to stabilization of the DNA replication licensing factor and CRL substrate CDT1169. MLN4924-treated cells accumulate DNA damage, which they fail to repair, and enter apoptosis169 or senescence168.
MLN4924 administration would also inhibit the neddylation of proposed non-cullin neddylation substrates. Although the consequences for the therapeutic efficacy remain unstudied, there are strong indications for both synergistic as well as counter-active effects. For example, neddylation is thought to inhibit the transcriptional activity of the well-established tumor suppressors TP53 and TP73, and stabilize HuR, which is important for the survival of hepatocellular carcinoma and colon cancer cells170,171, as well as to stabilize HIF transcription factors172. Hence, neddylation inhibition in the above examples will probably enhance the anti-tumorigenic properties of MLN4924. By contrast, non-cullin neddylation is thought to be tumor-suppressive in a number of cases, including TGF-β-induced G1/S cell cycle arrest through TGFβRII neddylation146, stimulation of DNA repair64, stimulation of VHL tumour suppressor-mediated involvement in fibronectin-dependent extracellular matrix assembly113,114, inhibition of E2F transcription133, inhibition of EGFR signaling90, and inhibition of caspase activity86. Consequently, inhibition of neddylation in the above cases might be counterproductive in cancer therapy and merits attention.
Neddylation of Ubiquitin E3 Ligases
The best-characterized and fully validated neddylation substrates are the cullin family members, which nucleate the largest class of RING ubiquitin E3 ligases, the cullin RING ligases. Cullin neddylation results in the activation of ubiquitin transfer activity by favoring alternative conformations of the cullin C-terminal domain and of RBX85,103–105, whereas CSN complex-mediated deneddylation of cullins enables the substrate receptor exchange factor CAND1 to bind to the cullin, removing CSN and triggering the exchange of the ubiquitylation-substrate specific receptor82,106–108. These functions of neddylation have been reviewed extensively elsewhere109.
Recently, several other ubiquitin E3 ligases have been reported to be neddylated, although further experiments will be required for validation using all of the criteria outlined in Box 1. The HECT-domain ubiquitin E3 ligase110 SMURF1, as well as its budding yeast orthologue Rsp5, has been reported to be activated by neddylation111. SMURF1 autoneddylates on multiple lysines through an active site cysteine that is distinct from the characteristic HECT domain ubiquitylation active site (Figure 3A). Moreover, SMURF1 hyper-activation through neddylation correlates with poor prognosis in colorectal cancer111. Other ubiquitin E3 ligases reportedly targeted by neddylation — although less well established by the criteria outlined in Box 1 — include Parkin (PARK2), VHL, BRAP2 and MDM2. All of these E3 ligases are members of the RING domain family. Several other HECT-domain E3 ligases have also been identified in large scale screens as neddylation targets59 but have not been studied further.
Figure 3. Proposed non-cullin Neddylation Substrates.
A Multiple auto-neddylation of the HECT domain ubiquitin E3 ligase SMURF1 on multiple lysines is mediated by a dedicated neddylation active site cysteine, which activates the ubiquitylation activity of SMURF1. B Both neddylation and ubiqutiylation of TP53 occur through MDM2 and together with NUB1 lead to the nuclear export and consequent inactivation of mono-ubiquitinated TP53. Neddylation of E2F1 (lower) in its DNA-binding domain is stimulated by methylation and leads to reduced protein levels and impaired transcriptional activity. C Differential regulation of receptor tyrosine kinase signaling by neddylation: EGF-induced c-CBL-mediated neddylation of the EGFR synergizes with ubiquitylation to target EGFR for lysosomal degradation. On the contrary, TGFβ-induced c-CBL-mediated neddylation of the TGFβRII counteracts the destructive effects of ubiquitylation and stabilizes TGFβRII. D Poly-neddylation of histone H4, recognized by RNF168, can function as a signal for amplification of the DNA damage response cascade. E Neddylation in nucleolar stress signaling. The large ribosomal subunit L11 can be neddylated by MDM2, following impaired ribosome assembly, such that it localizes to the nucleolus and evades proteasomal destruction. Deneddylation by DEN1 counteracts this process, but nucleoplasmic localization of L11 also leads to stabilization of TP53 and cell cycle arrest.
VHL, a substrate receptor of a CUL2-based CRL112, was found to be neddylated in prostate cancer cells113. However, as NEDD8 is overexpressed in certain cancer cell lines8,16, it remains unclear if VHL is also neddylated under non-pathological conditions. VHL is predominantly neddylated in a region that is important for incorporation into CRL2, and neddylation might thus impede complex formation114. VHL reportedly also has a CRL2-independent function as a positive regulator of fibronectin extracellular matrix assembly. Neddylated VHL is incapable of binding fibronectin but an intact neddylation pathway is required for the VHL–fibronectin interaction, as non-neddylatable VHL mutants have defects in promoting differentiated cell morphology113,114. It is therefore possible that transient neddylation of VHL is required as a switch between CUL2 and fibronectin binding.
Parkin is a RING-in-between-RING ubiquitin E3 ligase115 that promotes the selective degradation of damaged mitochondria by a process known as mitophagy4. Deregulation of this pathway is linked to the aetiology of Parkinson’s disease [G]116. Endogenous parkin has been found to be neddylated by endogenous NEDD8 in brain samples of patients with Parkinson’s disease117 as well as in cultured neuroblastoma cells, but UBC12 was not required for neddylation118. An aberrant ratio of NEDD8 to ubiquitin in these samples was, however, not excluded. Neddylation has been detected in aggregates from various neurodegenerative diseases117,119–122. Moreover, both NEDD8 and neddylated parkin were predominantly cytoplasmic in patients with Parkinson’s disease118, as observed for other neurodegenerative pathologies123. As NEDD8 is predominantly nuclear under normal conditions6, it is possible that in disease tissues NEDD8 is either overexpressed and/or aberrantly translocated to the cytoplasm, where it modifies the cytosolic pool of parkin, potentially through the ubiquitylation machinery. Parkin can be neddylated on multiple lysines distributed along the entire protein118. Parkin neddylation does not affect its solubility or subcellular localization but does enhance its ubiquitylation activity in vivo117,118. Parkin was recently discovered to adopt an auto-inhibited conformation115. It would therefore be of particular interest to reconstitute Parkin neddylation and ubiquitylation in vitro and to investigate whether Parkin ubiquitylation activity can be activated by specific neddylation. Indeed, whereas the ubiquitin E2-binding site in the Parkin structure is occluded in its unmodified and uncomplexed form115, Parkin neddylation resulted in increased affinity for its E2 and substrates in vivo118. A parallel can be drawn to the intriguing recent observation that auto-inhibition of the Ariadne subfamily of RING-in-between-RING E3 ligases can be relieved by binding to neddylated CRLs95.
Neddylation in Transcriptional Regulation
Transcription factors are among the most commonly reported putative neddylation targets. Neddylation seems to generally suppress their activity, albeit through different pathways, including altered protein stability, protein-protein and protein-DNA interactions, as well as subcellular localization. Multiple studies have examined neddylation of the transcription factor TP53 (Supp Table 1). TP53 helps cells to react to various stress signals such as nutrient deprivation, DNA damage, hypoxia and ribosomal stress by, for example, inhibiting cell cycle progression or triggering senescence or apoptosis and TP53 thus functions as a crucial tumour suppressor124. The activity of TP53 is inhibited under normal growth conditions by the RING-domain E3 ligase MDM2, which targets TP53 for proteasomal degradation through ubiquitylation. MDM2 has been reported to also function as a NEDD8–E3 ligase for TP53, although at this point the mechanisms of MDM2-mediated neddylation remain unclear. Neddylation is thought to inhibit TP53 transcriptional activity but not significantly affect its stability125 (Figure 3B). The ubiquitylation and neddylation activities of MDM2 towards TP53 are reported to have distinct regulatory mechanisms. The histone acetyl transferase TIP60 can inhibit MDM2-mediated neddylation of TP53, whereas the cyclin-dependent kinase inhibitor p14ARF inhibits MDM2-mediated ubiquitylation126. The balance between neddylation and ubiquitylation of TP53 is further regulated by NUB1, which decreases the former and promotes the latter127. Neddylation, ubiquitylation and NUB1 cooperate in the cytoplasmic translocation and inactivation of TP53 in a manner that is not fully understood but is mutually dependent127. Mono-ubiquitylation rather than neddylation of TP53, however, is the main signal triggering its nuclear export128. The TP53 family member TP73 also seems to be neddylated by MDM2, similarly resulting in cytoplasmic localization and down-regulated transcriptional activity129. A second E3 ligase, SCFFbxo11, was discovered to neddylate but not ubiquitylate TP53, thereby downregulating its transcriptional activity without affecting its stability130. It would be interesting to study whether CUL1 neddylation and/or DCNLs stimulate this activity.
E2F1
E2Fs are transcription factors that can both stimulate and repress cell cycle progression131. Two studies have reported that neddylation of E2Fs leads to down-regulation of their transcriptional activity132,133 (Figure 3B). E2F1 was observed to be neddylated mainly in the DNA-binding domain and its protein levels were reduced following neddylation133. DEN1 can deneddylate E2Fs and activate E2F-mediated transcription. Interestingly, E2Fs are known to be stabilized by DNA-damaging chemotherapeutics in which case they were observed to be less neddylated132,133, probably due to DEN1 overexpression134. There is evidence that both neddylation and deneddylation specifically regulate a subset of E2F target genes. In particular, E2F1 deneddylation follows DNA damage and enhances the interaction of E2F1 specifically with its cofactor MCPH1, which triggers transcription of pro-apoptotic factors132. Moreover, there are indications that E2F1 neddylation can be stimulated by prior methylation of E2F1 by Set7/9133. The latter occurs during DNA damage and, similar to neddylation, leads to a destabilization of E2F1135.
NF-κB signaling
The nuclear factor-κB (NF-κB) family comprises heterodimeric transcription factors that are primarily responsible for the inflammatory response and cell survival136. In the absence of extracellular signaling, NF-κB is located in the cytoplasm and is inhibited by IκB family members. Upon stimulation of a cell with inflammatory cytokines such as TNF, a heterotrimeric kinase composed of IKKα and IKKβ in complex with NEMO (also known as IKKγ) phosphorylates IκB. IκB is consequently ubiquitylated and targeted for degradation by SCFβTrCP, with the result that the NF-κB transcription factors enter the nucleus and stimulate gene expression. In this regard, the best-studied effect of NEDD8 on NF-κB signaling is related to its regulation of the cullin E3 ubiquitin ligases responsible for IκB ubiquitylation137.
Several recent reports have proposed non-cullin neddylation substrates with an effect on NF-κB-mediated transcription. For example, IKKγ is neddylated by the RING E3 ligase TRIM40, thus inhibiting NF-κB signaling138. Importantly, a comparison of endogenous IKKγ neddylation levels revealed a significant reduction in gastric epithelium samples from patients with gastric cancer relative to healthy controls, which indicates that IKKγ neddylation might have a tumour suppressive function138. In a further example, NF-κB-mediated transcription was inhibited by neddylated BCA3, which interacts with the NF-κB subunit RELA and further recruits the histone deacetylase and transcriptional repressor SIRT1139. However, a subsequent study challenged these conclusions arguing that BCA3 is only a repressor of NF-κB transcription at very high TNF concentrations, and that BCA3 behaves as a positive regulator of NF-κB at close to physiological levels of TNF140. As its neddylation status was not investigated in this latter study, the exact functions of BCA3 and the role of its neddylation in NF-κB regulation merit further investigation under physiological conditions. In a final study, the RING E3 ubiquitin ligase BRAP2, which has several reported functions in intracellular signaling, was found both to interact with NEDD8 and to be neddylated, albeit solely by over-expression studies141. Interestingly, BRAP2 was neddylated on a lysine residue in a sequence stretch that is homologous to the NEDD8 consensus site in cullins. TNF stimulation leads to BRAP2 neddylation and suppression of NF-κB nuclear translocation. However, a link between the two effects is lacking and it is possible that the inhibition of NF-kB is rather mediated through interaction with CUL1 by a neddylation-independent mechanism141. Thus, further work is required to assess whether BRAP2 neddylation has a physiological significance and a function in NF-κB signaling.
APP/AICD
The amyloid-beta precursor protein (APP) is a cell surface protein that is sequentially cleaved by α-, β- and γ-secretase to produce two fragments. One is the extracellular amyloid-beta peptide, which is associated with Alzheimer’s disease [G], and the other is APP intracellular domain (AICD). The AICD functions as a component of a transcription factor complex, further comprising the nuclear adaptor protein FE65 and the histone acetyltransferase TIP60142. Neddylation of AICD sterically blocks the interaction with FE65 and consequently attenuates the transcriptional activity of the complex143. The required E1, E2 and E3 enzymes for neddylation of AICD were not determined in this study, leaving open the question of whether the typical NEDD8 machinery is involved or whether neddylation occurs through the ubiquitylation pathway. In this particular case, a third more speculative possibility also exists. As APP binds to the NAE subunit APPBP1 close to the lysine residues reported to be neddylated, it is conceivable that the transfer of NEDD8 from the catalytic cysteine of NAE onto a lysine residue in AICD requires no E2 or E3 enzymes144.
Neddylation in Signaling Pathways
As summarized below, neddylation has been reported to potentially regulate numerous signaling pathways, including receptor tyrosine kinase signaling, apoptosis, DNA damage and nucleolar stress signaling.
Receptor Tyrosine Kinase signaling
There are two examples of neddylation regulating receptor tyrosine kinase signaling (Figure 3C). The transforming growth factor-β type II receptor (TGFβRII) is a receptor tyrosine kinase that inhibits cellular proliferation145. A recent study showed that TGFβRII phosphorylation activates the RING E3 ligase c-CBL, which can neddylate TGFβRII and thus stabilize and prolong its signalling by targeting it for clathrin-mediated endocytosis under endogenous conditions146. Deneddylation of TGFβRII by DEN1, by contrast, promotes TGFβRII ubiquitylation, which leads to its degradation through lipid raft-/caveolin-mediated endocytosis [G]146. Although this study146 reported that c-CBL activates UBC12 to neddylate TGFβRII, this seems to be a structurally unlikely event (Figure 2C, D). Therefore conclusive evidence that TGFβRII is conjugated by the neddylation machinery is currently missing.
The epidermal growth factor receptor (EGFR) is a receptor tyrosine kinase activated by binding to extracellular growth hormones that in turn activates numerous signaling cascades in the cell. As hyper-activation of this signaling cascade would be detrimental to the organism, the activated EGFR is rapidly internalized through endocytosis and degraded in the lysosome in a process regulated by phosphorylation and c-CBL, which ubiquitylates the EGFR147. c-CBL has also been reported to neddylate EGFR, resulting in its increased ubiquitylation and endocytic internalization90.
Apoptosis
Using in vivo RNAi screening followed by genetic validation, the deneddylase DEN1 was shown to have a proapoptotic role in D. melanogaster86. Indeed, the effector caspase [G] drICE, but not the initiator caspase [G] DRONC, was found to be neddylated under endogenous conditions by the RING ubiquitin E3 ligase dIAP1 and to be deneddylated by DEN1. Moreover, neddylation of drICE was shown to inhibit its ability to cleave its substrate poly ADP-ribose polymerase-1 (PARP1) in vitro, rationalizing its anti-apoptotic effect in vivo. Interestingly, in vitro ubiquitylation of drICE also inhibits its proteolyic activity but the underlying mechanisms are probably different, as neddylation seems to function through a non-competitive mechanism, whereas ubiquitylation partly decreases the affinity of drICE for PARP1. Convincing proof for neddylation under endogenous conditions of the human drICE orthologue, Caspase-7, is currently lacking86,148.
DNA damage
Both ubiquitin and SUMO have essential roles in the DNA damage response. Similarly, NEDD8 was observed to form nuclear foci that co-localize with sites of DNA damage64. In particular, following DNA damage the RING E3 ligase RNF111 together with UBC12 assembles NEDD8 chains on histone H4, which are then recognized by the UIM domains of another RING E3 ligase, RNF168 (Figure 3D). The latter amplifies the DNA damage response cascade by recruiting the homologous recombination repair factors BRCA1 and 53BP164. It is likely that ubiquitin and NEDD8 chains have very similar functions in the DNA damage response.
Nucleolar stress signaling
Ribosome biogenesis consumes a large percentage of the cellular energy and is tightly monitored and regulated. Impaired ribosome assembly can lead to cell cycle arrest, mediated by the binding of the large ribosomal protein L11 to MDM2, leading to competitive inhibition of the MDM2-TP53 interaction and stabilization of TP53149. This process is, in part, regulated by reversible neddylation of L11 (Figure 3E). L11 can be neddylated by MDM2, leading to localization to the nucleolus and stabilization, whereas DEN1-mediated deneddylation of L11 results in its nucleoplasmic relocalization and proteasomal degradation by an unknown E3 ligase61,150. Importantly, the deneddylated L11 also co-localizes with TP53 and MDM2 in the nucleoplasm, leading to the aforementioned TP53 stabilization and aiding the recruitment of TP53 co-activators151. Moreover, the ribosomal protein S14 has also been reported to trigger TP53 activation upon nucleolar stress152. Similarly to L11, S14 neddylation results in its nucleolar localization and increased stability, and S14 neddylation is reversed by DEN1 in a reaction that is stimulated by the DEN1 interactor hCINAP152. In contrast to L11, however, the neddylated form of S14 seems to inhibit MDM2 and thus stabilize TP53152. Further studies are needed to elucidate the potential interplay and/or feedback loops between these pathways as well as whether the requirement for NEDD8 to activate TP53 during nucleolar stress is due to additional factors.
Conclusions and future perspectives
Although as discussed above there is potential that non-cullin proteins are targeted for neddylation under homeostatic conditions, compelling evidence for the reported substrates is still lacking (Box 1 and Supp. Table 1). Numerous reports of non-cullin neddylation suggest that it might have a wider diversity of functions than the regulation of cullin RING ligases, potentially including regulation of protein stability through proteasomal and lysosomal pathways, altering subcellular localization, as well as protein-protein interactions. Although the physiological relevance of several reported NEDD8 substrates is unknown, neddylation may directly regulate receptor tyrosine kinase signalling, the DNA damage response, transcription and translation. Many of these targets are involved in human pathologies and the pharmacological inhibition of neddylation by MLN4924 (Box 2) should not be regarded as synonymous with inhibition of CRL-mediated ubiquitylation. Moreover, the cross-talk between the neddylation and ubiquitylation pathways under various stress and pathological conditions should be considered in the evaluation of neddylation as a drug target.
An important open question is whether neddylation is functionally distinct from ubiquitylation. For example, the yeast CUL4-orthologue Rtt101 can be neddylated or ubiquitinated, both leading to its catalytic activation70. Moreover, it is possible that poly-neddylation and poly-ubiquitylation at DNA damage sites or in response to other stress conditions are functionally redundant, as NEDD8 and ubiquitin can be recognized by the same interaction motifs. By contrast, ubiquitylation and neddylation of the TGFβRII elicit distinct biological responses. A research area in need of closer attention is the identification and functional characterization of specific NEDD8-interacting domains and proteins as well as molecular and structural understanding of the mechanisms through which they discriminate between NEDD8 and other UBLs, and decode and relay NEDD8-associated signals.
DEN1 has emerged as the primary deneddylase of the postulated non-cullin neddylation substrates and its expression seems to regulate NEDD8-based signalling in response to cellular stresses. However, DEN1 depletion does not lead to strong cellular phenotypes38, indicating either the non-essential nature of non-cullin deneddylation under the studied conditions or as yet unknown functionally redundant deneddylases. A further noteworthy observation is that in contrast to cullin neddylation, the reported neddylation sites in non-cullin substrates are often numerous and broadly distributed across the target’s surface-exposed lysines, hinting at the possibility that non-cullin neddylation signals are transmitted through multiple mono-neddylation of whole domains. Further functional, mechanistic and structural investigations into non-cullin neddylation are thus highly warranted.
As future searches for neddylation targets will require rigorous experimental validation, we anticipate that the increasingly accessible genome editing techniques, such as the CRISPR/Cas9 technology153, will be of great aid. Last but not least, the propensity of NEDD8 itself to be post-translationally modified merits closer attention. In addition to NEDD8- and ubiquitin-chain formation, proteomic studies have reported phosphorylation, acetylation and succinylation sites on NEDD8154–157*, whose functional significance remains unknown.
Supplementary Material
Online summary.
NEDD8 and ubiquitin have the highest sequence and structural similarity among all ubiquitin-like proteins
NEDD8-specific conjugation and de-conjugation pathways exist in all studied eukaryotes and they are capable of discriminating between NEDD8 and other ubiquitin-like proteins through NEDD8-specific interaction domains
Nevertheless, a perturbed ratio of free NEDD8 and ubiquitin or cellular stress can cause NEDD8-conjugation through the ubiquitylation machinery onto ubiquitylation substrates. This can lead to mis-assignments of neddylation targets and most published reports lack sufficient evidence to substantiate the discovery of genuine neddylation substrates.
We propose a list of necessary criteria for bona fide neddylation substrates and re-evaluate published studies in the light of these criteria. Cullins are the best-studied and only neddylation targets to date which fulfill all of these criteria.
We discuss potential examples of neddylation regulating non-cullin ubiquitin E3 ligases, transcription, ribosomal stress, as well as various signaling pathways.
Pharmacological inhibition of neddylation is a promising new direction for cancer therapy. We discuss the potential effects of inhibiting non-cullin, as well as cullin, neddylation.
Acknowledgments
We thank D. Xirodimas and A. Smith for critical reading of the manuscript. R.I.E. is supported through a Marie-Curie post-doctoral fellowship, and work in the Peter lab is supported by the European Research Council (ERC), the Swiss National Science Foundation (SNF), and the ETH Zürich. B.A.S. acknowledges the Howard Hughes Medical Institute and NIH R01GM069530, P30CA021765, and ALSAC for support.
Glossary
- cullin RING ligases
a large family of E3 ligases, nucleated by one of seven cullins, which bring together the ubiquitylation substrate through substrate-specific adaptors and a RING-domain RBX subunit, which bind an E2 enzyme and catalyzes the ubiquitin discharge.
- JAMM motif
a metaloprotease His-X-His-X(10)-Asp motif, which coordinates a zinc, present in multiple pro- and eukaryotic enzymes, including the deubiquitinating enzymes RPN11, AMSH-LP and CSN5.
- P97
an AAA-ATPase hexamer, involved in eukaryotic signaling and quality control pathways, delivering ubiquitylated proteins to the 26S proteasome through numerous adaptors.
- UBA domain
the ubiquitin-associated domain is structurally characterized by a three-helix bundle, which recognizes the Ile44 hydrophobic patch of ubiquitin.
- UIM domain
the ubiquitin-interacting motif domain comprises an amphipathic helix with conserved negatively charged residues at its N-terminus and contiguous hydrophobic patch in the middle and a C-terminal serine residue. A conserved alanine side chain of the UIM inserts in the Ile44 hydrophobic pocket of ubiquitin.
- MIU domain
the motif interacting with ubiquitin domain is similar to the UIM domain but the helix runs in the opposite direction. MIU domains are known to recognize Lys63-linked ubiquitin chains.
- Parkinson’s Disease
a neurodegenerative disease, characterised by the loss of dopaminergic neurons of the midbrain and accumulation of insoluble inclusions containing α-synuclein amyloid fibrils.
- Alzheimer’s disease
a neurodegenerative disease of the cerebral cortex characterised by accumulation of aggregated amyloid-Beta (Aβ).
Footnotes
References
- 1.van der Veen AG, Ploegh HL. Ubiquitin-like proteins. Annu Rev Biochem. 2012;81:323–357. doi: 10.1146/annurev-biochem-093010-153308. [DOI] [PubMed] [Google Scholar]
- 2.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin–proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–689. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schreiber A, Peter M. Substrate recognition in selective autophagy and the ubiquitin-proteasome system. Biochim Biophys Acta. 2014;1843:163–181. doi: 10.1016/j.bbamcr.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 5.Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 6.Kamitani T, Kito K, Nguyen HP, Yeh ET. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem. 1997;272:28557–28562. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- 7.Kim DY, et al. CBFβ stabilizes HIV Vif to counteract APOBEC3 at the expense of RUNX1 target gene expression. Mol Cell. 2013;49:632–644. doi: 10.1016/j.molcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hori T, et al. Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene. 1999;18:6829–6834. doi: 10.1038/sj.onc.1203093. [DOI] [PubMed] [Google Scholar]
- 9.Li T, Chen X, Garbutt KC, Zhou P, Zheng N. Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase. Cell. 2006;124:105–117. doi: 10.1016/j.cell.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 10.Kumar S, Yoshida Y, Noda M. Cloning of a cDNA which encodes a novel ubiquitin-like protein. Biochem Biophys Res Commun. 1993;195:393–399. doi: 10.1006/bbrc.1993.2056. [DOI] [PubMed] [Google Scholar]
- 11.Li T, Robert EI, van Breugel PC, Strubin M, Zheng N. A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery. Nat Struct Mol Biol. 2010;17:105–111. doi: 10.1038/nsmb.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar S, Tomooka Y, Noda M. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem Biophys Res Commun. 1992;185:1155–1161. doi: 10.1016/0006-291x(92)91747-e. [DOI] [PubMed] [Google Scholar]
- 13.Carrabino S, Carminati E, Talarico D, Pardi R, Bianchi E. Expression pattern of the JAB1/CSN5 gene during murine embryogenesis: colocalization with NEDD8. Gene Expr Patterns. 2004;4:423–431. doi: 10.1016/j.modgep.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 14.Noh EH, et al. Covalent NEDD8 conjugation increases RCAN1 protein stability and potentiates its inhibitory action on calcineurin. PLoS ONE. 2012;7:e48315. doi: 10.1371/journal.pone.0048315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salon C, et al. Altered pattern of Cul-1 protein expression and neddylation in human lung tumours: relationships with CAND1 and cyclin E protein levels. J Pathol. 2007;213:303–310. doi: 10.1002/path.2223. [DOI] [PubMed] [Google Scholar]
- 16.Chairatvit K, Ngamkitidechakul C. Control of cell proliferation via elevated NEDD8 conjugation in oral squamous cell carcinoma. Mol Cell Biochem. 2007;306:163–169. doi: 10.1007/s11010-007-9566-7. [DOI] [PubMed] [Google Scholar]
- 17.Tateishi K, Omata M, Tanaka K, Chiba T. The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J Cell Biol. 2001;155:571–579. doi: 10.1083/jcb.200104035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dharmasiri S, Dharmasiri N, Hellmann H, Estelle M. The RUB/Nedd8 conjugation pathway is required for early development in Arabidopsis. EMBO J. 2003;22:1762–1770. doi: 10.1093/emboj/cdg190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ou CY, Lin YF, Chen YJ, Chien CT. Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev. 2002;16:2403–2414. doi: 10.1101/gad.1011402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones D, Candido EP. The NED-8 conjugating system in Caenorhabditis elegans is required for embryogenesis and terminal differentiation of the hypodermis. Dev Biol. 2000;226:152–165. doi: 10.1006/dbio.2000.9847. [DOI] [PubMed] [Google Scholar]
- 21.Osaka F, et al. Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 2000;19:3475–3484. doi: 10.1093/emboj/19.13.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 1998;17:2208–2214. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lammer D, et al. Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitby FG, Xia G, Pickart CM, Hill CP. Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. J Biol Chem. 1998;273:34983–34991. doi: 10.1074/jbc.273.52.34983. Structural and biochemical characterization of NEDD8, defining specificity-determining residues. [DOI] [PubMed] [Google Scholar]
- 25.Rao-Naik C, et al. The rub family of ubiquitin-like proteins. Crystal structure of Arabidopsis rub1 and expression of multiple rubs in Arabidopsis. J Biol Chem. 1998;273:34976–34982. doi: 10.1074/jbc.273.52.34976. [DOI] [PubMed] [Google Scholar]
- 26.Choi YS, Jeon YH, Ryu KS, Cheong C. 60th residues of ubiquitin and Nedd8 are located out of E2-binding surfaces, but are important for K48 ubiquitin-linkage. FEBS Lett. 2009;583:3323–3328. doi: 10.1016/j.febslet.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 27.Walden H, et al. The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol Cell. 2003;12:1427–1437. doi: 10.1016/s1097-2765(03)00452-0. Structural and biochemical characterization of NAE, defining the residues that mediate specific interactions with NEDD8. [DOI] [PubMed] [Google Scholar]
- 28.Reverter D, et al. Structure of a complex between Nedd8 and the Ulp/Senp protease family member Den1. J Mol Biol. 2005;345:141–151. doi: 10.1016/j.jmb.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 29.Shen LN, et al. Structural basis of NEDD8 ubiquitin discrimination by the deNEDDylating enzyme NEDP1. EMBO J. 2005;24:1341–1351. doi: 10.1038/sj.emboj.7600628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 31.Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linghu B, Callis J, Goebl MG. Rub1p processing by Yuh1p is required for wild-type levels of Rub1p conjugation to Cdc53p. Eukaryotic Cell. 2002;1:491–494. doi: 10.1128/EC.1.3.491-494.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wada H, Kito K, Caskey LS, Yeh ET, Kamitani T. Cleavage of the C-terminus of NEDD8 by UCH-L3. Biochem Biophys Res Commun. 1998;251:688–692. doi: 10.1006/bbrc.1998.9532. [DOI] [PubMed] [Google Scholar]
- 34.Gan-Erdene T, et al. Identification and characterization of DEN1, a deneddylase of the ULP family. J Biol Chem. 2003;278:28892–28900. doi: 10.1074/jbc.M302890200. [DOI] [PubMed] [Google Scholar]
- 35.Mendoza HM, et al. NEDP1, a highly conserved cysteine protease that deNEDDylates Cullins. J Biol Chem. 2003;278:25637–25643. doi: 10.1074/jbc.M212948200. [DOI] [PubMed] [Google Scholar]
- 36.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu K, et al. DEN1 is a dual function protease capable of processing the C terminus of Nedd8 and deconjugating hyper-neddylated CUL1. J Biol Chem. 2003;278:28882–28891. doi: 10.1074/jbc.M302888200. [DOI] [PubMed] [Google Scholar]
- 38.Chan Y, et al. DEN1 deneddylates non-cullin proteins in vivo. J Cell Sci. 2008;121:3218–3223. doi: 10.1242/jcs.030445. [DOI] [PubMed] [Google Scholar]
- 39.Kurihara LJ, Semenova E, Levorse JM, Tilghman SM. Expression and functional analysis of Uch-L3 during mouse development. Mol Cell Biol. 2000;20:2498–2504. doi: 10.1128/mcb.20.7.2498-2504.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olsen SK, Capili AD, Lu X, Tan DS, Lima CD. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature. 2010;463:906–912. doi: 10.1038/nature08765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang DT, et al. Basis for a ubiquitin-like protein thioester switch toggling E1–E2 affinity. Nature. 2007;445:394–398. doi: 10.1038/nature05490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang DT, et al. Structural Basis for Recruitment of Ubc12 by an E2 Binding Domain in NEDD8’s E1. Mol Cell. 2005;17:341–350. doi: 10.1016/j.molcel.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 43.Huang DT, et al. A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8. Nat Struct Mol Biol. 2004;11:927–935. doi: 10.1038/nsmb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deshaies RJ, Joazeiro CAP. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 45.Huang DT, et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell. 2009;33:483–495. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 1999;13:2928–2933. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monda JK, et al. Structural Conservation of Distinctive N-terminal Acetylation-Dependent Interactions across a Family of Mammalian NEDD8 Ligation Enzymes. Structure. 2013;21:42–53. doi: 10.1016/j.str.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eletr ZM, Huang DT, Duda DM, Schulman BA, Kuhlman B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat Struct Mol Biol. 2005;12:933–934. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
- 49.Pruneda JN, Stoll KE, Bolton LJ, Brzovic PS, Klevit RE. Ubiquitin in motion: structural studies of the ubiquitin-conjugating enzyme∼ubiquitin conjugate. Biochemistry. 2011;50:1624–1633. doi: 10.1021/bi101913m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat Struct Mol Biol. 2012;19:876–883. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Plechanovová A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat Struct Mol Biol. 2006;13:491–499. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
- 53.Berndsen CE, Wiener R, Yu IW, Ringel AE, Wolberger C. A conserved asparagine has a structural role in ubiquitin-conjugating enzymes. Nat Chem Biol. 2013;9:154–156. doi: 10.1038/nchembio.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scott DC, et al. Structure of a RING E3 Trapped in Action Reveals Ligation Mechanism for the Ubiquitin-like Protein NEDD8. Cell. 2014;157:1671–1684. doi: 10.1016/j.cell.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Enchev RI, Schreiber A, Beuron F, Morris EP. Structural insights into the COP9 signalosome and its common architecture with the 26S proteasome lid and eIF3. Structure. 2010;18:518–527. doi: 10.1016/j.str.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 56.Gazdoiu S, et al. Proximity-induced activation of human Cdc34 through heterologous dimerization. Proc Natl Acad Sci USA. 2005;102:15053–15058. doi: 10.1073/pnas.0507646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ohki Y, Funatsu N, Konishi N, Chiba T. The mechanism of poly-NEDD8 chain formation in vitro. Biochem Biophys Res Commun. 2009;381:443–447. doi: 10.1016/j.bbrc.2009.02.090. [DOI] [PubMed] [Google Scholar]
- 58.Wu PY. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 2003;22:5241–5250. doi: 10.1093/emboj/cdg501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones J, et al. A targeted proteomic analysis of the ubiquitin-like modifier nedd8 and associated proteins. J Proteome Res. 2008;7:1274–1287. doi: 10.1021/pr700749v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jeram SM, et al. An improved SUMmOn-based methodology for the identification of ubiquitin and ubiquitin-like protein conjugation sites identifies novel ubiquitin-like protein chain linkages. Proteomics. 2010;10:254–265. doi: 10.1002/pmic.200900648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xirodimas DP, et al. Ribosomal proteins are targets for the NEDD8 pathway. EMBO Rep. 2008;9:280–286. doi: 10.1038/embor.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leidecker O, Matic I, Mahata B, Pion E, Xirodimas DP. The ubiquitin E1 enzyme Ube1 mediates NEDD8 activation under diverse stress conditions. Cell Cycle. 2012;11:1142–1150. doi: 10.4161/cc.11.6.19559. Together with Ref. 100, describes a stress-induced neddylation pathway through the ubiquitylation machinery, which can lead to artefacts in the search for neddylation substrates. [DOI] [PubMed] [Google Scholar]
- 63.Girdwood D, Xirodimas DP, Gordon C. The essential functions of NEDD8 are mediated via distinct surface regions, and not by polyneddylation in Schizosaccharomyces pombe. PLoS ONE. 2011;6:e20089. doi: 10.1371/journal.pone.0020089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ma T, et al. RNF111-dependent neddylation activates DNA damage-induced ubiquitination. Mol Cell. 2013;49:897–907. doi: 10.1016/j.molcel.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh RK, et al. Recognition and cleavage of related to ubiquitin 1 (Rub1) and Rub1-ubiquitin chains by components of the ubiquitin-proteasome system. Molecular & Cellular Proteomics. 2012;11:1595–1611. doi: 10.1074/mcp.M112.022467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kurz T, et al. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S cerevisiae. Nature. 2005;435:1257–1261. doi: 10.1038/nature03662. [DOI] [PubMed] [Google Scholar]
- 67.Kurz T, et al. Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol Cell. 2008;29:23–35. doi: 10.1016/j.molcel.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 68.Scott DC, et al. A Dual E3 Mechanism for Rub1 Ligation to Cdc53. Mol Cell. 2010;39:784–796. doi: 10.1016/j.molcel.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science. 2011;334:674–678. doi: 10.1126/science.1209307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rabut G, et al. The TFIIH subunit Tfb3 regulates cullin neddylation. Mol Cell. 2011;43:488–495. doi: 10.1016/j.molcel.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang G, Kaufman AJ, Ramanathan Y, Singh B. SCCRO (DCUN1D1) promotes nuclear translocation and assembly of the neddylation E3 complex. J Biol Chem. 2011;286:10297–10304. doi: 10.1074/jbc.M110.203729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu K, et al. Mono-ubiquitination drives nuclear export of the human DCN1-like protein hDCNL1. J Biol Chem. 2011;286:34060–34070. doi: 10.1074/jbc.M111.273045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meyer-Schaller N, et al. The human Dcn1-like protein DCNL3 promotes Cul3 neddylation at membranes. Proc Natl Acad Sci USA. 2009;106:12365–12370. doi: 10.1073/pnas.0812528106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Compe E, Egly JM. TFIIH: when transcription met DNA repair. Nat Rev Mol Cell Biol. 2012;13:343–354. doi: 10.1038/nrm3350. [DOI] [PubMed] [Google Scholar]
- 75.Cui J, et al. Glutamine deamidation and dysfunction of ubiquitin/NEDD8 induced by a bacterial effector family. Science. 2010;329:1215–1218. doi: 10.1126/science.1193844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yao Q, et al. Structural mechanism of ubiquitin and NEDD8 deamidation catalyzed by bacterial effectors that induce macrophage-specific apoptosis. Proc Natl Acad Sci USA. 2012;109:20395–20400. doi: 10.1073/pnas.1210831109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cope GA, et al. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 78.Echalier A, et al. Insights into the regulation of the human COP9 signalosome catalytic subunit, CSN5/Jab1. Proc Natl Acad Sci USA. 2013;110:1273–1278. doi: 10.1073/pnas.1209345110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sharon M, et al. Symmetrical modularity of the COP9 signalosome complex suggests its multifunctionality. Structure. 2009;17:31–40. doi: 10.1016/j.str.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 80.Birol M, et al. Structural and Biochemical Characterization of the Cop9 Signalosome CSN5/CSN6 Heterodimer. PLoS ONE. 2014;9:e105688. doi: 10.1371/journal.pone.0105688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lingaraju GM, et al. Crystal structure of the human COP9 signalosome. Nature. 2014;512:161–165. doi: 10.1038/nature13566. [DOI] [PubMed] [Google Scholar]
- 82.Enchev RI, et al. Structural basis for a reciprocal regulation between SCF and CSN. Cell Rep. 2012;2:616–627. doi: 10.1016/j.celrep.2012.08.019. Refs. 81 and 82 describe the structural and biochemical characterization of CSN and CSN-SCF complexes, indicating the interaction interfaces. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Emberley ED, Mosadeghi R, Deshaies RJ. Deconjugation of Nedd8 from Cul1 is directly regulated by Skp1-Fbox and substrate, and CSN inhibits deneddylated SCF by a non-catalytic mechanism. J Biol Chem. 2012;287:29679–29689. doi: 10.1074/jbc.M112.352484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fischer ES, et al. The Molecular Basis of CRL4DDB2/CSA Ubiquitin Ligase Architecture, Targeting, and Activation. Cell. 2011;147:1024–1039. doi: 10.1016/j.cell.2011.10.035. [DOI] [PubMed] [Google Scholar]
- 85.Duda DM, et al. Structural Insights into NEDD8 Activation of Cullin-RING Ligases: Conformational Control of Conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. Structural and biochemical characterization of neddylated cullin-RING E3 ligases, rationalizing the activatory effects of neddylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Broemer M, et al. Systematic in vivo RNAi analysis identifies IAPs as NEDD8-E3 ligases. Mol Cell. 2010;40:810–822. doi: 10.1016/j.molcel.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 87.Hemelaar J, et al. Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin-like proteins. Mol Cell Biol. 2004;24:84–95. doi: 10.1128/MCB.24.1.84-95.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ye Y, et al. Polyubiquitin binding and cross-reactivity in the USP domain deubiquitinase USP21. EMBO Rep. 2011;12:350–357. doi: 10.1038/embor.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kamitani T, Kito K, Fukuda-Kamitani T, Yeh ET. Targeting of NEDD8 and its conjugates for proteasomal degradation by NUB1. J Biol Chem. 2001;276:46655–46660. doi: 10.1074/jbc.M108636200. [DOI] [PubMed] [Google Scholar]
- 90.Oved S, et al. Conjugation to Nedd8 instigates ubiquitylation and down-regulation of activated receptor tyrosine kinases. J Biol Chem. 2006;281:21640–21651. doi: 10.1074/jbc.M513034200. [DOI] [PubMed] [Google Scholar]
- 91.Besten den W, Verma R, Kleiger G, Oania RS, Deshaies RJ. NEDD8 links cullin-RING ubiquitin ligase function to the p97 pathway. Nat Struct Mol Biol. 2012;19:511–6-S1. doi: 10.1038/nsmb.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bandau S, Knebel A, Gage ZO, Wood NT, Alexandru G. UBXN7 docks on neddylated cullin complexes using its UIM motif and causes HIF1α accumulation. BMC Biol. 2012;10:36. doi: 10.1186/1741-7007-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shamay M, Greenway M, Liao G, Ambinder RF, Hayward SD. De novo DNA methyltransferase DNMT3b interacts with NEDD8-modified proteins. J Biol Chem. 2010;285:36377–36386. doi: 10.1074/jbc.M110.155721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Antenos M, Casper RF, Brown TJ. Interaction with Nedd8, a ubiquitin-like protein, enhances the transcriptional activity of the aryl hydrocarbon receptor. J Biol Chem. 2002;277:44028–44034. doi: 10.1074/jbc.M202413200. [DOI] [PubMed] [Google Scholar]
- 95.Kelsall IR, et al. TRIAD1 and HHARI bind to and are activated by distinct neddylated Cullin-RING ligase complexes. EMBO J. 2013;32:2848–2860. doi: 10.1038/emboj.2013.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tanaka T, Kawashima H, Yeh ETH, Kamitani T. Regulation of the NEDD8 conjugation system by a splicing variant, NUB1L. J Biol Chem. 2003;278:32905–32913. doi: 10.1074/jbc.M212057200. [DOI] [PubMed] [Google Scholar]
- 97.Bohnsack RN, Haas AL. Conservation in the mechanism of Nedd8 activation by the human AppBp1-Uba3 heterodimer. J Biol Chem. 2003;278:26823–26830. doi: 10.1074/jbc.M303177200. [DOI] [PubMed] [Google Scholar]
- 98.Burch TJ, Haas AL. Site-Directed Mutagenesis of Ubiquitin. Differential Roles for Arginine in the Interaction with Ubiquitin-Activating Enzyme. Biochemistry. 1994;33:7300–7308. doi: 10.1021/bi00189a035. [DOI] [PubMed] [Google Scholar]
- 99.Walden H, Podgorski MS, Schulman BA. Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature. 2003;422:330–334. doi: 10.1038/nature01456. [DOI] [PubMed] [Google Scholar]
- 100.Hjerpe R, et al. Changes in the ratio of free NEDD8 to ubiquitin triggers NEDDylation by ubiquitin enzymes. Biochem J. 2012;441:927–936. doi: 10.1042/BJ20111671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Soucy TA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. Development and characterization of a specific NAE inhibitor. [DOI] [PubMed] [Google Scholar]
- 103.Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yamoah K, et al. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1’s C-terminal tail. Proc Natl Acad Sci USA. 2008;105:12230–12235. doi: 10.1073/pnas.0806155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Boh BK, Smith PG, Hagen T. Neddylation-induced conformational control regulates cullin RING ligase activity in vivo. J Mol Biol. 2011;409:136–145. doi: 10.1016/j.jmb.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 106.Pierce NW, et al. Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins. Cell. 2013;153:206–215. doi: 10.1016/j.cell.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zemla A, et al. CSN- and CAND1-dependent remodelling of the budding yeast SCF complex. Nat Commun. 2013;4:1641. doi: 10.1038/ncomms2628. [DOI] [PubMed] [Google Scholar]
- 108.Wu S, et al. CAND1 controls in vivo dynamics of the cullin 1-RING ubiquitin ligase repertoire. Nat Commun. 2013;4:1642. doi: 10.1038/ncomms2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lydeard JR, Schulman BA, Harper JW. Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep. 2013;14:1050–1061. doi: 10.1038/embor.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Scheffner M, Kumar S. Mammalian HECT ubiquitin-protein ligases: biological and pathophysiological aspects. Biochim Biophys Acta. 2014;1843:61–74. doi: 10.1016/j.bbamcr.2013.03.024. [DOI] [PubMed] [Google Scholar]
- 111.Xie P, et al. The covalent modifier Nedd8 is critical for the activation of Smurf1 ubiquitin ligase in tumorigenesis. Nat Commun. 2014;5:3733. doi: 10.1038/ncomms4733. Functional characterization of the neddylation of a HECT E3 ligase. [DOI] [PubMed] [Google Scholar]
- 112.Kaelin WG., Jr von Hippel-Lindau Disease. Annu Rev Pathol Mech Dis. 2007;2:145–173. doi: 10.1146/annurev.pathol.2.010506.092049. [DOI] [PubMed] [Google Scholar]
- 113.Stickle NH, et al. pVHL modification by NEDD8 is required for fibronectin matrix assembly and suppression of tumor development. Mol Cell Biol. 2004;24:3251–3261. doi: 10.1128/MCB.24.8.3251-3261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Russell RC, Ohh M. NEDD8 acts as a ‘molecular switch’ defining the functional selectivity of VHL. EMBO Rep. 2008;9:486–491. doi: 10.1038/embor.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Trempe JF, et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340:1451–1455. doi: 10.1126/science.1237908. [DOI] [PubMed] [Google Scholar]
- 116.Deas E, Wood NW, Plun-Favreau H. Mitophagy and Parkinson’s disease: The PINK1–parkin link. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2011;1813:623–633. doi: 10.1016/j.bbamcr.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Choo YS, et al. Regulation of parkin and PINK1 by neddylation. Hum Mol Genet. 2012;21:2514–2523. doi: 10.1093/hmg/dds070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Um JW, et al. Neddylation positively regulates the ubiquitin E3 ligase activity of parkin. J Neurosci Res. 2012;90:1030–1042. doi: 10.1002/jnr.22828. [DOI] [PubMed] [Google Scholar]
- 119.Dil Kuazi A, et al. NEDD8 protein is involved in ubiquitinated inclusion bodies. J Pathol. 2003;199:259–266. doi: 10.1002/path.1283. [DOI] [PubMed] [Google Scholar]
- 120.Mori F, et al. Accumulation of NEDD8 in neuronal and glial inclusions of neurodegenerative disorders. Neuropathol Appl Neurobiol. 2005;31:53–61. doi: 10.1111/j.1365-2990.2004.00603.x. [DOI] [PubMed] [Google Scholar]
- 121.Mori F, et al. Ubiquitin-related proteins in neuronal and glial intranuclear inclusions in intranuclear inclusion body disease. Pathol Int. 2012;62:407–411. doi: 10.1111/j.1440-1827.2012.02812.x. [DOI] [PubMed] [Google Scholar]
- 122.Odagiri S, et al. Immunohistochemical analysis of Marinesco bodies, using antibodies against proteins implicated in the ubiquitin-proteasome system, autophagy and aggresome formation. Neuropathology. 2012;32:261–266. doi: 10.1111/j.1440-1789.2011.01267.x. [DOI] [PubMed] [Google Scholar]
- 123.Chen Y, Liu W, McPhie DL, Hassinger L, Neve RL. APP-BP1 mediates APP-induced apoptosis and DNA synthesis and is increased in Alzheimer’s disease brain. J Cell Biol. 2003;163:27–33. doi: 10.1083/jcb.200304003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 125.Xirodimas DP, Saville MK, Bourdon J-C, Hay RT, Lane DP. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 2004;118:83–97. doi: 10.1016/j.cell.2004.06.016. TP53 transcriptional activity can be regulated by neddylation. [DOI] [PubMed] [Google Scholar]
- 126.Dohmesen C, Koeppel M, Dobbelstein M. Specific inhibition of Mdm2-mediated neddylation by Tip60. Cell Cycle. 2008;7:222–231. doi: 10.4161/cc.7.2.5185. [DOI] [PubMed] [Google Scholar]
- 127.Liu G, Xirodimas DP. NUB1 promotes cytoplasmic localization of p53 through cooperation of the NEDD8 and ubiquitin pathways. Oncogene. 2010;29:2252–2261. doi: 10.1038/onc.2009.494. [DOI] [PubMed] [Google Scholar]
- 128.Carter S, Bischof O, Dejean A, Vousden KH. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat Cell Biol. 2007;9:428–435. doi: 10.1038/ncb1562. [DOI] [PubMed] [Google Scholar]
- 129.Watson IR, Blanch A, Lin DCC, Ohh M, Irwin MS. Mdm2-mediated NEDD8 modification of TAp73 regulates its transactivation function. J Biol Chem. 2006;281:34096–34103. doi: 10.1074/jbc.M603654200. [DOI] [PubMed] [Google Scholar]
- 130.Abida WM, Nikolaev A, Zhao W, Zhang W, Gu W. FBXO11 promotes the Neddylation of p53 and inhibits its transcriptional activity. J Biol Chem. 2007;282:1797–1804. doi: 10.1074/jbc.M609001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.van den Heuvel S, Dyson NJ. Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol. 2008;9:713–724. doi: 10.1038/nrm2469. [DOI] [PubMed] [Google Scholar]
- 132.Aoki I, Higuchi M, Gotoh Y. NEDDylation controls the target specificity of E2F1 and apoptosis induction. Oncogene. 2013;32:3954–3964. doi: 10.1038/onc.2012.428. [DOI] [PubMed] [Google Scholar]
- 133.Loftus SJ, Liu G, Carr SM, Munro S, La Thangue NB. NEDDylation regulates E2F-1-dependent transcription. EMBO Rep. 2012;13:811–818. doi: 10.1038/embor.2012.113. Negative regulation of E2F transcription by neddylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Watson IR, et al. Chemotherapy induces NEDP1-mediated destabilization of MDM2. Oncogene. 2010;29:297–304. doi: 10.1038/onc.2009.314. [DOI] [PubMed] [Google Scholar]
- 135.Kontaki H, Talianidis I. Lysine methylation regulates E2F1-induced cell death. Mol Cell. 2010;39:152–160. doi: 10.1016/j.molcel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 136.Hayden MS, Ghosh S. NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Amir RE, Iwai K, Ciechanover A. The NEDD8 pathway is essential for SCF(beta -TrCP)-mediated ubiquitination and processing of the NF-kappa B precursor p105. J Biol Chem. 2002;277:23253–23259. doi: 10.1074/jbc.M200967200. [DOI] [PubMed] [Google Scholar]
- 138.Noguchi K, et al. TRIM40 promotes neddylation of IKKγ and is downregulated in gastrointestinal cancers. Carcinogenesis. 2011;32:995–1004. doi: 10.1093/carcin/bgr068. [DOI] [PubMed] [Google Scholar]
- 139.Gao F, Cheng J, Shi T, Yeh ETH. Neddylation of a breast cancer-associated protein recruits a class III histone deacetylase that represses NFkappaB-dependent transcription. Nat Cell Biol. 2006;8:1171–1177. doi: 10.1038/ncb1483. [DOI] [PubMed] [Google Scholar]
- 140.Gao N, Asamitsu K, Hibi Y, Ueno T, Okamoto T. AKIP1 enhances NF-kappaB-dependent gene expression by promoting the nuclear retention and phosphorylation of p65. J Biol Chem. 2008;283:7834–7843. doi: 10.1074/jbc.M710285200. [DOI] [PubMed] [Google Scholar]
- 141.Takashima O, et al. Brap2 regulates temporal control of NF-κB localization mediated by inflammatory response. PLoS ONE. 2013;8:e58911. doi: 10.1371/journal.pone.0058911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cao X, Südhof TC. A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- 143.Lee MR, Lee D, Shin SK, Kim YH, Choi CY. Inhibition of APP intracellular domain (AICD) transcriptional activity via covalent conjugation with Nedd8. Biochem Biophys Res Commun. 2008;366:976–981. doi: 10.1016/j.bbrc.2007.12.066. [DOI] [PubMed] [Google Scholar]
- 144.Chen Y, Neve RL, Liu H. Neddylation dysfunction in Alzheimer’s disease. J Cell Mol Med. 2012;16:2583–2591. doi: 10.1111/j.1582-4934.2012.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ikushima H, Miyazono K. TGFbeta signalling: a complex web in cancer progression. Nat Rev Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- 146.Zuo W, et al. c-Cbl-mediated neddylation antagonizes ubiquitination and degradation of the TGF-β type II receptor. Mol Cell. 2013;49:499–510. doi: 10.1016/j.molcel.2012.12.002. Neddylation could regulate protein stability through endocytosis- lysosomal pathway. [DOI] [PubMed] [Google Scholar]
- 147.Marmor MD, Yarden Y. Role of protein ubiquitylation in regulating endocytosis of receptor tyrosine kinases. Oncogene. 2004;23:2057–2070. doi: 10.1038/sj.onc.1207390. [DOI] [PubMed] [Google Scholar]
- 148.Nagano T, Hashimoto T, Nakashima A, Kikkawa U, Kamada S. X-linked inhibitor of apoptosis protein mediates neddylation by itself but does not function as a NEDD8-E3 ligase for caspase-7. FEBS Lett. 2012;586:1612–1616. doi: 10.1016/j.febslet.2012.04.056. [DOI] [PubMed] [Google Scholar]
- 149.Teng T, Thomas G, Mercer CA. Growth control and ribosomopathies. Curr Opin Genet Dev. 2013;23:63–71. doi: 10.1016/j.gde.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 150.Sundqvist A, Liu G, Mirsaliotis A, Xirodimas DP. Regulation of nucleolar signalling to p53 through NEDDylation of L11. EMBO Rep. 2009;10:1132–1139. doi: 10.1038/embor.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mahata B, Sundqvist A, Xirodimas DP. Recruitment of RPL11 at promoter sites of p53-regulated genes upon nucleolar stress through NEDD8 and in an Mdm2-dependent manner. Oncogene. 2012;31:3060–3071. doi: 10.1038/onc.2011.482. [DOI] [PubMed] [Google Scholar]
- 152.Zhang J, Bai D, Ma X, Guan J, Zheng X. hCINAP is a novel regulator of ribosomal protein-HDM2-p53 pathway by controlling NEDDylation of ribosomal protein S14. Oncogene. 2014;33:246–254. doi: 10.1038/onc.2012.560. [DOI] [PubMed] [Google Scholar]
- 153.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bian Y, et al. An enzyme assisted RP-RPLC approach for in-depth analysis of human liver phosphoproteome. Journal of Proteomics. 2014;96:253–262. doi: 10.1016/j.jprot.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 155.Kettenbach AN, et al. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci Signal. 2011;4:rs5. doi: 10.1126/scisignal.2001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Weinert BT, et al. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013;4:842–851. doi: 10.1016/j.celrep.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 157.Rigbolt KTG, et al. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal. 2011;4:rs3. doi: 10.1126/scisignal.2001570. [DOI] [PubMed] [Google Scholar]
- 158.Rabut G, Peter M. Function and regulation of protein neddylation. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:969–976. doi: 10.1038/embor.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jia J, et al. Mechanisms of drug combinations: interaction and network perspectives. Nat Rev Drug Discov. 2009;8:111–128. doi: 10.1038/nrd2683. [DOI] [PubMed] [Google Scholar]
- 162.Melchor L, et al. Comprehensive characterization of the DNA amplification at 13q34 in human breast cancer reveals TFDP1 and CUL4A as likely candidate target genes. Breast Cancer Res. 2009;11:R86. doi: 10.1186/bcr2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 164.Chang FM, et al. Inhibition of neddylation represses lipopolysaccharide-induced proinflammatory cytokine production in macrophage cells. J Biol Chem. 2012;287:35756–35767. doi: 10.1074/jbc.M112.397703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mathewson N, et al. Neddylation plays an important role in the regulation of murine and human dendritic cell function. Blood. 2013;122:2062–2073. doi: 10.1182/blood-2013-02-486373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Nawrocki ST, Griffin P, Kelly KR, Carew JS. MLN4924: a novel first-in-class inhibitor of NEDD8-activating enzyme for cancer therapy. Expert Opin Investig Drugs. 2012;21:1563–1573. doi: 10.1517/13543784.2012.707192. [DOI] [PubMed] [Google Scholar]
- 167.Brownell JE, et al. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell. 2010;37:102–111. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 168.Lin JJ, Milhollen MA, Smith PG, Narayanan U, Dutta A. NEDD8-targeting drug MLN4924 elicits DNA rereplication by stabilizing Cdt1 in S phase, triggering checkpoint activation, apoptosis, and senescence in cancer cells. Cancer Res. 2010;70:10310–10320. doi: 10.1158/0008-5472.CAN-10-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Milhollen MA, et al. Inhibition of NEDD8-activating enzyme induces rereplication and apoptosis in human tumor cells consistent with deregulating CDT1 turnover. Cancer Res. 2011;71:3042–3051. doi: 10.1158/0008-5472.CAN-10-2122. [DOI] [PubMed] [Google Scholar]
- 170.Embade N, et al. Murine double minute 2 regulates Hu antigen R stability in human liver and colon cancer through NEDDylation. Hepatology. 2012;55:1237–1248. doi: 10.1002/hep.24795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.McLarnon A. Cancer: Mdm2-regulated stabilization of HuR by neddylation in HCC and colon cancer--a possible target for therapy. Nat Rev Gastroenterol Hepatol. 2012;9:4. doi: 10.1038/nrgastro.2011.241. [DOI] [PubMed] [Google Scholar]
- 172.Ryu JH, et al. Hypoxia-inducible factor α subunit stabilization by NEDD8 conjugation is reactive oxygen species-dependent. J Biol Chem. 2011;286:6963–6970. doi: 10.1074/jbc.M110.188706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Artavanis-Tsakonas K, et al. Characterization and structural studies of the Plasmodium falciparum ubiquitin and Nedd8 hydrolase UCHL3. J Biol Chem. 2010;285:6857–6866. doi: 10.1074/jbc.M109.072405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21:873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 175.Sakata E, et al. Direct interactions between NEDD8 and ubiquitin E2 conjugating enzymes upregulate cullin-based E3 ligase activity. Nat Struct Mol Biol. 2007;14:167–168. doi: 10.1038/nsmb1191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.