Summary
The heterotetrameric Ndc80 complex establishes connectivity along the principal longitudinal axis of a kinetochore. Its two heterodimeric subcomplexes, each with a globular end and a coiled-coil shaft, connect end-to-end to create a ∼600 Å long rod spanning the gap from centromere-proximal structures to spindle microtubules. Neither subcomplex has a known function on its own, but the heterotetrameric organization and the characteristics of the junction are conserved from yeast to man. We have determined crystal structures of two shortened (“dwarf”) Ndc80 complexes that contain the full tetramer junction and both globular ends. The junction connects two α-helical coiled coils through regions of four-chain and three-chain overlap. The complexity of its structure depends on interactions among conserved amino-acid residues, suggesting a binding site for additional cellular factor(s) not yet identified.
Keywords: cell division, kinetochore structure, kinetochore assembly, coiled-coil junction, x-ray crystallography
Graphical abstract
Introduction
Kinetochores bridge centromeric DNA and spindle microtubules (MTs). They contain more than 50 distinct components in budding yeast and an even larger number in multi-cellular organisms, most associated into well-defined subassemblies (De Wulf et al., 2003; Santaguida and Musacchio, 2009). The heterotetrameric Ndc80 complex (Ndc80c), composed of Ndc80 (Hec1 in human cells), Nuf2, Spc24 and Spc25, contacts MTs (Cheeseman et al., 2006; DeLuca et al., 2006; Janke et al., 2001; Wei et al., 2007); its regulated interactions, with MTs and other kinetochore subassemblies, are the ultimate targets of the elaborate set of controls known as the spindle-assembly checkpoint (SAC) (Foley and Kapoor, 2013; Musacchio, 2011). The four Ndc80c subunits associate as two, independently stable, rod-like heterodimers (Ndc80:Nuf2 and Spc24:Spc25). The resulting complex is approximately 600 Å in length, with a parallel, α-helical, coiled-coil shaft and globular extremities (Ciferri et al., 2008; Wei et al., 2005). The globular regions are N-terminal to a long coiled-coil segment in Ndc80 and Nuf2 and C-terminal to a somewhat shorter coiled-coil segment in Spc24 and Spc25 (Wei et al., 2005). The coiled-coils of the two subassemblies join end-to-end; both parts of the shaft therefore have the same N-to-C polarity (Figure 1A).
Figure 1.
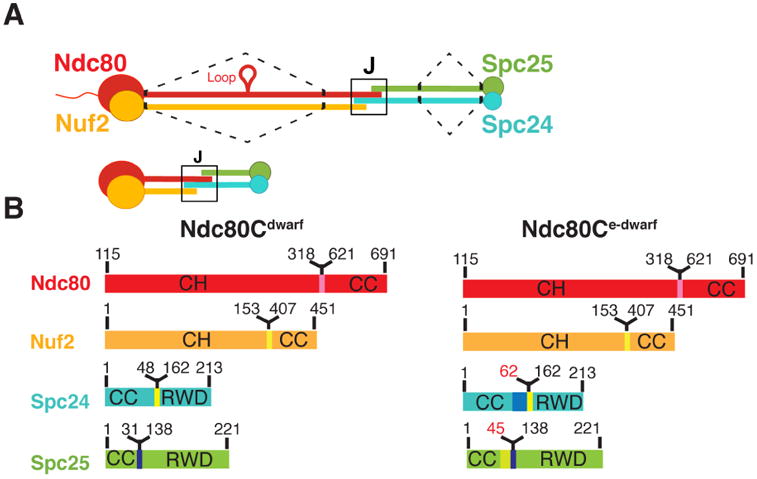
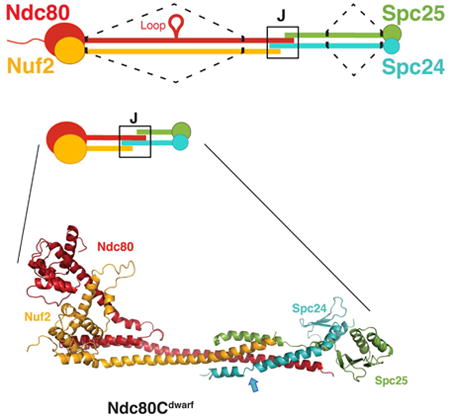
Ndc80c. (A) Schematic of Ndc80c. Upper diagram: Ndc80c was shortened by joining the globular ends of each heterodimer (Ndc80:Nuf2 or Spc24:Spc25) to the beginning of a heptad in the CC region, as illustrated by the dashed line. Lower diagram: The shortened Ndc80c retains the junction (J). Color code, used in all figures: Ndc80, red, Nuf2, orange, Spc24, green, Spc25, blue. (B) Polypeptide chains of Ndc80cdwarf (left) and Ndc80ce-dwarf (right). Numbers indicate boundaries determined from the crystal structures. The two additional heptads in Scp24e-dwarf and Spc25e-dwarf are in blue and yellow, respectively. Polypeptide chains of Ndc80ce-dwarf and Ndc80dwarf are otherwise identical. CH: calponin homology domains; RWD: RWD domains; CC: coiled-coil regions, including residues in junction.
The Ndc80 and Nuf2 globular regions are calponin-homology (CH) domains -- known MT-binding modules (Alushin et al., 2010; Ciferri et al., 2008; Wei et al., 2007). An approximately 100-residue, flexible, N-terminal segment of Ndc80 also participates in the Ndc80:MT interface (Alushin et al., 2012; Zaytsev et al., 2015). The coiled-coil forming segments appended to the C-terminal end of each CH domain pair to form about two-thirds of the shaft of Ndc80c; the coiled-coil of Spc24:Spc25 makes up the rest. The Spc24 and Spc25 globular regions at the C-termini of each chain are RWD domains, a module found in several other kinetochore proteins (Corbett et al., 2010; Kim et al., 2012; Petrovic et al., 2014; Schmitzberger and Harrison, 2012). The tightly associated RWD domains of Spc24 and Spc25 associate with the MIND (Mis12/Mtw1) complex, another conserved heterotetrameric kinetochore component.
The length of the Ndc80c shaft, measured from electron micrographs, and the lengths of the corresponding polypeptide chains, from the C-termini of CH-domains to the N-termini of the RWD domains, are consistent with an essentially continuous, α-helical coiled-coil, with only modest overlap where the two coiled-coils abut -- a structure we refer to here as the “junction” (Wei et al., 2005). Data from cross-linking mass spectrometry and analysis of heptad repeats in the amino-acid sequences of Ndc80 and Nuf2 suggest only one other specialized region between the paired CH domains of Ndc80 and Nuf2 and the paired RWD domains of Spc24 and Spc25 -- the “loop”, a ∼50-residue segment in Ndc80 midway between its CH domain and its C-terminus (Maiolica et al., 2007). The consequences of deleting it indicate that it interacts with the DASH-Dam1 complex (Maure et al., 2011). Images of negatively stained or rotary-shadowed Ndc80c molecules show no obvious globular specializations along the shaft, but images of negatively stained Ndc80c do show a relatively consistent kink about 160 Å from the Ndc80:Nuf2 end (Wang et al., 2008; Wei et al., 2005).
We know the structures of both globular ends of Ndc80c from x-ray crystallographic analyses of a C-terminal fragment of the Spc24:Spc25 heterodimer (Wei et al., 2006), an isolated Ndc80 CH domain (Wei et al., 2007), and a “designer protein”, called Ndc80cbonsai, that links (through a short shaft) the globular end of human Ndc80(Hec1):Nuf2 with the globular end of human Spc24:Spc25 (Ciferri et al., 2008). The design of Ndc80cbonsai fuses the first 286 residues of human Ndc80(Hec1) with the last 107 residues of Spc25 and the first 169 of Nuf2 with the last 76 of Spc24. One end binds MTs with presumably native contacts (Alushin et al., 2010; Ciferri et al., 2008); the other end binds MIND, also with presumably native contacts (Malvezzi et al., 2013; Petrovic et al., 2014). Why has evolution of the full-length protein never adopted a similar strategy? The four proteins function together in kinetochores, and we know of no separate role for either heterodimer. Yet the Ndc80c junction is a conserved joint in all eukaryotes (Janke et al., 2001; McCleland et al., 2004; Wei et al., 2005), suggesting some functional role beyond merely cementing the two halves of the complex together and posing the following questions. How do two α-helical coiled-coils join end-to-end firmly and rigidly? Can we build a more complete model of Ndc80c, by interpolating between the two globular extremities and the junction in the center, with heptad pairing constrained by cross-linking data? What hypotheses might explain why Ndc80c is always a heterotetramer?
To study the structure of the Ndc80c junction, we prepared constructs that remove central parts of the coiled-coil in both heterodimers, preserving the heptad registers while leaving the globular ends and the tetramerization region intact. We crystallized two such shortened complexes, which differed only in the number of residues between the junction and the deletions in Spc24 and Spc25, and determined their structures -- at 2.8 Å resolution for the shorter complex and at lower resolution (about 7.5 Å) for the longer one. The junction organizes the transition from one coiled coil to another through successive regions of four-chain overlap and three-chain overlap. The complexity of its structure and the conservation (among fungi) of particular features suggest that the Ndc80c junction interacts with additional factors that remain to be identified. In the structure of the longer of the two complexes, the junction includes the epitope for a single domain antibody (nanobody) included to assist crystallization. Thus, the exposed surface of the tetramer junction has sufficient specificity to bind a potential cellular factor. Comparison of the two shortened complexes shows a hinge between the globular heads of Ndc80:Nuf2 and the coiled-coil shaft that may facilitate successive binding and unbinding of the globular heads to MTs.
Results
Shortened Ndc80c tetramers
From published cross-linking data (Maiolica et al., 2007) and analysis of heptad repeats, we devised a strategy for excising the central shafts of Ndc80:Nuf2 and Spc24:Spc25, while keeping the register of their coiled-coils and retaining the domains at their ends (Figure 1). We fused the globular end of Ndc80 to a heptad C-terminal to the loop and the globular end of Nuf2 to the corresponding heptad on the apposed helix; we took a similar approach for Spc24:Spc25. We co-expressed various possible pairs of heterodimers in bacteria, to ask which combinations of shortened Ndc80:Nuf2 and Spc24:Spc25 heterodimers produced stable heterotetramers (Figure S1A). We assayed for tetramerization and stability by gel filtration and analytical ultracentrifugation or multi-angle light scattering and confirmed by negative-stain electron microscopy that the fusion had introduced no obvious distortions (data not shown). To check our assignment of the heptad register in the coiled-coil region, we made a short version of Ndc80:Nuf2 with an alternative heptad register, also compatible with cross linking data, in which the head of Nuf2 was fused to a heptad upstream of the equivalent heptad in Ndc80. This alternative heptad register yielded unstable products (Figure S1: Ndc80:Nuf2 deletant designated by Δ1°).
Structures of two shortened Ndc80 complexes
We determined the structures of two shortened versions of the Ndc80 complex from S. cerevisiae (Figure 2). By analogy with the “bonsai” Ndc80 complex (Ndc80cbonsai), we call these “dwarf Ndc80 complex” (Ndc80cdwarf) and “extended-dwarf Ndc80 complex” (Ndc80ce-dwarf), respectively. The protein compositions of the two differ from each other by only two heptads in the coiled-coil region of Scp24:Spc25. Crystals of Ndc80cdwarf gave measurable diffraction to a minimum Bragg spacing of 2.8 Å. We determined phases from single wavelength anomalous scattering by a selenomethionine substituted complex. The asymmetric unit contained one tetramer, approximately 150 Å long (Figure 2A). A residual N-terminal tag on Nuf2, left over from affinity purification and TEV cleavage, appeared to have determined crystal packing by contacting Spc24:Spc25 of a neighboring tetramer across a crystallographic dyad.
Figure 2.
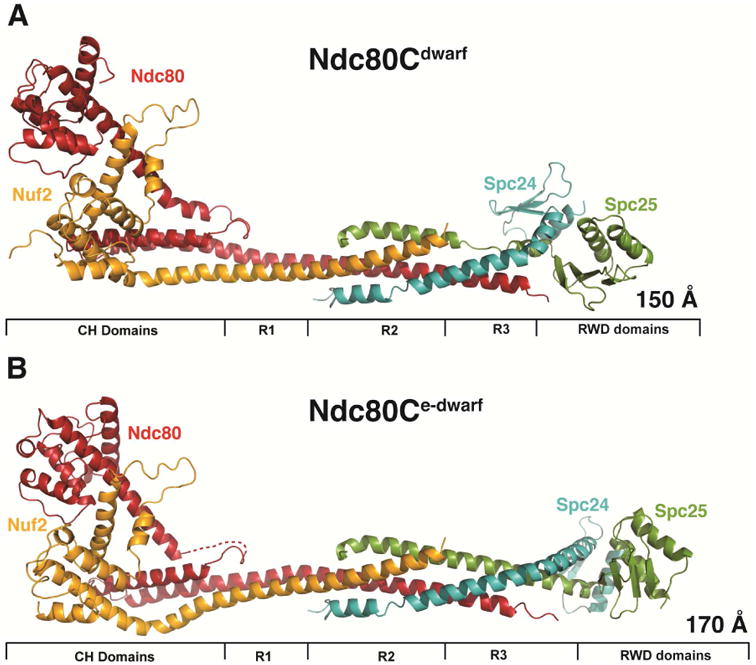
Crystal structures of Ndc80cdwarf and Ndc80ce-dwarf. (A) Ndc80cdwarf. Paired CH domains of Ndc80 and Nuf2 on the left; paired RWD domains on the right; tetramer junction in the middle. R1: region of two-chain coiled coil. R2: region of four-chain overlap. R3: region of three-chain overlap. (B) Ndc80ce-dwarf. Labels as in (A). Dashed line: Ndc80 residues 250-259, not resolved in electron density map. The structure was determined with a bound, single-chain antibody, omitted for clarity from the figure. See also Figure S2.
Because of our initial choice of excision points within the Spc24:Spc25 rod, a C-terminal segment of Ndc80, which extends beyond the tetramerization structure, reaches the Spc25 RWD domain. To ensure that we had not missed contacts of the C-terminal residues of Ndc80 with the Spc24:Spc25 coiled coil, we also determined the structure of Ndc80ce-dwarf, in which Scp24 and Spc25 each have two additional heptads of native sequence on the N-terminal side of the fusion (Figure 2B). The Ndc80:Nuf2 component of Ndc80ce-dwarf is the same as in the dwarf construct. We chose this way to extend the construct, because a disordered region immediately precedes the RWD domains of both Spc24 and Spc25 (see further discussion below) (Wei et al., 2006).
Ndc80ce-dwarf crystallized with assistance of an alpaca nanobody, isolated following immunization with the deletant combinding Δ1 and ΔA constructs (Figure S1). The crystals yielded measurable intensities to a minimum Bragg spacing of 7.5 Å. Failing to find a satisfactory molecular replacement solution with Ndc80cdwarf as a search model, we recorded anomalous diffraction data from crystals soaked in ethyl mercuric phosphate and potassium tetrachloroplatinate(II), from which we obtained excellent experimental maps. We adjusted the structure of Ndc80cdwarf to fit the density and built the two-heptad extension in the coiled-coil region of Scp24 and Spc25 using the constellation of mercury and platinum atoms as landmarks to guide placement of the polypeptide chains. There was continuous electron density for the tetramer in the solvent-flattened experimental maps, except for the unstructured loop in the Nuf2 head and residues 247 - 261 of Ndc80. We prepared a homology model of the nanobody and fit it into the map. The nanobody contacts the tetramerization region -- in particular, the C-terminal helical segment of Ndc80 and the N-terminal helix of Spc24 (Figure S2A). Qualitative binding studies with various constructs are consistent with this interpretation (Figure S2A,B), but the resolution of the map does not allow us to describe the interaction in detail (Figure S2C).
The globular ends and the tetramer junction in Ndc80ce-dwarf are essentially the same as in the shorter construct. As expected from the design, the contacts between Ndc80 and Spc25 seen in Ndc80cdwarf are absent in the extended structure. A kink in the heterodimeric rod leading into the RWD domains of Spc24 and Spc25 is present in both (Figure S3A); it is a consequence of the choice of deletion points in the two chains. The published crosslinking data were most consistent with the assumption that the heptad comprising residues 49 – 55 in Spc24 paired with the heptad comprising residues 32 – 38 in Spc25; the structure, in which formation of the junction dictated a correct pairing, showed that our assignment had added an extra six residues to Spc24; the kink that follows the fusion of N- and C-terminal heterodimeric fragments and the segment of Spc25 non-helical structure in the dwarf complex compensate for the extra residues. In the native complex, the RWD domains are likely to project axially, as in Ndc80cbonsai.
Structure of the junction of Ndc80:Nuf2 with Spc24:Spc25
The junction, where chains from both heterodimers interact, includes both a four-chain overlap (R2) and a three-chain overlap (R3)(Figure 3A). Leading into the junction, Ndc80 and Nuf2 chains pair in a regular coiled-coil (R1), except for an extra “stutter” residue at the N-terminal junction boundary. The four-chain overlap, which spans about 20 residues (three heptads), is a parallel four helix bundle with a solvent-inaccessible central axis characterized by van der Waals interactions among residues conserved in Ndc80c sequences from other budding yeast -- in particular, a stack of four aromatic side chains, one from each subunit (Figure 3B). At its N-terminal end, the four-helix bundle extends the heterodimeric coiled-coil interaction of Ndc80 and Nuf2, with the Spc24 and Spc25 helices packed into the grooves on either side. At its C-terminal end, the axes of the Ndc80 and Nuf2 helices diverge from each other, and the rod continues as a three-chain coiled coil (Ndc80, Spc24, Spc25) which extends about three heptads until the end of the Ndc80 helix.
Figure 3.
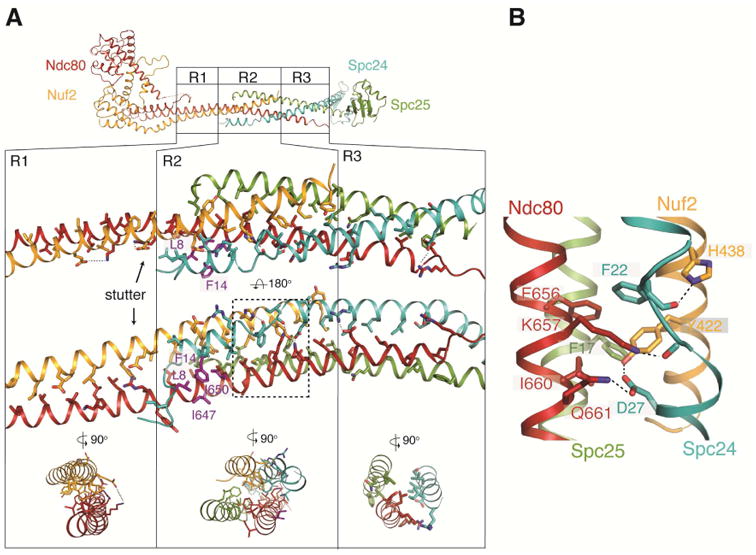
Central region of Ndc80cdwarf/e-dwarf. (A) Top: Diagram of Ndc80ce-dwarf; regions R1, R2 and R3 are identical in the Ndc80cdwarf and Ndc80e-dwarf structures (Cα rmsd = 0.8 Å). Middle: Expanded view of R1, R2 and R3. Arrow points to an additional, “stutter” residue in the third heptad in R1. Residues shown as purple sticks (Ndc80: I650 and I647; Spc24: L8 and F14) form a hydrophobic surface that contacts the nanobody (Figure S3). Bottom: cross section diagrams to show two-chain, four-chain, and three-chain packing in R1, R2 and R3, respectively. (B) Expanded view of a network of aromatic and ionic interactions, conserved in fungi. See also Figure S3.
In the middle of the junction, the Spc24 α-helix breaks and then resumes with three residues spanning an interval that would, in a continuous helix, require six (Figure 3A,B). A set of interactions, conserved in both budding and fission yeast, determines this interruption of the Spc24 helical path (Figure S3B). In particular, a hydrogen-bonding network, reinforced by a well-packed aromatic core, links the side chains of Asp27 (Spc24), Tyr442 (Nuf2), Lys657 (Ndc80) and Gln661 (Ndc80), and the main-chain carbonyl of Asp23 (Spc24) (Figure 3B).
The junction is also the nanobody epitope, which includes a conserved hydrophobic patch with contributions from Spc24 Leu8 and Phe14 and from Ndc80 Ile647 and Ile650 (Figure 3A). Although the nanobody is an exogenous ligand, this contact illustrates that the site has characteristics typical of protein-protein interaction surfaces. Conservation of both a unique interruption of otherwise continuous α-helix in all four chains and a patch of exposed hydrophobic residues suggests a functional contact point, perhaps for other kinetochore components.
We assessed contributions to stability of the junction by pairing Ndc80:Nuf2-dwarf with N-terminally truncated versions of Spc24:Spc25 and determining tetramer formation by size exclusion chromatography (Figure S4). Deleting up to 16 amino acid residues from the N-terminus of Spc24 did not affect tetramer formation; deleting to residue V18 of Spc24 weakened the tetramer interface, as shown by a shoulder on the tetramer peak and presence of a peak for unbound Spc24D18:Spc25. Deleting to residue F22, in the segment that interrupts the Spc24 helix, or mutating F22 to Pro eliminated the tetramer completely.
Ndc80:Nuf2 globular domains
The N-terminal “heads” of Ndc80 and Nuf2 are both calponin homology (CH) domains, compact clusters of six α-helices, about 100 residues in total polypeptide-chain length (Figure 4). The last helix of both domains (αG in Figure 4A,B and Figure S3C,D) is longer than in most other CH domains and connects to the principal coiled-coil of the complex, directly through a sharp turn in the case of Nuf2 but with a bend at the end of αG and an intervening helical hairpin (αH and αI) in the case of Ndc80.
Figure 4.
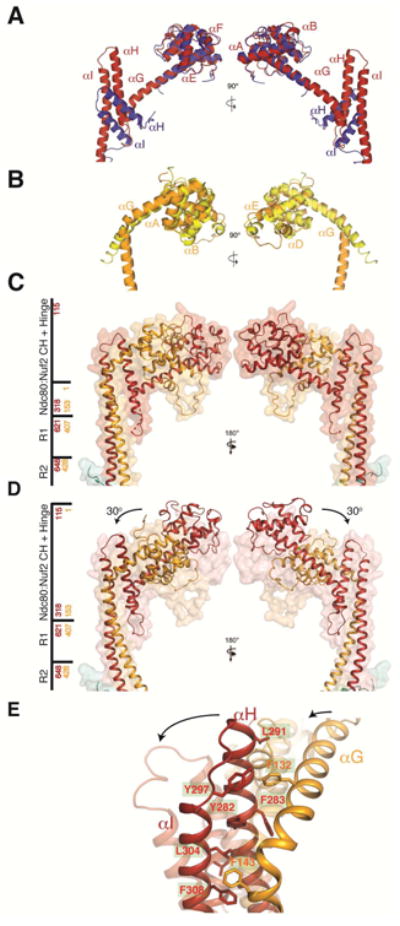
Comparison of yeast and human Ndc80 and Nuf2 CH domains and Ndc80c hinge. (A) Superposition of the Ndc80 CH domain from human (in blue; coordinates from chain B of PDB entry 2VE7) and yeast (in red, chain A of the Ndc80cdwarf crystal structure). (B) Superposition of Nuf2 CH domains from human (in yellow; chain D of PDB entry 2VE7) and yeast CH domain (in orange; chain B of the Ndc80cdwarf crystal structure). (C) Structure of CH domains and adjacent coiled coil from Ndc80ce-dwarf as both secondary-structure ribbons and transparent surface. Side bar marks regions of the structure; red and yellow numbers correspond to Ndc80 and Nuf2, respectively. (D) Structure of Ndc80cdwarf juxtaposed on transparent surface of Ndc80ce-dwarf. The arrows show the reorientation of the Ndc80:Nuf2 CH domains with respect to the coiled-coil axis. Structures aligned with respect to residues encompassing the R1 and R2 segments of the middle region with an RMSD of 0.8 Å2. (E) Fulcrum of hinge, showing cluster of aromatic residues. See also Figure S3E and Supplemental Movie 1.
The globular head of Ndc80c anchors it to a spindle MT. The CH domains in the yeast complex superpose well on their human counterparts (Figure 4C,D); the characteristics of their microtubule interface are therefore likely to be similar. Comparison of the Ndc80e-dwarf and Ndc80dwarf structures shows that the coiled-coil rod swings by about 30° with respect to the globular head (Figure 4C,D, Figure S3E, and Supplemental Movie 1). The intramolecular hinge point in both molecules is at the end of αG. The Ndc80e-dwarf and Ndc80dwarf constructs are identical at the Ndc80:Nuf2 end: crystal packing and a “soft” hinge must therefore account for the swing. A cluster of aromatic residues, conserved in budding yeasts, appears to create a “well-oiled” fulcrum for this hinge (Figure 4E). A capacity to swing by at least 30° may be a functionally relevant property of the full-length complex. (A further, somewhat smaller, packing induced bend in Ndc80ce-dwarf is at the point of fusion of the heads to the C-proximal shaft; because this response could be a property of the sequence discontinuity, the analysis here applies only to the hinge point within fully native sequence, at the C-terminal end of helix G.)
Discussion
The Ndc80cdwarf and Ndc80ce-dwarf structures augment our picture of the principal axial component of a kinetochore. First, they show conserved features in the tetramer junction between Ndc80:Nuf2 and Spc24:Spc25 that suggest it does more than simply hold the two heterodimers together. Second, by giving us a direct view of the overlap and coiled-coil interaction within the junction, they allow us to infer some characteristics of the shaft segments between the new structures and those already known from crystals of the Spc24:Scp25 heterodimer and from Ncd80Cbonsai. Third, they suggest a possible point of flexion where the Ndc80:Nuf2 globular regions connect with the coiled-coil shaft.
Tetramer junction
The tetramer junction has some of the features one might expect for an end-to-end joint between two α-helical coiled-coils of the same polarity -- particularly the way the two helical chains of one partner slide into the grooves on the surface of the helical pair in the other partner -- but its structure is more elaborate than required simply to oligomerize the complex. It lacks the symmetrical regularity of the Csm1-Lrs4 monopolin complex, for example, which creates a Y-shaped joint of three helical pairs rather than an end-to-end joint of two (Corbett et al., 2010). Three noteworthy features of the Ndc80c tetramer junction, all conserved among budding yeasts, are the non-polar surface contributing to the nanobody epitope, the aromatic side-chain stack at the center of the junction, and the network of polar contacts supporting the break in an otherwise continuous, α-helical conformation of Spc24. Sequence alignments suggest that similar features may also be present in mammalian Ndc80c.
Conservation of these structural characteristics correlates with the concentration of conserved residues in the tetramer junction. Moreover, Ndc80c is a heterotetramer in all eukaryotic species, and each of its two heterodimeric subcomplexes has essentially the same length and characteristics. The two heterodimeric components have no known independent functions, however, and the individual subunits are unstable on their own. These observations argue for recognition of the junction by some additional kinetochore-associated factor and potentially for regulation of Ndc80c assembly. The Spc24 truncation experiments also point to conserved residues on the N-terminal side of the junction that are not essential for tetramer formation but that might have a role in binding kinetochore factors. The only published candidate for an interaction partner other than known kinetochore structural components, such as Spc105 or Stu2, is Ybp2, which associates under the condition of affinity-capture assays with Ndc80, Nuf2 and Spc25, but not with Spc24, and which pulls down CEN DNA in chromatin immunoprecipitation experiments (Ohkuni et al., 2008). These observations suggest that Ybp2 could be an Ndc80 assembly chaperone, but there are no biochemical data from experiments with purified components.
Coiled-coil
Both sequence and mass per unit length calculations show that, except for the loop, the Ndc80:Nuf2 heterodimer is largely continuous coiled-coil from the point at which the heads join the shaft to the beginning of the tetramer junction. In Nuf2, a pattern of heptads with primarily non-polar residues at positions a and d continues throughout this 298-residue length of polypeptide chain, and the paired stretch of Ndc80 has a heptad pattern of comparable extent (assuming an extruded loop). Crosslinking data from the human ortholog are consistent with colinearity of the heptad repeats (Maiolica et al., 2007).
The Spc24:Spc25 sequences likewise show heptads extending from the junction toward the C-terminus, with an evident interruption N-terminal to the RWD domains. The latter observation is consistent with NMR spectra from N-terminally truncated Spc24:Scp25 heterodimers indicating unstructured backbone for residues 138–154 of Spc24 and 128–132 of Spc25 (Wei et al., 2006). This flexible connection allows the paired RWD domains to project in variable directions from the coiled-coil shaft. The Spc24 and Spc25 deletants in Ndc80ce-dwarf exclude about 10 heptads of coiled-coil in Spc24:Spc25 as well as the unstructured region. The mammalian and yeast Spc24 and Spc25 sequences diverge substantially, preventing useful correlation with the available crosslinking data.
Hinge at the Ndc80:Nuf2 globular head
The distinct relative orientations of the Ndc80:Nuf2 head with respect to its coiled-coil shaft, generated by different lattice contacts in our crystals of Ndc80dwarf and Ndc80e-dwarf, may reflect a functional role for CH domain flexion. Ndc80c appears to track along a depolymerizing microtubule because of preferential attachment to a lattice of straight, rather than curved protofilaments (Alushin et al., 2010). This postulated biased diffusion mechanism requires a local equilibrium, such that detachment of Ndc80c from the microtubule lattice allows one or more heterodimers of the underlying protofilaments to curl outwards followed by reattachment of Ndc80c to the adjacent straight zone. In budding yeast and in Candida albicans, both with just one microtubule attachment per centromere (Zhang et al., 2012), the DASH/Dam1C ring is an essential component for end-on microtubule attachment (Thakur and Sanyal, 2011). Several lines of evidence suggest that the Ndc80 loop, at about the midpoint in the coiled-coil shaft between the Ndc80:Nuf2 head and the heterotetramer junction, attaches Ndc80c to DASH/Dam1C in yeast (Maure et al., 2011) and to the Ska complex in mammalian cells (Zhang et al., 2012). If so, then some flexibility between the presumed anchor point on the ring and the globular head is necessary for the attachment/reattachment cycle postulated to accompany microtubule shortening. The angular shift we see when comparing the head-shaft connection in our two crystal structures could contribute to that flexibility.
Conclusion
We draw two principal conclusions from the two structures reported here. (1) The Ndc80c tetramer junction is an elaborately specialized, end-to-end connection between two heterodimeric coiled-coils with the same polarity. Successive regions of four-chain and three-chain overlap and irregular structural features with conserved amino-acid residues suggest conserved, functional interactions with other proteins, either to regulate assembly or to buttress kinetochore organization. (2) The position at which the CH-domain “heads” of Ndc80 and Nuf2 join the coiled-coil stalk has hinge-like characteristics. This flexibility imparts degrees of freedom likely to be required for dynamic end-on attachment during microtubule shortening and attendant sister-chromatid separation.
Experimental Procedures
Shortened versions of Ndc80, Nuf2, Spc24, and Spc25 (Figure S1) were cloned with a cleavable, N-terminal 6-His-tag. The proteins were co-expressed in E. coli strain Rosetta2 (DE3) pLys (Novagen), purified by Co2+ affinity, ion-exchange, and size exclusion chromatography (SEC), and crystallized (see Supplemental Experimental Procedures for details). Ndc80cdwarf at 15 mg/ml crystallized in space group C2221 (a=169.4 b=186.6, c=122.0) from 1.2 – 1.7 M magnesium sulfate, 0.1 M MES (2-(N-morpholino)ethanesulfonic acid), pH 6.0 – 6.7; the Ndc80ce-dwarf: nanobody complex (1:1 molar ratio) at 11 mg/ml crystallized in space group I4122 (a=226.282, 226.82, 237.26) from 12 – 16% polyethylene glycol 4,000, 0.1 M CHES (N-Cyclohexyl-2-aminoethanesulfonic acid), pH 9.0 – 9.4. Diffraction data were recorded from crystals cryopreserved in mother liquor supplemented with 25% glycerol on NE-CAT beamline 24-ID-C at the Advanced Photon Source. Phases were determined from anomalous scattering by Se atoms in crystals of selenomethionine substituted protein (Ndc80cdwarf) or by the Hg or Pt atoms in crystals soaked in ethyl mercuric phosphate or tetrachloroplatinate(II) (Ndc80ce-dwarf). For Ndc80cdwarf, iterative rounds of model building and density modification, starting with the map from single-wavelength anomalous dispersion phases (17 Se atoms), led to a model that could be refined at 2.8 Å resolution by standard methods (see Supplemental Experimental Procedures). For the Ndc80ce-dwarf structure, six Hg sites located from anomalous differences were used for single-wavelength anomalous dispersion phasing. A partial model from segments of the Ndc80cdwarf was placed in the initial map and adjusted to match the positions of cysteine residues and the Hg locations. Iterative rounds of model adjustment, density modification and refinement led to a model that was combined with one for most of the nanobody and further refined against the x-ray data to 7.5 Å. Pt sites from anomalous difference maps confirmed the placement of methionine residues. See Supplemental Experimental Procedures, for complete details of structure determination and refinement, and Table S1, for data collection and refinement statistics. Nanobodies isolated from an alpaca immunized with a shortened Ndc80c construct (Ndc80Δ1:Nuf2Δ1/Spc24ΔA:Spc25ΔA -- see Figure S1) were elicited and prepared as described in Supplemental Experimental Procedures.
Association of Ndc80:Nuf2 constructs with Spc24:Spc25 constructs was assessed by analytical SEC at 4 °C on Superdex-S200 Increase 10 /300 GL (GE Healthcare). Samples were mixed in a 1:1 molar ratio, equilibrated at room temperature for 30 minutes, and injected into the column in a final volume of 200 uL. The elution buffer was 300 mM NaCl, 30 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid), pH 7.6, and 1 mM tris(2-carboxyethyl)phosphine (TCEP). Fractions were collected and analyzed by SDS-PAGE.
Supplementary Material
Acknowledgments
We thank Y. Dimitrova, S. Jenni and S. Hinshaw for advice and comments on the manuscript, C. Shoemaker and J. Mukherjee (Tufts University Cummings Veterinary School) for assistance in alpaca immunization and H. Ploegh (Whitehead Institute for Biomedical Research) for sponsoring and directing nanobody production, supported by a NIH Director's Pioneer Award (to H. L. Ploegh). X-ray diffraction data were recorded at the Advanced Photon Source (APS) Northeastern Collaborative Access Team beam line 24-ID-C, funded by NIGMS P41 GM103403, with a Pilatus 6M detector funded by NIH-ORIP S10 RR029205. The APS is a U.S. Department of Energy (DOE) Office of Science User Facility, operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. S.C.H. is an Investigator in the Howard Hughes Medical Institute.
Footnotes
Accession Codes: Coordinates have been deposited in the Protein Data Bank under accession codes PDB 5TCS (Ndc80cdwarf) and 5TD8 (Ndc80ce-dwarf_Hg).
Author Contributions: R.V. and S.C.H designed experiments; R.V. conducted experiments, collected and analyzed diffraction data, with guidance from S.C.H.; J.I. isolated the nanobody; S.C.H and R.V. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alushin GM, Musinipally V, Matson D, Tooley J, Stukenberg PT, Nogales E. Multimodal microtubule binding by the Ndc80 kinetochore complex. Nat Struct Mol Biol. 2012;19:1161–1167. doi: 10.1038/nsmb.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin GM, Ramey VH, Pasqualato S, Ball DA, Grigorieff N, Musacchio A, Nogales E. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467:805–810. doi: 10.1038/nature09423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett KD, Yip CK, Ee LS, Walz T, Amon A, Harrison SC. The monopolin complex crosslinks kinetochore components to regulate chromosome-microtubule attachments. Cell. 2010;142:556–567. doi: 10.1016/j.cell.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wulf P, McAinsh AD, Sorger PK. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol. 2013;14:25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Ortiz J, Lechner J, Shevchenko A, Magiera MM, Schramm C, Schiebel E. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. Embo J. 2001;20:777–791. doi: 10.1093/emboj/20.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Sun H, Tomchick DR, Yu H, Luo X. Structure of human Mad1 C-terminal domain reveals its involvement in kinetochore targeting. Proc Natl Acad Sci U S A. 2012;109:6549–6554. doi: 10.1073/pnas.1118210109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiolica A, Cittaro D, Borsotti D, Sennels L, Ciferri C, Tarricone C, Musacchio A, Rappsilber J. Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Molecular & cellular proteomics : MCP. 2007;6:2200–2211. doi: 10.1074/mcp.M700274-MCP200. [DOI] [PubMed] [Google Scholar]
- Malvezzi F, Litos G, Schleiffer A, Heuck A, Mechtler K, Clausen T, Westermann S. A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors. Embo J. 2013;32:409–423. doi: 10.1038/emboj.2012.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maure JF, Komoto S, Oku Y, Mino A, Pasqualato S, Natsume K, Clayton L, Musacchio A, Tanaka TU. The Ndc80 loop region facilitates formation of kinetochore attachment to the dynamic microtubule plus end. Curr Biol. 2011;21:207–213. doi: 10.1016/j.cub.2010.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland ML, Kallio MJ, Barrett-Wilt GA, Kestner CA, Shabanowitz J, Hunt DF, Gorbsky GJ, Stukenberg PT. The vertebrate Ndc80 complex contains Spc24 and Spc25 homologs, which are required to establish and maintain kinetochore-microtubule attachment. Curr Biol. 2004;14:131–137. doi: 10.1016/j.cub.2003.12.058. [DOI] [PubMed] [Google Scholar]
- Musacchio A. Spindle assembly checkpoint: the third decade. Philos Trans R Soc Lond B Biol Sci. 2011;366:3595–3604. doi: 10.1098/rstb.2011.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuni K, Abdulle R, Tong AH, Boone C, Kitagawa K. Ybp2 associates with the central kinetochore of Saccharomyces cerevisiae and mediates proper mitotic progression. PLoS One. 2008;3:e1617. doi: 10.1371/journal.pone.0001617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic A, Mosalaganti S, Keller J, Mattiuzzo M, Overlack K, Krenn V, De Antoni A, Wohlgemuth S, Cecatiello V, Pasqualato S, et al. Modular assembly of RWD domains on the Mis12 complex underlies outer kinetochore organization. Mol Cell. 2014;53:591–605. doi: 10.1016/j.molcel.2014.01.019. [DOI] [PubMed] [Google Scholar]
- Santaguida S, Musacchio A. The life and miracles of kinetochores. Embo J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitzberger F, Harrison SC. RWD domain: a recurring module in kinetochore architecture shown by a Ctf19-Mcm21 complex structure. EMBO Rep. 2012;13:216–222. doi: 10.1038/embor.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur J, Sanyal K. The essentiality of the fungus-specific Dam1 complex is correlated with a one-kinetochore-one-microtubule interaction present throughout the cell cycle, independent of the nature of a centromere. Eukaryot Cell. 2011;10:1295–1305. doi: 10.1128/EC.05093-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HW, Long S, Ciferri C, Westermann S, Drubin D, Barnes G, Nogales E. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J Mol Biol. 2008;383:894–903. doi: 10.1016/j.jmb.2008.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- Wei RR, Schnell JR, Larsen NA, Sorger PK, Chou JJ, Harrison SC. Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure. 2006;14:1003–1009. doi: 10.1016/j.str.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci U S A. 2005;102:5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaytsev AV, Mick JE, Maslennikov E, Nikashin B, DeLuca JG, Grishchuk EL. Multisite phosphorylation of the NDC80 complex gradually tunes its microtubule-binding affinity. Mol Biol Cell. 2015;26:1829–1844. doi: 10.1091/mbc.E14-11-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Kelstrup CD, Hu XW, Kaas Hansen MJ, Singleton MR, Olsen JV, Nilsson J. The Ndc80 internal loop is required for recruitment of the Ska complex to establish end-on microtubule attachment to kinetochores. J Cell Sci. 2012;125:3243–3253. doi: 10.1242/jcs.104208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


