Abstract
Stomata are microscopic valves on plant surfaces that originated over 400 million years ago and facilitated the greening of Earth’s continents by permitting efficient shoot-atmosphere gas exchange and plant hydration1. However, the core genetic machinery regulating stomatal development in non-vascular land plants is poorly understood2–4 and their function has remained a matter of debate for a century5. Here, we show that genes encoding the two basic helix-loop-helix proteins PpSMF1 and PpSCRM1 in the moss Physcomitrella patens are orthologous to transcriptional regulators of stomatal development in the flowering plant Arabidopsis thaliana and essential for stomata formation in moss. Targeted knock-out P. patens mutants lacking either PpSMF1 or PpSCRM1 develop gametophytes indistinguishable from wild-type plants but mutant sporophytes lacking stomata. Protein-protein interaction assays reveal heterodimerisation between PpSMF1 and PpSCRM1 which, together with moss-angiosperm gene complementations6, suggests deep functional conservation of the heterodimeric SMF1 and SCRM1 unit required to activate transcription for moss stomatal development, as in A. thaliana7. Moreover, stomata-less sporophytes of ΔPpSMF1 and ΔPpSCRM1 mutants exhibited delayed dehiscence, implying stomata might have promoted dehiscence in the first complex land plant sporophytes.
Colonization of terrestrial environments by green plants approximately 500 million years ago (Ma) established the basis for the emergence of complex land-based ecosystems that fundamentally transformed the biogeochemical cycling of carbon, water and energy1,8. Fossils suggest stomata originated on the small leafless sporophytes of the earliest vascular land plants, such as Cooksonia, over 410 Ma, and predated the evolutionary appearance of leaves and roots9. Insight into the core developmental modules has emerged from studies on the evolution of roots10,11, shoots12, and land plant life cycles13,14. We know little, however, about the core regulatory genes governing the specialized differentiation of guard cells that formed stomatal pores in basal land plant lineages.
Here, we address the origin of stomata in land plants by elucidating the key genetic components controlling stomatal development in the moss Physcomitrella patens. Targeted molecular genetic studies with P. patens provide insight into the genetic toolkit adopted by early land plants because stomata evolved in the common ancestor of mosses and vascular plants15. P. patens belongs to an extant basal lineage of non-vascular land plants that develop stomata exclusively on the diploid sporophyte (Figures 1 a–c), although the major photosynthetic moss tissue is the haploid leafy gametophyte. Knowledge of the genetic controls on moss stomatal development is rudimentary2. In Arabidopsis, a representative of the dicot flowering plants, developmental stages leading to stomatal formation are controlled primarily by the action of three closely related Group Ia basic helix-loop-helix (bHLH) proteins (SPEECHLESS (SPCH), MUTE and FAMA)16. Each of these three bHLHs regulates a key successive step in stomatal lineage behaviour, and each requires heterodimerisation with either of the more broadly expressed Group IIIb bHLH proteins SCREAM1(SCRM1)/ICE1 or SCRM27,17. Evolutionary loss of stomatal bHLH developmental genes, including SPCH, MUTE, FAMA and SCRM2 orthologues, from the genome of the marine flowering plant eelgrass (Zostera marina) around 70–60 Ma ago correlates with a complete absence of stomata18.
Figure 1. The moss Physcomitrella patens genome encodes orthologues of the basic helix loop helix (bHLH) transcription factors regulating stomatal development in flowering plants.
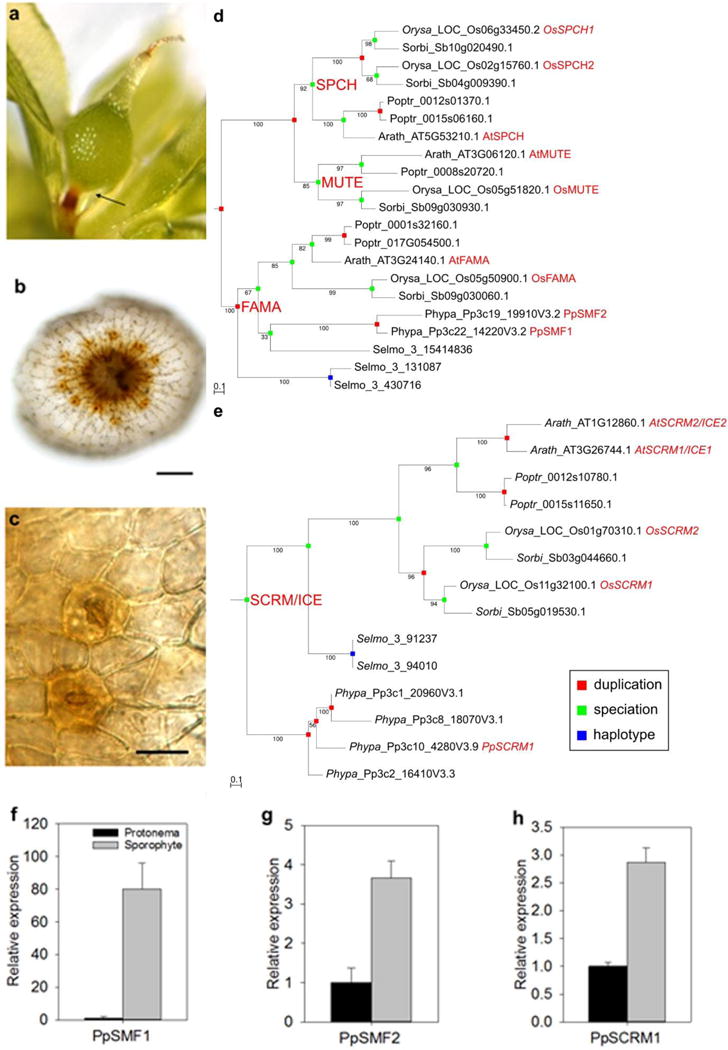
(a) Developing P. patens sporophyte, arrow indicating region of stomatal placement, and (b) excised sporophyte with stomata (orange/brown pores) forming a ring around the base. (c) Close-up of the sporophyte epidermis with single celled guard cells and central pores. (d and e) Bootstrapped Maximum Likelihood phylogenies of the SMF gene family comprising the FAMA, SPCH and MUTE subfamilies and the SCRM/ICE gene family in sequenced land plants. Internal node names in bold red indicate inferred subfamily ancestry. Internal nodes are coloured to indicate either duplication (red), speciation (green) or haplotype (blue) origin of the descendant nodes. Edge values represent bootstrap values. External node names comprise species abbreviations, original accession numbers of the protein sequences and accepted gene names of experimentally studied representatives in bold red. Species abbreviations in five-letter-code: Arabidopsis thaliana, Populus trichocarpa, Oryza sativa, Sorghum bicolor, Selaginella moellendorffii and Physcomitrella patens. (f, g and h) Relative expression of PpSMF1, PpSMF2 and PpSCRM1 in the developing sporophyte (grey bars) and protonema tissue (black bars) analysed by qRT-PCR. Error bars indicate standard error of the mean. Three replicates per tissue type were used. The scale bar in a = 100μm, in b = 100μm, in c = 25μm.
Phylogenetic analyses indicate that homologues of FAMA-like genes of Arabidopsis are found in lineages that diverged early in the evolution of land plants19. Group Ia genes have not been identified in the liverwort Marchantia polymorpha or in algae, both plant lineages lacking stomata, suggesting that Group Ia bHLHs are intimately linked to stomatal evolution. The P. patens genome harbours two Group Ia bHLH inparalogous genes, PpSMF1 and PpSMF26,19, and four SCRM1/SCRM2 Group 3b genes (PpSCRM1/Pp3c10_4260V3, Pp3c2_16410V3, Pp3c_20960V3 and Pp3c8_18070V3) (Figures 1 d,e). In line with a previous analysis with broader taxonomic sampling11, our phylogenetic inference robustly suggests that PpSMF1 and PpSMF2 are co-orthologous to AtFAMA which, in Arabidopsis, is essential for guard cell fate. Both analyses robustly reject a (co-)orthologous relationship of the SMF genes in Physcomitrella and Selaginella with the MUTE/SPCH clade, as suggested by our earlier phylogenetic analysis6. Reasoning that genes encoding stomatal regulators would be preferentially expressed in the stomatal-bearing sporophyte, we interrogated microarray datasets20 and P. patens transcriptome atlas results21 that identified PpSMF1, PpSMF2 and PpSCRM1 as strong candidates because of their up-regulation in the sporophyte relative to protonemal tissue, as supported by qRT-PCR (Figures 1 f–h; Supp. Info. Figures 1–2). Additionally, PpSCRM1 is the most highly expressed of the four PpSCRM paralogues across P. patens tissues including developing sporophytes21 (Supp. Info. Figure 2). Based on these analyses, we investigated the role of PpSMF1, PpSMF2 and PpSCRM1 in regulating stomatal formation in P. patens by generating targeted gene deletion mutants via homologous recombination. Altogether, we generated two independent knock-out lines for each of PpSMF1, PpSMF2, and PpSCRM1. Flow cytometry analyses verified gametophytes of all the mutants were haploid, as in the wild-type, and not polyploid transformants (Supp. Info. Figure 3).
Stomata of P. patens form exclusively during the sporophyte stage of the life cycle (Figure 1a) and are restricted to a small area around the base (Figure 1b). P. patens lacks the early meristematic lineage for stomata seen in A. thaliana. Instead, the formation of a cell equivalent to a guard mother cell (GMC) is specified3 which, in common with the closely related Funaria hygrometrica22, appears to undergo an incomplete symmetric division leading to the formation of a single guard cell and a central pore (Figure 1c). Strikingly, in both ΔPpSMF1 and ΔPpSCRM1 mutant lines, the stomatal developmental program is halted resulting in no mature guard cells. Instead, only pavement-like cells develop and in ΔPpSCRM1, very occasionally cells form that enter the stomatal lineage but fail to mature into stomata (Figures 2a, b). In contrast, ΔPpSMF2 mutants develop normal wild-type stomata (Figures 2a, b). We confirmed integration of the transgenes at the targeted loci and verified absence of gene expression in all mutant lines using genomic PCR and RT-PCR. (Figure 2c; Supp. Info. Figures 4–6). Closer anatomical inspection revealed a correlation between the presence of stomata and of sub-stomatal cavities, pointing to functional stomata: Sectioning of sporophytes revealed loss of stomata in ΔPpSMF1 and ΔPpSCRM1 was accompanied by the loss of sub-stomatal cavities, whereas in WT and in ΔPpSMF2 stomata and sub-stomatal cavities were present (Supp. Info. Figure 7). We found no differences in sporophyte sizes between the different mutants and WT lines (Supp. Info. Figure 8). These results establish PpSMF1 and PpSCRM1, but not PpSMF2, as essential for the formation of stomata in P. patens. Our targeted knock-out results are independently supported by cross-species gene complementation studies in which PpSMF1, but not PpSMF2, partially complemented A. thaliana mute and fama mutants6. Taken together, these data strengthen our hypothesis that a single ancestral PpSMF1-like gene and a SCRM partner were responsible for stomatal development in early land plants.
Figure 2. PpSMF1 and PpSCRM1 are required for stomatal development in the moss Physcomitrella patens.
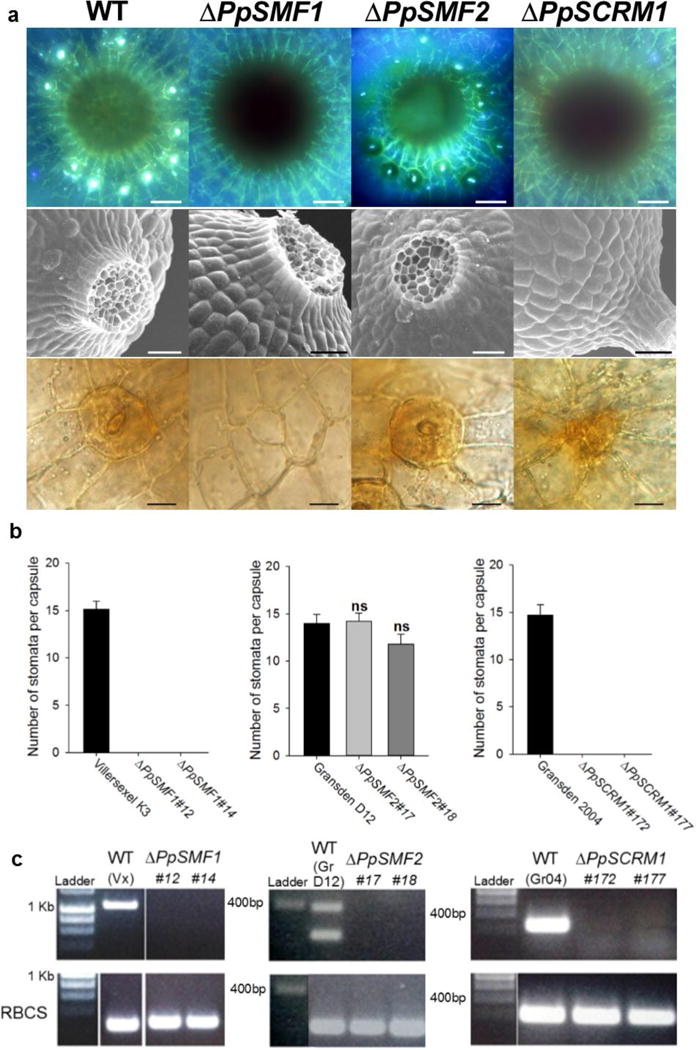
(a) Stacked UV fluorescence images (upper panel), scanning electron microscope images (middle panel) and bright field images (bottom panel) showing the spore capsule base and epidermal close-ups from P. patens wild-type, ΔPpSMF1, ΔPpSMF2 and ΔPpSCRM1 knock-out mutants, respectively. The top panel wild-type representative is from Villersexel K3 ecotype of P. patens, the middle panel wild-type representative is from the Gransden D12 ecotype and the bottom panel wild-type relates to the Gransden 2004 ecotype. There were no discernible differences between the sporophytes of the different background lines. For both of the ΔPpSCRM1 lines generated we observed one such instance of aborted stomata (see bottom right panel) in the 7 capsules of each line surveyed. (b) Number of stomata formed per sporophyte in two independent lines of each genotype versus wild-type controls. Error bars indicate one standard error of the mean. For ΔPpSMF1 and ΔPpSCRM1 and the corresponding wild-types, n = 7 capsules of each line were analysed. For ΔPpSMF2 and wild-type background, 5 capsules were surveyed. A One-way ANOVA was performed to test for differences between the wild-type and ΔPpSMF2 lines and no significant differences (denoted ns) were found. (c) RT-PCR to confirm loss of the respective transcript in each of the P. patens knock-out lines (top panel). A Rubisco (RBCS) control was run to verify the integrity of the produced cDNA (Bottom panel). For labelling purposes the wild-types Villersexel K3, Gransden D12 and Gransden 2004 are denoted Vx, GrD12 and Gr04. For PpSMF2 two bands were amplified in the control for which the smaller 239bp product represents the size expected for PpSMF2. Scale bars in a = 50 μm in the top and middle panels, in the bottom panel = 15 μm.
Because Group Ia bHLH proteins are obligate heterodimers with Group III bHLHs in A. thaliana, we next used bimolecular fluorescence complementation (BiFC) assays23 and yeast two-hybrid (Y2H) experiments24 to determine direct protein-protein interactions between PpSMF1 and PpSCRM1 in vivo. Transient co-expression of PpSMF1::YFPn and PpSCRM1::YFPc, as well as PpSMF1::YFPc and PpSCRM1::YFPn, resulted in strong YFP-fluorescence in the nuclei of Allium cepa cells, whereas no YFP-fluorescence was detected in controls (Figure 3a; Supp. Info. Figure 9). Specific interaction of PpSMF1 and PpSCRM1 was also demonstrated by Y2H experiments. PpSMF1 and PpSCRM1 fused with Gal4-DB alone showed no transcriptional activation, but strong activation was observed by using PpSMF1 as bait and PpSCRM1 as prey (Figures 3 b–d). These results support PpSMF1 and PpSCRM1 as physically-interacting heterodimeric partners. Furthermore, their nuclear localization is consistent with a role as DNA-binding transcription factors, reinforcing functional orthology to the A. thaliana Group Ia and IIIb bHLHs, respectively.
Figure 3. Bimolecular fluorescence complementation and Yeast 2-Hybrid assays demonstrating PpSMF1 and PpSCRM1 protein-protein interactions.
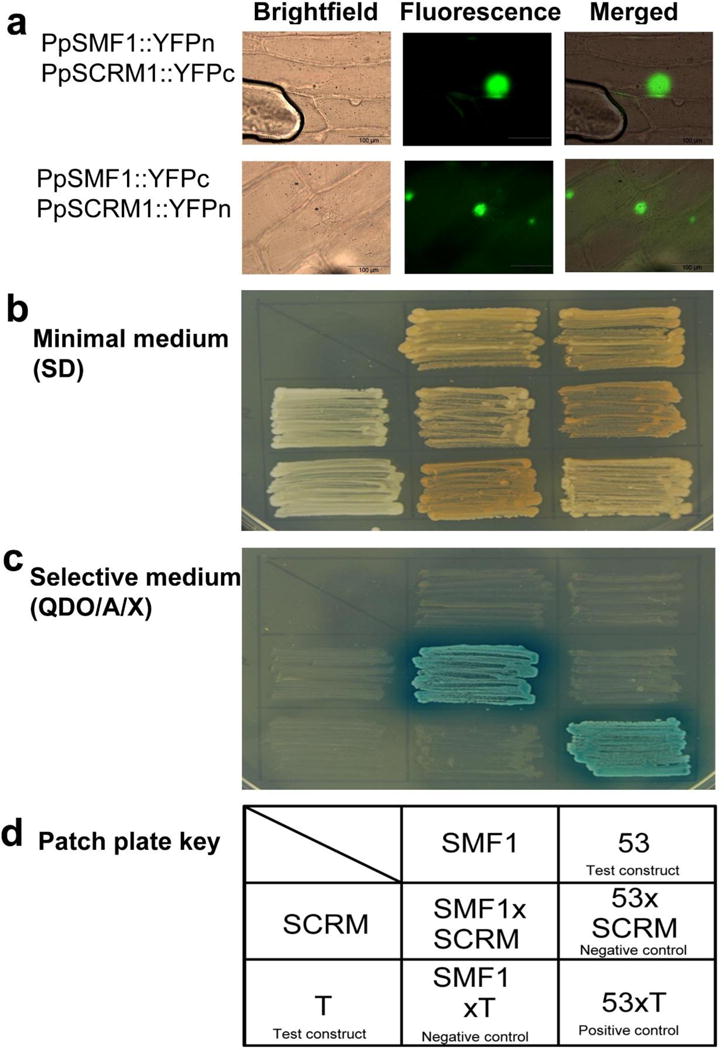
(a) Representative bright-field, fluorescence and overlay/merged images of BiFC analysis showing pairwise combinations of bHLH constructs, each fused with a complementary, half-YFP molecule (nYFPn fusion and YFPc fusions, respectively). In the intact Allium cepa epidermis using bimolecular fluorescent complementation (BiFC), PpSMF1 and PpSCRM1 showed strong heterodimerization in the nuclei. Controls are described in Supp. Info. Fig 9. Scale = 100 μm. (b–d) Yeast two-hybrid analysis: (b) Growth on minimal medium. (c) Growth on stringent selection medium. Blue indicates reporter activation. (d) Key to patch plate assays is shown in (b) and (c).
The BiFC and Y2H results suggest that PpSMF1-PpSCRM1 heterodimerisation could occur in P. patens cells due to highly conserved protein-protein interactions. In-silico analysis of the putative key domains involved in DNA binding during heterodimerisation suggests that an E-box binding domain (EBD) in PpSMF1 and PpSMF2, a corresponding DNA binding domain in PpSCRM1 and coiled-coil domains in both peptides are conserved between P. patens and A. thaliana (Supp. Info. Figure 10). However, PpSMF2 expression is very low compared to PpSMF1 and it is therefore unsurprising there is no aberrant phenotype in ΔPpSMF2 mutants despite key regulatory motifs being present. Conservation of functional motifs of PpSMF1 and PpSCRM1, which are both strongly expressed in the sporophyte21, taken together with our experimental data (Figures 1–3), suggests that a heterodimeric bHLH partnership first existed in the ancestor of mosses and flowering plants which could both initiate and complete stomatal development.
Having produced mosses with stomata-less sporophytes, we next addressed the long-standing mystery relating to stomatal function in an early diverging non-vascular land plant lineage5,25. Current opinion suggests moss stomata facilitate nutrient and water transport and gas exchange in the developing sporophyte26,27 and also assist dehiscence and release of spores during sporophyte maturation28, when pores become less able to close. We tested the function of stomata in P. patens in this context by tracking the development and subsequent dehiscence of the sporophytes in WT and mutants (Figure 4). Absence of stomata had no effect on spore development, morphology or viability in lines of ΔPpSMF1 and ΔPpSCRM1 as determined using SEM and bright-field microscopy and spore germination assays, respectively (Supp. Info. Figures 11 and 12). In contrast, observations of sporophyte development over time indicated that stomata-less ΔPpSMF1 and ΔPpSCRM1 mutants showed significantly (P < 0.01) delayed capsule dehiscence relative to WT during the late stages of development, as measured by the percentage of open capsules and timing of dehiscence (Figure 4; Supp. Info. Figures 13–14)., Although the reduced sporophyte of Physcomitrella is different to that of larger complex mosses, such as Funaria, our data suggest stomata during late stage sporophyte development may function in a similar manner aiding capsule dehiscence29. Intriguingly, delayed sporophyte dehiscence in P. patens seems to be decoupled from the browning of the sporophyte capsules, which is commonly assumed to be an indicator of capsule and spore maturation. As indicated by our quantitative analysis of the transition of capsule colouring (Supp. Info. Figure 15), ΔPpSMF1 capsules did not reveal any significant deviation from WT. However, in young green sporophytes of P. patens, and F. hygrometrica, stomata open and close in response to cues, such as light and abscisic acid, through molecular pathways co-opted from the gametophyte27,30, suggesting gas exchange functionality. A complex picture of stomatal function in early land plant lineage sporophytes is therefore emerging relating to age, and possibly environmental conditions, but with stomatal action ultimately linked to reproductive success.
Figure 4. Loss of PpSMF1 and the PpSCRM1 gene functions results in delayed dehiscence of spore capsules.
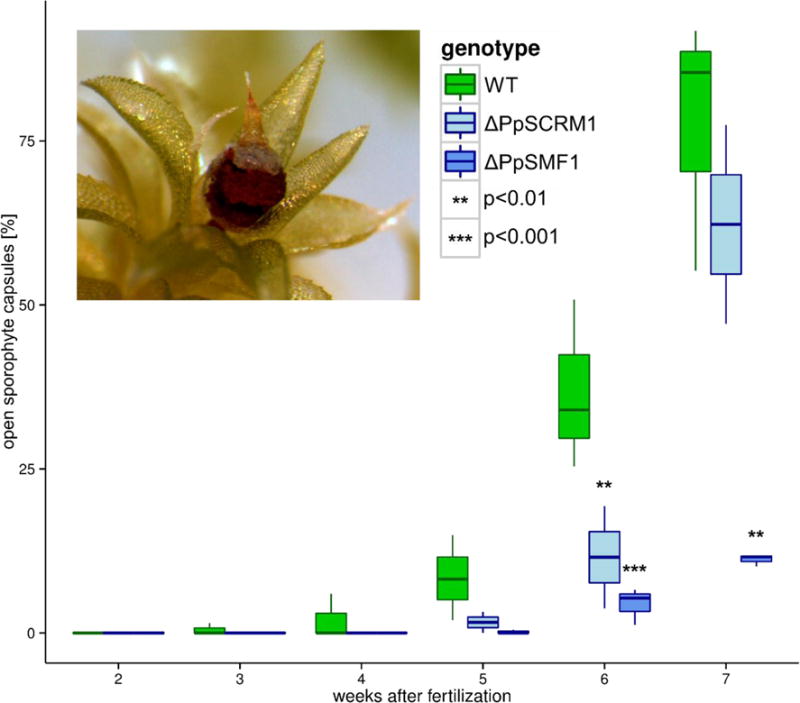
Box-whisker plots of the percentages of ruptured sporophyte capsules in the wild-type, ΔPpSMF1 and ΔPpSCRM1 lines over a developmental time series experiment ranging from second and seventh week after induction of fertilization. Vertical lines within boxes mark the median. The boxes indicate the upper (75 %) and lower (25 %) quartiles. Whiskers indicate the ranges of the minimal and maximal values. Inset photograph depicts an open/ruptured spore capsule in the Gransden wild-type strain. Significance of differences between mutants and the wild type was tested using a binomial model with a nested error term correcting for repeated measurements in the combined data set, and for each genetic background independently, with consistent results. Significant (P < 0.05) deviations from the wild type are indicated by asterisks.
We propose that an ancestral land plant possessed a multifunctional ancestral dimer, comprised of ancient variants of PpSMF1 and PpSCRM1, which was sufficient to initiate and drive stomatal development in the early sporophyte. Specifically, results from our experiments with knock-out mutants in the moss P. patens, belonging to an extant lineage of non-vascular land plants with stomata, and our protein-protein interaction evidence, support the notion that a MUTE-FAMA-like and SCRM1/SCRM-like bHLH partnership was responsible for the origin of stomata in the earliest vascular land plants over 400 Ma. Remarkably, the origin of this genetic system that gave rise to stomata, together with those for roots10,11 and leafy shoots12, ultimately helped facilitate the evolutionary radiation of plants on land leading to increases in terrestrial ecosystem complexity and primary production1,8,31 that supported a burgeoning diversity of life on the continents.
Methods
Plant material and culture conditions
Physcomitrella patens subspecies patens (Hedwig) Bruch & Schimp. WT strain “Gransden 2004”, used for genome sequencing32, provided the genetic background for the generation of ΔPpSCRM1 mutants (“Gransden 2004”, Freiburg) and ‘Villersexel’ the genetic background for the generation of ΔPpSMF1 mutants (Sheffield), and “Gransden D12” was the background for production of the ΔPpSMF2 mutants. P. patens was grown axenically on BCDAT medium33 supplemented with 1 mM calcium chloride and overlaid with cellophane discs (AA Packaging, UK), in 9 cm Petri dishes sealed with Micropore tape (3M) in Sanyo MLR incubators under continuous light (140 μmol m−2 s−1) at 25 °C34. P. patens (Freiburg) was grown in liquid or on solid (12 g/L purified agar (OXOID, Thermo Scientific, Waltham, MA, USA)) supplemented Knop medium35,36 and cultivated at 23 °C under a 16-hour light and 8-hour dark cycle37. Sporophyte development was induced according to ref38.
Generation of transgenic lines
To create the PpSMF1 and PpSMF2 knock-out (KO) constructs for gene targeting, 5′- and 3′-targeting sequences (coordinates Chr22: 9308333 -9307319 (5′) and Chr22: 9306131-9305111 (3′) for PpSMF1 and Chr19: 13226647-13227667 (5′)and Chr19: 13228404-13229099 (3′) for PpSMF2 were cloned on either side of a KanR(SMF1-KO) and HygR (SMF2-KO) selection cassette, respectively. The resulting constructs were amplified by PCR and used to transform P. patens. To produce the PpSCRM1 KO construct a 1,365 bp fragment of the PpSCRM1 gene (Pp3c10_4280) was PCR-amplified with the primers listed in Supp. Table 1 introducing EcoRI sites to the ends of the PCR product. After cloning to plasmid pJet1.2 (Thermo Fisher) an nptII selection cassette39 was inserted into this fragment via unique restriction sites for HincII and BcuI, respectively. Before moss transformation the KO construct was released from the vector backbone via EcoRI digest. Polyethylene glycol-mediated protoplast transformation of P. patens and analysis and confirmation of gene targeted loci, were conducted according to ref (40).
RNA was isolated from all tissues using the Spectrum Plant Total RNA Kit (Sigma-Aldrich) following the manufacturer’s protocol. RNA was quantified using a NanoDrop ND-8000 UV-Vis spectrophotometer (ThermoScientific). For RT-PCR, eluted RNA was DNase-treated with Ambion DNA-free™ DNA Removal Kit and then used as a template for cDNA synthesis with M-MLV Reverse Transcriptase (Life Technologies, New York) as per the manufacturer’s protocol. The resulting cDNA was used for PCR amplification (35 to 40 cycles) (Table S1 for primers). At the end of the PCR program samples were loaded into wells for agarose gel (1% w/v) electrophoresis and visualised by a UVItec (Cambridge, UK) digital camera. Primer sequences were designed and selected using Primer3 (http://frodo.wi.mit.edu/primer3/).
Molecular analysis
Three replicates of 7 day old protonemata grown on BCDAT, and 3 replicates of peat-pellet derived sporophyte capsules were used to compare the relative expression of PpSMF1, PpSMF2 and PpSCRM1. For protonemata, RNA was extracted from half a plate of tissue for each replicate. For sporophyte samples, early expanding sporophytes were harvested from 2 peat-pellets per replicate in order to generate sufficient RNA (approx. 300 capsules per replicate). RNA was extracted and processed using the above described methods. Prior to DNase treatment and cDNA synthesis the replicate RNA was assessed using the Nanodrop (Thermo Scientific) to ensure the same amount of RNA in all replicates prior to downstream applications. Relative qRT-PCR was performed using the Rotor-Gene SYBR Green PCR Kit (400) on a Corbett Rotor Gene 6000 (Qiagen, Venlo, Netherlands) following the manufacturer’s protocols. Relative quantification was performed by normalising the take-off value and amplification efficiency of the genes analysed relative to three housekeeping genes41.
Microscopic analysis
For epidermal phenotyping, 5–7 mature sporophytes of each line, and the corresponding WT, were removed from individual peat pellet-grown gametophores beneath a Leica MZCFLIII stereomicroscope. Capsules were stored in a modified Carnoy’s solution (2:1 ethanol: acetic acid) for a period of 2 weeks prior to dissection. Dissected sporophytes were viewed with an Olympus BX51 microscope and photomicrographs taken using an Olympus DD71 camera. Images were analysed using ImageJ software.
Sporophyte maturation and dehiscence
Gametophores were cultivated from spores on agar plates with Knop medium including microelements36. Individual three week old colonies were identified and transferred to Knop plates. Between 8 and 10 plants were isolated per plate and generating at least five plates per line. Plates were sealed with 7/8 of Parafilm and 1/8 of Micropore film and grown under long day conditions at 25 °C. After five weeks, plates were transferred into climate cabinets with short day conditions at 15 °C, sealed with Parafilm and grown for four weeks until formation of gametangia. Fertilization was initialized by soaking plates with sterilised water (re-closed with Parafilm), re-opening the plates after five days to remove the water, resealing with Micropore film and then cultured for three to six weeks at 15 °C short-day conditions. Developing sporophytes were recorded and traced by marking and numbering them on the plate lids as they appeared.
Supplementary Material
Acknowledgments
We thank Richard Haas and Tim Fulton for excellent technical assistance. R.C. was supported by a NERC studentship. D.C.B is a GBMF investigator of the Howard Hughes Medical Institute. D.J.B. acknowledges funding through an ERC Advanced Grant (CDREG, 322998). R.R. acknowledges funding through the Excellence Initiative of the German Federal and States Governments (EXC294). A.C.C. and Y.K. acknowledge support from BBSRC (Grant numbers BB/F001797/1 and BB/I006710/1).
Footnotes
Author contributions
C.C.C., R.C., R.R., W.F., J.E.G., A.F., D.J.B. and R.R. designed the study, C.C.C., R.C., D.L. and M.T. undertook the experiments with contributions from S.W., Y.K. and A.C.C., C.A.M., S.C., D.C.B., D.L., E.L.D and W.F. contributed materials and advice. A.C.C. constructed vectors for moss targeted knockout (SMF1 and SMF2), Y.K. carried out moss transformation (SMF2-KO) and yeast-2-hybrid analysis, A.C.C. and Y.K. carried out Southern blot hybridisation of the knockout mutant lines. D.J.B., R.R, C.C.C. and R.C. wrote the paper with contributions from D.C.B. All authors read, commented on and approved the final version of the manuscript.
Availability. All moss mutants described here were deposited in the International Moss Stock Center IMSC (Supplemental Table S2).
References
- 1.Berry JA, Beerling DJ, Franks PJ. Stomata: key players in the earth system, past and present. Current Opinion in Plant Biology. 2010;13:233–240. doi: 10.1016/j.pbi.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 2.Chater C, Gray JE, Beerling DJ. Early evolutionary acquisition of stomatal control and development gene signalling networks. Current Opinion in Plant Biology. 2013;16:638–646. doi: 10.1016/j.pbi.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 3.Vaten A, Bergmann DC. Mechanisms of stomatal development: an evolutionary view. Evodevo. 2012;3:9. doi: 10.1186/2041-9139-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pressel S, Goral T, Duckett JG. Stomatal differentiation and abnormal stomata in hornworts. Journal of Bryology. 2014;36:87–103. doi: 10.1179/1743282014y.0000000103. [DOI] [Google Scholar]
- 5.Haberlandt G. Physiologische Pflanzenanatomie. 5. Engelmann; 1918. [Google Scholar]
- 6.MacAlister CA, Bergmann DC. Sequence and function of basic helix-loop-helix proteins required for stomatal development in Arabidopsis are deeply conserved in land plants. Evolution & Development. 2011;13:182–192. doi: 10.1111/j.1525-142X.2011.00468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanaoka MM, et al. SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. Plant Cell. 2008;20:1775–1785. doi: 10.1105/tpc.108.060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beerling DJ. The emerald planet: how plants changed Earth’s history. Oxford University Press; 2007. [Google Scholar]
- 9.Edwards D, Kerp H, Hass H. Stomata in early land plants: an anatomical and ecophysiological approach. Journal of Experimental Botany. 1998;49:255–278. doi: 10.1093/jexbot/49.suppl_1.255. [DOI] [Google Scholar]
- 10.Menand B, et al. An ancient mechanism controls the development of cells with a rooting function in land plants. Science. 2007;316:1477–1480. doi: 10.1126/science.1142618. [DOI] [PubMed] [Google Scholar]
- 11.Tam THY, Catarino B, Dolan L. Conserved regulatory mechanism controls the development of cells with rooting functions in land plants. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E3959–E3968. doi: 10.1073/pnas.1416324112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrison CJ, et al. Independent recruitment of a conserved developmental mechanism during leaf evolution. Nature. 2005;434:509–514. doi: 10.1038/nature03410. [DOI] [PubMed] [Google Scholar]
- 13.Sakakibara K, Nishiyama T, Deguchi H, Hasebe M. Class 1 KNOX genes are not involved in shoot development in the moss Physcomitrella patens but do function in sporophyte development. Evolution & Development. 2008;10:555–566. doi: 10.1111/j.1525-142X.2008.00271.x. [DOI] [PubMed] [Google Scholar]
- 14.Horst NA, et al. A single homeobox gene triggers phase transition, embryogenesis and asexual reproduction. Nature Plants. 2016;2:6. doi: 10.1038/nplants.2015.209. [DOI] [PubMed] [Google Scholar]
- 15.Raven JA. Selection pressures on stomatal evolution. New Phytologist. 2002;153:371–386. doi: 10.1046/j.0028-646X.2001.00334.x. [DOI] [PubMed] [Google Scholar]
- 16.MacAlister CA, Ohashi-Ito K, Bergmann DC. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature. 2007;445:537–540. doi: 10.1038/nature05491. [DOI] [PubMed] [Google Scholar]
- 17.Chinnusamy V, et al. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes & Development. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olsen JL, et al. The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature. 2016;530:331–+. doi: 10.1038/nature16548. [DOI] [PubMed] [Google Scholar]
- 19.Ran JH, Shen TT, Liu WJ, Wang XQ. Evolution of the bHLH Genes Involved in Stomatal Development: Implications for the Expansion of Developmental Complexity of Stomata in Land Plants. Plos One. 2013;8:11. doi: 10.1371/journal.pone.0078997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Donoghue MT, et al. Genome-wide transcriptomic analysis of the sporophyte of the moss Physcomitrella patens. Journal of Experimental Botany. 2013;64:3567–3581. doi: 10.1093/jxb/ert190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ortiz-Ramirez C, et al. A Transcriptome Atlas of Physcomitrella patens Provides Insights into the Evolution and Development of Land Plants. Mol Plant. 2016;9:205–220. doi: 10.1016/j.molp.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Sack FD, Paolillo DJ. Incomplete cytokinesis in Funaria stomata. American Journal of Botany. 1985;72:1325–1333. doi: 10.2307/2443504. [DOI] [Google Scholar]
- 23.Weinthal D, Tzfira T. Imaging protein-protein interactions in plant cells by bimolecular fluorescence complementation assay. Trends in Plant Science. 2009;14:59–63. doi: 10.1016/j.tplants.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Ito T, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haig D. Filial mistletoes: the functional morphology of moss sporophytes. Annals of Botany. 2013;111:337–345. doi: 10.1093/aob/mcs295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merced A, Renzaglia KS. Moss stomata in highly elaborated Oedipodium (Oedipodiaceae) and highly reduced Ephemerum (Pottiaceae) sporophytes are remarkably similar. American Journal of Botany. 2013;100:2318–2327. doi: 10.3732/ajb.1300214. [DOI] [PubMed] [Google Scholar]
- 27.Chater C, et al. Regulatory Mechanism Controlling Stomatal Behavior Conserved across 400 Million Years of Land Plant Evolution. Current Biology. 2011;21:1025–1029. doi: 10.1016/j.cub.2011.04.032. [DOI] [PubMed] [Google Scholar]
- 28.Garner DLB, Paolillo DJJ. On the functioning of stomates in Funaria. Bryologist. 1973;76:423–427. doi: 10.2307/3241726. [DOI] [Google Scholar]
- 29.Merced A, Renzaglia KS. Patterning of stomata in the moss Funaria: a simple way to space guard cells. Annals of Botany. 2016;117:985–994. doi: 10.1093/aob/mcw029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lind C, et al. Stomatal Guard Cells Co-opted an Ancient ABA-Dependent Desiccation Survival System to Regulate Stomatal Closure. Current Biology. 2015;25:928–935. doi: 10.1016/j.cub.2015.01.067. [DOI] [PubMed] [Google Scholar]
- 31.Franks PJ, Beerling DJ. Maximum leaf conductance driven by CO2 effects on stomatal size and density over geologic time. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:10343–10347. doi: 10.1073/pnas.0904209106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rensing SA, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 33.Cove D. The moss, Physcomitrella patens. Journal of Plant Growth Regulation. 2000;19:275–283. doi: 10.1007/s003440000031. [DOI] [Google Scholar]
- 34.Wallace S, et al. Conservation of Male Sterility 2 function during spore and pollen wall development supports an evolutionarily early recruitment of a core component in the sporopollenin biosynthetic pathway. New Phytologist. 2015;205:390–401. doi: 10.1111/nph.13012. [DOI] [PubMed] [Google Scholar]
- 35.Reski R, Abel WO. Induction of budding on choloronemata and caulonemata of the moss, Physcomitrella patens, using isopentenyladenine. Planta. 1985;165:354–358. doi: 10.1007/bf00392232. [DOI] [PubMed] [Google Scholar]
- 36.Egener T, et al. High frequency of phenotypic deviations in Physcomitrella patens plants transformed with a gene-disruption library. BMC plant biology. 2002;2:6. doi: 10.1186/1471-2229-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frank W, Ratnadewi D, Reski R. Physcomitrella patens is highly tolerant against drought, salt and osmotic stress. Planta. 2005;220:384–394. doi: 10.1007/s00425-004-1351-1. [DOI] [PubMed] [Google Scholar]
- 38.Hohe A, Rensing SA, Mildner M, Lang D, Reski R. Day length and temperature strongly influence sexual reproduction and expression of a novel MADS-box gene in the moss Physcomitrella patens. Plant Biology. 2002;4:595–602. doi: 10.1055/s-2002-35440. [DOI] [Google Scholar]
- 39.Hohe A, et al. An improved and highly standardised transformation procedure allows efficient production of single and multiple targeted gene-knockouts in a moss, Physcomitrella patens. Curr Genet. 2004;44:339–347. doi: 10.1007/s00294-003-0458-4. [DOI] [PubMed] [Google Scholar]
- 40.Kamisugi Y, Cuming AC, Cove DJ. Parameters determining the efficiency of gene targeting in the moss Physcomitrella patens. Nucleic Acids Research. 2005;33:10. doi: 10.1093/nar/gnil172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luna E, et al. Plant perception of beta-aminobutyric acid is mediated by an aspartyl-tRNA synthetase. Nature Chemical Biology. 2014;10:450–456. doi: 10.1038/nchembio.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


