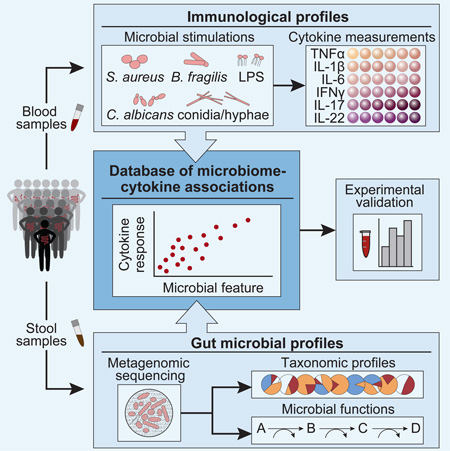
Summary
Gut microbial dysbioses are linked to aberrant immune responses, which are often accompanied by abnormal production of inflammatory cytokines. As part of the Human Functional Genomics Project (HFGP), we investigate how differences in composition and function of gut microbial communities may contribute to inter-individual variation in cytokine responses to microbial stimulations in healthy humans. We observe microbiome-cytokine interaction patterns that are stimulus-specific, cytokine-specific, and cytokine- and stimulus-specific. Validation of two predicted host-microbial interactions reveal that TNFα and IFNγ production are associated with specific microbial metabolic pathways: palmitoleic acid metabolism and tryptophan degradation to tryptophol. Besides providing a resource of predicted microbially-derived mediators that influence immune phenotypes in response to common microorganisms, these data can help to define principles for understanding disease susceptibility. The three HFGP studies presented in this issue lay the groundwork for further studies aimed at understanding the interplay between microbial, genetic, and environmental factors in the regulation of the immune response in humans.
Keywords: Database of microbiome-cytokine associations, microbiome-host interactions, human gut microbiome, inflammatory cytokine response, Human Functional Genomics Project, metagenomics, microbial profiles, immunological profiles, interindividual variation, healthy human cohort
Graphical Abstract
Introduction
The gut microbiome is a crucial factor for shaping and modulating immune system responses, with gut microbial dysbioses linked to several autoimmune and immune-mediated diseases (Gevers et al., 2014; Kosiewicz et al., 2011; Paun et al., 2016). In the case of rheumatoid arthritis, this dysbiosis can be partially resolved after treatment (Zhang et al., 2015). While responding to pathogenic organisms is a main function of the immune system, recognition and tolerance of commensal bacteria are equally important for host health (Kosiewicz et al., 2011). Commensal microbes calibrate innate and adaptive immune responses and impact the activation threshold for pathogenic stimulations, in large part by producing small molecules that mediate host-microbial interactions (Donia and Fischbach, 2015). Short-chain fatty acids (SCFAs) are a classic example of how bacterial-derived molecules contribute to gut immune homeostasis (Lee and Hase, 2014; Thorburn et al., 2014). SCFAs serve as energy sources for gut epithelial cells, modulate cytokine production, and induce expansion of regulatory T cells. Consistent with these functions, SCFAs were implicated in anti-inflammatory properties of a mix of 17 human-derived Clostridia strains (Atarashi et al., 2013; Smith et al., 2013). Gut microbial communities harboring Clostridia are additionally beneficial in that they regulate immune function and epithelial permeability to food antigens to protect against allergen sensitization (Stefka et al., 2014).
Commensal microbiota also modulate systemic immune responses (Lee and Hase, 2014). While the epithelial barrier ensures that microorganisms are largely confined to the gut, microbial metabolites can penetrate the epithelial barrier, allowing them to enter and accumulate in the host circulatory system where they are sensed by immune cells (Dorrestein et al., 2014). For example, dietary tryptophan is metabolized by different gut bacteria resulting in distinct derivatives, including indole propionic acid, which is absorbed through the intestinal epithelium and enters the bloodstream. The biological activity of indole propionic acid, as is the case for many other small molecules, is largely unknown (Wikoff et al., 2009).
Given the many examples of commensal microbiota modulating immune responses, we investigated the relationship between inter-individual variation in gut microbial community composition and the inflammatory cytokine response to microbial stimulation in healthy individuals (Figure 1). To this end, we assessed potential functional relationships in multi-omic data including microbial and cytokine profiles from approximately 500 healthy individuals of Western-European genetic background in the 500 Functional Genomics (500FG) cohort from the HFGP (Figure 1A; for exact sample numbers see Figures S1A and S1B). Several types of microbiome-cytokine interaction patterns were detected. We identified specific bacterial species and genera that are predicted to influence cytokine production capacity, and found putative interactions between microbial metabolism and tumor necrosis factor alpha (TNFα) and interferon gamma (IFNγ), a subset of which we experimentally validated. This work is complemented by two accompanying studies linking cytokine responses to host genetics (Li et al., 2016) and to host and environmental factors independent of the microbiome (ter Horst et al., 2016) yielding a comprehensive picture of the factors influencing human cytokine responses and host defense.
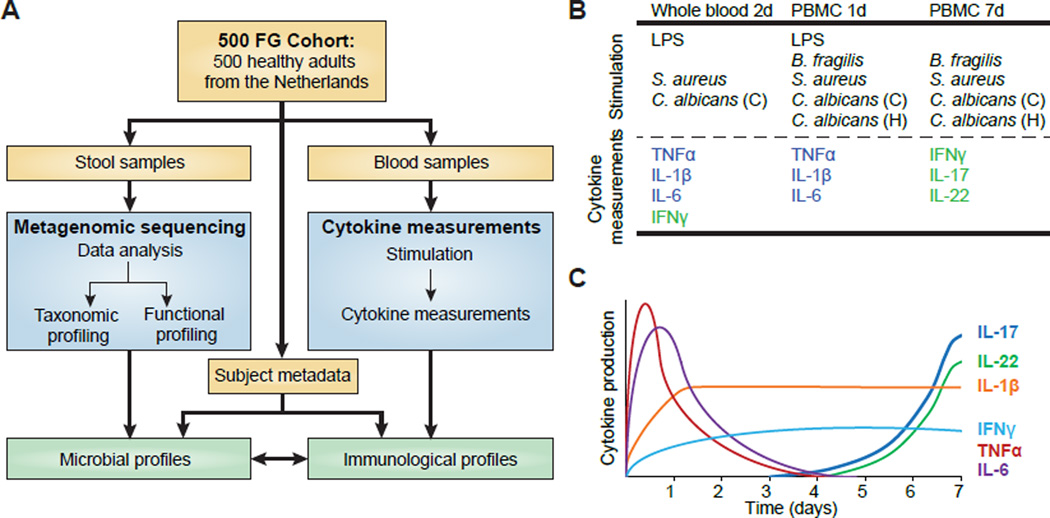
Figure 1. Linking inter-individual variation in immune response to the gut microbiome in the 500FG cohort.
(A) The 500FG cohort comprises 500 healthy adults from the Netherlands. Stool samples were collected and subjected to metagenomic sequencing to infer gut microbial profiles. Concurrent blood samples were collected to measure the inflammatory cytokine response in connection with various microbial stimulations.
(B) Monocyte- (blue) [IL-6, TNFα, IL-1β] and lymphocyte-derived cytokines (green) [IFNγ, IL-17, IL-22] were measured in connection with three bacterial (LPS, B. fragilis, and S. aureus) and two fungal (C. albicans conidia and hyphae) stimulations in whole blood and/or PBMCs.
(C) Schematic illustrating production kinetics of different cytokines in PBMCs. IL-17 and IL-22 increase steadily during a 7-day period, whereas IL-6 and TNFα production is maximal in the first 24 hr and decreases thereafter. IFNγ reaches its maximum at 4–5 days, and IL-1β reaches its maximum level at 24 hr, after which a plateau is attained. (Adapted from (Ruschen et al., 1992; van de Veerdonk et al., 2009))
See also Figure S1.
Results
Study Overview
To identify associations between the gut microbiome and stimulus-induced cytokine responses, we collected stool samples from 500 healthy individuals to generate microbial taxonomic and functional profiles. Blood samples were taken concurrently from each individual and the cytokine response was measured ex vivo in peripheral blood mononuclear cells (PBMCs) and whole blood under five different microbial stimulations. Three stimulations were bacteria-derived (purified Escherichia coli-derived LPS and Bacteroides fragilis representing Gram-negative bacteria, and Staphylococcus aureus representing Gram-positive bacteria) and two were fungal-derived (Candida albicans hyphae and conidia yeast) (Figure 1B and S1C). Although most stimulations were pathogens, we also included the common gut commensal organism Bacteroides fragilis, as this species can mediate development of the host immune system and was detected in 41% of all samples (Mazmanian et al., 2005). Three monocyte-derived cytokines (IL-1β, TNFα, IL-6) and three lymphocyte-derived cytokines (IFNγ, IL-17, IL-22) were measured at different times to capture their peak abundance (Figures 1B and 1C). We then used multivariate analyses to identify significant associations between microbial features and specific cytokine responses.
Between-Subject Variation Is Substantial Both in Cytokine Response to Stimulation and in Microbial Composition
We first analyzed the inter-individual variation in the inflammatory cytokine response and the gut microbiome composition, as this forms the basis for the identification of potential interactions between the two components (Figure 2A). The cytokine response to the stimulations was compared to the unstimulated state (RPMI) in whole blood and/or PBMCs (Figure 1B). While the unstimulated cells exhibited only a small degree of variation in cytokine levels, a high degree of inter-individual variation was observed in response to all stimulations. The difference in the degree of variation was significant for all stimulatory conditions compared to the unstimulated state for each cytokine and cell type, respectively (Fligner-Killeen test, all p < 1e-15). Generally, the intensity of the response to a particular stimulation was cytokine-dependent and in some cases also differed between whole blood and PBMCs. For most cytokines, the strongest response was induced by C. albicans; IL-6 was the only cytokine to show a stronger response to LPS stimulation.
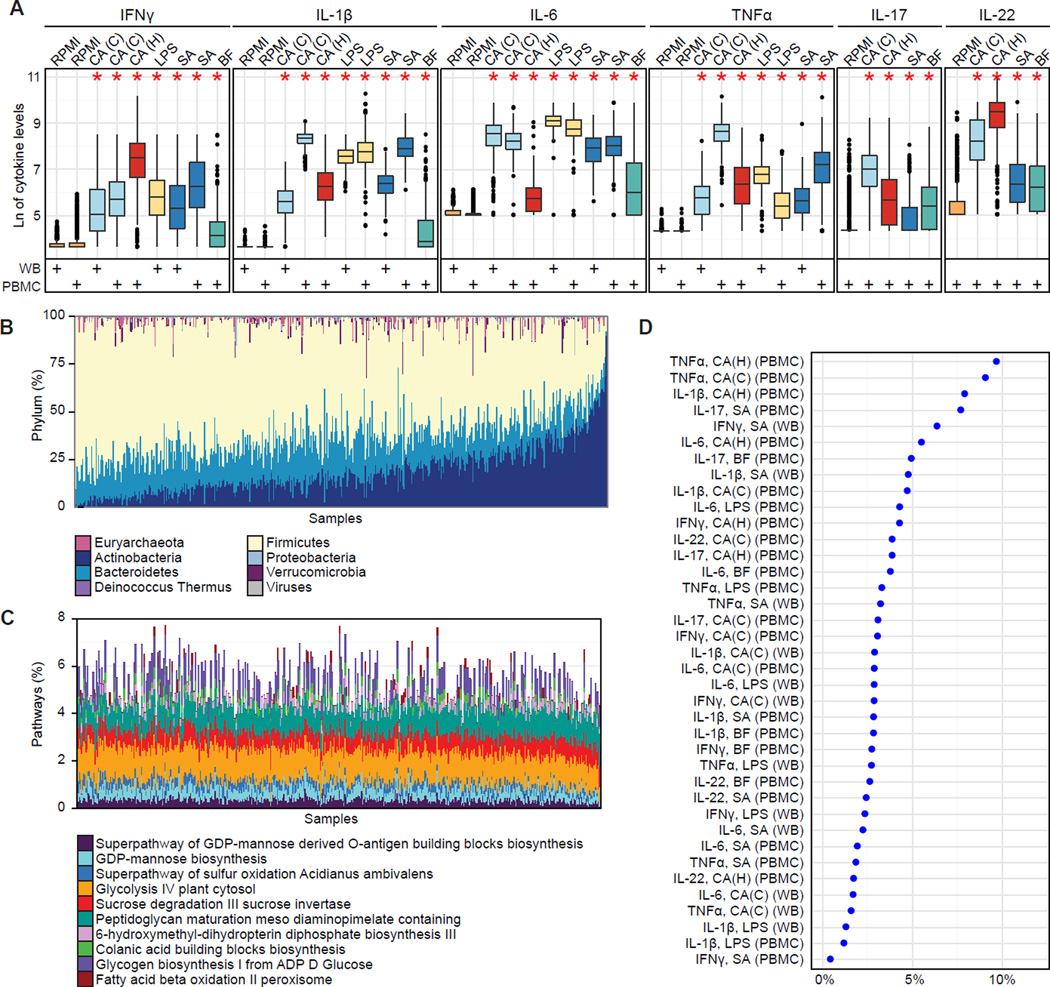
Figure 2. Healthy individuals show significant inter-individual variability in stimulated cytokine responses and in gut microbiota.
(A) Inter-individual variation in cytokine response. Each color represents a type of stimulation, also indicated on the upper x-axis: C. albicans conidia, CA(C); C. albicans hyphae, CA(H); E. coli-derived lipopolysaccharide 100 ng, LPS; S. aureus, SA; and B. fragilis, BF. Cell type is indicated below the x-axis (whole blood, WB; peripheral blood mononuclear cells, PBMC). The y-axis specifies the cytokine response. Each cytokine was measured in a non-stimulated control for each cell type (RPMI, orange). Stars indicate significant differences in variation for each stimulatory response compared to controls, respectively for each cell type (Fligner-Killeen test, all p < 1e-15). (Note that B. fragilis-induced TNFα measurements were not considered for further analyses due to a small degree of variation across individuals.) Whole blood measurements are based on 456 individuals. TNFα, IL-1β, and IL-6 measurements in PBMCs (1 d) were available for 401 individuals; IFNγ, IL-17, and IL-22 measurements in PBMCs (7 d) are based on 462 individuals.
(B) Taxonomic microbial profiles displaying phylum-level composition. Sample order was determined by the first principal component (PCA with Bray-Curtis distance).
(C) Functional microbial profiles displaying the abundance of the ten most variable MetaCyc pathways (based on variance). Samples are in the same order as in (B).
(D) The overall percentage of cytokine variation explained by species composition of the gut microbiome was 0.4–9.6%. To avoid overestimation due to species-species correlations, we represented the microbiota through the first 20 principal coordinates (PCoA with Bray-Curtis distance) which explain ∼50% of the variance. The cytokine variance explained by these principal coordinates was estimated through permutation ANOVA by summing over the significant contributions (p < 0.2).
See also Figure S2.
In addition to immunological profiles, we used metagenomic sequencing reads to identify the species-level composition of the communities and their functional potential in the form of encoded genes and pathways. As expected, inter-individual variation in taxonomic composition was high, with even high-level phylum composition varying greatly across individuals (Figure 2B) (Human Microbiome Project, 2012). At the functional level, we detected many MetaCyc pathways with comparable levels across samples, with a subset of generally less abundant pathways varying more substantially between individuals (Figure 2C). The microbial profiles from the 500FG cohort are similar to those of LifeLines-DEEP, another healthy Dutch cohort within the HFGP (Fu et al., 2015; Tigchelaar et al., 2015; Zhernakova et al., 2016) and appear to represent a single population (Figure S1D). In contrast, when these data were compared with the Human Microbiome Project cohort (consisting of 242 healthy adults from the United States (Human Microbiome Project, 2012)), the two Dutch cohorts had higher levels of Actinobacteria and lower levels of Bacteroidetes. However, a high degree of inter-individual variability was observed within all cohorts.
Cytokine Variation Explained by the Microbiome
For each of the cytokine measurements, we quantified how much of the overall variation could be attributed to the gut microbiota (Figure 2D). To avoid an overestimation of variability due to species-species correlations, we used the first 20 principal coordinates to represent the species composition, accounting for ∼50% of the variability in the microbial composition of the samples; thus the variation we observed is a conservative estimate. The largest percentage of variation in the cytokine response was explained for TNFα in connection with C. albicans hyphae (9.7%) and conidia (9.0%) stimulation in PBMCs. For other stimulations, the microbiome accounted for less than 5% of the variation in the TNFα response, highlighting the stimulus-specific effect of the predicted microbiome-cytokine interactions. In addition, we assessed variation due to functional features using MetaCyc metabolic pathways and gene ontology (GO) categories. A maximum of 7.7% and 8.6% of the cytokine variation was explained by metabolic pathways and GO categories, respectively (Figure S2).
Three Classes of Microbiome-Cytokine Interaction Patterns
Since we observed significant inter-individual variation in cytokine responses and gut microbial profiles, we next assessed whether these factors were correlated. We predicted three types of interaction patterns (IP), referred to as IP1 through IP3 (Figure 3A). The first interaction patterns (IP1) was stimulus-specific. In this case, gut microbial features were associated with the response of several cytokines, but only in connection with a particular type of stimulation. The second interaction pattern (IP2) describes cytokine-specific associations that are independent of the type of stimulation. In other words, a high/low response of a particular cytokine was connected to the abundance of a specific gut microbial species or function and observed in connection with several of the tested stimulations. Lastly, IP3 microbiome-cytokine interaction patterns were cytokine- and stimulus-specific. Here, a gut microbial component affected only the response of a particular cytokine and only in connection with a specific stimulation.
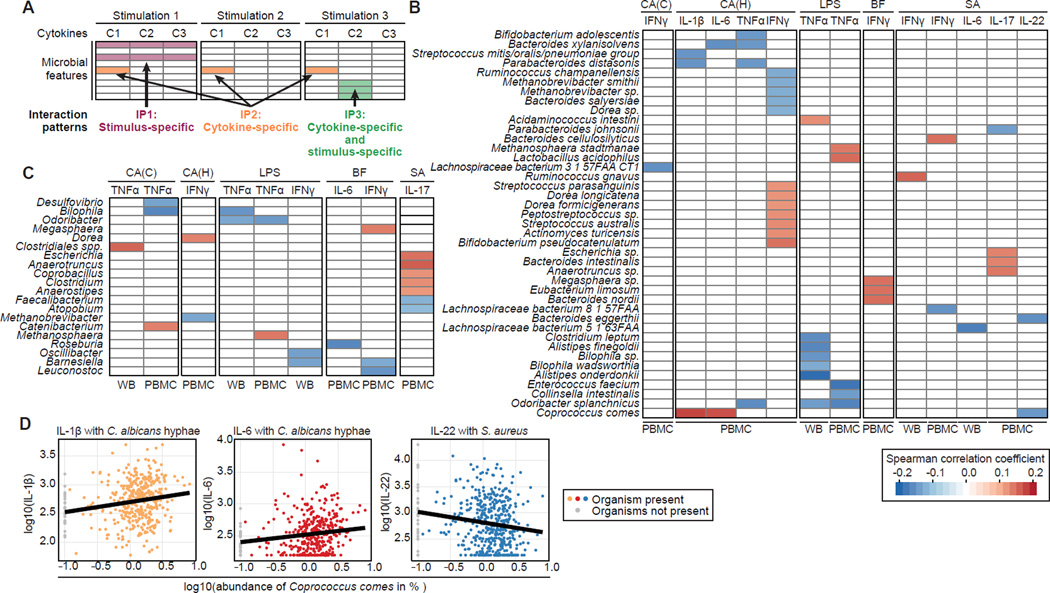
Figure 3. Significant correlations between gut microbial abundances and cytokine responses to stimulations.
(A) Three microbiome-cytokine interaction patterns were observed. Each colored cell represents a significant interaction between a specific microbial feature (y-axis) and a stimulus-induced cytokine response (x-axis). Interaction pattern 1 (IP1) refers to stimulus-specific interactions, where the same microbial feature is associated with several cytokines in connection with the same stimulation. IP2 describes cytokine-specific interactions, regardless of stimulation. IP3 refers to interactions that are cytokine- as well as stimulation-specific.
(B and C) Summary of species (B) and genus (C) associations with cytokine responses using Spearman correlation with Benjamini-Hochberg FDR correction (α ≤ 0.2). All species/genera were required to be detected in ≥ 3% of all samples (corresponding to ≥ 14 samples). Only species/genera significantly associated with at least one cytokine response are displayed. C. albicans conidia, CA(C); C. albicans hyphae, CA(H); lipopolysaccharide 100 ng, LPS; B. fragilis, BF; S. aureus, SA; whole blood, WB; peripheral blood mononuclear cells, PBMC.
(D) A stimulus-specific association of Coprococcus comes was observed for IL-1β and IL-6 in connection with C. albicans hyphae stimulation. C. comes was also negatively correlated with S. aureus-induced IL-22 production. All displayed cell types are PBMCs.
Gut Microbial Species and Genera are Significantly Associated with Inflammatory Cytokine Responses
We performed pairwise correlation tests between microbial taxonomic composition at the species and genus levels with each cytokine in connection with a specific stimulation (Spearman correlation with Benjamini-Hochberg FDR correction α ≤ 0.2, Figures 3B and 3C). A total of 41 species and 20 genera were significantly associated with at least one potential interaction and associations were identified for all of the tested cytokines. All three interaction patterns were observed (Figure 3A) with the majority of associations being cytokine- and stimulus-specific (IP3). Three stimulus-specific (IP1) associations were observed, with C. albicans hyphae stimulation in connection with Bacteroides xylanisolvens, Parabacteroides distasonis, and Coprococcus comes. Furthermore, three potential cytokine-specific interactions (IP2) were identified. Odoribacter splanchnicus (Figure 3B) and the genus Bilophila (Figure 3C) were negatively correlated with TNFα production for LPS and C. albicans stimulations. Barnesiella was negatively associated with LPS and B. fragilis-induced IFNγ production (Figure 3C). Below, we detail the associations encountered for each of the stimulation types.
Taxonomic Associations with Fungal-Induced Cytokine Responses
Taxonomic features associated with fungal-induced cytokine responses were often connected to an altered IFNγ response (Figures 3B and 3C). This included common gut commensals such as Dorea longicatena and Dorea formicigenerans, where higher species abundances were connected to higher IFNγ levels in response to C. albicans hyphae. Both Dorea species can metabolize sialic acids, which are commonly found at terminal ends of mucins; release of these acids is implicated in mucin degradation, potentially increasing gut permeability (Crost et al., 2013). In contrast, another unclassified Dorea species was negatively correlated with the IFNγ response. Thus, different species of the same genus may affect the IFNγ response in opposite ways. We also observed different species from the same genus affecting different cytokines. For example, Streptococcus parasanguinis and Streptococcus australis were associated with IFNγ production, but other species such as the Streptococcus mitis/oralis/pneumoniae group were associated with IL-1β. Examples of a microbial species predicted to affect multiple cytokines (IP1) include Bifidobacterium, which has been previously associated with IFNγ production (Lopez et al., 2011; Menard et al., 2008). Here, we observed a positive correlation between Bifidobacterium pseudocatenulatum and IFNγ. In contrast, Bifidobacterium adolescentis was inversely correlated with TNFα production, further emphasizing the potential cytokine- and species-specificity of these associations. Further, P. distasonis was negatively associated with TNFα and IL-1β in response to C. albicans hyphae stimulation (Figure 3B). In human biopsies, P. distasonis was significantly more abundant in uninflamed versus inflamed tissue (Zitomersky et al., 2013). Consistent with this observation, oral administration of P. distasonis antigens attenuates intestinal inflammation during colitis in mice (Kverka et al., 2011).
These associations can also be used to form hypotheses regarding principles of host-microbial interactions. For instance, C. comes exhibited stimulus-specific associations with IL-1β and IL-6 in connection with C. albicans hyphae stimulation, suggesting that C. comes potentially modulates acute phase responses (Figure 3D). The acute phase response is a non-specific defense mechanism against microorganisms, characterized by a cascade involving stimulation of IL-1β and subsequent IL-6 production followed by fever and synthesis of acute-phase proteins. Both cytokines were positively correlated with C. comes, suggesting immunostimulatory properties of this microorganism on the acute phase pathway. In an accompanying manuscript examining host factors that influence cytokine response, ter Horst et al. found correlations between IL-1β and the acute phase protein AAT (alpha-1-antitrypsin) (ter Horst et al., 2016). We therefore assessed the impact of C. comes on the circulating concentrations of AAT and found a positive correlation (r = 0.08, p = 0.08) (Figure S3) providing further evidence for the connection between C. comes and acute phase responses. Furthermore, C. comes demonstrated immunosuppressive properties in connection with IL-22 production induced by S. aureus (Figure 3D). The negative correlation of C. comes with IL-22 (in contrast to the positive correlations observed for IL-1β and IL-6) might at first appear contradictory, as IL-1β and IL-6 are both required for Th17 and IL-17/IL-22 production. However, complementary data from the 500FG and 200FG projects show an absence of correlations between monocyte-derived cytokines (e.g., IL-1β and IL-6) and Th-responses (e.g., IL-22) (Li et al., 2016).
Taxonomic Associations with Bacterial-Induced Cytokine Responses
Three bacterial stimulations were used: LPS, S. aureus, and B. fragilis. For TNFα, IL-17, and IL-22, associations were only observed in connection with a specific bacterial stimulus (Figures 3B and 3C). A differential IFNγ response was observed in connection with B. fragilis and S. aureus at the species level (Figure 3B) and LPS and B. fragilis at the genus level (Figure 3C). In contrast, microbial associations with TNFα were exclusively detected in connection with LPS stimulation, suggesting that the gut microbiome influences TNFα production in a stimulus-dependent manner (Figures 3B and 3C; Tables S1–S4). In total, three positive and nine negative associations were found at the species level for LPS-induced TNFα responses in whole blood and PBMCs. For example, negative associations between the microbiota and LPS-induced TNFα response involved multiple diet-sensitive bacteria, including Alistipes spp., Clostridium spp., and Bilophila spp. (in particular B. wadsworthia) (David et al., 2014; Devkota et al., 2012). This effect is particularly interesting given that anti-TNFα therapy is a common treatment for chronic inflammatory diseases such as inflammatory bowel disease and rheumatoid arthritis, and these associations motivate potential microbiome-based therapeutic approaches.
S. aureus was the only bacterial stimulus where a differential Th17 response was associated with the microbiota (IL-17 and IL-22; Figures 3B and 3C). While the IL-17 and IL-22 response to S. aureus was generally lower compared to fungal-induced responses, a significant amount of variation was observed for all stimulations (see Figure 2A). For IL-17, five positive genus associations were identified, including Clostridium. This genus includes many species from Clostridium clades IV and XIV, which enhance T regulatory cell abundance and induce anti-inflammatory molecules, including IL-17 (Atarashi et al., 2013). Two negative genus associations with IL-17 production involved Faecalibacterium and Atopobium (Figure 3C). A prominent member of the Faecalibacterium genus, F. prausnitzii, inhibits IL-17 production in rats (Zhang et al., 2014). Our finding suggests that this may also be true in humans.
In contrast to these stimulus-specific examples, a differential IFNγ response was observed for all bacterial stimulations (Figure 3B and 3C). For LPS, for example, two negative associations were detected with Oscillibacter and Barnesiella. Barnesiella was additionally implicated in the B. fragilis-induced IFNγ response, representing a cytokine-specific interaction (IP2). One of the species-level associations involving B. fragilis stimulation was another Bacteroides species, B. nordii. However, in general the associations observed in connection with B. fragilis, a commensal gut bacterium, were not unique compared to the pathogenic stimulations. For example, the S. aureus-induced IFNγ response had three microbial associations, one being a positive association with Bacteroides cellulosilyticus. This observation suggests that while the various stimulations yield different associations, there does not appear to be a unique preferential association of B. fragilis with other Bacteroides.
Microbial Metabolites Attenuate Pathogen-Induced TNFα and IFNγ Responses
The immune system recognizes and reacts to small molecules produced by gut microbes, such as the previously mentioned example of SCFAs. These microbial functional pathways are often driven by multiple microorganisms, and hence functional associations would not necessarily be detected in taxonomic pairwise association analyses. Therefore we investigated correlations between cytokine production and MetaCyc metabolic pathways as well as GO categories, including biological processes and molecular functions, to identified functional categories implicated in differential cytokine responses (Figures 4A and 4B, Tables S5–S8).
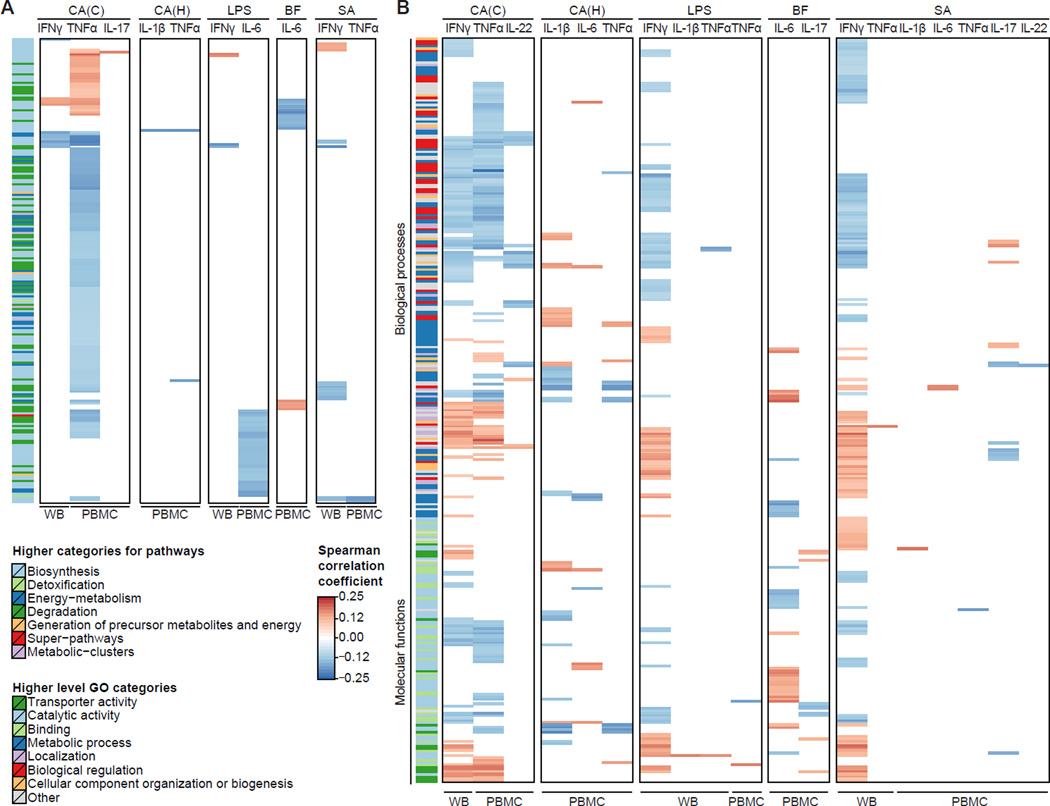
Figure 4. Significant correlations between functional potential of the gut microbiome and cytokine responses.
Functional summary for MetaCyc pathways (A) and Gene Ontology (GO) categories (B) using Spearman correlation with Benjamini-Hochberg FDR correction (α ≤ 0.2). All functional categories were required to occur in ≥ 3% of the samples (≥ 14 samples). C. albicans conidia, CA(C); C. albicans hyphae, CA(H); lipopolysaccharide 100ng, LPS; B. fragilis, BF; S. aureus, SA; whole blood, WB; peripheral blood mononuclear cells, PBMC. For MetaCyc pathways and GO categories, respectively, higher level functional categories are indicated on the left side.
Functional Associations with Fungal-Induced Cytokine Responses
Analogous to the taxonomic analysis, we first focused on functional associations in connection with fungal-induced cytokine responses. The majority of functional associations were detected for the C. albicans conidia-induced TNFα and IFNγ response (Figures 4A and 4B, Tables S5–S8). For TNFα, this observation was in contrast to the taxonomic analysis, where all associations were detected in connection with LPS stimulation (Figures 3B and 3C). Stimulus-specific interactions patterns involving TNFα and IFNγ are not surprising, as both are regulated by similar transcription factors in T cells, and IFNγ can further induce TNFα expression in dendritic cells.
Furthermore, 11 microbial functional features were predicted to affect the C. albicans conidia-induced production of TNFα and IL-22 in the same manner (Figures 4A and 4B). Both cytokines are produced by Th22 cells and can synergistically induce an effective innate immune response of epithelial cells during C. albicans infection, conferring protection in humans (Eyerich et al., 2011). Our results indicate that there are potential microbial factors that correlate with the production of both cytokines. In comparison to conidia, very few associations were detected for C. albicans hyphae, suggesting that the developmental state of the fungal pathogen is an important factor in determining host-microbial interactions.
Functional Associations with Bacterial-Induced Cytokine Responses
Microbial associations with the IL-6 response were detected for all bacterial stimulations (Figures 4A and 4B). For example, three MetaCyc pathways (Figure 4A) associated with the LPS-induced IL-6 response were involved in arginine degradation: cycloserine biosynthesis (PWY-7274), L-arginine and L-ornithine degradation (ORNARGDEG-PWY), and L-arginine, putrescine and 4-aminobutanoate degradation (ARGDEG-PWY) (Table S5). L-arginine depletion mediated by myeloid cell arginase (ARG) has emerged as a fundamental mechanism of inflammation-associated immunosuppression (Bronte and Zanovello, 2005). Our data suggest that the microbiome may play an important role in arginine depletion and may need to be taken into consideration to address the immunomodulatory role of ARG. We also found a connection to the fungal-induced TNFα response, which was negatively correlated with two of these pathways (ORNARGDEG-PWY and ARGDEG-PWY).
Immunomodulatory Effects of Tryptophan and Tryptophol
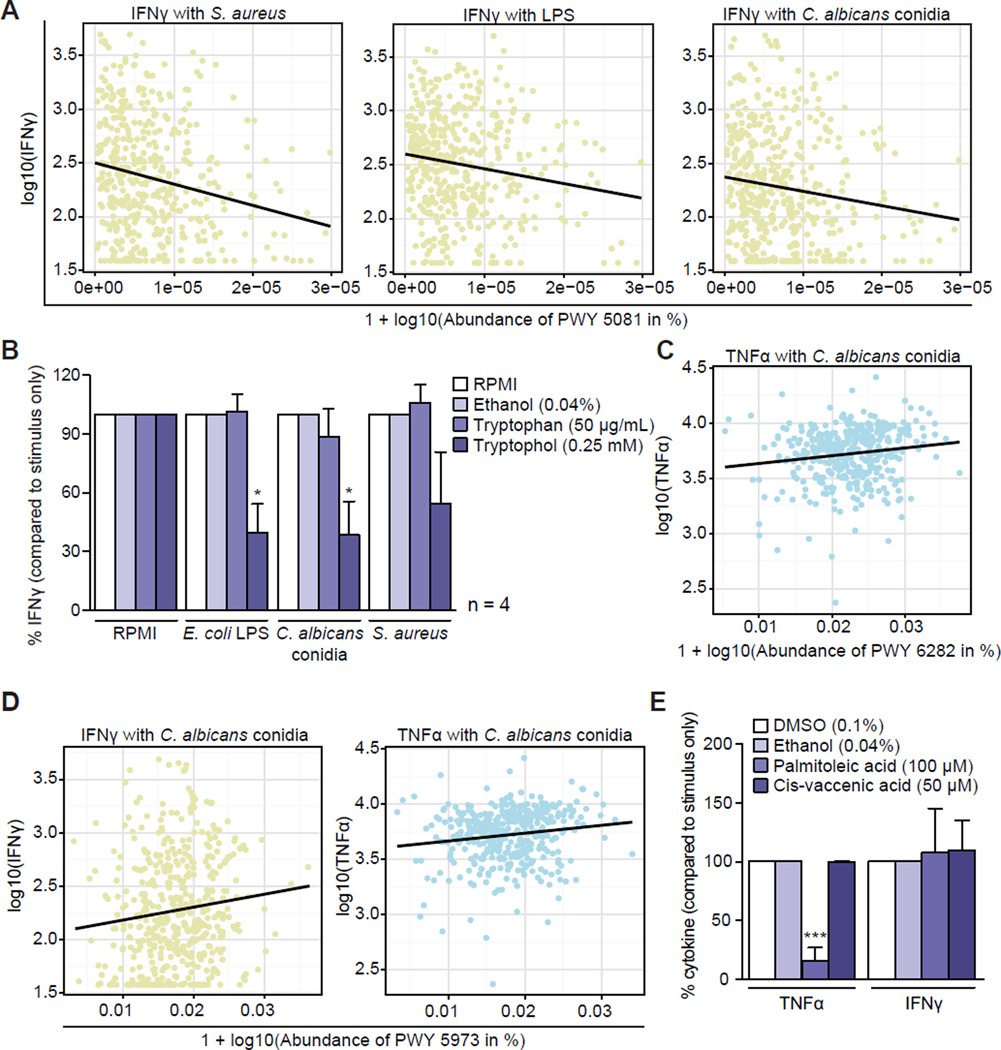
Most functional associations in connection with bacterial stimulations were detected for IFNγ, involving a large number of cytokine-specific interactions (IP2) (Figure 4B). For example, microbial tryptophan degradation to tryptophol (MetaCyc pathway 5081) was negatively associated with IFNγ production in whole blood for S. aureus, LPS, and C. albicans conidia (Figure 5A). Tryptophan is metabolized in the gut into several molecules that play an important role in immune regulation, such as limiting inflammation by interacting with the aryl hydrocarbon receptor (Lamas et al., 2016; Rothhammer et al., 2016) and both tryptophan depletion and downstream tryptophan metabolites can confer protection against excessive inflammation by immune inhibition (Opitz et al., 2007). To determine whether this observed decrease in IFNγ levels was due to a decrease in tryptophan or an increase in the resulting metabolite tryptophol, we experimentally validated these findings using a whole blood stimulation assay in healthy human donors. When tryptophan was added to whole blood, there was no effect on IFNγ production induced by LPS, S. aureus, or C. albicans conidia (Figure 5B). In contrast, tryptophol significantly decreased C. albicans- and LPS-induced production of IFNγ. Stimulation with S. aureus also showed a trend toward decreased IFNγ production when tryptophol was added (Figure 5B). These results show that microbially produced tryptophol has a strong effect on IFNγ production.
Figure 5. Immunomodulatory effects of tryptophol and palmitoleic acid.
(A) L-tryptophan degradation to tryptophol (MetaCyc PWY 5081) was negatively associated with IFNγ in connection with S. aureus, LPS, and C. albicans conidia in whole blood.
(B) Whole blood was stimulated with LPS, C. albicans conidia, or S. aureus in the presence or absence of tryptophan (50 µg/mL) or tryptophol (0.25 mM) and IFNγ concentrations were measured in supernatants for 4 donors. Group differences are expressed as percentage change compared to RPMI (tryptophan) or ethanol (tryptophol) and analyzed by paired t-test. *p < 0.05. Mean values for RPMI controls in pg/mL: RPMI: 8, LPS: 370, C. albicans conidia: 115, S. aureus: 405. Mean values for ethanol controls in pg/mL: RPMI: 8, LPS: 219, C. albicans conidia: 131, S. aureus: 91.
(C) Palmitoleic acid biosynthesis (MetaCyc PWY 6282) in PBMCs was positively correlated with C. albicans conidia-induced TNFα production.
(D) Cis-vaccenate biosynthesis (MetaCyc PWY 5973) was positively correlated with IFNγ (in whole blood) and TNFα (in PBMCs) in connection with C. albicans conidia stimulation.
(E) PBMCs from 5–6 healthy volunteers were stimulated with C. albicans conidia in the presence or absence of palmitoleic acid and cis-vaccenate (cis-vaccenic acid). Cytokine concentrations were measured in supernatants. Group differences are expressed as percentage change compared to ethanol or DMSO measurements and analyzed by paired t-test. ***p < 0.001. Mean values in pg/mL: TNFα (DMSO: 10,000, ethanol: 10,000, palmitoleic acid: 1,504, cis-vaccenate: 9,957; IFNγ (DMSO: 1,370, ethanol: 1,571, palmitoleic acid: 976, cis-vaccenate: 2,241).
See also Figure S4.
Palmitoleic Acid Can Inhibit the Pro-Inflammatory Cytokine Response
Two related MetaCyc pathways showed positive correlations with TNFα in response to C. albicans conidia stimulation: palmitoleic acid biosynthesis (MetaCyc PWY 6282) and cis-vaccenate biosynthesis (MetaCyc PWY 5973) (Figures 5C and 5D). Cis-vaccenate biosynthesis was also positively associated with IFNγ production. Palmitoleic acid biosynthesis has a branch point from a palmitoleoyl [acp] to the cis-vaccenate pathway (Figure S4A). Thus, increased activity in the palmitoleic acid biosynthesis pathway could result in either more palmitoleic acid or more cis-vaccenate if the cis-vaccenate pathway were also active. In the second scenario, an increase of cis-vaccenate would utilize the substrate for palmitoleic acid biosynthesis, resulting in less palmitoleic acid production. Given that our associations with cis-vaccenate biosynthesis have a positive correlation with TNFα and IFNγ, it is possible that increased palmitoleic acid corresponds to decreased cytokine production. We tested how the presence of these molecules affects C. albicans conidia-induced cytokine production in human PBMCs (Figure 5E). Palmitoleic acid decreased TNFα production but had no effect on IFNγ. However, cis-vaccenate (cis-vaccenic acid) did not influence cytokine production under any of the tested stimulations (Figure 5E). Interestingly, palmitoleic acid displayed an antiinflammatory effect in connection with all of the monocyte-derived cytokines (IL-1β, TNFα, IL-6), but had no effect on lymphocyte-derived cytokines (IFNγ, IL-17, IL-22) (Figures S4B and S4C).
Discussion
The HFGP is a comprehensive study that aims to identify the most important factors influencing cytokine production capacity in healthy humans. In three complementary studies, we report the environmental, genetic, and microbiome factors associated with cytokine production induced by different microbial stimulations in a cohort of 500 healthy volunteers. While accompanying studies demonstrate the impact of environmental and host factors (genetic and non-genetic) for the modulation of cytokine production (Li et al., 2016; ter Horst et al., 2016), in this study we provide evidence that the gut microbiome also has biologically relevant effects.
Our analysis indicates that inter-individual variation in cytokine response is linked to specific microbial organisms as well as microbial functions. The majority of detected associations were both cytokine- and stimulus-specific (IP3), suggesting that the immune system recognizes and interacts with microbial organisms and products with high specificity and that these microbial factors are associated with a particular immunological phenotype. In particular, a broad range of microbial functional features was associated with cytokine responses. This observation suggests that modulation of host defense by the microbiota may be exerted mainly through the release of intermediary common mediators (such as metabolites) rather than direct interaction between specific microorganisms and immune cells. This finding constitutes the first important conclusion that can be drawn from our data.
An important role for metabolites in microbiome-cytokine interactions is supported by the fact that a large proportion of the metabolites in the blood originate from the gut (Sridharan et al., 2014; Wikoff et al., 2009), as well as our findings of the strong impact of microbial metabolic processes on cytokine production (Figure 4). Among these metabolic processes, we experimentally validated the important impact of microbial tryptophan metabolism, a pathway known to modulate cytokine production at the level of host metabolism (Moffett and Namboodiri, 2003; Nowak et al., 2012). Here, we demonstrate that microbial tryptophan metabolism strongly influences cytokine production. Interestingly, this effect is mainly mediated by the tryptophan metabolite tryptophol, which has strong inhibitory effects on the TNFα response. However, the inhibitory effect depends on the particular pathogenic stimulation. We also validated a predicted interaction involving palmitoleic acid metabolism. Palmitoleic acid inhibits apoptosis induced by a combination of IL-1β and IFNγ (Welters et al., 2004), but a specific effect on IFNγ synthesis has not been previously reported. Interestingly, we not only validated this relationship in human cell stimulation assays, but also demonstrated a specific effect of palmitoleic acid on monocyte-derived cytokines (TNFα, IL-1β, IL-6) but not lymphocyte-derived cytokines (IFNγ, IL-17, IL-22). The molecular mechanisms responsible for this specificity remain to be elucidated in future studies. These validations serve as proof-of-principle of how predicted associations can be used to provide insight into host-microbial interactions.
In addition to the observations concerning modulation of cytokine production capacity by microbial functions, we also predicted interactions of specific microbial species and genera, including the effect of C. comes on cytokines of the acute phase pathway (IL-1β and IL-6). The production of IL-1β and IL-6 is regulated largely at a genetic level (Li et al., 2016) and less influenced by microorganisms. The physiological relevance of this finding is supported by our subsequent analysis showing that C. comes is associated with circulating concentrations of the acute phase protein AAT. This connection was observed despite the fact that AAT was measured during health, not after microbial challenge or infection.
Insight into gut microbiota and the immune system interactions is a crucial step in understanding immune-mediated and infectious diseases. For example, differential abundance of specific gut bacteria was associated with cytokine responses and may impact disease susceptibility. This effect was exerted directly on the intrinsic cytokine production capacity of the immune cells, rather than by influencing the number of cells in circulation: no effect of microbiota composition on the most important immune cell populations (T/B-lymphocytes, monocytes, neutrophils, NK-cells) was detected in the present study (data not shown).
Evaluating the results of the three studies presented in this issue, it appears that variation at many levels affects the inflammatory response. An earlier study showed that host genetic variants modulate pathogen sensing in dendritic cells stimulated with microbial ligands (Lee et al., 2014), and we now present a much broader assessment of the genetic (Li et al., 2016) and environmental factors (ter Horst et al., 2016) that modulate cytokine production. Non-genetic factors such as age, gender, and season have a strong influence on cytokine production capacity, and genome-wide analyses have identified 17 independent loci that influence cytokine production at a genome-wide level. Here, we demonstrate that the microbiome also plays a key role in the regulation of cytokine production. This conclusion is supported by studies showing that perturbations such as antibiotic treatment greatly impact development of autoimmune diseases such as psoriasis (Zanvit et al., 2015) and are associated with allergic asthma susceptibility (Russell et al., 2012). In addition, the use of antibiotics is associated with a more pronounced dysbiosis in children with Crohn’s disease, implicating perturbations of the gut microbiota in disease (Gevers et al., 2014).
The HFGP provides the opportunity to evaluate the importance of these host-dependent factors. Although host genetic variability and microbiome composition both influence cytokine production, our analyses suggest a greater impact of host genetics, explaining 25–50% of the variability of some cytokine responses, compared to the microbiome, explaining only up to 10% of cytokine variability.
Several limiting factors can introduce noise to the microbiome data, resulting in smaller correlation coefficients. First, any experimentally obtained data contains noise; for example, the immune cells were stimulated ex vivo, outside the environment where the cells were exposed to microbial factors, potentially weakening the detectable effects of host-microbial interactions. Second, simultaneously occurring host-microbial associations may affect each other adversely. Importantly, the effect size does not necessarily provide knowledge about biological significance, as illustrated by the experimental validation of two functional associations, providing conclusive evidence for the specificity of these host-microbial interactions.
The HFGP studies provide a comprehensive assessment of the effects of host and environmental factors on cytokine production capacity at a population level. Integrated approaches initiated in HFGP enabled us to identify important general interaction characteristics of different biological traits in the human hosts (Figure 6). First, general characteristics of the host such as age and gender have an important impact on inflammation. Men display a higher production of monocyte-derived proinflammatory cytokines, whereas women have a greater Th17 response (ter Horst et al., 2016). These differences may provide a partial explanation for differences in pathologies between men and women: men are more susceptible to metabolic and cardiovascular diseases, whereas women more often have autoimmune diseases (Regitz-Zagrosek, 2006; Regitz-Zagrosek et al., 2006). In addition, a defective IFNγ production capacity has been observed in elderly people (Ouyang et al., 2002), which may explain their increased susceptibility to infections. Second, genomic approaches in HFGP demonstrate the importance of host genetics for determining cytokine responses. Although this concept is not new, here we have analyzed a large healthy population in a genome-wide fashion, and report 17 new genetic loci associated with specific responses (Li et al., 2016). Third, we found that the effects of both genetic and microbiome factors are mainly stimulus- and cytokine-specific, and interesting new patterns emerged: TNFα and IFNγ production capacity appear to be more strongly influenced by the microbiome, whereas other cytokines such as IL-1β, IL-6, and Th17-derived IL-17 and IL-22 exhibit fewer, but more specific, associations with the gut microbiota. This finding is in line with genetic data showing an opposite trend for monocyte- and Th-derived cytokines (Li et al., 2016). These findings may have important consequences for approaches to immunotherapy targeting specific cytokines to alter immune responses: while inhibition of cytokines that are strongly influenced by the microbiome can potentially be modulated through diet, elimination of specific species, or fecal microbiota transplantation, host genomics-modulated cytokines may be more effectively targeted through inhibitory pharmacological approaches (e.g., neutralizing monoclonal antibodies).
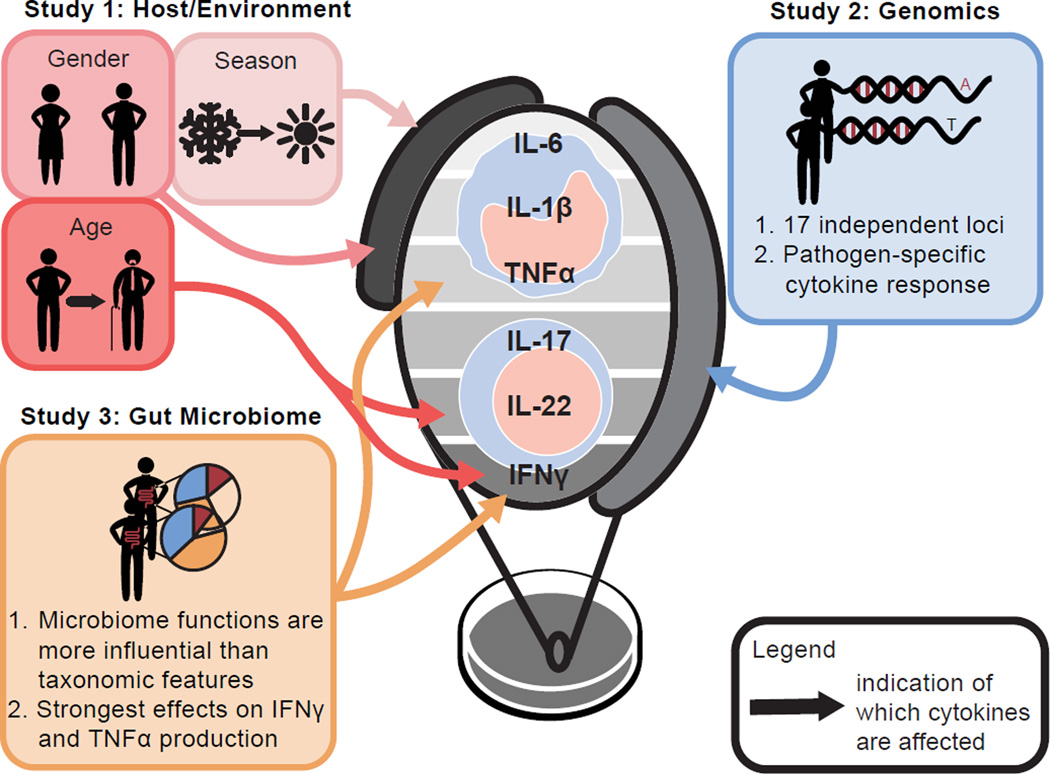
Figure 6. Environmental, host genetic and gut microbial factors impact human cytokine responses.
The impact of host environmental factors (ter Horst et al., 2016), host genetics (Li et al., 2016), and the gut microbiome (this study) on stimulus-induced cytokine responses was assessed in three complementary studies of the HFGP. While gender and seasonality were the main environmental factors affecting the response of monocyte-derived cytokines, age was associated with IL-22 and IFNγ production. Further, 17 independent loci were implicated in specific differential cytokine responses of monocyte- and Th-derived cytokines. Lastly, gut microbial functions were more influential on cytokine production than taxonomic features, where the strongest effects were observed for the stimulus-induced IFNγ and TNFα responses.
Through these three complementary HFGP studies assessing the impact of the host microbiome (this study), environmental (ter Horst et al., 2016), and host genetic factors (Li et al., 2016), we are able to glimpse the complexity of the mechanisms driving the variability of human cytokine responses. These studies open the possibility for future similar investigations in patient groups to identify the variation and disturbances responsible for their diseases in a personalized fashion, thereby providing the basis for precision medicine.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to and will be fulfilled by corresponding authors Ramnik J. Xavier (xavier@molbio.mgh.harvard.edu) and Mihai G. Netea (mihai.netea@radboudumc.nl).
Experimental Model and Subject Details
Human Cohort
The 500 Functional Genomics (500FG) project consists of 534 adult healthy volunteers sampled between July 2013 and December 2014. Inclusion criteria were >18 years of age and Western European descent. Exclusion criteria were pregnancy/breastfeeding, chronic or acute disease at the time of assessment, and use of chronic or acute medication during the last month before the study. After visiting the hospital to donate blood, volunteers received an extensive online questionnaire about lifestyle, diet, and disease history. Upon analyzing the questionnaire data we excluded 45 volunteers as they were under medication, non-European descent, or had kidney disease or diabetes mellitus (Figure S1A).
Ethics Statement
The 500FG study was approved by the Ethical Committee of Radboud University Nijmegen (NL42561.091.12, 2012/550). Inclusion of volunteers and experiments were conducted according to the principles expressed in the Declaration of Helsinki. All volunteers gave written informed consent before any material was taken.
Method Details
Stimulation of Whole Blood and Peripheral Blood Mononuclear Cells
Whole blood experiments
100 µl heparin blood was added to a 48-well plate and stimulated (Figure S1C) in a total volume of 500 µl for 48 hr at 37°C and 5% CO2. Supernatants were collected and stored at −20°C until used for ELISA.
Peripheral blood mononuclear cell (PBMC) experiments
PBMCs were obtained by density centrifugation of diluted blood (1 part blood to 1 part pyrogen-free saline) over Ficoll-Paque (Pharmacia Biotech, Uppsala, Sweden). Cells were washed twice in saline and suspended in medium (RPMI 1640) supplemented with gentamicin (10 mg/mL), L-glutamine (10 mM), and pyruvate (10 mM). PBMCs were counted in a Coulter counter (Coulter Electronics, Buckinghamshire, England) and their concentrations were adjusted to 5 × 106 cells/mL. 5 × 105 PBMCs in a total volume of 200 µL per well were stimulated in round-bottom 96-well plates (Greiner) with different stimulations (Figure S1C) for either 24 hr or 7 days at 37°C and 5% CO2. Seven-day stimulations were performed in the presence of 10% human pooled serum. Supernatants were collected and stored in −20°C until used for ELISA.
Cytokine Measurements
The concentrations of IL-1β, IL-6, IL-17, IL-22, IFNγ, and TNFα were measured in cell culture supernatants using enzyme-linked immunosorbent assay (ELISA) (IL-1β, IL-17, IL-22, TNFα: R&D Systems; IL-6, IFNγ: Sanquin), according to the manufacturer’s instructions. The circulating concentration of alpha-1-antitrypsin (AAT) was measured in EDTA plasma using an R&D Systems DuoSet ELISA kit following the manufacturer’s standard protocol.
Stool Samples and Sequencing
Participants collected stool samples at home the day before, or on the day of blood collection. Participants were asked to store their stool sample in the refrigerator before delivering the sample to the hospital where it was aliquoted and stored at −80°C until DNA extraction. DNA was isolated with the AllPrep DNA/RNA Mini Kit (Qiagen) with the addition of mechanical lysis. DNA samples were subsequently quantified by Quant-iT PicoGreen dsDNA Assay (Life Technologies) and normalized to a concentration of 50 pg/µL. Whole-genome shotgun sequencing libraries were prepared according to the manufacturer’s instructions using the Nextera XT DNA Library Preparation kit (Illumina) with 100–250 pg input DNA. Libraries were pooled by transferring equal volumes of each library using a Labcyte Echo 550 liquid handler. The concentrations and insert size ranges for each pooled library were checked using an Agilent Bioanalyzer DNA 1000 kit (Agilent Technologies). Libraries were subsequently sequenced on the Illumina HiSeq 2000 platform in paired-end mode (2×101bp) targeting ∼2.5Gb of sequences per sample.
Functional Validation Experiments
Cohort
PBMCs were isolated from buffy coats isolated from healthy volunteers (Netherlands).
Ethics statement
All studies with human blood samples were conducted in the Radboud University Nijmegen Medical Center and the use of healthy volunteers was approved by the institutional ethics review board. Peripheral venous blood samples from healthy volunteers were obtained after written informed consent was provided.
Reagents
The following reagents were used for cell isolation: Ficoll-Paque (GE Healthcare, Diegem, Belgium), RPMI 1640 Dutch modifications culture medium (Sigma, Zwijndrecht, the Netherlands). RPMI 1640 medium was supplemented with 1% gentamicin, 1% L-glutamine, and 1% pyruvate (Life Technologies, Nieuwerkerk, the Netherlands). For cell stimulation experiments, LPS (Escherichia coli serotype 055:B5) was purchased from Sigma and an extra purification step was performed as described previously (Hirschfeld et al., 2000). Purified LPS was tested in Toll-like receptor (TLR) 4−/− mice for the presence of contaminants and did not have any TLR4-independent activity (Sutmuller et al., 2006). C. albicans ATCC MYA-3573 (UC 820) (Lehrer and Cline, 1969) was grown overnight in Sabouraud broth at 37°C; cells were harvested by centrifugation, washed twice, and resuspended in culture medium. C. albicans conidia or hyphae were heat-killed for 1 hr at 100°C. S. aureus (ATCC 25923) and B. fragilis (NCTC 10584) were heat-killed for 30 min at 100°C a nd 95°C, respectively. L-tryptophan was purchased from Sigma. Palmitoleic acid, tryptophol, palmitic acid, and cis-vaccenic acid were purchased from Sigma (P9417, T90301, P0500, and V0384, respectively). Tryptophol and cis-vaccenic acid were dissolved in ethanol (Merck 1.00983.1000) and therefore ethanol was used as a control.
Human peripheral blood mononuclear cells
PBMCs were obtained by density centrifugation of diluted blood (1 part blood to 1 part pyrogen-free saline) over Ficoll-Paque (Pharmacia Biotech, Uppsala, Sweden). PBMCs were washed twice in saline and suspended in culture medium. The PBMCs were counted in a Coulter counter (Coulter Electronics, Buckinghamshire, England) and their concentration was adjusted to 5 × 106 cells/mL.
Stimulations
5 × 105 PBMCs in a total volume of 200 µL per well were incubated at 37°C in round-bottomed, 96-well plates (Greiner) with different stimulations. The effects of various metabolites (tryptophan, tryptophol, palmitoleic acid, cis-vaccenate) were studied by adding them 10 min before microbial stimulation. RPMI and DMSO medium were used as controls. After 24 hr, 48 hr, or 7 days of incubation with or without the various metabolites, supernatants were collected and stored at −20°C until assayed. S even-day cultures were performed in the presence of 10% human pooled serum.
Cytokine measurements
The concentrations of IL-1β, IL-6, IL-17, IL-22, IFNγ, and TNFα were measured in cell culture supernatants using enzyme-linked immunosorbent assay (ELISA) (R&D Systems & Sanquin) according to the manufacturer’s instructions.
Quantification and Statistical Analysis
Metagenomic Data Analysis
Samples that were included in the analysis had at least 4 million sequencing reads. Reads were first processed using KneadData (http://huttenhower.sph.harvard.edu/kneaddata). This included quality-trimming (trimmomatic parameters: MAXINFO:90:0.5), read-filtering based on a minimum read length of 60 bp, and removal of potential human contamination by filtering reads that aligned to the human genome (reference genome hg19). Quality-controlled, paired-end reads were aligned against a database of unique clade-specific marker genes using Bowtie2 and taxonomic profiles were inferred with MetaPhlAn 2.2 (Segata et al., 2012). For subsequent analysis, we considered species as well as genus composition of the samples. Functional profiling was performed using HUMAnN2 (http://huttenhower.sph.harvard.edu/humann2). Briefly, reads are mapped against a customized database of functionally annotated pangenomes, only considering organisms that were identified during the taxonomic profiling step. Functional annotation of the protein sequences in the pangenomes to their respective UniRef50 family is provided with the software. Reads that cannot be mapped are subsequently aligned against the complete UniRef50 database. The community totals are computed for each protein family (RPK) and converted into relative abundances. For subsequent downstream analysis these tens of thousands of gene families were further grouped into broader functional categories: MetaCyc metabolic pathways and informative GO categories, focusing on molecular functions and biological processes as previously described (Vatanen et al., 2016). Specifically, the selected GO terms were each annotated to > 2,000 proteins in UniRef50, while all their descendant (more specific) terms were annotated to < 2,000 proteins. Taxonomic as well as functional features were required to occur in at least 3% of the samples (at least 14 samples) to be considered for association testing. Overall, this resulted in 219 species, 83 genera, 639 MetaCyc pathways, and 520 GO categories (213 molecular functions and 307 biological processes).
Cytokine Data Analysis
Inter-individual variation of cytokine responses was evaluated using Fligner-Killeen test of homogeneity of variances. The variance for each of the stimulation conditions was compared to the unstimulated state (RPMI medium). For all stimulation conditions the level of variation was significantly increased (all p < 1e-15).
Correlation Analysis
Associations for each cytokine measurement in connection with a particular stimulus were analyzed using Spearman correlation coefficient in combination with Benjamini-Hochberg correction to account for multiple hypothesis testing (significance threshold α ≤ 0.2). For whole blood samples, cytokine measurements were obtained from 456 samples. For PBMCs, samples from 401 and 466 individuals were available for cytokine measurements after 1 day and 7 days, respectively.
Functional Validation Experiments
Significance testing
Experiments were performed in duplicate, and supernatants were pooled. When values were below or above the detection limit of the ELISA, the corresponding detection limit was used. For each experiment, 4–8 biological replicates were obtained. Differences between groups were analyzed using a paired t-test. The level of significance between groups was set at p < 0.05 (*), p < 0.01 (**), p < 0.001 (***) or p < 0.0001 (****). Data are presented as percentage change compared to control ± standard error.
Data and Software Availability
Data Resources
The data from the Human Functional Genomics Project are available online at http://hfgp.bbmri.nl/. The microbiome data are also available through SRA (Bioproject ID: PRJNA319574).
Supplementary Material
(A) For the 500FG project, samples from 534 individuals were collected and all volunteers were asked to complete a detailed questionnaire. Only samples from healthy adults were further processed and the filtering criteria included several diseases as well as medication use. Further, only a very small number of the volunteers were from non-European descent and therefore these samples were excluded from the subsequent analysis.
(B) The 500FG cohort consists of 489 volunteers. Stool samples were collected from all individuals and subjected to metagenomic shotgun sequencing. Filtering samples with less than 4 million reads resulted in a subset of 471 samples. Cytokine data is available for a subset of 456/401/466 of these samples for measurements in whole blood/PBMC 1d/PBMC 7d.
(C) Experiments were performed in whole blood and peripheral blood mononuclear cells (PBMCs). Cytokines were measured in RPMI media (control) as well as in connection with lipopolysaccharide (LPS), Candida albicans conidia/hyphae and Staphylococcus aureus stimulation.
(D) To determine the generalizability of the microbial profiles from our cohort, we compared the profiles of the 500FG cohort to those of LifeLines-DEEP, another cohort within the HFGP. As part of LifeLines-DEEP, metagenomic sequencing was performed on 1,326 fecal samples from healthy adults from the Netherlands. Taxonomic profiles of the samples were generated for both cohorts (see STAR Methods) and samples were compared using Principal Coordinate Analysis (PCoA) with Bray-Curtis distance at species level. The 500FG and LLDeep cohort exhibit similar gut microbial profiles and appear to represent a single population.
Analogous to Figure 2D, we computed the overall percentage of cytokine variation explained by microbial pathways (A) and GO categories (B). To avoid overestimation due to correlations of the microbial features, we represented the microbiome data through the first 20 principle coordinates (PCoA analysis with Bray-Curtis distance) for pathways and GO categories, respectively. The cytokine variance explained by these principle coordinates was estimated through permutation ANOVA by summing over the significant contributions (p < 0.2).
Validation of the interaction of C. comes with the acute phase response pathway. A positive correlation was observed for C. comes abundance and concentration of the acute phase protein AAT (r = 0.08, p = 0.08).
(A) Palmitoleic acid (palmitoleate) biosynthesis (MetaCyc pathway 6283) converts a (5Z)-dodec-5-enoyl-[acp] into palmitoleic acid. One of the intermediates of this pathway, a palmitoleoyl-[acp], constitutes a branching point of this pathway leading to cis-vaccenate biosynthesis (MetaCyc pathway 5973).
(B) and (C) PBMCs were stimulated for 24 hours (IL-1β and IL-6) or 7 days (IL-17 and IL-22) with HK C. albicans conida in the absence or presence of palmitoleic acid (100 µM) or cis-vaccenic acid (50 µM). IL-1β, IL-6, IL-17, and IL-22 were measured in cell culture supernatants using ELISA. Cytokine production is expressed as percentage compared to DMSO (control palmitoleic acid) or ethanol (control cis-vaccenic acid). Groups were compared using paired-t-test.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| No specific antibodies were used in this study | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| L-tryptophan | Sigma | H9772 |
| Palmitoleic acid | Sigma | P9417 |
| Palmitic acid | Sigma | P0500 |
| Tryptophol | Sigma | T90301 |
| Cis-vaccenic acid | Sigma | V0384 |
| Critical Commercial Assays | ||
| Human IL-1β ELISA Kit | RD Systems | DY201 |
| Human IL-6 ELISA Kit | PeliKine Compact | M9316 |
| Human TNFα ELISA Kit | RD Systems | DY210 |
| Human IL-22 ELISA Kit | RD Systems | DY782 |
| Human IL-17 ELISA Kit | RD Systems | DY317 |
| Human IFNγ ELISA Kit | PeliKine Compact | M9333 |
| Human AAT ELISA Kit | RD Systems | DY1268 |
| Deposited Data | ||
| ELISA cytokine measurements and other cohort data | This paper | http://hfgp.bbmri.nl/ |
| Metagenomic shotgun sequencing data | This paper | SRA: PRJNA319574, http://hfgp.bbmri.nl/ |
| Experimental Models: Cell Lines | ||
| No cell lines were used in this study | ||
| Experimental Models: Organisms/Strains | ||
| Human PBMCs and Human Monocytes | Primary/Healthy volunteers |
N/A |
| Candida albicans conidia/hyphae | ATCC | UC820 |
| Staphylococcus aureus | ATCC | ATCC 29213 |
| Bacteroides fragilis | ATCC | ATCC 25285 |
| Escherichia coli LPS | Sigma |
E. coli serotype 055:B5 |
| Recombinant DNA | ||
| No recombinant DNA was used in this study | ||
| Sequence-Based Reagents | ||
| AllPrep DNA/RNA Mini Kit | Qiagen | 80204 |
| Quant-iT PicoGreen dsDNA Assay | Life Technologies | P7589 |
| Nextera XT DNA Library Preparation kit | Illumina | FC-131-1096 |
| Software and Algorithms | ||
| KneadData | http://huttenhower.sph.harvard.edu/knead data | http://huttenhower.sph.harvard.edu/knead data |
| MetaPhlAn 2.2 | Segata et al., 2012 | http://huttenhower.sph.harvard.edu/metaphlan |
| HUMAnN2 | http://huttenhower.sph.harvard.edu/humann2 | http://huttenhower.sph.harvard.edu/humann2 |
| R | The R Project for Statistical Computing |
http://www.r-project.org/ |
| Other | ||
Acknowledgments
The authors thank all volunteers from the 500FG cohort for participating. The HFGP is supported by grants to MGN [ERC Consolidator Grant (3310372), Spinoza Prize (NOW SPI 94-212), and IN-CONTROL CVON grant] and CW [ERC Advanced Grant (FP/2007-2013/ERC grant 2012-322698), Spinoza Prize (NWO SPI 92-266), the Dutch Digestive Diseases Foundation (MLDS WO11-30), and the European Union’s Seventh Framework Programme (EU FP7) TANDEM project (HEALTH-F3-2012-305279)]. RJX was supported by grants from NIH (U54DK102557 and R01DK092405), Helmsley Charitable Trust, JDRF (17-2011-529), and CCFA (20144126). AZ, JF, and AK were supported by a CVON grant. SPS was financially supported by STW grant STW13546. AZ holds a Rosalind Franklin Fellowship (University of Groningen). JF is funded by the Netherlands Organization for Scientific Research (NWO-VIDI 864.13.013). We thank Tiffany Poon for help in sequence production and sample management and Natalia Nedelsky (Massachusetts General Hospital) for editorial help and help with figure generation. We also thank Edith Adriaanse, Marije van der Geest, and Marieke Bijlsma for structuring the online database and the MOLGENIS open source team, Dennis Hendriksen, Erwin Winder, Bart Charbon, Fleur Kelpin, Jonathan Jetten, Mark de Haan, Tommy de Boer, David van Enckevort, Chao Pang, Joeri van der Velde, and Morris Swertz for designing and implementing the database software. The creation and hosting of the online database was supported by BBMRI-NL, a research infrastructure financed by the Netherlands Organization for Scientific Research (NWO), grant number 184.021.007. We thank Maria Carmen Cenit, Astrid Maatman, Mathieu Plateel, and Jody Arends for technical support, and Dan Graham for helpful advice and discussions. We also thank thenounproject.com for providing the figure icons “Male Aging”/”Female Aging” by Marie Van den Broeck.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
MS performed the metagenomics data analyses and correlation analyses. SPS, MJ, MO, AK, and MJB performed the immunological assays and processed and analyzed the data. MS, HV, EAF, CH, MGN, and RJX interpreted the results. MS, EAF, and CH led the computational methods and research development. SPS, MJ, and MO collected clinical samples. SPS, TJ, and LJ performed functional validation experiments. MS, SPS, HV, CH, MGN, and RJX assembled and wrote the paper. MS, HV, MGN, and RJX served as project leaders. JF, AZ, LABJ, CW, MGN, and RJX designed the cohort study. CH, CW, MGN, and RJX served as principal investigators.
Table S1. Related to Figure 3. Correlation coefficients for all significant species associations
Table S2. Related to Figure 3. FDR values for all significant species associations
Table S3. Related to Figure 3. Correlation coefficients for all significant genus associations
Table S4. Related to Figure 3. FDR values for all significant genus associations
Table S5. Related to Figure 4. Correlation coefficients for all significant pathway associations
Table S6. Related to Figure 4. FDR values for all significant pathway associations
Table S7. Related to Figure 4. Correlation coefficients for all significant GO category associations
Table S8. Related to Figure 4. FDR values for all significant GO category associations
References
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- Bronte V, Zanovello P. Regulation of immune responses by L-arginine metabolism. Nat. Rev. Immunol. 2005;5:641–654. doi: 10.1038/nri1668. [DOI] [PubMed] [Google Scholar]
- Crost EH, Tailford LE, Le Gall G, Fons M, Henrissat B, Juge N. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS ONE. 2013;8:e76341. doi: 10.1371/journal.pone.0076341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia MS, Fischbach MA. HUMAN MICROBIOTA. Small molecules from the human microbiota. Science. 2015;349:1254766. doi: 10.1126/science.1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrestein PC, Mazmanian SK, Knight R. Finding the missing links among metabolites, microbes, and the host. Immunity. 2014;40:824–832. doi: 10.1016/j.immuni.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyerich S, Wagener J, Wenzel V, Scarponi C, Pennino D, Albanesi C, Schaller M, Behrendt H, Ring J, Schmidt-Weber CB, et al. IL-22 and TNF-alpha represent a key cytokine combination for epidermal integrity during infection with Candida albicans. Eur. J. Immunol. 2011;41:1894–1901. doi: 10.1002/eji.201041197. [DOI] [PubMed] [Google Scholar]
- Fu J, Bonder MJ, Cenit MC, Tigchelaar EF, Maatman A, Dekens JA, Brandsma E, Marczynska J, Imhann F, Weersma RK, et al. The Gut Microbiome Contributes to a Substantial Proportion of the Variation in Blood Lipids. Circ. Res. 2015;117:817–824. doi: 10.1161/CIRCRESAHA.115.306807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschfeld M, Ma Y, Weis JH, Vogel SN, Weis JJ. Cutting edge: repurification of lipopolysaccharide eliminates signaling through both human and murine toll-like receptor 2. J. Immunol. 2000;165:618–622. doi: 10.4049/jimmunol.165.2.618. [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project, C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosiewicz MM, Zirnheld AL, Alard P. Gut microbiota, immunity, and disease: a complex relationship. Front Microbiol. 2011;2:180. doi: 10.3389/fmicb.2011.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kverka M, Zakostelska Z, Klimesova K, Sokol D, Hudcovic T, Hrncir T, Rossmann P, Mrazek J, Kopecny J, Verdu EF, et al. Oral administration of Parabacteroides distasonis antigens attenuates experimental murine colitis through modulation of immunity and microbiota composition. Clin. Exp. Immunol. 2011;163:250–259. doi: 10.1111/j.1365-2249.2010.04286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JM, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016;22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MN, Ye C, Villani AC, Raj T, Li W, Eisenhaure TM, Imboywa SH, Chipendo PI, Ran FA, Slowikowski K, et al. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 2014;343:1246980. doi: 10.1126/science.1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WJ, Hase K. Gut microbiota-generated metabolites in animal health and disease. Nat. Chem. Biol. 2014;10:416–424. doi: 10.1038/nchembio.1535. [DOI] [PubMed] [Google Scholar]
- Lehrer RI, Cline MJ. Interaction of Candida albicans with human leukocytes and serum. J. Bacteriol. 1969;98:996–1004. doi: 10.1128/jb.98.3.996-1004.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Oosting M, Smeekens SP, Jaeger M, Aguirre-Gamboa R, Le KTT, Deelen P, Ricano-Ponce I, Schoffelen T, Jansen AFM, et al. A Functional Genomics Approach to Understand Variation in Cytokine Production in Humans. Cell. 2016 doi: 10.1016/j.cell.2016.10.017. In Press. [DOI] [PubMed] [Google Scholar]
- Lopez P, Gonzalez-Rodriguez I, Gueimonde M, Margolles A, Suarez A. Immune response to Bifidobacterium bifidum strains support Treg/Th17 plasticity. PLoS ONE. 2011;6:e24776. doi: 10.1371/journal.pone.0024776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Menard O, Butel MJ, Gaboriau-Routhiau V, Waligora-Dupriet AJ. Gnotobiotic mouse immune response induced by Bifidobacterium sp. strains isolated from infants. Appl. Environ. Microbiol. 2008;74:660–666. doi: 10.1128/AEM.01261-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett JR, Namboodiri MA. Tryptophan and the immune response. Immunol. Cell Biol. 2003;81:247–265. doi: 10.1046/j.1440-1711.2003.t01-1-01177.x. [DOI] [PubMed] [Google Scholar]
- Nowak EC, de Vries VC, Wasiuk A, Ahonen C, Bennett KA, Le Mercier I, Ha DG, Noelle RJ. Tryptophan hydroxylase-1 regulates immune tolerance and inflammation. J. Exp. Med. 2012;209:2127–2135. doi: 10.1084/jem.20120408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz CA, Wick W, Steinman L, Platten M. Tryptophan degradation in autoimmune diseases. Cell. Mol. Life Sci. 2007;64:2542–2563. doi: 10.1007/s00018-007-7140-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Q, Wagner WM, Wikby A, Remarque E, Pawelec G. Compromised interferon gamma (IFN-gamma) production in the elderly to both acute and latent viral antigen stimulation: contribution to the immune risk phenotype? Eur. Cytokine Netw. 2002;13:392–394. [PubMed] [Google Scholar]
- Paun A, Yau C, Danska JS. Immune recognition and response to the intestinal microbiome in type 1 diabetes. J. Autoimmun. 2016;71:10–18. doi: 10.1016/j.jaut.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Regitz-Zagrosek V. Therapeutic implications of the gender-specific aspects of cardiovascular disease. Nat. Rev. Drug Discov. 2006;5:425–438. doi: 10.1038/nrd2032. [DOI] [PubMed] [Google Scholar]
- Regitz-Zagrosek V, Lehmkuhl E, Weickert MO. Gender differences in the metabolic syndrome and their role for cardiovascular disease. Clin. Res. Cardiol. 2006;95:136–147. doi: 10.1007/s00392-006-0351-5. [DOI] [PubMed] [Google Scholar]
- Rothhammer V, Mascanfroni ID, Bunse L, Takenaka MC, Kenison JE, Mayo L, Chao CC, Patel B, Yan R, Blain M, et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 2016;22:586–597. doi: 10.1038/nm.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruschen S, Stellberg W, Warnatz H. Kinetics of cytokine secretion by mononuclear cells of the blood from rheumatoid arthritis patients are different from those of healthy controls. Clin. Exp. Immunol. 1992;89:32–37. doi: 10.1111/j.1365-2249.1992.tb06873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, Gill N, Blanchet MR, Mohn WW, McNagny KM, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13:440–447. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods. 2012;9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan GV, Choi K, Klemashevich C, Wu C, Prabakaran D, Pan LB, Steinmeyer S, Mueller C, Yousofshahi M, Alaniz RC, et al. Prediction and quantification of bioactive microbiota metabolites in the mouse gut. Nat. Commun. 2014;5:5492. doi: 10.1038/ncomms6492. [DOI] [PubMed] [Google Scholar]
- Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, Tjota MY, Seo GY, Cao S, Theriault BR, et al. Commensal bacteria protect against food allergen sensitization. Proc. Natl. Acad. Sci. U. S. A. 2014;111:13145–13150. doi: 10.1073/pnas.1412008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutmuller RP, den Brok MH, Kramer M, Bennink EJ, Toonen LW, Kullberg BJ, Joosten LA, Akira S, Netea MG, Adema GJ. Toll-like receptor 2 controls expansion and function of regulatory T cells. J. Clin. Invest. 2006;116:485–494. doi: 10.1172/JCI25439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter Horst R, Jaeger M, Smeekens SP, Oosting M, Swertz MA, Li Y, Kumar V, Diavatopoulos DA, Jansen AFM, Lemmers H, et al. Host and environmental factors influencing individual human cytokine responses. Cell. 2016 doi: 10.1016/j.cell.2016.10.018. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn AN, Macia L, Mackay CR. Diet, metabolites, and “western-lifestyle” inflammatory diseases. Immunity. 2014;40:833–842. doi: 10.1016/j.immuni.2014.05.014. [DOI] [PubMed] [Google Scholar]
- Tigchelaar EF, Zhernakova A, Dekens JA, Hermes G, Baranska A, Mujagic Z, Swertz MA, Munoz AM, Deelen P, Cenit MC, et al. Cohort profile: LifeLines DEEP, a prospective, general population cohort study in the northern Netherlands: study design and baseline characteristics. BMJ Open. 2015;5:e006772. doi: 10.1136/bmjopen-2014-006772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Veerdonk FL, Marijnissen RJ, Kullberg BJ, Koenen HJ, Cheng SC, Joosten I, van den Berg WB, Williams DL, van der Meer JW, Joosten LA, et al. The macrophage mannose receptor induces IL-17 in response to Candida albicans. Cell Host Microbe. 2009;5:329–340. doi: 10.1016/j.chom.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, Yassour M, Kolde R, Vlamakis H, Arthur TD, Hamalainen AM, et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell. 2016;165:842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welters HJ, Tadayyon M, Scarpello JH, Smith SA, Morgan NG. Mono-unsaturated fatty acids protect against beta-cell apoptosis induced by saturated fatty acids, serum withdrawal or cytokine exposure. FEBS Lett. 2004;560:103–108. doi: 10.1016/S0014-5793(04)00079-1. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanvit P, Konkel JE, Jiao X, Kasagi S, Zhang D, Wu R, Chia C, Ajami NJ, Smith DP, Petrosino JF, et al. Antibiotics in neonatal life increase murine susceptibility to experimental psoriasis. Nat. Commun. 2015;6:8424. doi: 10.1038/ncomms9424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Qiu X, Zhang H, Yang X, Hong N, Yang Y, Chen H, Yu C. Faecalibacterium prausnitzii inhibits interleukin-17 to ameliorate colorectal colitis in rats. PLoS ONE. 2014;9:e109146. doi: 10.1371/journal.pone.0109146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, Wu X, Li J, Tang L, Li Y, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 2015;21:895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, Mujagic Z, Vila AV, Falony G, Vieira-Silva S, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitomersky NL, Atkinson BJ, Franklin SW, Mitchell PD, Snapper SB, Comstock LE, Bousvaros A. Characterization of adherent bacteroidales from intestinal biopsies of children and young adults with inflammatory bowel disease. PLoS ONE. 2013;8:e63686. doi: 10.1371/journal.pone.0063686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) For the 500FG project, samples from 534 individuals were collected and all volunteers were asked to complete a detailed questionnaire. Only samples from healthy adults were further processed and the filtering criteria included several diseases as well as medication use. Further, only a very small number of the volunteers were from non-European descent and therefore these samples were excluded from the subsequent analysis.
(B) The 500FG cohort consists of 489 volunteers. Stool samples were collected from all individuals and subjected to metagenomic shotgun sequencing. Filtering samples with less than 4 million reads resulted in a subset of 471 samples. Cytokine data is available for a subset of 456/401/466 of these samples for measurements in whole blood/PBMC 1d/PBMC 7d.
(C) Experiments were performed in whole blood and peripheral blood mononuclear cells (PBMCs). Cytokines were measured in RPMI media (control) as well as in connection with lipopolysaccharide (LPS), Candida albicans conidia/hyphae and Staphylococcus aureus stimulation.
(D) To determine the generalizability of the microbial profiles from our cohort, we compared the profiles of the 500FG cohort to those of LifeLines-DEEP, another cohort within the HFGP. As part of LifeLines-DEEP, metagenomic sequencing was performed on 1,326 fecal samples from healthy adults from the Netherlands. Taxonomic profiles of the samples were generated for both cohorts (see STAR Methods) and samples were compared using Principal Coordinate Analysis (PCoA) with Bray-Curtis distance at species level. The 500FG and LLDeep cohort exhibit similar gut microbial profiles and appear to represent a single population.
Analogous to Figure 2D, we computed the overall percentage of cytokine variation explained by microbial pathways (A) and GO categories (B). To avoid overestimation due to correlations of the microbial features, we represented the microbiome data through the first 20 principle coordinates (PCoA analysis with Bray-Curtis distance) for pathways and GO categories, respectively. The cytokine variance explained by these principle coordinates was estimated through permutation ANOVA by summing over the significant contributions (p < 0.2).
Validation of the interaction of C. comes with the acute phase response pathway. A positive correlation was observed for C. comes abundance and concentration of the acute phase protein AAT (r = 0.08, p = 0.08).
(A) Palmitoleic acid (palmitoleate) biosynthesis (MetaCyc pathway 6283) converts a (5Z)-dodec-5-enoyl-[acp] into palmitoleic acid. One of the intermediates of this pathway, a palmitoleoyl-[acp], constitutes a branching point of this pathway leading to cis-vaccenate biosynthesis (MetaCyc pathway 5973).
(B) and (C) PBMCs were stimulated for 24 hours (IL-1β and IL-6) or 7 days (IL-17 and IL-22) with HK C. albicans conida in the absence or presence of palmitoleic acid (100 µM) or cis-vaccenic acid (50 µM). IL-1β, IL-6, IL-17, and IL-22 were measured in cell culture supernatants using ELISA. Cytokine production is expressed as percentage compared to DMSO (control palmitoleic acid) or ethanol (control cis-vaccenic acid). Groups were compared using paired-t-test.