Abstract
The oligosaccharyltransferase is the signature enzyme for N-linked glycosylation in all domains of life. In Archaea, this enzyme termed AglB, is responsible for transferring lipid carrier-linked glycans to select asparagine residues in a variety of target proteins including archaellins, S-layer proteins and pilins. This study investigated the ability of a variety of AglBs to compensate for the oligosaccharyltransferase activity in Methanococcus maripaludis deleted for aglB, using archaellin FlaB2 as the reporter protein since all archaellins in Mc. maripaludis are modified at multiple sites by an N-linked tetrasaccharide and this modification is required for archaellation. In the Mc. maripaludis ΔaglB strain FlaB2 runs as at a smaller apparent molecular weight in western blots and is nonarchaellated. We demonstrate that AglBs from Methanococcus voltae and Methanothermococcus thermolithotrophicus functionally replaced the oligosaccharyltransferase activity missing in the Mc. maripaludis ΔaglB strain, both returning the apparent molecular weight of FlaB2 to wildtype size and restoring archaellation. This demonstrates that AglB from Mc. voltae has a relaxed specificity for the linking sugar of the transferred glycan since while the N-linked glycan present in Mc. voltae is similar to that of Mc. maripaludis, the Mc. voltae glycan uses N-acetylglucosamine as the linking sugar. In Mc. maripaludis that role is held by N-acetylgalactosamine. This study also identifies aglB from Mtc. thermolithotrophicus for the first time by its activity. Attempts to use AglB from Methanocaldococcus jannaschii, Haloferax volcanii or Sulfolobus acidocaldarius to functionally replace the oligosaccharyltransferase activity missing in the Mc. maripaludis ΔaglB strain were unsuccessful.
Introduction
N-glycosylation refers to the covalent attachment of glycans to target proteins at asparagine residues located within a conserved sequon (Asn-X-Ser/Thr, where X can be any amino acid except proline). The oligosaccharyltransferase (OST) is the signature enzyme of the N-glycosylation pathways in all three domains of life [1–3]. In both prokaryotic domains, Bacteria and Archaea, the OST exists as a single subunit designated PglB [4] and AglB [5], respectively. In higher Eukaryotes, the OST is a multi-subunit complex with the Stt3 subunit identified as the catalytic subunit [6]. Both PglB and AglB are homologs of Stt3 [1,4].
The N-glycosylation pathway in Archaea has been best studied, in terms of both glycan structure and genetic analysis of the biosynthesis and assembly of the glycan, in three distinctive model organisms, namely the methanogen Methanococcus maripaludis, the extreme halophile Haloferax volcanii and the thermoacidophile Sulfolobus acidocaldarius [1,7,8]. Glycan structures alone are published for several other varied members of this domain [1,9,11]. The most commonly studied proteins modified by N-linked glycans in archaea are S-layer proteins [12–14], archaellins [15–19] and pilins [20–22]. The current model of the archaeal N-glycosylation pathway involves the sequential addition of sugar monomers onto a dolichol-type lipid carrier embedded in the cytoplasmic membrane, followed by a flipping of the lipid-linked glycan across the membrane. Finally, on the external face side of the cytoplasmic membrane, AglB transfers the glycan from the lipid carrier onto the acceptor protein en bloc [1,23]. Further addition of sugar monomers onto the protein-bound N-glycan has been shown to occur in a very limited number of archaeal species [24,25].
Among the three domains, it appears that glycan structures as well as the nature of the linking sugar and the lipid carrier are most variable in Archaea [23]. In different archaea, the sugar linking the glycan to the asparagine of the target protein can be N-acetyl-glucosamine, N-acetyl-galactosamine or a simple hexose [1]. The lipid carrier has been identified in several archaea and can be dolichol phosphate or dolichol diphosphate and the dolichol can vary in length as well as degree of saturation [2,24,26–30]. These are all aspects that may affect AglB activity. In both Mc. maripaludis and Hfx. volcanii, aglB deletion mutants have been isolated [5,12,31] while the enzyme is essential for S. acidocaldarius [32].
The specificity of AglB for different sugar-donor substrates was first reported for enzymes from Archeoglobus fulgidus and Pyrococcus furiosus. In in vitro experiments, neither enzyme could process the lipid-linked glycan of the other organism [33], indicating specificity of the enzyme but it is unclear whether this is due to the structure of the different glycans or other factors. More recently, Eichler’s group showed substrate promiscuity of AglB from various extreme halophiles [34]. Specifically, AglB from Haloarcula marismortui, Halobacterium salinarum and Haloferax mediterranei could all functionally replace the oligosaccharyltransferase activity in a Hfx. volcanii ΔaglB strain. While all of these species are extreme halophiles, their respective AglBs, though structurally similar, all transfer lipid-linked glycans in their native cells that are distinct from that found in Hfx. volcanii. In addition, in at least some cases, the heterologous AglB had to accommodate a different linking sugar or dolichol length to be effective, demonstrating a relaxed specificity of the enzymes.
The overwhelming majority of archaea (166/168 sequenced genomes examined) contain an identifiable aglB [35], including a significant number of organisms (>30) that appear to possess multiple copies of the gene. Examination of the multiple copies of aglB within a given organism indicated that, at least in some cases, distinct versions of the enzyme are present that possess, for example, variations in the catalytic motif WWDXG. Other studies have indicated that not all versions of AglB are constitutively expressed in organisms that have multiple copies [10,24]. In the case of Hfx. volcanii, two distinct N-linked glycans have been reported, depending on the salt concentration of the medium [36]. The transfer of the low salinity glycan still occurs in an aglB mutant even though only a single aglB gene is detected in the sequenced genome [35] suggesting the existence of either an additional aglB with such low sequence similarity to known aglBs that it escaped detection or a novel mechanism/enzyme for the transfer of the low salinity glycan. The presence of multiple AglBs in a single organism, some not constitutively expressed, some with variations in conserved motifs, suggest that they may be involved in different transfer reactions with different substrates. Such is the case in Trypanosoma brucei, where there are three single subunit OSTases, all with distinct donor and acceptor specificities [37].
In Mc. maripaludis, N-glycosylation of archaellins (the structural proteins of the motility apparatus archaella [38,39]) is critical for their assembly into archaella [31,40,41]. In wildtype cells, the archaellins are decorated at multiple locations with a tetrasaccharide [17]. In mutants in which the resulting glycan is truncated, the shortened glycan, even a monosaccharide version, is still efficiently transferred to the target archaellins indicating that AglB does not require the complete glycan structure for transfer [31]. In vitro assays using purified Mc. voltae AglB showed efficient transfer of a truncated disaccharide glycan but not if Dol-P with a monosaccharide was used as the donor [2]. While cells can assemble functional archaella with only a truncated disaccharide attached to archaellins, if a mutation occurs that results in the glycan being truncated to a single sugar or in archaellins that are completely non-glycosylated, then the cells are nonarchaellated [31]. The latter case occurs if aglB is deleted. Interestingly, in cases where genes are deleted that result in nonarchaellated cells, such as the ΔaglB deletion mutant, these mutants subsequently stop transcription of the fla operon, a series of genes which includes the three archaellin genes as well as a number of accessory genes (flaC-J) also required for archaella assembly [42]. This occurs, at least in some cases, due to a second mutation in a recently described transcriptional activator for the fla operon, EarA [43].
In this contribution, we examine the ability of several heterologous AglBs to functionally compensate for the oligosaccharyltransferase activity in a Mc. maripaludis strain deleted for aglB. This information contributes to the substrate specificity/variability of AglB and may aid in studies designed to use Archaea for glyco-engineering purposes [44,45].
Materials and Methods
Strains and growth conditions
Methanococcus maripaludis S2 Δhpt (Mm900) [46], Methanococcus voltae PS, Methanothermococcus thermolithotrophicus DSM2095 were all grown in Balch Medium III [47] under a headspace of CO2/H2 (20:80). Methanocaldococcus jannaschii JAL-1 was grown in the minimal medium described by Ferrante et al. [48]. Mc. maripaludis and Mc. voltae were incubated at 35°C, Mtc. thermolithotrophicus at 60°C and Mcc. jannaschii at 80°C. For complementation studies, Mc. maripaludis ΔaglB harbouring the various complementation vectors were grown in nitrogen-free medium containing puromycin (2.5μg/ml) for plasmid selection and supplemented with either L-alanine (10mM) or NH4Cl (10mM) as sole nitrogen source [49]. Escherichia coli Top10 cells (Invitrogen, Burlington ON, Canada), used for various cloning steps, were grown at 37°C in Luria-Bertani (LB) broth or agar with ampicillin added (100μg/ml) for plasmid selection when required.
Isolation of a Mc. maripaludis ΔaglB mutant
An in-frame deletion of aglB in Mc. maripaludis was re-created using pKJ574 and methodology previously reported [31]. Following confirmation of the inframe deletion by PCR screening and sequencing this strain was designated as ΔaglB-14-9.
Cloning of various aglBs
aglB, from Mc. voltae (GenBank accession ABD17750), was amplified by PCR (50°C annealing temperature, 3 min extension, 30 cycles) using primers listed in Table 1 and washed Mc. voltae cells as template. The forward and reverse primers had either NsiI or MluI restriction sites added, respectively. For Mtc. thermolithotrophicus, the aglB sequence has not been previously reported. A BLAST search using the AglB protein sequence from Mc. maripaludis as bait, retrieved WP_018154595 in GenPept as the only hit (100% coverage, 59% identity, 77% similarity) in Mtc. thermolithotrophicus DSM 2095. The gene (GenBank accession NZ_AQXV01000054.1) was amplified by PCR (50°C annealing temperature, 3 min extension, 30 cycles) using the primers listed in Table 1 and washed cells of M. thermolithotrophicus as template. The aglB gene from Mcc. jannaschii (MJ_RS08150) was amplified by PCR (50°C annealing temperature, 3 min extension, 30 cycles) using the primers listed in Table 1 and washed cells of Mcc. jannaschii as template. The forward primer in this case included a single nucleotide change to remove an internal NsiI restriction site while leaving the amino acid sequence unchanged. The aglB gene from Hfx. volcanii (HVO_RS12050) was amplified by PCR (62.5°C annealing temperature, 3 min extension, 30 cycles) with the primers listed in Table 1 and genomic DNA (gift of Jerry Eichler) as template. Hfx. volcanii aglB and Sulfolobus acidocaldarius aglB (NC_007181.1) were also synthesized with a C-terminal FLAG-tag using Mc. maripaludis codon preferences while avoiding NsiI and MluI sites (GenScript, NJ). Each of the aglB genes was digested with NsiI and MluI and cloned into pHW40 previously digested with the same restriction enzymes to generate the complementation plasmids used in this study (Table 2). Transcription of the cloned aglB is under the control of a regulatable nif promoter in this vector. The complementation plasmid carrying the Mc. maripaludis aglB, pKJ677, was previously constructed [31]. A site directed mutagenesis step to remove an internal NsiI restriction site was performed prior to the cloning of the Mc. maripaludis aglB into pHW40 as an NsiI/XbaI fragment (see Table 1 for primer pair used).
Table 1. Primers used in this study.
| Primers | Sequence (5’ to 3’) | Restriction site (underlined) |
|---|---|---|
| AglB complementation | ||
| MaraglB-F | CCCATGCATGGGTGAATTTTTAAATAAAGTC | NsiI |
| MaraglB-R | GCTCTAGATTAGTGATGGTGGTGATGATGATTGAGATAGTCAGTTCCA | XbaI |
| VoltaglB-F | CGTAATGCATGACTGAAAACAACGAAAAAGTCAAAAATTCCGATTCTGC | NsiI |
| VoltaglB-R | GACTACGCGTTTATTTTGAGTAATTACCGTAATCCACGC | MluI |
| ThermaglB-F | CCAATGCATGGGGGAATTCCTGAACAAATTTTCC | NsiI |
| ThermaglB-R | AGCACGCGTTTAGTTAAGATAATCTATTCCATAATC | MluI |
| JannaglB-F* | CCAATGCATGTATATAAAGGTGAAACTTATGAGTAATGCTTTAG | NsiI |
| JannaglB-R | AGCACGCGTTATTTTAGATAATCAGTTCCATAATC | MluI |
| HvaglB-F | CAGTATGCATGAGTGACGAGCAAACAAAGTATTCG | NsiI |
| HvaglB-R | TACGTCTAGATTACGCGGCGGCCGAGACCG | XbaI |
| SDM** | ||
| MaraglBSDM-F | GAAGTTGAAAACGCATGCCCAACC | |
| MaraglBSDM-R | GGTTGGGCATGCGTTTTCAACTTC | |
| RT-PCR | ||
| SaaglB-RT-F | CACACTTACTCTTGTTTCACC | |
| SaaglB-RT-R | GCATCTCGAGCCAGTTATCGGCAGAATCGTAG | |
| HvaglB-RT-F | GATTCGCAGACCACAACATCG | |
| HvaglB-RT-R | GAAGTTTCTTGCGATTGAGTCG |
*T shown in bold underline removes and internal NsiI site
**SDM primers remove internal NsiI site in Mc. maripaludis aglB. Change is underlined
Table 2. Plasmids used in this study.
| Plasmids | Description | Reference/Source |
|---|---|---|
| pHW40 | nif promoter-lacZ fusion plus Purr cassette;Ampr | John Leigh |
| pCRPrtNeo | hmv promoter-hpt fusion plus Neor cassette in pCR2.1Topo; Ampr | [46] |
| pKJ574 | pCRPrtNeo harboring an inframe deletion of aglB | [31] |
| pKJ677 | pHW40 harboring Mc. maripaludis aglB | This study |
| pKJ1229 | pHW40 harboring Mc. voltae aglB | This study |
| pKJ1240* | pHW40 harboring S. acidocaldarius aglB | This study |
| pKJ1248 | pHW40 harboring Mtc. thermolithotrophicus aglB | This study |
| pKJ1251* | pHW40 harboring Hfx. volcanii aglB | This study |
| pKJ1267 | pHW40 harboring Mcc. jannaschii aglB | This study |
*gene synthesized using Mc. maripaludis codon preferences
Complementation of the Mc. maripaludis ΔaglB mutant
Mc. maripaludis ΔaglB 14–9 was transformed with the various pHW40-derived plasmids containing different aglB genes using the PEG precipitation method [50]. The complementation strains were subsequently grown in the presence of puromycin (2.5 μg/ml) in nitrogen-free medium supplemented with either 10 mM NH4Cl (nif promoter repressed) or alanine (nif promoter induced). At least three transfers in nitrogen-free medium supplemented with alanine were done prior to analysis.
Reverse transcription polymerase chain reaction (RT-PCR)
Total RNA from ΔaglB mutant cells carrying either pKJ1240 (with the Mc. maripaludis codon-optimized aglB from S. acidocaldarius, aglBsa) or pKJ1251 (with the Mc. maripaludis codon-optimized aglB from Hfx. volcanii, aglBhv) was extracted using a High Pure RNA Isolation Kit (Roche Life Science), followed with an additional DNase digest using an Ambion™ TURBO DNA-free Kit (Invitrogen). The presence of the aglBsa or aglBhv transcript in the RNA extract was detected using a OneStep RT-PCR Kit (Qiagen) with 10 ng of the total RNA extract as template and the corresponding gene-specific primers listed in Table 1. After a 30 min reverse transcription step at 50°C and a 15 min initial PCR activation step at 94°C, the polymerase chain reaction consisted of 30 cycles of 30 sec denaturation at 94°C, 30 sec annealing at 48°C, and 30 sec extension at 72°C. Amplification experiments using RNA extracts from ΔaglB mutant cells carrying either pKJ1240 or pKJ1251 as template and the corresponding gene-specific primer pair, but omitting the reverse transcription step were conducted to rule out the possibility of DNA contamination of the RNA template.
PCR experiments using pKJ1240 or pKJ1251 as template and the corresponding gene-specific primer pairs were performed to confirm the amplicon size and primer specificity in the RT-PCR. In addition, RT-PCR experiments were also conducted using RNA extract from ΔaglB mutant cells carrying plasmid-borne aglBsa (pKJ1240) as template and the aglBhv primer pair and RNA extract from ΔaglB mutant cells carrying plasmid-borne algBhv (pKJ1251) as template and the aglBsa primer pair to further exclude the possibility of non-specific amplification of the two primer pairs.
Western blotting
Whole cell lysates of Mc. maripaludis Mm900, Mc. maripaludis ΔaglB-14-9 and the various Mc. maripaludis ΔaglB-14-9 strains carrying complementation plasmids were separated by SDS-PAGE (12.5% acrylamide gels) and transferred to Immobilon-P membrane (Millipore, MA) and examined by western blotting using anti-FlaB2 specific antibodies, as previously described [42]. Complemented strains were transferred three times in nitrogen-free medium supplemented with either NH4Cl or alanine prior to the western blotting experiments. Detection of FLAG-tagged versions of Hfx. volcanii and S. acidocaldarius AglB in whole cell lysates of the appropriate complementation strains was attempted using anti-FLAG antibodies (mouse monoclonal, Sigma).
Electron microscopy
Transmission electron microscopy was performed on the various Mc. maripaludis strains after staining with 2% (w/v) phosphotungstic acid, pH 7.0, as previously described [43].
Bioinformatics
The presence of transmembrane domains in AglB proteins selected for this study was predicted using HMMTOP (http://www.enzim.hu/hmmtop/) [51].
Results and Discussion
AglB from Mc. maripaludis restores N-glycan modification of FlaB2 and archaellation in the ΔaglB-14-9 mutant
AglB is the oligosaccharyltransferase that performs the most conserved and critical terminal step in the N-glycosylation pathway in Archaea, namely the transfer of a glycan from its lipid carrier to the target protein [5]. Following the initial isolation of the first aglB deletion strain in Mc. maripaludis, attempts were made to complement that strain with a wildtype copy of aglB expressed in trans. However, it was not possible to confirm that the expressed wildtype copy of aglB could rescue the deletion strain since the aglB mutant quickly stopped synthesis of FlaB2 upon repeated serial transfers [31]. This turned out to be a common occurrence for any mutant carrying a deletion in any gene that resulted in nonarchaellated cells [43].
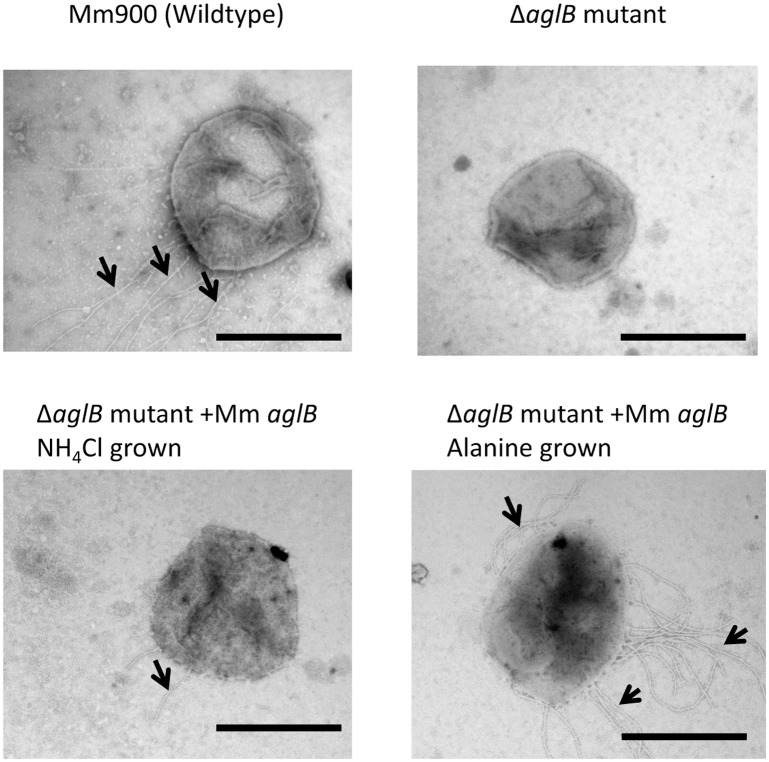
Thus, for this study, we initially had to recreate the aglB deletion and immediately store aliquots of the mutant, designated ΔaglB-14-9, at -80°C while it was still synthesizing FlaB2 that could be detected in western blots. Using pKJ677 expressing wildtype Mc. maripaludis aglB from a nif promoter, it was possible to show complementation of the deleted aglB in the ΔaglB-14-9 mutant. This was initially demonstrated by western blotting where the faster migrating version of the reporter glycoprotein FlaB2 in ΔaglB-14-9 was fully restored to wildtype size following growth of the complemented strain in nitrogen-free medium supplemented with alanine, where transcription from the nif promoter is induced (Fig 1). A small amount of the wildtype size FlaB2, in addition to minor amounts of intermediate sized FlaB2, was also observed when complemented cells were grown in nitrogen-free medium supplemented with NH4Cl, conditions in which transcription from the nif promoter is repressed. This has been observed previously in complementation experiments and attributed to very small amounts of transcription from the nif promoter even under NH4Cl growth conditions [31,40,52,53]. aglB deletion mutants are nonarchaellated due to a requirement for at least a truncated disaccharide version of the wildtype tetrasaccharide N-linked glycan to be attached to the archaellins for their assembly into archaella [31]. Thus, complemented cells were also examined by electron microscopy for the presence of archaella. Such examination revealed that under alanine growth conditions the complemented ΔaglB-14-9 cells were now archaellated (Fig 2).
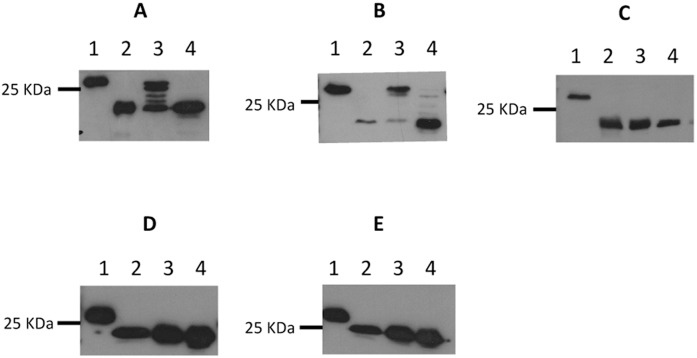
Fig 1. Western blot analysis of FlaB2 in an Mc. maripaludis ΔaglB-14-9 mutant complemented in trans with homologous aglB.
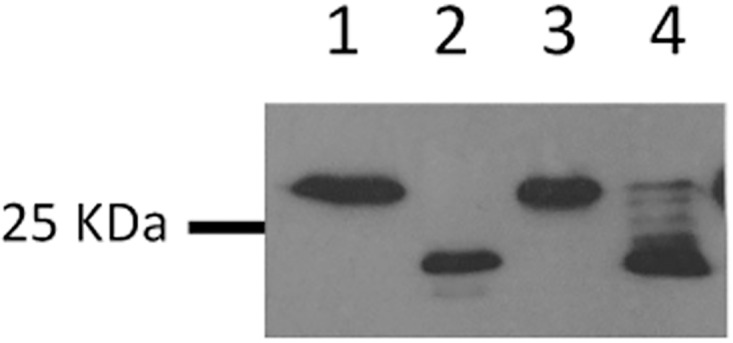
The ΔaglB-14-9 mutant was complemented with a plasmid borne version of Mc. maripaludis aglB under the control of the nif promoter. The complemented ΔaglB-14-9 mutant was grown in nitrogen-free medium supplemented with alanine or NH4Cl which results in transcription from the nif promoter being on (alanine) or off (NH4Cl). Lane 1 is Mm900 (WT), lane 2 is ΔaglB-14-9 mutant, lane 3 is ΔaglB-14-9 mutant complemented cells grown in nitrogen-free medium supplemented with alanine, lane 4 is ΔaglB-14-9 mutant complemented cells grown in nitrogen-free medium supplemented with NH4Cl.
Fig 2. Electron micrographs of the Mc. maripaludis ΔaglB-14-9 mutant complemented in trans with homologous aglB.
The ΔaglB-14-9 mutant was complemented with a plasmid borne versions of Mc. maripaludis aglB under the control of the nif promoter. Complemented cells were examined after a minimum of three transfers in nitrogen-free medium supplemented with NH4Cl or alanine by negative staining (2% phosphotungstic acid, pH 7.0). Arrows indicate archaella. Bar equals 1μm.
Comparison of selected AglBs used in this study
To study the promiscuous nature of archaeal AglBs, the ΔaglB-14-9 strain was also complemented with various heterologous aglBs, including from other methanogens (Mc. voltae, Mtc. thermolithotrophicus and Mcc. jannaschii) and from other more distantly related archaeal species in which the N-glycosylation system has been best studied, namely S. acidocaldarius and Hfx. volcanii. Successful complementation would also indicate, in cases where it was not yet known (Mtc. thermolithotrophicus and Mcc. jannaschii), that the putative aglB gene did encode an active oligosaccharyltransferase. The topology of AglB proteins include a typical 13 transmembrane helices in the N-termini with extracellular loops between transmembrane helixes, and a soluble C-terminal domain located in the extracellular side of the cytoplasmic membrane where the catalytic site is located [54–56]. A pairwise comparison of the Mc. maripaludis AglB to the other AglBs used in this study is presented in Table 3 (see also S1–S5 Figs for EMBOSS Needle pairwise alignments). The three methanogen AglBs used in this study were from the mesophilic Mc. voltae, the thermophilic Mtc. thermolithotrophicus (optimal growth at 60–65°C) and the hyperthermophilic Mcc. jannaschii (optimal growth at 80–85°C). These methanogens, as well as Mc. maripaludis, are all stringent anaerobes that grow optimally at neutral pH with 1–4% NaCl. Hfx. volcanii is an extreme halophile growing optimally in 1.5–2.5M NaCl, at pH 7 and 45°C. S. acidocaldarius, on the other hand, is a thermoacidophile, growing optimally at 70–75°C and pH 2–3. Since the catalytic site in AglB is predicted to be orientated extracellularly [34], the enzymes from Hfx. volcanii and S. acidocaldarius may have specific adaptations to function in high salt or low pH that would not be required for the methanogen AglBs.
Table 3. Comparison of Mc. maripaludis AglB to other AglBs used in this study (pairwise EMBOSS Needle).
| Organism | Length (aa) | TMDs | % identity | % similarity | Gaps |
|---|---|---|---|---|---|
| Mc. maripaludis | 852 | 13 | |||
| Mc. voltae | 917 | 13 | 49.2 | 66.3 | 8.8 |
| Mtc. thermolithotrophicus | 860 | 13 | 59.4 | 77.5 | 1.4 |
| Mcc. jannaschii | 933 | 15 | 46.9 | 62.6 | 13.7 |
| Hfx. volcanii | 1054 | 14 | 17.9 | 31.0 | 39.7 |
| S. acidocaldarius | 754 | 15 | 19.9 | 34.6 | 30.1 |
AglB of Mtc. thermolithotrophicus was most similar to AglB of Mc. maripaludis, being almost 60% identical (and 77.5% similar) and almost identical in length (860 amino acids vs 852 for Mc. maripaludis AglB). AglBs from Mc. voltae and Mcc. jannaschii were larger by 65 and 81 amino acids, respectively and slightly less than 50% identical to the Mc. maripaludis enzyme. Most of the extra length in the Mcc. jannaschii enzyme is found in one large insertion of about 70 amino acids (between amino acids 227 and 299), when compared to the Mc. maripaludis AglB. This insertion is predicted to contain 2 additional transmembrane helices, resulting in a total of 15, compared to 13 for AglBs of Mc. maripaludis, Mc. voltae and Mtc. thermothithotrophicus. The AglBs from the two non-methanogens tested, Hfx. volcanii and S. acidocaldarius, were much less similar to the Mc. maripaludis AglB, being only 17.9 and 19.9% identical, respectively. In addition, these two oligosaccharyltransferases were much different in length compared to the Mc. maripaludis AglB. The Hfx. volcanii AglB was almost 200 amino acids larger than the Mc. maripaludis enzyme while that of S. acidocaldarius was almost 100 amino acids smaller.
Examination of the 6 AglBs revealed differences in conserved motifs previously shown to be important for catalysis (Fig 3). The xxD motif (sometimes referred to as the ExD or DxD motif) is thought to bind the dolichol carrier (DolP or DolPP). In all four methanogen AglBs, this motif is ALD, while in Hfx. volcanii it is GND and in S. acidocaldarius it is GFD. In all cases, this motif is found in the first extracellular loop in the N-terminus of the protein. The second motif, WWDXG, is necessary for catalysis and found in the extracellular C-terminal domain. In the four methanogens, this motif is identical, WWDNG, while in both Hfx. volcanii and S. acidocaldarius the sequence of this motif is WWDYG. Most variation is found in the third motif, the DK (DXXK or the relaxed version DXXMXXX(M/I)) or MI (MXXIXXX(I/V/W) motif [57]. This motif helps to form the pocket that recognizes the serine or threonine residue found in the N-linked glycosylation sequon (N-X-S/T) and is found adjacent to the WWDXG motif in the tertiary structure of the enzymes. The DK motif is found in most AglB proteins (as well as in eukaryotic Stt3) although variations in the motif have been reported in terms of an insertion of 4–14 amino acids that interrupts the DK motif in some archaea (DXXMX(4–14)K), including in certain extreme halophiles. Hfx. volcanii AglB has a 5 amino acid insertion here. S. acidocaldarius AglB has the DK motif (DIAK) which is typical of Crenarchaeota. In lieu of the DK motif, some archaea (as well as bacterial PglBs) have an MI motif at the same position which is thought to perform the same function as the DK motif. This is the case for the four methanogens in this study. The three key amino acids are conserved in all four cases (MXXIXXXW) although there is some variation in the X positions (Fig 3). Since the side chains of the amino acids that comprise the DK and MI motif are very different, it is unclear what differences in substrate recognition this variation may have [57].
Fig 3. Comparison of the conserved motifs in the various AglBs used in this study.

The putative xxD, WWDXG and DK/MI motifs for the 6 AglBs used in this study are indicated in bold.
AglB homologues from Mc. voltae and Mtc. thermolithotrophicus but not Mcc. jannaschii could functionally replace the OST activity in Mc. maripaludis ΔaglB-14-9
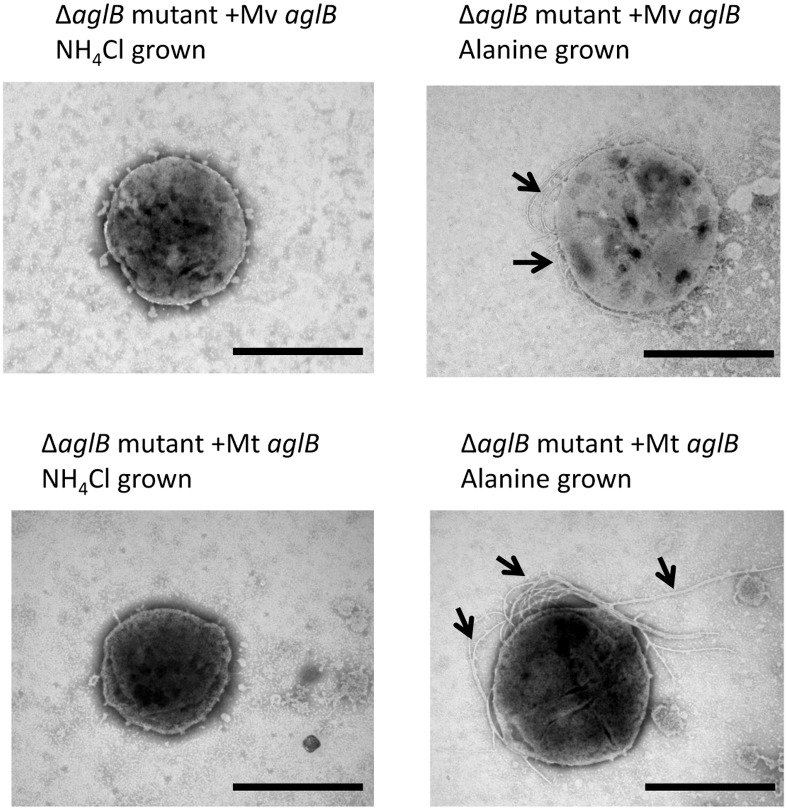
In all attempted cases of heterologous complementation of the Mc. maripaludis ΔaglB mutant, the initial screen to indicate successful complementation was by western blotting after growth of the complemented ΔaglB-14-9 strain in nitrogen-free medium supplemented with alanine or NH4Cl. Only a limited number of archaeal N-linked glycan structures are known [1] and of the three other methanogens tested in this study only in Mc. voltae has an N-linked glycan structure been reported. It is a trisaccharide with significant similarity to the tetrasaccharide of Mc. maripaludis [15,17]. Interestingly, however, the linking sugar is different, though both are N-acetylated. In the case of Mc. voltae, this position is held by an N-acetyl-glucosamine while in Mc. maripaludis the linking sugar in the glycan is N-acetyl-galactosamine. Transformation of the ΔaglB-14-9 strain with pKJ1229 containing Mc. voltae aglB, followed by growth in alanine supplemented nitrogen-free medium indicated that the Mc. voltae enzyme was capable of restoring the lost oligosaccharyltransferase activity leading to the production of wildtype sized FlaB2s (Fig 4A) which were assembled into archaella (Fig 5). Clearly, the Mc. voltae enzyme is active on glycan structures with either GlcNAc or GalNAc as the linking sugar. The promiscuity towards glycans with different linking sugars is similar to reports in the halophile system where AglBs from different halophiles can transfer glycans with hexose or N-acetylhexuronic acid acting as the linking sugar [34]. This confirms by enzyme activity the aglB gene of Mc. voltae previously identified only by deletion analysis [5].
Fig 4. Western blot analysis of FlaB2 in an Mc. maripaludis ΔaglB-14-9 mutant complemented in trans with heterologous aglBs.
The ΔaglB-14-9 mutant was complemented with plasmid borne versions of heterologous aglB from Mc. voltae (A), Mtc. thermolithotrophicus (B), Mcc. jannaschii (C), Hfx. volcanii (D) or S. acidocaldarius (E). For D and E the aglB was synthesized using Mc. maripaludis codon preferences. The complemented ΔaglB-14-9 mutant was grown in nitrogen-free medium supplemented with alanine or NH4Cl which results in transcription from the nif promoter being on (alanine) or off (NH4Cl). In each panel, lane 1 is Mm900 (WT), lane 2 is ΔaglB-14-9 mutant, lane 3 is ΔaglB-14-9 mutant complemented cells grown in nitrogen-free medium supplemented with alanine, lane 4 is ΔaglB-14-9 mutant complemented cells grown in nitrogen-free medium supplemented with NH4Cl.
Fig 5. Electron micrographs of the Mc. maripaludis ΔaglB-14-9 mutant complemented in trans with heterologous aglBs.
The ΔaglB-14-9 mutant was complemented with plasmid borne versions of aglB from Mc. voltae (Mv aglB) or Mtc. thermolithotrophicus (Mt aglB). Complemented cells were examined after a minimum of three transfers in nitrogen-free medium supplemented with NH4Cl or alanine by negative staining (2% phosphotungstic acid, pH 7.0). Arrows indicate archaella. Bar equals 1μm.
aglB from two other genera of Methanococcales with higher optimal growth temperatures was also tested for their ability to complement the ΔaglB-14-9 strain. For both Mtc. thermolithotrophicus and Mcc. jannaschii, no N–linked glycan structures have been reported, although the archaellins of Mtc. thermolithotrophicus contain many potential glycosylation sequons [58] and were reported to stain weakly with glycoprotein stains [59]. Mtc. thermolithotrophicus was not one of the 168 species of archaea examined for the presence of aglB by the Eichler group [35] since a complete, annotated genome is not available. However, a BLAST search revealed a single Mtc. thermolithotrophicus gene (GenBank accession No. NZ_AQXV01000054.1) from a whole genome shotgun sequence (GenBank accession No. NZ_AQXV01000019) with high sequence identity to aglB from Mc. maripaludis. Despite being a thermophile with optimal growth near 60°C, this gene from Mtc. thermolithotrophicus was able to very effectively restore wildtype size FlaB2 (Fig 4B) as well as archaellation (Fig 5) to the ΔaglB-14-9 strain grown at 35°C when used in complementation studies, thereby confirming the identity of the Mtc. thermolithotrophicus OST by activity.
All attempts to show complementation of the ΔaglB-14-9 strain with the Mcc. jannaschii aglB homolog were unsuccessful (Fig 4C). The failure of the Mcc. jannaschii AglB to complement is intriguing. This AglB has very high sequence identity and similarity to that of Mc. maripaludis. Furthermore, the three conserved motifs examined had identical sequences in the Mcc. jannaschii AglB to that found in AglB from Mtc. thermolithotrophicus, which did complement the aglB deletion strain of Mc. maripaludis. Strikingly though, there is a large insertion (approximately 70 amino acids) found in the Mcc. jannaschii AglB lacking in the remaining three methanogen AglBs. The lack of complementation may be due simply to the activity profile of the Mcc. jannaschii AglB in regards to temperature. Complementations of the aglB mutant of Mc. maripaludis were done at 35°C and 40°C while Mcc. jannaschii grows optimally above 80°C. Growth of Mc. maripaludis can occur to 45°C but above 42°C the reporter protein we use, FlaB2 archaellin, is not made [60]. However, other possible reasons for the failure of this complementation can also be considered. While all aglBs were cloned into the same site of the same vector and transcribed from the same nif promoter, we cannot rule out that the lack of activity demonstrated was due to degradation/instability of the expressed Mcc. jannaschii AglB. Furthermore, the lipid composition of the cytoplasmic membrane of Mcc. jannaschii is known to vary considerably depending on the growth temperature. At the low end of its growth range (45°C), the lipids are mostly typical diethers, but as the growth temperature is raised to 75°C the proportion of tetraether lipids and a novel macrocyclic diether lipid dramatically increase [61]. The inability of the Mcc. jannaschii aglB to complement in Mc. maripaludis may thus be due to faulty insertion of the AglB into the strictly diether lipids in the cytoplasmic membrane of Mc. maripaludis [62]. This would not have been a problem for the AglBs of Mc. voltae or Mtc. thermolithotrophicus since both these organisms have membrane lipids that are also diethers [63]. As with Mtc. thermolithotrophicus, no N-linked glycan structures have been reported for Mcc. jannaschii and, likewise, there are no reports on the structures of dolichol phosphate carriers so it is possible that AglB in Mcc. jannaschii would naturally recognize a unique glycan structure possibly with a different linking sugar or a different lipid carrier and was not able to utilize the Mc. maripaludis versions.
AglB homologues from S. acidocaldarius and Hfx. volcanii could not functionally replace the OST activity in Mc. maripaludis ΔaglB-14-9
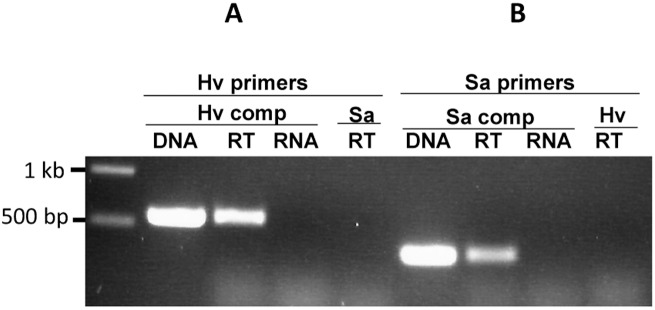
The other two failed complementations used the aglBs from S. acidocaldarius and Hfx. volcanii (Fig 4D and 4E). These two organisms are much more distantly related to Mc. maripaludis than Mc. voltae and Mtc. thermolithotrophicus, with S. acidocaldarius being a member of a different phylum, the Crenarchaeota. These organisms were chosen as, along with Mc. maripaludis, they are two of the best studied in terms of various aspects of the N-glycosylation pathways with information available on glycan structure, linking sugars, dolichol carriers and AglB [1]. For these complementation studies, the Hfx. volcanii and S. acidocaldarius aglB were synthesized using Mc. maripaludis codon preferences and with the addition of a C-terminal FLAG tag. Using the same conditions that resulted in successful complementation as reported above, no indication of successful complementation of the ΔaglB-14-9 strain was observed by western blot using either the S. acidocaldarius or Hfx. volcanii aglB. While attempts to detect synthesis of the AglBs from Hfx. volcanii and S. acidocaldarius with anti-FLAG antiserum in the complemented ΔaglB-14-9 strain lysates were unsuccessful (not shown), RT-PCR demonstrated that the mRNA for each aglB was present in the respective complementations (Fig 6). Expression of archaeal AglBs in foreign hosts can be problematic. While good expression of Mc. voltae AglB in E.coli was reported [2], Taguchi et al. (2016) were unable to produce more than a trace of AglB from Sulfolobus solfataricus or Pyrobaculum calidifontis in E.coli [24]. Expression of active AglB from three different extreme halophiles in Hfx. volcanii was achieved although to greatly varying degrees [34]. The aglB from Hfx. volcanii was also cloned directly from genomic DNA without Mc. maripaludis codon optimization but it too could not rescue oligosaccharyltransferase activity in the ΔaglB-14-9 strain (not shown).
Fig 6. Detection of transcription of Hfx. volcanii and S. acidocaldarius aglB in complemented Mc. maripaludis ΔaglB-14-9 mutant cells.
Total RNA was purified from complemented cells grown for three successive transfers in nitrogen-free medium supplemented with alanine to allow transcription of the aglB gene from the nif promoter in the complementation vector. In both cases, the aglB was synthesized using Mc. maripaludis codon preferences. A. Mc. maripaludis ΔaglB-14-9 mutant complemented with Hfx. volcanii aglB. Purified RNA with (RT) or without (RNA) a reverse transcription step was used as template using RT primers for Hfx. volcanii aglB (Table 2). Standard PCR with the same primers was used with pKJ1251 as template for amplicon size verification (DNA). The Hfx. volcanii aglB RT-PCR primers were also used in reactions with RNA isolated from Mc. maripaludis ΔaglB-14-9 mutant complemented with S. acidocaldarius aglB (Sa RT). B. Mc. maripaludis ΔaglB-14-9 mutant complemented with S. acidocaldarius aglB. Purified RNA with (RT) or without (RNA) a reverse transcription step was used as template using RT primers for S. acidocaldarius aglB (Table 2). Standard PCR with the same primers was used with pKJ1240 as template for amplicon size verification (DNA). The S. acidocaldarius aglB RT-PCR primers were also used in reactions with RNA isolated from Mc. maripaludis ΔaglB-14-9 mutant complemented with Hfx. volcanii aglB (Hv RT).
In Hfx. volcanii, the N-linked glycosylation system is very complicated. Completely different glycans are found depending on the salt content of the growth medium. At high salt (3.4M NaCl), a pentasaccharide (mannose-1,2-[methyl-O-4-]GlcA-β-1,4-galacturonic acid-α1,4-GlcA-β1,4-glucose-β1-Asn) is made [9]. The first four sugars are assembled on a single dolichol carrier and transferred by AglB while the terminal mannose loaded onto a separate dolichol carrier is transferred to the Asn-bound tetrasaccharide by AglS [23,25]. Here, the linking sugar is unusual in being glucose, rather than an acetylated sugar. At growth in low salt (1.75M NaCl), a tetrasaccharide consisting of a sulfated hexose, two hexoses and a rhamnose is found [64]. Intriguingly, only a single aglB gene is reported for Hfx. volcanii but this AglB is only required for the transfer of the pentasaccharide but not the low salt tetrasaccharide [36]. The oligosaccharyltransferase responsible for transfer of the low salt glycan is not yet identified. The percent identity and similarity of the Hfx. volcanii AglB to that of Mc. maripaludis is much lower (17.9/ 31.0%) than that shown by the Mc. voltae and Mtc. thermolithotrophicus enzymes and the halophile enzyme is much longer. In addition, the DK/MI motif is completely different in halophiles and methanogens. The DK motif has been shown to be catalytically important both in yeast Stt3 [65] as well as the AglBs of P. furiosus and A. fulgidus [33,56]. In Hfx. volcanii, the sequence is the modified DM version of the motif, DWQMASTDAK. Methanococci have the MI motif at the corresponding position in AglB believed to perform the same role as the DK/DM motif and in Mc. maripaludis the MI motif is MTSIASVW. In PglB of Campylobacter jejuni, the MI motif (MSLIFSTV) has been shown by mutational analysis to be important for catalytic activity with only chemically similar amino acids tolerated at the conserved positions [66]. Given that the chemical properties of the DK, DM and MI side chains are considerably different and the effect this has on enzyme function is not understood [57], it is possible the replacement of the DK/DM motif with the MI motif affects recognition of substrates by the different AglBs. Furthermore, the catalytic site of all AglBs is in the C-terminal domain predicted to be located external to the cell. Consequently, this region may be specifically adapted to the high external salt environment of Hfx. volcanii and not properly folded in the 2% NaCl-containing medium used for Mc. maripaludis. It is also possible that the halophile AglB was misfolded and either degraded or did not insert properly into the cytoplasmic membrane, although the membrane lipids of Hfx. volcanii are diether types like that in Mc. maripaludis [67]. Other less likely, in our opinion, possibilities for the failure of the Hfx. volcanii AglB complementation can also be envisioned. One is the difference in the linking sugar in the glycans of Hfx. volcanii and Mc. maripaludis. However, it is known that the Hfx. volcanii aglB can be replaced with aglB from different extreme halophiles that can transfer glycans with either a hexose or N-acetylhexuronic acid as a linking sugar so one would expect that the AglB from Hfx. volcanii would accommodate a GalNAc linking sugar. Another unlikely reason for the failure of the Hfx. volcanii AglB to complement could lie in the nature of the dolichol carrier, but the Hfx. volcanii carrier is C55-C60 dolichol phosphate with saturated isoprenes at the α and ω positions [27] and in Mc. maripaludis it is a C55 dolichol phosphate that has two sites of saturation, presumed to be the α and ω positions too [2].
The other failed complementation occurred with the S. acidocaldarius AglB. In S. acidocaldarius, the N-linked glycan is a complex tri-branched hexasaccharide containing the unusual sulfated sugar, sulfoquinovose and with a linking sugar of N-acetylglucosamine [14]. Many possible reasons for the failure can be presented. Obviously, the active site of this AglB would be facing an extremely acidic environment and the enzyme would need to be adapted for the high growth temperature as well, both not found in the growth conditions of Mc. maripaludis. Even with an AglB from the much more closely related hyperthermophile Mcc. jannaschii, we were unable to show complementation. Furthermore, in its native environment AglB of S. acidocaldarius would be inserted into a cytoplasmic membrane comprised of tetraether lipids, including ones that could contain up to 4 cyclopentane rings [67,68]. Cytoplasmic membranes composed solely of tetraether lipids are thought to form a lipid monolayer and not the lipid bilayer seen in bacteria or in archaea which have diether lipids [69,70]. As with the Hfx. volcanii AglB, the one from S. acidocaldarius shows low sequence identity and similarity to that of Mc. maripaludis (19.9%/34.1%). The S. acidocaldarius AglB has the DK motif (DIAK) rather than the MI motif of Mc. maripaludis AglB. Furthermore, there are potentially significant differences in the substrates encountered. The linking sugar in the glycan of S. acidocaldarius is GlcNAc and not GalNAc as in Mc. maripaludis. In addition, the dolichol carrier normally recognized by AglB in S. acidocaldarius was recently shown to be an unusual, short (C45) dolichol pyrophosphate [30] and not the dolichol phosphate initially believed to serve as the glycan lipid carrier [28] and this could cause recognition problems for the S. acidocaldarius AglB towards the Mc. maripaludis dolichol carrier, identified as C55 dolichol phosphate [2].
Conclusions
This study extends to methanogens the examination of AglB promiscuity previously reported in in vivo experiments with extreme halophiles [34] and in vitro with the hyperthermophiles A. fulgidus and P. furiosus [33]. We have shown the ability of Mc. voltae AglB to transfer glycans of similar but different composition, including glycans with either GlcNAC or GalNAc as the linking sugar. We have also identified for the first time the AglB from the thermophilic methanogen Mtc. thermolithotrophicus and demonstrate that it can functionally replace the oligosaccharyltransferase activity in the Mc. maripaludis ΔaglB mutant. As there are currently no published data on the nature of N-linked glycans in Mtc. thermolithotrophicus, it is not possible to state how different the Mc. maripaludis glycan is from that naturally transferred by the Mtc. thermolithotrophicus AglB. We have begun studies to determine the N-glycan attached to archaellins in this thermophilic methanogen to address this issue. Attempts to complement using AglBs from more distantly related and more extremophilic archaea for which data on N-linked glycosylation systems is well known were unsuccessful, although the possible reasons for this are many. The accumulation of data on the relaxed nature of substrates accepted by various AglBs will aid in any efforts to develop archaea as platforms for glycoengineering [44].
Supporting Information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Funding for the project was supplied by two separate grants from Natural Sciences and Engineering Research Council of Canada (NSERC), one to KFJ and the other to CMK. NSERC website http://www.nserc-crsng.gc.ca/index_eng.asp. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jarrell KF, Ding Y, Meyer BH, Albers SV, Kaminski L, Eichler J. N-Linked glycosylation in Archaea: a structural, functional, and genetic analysis. Microbiol Mol Biol Rev 2014;78:304–341. 10.1128/MMBR.00052-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larkin A, Chang MM, Whitworth GE, Imperiali B. Biochemical evidence for an alternate pathway in N-linked glycoprotein biosynthesis. Nature Chem Biol 2013;9:367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwarz F, Aebi M. Mechanisms and principles of N-linked protein glycosylation. Curr Opin Struct Biol 2011;21:576–582. 10.1016/j.sbi.2011.08.005 [DOI] [PubMed] [Google Scholar]
- 4.Szymanski CM, Logan SM, Linton D, Wren BW. Campylobacter—a tale of two protein glycosylation systems. Trends Microbiol 2003;11:233–238. [DOI] [PubMed] [Google Scholar]
- 5.Chaban B, Voisin S, Kelly J, Logan SM, Jarrell KF. Identification of genes involved in the biosynthesis and attachment of Methanococcus voltae N-linked glycans: insight into N-linked glycosylation pathways in Archaea. Mol Microbiol 2006;61:259–268. 10.1111/j.1365-2958.2006.05226.x [DOI] [PubMed] [Google Scholar]
- 6.Kelleher DJ, Gilmore R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 2006;16:47R–62R. 10.1093/glycob/cwj066 [DOI] [PubMed] [Google Scholar]
- 7.Meyer BH, Albers SV. Hot and sweet: protein glycosylation in Crenarchaeota. Biochem Soc Trans 2013;41:384–392. 10.1042/BST20120296 [DOI] [PubMed] [Google Scholar]
- 8.Kaminski L, Naparstek S, Kandiba L, Cohen-Rosenzweig C, Arbiv A, Konrad Z, et al. Add salt, add sugar: N-glycosylation in Haloferax volcanii. Biochem Soc Trans 2013;41:432–435. 10.1042/BST20120142 [DOI] [PubMed] [Google Scholar]
- 9.Kandiba L, Lin CW, Aebi M, Eichler J, Guerardel Y. Structural characterization of the N-linked pentasaccharide decorating glycoproteins of the halophilic archaeon Haloferax volcanii. Glycobiology 2016;26:745–756. 10.1093/glycob/cww014 [DOI] [PubMed] [Google Scholar]
- 10.Fujinami D, Nyirenda J, Matsumoto S, Kohda D. Structural elucidation of an asparagine-linked oligosaccharide from the hyperthermophilic archaeon, Archaeoglobus fulgidus. Carbohydr Res 2015;413:55–62. 10.1016/j.carres.2015.05.010 [DOI] [PubMed] [Google Scholar]
- 11.Fujinami D, Matsumoto M, Noguchi T, Sonomoto K, Kohda D. Structural elucidation of an asparagine-linked oligosaccharide from the hyperthermophilic archaeon, Pyrococcus furiosus. Carbohydr Res 2014;387:30–36. 10.1016/j.carres.2014.01.021 [DOI] [PubMed] [Google Scholar]
- 12.Abu-Qarn M, Eichler J. Protein N-glycosylation in Archaea: defining Haloferax volcanii genes involved in S-layer glycoprotein glycosylation. Mol Microbiol 2006;61:511–525. 10.1111/j.1365-2958.2006.05252.x [DOI] [PubMed] [Google Scholar]
- 13.Jarrell KF, Jones GM, Nair DB. Role of N-linked glycosylation in cell surface structures of Archaea with a focus on flagella and S layers. Int J Microbiol 2010. Article ID 470138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peyfoon E, Meyer B, Hitchen PG, Panico M, Morris HR, Haslam SM, et al. The S-layer glycoprotein of the crenarchaeote Sulfolobus acidocaldarius is glycosylated at multiple sites with the chitobiose-linked N-glycans. Archaea 2010. Article ID 754101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voisin S, Houliston RS, Kelly J, Brisson JR, Watson D, Bardy SL, et al. Identification and characterization of the unique N-linked glycan common to the flagellins and S-layer glycoprotein of Methanococcus voltae. J Biol Chem 2005;280:16586–16593. 10.1074/jbc.M500329200 [DOI] [PubMed] [Google Scholar]
- 16.Meyer BH, Birich A, Albers SV. N-Glycosylation of the archaellum filament is not important for archaella assembly and motility, although N-glycosylation is essential for motility in Sulfolobus acidocaldarius. Biochimie 2015;118(294):301. [DOI] [PubMed] [Google Scholar]
- 17.Kelly J, Logan SM, Jarrell KF, Vandyke DJ, Vinogradov E. A novel N-linked flagellar glycan from Methanococcus maripaludis. Carbohydr Res 2009;344:648–653. 10.1016/j.carres.2009.01.006 [DOI] [PubMed] [Google Scholar]
- 18.Tripepi M, You J, Temel S, Önder Ö, Brisson D, Pohlschröder M. N-glycosylation of Haloferax volcanii flagellins requires known Agl proteins and is essential for biosynthesis of stable flagella. J Bacteriol 2012;194:4876–4887. 10.1128/JB.00731-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wieland F, Paul G, Sumper M. Halobacterial flagellins are sulfated glycoproteins. J Biol Chem 1985;260:15180–15185. [PubMed] [Google Scholar]
- 20.Nair DB, Jarrell KF. Pilin processing follows a different temporal route than that of archaellins in Methanococcus maripaludis. Life (Basel) 2015;5:85–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng SYM, Wu J, Nair DB, Logan SM, Robotham A, Tessier L, et al. Genetic and mass spectrometry analysis of the unusual type IV-like pili of the archaeon Methanococcus maripaludis. J Bacteriol 2011;193:804–814. 10.1128/JB.00822-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esquivel RN, Schulze S, Xu R, Hippler M, Pohlschroder M. Identification of Haloferax volcanii pilin N-glycans with diverse roles in pilus biosynthesis, adhesion, and microcolony formation. J Biol Chem 2016;291:10602–10614. 10.1074/jbc.M115.693556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eichler J. Extreme sweetness:protein glycosylation in Archaea. Nature Rev. Microbiol. 2013;11:151–156. [DOI] [PubMed] [Google Scholar]
- 24.Taguchi Y, Fujinami D, Kohda D. Comparative analysis of archaeal lipid-linked oligosaccharides that serve as oligosaccharide donors for Asn glycosylation. J Biol Chem 2016;291:11042–11054. 10.1074/jbc.M115.713156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen-Rosenzweig C, Yurist-Doutsch S, Eichler J. AglS, a novel component of the Haloferax volcanii N-glycosylation pathway, is a dolichol phosphate-mannose mannosyltransferase. J Bacteriol 2012;194:6909–6916. 10.1128/JB.01716-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang MM, Imperiali B, Eichler J, Guan Z. N-linked glycans are assembled on highly reduced dolichol phosphate carriers in the hyperthermophilic Archaea Pyrococcus furiosus. PloS one 2015;10:e0130482 10.1371/journal.pone.0130482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guan Z, Eichler J. Liquid chromatography/tandem mass spectrometry of dolichols and polyprenols, lipid sugar carriers across evolution. Biochim Biophys Acta 2011;1811:800–806. 10.1016/j.bbalip.2011.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guan Z, Meyer BH, Albers SV, Eichler J. The thermoacidophilic archaeon Sulfolobus acidocaldarius contains an unusually short, highly reduced dolichyl phosphate. Biochim Biophys Acta 2011;1811:607–616. 10.1016/j.bbalip.2011.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lechner J, Sumper M. Structure and biosynthesis of prokaryotic glycoproteins. Annu Rev Biochem 1989;58:173–194. 10.1146/annurev.bi.58.070189.001133 [DOI] [PubMed] [Google Scholar]
- 30.Guan Z, Delago A, Nußbaum P, Meyer B, Albers SV, Eichler J. N-glycosylation in the thermoacidophilic archaeon Sulfolobus acidocaldarius involves a short dolichol pyrophosphate carrier. FEBS Lett 2016; 590:3168–3178. 10.1002/1873-3468.12341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vandyke DJ, Wu J, Logan SM, Kelly JF, Mizuno S, Aizawa SI, et al. Identification of genes involved in the assembly and attachment of a novel flagellin N-linked tetrasaccharide important for motility in the archaeon Methanococcus maripaludis. Mol Microbiol 2009;72:633–644. 10.1111/j.1365-2958.2009.06671.x [DOI] [PubMed] [Google Scholar]
- 32.Meyer BH, Albers SV. AglB, catalyzing the oligosaccharyl transferase step of the archaeal N-glycosylation process, is essential in the thermoacidophilic crenarchaeon Sulfolobus acidocaldarius. Microbiology Open 2014;3:531–543. 10.1002/mbo3.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsumoto S, Igura M, Nyirenda J, Matsumoto M, Yuzawa S, Noda N, et al. Crystal structure of the C-terminal globular domain of oligosaccharyltransferase from Archaeoglobus fulgidus at 1.75 Å resolution. Biochemistry 2012;51:4157–4166. 10.1021/bi300076u [DOI] [PubMed] [Google Scholar]
- 34.Cohen-Rosenzweig C, Guan Z, Shaanan B, Eichler J. Substrate promiscuity: AglB, the archaeal oligosaccharyltransferase, can process a variety of lipid-linked glycans. Appl Environ Microbiol 2014;80:486–496. 10.1128/AEM.03191-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaminski L, Lurie-Weinberger MN, Allers T, Gophna U, Eichler J. Phylogenetic- and genome-derived insight into the evolution of N-glycosylation in Archaea. Mol Phylogenet Evol 2013;68:327–339. 10.1016/j.ympev.2013.03.024 [DOI] [PubMed] [Google Scholar]
- 36.Kaminski L, Guan Z, Yurist-Doutsch S, Eichler J. Two distinct N-glycosylation pathways process the Haloferax volcanii S-layer glycoprotein upon changes in environmental salinity. Mbio 2013;4:e00716–13. 10.1128/mBio.00716-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Izquierdo L, Schulz BL, Rodrigues JA, Güther ML, Procter JB, Barton GJ, et al. Distinct donor and acceptor specificities of Trypanosoma brucei oligosaccharyltransferases. EMBO J 2009;28:2650–2661. 10.1038/emboj.2009.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jarrell KF, Albers SV. The archaellum: an old motility structure with a new name. Trends Microbiol 2012;20:307–312. 10.1016/j.tim.2012.04.007 [DOI] [PubMed] [Google Scholar]
- 39.Albers SV, Jarrell KF. Archaellum moves Archaea with distinction. Microbe 2015;10:283–288. [Google Scholar]
- 40.Siu S, Robotham A, Logan SM, Kelly JF, Uchida K, Aizawa SI, et al. Evidence that biosynthesis of the second and third sugars of the archaellin tetrasaccharide in the archaeon Methanococcus maripaludis occurs by the same pathway used by Pseudomonas aeruginosa to make a di-N-acetylated sugar. J Bacteriol 2015;197:1668–1680. 10.1128/JB.00040-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding Y, Uchida K, Aizawa SI, Murphy K, Berezuk A, Khursigara CM, et al. Effects of N-glycosylation site removal in archaellins on the assembly and function of archaella in Methanococcus maripaludis. PloS one 2015;10:e0116402 10.1371/journal.pone.0116402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaban B, Ng SY, Kanbe M, Saltzman I, Nimmo G, Aizawa SI, et al. Systematic deletion analyses of the fla genes in the flagella operon identify several genes essential for proper assembly and function of flagella in the archaeon, Methanococcus maripaludis. Mol Microbiol 2007;66:596–609. 10.1111/j.1365-2958.2007.05913.x [DOI] [PubMed] [Google Scholar]
- 43.Ding Y, Nash J, Berezuk A, Khursigara CM, Langelaan DN, Smith SP, et al. Identification of the first transcriptional activator of an archaellum operon in a euryarchaeon. Mol Microbiol 2016;102:54–70 10.1111/mmi.13444 [DOI] [PubMed] [Google Scholar]
- 44.Calo D, Guan Z, Eichler J. Glyco-engineering in Archaea: differential N-glycosylation of the S- layer glycoprotein in a transformed Haloferax volcanii strain. Microb Biotechnol 2011;4:461–470. 10.1111/j.1751-7915.2011.00250.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calo D, Eilam Y, Lichtenstein RG, Eichler J. Towards glycoengineering in archaea: replacement of Haloferax volcanii AglD with homologous glycosyltransferases from other halophilic archaea. Appl Environ Microbiol 2010;76:5684–5692. 10.1128/AEM.00681-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore BC, Leigh JA. Markerless mutagenesis in Methanococcus maripaludis demonstrates roles for alanine dehydrogenase, alanine racemase, and alanine permease. J Bacteriol 2005;187:972–979. 10.1128/JB.187.3.972-979.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. Methanogens: reevaluation of a unique biological group. Microbiol Rev 1979;43:260–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferrante G, Richards JC, Sprott GD. Structures of polar lipids from the thermophilic, deep-sea archaeobacterium Methanococcus jannaschii. Biochem Cell Biol 1990;68:274–283. [DOI] [PubMed] [Google Scholar]
- 49.Lie TJ, Leigh JA. A novel repressor of nif and glnA expression in the methanogenic archaeon Methanococcus maripaludis. Mol Microbiol 2003;47:235–246. [DOI] [PubMed] [Google Scholar]
- 50.Tumbula DL, Makula RA, Whitman WB. Transformation of Methanococcus maripaludis and identification of a PstI-like restriction system. FEMS Microbiol Lett 1994;121:309–314. [Google Scholar]
- 51.Tusnády GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics 2001;17:849–850. [DOI] [PubMed] [Google Scholar]
- 52.Lie TJ, Wood GE, Leigh JA. Regulation of nif expression in Methanococcus maripaludis: roles of the euryarchaeal repressor NrpR, 2-oxoglutarate, and two operators. J Biol Chem 2005;280:5236–5241. 10.1074/jbc.M411778200 [DOI] [PubMed] [Google Scholar]
- 53.Ding Y, Jones GM, Uchida K, Aizawa SI, Robotham A, Logan SM, et al. Identification of genes involved in the biosynthesis of the third and fourth sugars of the Methanococcus maripaludis archaellin N-linked tetrasaccharide. J Bacteriol 2013;195:4094–4104. 10.1128/JB.00668-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matsumoto S, Shimada A, Kohda D. Crystal structure of the C-terminal globular domain of the third paralog of the Archaeoglobus fulgidus oligosaccharyltransferases. BMC Struct Biol 2013;13:11 10.1186/1472-6807-13-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matsumoto S, Shimada A, Nyirenda J, Igura M, Kawano Y, Kohda D. Crystal structures of an archaeal oligosaccharyltransferase provides insights into the catalytic cycle of N-linked protein glycosylation. Proc Natl Acad Sci U S A 2013; 110:17868–17873. 10.1073/pnas.1309777110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Igura M, Kohda D. Selective control of oligosaccharide transfer efficiency for the N-glycosylation sequon by a point mutation in oligosaccharyltransferase. J Biol Chem 2011;286:13255–13260. 10.1074/jbc.M110.213900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maita N, Nyirenda J, Igura M, Kamishikiryo J, Kohda D. Comparative structural biology of eubacterial and archaeal oligosaccharyltransferases. J Biol Chem 2010;285:4941–4950. 10.1074/jbc.M109.081752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ng SY, Chaban B, Jarrell KF. Archaeal flagella, bacterial flagella and type IV pili: a comparison of genes and posttranslational modifications. J Mol Microbiol Biotechnol 2006;11:167–191. 10.1159/000094053 [DOI] [PubMed] [Google Scholar]
- 59.Kostyukova AS, Gongadze GM, Obraztsova AY, Laurinavichus KS, Fedorov OV. Protein composition of Methanococcus thermolithotrophicus flagella. Can J Microbiol 1992;38:1162–1166. [Google Scholar]
- 60.Ding Y, Lau Z, Logan SM, Kelly JF, Berezuk A, Khursigara CM, et al. Effects of growth conditions on archaellation and N-glycosylation in Methanococcus maripaludis. Microbiology (Reading, England) 2016;162:339–350. [DOI] [PubMed] [Google Scholar]
- 61.Sprott GD, Meloche M, Richards JC. Proportions of diether, macrocyclic diether, and tetraether lipids in Methanococcus jannaschii grown at different temperatures. J Bacteriol 1991;173:3907–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hedrick DB, Guckert JB, White DC. Archaebacterial ether lipid diversity analyzed by supercritical fluid chromatography: integration with a bacterial lipid protocol. J Lipid Res 1991;32:659–666. [PubMed] [Google Scholar]
- 63.Grant WD, Pinch G, Harris JE, De Rosa M, Gambacorta A. Polar lipids in methanogen taxonomy. J Gen Microbiol 1985;131:3277–3286. [Google Scholar]
- 64.Guan Z, Naparstek S, Calo D, Eichler J. Protein glycosylation as an adaptive response in Archaea: growth at different salt concentrations leads to alterations in Haloferax volcanii S-layer glycoprotein N-glycosylation. Environ Microbiol 2012;14:743–753. 10.1111/j.1462-2920.2011.02625.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Igura M, Maita N, Kamishikiryo J, Yamada M, Obita T, Maenaka K, et al. Structure-guided identification of a new catalytic motif of oligosaccharyltransferase. EMBO J 2008;27:234–243. 10.1038/sj.emboj.7601940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ihssen J, Kowarik M, Wiesli L, Reiss R, Wacker M, Thöny-Meyer L. Structural insights from random mutagenesis of Campylobacter jejuni oligosaccharyltransferase PglB. BMC Biotechnol 2012;12:67 10.1186/1472-6750-12-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Langworthy TA. Lipids of archaebacteria In: Woese CR, Wolfe RS, editors. The Bacteria: A treatise on structure and function. Volume VIII: Archaebacteria Orlando, Fl: Academic Press, Inc; 1985. p. 459–497. [Google Scholar]
- 68.Chong PL, Ayesa U, Daswani VP, Hur EC. On physical properties of tetraether lipid membranes: effects of cyclopentane rings. Archaea 2012. Article ID 138439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jain S, Caforio A, Driessen AJ. Biosynthesis of archaeal membrane ether lipids. Front Microbiol 2014;5:641 10.3389/fmicb.2014.00641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van de Vossenberg JL, Driessen AJ, Konings WN. The essence of being extremophilic: the role of the unique archaeal membrane lipids. Extremophiles 1998;2:163–170. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.