Abstract
The consistent appearance of specific chromosomal translocations in multiple myeloma has suggested that the positioning of chromosomes in the interphase nucleus might play a role in the occurrence of particular chromosomal rearrangements associated with malignant transformation. Using fluorescence in situ hybridization, we have determined the positions of selected chromosome pairs (18 and 19, 9 and 22, 4 and 14, 14 and 16, 11 and 14) in interphase nuclei of myeloma cells compared to normal lymphocytes of treatment‐naïve patients. All chromosome pairs were arranged in a nonrandom pattern. Chromosomes commonly involved in myeloma‐associated translocations (4 and 14, 14 and 16, 11 and 14) were found in close spatial proximity, and this is correlated with the occurrence of overlapping chromosome territories. The spatial distribution of chromosomes may increase the possibility of chromosomal translocations in multiple myeloma.
Keywords: 3D‐SIM, chromosome territory, myeloma, monoclonal gammopathy of undetermined significance
Short abstract
What's new?
The positioning of chromosomes in the interphase nucleus might play a role in the occurrence of chromosomal rearrangements associated with malignant transformation. Here, the authors examined the nuclei of myeloma cells and control lymphocytes from treatment‐naïve patients using conventional 3D imaging and super‐resolution microscopy. Common and distinct chromosome positions were found in lymphocytes and myeloma cells with respect to neighborhood relationships and tendency to overlap. Chromosomes commonly involved in myeloma‐associated translocations were found in close spatial proximity, which correlated with the occurrence of overlapping chromosome territories. The spatial distribution of chromosomes may increase the possibility of translocations in multiple myeloma.
Multiple myeloma (MM) is a malignant plasma cell proliferative disorder, which accounts for 1% of all cancers and ∼10% of all hematologic malignancies.1, 2 MM is almost always preceded by an asymptomatic premalignant stage known as monoclonal gammopathy of undetermined significance (MGUS).3 The risk of progression from MGUS to symptomatic MM is ∼1% per year.4, 5
The pathogenesis of MM and MGUS is a complex process that includes the progressive occurrence of multiple structural chromosomal changes.1 These alterations involve a combination of gains and losses of whole chromosomes, nonrandom chromosomal translocations and point mutations.6 One of the common abnormalities is hyperdiploidy, which is associated with the odd numbered chromosomes including 3, 5, 7, 9, 11, 15, 19 and 21.7, 8 Translocations involving the immunoglobulin heavy‐chain (IgH) locus are predominately found in the nonhyperdiploid group.8 The most common translocations involve the following chromosomes: t(11;14)(q13;q32), t(4;14)(p16;q32) and t(14;16)(q32;q23)1 at the following frequencies; 20%, 15–20% and 5% of the cases, respectively.9
Previous studies revealed the consistent association of specific chromosomal abnormalities with specific types of cancer, supporting the hypothesis that these nonrandom structural chromosomal abnormalities are the cause of malignant transformation in a particular cancer type.10 Advances in molecular cytogenetics have provided knowledge about chromatin organization in interphase nuclei at molecular resolution.11 It was proposed for a long time that neighboring chromosomes/gene loci may engage in translocations more frequently than chromosomes/loci that are found at larger distances to each other.12 In fact, studies by several groups indicated that neighborhood relationships appear critical for translocations to occur.10, 13, 14, 15, 16, 17, 18, 19, 20 Changes in the spatial position of chromosomes may also predispose chromosomes in close proximity to translocations that are typical of certain cancer types.10, 15, 21
Chromosome territories (CTs) were first postulated by Theodor Boveri.22, 23 His pivotal work was recently translated and annotated by Henri Harris.24 Interphase chromosomes are organized in a nonrandom fashion within the three‐dimensional (3D) nuclear space.21, 25, 26, 27, 28 The organization of chromatin and chromosomes in the nucleus influences gene expression and nuclear function.27, 29, 30 Several studies indicate that interphase positioning of CTs may not only be tissue‐specific, but is subject to changes during differentiation.21, 28, 31, 32, 33
Fluorescence in situ hybridization on 3D preserved nuclei (3D‐FISH) allows visualization of chromosomes as volume structures, providing detailed information about chromosomal positioning relative to nuclear structure.34, 35, 36 The combination of 3D‐FISH, 3D‐microscopy and image reconstruction is an effective way to analyze the spatial arrangement of chromosomes in the nucleus.37 The conventional, widely used epifluorescence light microscopy has a resolution limited by the diffraction limit of the objective lens. The development of super‐resolution fluorescence microscopy techniques, such as structured illumination microscopy (SIM), stimulated emission depletion (STED) and single molecule localization approaches (SMLM), have provided a great improvement in spatial resolution beyond the diffraction barrier.38, 39, 40, 41 These methods yield the ability of accurate measurement of subcellular structures at the nanometer scale in all three dimensions.35, 41, 42
In this study, we have used 3D‐FISH with fluorescent‐labeled whole chromosome‐specific paints and examined whether we could (1) determine the 3D arrangement of interphase CTs in myeloma nuclei of treatment‐naïve patients; (2) identify distinct characteristics of CTs in myeloma nuclei compared to control lymphocytes; and (3) elucidate possible interaction(s) between CTs in interphase nuclei, especially of myeloma cells, using 3D‐SIM and quantitative measurement techniques.
Materials and Methods
Patients
The pilot study population consisted of 20 treatment‐naïve patients, which where subdivided into two groups: MM (N = 10) and MGUS (N = 10) (Table 1). Diagnosis criteria were based on the International Myeloma Working Group (IMWG).43 Control lymphocytes were examined from the same patients. Ethics approval was obtained from CancerCare Manitoba Research Resource Impact Committee and Research Ethics Board on human studies of University of Manitoba, Manitoba, Canada (Ethics Reference No. H2010:170).
Table 1.
Clinical characteristics of the patients included in this study
| Clinical characteristic | MGUS patients | MM patients |
|---|---|---|
| Mean age (year) | 71.7 ± 10.2 | 63.1 ± 8.9 |
| BMPC (%) | 6.5 ± 2.0 | 49.2 ± 25.0 |
| Immunoglobulin isotype (mg/dL) | ||
| IgG | 15.0 ± 7.8 | 23.7 ± 27.8 |
| IgA | 3.4 ± 3.9 | 0.4 ± 0.4 |
| IgM | 12.7 ± 24.9 | 0.2 ± 0.1 |
| Serum free light chain (mg/l) | ||
| Kappa | 20.9 ± 12.7 | 323.2 ± 540.3 |
| Lambda | 34.3 ± 36.9 | 509.4 ± 1,417.4 |
| Albumin (g/l) | 36.1 ± 4.0 | 32.5 ± 5.1 |
| M protein (g/l) | 11.2 ± 10.1 | 31.0 ± 22.6 |
| β2 microglobulin (μg/ml) | 2.4 ± 0.4 | 7.7 ± 8.2 |
BMPC indicates bone marrow plasma cells.
Isolation of lymphocytes and myeloma cells
White blood cells were isolated as previously described.44 Briefly, 10 ml peripheral blood from each patient was collected in EDTA‐treated tubes. Mononuclear cells were separated using Ficoll–Paque (GE Healthcare Life Sciences, Quebec, Canada) by a 30 min centrifugation at 200g. The buffy coat was washed with 10 ml of a 1× phosphate buffered saline (PBS) solution.
Fluorescence in situ hybridization
Pretreatment of interphase nuclei
Cells were subsequently placed onto slides. The slides were equilibrated in 2 × saline‐sodium citrate (SSC) buffer solution for 10 minutes at room temperature (RT). The slides were incubated with 100 μg/ml ribonuclease A (RNase A) at 37°C for 1 hr and washed three times in 2 × SSC for 5 min each while shaking at RT followed by 10‐min pepsin/HCl treatment at 37°C. The slides were washed twice in 1 × PBS for 5 min each and then once in 50 mM MgCl2 in 1 × PBS for 5 min while shaking before incubated in 1% formaldehyde/50 mM MgCl2 in 1 × PBS for 10 min. Slides were washed once in 1 × PBS for 5 min while. The denaturation of slides was performed in 70% deionized formamide in 2 × SSC (pH 7.0) at 70°C for 2 min and then transferred immediately to a series of cold ethanol (70%, 90% and 100% ethanol).
Chromosome paints and labeling
Whole human chromosome‐specific paints for chromosome 4, 9, 11, 14, 16, 18, 19 and 22 were purchased from Applied Spectral Imaging Ltd (Carlsbad, CA). Cy3‐labeled and FITC‐labeled whole chromosome painting probes were used for hybridization, simultaneously. The probe were mixed and then denatured at 85–90°C for 7 min followed by a 30 min incubation at 37°C. The pre‐annealed probe was applied to the denatured nuclei. Slides were sealed with rubber cement and incubated in a humidified chamber at 37°C. After overnight hybridization, the slides were washed three times in 50%formamide/2 × SSC (5 min each, at 45°C), twice in 0.1% Tween‐20/4xSSC (5 min each, at 45°C) and once in 1 × SSC (2 min, at 45°C) while shaking. The slides were counterstained with 4′,6‐diamidino‐2‐phenylindole (DAPI) (0.1 μg/ml) and incubated in the dark for 3 min. Excess DAPI was removed with ddH2O. The slides were then mounted in fluorescence‐antifade solution (Vector Laboratories, Burlington, Ontario, Canada), using high performance cover glasses (No. 1 1/2, Schott, Mainz, Germany). The slides were sealed with nail polish and stored at 4°C until imaging.
Identification of myeloma cells
We previously identified myeloma cells based on green fluorescence signals emitted by the fluorescein isothiocyanate (FITC)–labeled CD138 antibody and on the size and intensity of the DAPI counterstained nucleus.44, 45 In comparison, normal lymphocyte nuclei have a smaller, rounder shape and emit a brighter DAPI signal than myeloma nuclei. In this study, we have identified myeloma and lymphocyte nuclei based on size and intensity of the DAPI staining.
Microscopy and Image Acquisition
All image processing was performed in MATLAB (MathWorks, Natick, MA) with the DIPimage toolbox.46
Conventional epifluorescence microscopy
Epiluorescence imaging was performed with an AxioImager Z1 microscope (Carl Zeiss Canada, Toronto, Canada) and an AxioCam HR charge‐coupled device (Carl Zeiss). A 63×/1.40 oil immersion objective lens (Carl Zeiss) was used with a DAPI filter, for nuclear DNA staining detection, a cyanine 3 (Cy3) filter and a fluorescein isothiocyanate (FITC) filter for detection of the whole chromosome paint signals. All imaging of interphase nuclei used the same protocol and tricolor beads were used for standardization.47 Eighty z‐stacks were acquired at a sampling distance of Δx = Δy = 102 nm and Δz = 200 nm for each slice of the stack. The acquired images were deconvolved using AxioVision 4.8 software (Carl Zeiss) using a constrained iterative algorithm.48 Deconvolved images were exported as TIFF files for analysis.
Image segmentation and 3D image analysis of conventional epifluorescence microscopy
The chromosome position was analyzed in two steps. First, the objects of interest were automatically identified (segmentation). Second, the relevant features were measured. The nucleus was segmented as described by Righolt et al. (2015) with some minor modifications for these cells. The chromosome channels are individually segmented from their respective images.49 The image is first band‐filtered with a difference of Gaussians filter, the negative values arising from this filter are clipped to zero. The product of the original image and the clipped band‐pass filtered image is subsequently thresholded over the nuclear mask with the isodata algorithm.50 Holes within connected regions are filled and the largest connected, detected regions are the detected chromosomes. The relevant volume and distance measures follow straightforward from these detected regions. The relative radial position was measured from the center of mass of the nucleus as described in Kuzyk et al. (2015).51 These measures were based on the full CT rather than just the center of mass.51
Super‐resolution microscopy (3D structured illumination microscopy, 3D‐SIM)
Super‐resolution imaging (3D‐SIM) was performed on a Zeiss Elyra PS1 equipped with a Zeiss Plan Apochromat inverted 63×/1.40 oil immersion objective lens using an Andor EM‐CCD iXon 885 camera and a 1.6× tube lens at room temperature. The DAPI channel was obtained with 405 nm laser excitation, 23 mm diffraction grating and filter cube SR Cube 07. The Cy3 channel was obtained with 561 nm laser excitation. The FITC channel was obtained with 488 nm laser excitation. The lateral pixel size, Δx and Δy, was 79 nm in the recorded images and 40 nm in the reconstructed image. The z‐stacks were acquired by capturing slices taken at 91 nm intervals through each nucleus, and consisted of 60–70 slices collected sequentially. A field of view was selected and the z‐stack boundaries were defined manually. The 3D‐SIM images were reconstructed using ZEN 2012 black edition (Carl Zeiss, Jena, Germany). Image stacks were exported as 16‐bit tiff image sequences.
The full 3D image was used for analysis. The features described below are based on measuring spatial proximity and overlap between the regions at the edges of CTs. The nucleus was segmented using an isodata threshold. The chromosomal DNA was segmented by applying an isodata threshold to the chromosome image within the nucleus. All detected objects with a volume smaller than a sphere with a 300 nm radius were discarded. Two features were measured in each cell. The first measure is based on the Manders colocalization coefficient (MCC). First the MCC are calculated based on the detected CTs.52 The mean of these two coefficients is then calculated and then taken as the MCC for the cell, MCC will refer to this mean of the two coefficients in this article. The second measure is based on the detected volumes in both CT channels, and , both binary images. This feature is called the percentage of intermingling and is the ratio of the intersection to the union of both volumes:
Statistical analysis
Group data were expressed as mean ± SD. Statistical analysis was performed using two‐sided, two‐sample Kolmogorov–Smirnov (KS) tests to determine significant differences in all imaging data, and the results were expressed as cumulative distribution plots. Histograms and scatter plots were generated using Microsoft Excel. We requested p‐values of <0.01 for a statistically significant and p < 0.001 for a highly significant difference in all analysis.
Results
Clinical data were available for all patients. The patients included 10 treatment‐naïve MGUS patients and 10 treatment‐naïve MM patients with a median age of 67.4 years (range, 45–80 years). Patient characteristics are given in Table 1.
FISH was performed on 3D preserved lymphocytes and myeloma nuclei. The nuclei were identified and imaged using fluorescence microscopy and 3D‐SIM. After acquisition and image reconstruction (see Materials and Methods), we determined the CT characteristics and positioning within the nuclear space. Hybridized chromosome signals for each chromosome paint were examined in at least 30 nuclei per sample. We analyzed a total of 5,518 lymphocytes, 2,917 MGUS and 2,906 MM nuclei for 5 pairs of chromosomes, including chromosome (4 and 14), (9 and 22), (11 and 14), (14 and 16) and (18 and 19). Chromosome 9 and 22 were chosen since they are neighbors in lymphocytes.18, 53 Chromosomes 18 and 19 were chosen as they are arranged based on their gene densities in human cells.54 All other listed chromosome pairs represent possible translocation partners in myeloma cells.9
Chromosome territories and nuclear volumes measurements after conventional epifluorescence microscopy
We analyzed the above chromosomes in interphase nuclei. Figure 1 a represents a screenshot of the program showing a segmentation example for chromosomes 4 and 14 in control lymphocytes (A), MGUS (B) and MM (C). A 2D image is displayed in pre‐segmentation mode (Fig. 1 a, Ai, Bi, Ci, left) and following segmentation of chromosomes and nuclei (Fig. 1 a, Ai, Bi, Ci right). The 3D reconstructed chromosomes are shown in the right panel of this figure (Fig. 1 a, Aii, Bii and Cii).
Figure 1.
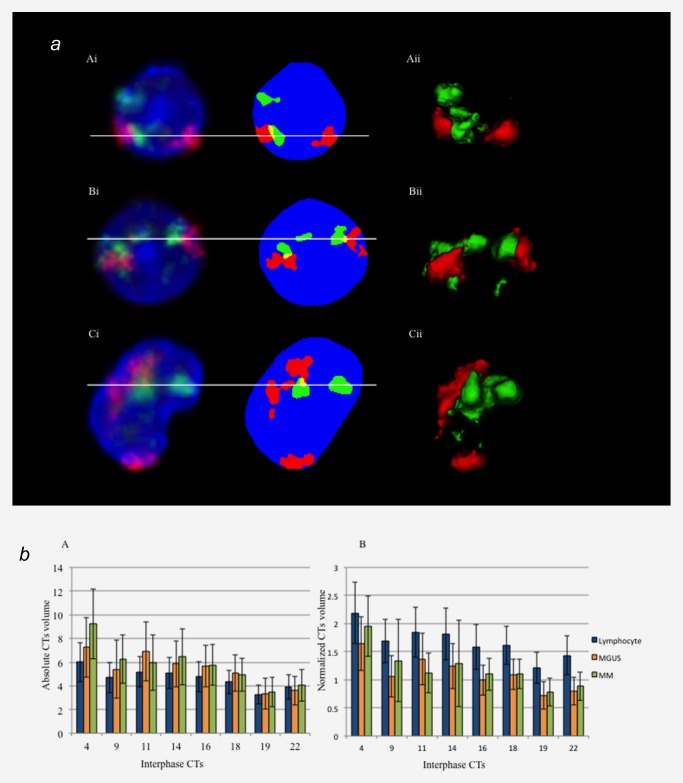
(a) Representative screenshot of chromosome segmentation and 3D reconstruction data of chromosome 4 and 14 territories in control lymphocyte (A), MGUS (B) and MM nuclei (C). Left panel (Ai, Bi and Ci) represents the 2D and pre‐segmentation image. Right panel (Aii, Bii and Cii) represents 3D reconstruction of each nucleus (DAPI is not shown in 3D‐image). Nuclei are counterstained with DAPI (blue), chromosome 4 territories are in red (Cyanine 3) and chromosome 14 territories are in green (FITC). Conventional 3D epifluorescence microscopy was used. Note the overlapping regions within CTs are shown in yellow. (b) Mean nuclear and CTs volume measurements in lymphocytes (blue), MGUS (orange) and MM nuclei (green). The error bar represents one standard error of the mean. The absolute CT volumes in myeloma nuclei are larger than in control lymphocytes but the normalized CTs volumes are largest in lymphocyte nuclei, and then followed by MM and MGUS nuclei.
Our automatic segmentation approach measured cell properties for each cell: the volume of CTs, the distance between them and their distances to the nuclear center (Materials and Methods). Total volumes were calculated for both the chromosomes and nuclei. The mean nuclear volumes are 281.98 ± 67.24 μm3 for lymphocytes, 478.68 ± 148.84 μm3 for MGUS cells and 476.72 ± 152.75 μm3 for MM nuclei. As expected the nuclear volumes were significantly larger in myeloma nuclei compared to control lymphocytes (p < 0.001). Among myeloma nuclei, the nuclear volume of MM nuclei and MGUS nuclei was not significantly different (p = 0.215). These results supported our previous findings demonstrating increased nuclear volumes in myeloma nuclei.45 Correspondingly, the absolute CT volumes in myeloma nuclei were also larger than in control lymphocytes (Fig. 1 b).
To avoid the effects of size increases of nuclear volumes and altered chromosome numbers in malignant nuclei, we normalized the CTs volumes and expressed them as a percentage of the nuclear volume. Nuclear volumes were calculated for the same nuclei used for CTs volume measurements. The calculated ratio of individual CT volumes to nuclear volumes varies depending on the chromosome and cell types (Fig. 1 b). All normalized CT volumes were largest in lymphocyte nuclei, and then followed by MM and MGUS nuclei. The absolute and normalized CTs volumes of the chromosomes studied in each cell type are shown in Table 2.
Table 2.
Mean CTs volume of each chromosome in lymphocyte, MGUS and MM nuclei
| Mean absolute CTs volumes | Normalized CTs volumes | |||||
|---|---|---|---|---|---|---|
| Chromosome | Lymphocyte | MGUS | MM | Lymphocyte | MGUS | MM |
| 4 | 6.01 ± 3.32 |
7.26 ± 5.05 (0.009) |
9.23 ± 5.89 (<0.001) |
2.19 ± 1.09 |
1.64 ± 0.96 (<0.001) |
1.96 ± 1.08 (<0.001) |
| 9 | 4.70 ± 2.55 |
5.41 ± 4.92 (<0.001) |
6.25 ± 4.05 (<0.001) |
1.69 ± 0.77 | 1.07 ± 0.74 (<0.001) |
1.34 ± 0.74 (<0.001) |
| 11 | 5.20 ± 2.62 |
6.92 ± 5.01 (<0.001) |
5.95 ± 4.66 (0.048) |
1.85 ± 0.89 |
1.37 ± 0.92 (<0.001) |
1.12 ± 0.70 (<0.001) |
| 14 | 5.08 ± 2.68 |
5.87 ± 3.90 (<0.001) |
6.46 ± 4.73 (<0.001) |
1.82 ± 0.92 |
1.24 ± 0.81 (<0.001) |
1.29 ± 0.77 (<0.001) |
| 16 | 4.78 ± 2.54 |
5.66 ± 3.47 (0.006) |
5.77 ± 3.47 (0.003) |
1.58 ± 0.79 |
0.99 ± 0.53 (<0.001) |
1.10 ± 0.57 (<0.001) |
| 18 | 4.33 ± 1.99 |
5.06 ± 3.08 (<0.001) |
4.91 ± 2.77 (0.003) |
1.61 ± 0.68 |
1.10 ± 0.54 (<0.001) |
1.11 ± 0.53 (<0.001) |
| 19 | 3.26 ± 1.55 |
3.33 ± 2.58 (<0.001) |
3.48 ± 2.47 (0.004) |
1.21 ± 0.55 |
0.73 ± 0.50 (<0.001) |
0.78 ± 0.49 (<0.001) |
| 22 | 3.91 ± 2.04 |
3.60 ± 2.42 (<0.001) |
4.06 ± 2.69 (0.128) |
1.43 ± 0.69 |
0.80 ± 0.49 (<0.001) |
0.89 ± 0.50 (<0.001) |
The CT volume was normalized for nuclear size by dividing by the nuclear volume. Note that values in parentheses represented KS p‐values when compared to control lymphocytes.
Chromosome positioning in myeloma and lymphocyte nuclei after conventional epifluorescence microscopy
To examine spatial chromosome organization, we have used two differentially labeled chromosome paints in combination. The results revealed that the chromosome positions were nonrandom in normal lymphocytes, MGUS and MM nuclei (Fig. 2 a). To evaluate position differences between CTs, the relative radial distribution of each CT was measured. The average radial distance of the CTs to the center of the nuclei was determined. The distances were normalized using the radius of the nucleus. Values of 0 and 1 correspond to theoretical positions at the center or edge of the nucleus, respectively (see Kuzyk et al., 2015 for more details of this method).51
Figure 2.
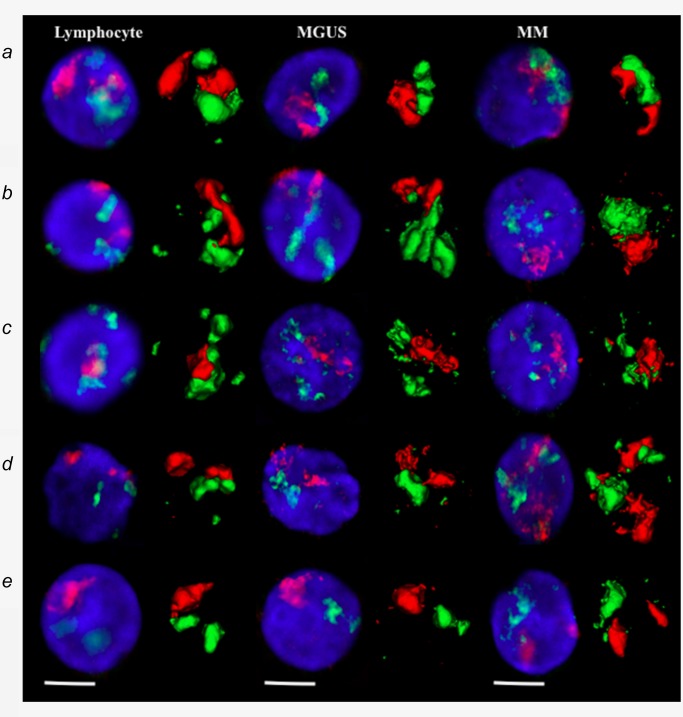
(a) Representative data of chromosome positions in interphase nuclei of lymphocytes, MGUS and MM after conventional 3D epifluorescence microscopy. Visualization of the 2D image and 3D reconstruction of specific CTs after 3D‐FISH using human whole chromosome painting probes on lymphocytes (left), MGUS cells (middle) and MM cells (right panel). (A) chromosome 4 (red) and 14 (green); (B) chromosome 11 (red) and 14 (green); (C) chromosome 14 (green) and 16 (red); (D) chromosome 9 (red) and 22 (green); and (E) chromosome 18 (red) and 19 (green). DNA counterstain is shown in blue (scale bar = 7 μm).
Our data show that chromosomes 4, 9, 11 and 18 were preferentially located toward the nuclear periphery, while chromosomes 19 and 22 were more centrally located. Chromosomes 14 and 16 occupied an intermediate position. The distributions of particular chromosomes were observed for both, the myeloma cells as well as the normal B‐lymphocytes. The two‐sided, two‐sample Kolmogorov–Smirnov (KS) analysis showed significant changes in the relative radial positions of chromosome 4, 9, 11, 14, 16 and 18 between lymphocyte and myeloma nuclei (Table 3). In contrast, there were no significant alterations in the relative radial position of chromosome 19 and 22, which were located toward a more central position within the cell nucleus.
Table 3.
The percentage of relative radial positioning (distance to the nuclear center) of each CTs distributed radially in lymphocyte and myeloma nuclei
| Chromosome | Median relative radial position (%) | KS test | |
|---|---|---|---|
| Lymphocyte | Myeloma | ||
| 4 | 61.37 | 58.18 | < 0.001 |
| 9 | 54.04 | 50.33 | < 0.001 |
| 11 | 51.43 | 52.06 | < 0.001 |
| 14 | 49.92 | 47.66 | < 0.001 |
| 16 | 42.94 | 43.42 | 0.0018 |
| 18 | 58.17 | 52.67 | < 0.001 |
| 19 | 30.08 | 33.02 | 0.0963 |
| 22 | 35.57 | 36.70 | 0.0786 |
We next analyzed the relationship of CTs volumes to the relative radial position within nuclei. Figure 3 a revealed that the CTs of the larger chromosomes tended to be further from the center than those of the small chromosomes in both lymphocyte and myeloma nuclei. Our results were consistent with previous studies that showed CTs size is one of the factors that influences radial position of chromosomes in interphase nucleus.26, 54
Figure 3.
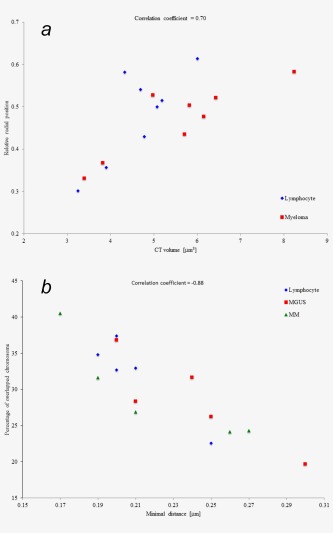
(a) Relationship between the size of CTs and their relative radial position within the nucleus of lymphocytes and myeloma. Blue data points represent lymphocytes and red data points represent myeloma cell. Each data point is the average for a patient. Position measurements are taken from CTs to the center of the nuclei and then normalized by dividing by the radius of the nucleus. The scatterplot shows a correlation between the size of CTs and their radial position within the nucleus (correlation coefficient = 0.70). Large chromosomes are located more distal to the nuclear center (larger relative radial position) than smaller chromosomes. (b) Relationship between the percentage of overlapping CTs and their minimal mutual distance in lymphocytes, MGUS and MM. These two measures correlate (correlation coefficient = −0.88). When chromosomes are located closer together they overlap more. Each data point represents the averages for a patient.
Relative proximity of CTs and overlapping CTs after conventional epifluorescence microscopy
To investigate localization and relationship between different CTs, we examined the relative position between CTs. The CT‐to‐CT distances were measured between CTs, and then expressed as a fraction of the nuclear radius. We observed that some nuclei had visibly touching neighboring CTs boundaries, distinctly pairs of chromosomes 4 and 14, 11 and 14 and 14 and 16. Minimal distances between CTs were measured as well. A value of zero in minimal distance between two CTs indicated that they were touching or sharing the same boundary. The minimal distance between CTs was highest for chromosome 18 and 19, which corresponds to their positions within nuclei. Chromosome 19 is located in the center while chromosome 18 is located toward nuclear periphery; thus they are physically far away from each other.
We also calculated the percentage of overlapping CTs between two different chromosomes. The most common overlapping pairs were CTs were 4 and 14, 11 and 14 and 14 and 16 for all three cells types, followed by 9 and 22 and finally 18 and 19 (Table 4). This Table shows which chromosomes edges are close together, overlapping or intermingling.
Table 4.
The minimal distance between CTs and percentage of overlapped CTs of lymphocyte and myeloma nuclei
| Minimal distance between CTs (μm) | Overlapped CTs (% of nuclei) | |||||
|---|---|---|---|---|---|---|
| Chromosome | Lymphocyte | MGUS | MM | Lymphocyte | MGUS | MM |
| 4 and 14 | 0.20 ± 0.26 |
0.21 ± 0.24 (0.06) |
0.19 ± 0.22 (0.52) |
37.34 | 28.35 | 31.63 |
| 9 and 22 | 0.20 ± 0.23 |
0.30 ± 0.31 (<0.001) |
0.27 ± 0.29 (0.002) |
32.67 | 19.65 | 24.30 |
| 11 and 14 | 0.21 ± 0.25 |
0.24 ± 0.30 (0.47) |
0.21 ± 0.22 (0.42) |
32.94 | 31.65 | 26.88 |
| 14 and 16 | 0.19 ± 0.23 |
0.20 ± 0.24 (1.00) |
0.17 ± 0.24 (0.37) |
34.80 | 36.77 | 40.50 |
| 18 and 19 | 0.25 ± 0.24 |
0.25 ± 0.28 (0.29) |
0.26 ± 0.27 (0.50) |
22.57 | 26.22 | 24.17 |
We next analyzed the relationship of the proximity of CTs to the percentage of overlap by combining the pairs of chromosomes by measuring the minimal distance between the CTs and the percentage of overlapping CTs. We found that the distance between chromosomes corresponded to the decrease of overlapping CTs (Table 4). Scatterplots showed an inverse relationship between minimal distances and overlapping CTs in all cell types, since nearby CTs exhibited an increased tendency to overlap compared to remote ones (Fig. 3 b).
Super‐resolution insights into CTs intermingling
Using 3D‐SIM to evaluate interterritorial relationships between selected CTs, we looked into the correlation between pairs of CTs within nuclei (Fig. 2 b). Pairs of CTs refer to two or more, if applicable CTs of two different types of chromosomes. We have used quantitative colocalization analysis to examine the degree of overlap of fluorescence signals representing each CTs. The Manders colocalization coefficient (MCC, Materials and Methods) was lowest in chromosomes 18 and 19, corresponding to their highest minimal distances between each other. For other pairs of CTs, the MCC of chromosomes 4 and 14, 11 and 14 and 14 and 16 were similar to 9 and 22, which are known to be neighbors (Table 5).18, 53 We analyzed the percentage of intermingling CT. The intermingling volumes were the lowest for chromosomes 18 and 19 and higher in other pairs of chromosomes, including 4 and 14, 9 and 22, 11 and 14 and 14 and16. These findings indicate that the distances between CT are correlated with the percentage of overlapping CTs and the interaction between them.
Figure 2.
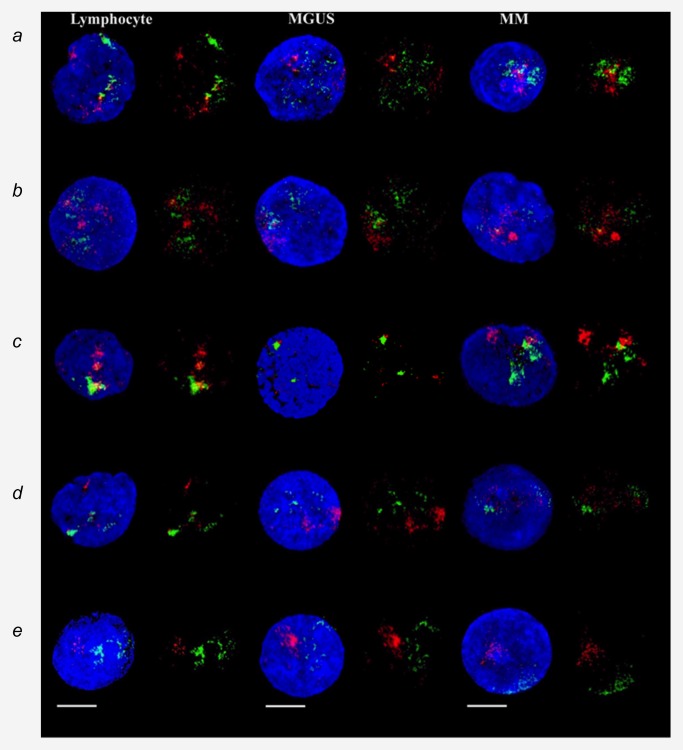
(b) Representative data of chromosome positions in interphase nuclei of lymphocytes, MGUS and MM after 3D‐SIM. Visualization of the 2D image and 3D reconstruction of specific CTs after 3D‐FISH using human whole chromosome painting probes on lymphocytes (left), MGUS cells (middle) and MM cells (right panel). (A) chromosome 4 (red) and 14 (green); (B) chromosome 11 (red) and 14 (green); (C) chromosome 14 (green) and 16 (red); (D) chromosome 9 (red) and 22 (green); and (E) chromosome 18 (red) and 19 (green). DNA counterstain is shown in blue (scale bar = 7 μm).
Table 5.
Mean Mander colocalization coefficient (MCC) and percentage of intermingling CTs of lymphocyte and myeloma nuclei studied by 3D‐SIM
| Mean Mander colocalization Coefficient (MCC) | Intermingling volume | |||||
|---|---|---|---|---|---|---|
| Chromosome | Lymphocyte | MGUS | MM | Lymphocyte | MGUS | MM |
| 4 and 14 | 0.12 ± 0.07 |
0.12 ± 0.05 (<0.001) |
0.11 ± 0.06 (0.001) |
6.28 ± 3.87 |
5.68 ± 2.45 (<0.001) |
5.17 ± 2.78 (<0.001) |
| 9 and 22 | 0.12 ± 0.07 |
0.12 ± 0.06 (0.002) |
0.11 ± 0.06 (0.12) |
5.81 ± 3.64 |
5.61 ± 2.69 (<0.001) |
5.14 ± 3.22 (0.09) |
| 11 and 14 | 0.10 ± 0.07 |
0.09 ± 0.04 (0.06) |
0.10 ± 0.06 (0.07) |
5.05 ± 3.61 |
4.32 ± 2.20 (0.06) |
4.71 ± 2.90 (0.09) |
| 14 and 16 | 0.11 ± 0.09 |
0.11 ± 0.06 (0.73) |
0.09 ± 0.06 (0.02) |
5.42 ± 4.55 |
4.97 ± 2.75 (0.69) |
3.99 ± 3.28 (0.02) |
| 18 and 19 | 0.05 ± 0.06 |
0.08 ± 0.06 (<0.001) |
0.07 ± 0.07 (<0.001) |
2.21 ± 2.94 |
3.73 ± 2.78 (<0.001) |
3.23 ± 3.41 (<0.001) |
Discussion
This study revealed the relative localization of CTs in human interphase nuclei of MM and MGUS and control lymphocytes. The eukaryotic interphase nucleus is a highly compartmentalized structure.27 The ability of FISH to reveal chromosomes and genes within nuclei demonstrated that gene‐rich chromosomes occupy predominately the nuclear interior whereas gene‐poor chromosomes locate toward the nuclear periphery.20, 25 Gene‐rich chromosomes such as 17 and 19 were found to locate in internal positions in the nucleus, while gene‐poor chromosomes such as 4, 13, 18 and inactive X (Xi) were typically found at the nuclear periphery.10, 55 Apart from the gene function, chromosome size and nuclear shape also correlated with the distribution of chromosomes within the nucleus. Small chromosomes are usually located more centrally and large chromosomes toward the periphery.26, 54 This preferential radial arrangement results in preferred clusters of neighboring chromosomes and might contribute to the control of certain sets of genes in a coordinated manner.12, 36
Previous studies showed that chromosome 9 and 22 are neighbors in lymphocyte nuclei; this proximity is thought to contribute to the t(9;22) translocation, a hallmark translocation found in chronic myeloid leukemia and adult acute lymphoblastic leukemia.18, 53 Here, we confirmed that chromosome 9 and 22 territories in lymphocyte are located in close proximity to each other. Pairs of chromosomes 4 and 14, 11 and 14 and 14 and 16 are also arranged close together, even closer than chromosomes 9 and 22 in both lymphocyte and myeloma nuclei. This leads to our assumption that chromosomes 4 and 14, 11 and 14 and 14 and 16 are neighboring chromosomes and that their very close association favors myeloma‐associated translocations. In contrast, chromosome 18 and 19 territories have the largest distance among the five analyzed pairs of chromosomes and are not usually engaged in translocations in myeloma.
Large variations in CTs sizes are characteristic for the eight chromosomes tested in this study. We observed a positive correlation between chromosome size and relative radial position. Large CTs tend to locate at the nuclear periphery and smaller CTs locate in more central positions. Despite similarities in the location of CTs between lymphocytes, MGUS and MM cell nuclei, we have demonstrated significant changes of relative radial positions among them. Compared to lymphocytes, chromosomes 4, 9, 14 and 18 in myeloma nuclei locate toward the nuclear center, whereas chromosomes 11 and 16 position toward the nuclear rim. This might result in alterations of gene expression and function in myeloma nuclei. Our data suggest significant changes of 3D chromosome positioning in interphase nuclei. In contrast, there are no differences in relative radial positions for chromosome 19 and 22, which locate in a central part of the nucleus.
Unbalanced chromosome translocation involving IgH loci are one of the most common pathogenesis of myeloma development. The three main common translocations are t(11;14), t(4;14) and t(14;16).1 Our results revealed that those pairs of chromosomes are neighboring chromosomes, which locate in close proximity and exhibit overlapping boundaries in both lymphocyte and myeloma nuclei. Increasing evidence suggests that chromosome organization is specific to the cell and tissue type.12, 21 Myeloma cells and lymphocytes originate from a common lymphoid progenitor, thus they are lineage‐related which probably contributes to the common structural arrangement of the chromosome within their nuclei. The results obtained in our experiments show that the preferential radial position of all CTs is maintained in myeloma nuclei when compared to lymphocytes. Additionally, we observed a relationship between minimal distance and chromosome overlap. We hypothesize that the close proximity of CTs increases their chance of overlap in the interphase nucleus. Our study showed that the highest percentage of overlapping occurred between chromosomes 4 and 14, 11 and 14 and 14 and 16, which correlated with the least minimal distance among all five pairs of chromosomes. Therefore, we conclude that the pairs of chromosomes potentially involved in a specific translocation event are located in close proximity in both normal and malignant nuclei. This spatial proximity of neighboring CTs appears to increase their chance to engage in translocations.
In accordance with the above findings, we expected differences in distribution of CTs in translocation‐containing nuclei; however, cytogenetic testing is not routinely done for all samples at our institution (cytogenetic testing was reported in 4/20 samples; t(11;14) in 2 samples and a hyperdiploid clone in 2 samples). Therefore, a subgroup analysis of chromosome positions in translocation‐positive myeloma was not performed here. Further studies are warranted to detail chromosomal organization in translocation‐positive compared to translocation‐negative myeloma.
Our findings of a larger size of CTs and nuclear volume in myeloma are consistent with our previous work that revealed a significant increase in the intranuclear submicron structure in myeloma interphase nuclei by using 3D‐SIM as well as increased DNA‐free/poor nuclear space compared to control lymphocyte.45 DNA‐free space refers to areas within the nucleus that have a low DNA density or no DNA at all.56
In the current study, we observed a direct relationship between changes in structural DNA content and the volumes of CTs in the interphase nucleus. Despite prominent larger sizes of interphase CTs volumes, myeloma nuclei have smaller normalized CTs volumes—when corrected for changes in the nuclear volumes—compared to lymphocyte nuclei, suggesting that the increases of CTs volume and nuclear volume are not proportional. CT volume enlarges at a lower ratio than the nuclear volume.
Taken together, this study has defined the 3D chromosome positions of translocation‐prone chromosomes in lymphocytes, MGUS and myeloma. Chromosomes in close spatial proximity have an increased likelihood of rearrangement and translocation.
Acknowledgements
The authors would like to thank all the patients who contributed blood samples to this study. We thank Donna Hewitt for obtaining patient consent and Shannon Kornelsen for providing clinical information.
References
- 1. Dimopoulos MA, Terpos E. Multiple myeloma. Ann Oncol 2010;21:vii143–50. [DOI] [PubMed] [Google Scholar]
- 2. Rajkumar SV, Kumar S. Multiple myeloma: diagnosis and treatment. Mayo Clin Proc 2016;91:101–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rajkumar SV. Multiple myeloma: 2012 update on diagnosis, risk‐stratification, and management. Am J Hematol 2012;87:79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rajkumar SV. MGUS and smoldering multiple myeloma: update on pathogenesis, natural history, and management. Hematol Am Soc Hematol Educ Program 2005;1:340–5. [DOI] [PubMed] [Google Scholar]
- 5. Korde N, Kristinsson SY, Landgren O. Monoclonal gammopathy of undetermined significance (MGUS) and smoldering multiple myeloma (SMM): novel biological insights and development of early treatment strategies. Blood 2011;17:5573–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hideshima T, Mitsiades C, Tonon G, et al Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat Rev Cancer 2007;7:585–98. [DOI] [PubMed] [Google Scholar]
- 7. Kuehl WM, Bergsagel PL. Molecular pathogenesis of multiple myeloma and its premalignant precursor. J Clin Invest 2012;122:3456–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer 2012;12:335–48. [DOI] [PubMed] [Google Scholar]
- 9. Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol 2005;23:6333–8. [DOI] [PubMed] [Google Scholar]
- 10. Bridger JM, Arican‐Gotkas HD, Foster HA , et al The non‐random repositioning of whole chromosomes and individual gene loci in interphase nuclei and its relevance in disease, infection, aging, and cancer. Adv Exp Med Biol. 2014;773:263–79. [DOI] [PubMed] [Google Scholar]
- 11. Iourov I. To see an interphase chromosome or: how a disease can be associated with specific nuclear genome organization. BioDiscovery 2012;4:1–5. [Google Scholar]
- 12. Meaburn KJ, Misteli T. Chromosome territories. Nature 2007;445:379–81. [DOI] [PubMed] [Google Scholar]
- 13. Rätsch A, Joos S, Kioschis P, et al Topological organization of the MYC/IGK locus in Burkitt's lymphoma cells assessed by nuclear halo preparations. Exp Cell Res 2002;273:12–20. [DOI] [PubMed] [Google Scholar]
- 14. Roix JJ, McQueen PG, Munson PJ, et al Spatial proximity of translocation‐prone gene loci in human lymphomas. Nat Genet 2003;34:287–91. [DOI] [PubMed] [Google Scholar]
- 15. Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription‐dependent associations. PLoS Biol 2006;4:e138 Doi:10.1371/journal.pbio.0040138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kozubek S, Lukásová E, Rýznar L, et al Distribution of ABL and BCR genes in cell nuclei of normal and irradiated lymphocytes. Blood 1997;89:4537–45. [PubMed] [Google Scholar]
- 17. Osborne CS, Chakalova L, Mitchell JA, et al Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol 2007;5:e192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guffei A, Sarkar R, Klewes L, et al Dynamic chromosomal rearrangements in Hodgkin's lymphoma are due to ongoing three‐dimensional nuclear remodeling and breakage‐bridge‐fusion cycles. Haematologica 2010;95:2038–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Righolt CH, Wiener F, Taylor‐Kashton C, et al Translocation frequencies and chromosomal proximities for selected mouse chromosomes in primary B lymphocytes. Cytometry A 2011;79A:276–83. [DOI] [PubMed] [Google Scholar]
- 20. Cremer T, Cremer C. Rise, fall and resurrection of chromosome territories: a historical perspective. Part II. Fall and resurrection of chromosome territories during the 1950s to 1980s. Part III. Chromosome territories and the functional nuclear architecture: experiments and models from the 1990s to the present. Eur J Histochem 2006;50:223–72. [PubMed] [Google Scholar]
- 21. Parada LA, McQueen PG, Misteli T. Tissue‐specific spatial organization of genomes. Genome Biol 2004;5:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boveri, T. Zur Frage der Entstehung maligner Tumoren. Jena: Gustav Fischer, 1914. 64. [Google Scholar]
- 23. Boveri, T. The origin of malignant tumors. Translated by Marcella Boveri. Baltimore: The Williams & Wilkens Company, 1929. 119. [Google Scholar]
- 24. Boveri T. Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. J Cell Sci 2008;121:1–84. [DOI] [PubMed] [Google Scholar]
- 25. Boyle S, Gilchrist S, Bridger JM, et al The spatial organization of human chromosomes within the nuclei of normal and emerin‐mutant cells. Hum Mol Genet 2001;10:211–19. [DOI] [PubMed] [Google Scholar]
- 26. Bolzer A, Kreth G, Solovei I, et al Three‐dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol 2005;3:826–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kumaran RI, Thakar R, Spector DL. Chromatin dynamics and gene positioning. Cell 2008;132:929–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cremer T, Cremer C. Chromosome territories. Cold Spring Harb Perspect Biol 2010;2:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sproul D, Gilbert N, Bickmore WA. The role of chromatin structure in regulating the expression of clustered genes. Nat Rev Genet 2005;6:775–81. [DOI] [PubMed] [Google Scholar]
- 30. Solovei I, Kreysing M, LanctÔt C, et al Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 2009;137:356–68. [DOI] [PubMed] [Google Scholar]
- 31. Marella NV, Bhattacharya S, Mukherjee L, et al Cell type specific chromosome territory organization in the interphase nucleus of normal and cancer cells. J Cell Physiol 2009;221:130–8. [DOI] [PubMed] [Google Scholar]
- 32. Kuroda M, Tanabe H, Yoshida K, et al Alteration of chromosome positioning during adipocyte differentiation. J Cell Sci 2004;15:5897–903. [DOI] [PubMed] [Google Scholar]
- 33. Hübner B, Lomiento M, Mammoli F, et al Remodeling of nuclear landscapes during human myelopoietic cell differentiation maintains co‐aligned active and inactive nuclear compartments. Epigenetics Chromatin 2015; 8:47.doi: 10.1186/s13072‐015‐0038‐0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rouquette J, Cremer C, Cremer T, et al Functional nuclear architecture studied by microscopy: present and future. Int Rev Cell Mol Biol 2010;282:1–90. [DOI] [PubMed] [Google Scholar]
- 35. Cremer T, Cremer M, Hübner B , et al The 4D nucleome: evidence for a dynamic nuclear landscape based on co‐aligned active and inactive nuclear compartments. FEBS Lett 2015;589:2931–43. [DOI] [PubMed] [Google Scholar]
- 36. Fritz AJ, Barutcu AR, Martin‐Buley L, et al Chromosomes at work: organization of chromosome territories in the interphase nucleus. J Cell Biochem 2016;117:9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cremer M, Grasser F, Lanctôt C , et al Multicolor 3D fluorescence in situ hybridization for imaging interphase chromosomes In: Hancock R, ed. The nucleus: Volume 1: nuclei and subnuclear components. New Jersey: Humana Press, 2008. 205–39. [DOI] [PubMed] [Google Scholar]
- 38. Heilemann M. Fluorescence microscopy beyond the diffraction limit. J Biotechnol 2010;149:243–51. [DOI] [PubMed] [Google Scholar]
- 39. Leung BO, Chou KC. Review of super‐resolution fluorescence microscopy for biology. Appl Spectrosc 2011;65:967–80. [DOI] [PubMed] [Google Scholar]
- 40. Schermelleh L, Heintzmann R, Leonhardt H. A guide to super‐resolution fluorescence microscopy. J Cell Biol 2010;190:165–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Żurek‐Biesiada D, Szczurek AT, Prakash K, et al Localization microscopy of DNA in situ using Vybrant® DyeCycle™ Violet fluorescent probe: a new approach to study nuclear nanostructure at single molecule resolution. Exp Cell Res 2015;343:97–106. [DOI] [PubMed] [Google Scholar]
- 42. Huang B, Bates M, Zhuang X. Super resolution fluorescence microscopy. Annu Rev Biochem 2009;78:993–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kyle RA, Rajkumar SV. Criteria for diagnosis, staging, risk stratification and response assessment of multiple myeloma. Leukemia 2009;23:3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Klewes L, Vallente R, Dupas E, et al Three‐dimensional nuclear telomere organization in multiple myeloma. Transl Oncol 2013;6:749–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sathitruangsak C, Righolt CH, Klewes L, et al Quantitative superresolution microscopy reveals differences in nuclear DNA organization of multiple myeloma and monoclonal gammopathy of undetermined significance. J Cell Biochem 2015;116:704–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Luengo Hendriks CL, van Vliet LJ, Rieger B , et al DIPimage: a scientific image processing toolbox for MATLAB Delft: Quantitative Imaging Group, 1999. The Netherlands: Delft University of Technology. [Google Scholar]
- 47. Poon SS, Lansdorp PM. Quantitative fluorescence in situ hybridization (Q‐FISH) In: Bonifacino JS, Dasso M, Harford JB, Lippincott‐Schwartz J, Yamada KM, eds. Current protocols in cell biology. New York: Wiley, 2001. 18.4.1–21. [DOI] [PubMed] [Google Scholar]
- 48. Schaefer LH, Schuster D, Herz H. Generalized approach for accelerated maximum likelihood based image restoration applied to three‐dimensional fluorescence microscopy. J Microsc 2001;204:99–107. [DOI] [PubMed] [Google Scholar]
- 49. Righolt CH, Schmalter AK, Kuzyk A, et al Measuring murine chromosome orientation in interphase nuclei. Cytometry A 2015;87:733–40. [DOI] [PubMed] [Google Scholar]
- 50. Ridler TW, Calvard S. Picture thresholding using an iterative selection method. IEEE Trans Syst Man Cybern 1978;8:630–2. [Google Scholar]
- 51. Kuzyk A, Booth S, Righolt C, et al MYCN overexpression is associated with unbalanced copy number gain, altered nuclear location, and overexpression of chromosome arm 17q genes in neuroblastoma tumors and cell lines. Genes Chromosomes Cancer 2015;54:616–28. [DOI] [PubMed] [Google Scholar]
- 52. Manders EMM, Verbeek FJ, Aten JA. Measurement of co‐localization of objects in dual‐colour confocal images. J Microsc 1993;169:375–82. [DOI] [PubMed] [Google Scholar]
- 53. Kozubek S, Lukášová E, Marečková A, et al The topological organization of chromosomes 9 and 22 in cell nuclei has a determinative role in the induction of t(9,22) translocations and in the pathogenesis of t(9,22) leukemias. Chromosoma 1999;108:426–35. [DOI] [PubMed] [Google Scholar]
- 54. Cremer M, von Hase J, Volm T, et al Chromosome Res 2001;9:541–67. [DOI] [PubMed] [Google Scholar]
- 55. Croft JA, Bridger JM, Boyle S, et al Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol 1999;145:1119–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Righolt CH, Guffei A, Knecht H, et al Differences in nuclear DNA organization between lymphocytes, Hodgkin and Reed–Sternberg cells revealed by structured illumination microscopy. J Cell Biochem 2014;115:1441–8. [DOI] [PMC free article] [PubMed] [Google Scholar]


