ABSTRACT
Neurogenesis is the process of neuron generation, which occurs not only during embryonic development but also in restricted niches postnatally. One such region is called the subventricular zone (SVZ), which gives rise to new neurons in the olfactory bulb (OB). Neurons that are born postnatally migrate through more complex territories and integrate into fully functional circuits. Therefore, differences in the differentiation of embryonic and postnatally born neurons may exist. Dendritogenesis is an important process for the proper formation of future neuronal circuits. Dendritogenesis in embryonic neurons cultured in vitro was shown to depend on the mammalian target of rapamycin (mTOR). Still unknown, however, is whether mTOR could regulate the dendritic arbor morphology of SVZ‐derived postnatal OB neurons under physiological conditions in vivo. The present study used in vitro cultured and differentiated SVZ‐derived neural progenitors and found that both mTOR complex 1 and mTOR complex 2 were required for the dendritogenesis of SVZ‐derived neurons. Furthermore, using a combination of in vivo electroporation of neural stem cells in the SVZ and genetic and pharmacological inhibition of mTOR, it was found that mTOR was crucial for the growth of basal and apical dendrites in postnatally born OB neurons under physiological conditions and contributed to the stabilization of their basal dendrites. © 2016 Wiley Periodicals, Inc. Develop Neurobiol 76: 1308–1327, 2016
Keywords: postnatal neurogenesis, mTOR, dendritogenesis, olfactory bulb, in vivo electroporation
INTRODUCTION
Neurogenesis is a process of neuron generation (Ming and Song, 2011), which occurs mainly during early embryonic development and in restricted form also occurs postnatally, including in adulthood (Götz and Huttner, 2005; Paridaen and Huttner, 2014). One postnatal neurogenic region is the subventricular zone (SVZ), from where neural progenitors migrate along the rostral migratory stream (RMS) toward the olfactory bulb (OB) where they differentiate into neurons (Carlén et al., 2002; Lledo et al., 2008) that are involved in OB‐dependent forms of plasticity (Nissant et al., 2009). Neurogenesis includes neuron birth, migration, differentiation, maturation, and integration. Many molecular mechanisms that underlie neuronal development are shared in the embryonic and postnatal periods (Urban and Guillemot, 2014). However, neurons that are born postnatally need to migrate through more complex territories and integrate into fully functional circuits (Petreanu and Alvarez‐Buylla, 2002). Although substantial progress has been made in understanding postnatal neurogenesis and subsequent neuron maturation, several questions remain unanswered. For example, the precise molecular mechanisms that regulate newborn neuron differentiation, maturation, and integration into pre‐existing circuits are still not fully known. Moreover, the existence of important differences between embryonic and postnatal neurogenesis have only just begun to be revealed (Ming and Song, 2011; Urban and Guillemot, 2014).
An important step of neuron maturation is dendritogenesis. The shape of the dendritic arbor reflects the function of a neuron within a particular neuronal circuit. Thus, its proper development is key for accurate connectivity and synaptic organization within any given neuronal ensemble (McAllister, 2000; Wong and Ghosh, 2002; Urbanska et al., 2008). The development of dendritic trees is a complex process that requires tight molecular regulation and depends on both the genetic program and various local environmental cues. Among the intrinsic factors that regulate the dendritogenesis of postnatally born neurons is the transcription factor cyclic adenosine monophosphate response element‐binding protein (CREB; Giachino et al., 2005). Examples of extrinsic cues include γ‐aminobutyric acid (GABA) and vascular endothelial growth factor (VEGF) (Gascon et al., 2006; Licht et al., 2010). However, considering the numerous factors that regulate the dendritic arbor development of embryonic neurons, the molecular aspects of postnatally born OB neuron dendritogenesis have only been sparsely investigated. mTOR is a large (259 kDa) serine/threonine kinase. In mammalian cells, mTOR forms two distinct protein complexes, mTORC1 and mTORC2, which are defined by the presence of unique mTOR protein partners (i.e., regulatory‐associated protein of mTOR [Raptor, mTORC1] and rapamycin‐insensitive companion of mTOR [Rictor, mTORC2]; Kim et al., 2002; Jacinto et al., 2004; Malik et al., 2013; Sarbassov et al., 2004). In addition to differences in protein composition, these two complexes differ in their substrates, cellular localization, function, and sensitivity to the mTOR inhibitor rapamycin (Malik et al., 2013). Studies of in vitro cultured neurons showed that mTOR plays important roles in neuronal development, including dendritogenesis (Jaworski et al., 2005; Kumar et al., 2005; Swiech et al., 2008; Urbanska et al., 2012a). In vivo studies are less numerous (Thomanetz et al., 2013). However, work from the Bordey group using a conditional knockout of TSC1 (Tuberous Sclerosis Complex 1), a well‐known mTORC1 inhibitor, in neural stem cells (NSCs) postnatally in SVZ, shows that mTOR hyperactivity results in dendritic arbor hypertrophy (Feliciano et al., 2012). Similarly, hyperactivation of mTOR in adult born neurons in dentate gyrus, due to loss of Disc1, leads to excessive dendritic branching (Kim et al., 2009; Zhou et al., 2013). Importantly, the role of mTOR in SVZ‐derived neurons under physiological conditions has not been studied in vivo. Chemical screening performed by Khodosevich and Monyer (2010) questioned the importance of mTOR activity for neurite formation by RMS‐derived neuroblasts cultured in vitro. This raises the question of whether mTOR can regulate the dendritic arbor morphology of SVZ‐derived GABAergic postnatal OB neurons under physiological conditions.
In the present study, we tested whether mTOR contributes to regulating the dendritic arbor formation of OB neurons that are generated early postnatally. Using in vitro cultured and differentiated SVZ‐derived neuroprecursors, we found that both mTORC1 and mTORC2 are required for the dendritogenesis of SVZ‐derived neurons. Furthermore, using a combination of in vivo electroporation of NSCs in the SVZ and genetic and pharmacological inhibition of mTOR, we found that mTOR is crucial for the growth of basal and apical dendrites of postnatally born OB neurons under physiological conditions, and it contributes to the stabilization of their basal dendrites.
METHODS
Drugs and Antibodies
The following inhibitors were purchased from commercial sources: rapamycin (LC Laboratories, Woburn, MA) and Ku‐0063794 (Chemdea, Ridgewood, NJ). Primary antibodies are listed in Table 1. Secondary anti‐rabbit and anti‐mouse antibodies conjugated to horseradish peroxidase (HRP; Jackson ImmunoResearch, West Grove, PA; 1:10,000) were used for Western blot. Donkey anti‐rabbit and anti‐mouse and goat anti‐chicken secondary antibodies conjugated to Alexa Fluor dyes (Invitrogen, Eugene, OR) were used for immunofluorescence both in cultured cells (1:300) and brain sections (1:500).
Table 1.
List of Primary Antibodies Used
| No. | Antigen | Catalog No. | Host | Application and Dilution | Vendor |
|---|---|---|---|---|---|
| 1 | GFAP | AB5804 | Rabbit |
IF 1:1000 (tissue culture) |
Millipore |
| 2 | GFP | 13970 | Chicken |
IF 1:1000 (brain sections) |
Abcam |
| 3 | GFP | 598 | Rabbit |
IF 1:1000 (tissue culture) |
MBL |
| 4 | GFP | NB100‐1770 | Goat |
IF 1:500 (tissue culture) |
Novus Biologicals |
| 5 | Lectin FITC | 128K1065 | Mouse |
IF 1:300 (tissue culture) |
Sigma‐Aldrich |
| 6 | MAP2 | M4403 | Mouse |
IF 1:300 (tissue culture) |
Sigma‐Aldrich |
| 7 | MAP2 | 4542 | Rabbit |
IF 1:1000 (tissue culture) |
Cell Signaling Technology |
| 8 | mTOR (7c10) | 2983 | Rabbit | WB 1:500 | Cell Signaling Technology |
| 9 | Nestin | Ab6142 | Mouse |
IF 1:300 (tissue culture) |
Abcam |
| 10 | NeuN | ab104225 | Mouse |
IF 1:1000 (tissue culture) |
Abcam |
| 11 | Oligodendrocytes (RIP) | MAB158 | Mouse |
IF 1:300 (tissue culture) |
Millipore |
| 12 | P‐S6 (Ser235/236) | 4858 | Rabbit |
IF 1:300 (tissue culture); WB 1:500 |
Cell Signaling Technology |
| 13 | P‐S6 (Ser240/Ser244) | 5364 | Rabbit |
IF 1:500 (brain sections) |
Cell Signaling Technology |
| 14 | S6 | 2217 | Rabbit | WB 1:500 | Cell Signaling Technology |
| 15 | P‐Akt (Ser473) | 4060 | Rabbit |
WB 1:1000 IF 1:300 |
Cell Signaling Technology |
| 16 | Akt | 2920 | Mouse | WB 1:1000 | Cell Signaling Technology |
| 17 | Tuj 1 (neuron‐specific class III β‐tubulin) | ab18207 | Mouse |
IF 1:500 (tissue culture) |
Abcam |
| 18 | α‐tubulin | T5168 | Mouse | WB 1:5000 | Sigma‐Aldrich |
IF, immunofluorescence; WB, Western blot.
DNA Constructs
The following mammalian expression plasmids, described previously, were obtained from Addgene or directly from other researchers: pCx‐EGFP‐N1 (Boutin et al., 2008), pCAG‐GFP (Matsuda and Cepko, 2004), pCALNL‐DsRED (Matsuda and Cepko, 2007), pCAG‐Cre (Matsuda and Cepko, 2007), pSUPER (Brummelkamp et al., 2002), pSUPER‐shRaptor#1 (Urbanska et al., 2012a), pSUPER‐shRictor#2 (Urbanska et al., 2012a), pSUPER‐scrRaptor#1 (Urbanska et al., 2012a), pSUPER‐scrRictor#2 (Urbanska et al., 2012a), and pSUPER‐mTOR7513 (Jaworski et al., 2005). The empty vector pCAG was obtained by excising the Cre coding sequence from pCAG‐Cre with EcoRI/NotI.
Animals Used for the Studies
To prepare primary in vitro cultures of neural progenitors, Wistar rat (Rattus norvegicus) pups (postnatal day [P] 1 or 2) were used. The animals were sacrificed by decapitation according a procedure approved by the First Local Ethics Committee in Warsaw (decision no. 987/2009), which is in compliance with the European Community Council Directive (86/609/EEC). For in vivo electroporation, wildtype (C57BL/6 strain) P1 or P2 mice or Mtorfl/fl mice (on a C57BL/6 background; Gangloff et al., 2004) were used in accordance with procedures approved by the First Local Ethics Committee in Warsaw (Decisions 189/2011), which are in compliance with the European Community Council Directive (86/609/EEC). All of the procedures that were performed at McGill University were in compliance with the Canadian Council on Animal Care guidelines and approved by McGill University.
Primary In Vitro Culture of Neural Progenitors and Neurons
Primary neural progenitor cultures were prepared from rat neonates 24 h after birth (P1) according to the protocol adapted from Giachino et al. (2009). A single rat pup (P1) was decapitated, the brain was removed and placed into cold Hank's balanced salt solution (HBSS). The meninges, OBs [Supporting Information Fig. 1(A), line #1], and cerebellum [Supporting Information Fig. 1(A), line #2] were removed. The brain was cut coronally, approximately in the middle [Supporting Information Fig. 1(A), line #3]. The SVZ was microdissected [Supporting Information Fig. 1(A), red dotted line], and the tissue was minced. The minced tissue was incubated for 30 min at 37°C with Papain mix composed of 30 U/mL papain (Sigma‐Aldrich, St. Louis, MO), 0.24 mg/mL cysteine (Sigma‐Aldrich), 40 mg/mL DNAseI Type IV (Sigma‐Aldrich) and Ovomucoid mix: 1 mg/mL trypsin inhibitor (Sigma‐Aldrich), 0.5 mg/mL BSA (Sigma‐Aldrich), 40 mg/mL DNAse I Type IV (Sigma‐Aldrich) in HBSS in the ratio 1:1. The enzymatic reaction was stopped by the addition of an equal volume of Ovomucoid mix followed by an additional 5–10 min incubation at room temperature. Next, the tissue was dissociated using a 1 mL filter tip. The obtained cell suspension was washed by adding and resuspending the dissociated tissue in 9 mL Dulbecco's Modified Eagle Medium (DMEM)/F12. The cells from the debris were separated by centrifugation 5 min at 100g. The supernatant was discarded and the cell pellet was resuspended in proliferation medium that contained DMEM/F12 with GlutaMax supplement (Invitrogen, Carlsbad, CA), 2% B27 (Invitrogen), 20 nM recombinant epidermal growth factor (EGF; Alomone Laboratories, Jerusalem, Israel), 20 nM recombinant basic fibroblast growth factor (bFGF; Alomone Laboratories), and 1% penicillin/streptomycin mixture (Sigma‐Aldrich) and plated as a single‐cell suspension on an uncoated plastic dish.
Figure 1.
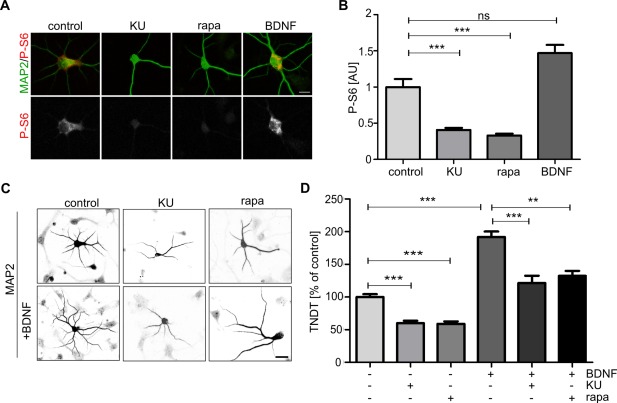
mTOR activity is necessary for proper dendritic arborization of SVZ‐derived neurons cultured in vitro. (A) Representative confocal images of SVZ‐derived neurons immunofluorescently stained for MAP2 and ribosomal protein S6 phosphorylated at Ser235/236 (P‐S6). Scale bar = 25 µm. Two days after plating, brain‐derived neurotrophic factor (BDNF; 50 ng/µL) and DMSO or mTOR inhibitors (Ku‐0063794 [KU], 30 nM; rapamycin [rapa], 20 nM) were added to the culture medium. Four days after plating, the cells were fixed and immunostained. (B) Quantitative analysis of average intensity of P‐S6 immunofluorescence of neurons treated as in A. Number of experiments (N) = 3. Number of analyzed cells (n) = 24 (control), 31 (KU), 32 (rapa), and 26 (BDNF). The data are expressed as mean values normalized to the control. Error bars indicate SEM. ***p < 0.001; ns, not significant (Kruskal–Wallis test with Dunn's post hoc test). AU, arbitrary units. (C) Representative confocal images of neurons treated as indicated in A and immunofluorescently labeled with an antibody against MAP2. Scale bar = 25 µm. (D) Quantitative analysis of number of dendrites based on MAP2 immunostaining of cells treated as in A. The bar graph shows the analysis of the number of dendritic tips as a percentage of the value normalized to the BDNF untreated control in a given experiment. Error bars indicate SEM. Number of experiments (N) = 3. Number of analyzed cells (n) = 123 (control), 72 (KU), 69 (rapa), 105 (control + BDNF), 53 (KU + BDNF), and 72 (rapa + BDNF). ***p < 0.001, **p < 0.01 (Kruskal–Wallis test with Dunn's post hoc test).
Neural progenitors were cultured as free‐floating neurospheres. The first passage was performed 5 days post‐isolation. The neurospheres were centrifuged at 100g for 5 min, resuspended in prewarmed 0.05% trypsin‐EDTA (Sigma‐Aldrich) in HBSS and incubated at 37°C for 10 min. Next, an equal volume of Ovomucoid mix was added and the neurosphere suspension was incubated for an additional 5 min at room temperature. Then the neurospheres were dissociated to a single cell using a 1 mL filter tip. The obtained cell suspension was washed by adding 9 mL DMEM/F12 and was centrifuged at 100g for 5 min. The supernatant was then discarded, and proliferation medium was added to the cell pellet. The cells were reseeded at a density of 1 × 104 cells/cm2 in proliferation medium and incubated at 37°C with 5% CO2 until the next passage. Neural progenitors were cultured as neurospheres for three to four passages before differentiation.
For the differentiation of neural progenitors to neurons, the neurospheres were first trypsinized and plated in neuron growth medium that contained Neurobasal medium (Invitrogen), 2% B27 (Invitrogen), 0.5 mM glutamine (Sigma‐Aldrich), 12.5 µM glutamate (Sigma‐Aldrich), 1% penicillin/streptomycin mixture (Sigma‐Aldrich) at a density of 500 cells/mm2 on glass coverslips coated with poly‐L‐ornithine (15 µg/mL, Sigma‐Aldrich), and laminin (5 µg/mL, Roche, Basel, Switzerland). Differentiating neural progenitors were kept in an incubator under standard culture conditions.
Transfection and Drug Treatment of Differentiating SVZ‐Derived Neural Progenitors
After three to four passages, the neural progenitors were first plated under differentiating conditions. The next day, they were transfected with Lipofectamine2000 as described previously for hippocampal neurons cultured in vitro (Jaworski et al., 2009). To accelerate dendritogenesis and activate mTOR signaling on day 2 after plating, differentiating neural progenitors were treated with brain‐derived neurotrophic factor (BDNF; 50 ng/µL, Sigma Aldrich). To inhibit mTOR activity, either rapamycin (20 nM) or Ku‐0063794 (30 nM) were added to the culture medium alone or in combination with BDNF 48 h post‐plating.
In Vivo Electroporation and Pharmacological Treatment
The protocol and electroporation parameters were adapted from Boutin et al. (2008). Neonates (C57BL/6, P1/P2) were anesthetized on an ice‐filled glass petri dish for approximately 5 min. Subsequently, the animals were placed on a custom‐made support in a stereotaxic rig under a Hamilton syringe with a manually pulled glass capillary (<50 μm diameter). The DNA (3 μg/μL) for electroporation was diluted in phosphate‐buffered saline (PBS)‐I (147 mM NaCl, 2.7 mM KCl, and 20 mM phosphate buffer [pH 7.4]) that contained 0.1% Fast Green. The syringe filled with the DNA solution was positioned at the level of the right lateral ventricle [Supporting Information Fig. 2(A)] and introduced 2 mm deep into the lumen. An injection was considered correct when Fast Green dye was visible under white light in the lateral ventricle. Successfully injected animals were subjected to five poring electrical pulses (50 ms, 95 V, pulses at 950 ms intervals) followed by five transfer pulses (50 ms, 20 V, pulses at 50 ms intervals) using a CUY21 device (Nepagene, Chiba, Japan) and 10 mm tweezer electrodes (CUY650P10, Nepagene). The tweezer electrodes were oriented to target plasmid DNA to the dorsal wall of the ventricles [Supporting Information Fig. 2(A)]. Afterward, electroporated animals were warmed on a heating pad for several minutes and returned to their dams. To inhibit mTOR activity, electroporated pups were treated with rapamycin. Rapamycin treatment was adapted from Anderl et al. (2011). Rapamycin was initially dissolved in 100% ethanol at a concentration of 0.1 mg/mL and stored at −20°C. Immediately before the injection, rapamycin was diluted in a sterile vehicle solution that contained 0.25% Tween 80 and 0.25% PEG 400 (low‐molecular‐weight grade of polyethylene glycol). Mouse neonates were intraperitoneally (IP) injected with either two or three doses of 1 mg/kg rapamycin beginning on P8 or P10, respectively. The doses were administered at 48 h intervals. A control group received ethanol in vehicle solution.
Figure 2.
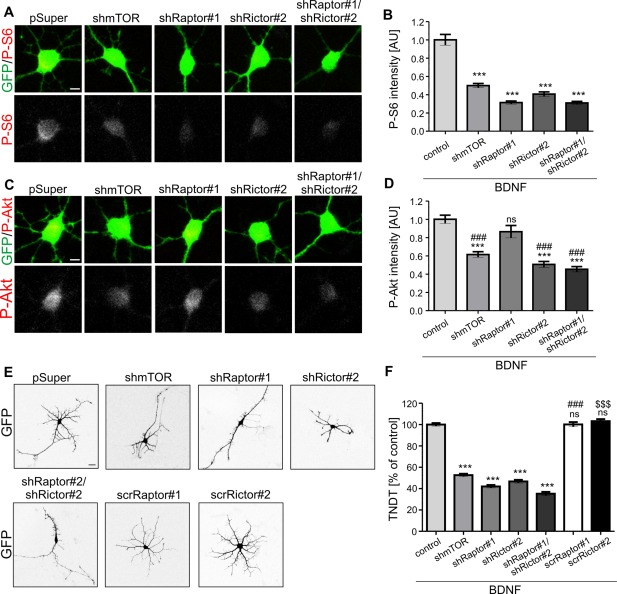
Proper dendritic arborization of SVZ‐derived neurons cultured in vitro requires both mTORC1 and mTORC2. (A) Representative confocal images of SVZ‐derived neurons transfected as indicated and immunofluorescently stained for ribosomal protein S6 phosphorylated at Ser235/236 (P‐S6) and GFP. Scale bar = 5 µm. Neural progenitors were isolated from the SVZ, dissociated, and plated in differentiation‐promoting conditions. On day 1, the cells were transfected with pSUPER (control), pSUPER‐mTOR7513 (shmTOR), pSUPER‐shRaptor#1 (shRaptor#1), pSUPER‐shRictor#2 (shRictor#2) or pSUPER‐shRaptor#1 and pSUPER‐shRictor#2 (shRaptor#1/shRictor#2). A GFP‐encoding plasmid was co‐transfected in all of the variants to visualize neuron morphology. Brain‐derived neurotrophic factor (BDNF) was added 24 h postransfection. Cells were fixed 4 days after plating. (B) Quantitative analysis of average intensity of P‐S6 immunofluorescence of neurons transfected and treated as in A. Number of experiments (N) = 2. Number of analyzed cells (n) = 59 (pSUPER, control), 37 (shmTOR), 44 (shRaptor#1), 38 (shRictor#2), 44 (shRaptor#1/shRictor#2). The data are expressed as mean values normalized to the control. Error bars indicate SEM. ***p < 0.001; ns, not significant vs. control (Kruskal–Wallis test with Dunn's post hoc test). AU, arbitrary units. (C) Representative confocal images of SVZ‐derived neurons transfected and treated as in A and stained for Akt phosphorylated at Ser473 (P‐Akt) and GFP. Scale bar = 5 µm. Progenitors were isolated from the SVZ, dissociated, and plated in differentiation‐promoting conditions with brain‐derived neurotrophic factor (BDNF). On day 1 cells were transfected as in A. (D) Quantitative analysis of average intensity of P‐Akt immunofluorescence of neurons treated as in A. Number of experiments (N) = 2. Number of analyzed cells (n) = 31 (pSUPER, control), 28 (shmTOR), 31 (shRaptor#1), 35 (shRictor#2), 31 (shRaptor#1/shRictor#2). The data are expressed as mean values normalized to the control. Error bars indicate SEM. ***p < 0.001; ns, not significant vs. control; ### p < 0.001 vs. shRaptor#1 (Kruskal–Wallis test with Dunn's post hoc test). AU, arbitrary units. (E) Representative confocal images of SVZ‐derived neurons transfected as indicated and immunofluorescently stained for GFP. Scale bar = 20 µm. Progenitors were isolated from the SVZ, dissociated, and plated in differentiation‐promoting conditions. On day 1, the cells were transfected with pSUPER (control), pSUPER‐mTOR7513 (shmTOR), pSUPER‐shRaptor#1 (shRaptor#1), pSUPER‐shRictor#2 (shRictor#2), pSUPER‐shRaptor#1 and pSUPER‐shRictor#2 (shRaptor#1/shRictor#2), pSUPER‐scrRaptor#1 (scrRaptor#1) or pSUPER‐scrRictor#2 (scrRictor#2). A GFP‐encoding plasmid was co‐transfected in all of the variants to visualize neuron morphology. BDNF was added 24 h postransfection. Cells were fixed 4 days after plating. (F) Quantitative analysis of the number of dendrites of neurons transfected and treated as in E. The bar graph shows the analysis of the number of dendritic tips as a percentage of the value normalized to the control in a given experiment. Error bars indicate SEM. Number of experiments (N) = 3. Number of analyzed cells (n) = 34 (pSUPER), 52 (shmTOR), 48 (shRaptor#1), 57 (shRictor#2), 56 (shRaptor#1; shRictor#2), 30 (scrRaptor#1), 30 (scrRictor#2). ***p < 0.001, ns ‐ not significant vs. control, ### p < 0.001 vs. shRaptor#1, $$$ p < 0.001 vs. shRictor#2 (Kruskal–Wallis test with Dunn's post hoc test).
Immunofluorescent Staining of In Vitro Cultured Cells
For immunofluorescence staining, cells that were cultured on glass coverslips were fixed with 4% paraformaldehyde (PFA) and 4% sucrose in PBS‐II (135 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, and 1.4 mM KH2PO4; pH 7.4) for 10 min at room temperature. After fixation, the cells were washed three times for 10 min each with PBS‐II and incubated with primary antibodies diluted in GDB (30 mM phosphate buffer [pH 7.4], 0.2% gelatin, 0.5% Triton X‐100, and 450 mM NaCl) overnight at 4°C. The next day, the cells were washed three times for 10 min each with PBS‐II and incubated for 1 h with appropriate Alexa Fluor‐conjugated secondary antibodies diluted in GDB at room temperature, followed by three washes for 10 min each with PBS‐II. During the second wash, Hoechst 33258 (1:10,000; Invitrogen) was added. In the last step, each coverslip was rinsed with purified, sterile water and closed with VECTASHIELD mounting medium (Vector Laboratories, Burlingame, CA) on a glass slide.
Immunofluorescent Staining of Mouse Brain Sections
The mice were deeply anesthetized with an overdose of pentobarbital (300 mg/kg body weight, IP) and transcardially perfused with PBS‐I followed by 4% PFA in 0.1M phosphate buffer (pH 7.4). Brains were isolated and postfixed for 2 h in 4% PFA in 0.1M phosphate buffer (pH 7.4). Postfixed brains were cryopreserved with 30% sucrose in 0.1M phosphate buffer for at least 72 h. Afterward, the brains were frozen and cut into 40 or 100 μm thick sections on a cryostat and collected in antifreeze medium. Immunofluorescence was performed on free‐floating sagittal or coronal brain sections. Prior to immunostaining, the brain sections were washed three times for 5 min each with PBS‐I. For 100‐µm thick sections, an additional step was included in the procedure. Specifically, PBS‐I was replaced with an antigen retrieval buffer (10 mM Trisodium citrate, 0.05% Tween 20), and the sections were incubated for an additional 15 min at 60°C. Next, the brain sections were washed three times for 5 min each with PBS‐I. Both 100 and 40 μm brain sections were next incubated for 1 h at room temperature with blocking buffer (PBS‐I that contained either 1% donkey or normal goat serum and 1% or 0.2% Triton‐X 100 for 100 and 40 μm thick brain sections, respectively). The brain slices were then incubated overnight at 4°C with primary antibodies in an appropriate blocking buffer. After this overnight incubation, the sections were washed three times with PBS‐I and incubated at room temperature for 1 or 2 h, depending on the section thickness, with fluorochrome‐conjugated secondary antibodies diluted in an appropriate blocking buffer. In the next step, the slices were rinsed three times with PBS‐I for 5 min each. Hoechst 33258 was added (1:10,000) to the second PBS‐I wash. Stained brain slices were mounted on microscope slides, dried for 15 min at room temperature, and then closed under glass coverslips using VECTASHIELD mounting medium.
Preparation of Brain Protein Extracts
The brains were removed immediately after decapitation. The OBs were dissected, frozen on dry ice, and stored at −80°C until use. The brain tissue was homogenized in homogenization buffer, followed by centrifugation at maximum speed (12,000g) for 15 min. Next, the supernatants were collected, and protein concentrations were measured using the BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA). Afterward, the proteins were denaturated in Laemmli buffer by heating at 95°C for 10 min. Protein extracts were then analyzed using standard Western blot techniques.
Image Acquisition and Analysis
Non‐confocal images of immunofluorescently labeled cells in culture were collected using a fluorescence microscope (Nikon Eclipse 80i) equipped with a 20× objective, digital camera (Media Cybernetics, Silver Spring, MD), and ImagePro Plus software (Media Cybernetics, Silver Spring, MD). Confocal images of immunofluorescently labeled in vitro cultured cells were collected using a Zeiss LSM 710NLO microscope and 20× dry objective. Images were collected at 1024 × 1024 pixel resolution as a series of z‐sections, each averaged twice per line. The obtained stack was flattened into a single image using maximum intensity projections. For all of the experiments that involved fluorescence intensity analysis, the acquisition settings were kept constant for all of the experimental variants. The fluorescence intensity of P‐S6 or P‐Akt immunostained culture neurons was measured using the native region measurement function of the Fiji software package. Neuronal somata were first freehand‐traced based on green fluorescent protein (GFP) expression. The intensity of the analyzed immunofluorescence at an appropriate emission wavelength was then measured.
Confocal images of immunolabeled brain sections were collected using a Zeiss LSM 710 NLO microscope with 10× and 20× dry objectives or 40× and 63× oil objectives, depending on the analyzed parameter. Images were acquired at a 1024 × 1024 pixel resolution as a set of z‐sections at either 4 or 1.2 µm intervals, depending on the type of analysis. For all of the experiments that involved fluorescence intensity analysis, the acquisition settings were kept constant for all of the experimental variants. For initial processing of the collected pictures, Zen 2010 software (Zeiss, Jena, Germany) was used.
Morphometric Analysis
For the morphometric analysis of in vitro cultured SVZ‐derived neurons, each z‐stack was flattened into a single image using the maximum intensity projection function of ZEN2010 software. To quantitatively describe changes in dendritic arbors, the total number of dendritic tips (TNDT) was analyzed. The analysis of TNDT of SVZ‐derived neurons was performed manually using the cell counter module of the Fiji software package.
The morphometric analysis of dendritic arbors and cell volume of neurons in vivo was performed using images that were acquired from 100‐µm thick brain slices. Three different square fields of view were taken for each slice. For dendritic arbor analysis, each image was a z‐stack of 10–20 images, depending on the neuron's location in the brain slice and slice thickness. For data analysis, only microscopic images with sufficient GFP expression and a good signal‐to‐noise ratio were included. Images of cells with incomplete dendritic arbors were excluded from the analysis. To quantitatively describe changes in dendritic arbors of neurons in vivo, neural arbors were first manually traced on a three‐dimensional reconstruction of collected images using Simple Neurite Tracer software, Fiji (Longair et al., 2011). On these traces, we analyzed TNDT, total dendrite length (TDL; sum of all dendrite lengths), and dendrite density distribution as a function of distance from the neuron cell soma (Sholl analysis; Sholl, 1953). The analyses were performed separately for apical and basal dendrites. To measure neuron cell soma volume, 100‐µm thick brain slices were used. For the analysis, three different square fields of view were taken for each slice. Each image was a z‐stack of five images. Neuron soma volume was measured using Imaris software (Bitplane, Zurich, Switzerland). Z‐stack images with visible GFP‐positive cells were uploaded to Imaris software, and the cell body volume was obtained using its native Surface mode function.
Statistical Analysis
For the in vitro culture experiments, data were collected from at least two independent primary cultures. For each experimental condition, images of cells from two coverslips were collected. The results are presented as the arithmetic mean ± standard error of the mean (SEM) and analyzed for statistically significant changes using the Kruskal–Wallis test and Dunn's post hoc test using Prism 5.03 software (GraphPad, San Diego, CA).
To analyze the results of the in vivo experiments, we used one of the following statistical tests, depending on the parameter under study: Mann–Whitney test, one‐way analysis of variance (ANOVA), or two‐way repeated‐measures ANOVA with appropriate within‐ and between‐subjects factors. For the Mann–Whitney test and ANOVA, Prism 5.03 software and SPSS 21 software (IBM) were used, respectively.
RESULTS
mTOR Activity is Necessary for Dendritogenesis of SVZ‐Derived Neurons In Vitro
mTOR plays an important role in regulating the dendritogenesis of embryonic neurons cultured in vitro. The question that arises, however, is whether mTOR is also needed for the dendritogenesis of postnatally born neurons (e.g., in the SVZ). To answer this question prior to laborious in vivo experimentation, we used primary tissue cultures of postnatal neural progenitors that were obtained from the SVZ and cultured under differentiating conditions. As shown in Supporting Information Figure 1(B), cells that were obtained from dissociated neurospheres and plated under differentiation conditions differentiated mainly into neurons (MAP2‐immunopositive cells) and astrocytes (GFAP‐immunopositive cells) after 4 days in culture. Cells that were positive for lectin and receptor interacting protein (RIP) were also present [Supporting Information Fig. 1(B)], suggesting that our preparations contained microglia and oligodendrocytes. Additional immunofluorescent staining for two independent neuronal markers (neuron‐specific β‐tubulin isotype III [Tuj1] and anti‐NeuN) confirmed that neural progenitors became neurons after 4 days in culture [Supporting Information Fig. 1(C)]. Cells that are derived from the SVZ are known to differentiate mainly into axonless, GABAergic granule neurons. MAP2‐positive cells stained negative for Tau‐1, an axonal marker (not shown).
Since our major aim was to study the development of dendritic arbors, we first established the precise timing of this process in our preparations. Depending on the neuron type and in vitro culture conditions, particular phases of development, including dendritogenesis, may occur with different timing. To this end, progenitors were cultured under defined differentiation conditions for 4 days. Each day, the cells were immunofluorescently labeled for nestin (neural progenitor marker) and MAP2 (mature neuron marker). MAP2 staining was additionally used to assess dendritic arbor morphology. As shown in Supporting Information Figure 1(D), on day 0 (day of cell plating) most of the cells that stained positive for nestin, were negative for MAP2. Over the following days, the number of nestin‐positive neural progenitors progressively declined and disappeared completely by the third day. Conversely, the number of MAP2‐positive cells began to increase on day 1. With regard to the development of proper morphology, progenitors began to grow neurites just a few hours after plating. On day 1, structures that resembled growth cones were clearly visible at the tip of the principal processes [Supporting Information Fig. 1(D)]. Cell protrusions progressively differentiated into dendrites, with numerous filopodia [Supporting Information Fig. 1(D)] from the second day onward. During the first 2 days in vitro, dendrites grew mainly in length. From the third day onward, the cells began to form secondary dendrites and develop more complex dendritic arbors, which were established by day 4 [Supporting Information Fig. 1(D)].
Having established the time course of dendritogenesis of SVZ‐derived neurons, we proceeded to study the role of mTOR during this process. We began the analysis using pharmacological inhibitors of mTOR. Two inhibitors of mTOR with different mechanisms of action were used, rapamycin and Ku‐0063794. We first tested whether both inhibitors effectively inhibit mTOR in our preparations at concentrations described previously (Jaworski et al., 2005; Urbanska et al., 2012a). Neural progenitor cells that were derived from the SVZ were first differentiated into neurons for 2 days, and then mTOR inhibitors were added to the culture for the next 48 h. Afterward, the cells were immunostained for MAP2 and the ribosomal protein S6 phosphorylated on serines 235/236 (P‐S6), a positive indicator of mTORC1 activity. As a positive control, cells were treated with BDNF (50 ng/µL), a known inducer of mTORC1 activity (Takei et al., 2004). As shown in Figure 1(A,B), the P‐S6 immunofluorescence intensity significantly decreased with both mTOR inhibitors, confirming their efficacy under the tested culture conditions. BDNF slightly but nonsignificantly increased P‐S6 immunofluorescence intensity compared with the control. Next, we tested the effects of the inhibitors on dendritic arborization under basal culture conditions adding them as described above. Additionally, we asked question whether BDNF, which was able to activate mTOR, can induce mTOR‐dependent increase in dendritic tree arborization of SVZ‐derived neurons. Although there were no previous reports on positive effects of BDNF on dendritogenesis of SVZ‐derived neurons cultured in vitro we decided to use this trophic factor because it has been shown previously in vivo to be critical for proper dendritic arbor development of different classes of neurons, including those present in the OB (Berghuis et al., 2006). In the case of BDNF treatment, the trophic factor was added with DMSO (as a control) or the inhibitor after 2 days in culture for another 48 h. Next, the cells were immunofluorescently labeled for MAP2, and the number of MAP2‐positive processes was counted [Fig. 1(C,D)]. Both rapamycin and Ku‐0063794 significantly reduced the number of dendrites, as previously described for embryonic hippocampal neurons cultured in vitro (Jaworski et al., 2005; Kumar et al., 2005; Urbanska et al., 2012a). The 2‐day treatment with BDNF caused a robust increase in the number of MAP2‐positive protrusions. In contrast, the application of either mTOR inhibitor significantly diminished the pro‐growth effect of BDNF [Fig. 1(C,D)]. Thus, we concluded that mTOR activity is needed for the proper arborization of SVZ‐derived neurons cultured in vitro, both under basal conditions and upon BDNF treatment.
mTORC1 and mTORC2 are Needed for Dendritogenesis of SVZ‐Derived Neurons In Vitro
The data presented above point to an important role for BDNF and mTOR in the dendritic arbor development of postnatally born SVZ‐derived neurons. However, both long‐term rapamycin treatment (>4 h; Urbanska et al., 2012a) and Ku‐006379 (allosteric inhibitor) blocked the activity of both mTOR complexes. Thus, our observations did not indicate which mTOR complex is involved in the regulation of SVZ‐derived neuron dendritogenesis upon BDNF treatment. To dissect the differential contributions of mTORC1 and mTORC2 to this process, we took advantage of plasmids that encoded shRNAs against Raptor and Rictor, which have been thoroughly characterized in our laboratory previously on hippocampal neurons (Urbanska et al., 2012a). First however, we decided to test if those shRNAs affect signaling downstream mTORC1 and mTORC2 in the SVZ‐derived preparations the same way as in hippocampal ones. The plasmids that encoded specific shRNAs against Raptor and Rictor were transfected to SVZ‐derived neurons individually or as a mix 1 day after plating. Transfection of the plasmid that encoded mTOR shRNA served as a positive control, and the empty vector pSUPER was used as a negative control, which was previously described to be indistinguishable from several tested scrambled shRNAs (Urbanska et al., 2012a). In all of the variants, a plasmid that encoded GFP was added to the transfection mixtures to help identify transfected cells. The next day after transfection (day 2 after plating), BDNF was added to the culture medium to enhance activation of both mTORCs. Two days later (day 4 after plating), the cells were fixed and immunofluorescently labeled for P‐S6 and P‐Akt (serine 473), which served as indicators of mTORC1 and mTORC2 activity, respectively. Knockdown of Raptor affected level of P‐S6 but not P‐Akt, whereas knockdown of Rictor affected both P‐S6 and P‐Akt [Fig. 2(A–D)], as previously shown for hippocampal neurons (Urbanska et al., 2012a). mTOR shRNA as well as mix of Raptor and Rictor shRNAs decreased levels of both P‐S6 and P‐Akt. This suggests that in SVZ‐derived neurons similar to hippocampal ones, mTORC2 may act upstream of mTORC1. We also noted that mTOR shRNA decreased P‐S6 and P‐Akt IF levels less efficiently than shRaptor and shRictor, respectively. Fact that it was also less potent than mix of Raptor and Rictor shRNAs indicates that shmTOR had relatively mild efficiency. This is consistent with results obtained previously with use of this mTOR shRNA (Jaworski et al., 2005).
In the next series of experiments, SVZ‐derived neurons were transfected and treated with BDNF, as described above, but instead of signaling pathway analysis we counted the total number of dendritic tips (TNDT) of transfected neurons based on GFP fluorescence [Fig. 2(E,F)]. As shown in Figure 2, transfection with mTOR shRNA reduced TNDT compared with pSUPER‐transfected cells. The knockdown of either Raptor or Rictor or both also effectively blocked dendritic arborization [Fig. 2(E,F)]. In contrast, overexpression of scrambled sequences of shRNAs against Raptor (scrRaptor#1) or Rictor (scrRictor#2) did not affect BDNF‐induced dendritic arborization [Fig. 2(E,F)]. Thus, we concluded that in the case of cultured in vitro postnatally born neurons derived from SVZ, similarly to embryonic hippocampal neurons, both mTORCs are indispensible for extracellular factor‐induced dendritogenesis. This result encouraged us to further study this process in vivo.
mTOR Knockout Reduces the Complexity of Dendritic Arbors of Postnatally Born OB Granular Neurons
To determine the involvement of mTOR in the dendritogenesis of SVZ‐born neurons that mature in the OB, we took advantage of the postnatal in vivo electroporation of neural stem cells that reside in the SVZ. We focused on the SVZ‐RMS‐OB system because the role of mTOR in the physiological dendritic arbor development of postnatally born OB neurons has not been previously demonstrated. Previous studies suggested that the hyperactivation of mTOR in this cells caused by knockout of TSC1 leads to the aberrant dendritic growth of OB neurons (Feliciano et al., 2012). To selectively decrease mTOR activity in postnatally born neurons, neonatal (P1‐P2) Mtorfl/fl mice were co‐electroporated with a mixture of pCAG‐Cre, pCALNL‐DsRed and pCAG‐GFP (experimental variant), or pCAG (empty vector), pCALNL‐DsRed and pCAG‐GFP (control variant) [Fig. 3(A)]. pCAG‐Cre encodes constitutively expressed Cre recombinase, and pCALNL‐DsRed causes DsRed expression only in cells that express Cre, serving as a positive marker of recombination [Fig. 3(B)]. Electroporated Mtorfl/fl mice were sacrificed on either 5 or 14 days post‐electroporation (dpe), and cryostat brain sections were immunostained for GFP and P‐S6. In control animals, electroporated cells in the RMS (5 dpe) and OB (14 dpe) stained positively for GFP [Fig. 3(C–E)] and were negative for DsRed expression. In animals from experimental variant (Cre) the expression of inducible DsRed was already visible in GFP‐positive RMS neuroblasts at 5 dpe [Fig. 3(D)]. At 14 dpe, approximately 40% of GFP‐positive neurons in the mouse OBs was DsRed‐positive [Fig. 3(E)]. The analysis of mTOR activity using P‐S6 immunofluorescence revealed lower level of P‐S6 in GFP‐ and DsRed‐positive neuroblasts at 5 dpe and OB neurons at 14 dpe compared with controls [Fig. 3(D,E)]. In addition to examining P‐S6 immunofluorescence, the cell volume of electroporated cells was measured as a known morphological hallmark of mTOR pathway inhibition in developing neurons. As shown in Figure 3(G), the cell volume of Cre‐expressing DsRed‐positive OB neurons decreased by 30% compared with controls. Unexpectedly, migrating neuroblasts lacking mTOR presented significant increase of the volume of cell soma [Fig. 3(F)]. This cell body swelling may represent a first hallmark of a cell death, as in some animals sacrificed at 14 dpe we observed less GFP‐positive cells in the OB of Cre‐electroporated mice than in control plasmid electroporated ones (not shown).
Figure 3.
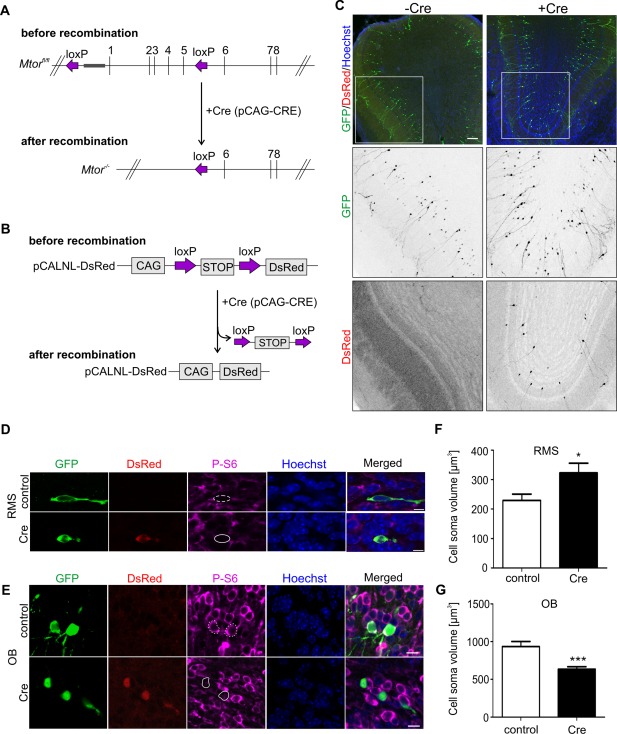
Overexpression of Cre recombinase in neural stem cells in Mtorfl/fl mice leads to mTORC1 inhibition in RMS neuroblasts and mature neurons in the OB. (A) Diagram illustrating genomic changes in cells of Mtorfl/fl mice electroporated with pCAG‐CRE encoding Cre recombinase. (B) principle of Cre‐dependent expression of DsRed in electroporated cells. (C) Representative confocal images of OB in Mtorfl/fl mice electroporated with pCAG‐GFP, pCALNL‐DsRed, and pCAG (control) or pCAG‐GFP, pCALNL‐DsRed, and pCAG‐Cre (experimental variant) on P1 and sacrificed at 14 dpe. Olfactory bulb sections were immunostained for GFP (green). DsRed was visualized solely by its fluorescence (red). Brain sections were additionally counterstained with Hoechst 33258 (Hoechst; blue). Scale bar = 100 µm. (D) Representative confocal images of migrating neuroblasts. Mice were electroporated as in C at the age of P1 and sacrificed at 5 dpe. Brain sections were immunostained for GFP (green) and the phosphorylated form of the ribosomal protein S6 (P‐S6, Ser 240/244) (magenta). DsRed was visualized solely by its fluorescence (red). White lines outline cell bodies that were positive for GFP/DsRed or GFP. Brain sections were additionally counterstained with Hoechst 33258 (Hoechst; blue) to visualize nuclei. Scale bar = 5 µm. (E) Representative confocal images of neurons in the olfactory bulb. Mice were electroporated as described in C and sacrificed at 14 dpe. Olfactory bulb sections were immunostained for GFP (green) and P‐S6 (magenta). DsRed was visualized solely by its fluorescence (red). All of the OB sections were additionally counterstained with Hoechst 33258 (Hoechst; blue) to visualize nuclei. White lines outline cell bodies that were positive for GFP/DsRed or GFP. Scale bar = 5 µm. (F) Quantitative analysis of neuroblast cell soma volume in mice electroporated as in C, presented as a mean value ± SEM. Number of animals (N) = 2 per group. Number of analyzed cells (n) = 20 (control), 20 (Cre). *p < 0.05 (Mann–Whitney test). (G) Quantitative analysis of OB neuron cell soma volume in mice electroporated as in C, presented as a mean value ± SEM. Number of animals (N) = 2 per group. Number of analyzed cells (n) = 20 (control), 20 (Cre). ***p < 0.001 (Mann–Whitney test).
Next, we repeated the experiment described above but this time focused on analyzing the effect of mTOR knockout on dendritic trees of OB neurons 14 dpe using three‐dimensional reconstructions of GFP‐positive cells [Fig. 4(A,B)]. Newly born neurons in the OB bear several short basal dendrites and one long primary apical dendrite, which next branches to distal dendrites of a higher order [Fig. 4(B)]. To quantitatively describe the morphology of the dendritic trees, we calculated the TNDT, the total dendritic length (TDL) and performed a Sholl analysis. As shown in Figure 4(C,E), neurons with active Cre recombinase exhibited a significant reduction of TNDT of both basal and apical dendrites, as compared with control. Moreover, the analysis of both basal and apical dendrites showed a decrease in TDL [Fig. 4(D,F)]. Thus, mTOR knockout significantly decreased both the number and total length of dendrites and shrunk both apical and basal dendritic arbors.
Figure 4.

mTOR knockout affects postnatally born OB neuron dendritic arbors. (A) Diagram illustrating the design of the experiment. Mice were electroporated with pCAG‐GFP, pCALNL‐DsRed, and pCAG (control) or pCAG‐GFP, pCALNL‐DsRed, and pCAG‐Cre (experimental variant) on P1‐P2 and sacrificed after 14 dpe. (B) Representative three‐dimensional reconstructions of GFP‐positive neurons after mTOR knockout vs. control in the olfactory bulb. Scale bar = 50 µm. (C, D) Quantification of the total number of dendritic tips (TNDT, C) and total dendritic length (TDL, D) of basal dendrites after mTOR knockout. (E, F) Quantification of the total number of dendritic tips (TNDT, E) and total dendritic length (TDL, F) of apical dendrites after mTOR knockout. The results are presented as a mean value ± SEM. Number of individual experiments = 4. Number of animals (N) = 11 (control), 10 (Cre). Number of analyzed cells (n) = 89 (control), 49 (Cre). ***p < 0.001 (Mann–Whitney test). (G, H) Sholl analysis of basal and apical dendrites in OB neurons. Significance for particular radii was assessed using one‐way ANOVA. *p < 0.05, **p < 0.01, ***p < 0.001. For all graphs: number of individual experiments = 4; number of animals (N) = 11 (control), 10 (Cre); number of cells (n) = 89 (control), 49 (Cre). [Color figure can be viewed at wileyonlinelibrary.com.]
Changes in dendritic arbor complexity were further assessed using Sholl analysis separately for basal [Fig. 4(G)] and apical [Fig. 4(H)] dendrites. The analyses for both basal and apical parts of dendritic trees showed the same significant radius effect (p < 0.001, two‐way repeated‐measures ANOVA with interaction), a significant reduction of the number of crossings due to the mTOR knockout effect (p < 0.001; two‐way repeated‐measures ANOVA with interaction), and a significant interaction between radius and the mTOR knockout effect (p < 0.001). Thus we concluded that in both groups, the number of crossings of basal and apical dendrites evolves with the radius. In addition, the leftward and downward shift in the knockout (Cre) profile plot is statistically significant, as compared with control. Moreover, the comparison (one‐way ANOVA) of corresponding radii showed a statistically significant difference in basal radii in the 30–90 µm range [Fig. 4(G)] and apical radii in the 20–150 µm range [Fig. 4(H)] from the center of the cell soma. This data shows that knockout of mTOR severely impairs complexity of both basal and apical dendritic arbors.
mTOR is Needed for the Stabilization of Basal Dendrites of Postnatally Born OB Granular Neurons
In the experiments described above, mTOR was inhibited in neuroblasts that migrated through the RMS and consequently also in the early stages of newly born neurons' development in the OB. To evaluate the role of mTOR during later stages of OB neuron dendritic tree formation in vivo, we used systemic administration of the mTORC1 inhibitor rapamycin in wildtype mice 10 days after electroporation in an effort to target neurons after they reached the OB and began dendritogenesis (Mizrahi, 2007). We focus on mTORC1 for two reasons. First, intraperitoneal injections of rapamycin have been previously extensively used to study mTOR in the brain. Second, our results from different types of in vitro cultured neurons suggested that mTORC1 may act downstream of mTORC2 (Fig. 2 and Urbanska et al., 2012a).
Our previous results suggested that chronic rapamycin treatment (4–6 weeks) in wildtype rodents can significantly affect the morphology of the brain through the induction of hydrocephaly (Macias et al., 2013). Thus, we first searched for the minimal dosage of the drug that is required for mTOR inhibition in the OB in neonates. Mouse neonates received either two or three IP injections of rapamycin (1 mg/kg rapamycin beginning on P8 or P10 at 48 h intervals) or ethanol in vehicle solution, and protein lysates were prepared from P14 mouse OBs to check for the levels of P‐S6 and P‐Akt. Western blot analysis of obtained protein lysates revealed a decrease in the levels of P‐S6 in the OB of rapamycin‐treated mice compared with vehicle‐injected control animals. Either two or three doses of rapamycin effectively decreased P‐S6 to a similar extent [Fig. 5(A)] without causing gross changes in brain morphology (data not shown). We also verified if such multiple rapamycin injections affect mTORC2 activity in OB in vivo, since it has been shown that prolonged exposure of in vitro cultured neurons to rapamycin may affect both complexes (Urbanska et al., 2012a). As shown in Figure 5(A), neither two nor three doses or rapamycin significantly decreased phosphorylation of Ser473 of Akt. In fact, slight increase in phosphorylation of P‐Akt was observed that is consistent with previous observation that rapamycin may block negative feedback loops from mTOR to the receptor tyrosine kinases. Therefore, two IP injections of the inhibitor were used to block mTORC1 in subsequent experiments focusing on neuronal morphology.
Figure 5.

Rapamycin treatment affects basal dendrites of postnatally born OB neurons. (A) Western blot analysis of phosphorylated ribosomal protein S6 (P‐S6, Ser235/236), total S6, phosphorylated Akt (P‐Akt, Ser473) and total Akt levels in protein lysates obtained from olfactory bulbs from animals treated with rapamycin (rapa) or vehicle solution (control). Mouse pups were treated with two or three doses of 1 mg/kg rapamycin from 8 (3 doses) or 10 days (2 doses) after birth. Doses were administered at 48 h intervals. Tubulin (Tub) was used as a loading control. (B) Quantitative analysis of cell soma volume of GFP‐positive neuronal cell body in olfactory bulb (OB). Mice were electroporated with a GFP‐encoding plasmid (pCx‐EGFP‐N1) on P1‐P2, treated with 2 doses of rapamycin or vehicle as described in A, and sacrificed at 14 dpe. All OB slices were immunostained for GFP. Neuron cell somas were three‐dimensionally reconstructed. The results are presented as a mean value ± SEM. Number of animals (N) = 5 (control), 5 (rapa). Number of analyzed cells (n) = 47 (control), 42 (rapa). ns, not significant (Mann‐Whitney test). (C) Representative three‐dimensional reconstructions of GFP‐positive neurons in olfactory bulb after IP injection of rapamycin vs. control. Scale bar = 50 µm. Mice were electroporated with pCx‐EGFP‐N1 on P1‐P2, intraperitoneally (IP) injected with two doses of rapamycin starting 10 dpe, and sacrificed at 14 dpe. (D, E) Quantification of the total number of dendritic tips (TNDT, D) and total dendritic length (TDL, E) of basal dendrites after IP rapamycin injections. (F, G) Quantification of the total number of dendritic tips (TNDT, F) and total dendritic length (TDL, G) of apical dendrites after IP rapamycin injections. The results are presented as a mean value ± SEM. For all graphs: number of individual experiments = 2; number of animals (N) = 5 (control), 5 (rapa); number of analyzed cells (n) = 52 (control), 57 (rapa). **p ≤ 0.01; ns, not significant (Mann–Whitney test). (H, I) Sholl analysis of basal and apical dendrites in OB neurons. Significance for particular radii was assessed using one‐way ANOVA. For all graphs: number of individual experiments = 2; number of animals (N) = 5 (control), 5 (rapamycin); number of analyzed cells (n): 52 (control), 57 (rapamycin). *p ≤ 0.05. [Color figure can be viewed at wileyonlinelibrary.com.]
Next, P1–P2 wildtype mice were electroporated with a GFP‐encoding plasmid (pCx‐EGFP‐N1) and treated with a total of two IP injections of rapamycin at 10 and 12 dpe. The animals were sacrificed at 14 dpe, and OB sections were immunostained for GFP followed by soma volume measurements and dendritic arbor analysis. As shown in Figure 5(B), no significant difference was found in soma volume between rapamycin and control mice. The analysis of dendritic trees, performed with three‐dimensional reconstructions of GFP‐positive neurons in the OB [Fig. 5(C)], revealed that in vivo rapamycin treatment altered the TNDT and TDL of neurons in the OB. But in contrast to Cre‐mediated knockout, rapamycin treatment specifically decreased the TNDT and TDL of basal dendrites [Fig. 5(D,E)]. The TNDT and TDL for apical dendrites did not differ significantly [Fig. 5(F,G)].
Finally, to more precisely analyze the pattern of dendritic arborization and assess the coverage of these dendritic fields, Sholl analysis was performed. The number of crossings for the arborescence of a single neuron at various radial distances from the cell soma was tested. A two‐way repeated‐measure ANOVA with interaction was used to analyze the data for basal and apical arbors separately. The analyses of basal dendrites [Fig. 5(H)] showed a significant radius effect (p < 0.001), no significant reduction of the number of crossings due to mTOR inhibition by rapamycin (p = 0.0828), and no significant interaction between radius and the rapamycin effect (p < 0.369). Analysis of the number of crossings, determined by the Sholl analysis, revealed small but nonsignificant effects on basal branching. Comparisons of corresponding radii showed significant differences only for radii that were 70 µm from the cell center.
For apical dendrites [Fig. 5(I)], the analysis showed a significant radius effect (p < 0.001), no significant reduction of the number of crossings due to mTOR inhibition (p = 0.198), and no significant interaction between radius and the rapamycin effect (p = 0.933). For apical dendrites, the number of crossings changed with the radius, but no changes were observed in the complexity or the size of apical dendrites in rapamycin‐treated animals, as compared with controls. In summary, rapamycin treatment led to a small but significant decrease in the TNDT and TDL of basal dendrites of neurons between rapamycin‐treated and control animals. The Sholl analysis confirmed a trend toward basal dendritic arbor simplification, but this effect did not reach statistical significance.
DISCUSSION
mTOR complexes were previously shown to modulate the dendritic arbor growth of different types of embryonic neurons cultured in vitro (for review, see Jaworski and Sheng, 2006; Swiech et al., 2008). However, its involvement in in vivo dendritic arbor development was studied almost exclusively under conditions of hyperactivation of the mTORC1 signaling pathway (Kwon et al., 2006; Chow et al., 2009; Kim et al., 2009; Feliciano et al., 2012; Zhou et al., 2013). Using in vitro cultures of differentiated SVZ‐derived neural progenitors and in vivo postnatal electroporation of cells in the SVZ, the present study found that mTOR is indeed important for the dendritogenesis of postnatally born neurons both in vitro and in vivo. Furthermore, the in vitro model revealed that both mTOR complexes are important for postnataly‐born neuron development. In the in vivo studies, we used two independent approaches to silence mTOR during neurogenesis, including the differentiation of postnatally born OB neurons. Each of these approaches yielded slightly different results, potentially revealing differences between mTOR complexes during dendritogenesis and/or the larger importance of mTOR signaling for basal dendritic arbor development than for apical dendritic arbor development and/or stability.
mTORCs Control the Formation of Dendritic Arbors of Postnatally Born Neurons Cultured In Vitro
In the in vitro experiments, we showed that the inhibition of mTOR using rapamycin, both under standard culturing conditions and upon cell stimulation with BDNF, prevented dendritic growth. This observation, although proving the importance of mTOR for the proper dendritic arborization of neurons born postnatally, is discordant with the observations of Khodosevich and Monyer (2010). In a large high‐throughput screening of signaling pathways that are involved in the neurite outgrowth of postnatally born neurons, these authors suggested that rapamycin does not affect this process. However, additional evidence presented herein further supports an important role for mTOR in the dendritic arbor formation of postnatally born neurons. First, treatment of postnatally born neurons with Ku‐0063794, a different mTOR inhibitor, also led to simplification of dendritic arbors of the analyzed neurons. Second, BDNF, a known inducer of mTOR, induced the dendritic growth of SVZ‐born neurons. Third, a validated shRNA against mTOR effectively blocked BDNF‐induced dendritogenesis of these cells. Fourth, the removal of either Rictor or Raptor yielded comparable results. The major technical difference between the present study and Khodosevich and Monyer (2010) is the doses of rapamycin used in both studies. Khodosevich and Monyer (2010) used rapamycin at a concentration of 50 pM. In the present study, we used 20 nM rapamycin, which is one of the lowest doses that is efficiently used in most long‐term studies on dendritic arbor development of neurons cultured in vitro (Jaworski et al., 2005; Kumar et al., 2005). Therefore, we believe that the dose of rapamycin that was used by Khodosevich and Monyer (2010) was insufficient.
Our in vitro studies also provided information concerning the potential mechanisms downstream of mTOR that may be involved in the dendritic arbor development of postnatally born neurons in response to BDNF. We found that both mTOR complexes (mTORC1 and mTORC2) are important for this process and mTORC2 likely acts upstream mTORC1. This observation is consistent with Urbanska et al. (2012a), who showed that both complexes are needed for basal and induced dendritic arborization in embryonic hippocampal neurons. However, further experiments are needed to determine the extent to which the development of SVZ‐derived neurons mimics the development of embryonic hippocampal neurons with regard to mTORC1 downstream effectors (e.g., Cytoplasmic Linker Protein‐170, Swiech et al., 2011).
mTOR Controls the Morphology of Postnatally Born Granule Neurons In Vivo
In the next step of the present study, we evaluated whether mTOR controls the morphology of dendritic arbors of postnatally born OB neurons in vivo. We used electroporation of NSCs in the SVZ in vivo and focused on postnatally born granule cells (GCs) because they form a relatively homogeneous group of GABAergic neurons. The results demonstrated the need for the presence of mTOR or its activity for the proper development of dendritic arbors of postnatally born GCs in the OB. Which regulators of dendritogenesis of postnatally born neurons act upstream mTOR? At least three such factors, namely VEGF, BDNF and GABA have been previously shown to activate mTOR in different types of neurons (Takei et al., 2004; Kim et al., 2008, 2009). However, thus far only GABA, which acts as an excitatory neurotransmitter at early steps of neurogenesis, was proven to indirectly, via increase in calcium influx, activate mTOR during dendritogenesis of adult born neurons in dentate gyrus of hippocampus in vivo (Kim et al., 2009). Whether similar mechanism exist in OB neurons needs to be established, but Gascon et al. (2006) showed that GABA acts as an excitatory neurotransmitter also on developing postnatally born OB neurons to stabilize newly formed dendrites. Another important regulator of mTOR‐dependent dendritogenesis in hippocampal and cortical neurons is reelin (Jossin and Goffinet, 2007; Matsuki et al., 2008). Since reelin is also expressed in OB postnatally (Hellwig et al., 2012) one could speculate that reelin controls dendritogenesis of postnatally born neurons in mTOR‐dependent fashion. However, thus far, there is no evidence that reelin is needed for dendritic arbor development in postnatally born OB neurons.
mTOR Knockout and Rapamycin Treatment Differentially Affect the Morphology of Postnatally Born Granule Neurons In Vivo
The data presented herein suggest that although mTOR is important for the growth of both basal and apical dendrites during the initial steps of dendritogenesis, it may be selectively involved in the growth and/or maintenance of basal dendrites at later stages of this process. In the present study, mTOR activity was modified in postnatally born OB neurons in two different ways: (i) electroporation of NSCs in the SVZ of Mtorfl/fl mice with constitutively active Cre and (ii) IP injection of rapamycin in wildtype mice. Methodological limitations did not allow us to directly prove the effectiveness of mTOR knockout in vivo. However, we provided evidence at the functional level, showing that both approaches decreased the level of the phosphorylated ribosomal S6 protein, a commonly used marker of mTORC1 activity (Hartman et al., 2013; Lafourcade et al., 2013; Macias et al., 2013). However, although both approaches used herein relatively efficiently decreased the activity of mTOR in the brain in vivo, they differentially affected dendritic morphology. Cre electroporation to NSCs in the SVZ of Mtorfl/fl mice led to the simplification of both apical and basal dendritic arbors of OB neurons, whereas rapamycin affected only basal dendritic arbors. This difference may be related to the scope and/or timing of mTOR inhibition as well as population of cells, which experience mTOR inhibition.
Rapamycin is a widely known inhibitor of mTOR, but it does not equally affect all mTOR activities (Dowling et al., 2010). Short‐term rapamycin treatment blocks, mTORC1 activity for some but not all targets and does not affect mTORC2. Prolonged treatment of in vitro cultured cells may affect a broader array of mTORC1 effectors, but some may remain insensitive to rapamycin treatment, depending on cell type (Choo et al., 2008). Prolonged rapamycin treatment of neurons cultured in vitro was shown to decrease mTORC2 activity (Urbanska et al., 2012a). These phenomena have not been comprehensively studied in the mammalian brain in vivo. Data presented in Figure 5(A) show, that our protocol of rapamycin application does not inhibit mTORC2 activity in OB. In contrast, Cre‐driven mTOR knockout results in the loss of both mTORC1 and mTORC2 activity. Thus, differences in the mode of action of our analytical tools (i.e., inhibition of a different set of mTOR targets) could explain the different phenotypes of apical dendrites of newly born OB neurons in response to Cre knockout and rapamycin treatment. Although we showed that mTORC1 acts downstream mTORC2, it is not a canonical effector of mTORC2. In fact, mTORC2 is widely known for regulation of small GTPases of Rho family (Jacinto et al., 2004), which are important regulators of actin dynamics and dendritic growth (Urbanska et al., 2008, 2012b). Thus, it is possible that canonical mTORC2 effectors are important for apical dendrite growth while mTORC1 is critical for basal dendritic arbor development.
Another explanation of the observed differences, not mutually exclusive with the above one, may involve the timing of mTOR inhibition/knockout. Based on our results, we can conclude that Cre electroporation of NSCs in the SVZ in Mtorfl/fl mice led to mTOR knockout by 5 dpe (i.e., before neuroblasts enter the OB at ∼9–10 dpe and start to grow dendrites). Rapamycin was administered at 10 dpe (i.e., after the majority of neuroblasts enter the OB and begin intensive dendritic arborization). Thus, using different means of mTOR inhibition, we targeted early (Cre‐driven knockout) and late dendritogenesis. Consequently, our results may be explained by the differential sensitivity of basal and apical dendrites to a lack of mTOR (or more specifically mTORC1) at later stages of development (i.e., during intensive remodeling and the stabilization of dendritic arbors between day 10 and 14) but not at earlier stages (before day 10). This possibility could be tested in the future with use of inducible Cre‐driven knockouts.
The apical and basal parts of dendritic trees are functionally specialized and differentially regulated (Cubelos et al., 2015). Little is known about the mechanisms that selectively maintain basal dendrites. One example is a study that was performed on Epac2 (exchange protein directly activated by cAMP 2), which helps regulate Rap, a Ras‐like small GTPase, that is highly enriched in the adult brain and dendrites (Srivastava et al., 2012). Epac2 knockdown robustly and selectively impaired basal dendrite maintenance in cortical pyramidal neurons in vivo and in culture (Srivastava et al., 2012). Another example was recently published by Wu et al. (2015) who showed that stability of basal dendrites of granular hippocampal neurons relies specifically on extrinsic cues. In this context it is worth noting that rapamycin, but not selective mTOR KO, blocks mTORC1 activity not exclusively in postnatally born neurons reaching OB but also in surrounding cells, which serve as a source of extrinsic cues. Thus, mTORCs may participate in such specific regulation of basal or apical maintenance.
Alternatively, one could speculate that apical and basal dendritic arbors become stabilized at different time points. At the time point at which we reached sufficient mTOR inhibition, apical dendrites were relatively stable, whereas basal dendrites were still undergoing intensive remodeling. Mizrahi (2007) showed that apical tufts of adult‐born GCs were relatively stable between 10 and 14 days after birth. However, his observations were restricted only to apical dendrites, and the level of basal dendrite stability remains unknown. The dynamics of the dendritic arborization of neonatally born GCs also remains to be determined. Therefore, further investigations are needed to fully explain the differences in sensitivity between basal and apical dendrites of postnatally born GCs to mTOR inhibition near the end of dendritogenesis.
Supporting information
Supporting Information Figure 1.
Supporting Information Figure 2.
ACKNOWLEDGMENTS
We thank Aimee K. Ryan, Krishnendu Ganguly, Leszek Kaczmarek, Angelique Bordey, David Feliciano, and Harald Cremer for sharing with us in vivo electroporation expertise and equipment and Harald Cremer for the pCx‐EGFP‐N1 plasmid. We are very grateful to Matylda Macias for sharing her knowledge and help with the experiments, Alina Zielinska and Anna Sylvestre for technical assistance. JJ is a recipient of a “Mistrz” Professorial subsidy of the Foundation for Polish Science.
Agnieszka Skalecka and Ewa Liszewska have contributed equally to this work.
REFERENCES
- Anderl S, Freeland M, Kwiatkowski DJ, Goto J. 2011. Therapeutic value of prenatal rapamycin treatment in a mouse brain model of tuberous sclerosis complex. Hum Mol Genet 20:4597–4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghuis P, Agerman K, Dobszay MB, Minichiello L, Harkany T, Ernfors P. 2006. Brain‐derived neurotrophic factor selectively regulates dendritogenesis of parvalbumin‐containing interneurons in the main olfactory bulb through the PLCgamma pathway. J Neurobiol 66:1437–1451. [DOI] [PubMed] [Google Scholar]
- Boutin C, Diestel S, Desoeuvre A, Tiveron MC, Cremer H. 2008. Efficient in vivo electroporation of the postnatal rodent forebrain. PloS One 3:e1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550–553. [DOI] [PubMed] [Google Scholar]
- Carlén M, Cassidy RM, Brismar H, Smith GA, Enquist LW, Frisén J. 2002. Functional integration of adult‐born neurons. Curr Biol 12:606–608. [DOI] [PubMed] [Google Scholar]
- Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. 2008. Rapamycin differentially inhibits S6Ks and 4E‐BP1 to mediate cell‐type‐specific repression of mRNA translation. Proc Natl Acad Sci 105:17414–17419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow DK, Groszer M, Pribadi M, Machniki M, Carmichael ST, Liu X, Trachtenberg JT. 2009. Laminar and compartmental regulation of dendritic growth in mature cortex. Nat Neurosci 12:116–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubelos B, Briz CG, Esteban‐Ortega GM, Nieto M. 2015. Cux1 and Cux2 selectively target basal and apical dendritic compartments of layer II‐III cortical neurons. Dev Neurobiol 75:163–172. [DOI] [PubMed] [Google Scholar]
- Dowling RJO, Topisirovic I, Fonseca BD, Sonenberg N. 2010. Dissecting the role of mTOR: Lessons from mTOR inhibitors. Biochim Biophys Acta 1804:433–439. [DOI] [PubMed] [Google Scholar]
- Feliciano DM, Quon JL, Su T, Taylor MM, Bordey A. 2012. Postnatal neurogenesis generates heterotopias, olfactory micronodules and cortical infiltration following single‐cell Tsc1 deletion. Hum Mol Genet 21:799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff YG, Mueller M, Dann SG, Svoboda P, Sticker M, Spetz J‐F, Um SH, et al. 2004. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Mol Cell Biol 24:9508–9516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascon E, Dayer AG, Sauvain M‐O, Potter G, Jenny B, De Roo M, Zgraggen E, et al. 2006. GABA regulates dendritic growth by stabilizing lamellipodia in newly generated interneurons of the olfactory bulb. J Neurosci 26:12956–12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giachino C, De Marchis S, Giampietro C, Parlato R, Perroteau I, Schütz G, Fasolo A, et al. 2005. cAMP response element‐binding protein regulates differentiation and survival of newborn neurons in the olfactory bulb. J Neurosci 25:10105–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giachino C, Basak O, Taylor V. 2009. Isolation and manipulation of mammalian neural stem cells in vitro. Meth Mol Biol Clifton NJ 482:143–158. [DOI] [PubMed] [Google Scholar]
- Götz M, Huttner WB. 2005. The cell biology of neurogenesis. Nat Rev Mol Cell Biol 6:777–788. [DOI] [PubMed] [Google Scholar]
- Hartman NW, Lin TV, Zhang L, Paquelet GE, Feliciano DM, Bordey A. 2013. mTORC1 targets the translational repressor 4E‐BP2, but not S6 kinase 1/2, to regulate neural stem cell self‐renewal in vivo. Cell Rep 5:433–444. [DOI] [PubMed] [Google Scholar]
- Hellwig S, Hack I, Zucker B, Brunne B, Junghans D. 2012. Reelin Together with ApoER2 Regulates Interneuron Migration in the Olfactory Bulb. PLoS ONE 7:e50646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN. 2004. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 6:1122–1128. [DOI] [PubMed] [Google Scholar]
- Jaworski J, Sheng M. 2006. The growing role of mTOR in neuronal development and plasticity. Mol Neurobiol 34:205–219. [DOI] [PubMed] [Google Scholar]
- Jaworski J, Spangler S, Seeburg DP, Hoogenraad CC, Sheng M. 2005. Control of dendritic arborization by the phosphoinositide‐3′‐kinase‐Akt‐mammalian target of rapamycin pathway. J Neurosci 25:11300–11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski J, Kapitein LC, Gouveia SM, Dortland BR, Wulf PS, Grigoriev I, Camera P, et al. 2009. Dynamic microtubules regulate dendritic spine morphology and synaptic plasticity. Neuron 61:85–100. [DOI] [PubMed] [Google Scholar]
- Jossin Y, Goffinet AM. 2007. Reelin signals through phosphatidylinositol 3‐kinase and Akt to control cortical development and through mTor to regulate dendritic growth. Mol Cell Biol 27:7113–7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodosevich K, Monyer H. 2010. Signaling involved in neurite outgrowth of postnatally born subventricular zone neurons in vitro. BMC Neurosci 11:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument‐Bromage H, Tempst P, et al. 2002. mTOR interacts with raptor to form a nutrient‐sensitive complex that signals to the cell growth machinery. Cell 110:163–175. [DOI] [PubMed] [Google Scholar]
- Kim BW, Choi M, Kim Y‐S, Park H, Lee H‐R, Yun C‐O, Kim EJ, et al. 2008. Vascular endothelial growth factor (VEGF) signaling regulates hippocampal neurons by elevation of intracellular calcium and activation of calcium/calmodulin protein kinase II and mammalian target of rapamycin. Cell Signal 20:714–725. [DOI] [PubMed] [Google Scholar]
- Kim JY, Duan X, Liu CY, Jang M‐H, Guo JU, Pow‐anpongkul N, Kang E, et al. 2009. DISC1 regulates new neuron development in the adult brain via modulation of akt‐mtor signaling through KIAA1212. Neuron 63:761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Zhang MX, Swank MW, Kunz J, Wu GY. 2005. Regulation of dendritic morphogenesis by Ras‐PI3K‐Akt‐mTOR and Ras‐MAPK signaling pathways. J Neurosci 25:11288–11299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CH, Luikart BW, Powell CM, Zhou J, Matheny SA, Zhang W, Li Y, et al. 2006. Pten regulates neuronal arborization and social interaction in mice. Neuron 50:377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafourcade CA, Lin TV, Feliciano DM, Zhang L, Hsieh LS, Bordey A. 2013. Rheb activation in subventricular zone progenitors leads to heterotopia, ectopic neuronal differentiation, and rapamycin‐sensitive olfactory micronodules and dendrite hypertrophy of newborn neurons. J Neurosci 33:2419–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licht T, Eavri R, Goshen I, Shlomai Y, Mizrahi A, Keshet E. 2010. VEGF is required for dendritogenesis of newly born olfactory bulb interneurons. Dev Camb Engl 137:261–271. [DOI] [PubMed] [Google Scholar]
- Lledo P‐M, Merkle FT, Alvarez‐Buylla A. 2008. Origin and function of olfactory bulb interneuron diversity. Trends Neurosci 31:392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longair MH, Baker DA, Armstrong JD. 2011. Simple Neurite Tracer: Open source software for reconstruction, visualization and analysis of neuronal processes. Bioinform Oxf Engl 27:2453–2454. [DOI] [PubMed] [Google Scholar]
- Macias M, Blazejczyk M, Kazmierska P, Caban B, Skalecka A, Tarkowski B, Rodo A, et al. 2013. Spatiotemporal characterization of mTOR kinase activity following kainic acid induced status epilepticus and analysis of rat brain response to chronic rapamycin treatment. PLoS One 8:e64455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik AR, Urbanska M, Macias M, Skalecka A, Jaworski J. 2013. Beyond control of protein translation: What we have learned about the non‐canonical regulation and function of mammalian target of rapamycin (mTOR). Biochim Biophys Acta 1834:1434–1448. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. 2004. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A 101:16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. 2007. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci U S A 104:1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuki T, Pramatarova A, Howell BW. 2008. Reduction of Crk and CrkL expression blocks reelin‐induced dendritogenesis. J Cell Sci 121:1869–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister AK. 2000. Cellular and molecular mechanisms of dendrite growth. Cereb Cortex 10:963–973. [DOI] [PubMed] [Google Scholar]
- Ming G, Song H. 2011. Adult neurogenesis in the mammalian brain: Significant answers and significant questions. Neuron 70:687–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrahi A. 2007. Dendritic development and plasticity of adult‐born neurons in the mouse olfactory bulb. Nat Neurosci 10:444–452. [DOI] [PubMed] [Google Scholar]
- Nissant A, Bardy C, Katagiri H, Murray K, Lledo P‐M. 2009. Adult neurogenesis promotes synaptic plasticity in the olfactory bulb. Nat Neurosci 12:728–730. [DOI] [PubMed] [Google Scholar]
- Paridaen JTML, Huttner WB. 2014. Neurogenesis during development of the vertebrate central nervous system. EMBO Rep 15:351–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petreanu L, Alvarez‐Buylla A. 2002. Maturation and death of adult‐born olfactory bulb granule neurons: Role of olfaction. J Neurosci 22:6106–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument‐Bromage H, Tempst P, et al. 2004. Rictor, a novel binding partner of mTOR, defines a rapamycin‐insensitive and raptor‐independent pathway that regulates the cytoskeleton. Curr Biol 14:1296–1302. [DOI] [PubMed] [Google Scholar]
- Sholl DA. 1953. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat 87:387–406. [PMC free article] [PubMed] [Google Scholar]
- Srivastava DP, Woolfrey KM, Jones KA, Anderson CT, Smith KR, Russell TA, Lee H, et al. 2012. An autism‐associated variant of Epac2 reveals a role for Ras/Epac2 signaling in controlling basal dendrite maintenance in mice. PLoS Biol 10:e1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiech L, Perycz M, Malik A, Jaworski J. 2008. Role of mTOR in physiology and pathology of the nervous system. Biochim Biophys Acta 1784:116–132. [DOI] [PubMed] [Google Scholar]
- Swiech L, Blazejczyk M, Urbanska M, Pietruszka P, Dortland BR, Malik AR, Wulf PS, et al. 2011. CLIP‐170 and IQGAP1 cooperatively regulate dendrite morphology. J Neurosci 31:4555–4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei N, Inamura N, Kawamura M, Namba H, Hara K, Yonezawa K, Nawa H. 2004. Brain‐derived neurotrophic factor induces mammalian target of rapamycin‐dependent local activation of translation machinery and protein synthesis in neuronal dendrites. J Neurosci 24:9760–9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomanetz V, Angliker N, Cloëtta D, Lustenberger RM, Schweighauser M, Oliveri F, Suzuki N, et al. 2013. Ablation of the mTORC2 component rictor in brain or Purkinje cells affects size and neuron morphology. J Cell Biol 201:293–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban N, Guillemot F. 2014. Neurogenesis in the embryonic and adult brain: Same regulators, different roles. Front Cell Neurosci 8:396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanska M, Blazejczyk M, Jaworski J. 2008. Molecular basis of dendritic arborization. Acta Neurobiol Exp (Wars) 68:264–288. [DOI] [PubMed] [Google Scholar]
- Urbanska M, Gozdz A, Swiech LJ, Jaworski J. 2012a. Mammalian target of rapamycin complex 1 (MTORC1) and 2 MTORC2) control the dendritic arbor morphology of hippocampal neurons. J Biol Chem 287:30240–30256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanska M, Swiech L, Jaworski J. 2012b. Developmental plasticity of the dendritic compartment: Focus on the cytoskeleton. Adv Exp Med Biol 970:265–284. [DOI] [PubMed] [Google Scholar]
- Wong RO, Ghosh A. 2002. Activity‐dependent regulation of dendritic growth and patterning. Nat Rev Neurosci 3:803–812. [DOI] [PubMed] [Google Scholar]
- Wu YK, Fujishima K, Kengaku M. 2015. Differentiation of apical and basal dendrites in pyramidal cells and granule cells in dissociated hippocampal cultures. Plos One 10:e0118482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Li W, Huang S, Song J, Kim JY, Tian X, Kang E, et al. 2013. mTOR inhibition ameliorates cognitive and affective deficits caused by disc1 knockdown in adult‐born dentate granule neurons. Neuron 77:647–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure 1.
Supporting Information Figure 2.


