Abstract
The utilization of scaffolds for enzyme immobilization involves advanced bionanotechnology applications in biorefinery fields, which can be achieved by optimizing the function of various enzymes. This review presents various current scaffolding techniques based on proteins, microbes and nanomaterials for enzyme immobilization, as well as the impact of these techniques on the biorefinery of lignocellulosic materials. Among them, architectural scaffolds have applied to useful strategies for protein engineering to improve the performance of immobilized enzymes in several industrial and research fields. In complexed enzyme systems that have critical roles in carbon metabolism, scaffolding proteins assemble different proteins in relatively durable configurations and facilitate collaborative protein interactions and functions. Additionally, a microbial strain, combined with designer enzyme complexes, can be applied to the immobilizing scaffold because the in vivo immobilizing technique has several benefits in enzymatic reaction systems related to both synthetic biology and metabolic engineering. Furthermore, with the advent of nanotechnology, nanomaterials possessing ideal physicochemical characteristics, such as mass transfer resistance, specific surface area and efficient enzyme loading, can be applied as novel and interesting scaffolds for enzyme immobilization. Intelligent application of various scaffolds to couple with nanoscale engineering tools and metabolic engineering technology may offer particular benefits in research.
Keywords: Cell surface anchoring, Designer enzyme, Nanoparticle, Scaffold, Whole‐cell biocatalyst
Abbreviations
- CBM
carbohydrate‐binding module
- GFP
Green fluorescence protein
- HLD
hydrophilic domain
- SLH
surface layer homology
1. Introduction
The synthesis of valuable products such as chemical building blocks and fuels from renewable biomasses is necessary to develop a biobased economy for sustainable economic growth 1. In recent years, waste residue of biomass materials, such as nonfood energy crops, e.g. switchgrass, have been regarded as inexpensive sugar sources to substitute starchy‐based glucose sources in microbial processes 2. As lignocellulosic materials are the most abundant, low‐cost and utilizable natural resources, these feedstocks are attractive options for use in microbial bioprocessing 3, 4, 5. Biorefinery research is a field of study for the bioconversion of fermentable sugars by degrading of lignocellulose in biomass (Fig. 1) 6. As the enzymatic hydrolysis of lignocellulosic biomass does not need high levels of energy cost in many commercial and industrial applications, enzymatic methods could replace those that use chemical catalysts 7. Enzymatic degradation methods showed simple facilities, excellent efficiency, mild reaction conditions, controllable products, low energy cost and minimal environmental pollution as advantages 8. Because lignocellulosic biomass consists of materials with different properties, various types of degrading and hydrolysis enzymes are required for its enzymatic hydrolysis (Fig. 2) 9. Although many stable cellulases have been identified for cellulose degradation, they are still not considered practical because these biological processes have had slow enzymatic degradation rates 10. Enzymes are effective and specific biocatalysts that are widely used in industry, but their application in industrial processes often requires desirable functions not found in naturally sourced enzymes 8. However, the hydrolysis process using cellulolytic enzymes by conventional expression from cellulolytic microbes is regarded as not cost effective 10. To obtain the desired enzymes, scientists have developed both rational and non‐rational design methods to enhance enzyme properties 8. Developing new, effective and economical enzymes is necessary to promote the broad application of biological hydrolysis processes 11.
Figure 1.
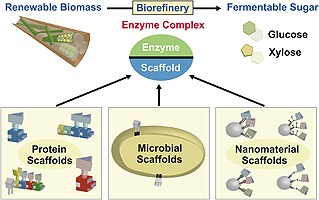
Diagram illustrates the background of renewable biomass utilization and biorefineries using enzyme complexes. The enzyme complex is composed of hydrolysis enzymes and various types of scaffolds such as protein scaffolds, microbial scaffolds and nanomaterial scaffolds. The scaffold‐assembled enzymes have a high level of hydrolysis activity and can convert the various biomasses to valuable biomaterials.
Figure 2.
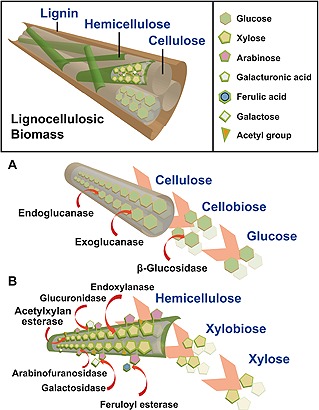
The Diagram illustrates the structural organization of the plant cell wall. Enzymatic degradation of cellulose is protected by hemicelluloses and lignin. (A) Simple chemical composition of cellulose as glucose polymer and three types of cellulases for its enzymatic degradation. (B) Various components of hemicellulose with xylose backbone and multiple sets enzymes for its enzymatic hydrolysis.
Many approaches containing immobilization systems on cell surface or extracellular secretion systems, have been implemented for the utilization of biomass through biological enzyme processing 12. The use of a scaffold is one strategy that can be used for effective lignocellulosic biomass hydrolysis 13. The term ”scaffold“ implies the formation of stable complexes, which are further reinforced by highly specific localization 14. Tight interactions between substrates and enzymes as well as working together with different enzymes are necessary to increase the enzymatic degradation of insoluble polysaccharides 15, 16. The use of a complexed enzyme system with a supporting scaffold is one strategy for producing cooperative enzymes with tight interactions. Scaffolds offer highly ordered structural organization as well as considerably greater degradative potential through enzyme proximity synergy. Also, substrate channeling is a main purpose for scaffolding enzymes. Substrate channeling is the phenomenon that yields and efficiencies in enzymatic reactions with diffusion processes are increased by proximity effects of enzyme complex modules between cascade enzymatic steps 17. Because the product of one enzyme is transferred to an adjacent cascade enzyme, mixing of intermediates with the bulk phase does not occur in the substrate channeling process 18. Various mechanisms of substrate channeling using natural and examples of constructing synthetic complexes for substrate channeling were presented with biotechnological potentials in synthetic biology 17, 18. For example, a synthetic metabolon containing three enzymes, triosephosphate isomerase, aldolase, and fructose 1,6‐bisphosphatase, was self‐assembled through the protein‐protein interactions with the protein scaffold 19, 20, 21. The synthetic metabolon accelerated reaction rate 19, decreased protein purification labor 20 compared to the noncomplexed enzymes. Thus, scaffolds help to provide more substantial and rapid degradation systems. 11 Also, DNA can be applied as scaffolds to generate DNA‐protein complexes, DNA nanodevices, for engineering enzyme pathways 22. Structural DNA and protein nanotechnologies can be introduced by engineering designed and programmable nanostructures as scaffolds.
For optimizing the function of target enzymes, the immobilization technique is served in industrial and research fields. Several different materials for improving the performance of immobilized enzymes have been described in the literature. Scaffold utilization involves advanced biorefinery applications with renewable biomass. These architectural scaffolds have been applied to useful strategies for protein engineering in several industrial and research fields 23. This review discusses these developments with emphasis on protein‐protein inter actions, cell surface display, and nanomaterials. Possible future directions of scaffold use are also summarized herein.
2. Protein scaffolds for biomass conversion and immobilization
Scaffolding proteins have critical roles to facilitate their concerted interactions and functions by bringing together multiple binding partners. Scaffolding proteins in complexed enzyme systems assemble different proteins in relatively durable configurations 14. Numerous scaffolding proteins found in nature have specific modules for multiple protein–protein interactions. We became aware of this feature of scaffolding proteins while researching lignocellulosic biomass utilization by microbes, which focused on the biomass contributions to the microbial carbon metabolism 14. Although various bacterial strains have been evaluated for their cellulase production potential, relatively few microorganisms have been reported to be able to degrade cellulosic substrates into glucose 6. These cellulolytic microbes have been found to produce cellulases as free enzymes or enzyme complexes that work synergistically 24. Among them, certain cellulolytic anaerobic bacteria have evolved intricate multi‐enzyme complexes with scaffolding proteins known as cellulosomes 25. Cellulosomes facilitate the synergistic breakdown of complex polymers in lignocellulose with the aid of scaffoldin, which is a noncatalytic scaffolding protein 26. Cellulosomes are a promising approach for solving the problems related to slow enzymatic degradation rates because complexed system showed degradative potential 11.
Cellulosomes have been researched in anaerobic microbes, i.e. Clostridium thermocellum 27, Clostridium cellulovorans 28, and Clostridium cellulolyticum 29. Cellulosome action involves scaffoldin binding to a variety of cellulolytic and hemicellulolytic subunits, including cellulases, hemicellulases, chitinase, pectinase, and other auxiliary enzymes, via interactions between the scaffoldin cohesin modules and the enzymatic dockerins 30. Cellulosomal cellulases tightly bind to scaffoldin, a large and nonenzymatic scaffolding protein. Although scaffoldin has no enzymatic ability, it helps to degrade polysaccharides of plant cell walls through substrate binding and structural organizing ability 15. Different cellulosomal and non‐cellulosomal enzymes have been reported to function synergistically and cooperatively 26. Additionally, cellulosomes bringing cellulases and polysaccharides into closer proximity via the strong binding of polysaccharides to the carbohydrate‐binding module (CBM) in scaffoldin 31. Thus, essentially, the cellulosome system of anaerobic microorganism showed higher hydrolytic activity than non‐cellulosomal systems of aerobic microorganisms 26, 32.
2.1. Protein interactions of complex system via binding modules
Scaffolding proteins achieve their concerted interactions and functions by being composed of several protein–protein interaction modules 14. In the same manner, cellulosomes are complexed by protein‐protein interactions with high affinity 15. Specific protein interactions could produce a cellulosome through several repeated sequences present on the non‐catalytic scaffoldin. These repeated sequences form the cohesin module, which is highly and moderately conserved within the same scaffolding protein and among different proteins, respectively 33. The hydrophobic cohesin modules of scaffoldin bind to the dockerin module, which is a duplicated sequence located on all cellulosomal enzymes and not found on non‐cellulosomal enzymes 27. Dockerin modules are 22‐amino‐acid residues separated by a linker sequence, which is a well‐conserved segment among bacterial species 33. Specific interactions between the dockerin module and one of several cohesin modules dictate the assembly of dockerin‐containing subunits into complexes with scaffoldin 34. Cohesin and dockerin modules and their interactions can be classified as type I or type II by sequence homology 35. Dockerin module in cellulosomal subunits and the cohesin module in the scaffoldin are interacted by type I interactions,. whereas the counter parts of type I interactions are type II dockerin‐cohesin interactions 36. Type I and type II interactions are type‐specific because cross‐interactions between type I and type II have been not reported. As the dockerin modules in each strain can distinguish the cohesin modules in other strains, interspecies specificity are found in diverse Clostridia microbes 37. The cohesin–dockerin interaction is the high‐affinity protein–protein interaction between cellulosomal subunits with dockerin modules and non‐catalytic scaffoldin with cohesin modules 38. As these high‐affinity interactions in comparison with other interactions of proteins showed a K D value ranging from 1.9 × 10−9 to 2.4 × 10−10, they could be applied to a various biological processes as the scaffold is based on affinity 37. Moreover, the cohesin‐dockerin interactions derived from anaerobic strains are considered to be broadly usable as advanced biotechnology tools.
2.2. Design of protein scaffolds for containing useful modules
Scaffoldin usually contains several cohesin modules, which have the affinity with the dockerin modules of each cellulosomal subunit 39. Because native scaffoldin is too large for expression by industrial microbe hosts, scaffoldin was spliced into a small recombinant scaffoldin (Fig. 3). For example, the scaffolding protein CbpA from C. cellulovorans was modified to the recombinant scaffoldin, which is called as the miniCbpA 15. Instead of nine cohesin modules in normal CbpA, a small miniCbpA contains only two cohesin modules as a small derivative. Although the small recombinant scaffoldins have only a few cohesin modules, these modules retain their complementary binding properties 40. Thus, small recombinant scaffoldin could be applied for the construction of useful enzyme complexes 15. Moreover, chimeric constructs of cellulosomal modules was applied in various research such as biorefinery, medicine and industry 41.
Figure 3.
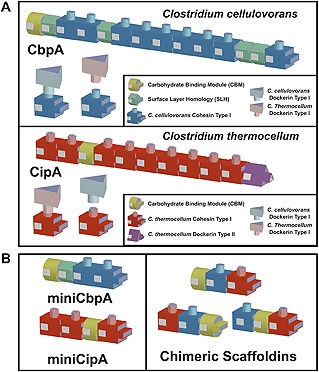
Schematic illustrations of protein scaffolds based on the cellulosome system. (A) Native protein scaffold CbpA and CipA from C. cellulovorans and C. thermocellum, respectively. Dockerin‐cohesin interactions from different Clostridia strains have interspecies specificity. Thus, the cohesin modules in each strain were able to discriminate the dockerins in other strains. (B) Small recombinant protein scaffolds, miniCbpA and miniCipA, derived from native protein scaffolds CbpA and CipA, respectively. In addition, chimeric protein scaffolds combined with various cohesin modules from different Clostridia strains. Because the native protein scaffold is too large for expression by industrial microbe hosts, scaffoldin was spliced into a small recombinant protein scaffolds.
Additionally, small recombinant scaffoldins derived from native scaffolding proteins were designed to have a CBM, which brings substrates into proximity with the cellulosomal enzymes 42. For example, normal CbpA consists of eight cohesin modules and the CBM has two X2 domains. Also, the recombinant miniCbpA (CBM‐Coh1‐Coh2) is a scaffolding protein containing two cohesin modules and a CBM. The cohesin modules and CBM enable complex formation and affinity applications through specific and high affinity modules, respectively 43. Because cellulose has several advantages such as inexpensive matrix, outstanding physical properties and low nonspecific binding with contaminants, it could be applied for affinity purification applications as large‐scale 15. By using the CBM in the scaffoldin, dockerin‐fused proteins could be recycled through the CBM‐based recycling method 44. By replacing the multi‐step process with single‐step CBM‐based recycling methods, accumulated loss of product can be avoided 43. Therefore, the single‐step CBM‐based recycling methods could remarkably enhance the cost effectiveness of protein purification for enzyme recycling.
2.3. Multi‐functional protein complexes for application on biorefinery
The assembly of multi‐functional protein complexes showed efficient enzymatic processes by enhancing hydrolysis as well as interactions between enzymes and their substrates 45. Enzyme complexes with scaffoldin, based on the cellulosome system, could convert the insoluble lignocellulosic materials to fermentable sugars by multi‐step reactions. Designed enzyme complexes with scaffoldin are constructed to contain substrate‐binding modules, such as CBM. As mentioned above, scaffoldin has been spliced into small recombinant scaffoldins, such as miniCbpA, for the construction of complexed enzyme systems. Additionally, chimeric enzymes with dockerin domains have also been used to assemble complexes via cohesin‐dockerin interactions 15. Previous reports have described the use of this interaction between dockerin and cohesin for the construction of protein complexes composed of endoglucanase or xylanase with increased hydrolytic ability with cellulose or hemicellulose as substrates, respectively 13, 44.
Hydrolytic enzyme complexes have been constructed to achieve improved lignocellulosic biomass degradation efficiency. The dockerin module of the cellulosomal subunits needs to be fused with the target proteins at the C‐terminus for assembly of complexed system 46. The multi‐step PCR strategy was used for the genetic fusion of target proteins with the dockerin module using overlapping primers. Designed proteins fused with the dockerin module could be connected to the small recombinant scaffolding proteins containing two cohesin modules through type I interactions 44. The designer cellulosomes, which are minicellulosomes containing only a few cohesin modules of native scaffoldin, showed higher hydrolysis activity of insoluble cellulose substrate than that of the free enzyme system 47. Previous reports have demonstrated that enzyme complexes enabled the effective targeting and concentration of the hydrolytic action through enzyme proximity effect by a coordinated organization 43. Additionally, these minicellulosomes could be recycled by single‐step CBM‐based recycling methods 13.
For example, the combination of endoglucanase with different hydrolysis enzymes had a synergistic effect on the degradation of lignocellulosic biomass 48. Additionally, chimeric endoglucanase cCelE and xylanase XynB were constructed as a complexed system with miniCbpA as scaffoldin. These enzyme complexes showed the effective degradation of lignocellulosic biomass with the aid of the CBM for efficient substrate affinity 13. As the construction of multi‐enzyme complexes has been shown to affect cellulose hydrolysis, the combination of cellulases with scaffolding subunits may be an effective means of achieving efficient degradation 49.
Artificial enzyme complexes could facilitate the development of biocatalysts for efficient utilization of lignocellulosic biomass 50. The strategy to design scaffoldin for the construction of enzyme complexes is an attractive concept that could be linked to a suitable host cell system with adequate configurations. This strategy provides evident benefits to biological applications for highly active enzymes with large quantities 49. Thus, construction of these artificial cellulosomes based on designer scaffoldin is very important to apply for the industrial and research fields.
3. Microbial scaffolds for biomass conversion and immobilization
A microbial strain can be applied to the immobilizing scaffold because the in vivo immobilizing technique has several benefits in enzymatic reaction systems related to both synthetic biology and metabolic engineering 51. For example, during batch fermentation, fixed hydrolysis activity was shown in displayed cellulases at each cycle via the induction of variant proteins 52. In addition, the in vivo immobilization system of cell surface display showed continuous hydrolysis ability without additional inducing agents and protein stabilization effect 53. Moreover, other research fields such as synthetic biology and metabolic engineering could be easily linked to cell surface display techniques using the microbial strain as a scaffold for fabricating valuable products 54. In the same manner, there are several advantages of whole‐cell biocatalysts that display lignocellulosic biomass hydrolytic enzymes with the ability to degrade cellulose. Although secreted free enzyme system directly saccharify the substrate far from cells, displayed enzymes directly consume the sugars after polysaccharide degradation in nearby cells (Fig. 4) 55. As glucose is instantly consumed by cells, such systems do not need excessive sterilization and multi‐type reactors. The immediate consumption of glucose also facilitates enzymatic activity because the activity of cellulases including β‐glucosidase and endoglucanase is inhibited by glucose and cellobiose 53. Furthermore, the various cellulases in enzyme complexes on the cell surface react in the similar location of the substrate 56. These types of displayed cellulase complexes have exhibited production of reducing sugars at high yields by direct degradation of lignocellulose with synergistic effects 53. These results suggest that the enzyme‐displayed whole‐cell biocatalysts with microbial scaffolds may be promising strategies for utilizing renewable biomass resources 54.
Figure 4.
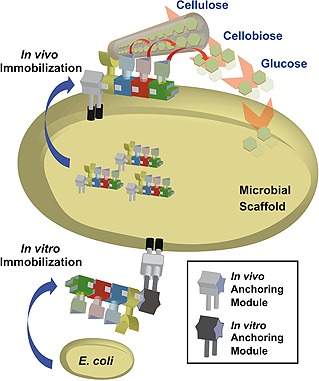
Schematic illustrations of advanced application of the microbial scaffold as surface display tool in bionanotechnology. Surface display based on anchoring modules for in vivo and in vitro immobilizations is shown as a diagram. The replacement of anchoring domains in protein scaffolds also leads to immobilization of enzymes on cell surface.
3.1. Anchoring modules to display scaffoldin on the cell surface
The scaffolding proteins of microbial cell surface display systems such as cellulosomes also possess a hydrophilic domain (HLD) or surface layer homology (SLH) domain as an anchoring module 57, 58. For example, cellulosomes bind to cell surface of C. cellulovorans via the HLD in the scaffoldin CbpA 57. However, this type of anchoring module is not suitable for the cell wall of yeast. Thus, the cell surface display of foreign proteins on industrial strains requires proper different anchoring modules 58.
In the case of yeast, a glycosylphosphatidylinositol is used as the anchor and fused with the scaffolding protein at the C‐terminus 58. Transmembrane proteins such as mechanosensitive channels (Msc) related to amino acid secretion and glucose consumption are used as anchoring modules in the host C. glutamicum. The macromolecular enzyme complexes are secreted by the Msc and attached to the cell surface 59. In particular, type IV pili (T4P), which are polymers of the major component pilin, were used as anchoring proteins for in vitro immobilization of enzymes on the cell surface of many gram‐negative bacteria 60.
3.2. Design of cell surface display systems by fusion of protein modules
For whole‐cell biocatalyst applications, target enzymes or scaffolding proteins need to be fused with cell surface‐anchoring modules 54. The cell surface display of the anchoring protein can be observed using fluorescence microscopy. Green fluorescence protein (GFP) can be used to label the anchoring module 53. Observing the anchoring module fused with GFP on the cell surface as a display state indicates that the scaffoldin fused with the anchoring module is also successfully displayed on the cell surface of target microbes 54. Additionally, by comparing enzyme activity between the fractions in the supernatant and the cell pellet, successful association of the enzymes with the cell surface of C. glutamicum can be confirmed 53.
The microbial cell‐surface display enzyme systems that have been designed successfully increase conversion efficiency and thermostability through the substrate‐binding‐affinity effect and intensive glycosylation, respectively 54. The rigid maintenance on the conformation of the immobilized cellulase complexes improved their thermostability. In addition, the formation of macromolecular complexes via dockerin‐cohesin interactions results in the rigid maintenance on the conformation that prevents protein denaturation in thermal conditions 61. For example, conjugating the scaffolding protein with Msc containing many hydrophobic residues (48.0%) enhanced the hydrophobicity of the scaffolding protein from 41.5 to 44.7% and stabilized the immobilized enzymes in the compact structure 53. Increasing cellulose degradation temperature showed no cooling, low contamination and high enzyme activity in order to reduce enzymatic process costs while taking its biological aspects into account. Over the past two decades, many methodologies have been used to improve protein thermostability, including mimicking naturally occurring homologous proteins and rational design through the introduction of mutations or directed evolution 11. Thus, the use of thermostable enzymes with microbial scaffolds is one strategy for reducing the contaminants in biorefinery process and decreasing the cooling step because they maintain the stable structure in high temperatures after pretreatment step for lignin degradation 62. These constructs can exhibit synergy, highly efficient saccharification and thermostability through simple cell surface immobilization via anchoring modules 53.
3.3. Whole‐cell biocatalysts for direct hydrolysis of polysaccharides
The manufacturing of valuable materials using plant‐based lignocellulosic biomass as feedstock requires useful whole‐cell biocatalysts with cellulolytic activity 2. Whole‐cell biocatalysts capable of cellulose degradation have been shown to directly consume the simple sugars such as glucose and cellobiose to prevent the inhibition of the activity of cellulolytic enzymes such as β‐glucosidase and cellulase in the same process 5. In addition, similar to designer enzyme complexes with a CBM module, enzyme proximity effects were induced by displayed scaffoldin. Subsequently, the development of industrial strains displaying functional enzyme complexes showed increased enzyme activity via enzyme proximity to the cell surface as a whole‐cell biocatalyst 53, 63, 64.
For example, C. glutamicum displaying cellulase complexes leads to enhanced direct hydrolysis of polysaccharides from low‐cost biomass. By using the Msc as anchoring module of C. glutamicum, cellulase complexes containing two different types of cellulases have been successfully co‐displayed on the cytoplasmic membrane 53. Conjugating endoglucanase and β‐glucosidase to scaffoldin and in turn, conjugating scaffoldin to an anchor protein, induced the synergistic production of high yields of reducing sugars 56. Spontaneous saccharification of lignocellulosic biomass was performed by the displayed endoglucanase and β‐glucosidase in C. glutamicum without any additional β‐glucosidase solution 53. The successful display of the endoglucanase and β‐glucosidase complexes allowed the efficient hydrolysis of biomass to glucose through enzyme proximity synergy. The synergistic effect of the displayed cellulase complexes led to direct conversion of rice straw, miscanthus and rape stem to reducing sugars that was 6.0‐, 3.1‐ and 3.3‐fold greater than that of the secreted cellulase complexes, respectively 53.
Additionally, the displayed cellulase complexes showed increased thermostability at 70°C with synergy ratios for endoglucanase and β‐glucosidase 2.3‐ and 3.4‐folder higher, respectively, than those of the secreted cellulase complexes 53. At 80°C, the displayed β‐glucosi dase showed a remaining activity up to 76.9%, but the anchored endoglucanase exhibited relative activity that had decreased to 33.6%. These differences indicated that the surface display effect is more sensitive to β‐glucosidase due to the increasing hydrophobicity after assembling into the complexed system (39.7–41.0%) and immobilizing by surface display techniques (39.7–43.5%) 53. In case of xylanase, the enzyme displayed by anchoring modules such as a polyglutamate synthetase (PgsA) retains up to 77 and 25% relative activity at 65°C and 70°C, respectively 65. However, xylanase anchored by the PgsA anchor protein showed a lower thermally stable synergy than cellulase anchored by the Msc anchor module.
4. Nanomaterial scaffolds for biomass conversion and immobilization
Nanoscale supports with large surface and small size including nanofibers, nanoparticles, sol‐gel silica, mesopo rous materials, crosslinked enzyme aggregates (CLEAs)/crystals (CLECs) and alginate‐based microspheres have emerged as useful scaffolds being excellent enzyme immobilization supports 66. The use of solid supports at the nanoscale offers the inherent benefit of being able to load large quantities of catalytic molecules onto a matrix 67. Cellulase enzymes immobilized on nanomaterials have been reported by various investigators 68. Nano scale supports have attracted increasing amounts of attention as novel materials for protein immobilization because of their unique physicochemical properties 69. Nanoscale supports show advantages such as effective enzyme loading, mass transfer resistance and specific surface area for determination of biocatalyst efficiency. The stability and activity of enzymes has been observed to increase when they are immobilized on such materials. In particular, several studies on enzyme immobilization on nanomaterials have reported generally enhanced enzyme stability under harsh conditions and improved enzyme reusability 70.
4.1. Nanomaterial scaffold synthesis methods for enzyme immobilization
The synthesis of noble nanoscale materials as scaffolds for enzyme immobilization has recently been gaining attention for various applications 71. A number of methodologies such physical, chemical or biological methods as for the synthesis of nanoscale scaffolds have been described in the literature 72. The selection of an immobilization method is very important to avoid introducing reactive groups that may lead to a loss of the desired enzyme activity. Weak enzyme‐matrix interactions and covalent bonds forming between the support and the enzyme are common problems of physical and chemical methods, respectively. Physical and chemical methods generally consume energy and require toxic ingredients or hazardous materials, rendering these unfavorable synthesis methods 73. Biological nanoparticle synthesis methods are relatively simple, cost effective and environment friendly compared with conventional chemical/physical synthesis methods, which makes them more desirable 74. In addition, biological methods can be applied for important applications due to site‐specific, powerful and efficient protein anchoring principles. The synthesis of nanoparticles using pure enzymes can serve as an example of the biochemical reactions involved in these biosynthetic pathways 72. The interaction time of enzymes and reducing agents with metal ions plays an important role. The synthesized nanoparticles effectively adsorb the enzyme molecules and thus serve as the immobilization matrix. Furthermore, biosynthesized nanoparticles can be also exploited as an immobilization matrix for other enzymes or enzyme complexes containing cellulases.
4.2. Nanoparticles for cellulase immobilization to improve various properties
Among other nanomaterial scaffolds used for enzyme immobilization, nanoparticles act as very efficient support materials. Nanoparticles with immobilized enzymes can lead to improved stability, performance, and protein folding. Simple separation can also be accomplished via an external magnetic field 75. Four main methodologies can be applied to anchor enzymes to nanoparticles, including adsorption by electrostatics, attachment by covalent reactions to functionalized nanoparticles, conjugation using protein with a specific affinity, and direct conjugation to the nanoparticle surface (Fig. 5). Among all the immobilization methods, adsorption is the simplest, and there have been many reports regarding cellulase immobilization via adsorption 69. Various studies on enzyme immobilization have been performed using various types of nanoparticles, such as porous and polymeric nanoparticles, metal nanoparticles and magnetic nanoparticles 76. Enzymatic immobilization on nanoparticles has been studied using both whole cells and isolated enzymes 77. Many researchers have attempted to immobilize cellulase through encapsulation in nanofibrous poly(vinyl alcohol) (PVA) membranes via electrospinning, prepared chitosan microspheres, modified PVA‐coated chitosan beads and mesoporous silica (SBA‐15) 78. Also, the cellulose‐containing nanoparticles were used as new supports for immobilizing and recycling of CBM tagged enzymes by binding affinity and magnetic force, respectively 79. To date, metal nanoparticles with diverse shapes and sizes have been prepared with wet chemistry approaches. The unique structural and functional features of metal nanoparticles make them excellent candidates for the development of electrochemical catalysts, electrical immunosensors, and supports for enzyme immobilization 80. These types of immobilized enzymes have shown higher activity and thermal stability than their free counterparts 81.
Figure 5.
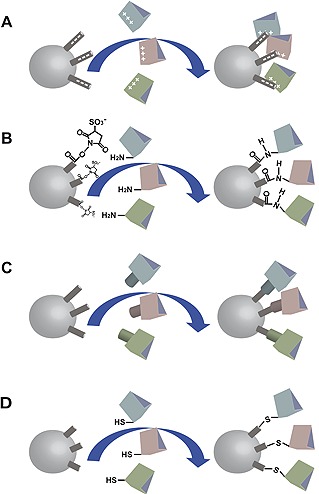
Four main methodologies for linking enzymes to nanoparticles, including (A) electrostatic adsorption, (B) covalent attachment to functionalized nanoparticles, (C) conjugation using proteins with a specific affinity, and (D) direct conjugation to the nanoparticle surface.
The cellulase was immobilized on biologically synthesized silver and gold nanoparticles by a simple adsorption process and showed greater thermal stability and reusability than did the free cellulase. The immobilized cellulase on the nanoparticles maintains 77–80% activity of the free enzyme at 75°C after 60 min 74. While the cellulase immobilization permit recycling for six times with a 22–27% activity loss of enzymes 74. This gradual activity loss of cellulase immobilized on silver and gold nanoparticles may be due to various factors, such as end‐product inhibition, protein denaturation and removal of several components of the complex 82.
Considering biocompatibility, silicon is also a good material for nanoparticle biosensors and electrochemical immunosensors. In addition, silicon can be hybridized with metal nanoparticles to obtain core–shell magnetic nanoparticles for enzyme immobilization and for the fabrication of enzyme devices 83. Supermagnetic nanoparticles have been coated with silicon via a simple molecular imprinting technique and employed as the support for cellulase immobilization. A high immobilization yield was achieved, and the immobilized cellulase exhibited high thermostability and pH stability, easy reusability and low K m 69. Compared with free enzyme, the immobilized cellulase displayed a relatively high activity at temperatures ranging from 30 to 80°C. When the enzyme is specifically adsorbed onto the imprinted nanoparticles, the immobilization limits the thermal vibration of cellulase. The immobilized enzyme also showed relatively higher activity at pH 3.0–6.0, although the optimal pH was not changed. Typically, the optimal pH of an immobilized enzyme changes with the charge of the scaffold. When the surface of prepared support is not charged, the immobilized enzyme showed that optimal pH does not differ from that of free enzyme 69. The anchored cellulase displayed a higher catalytic affinity for CMC than did free enzyme, possibly because immobilization via molecular imprinting fixes the proper figuration of cellulase, allowing substrates to enter the active site more easily 69. Although free cellulase is thermally stable, there is a 3.3‐fold improvement over that of free enzyme because immobilization improves enzyme rigidity 84. In addition, only 30% activity was lost after 10 cycles of reuse. Reusing these immobilized enzymes is very easy because of the superparamagnetic support 69. Thus, various nanoparticles can be successfully prepared as a scaffold for cellulase immobilization. The preparation process is very simple and yields high immobilization efficiency. Immobilized cellulase has higher affinity for cellulosic substrates than the free enzyme. These results suggest that nanoparticles have the potential for application in the purification and immobilization of cellulase from crude enzyme solutions and various industrial processes involving bioethanol production, paper and pulp, as well as other applications in the pharmaceutical industry 69.
5. Concluding remarks
In addition to industrial applications, immobilized enzymes can be used in laboratory‐scale organic synthesis processes as well as in various industrial fields. Also, displayed enzymes can link the reactions for organic synthesis because enzymes can perform the reactions both in inorganic and aqueous solutions to enhance the activity and stability of enzymes. Most immobilization research has focused on the design, preparation, and modification of immobilization supports. This review described the development of scaffolds for enzyme immobilization, which includes efficient, artificial, complexed enzyme systems. Designer enzyme complexes based on various scaffolds have the potential to efficiently produce valuable materials from lignocellulosic biomass. Intelligent application of protein scaffolds may deserve particular attention in research for rational design of enzyme complexes via nanoscale engineering tools with computational modelling 85. Also, cell surface display technique will be easily extended to be coupled with metabolic engineering technology such as substrate channeling to achieve higher efficiency of enzymes by synergy 86.
Acknowledgement
This work was supported by the Industrial Strategic Technology Development Program (10051513) funded by the Ministry of Trade, Industry & Energy (MI, Korea). We would like to express gratitude to Brain Korea 21 Plus (Department of Biotechnology, Korea University) for processing charges in Open Access journals.
The authors declare no financial or commercial conflict of interest.
Biographies
Jeong Eun Hyeon is a Research Professor and Postdoctoral Research Scientist at the department of Biotechnology at the Korea University (Republic of Korea). He obtained his M.Sc. and Ph.D. in biochemistry with the area of designer microbes for biorefinery of various bioresources in October 2015 under the guidance of Prof. Sung Ok Han at the Korea University (Republic of Korea). He has a vast research background in the field of biotechnology. His current research interests focus on the application of multi enzyme complex system in biocatalysis, biomass degradation, C1 gas conversion, bioactive compound synthesis, and bioplastic polymerization.

Sang Kyu Shin is integrated M.S. and Ph. D course student at department of biotechnology under the guidance of Prof. Sung Ok Han at the Korea University (Republic of Korea). He received his B.Sc. in chemical and biological engineering from Korea Polytechnic University (Republic of Korea). He researched efficient lignocellulosic biomass utilization to biofuel with multi enzyme complexes which consist of cellulase and hemicellulase. His current research interests focus on the application of C1 gas conversion to valuable products through expression inhibitor inspection.

Sung Ok Han is a Professor in the department of Biotechnology at Korea University (Republic of Korea). He received his Ph.D. in Microbiology at the University of Sydney (Australia) in 1998. His first appointments were as a postdoctoral researcher at the University of California, Davis (USA) and then as a senior researcher at Research Institute of Innovative Technology for the Earth (Japan). His research interests are focused on industrial microbiology and biomolecular engineering. He has been working on utilization of C1/3/5/6 bioresources for biorefinery using genetic, protein and metabolic engineering technology.

References
- 1. Yu, K. O. , Kim, S. W. , Han, S. O. , Engineering of glycerol utilization pathway for ethanol production by Saccharomyces cerevisiae . Bioresour. Technol. 2010, 101, 4157–4161. [DOI] [PubMed] [Google Scholar]
- 2. Fujita, Y. , Ito, J. , Ueda, M. , Fukuda, H. , Kondo, A. , Synergistic saccharification, and direct fermentation to ethanol, of amorphous cellulose by use of an engineered yeast strain codisplaying three types of cellulolytic enzyme. Appl. Environ. Microbiol. 2004, 70, 1207–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Galbe, M. , Zacchi, G. , A review of the production of ethanol from softwood. Appl Microbiol. Biotechnol. 2002, 59, 618–628. [DOI] [PubMed] [Google Scholar]
- 4. Jorgensen, H. , Kutter, J. P. , Olsson, L. , Separation and quantification of cellulases and hemicellulases by capillary electrophoresis. Anal. Biochem. 2003, 317, 85–93. [DOI] [PubMed] [Google Scholar]
- 5. Tokuhiro, K. , Ishida, N. , Kondo, A. , Takahashi, H. , Lactic fermentation of cellobiose by a yeast strain displaying beta‐glucosidase on the cell surface. Appl. Microbiol. Biotechnol. 2008, 79, 481–488. [DOI] [PubMed] [Google Scholar]
- 6. Das, M. , Royer, T. V. , Leff, L. G. , Diversity of fungi, bacteria, and actinomycetes on leaves decomposing in a stream. Appl. Environ. Microbiol. 2007, 73, 756–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Erickson, B. , Nelson, J. E. , Winters, P. , Perspective on opportunities in industrial biotechnology in renewable chemicals. Biotechnol. J. 2012, 7, 176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hong, J. , Wang, Y. , Kumagai, H. , Tamaki, H. , Construction of thermotolerant yeast expressing thermostable cellulase genes. J. Biotechnol. 2007, 130, 114–123. [DOI] [PubMed] [Google Scholar]
- 9. Shill, K. , Padmanabhan, S. , Xin, Q. , Prausnitz, J. M. et al., Ionic liquid pretreatment of cellulosic biomass: Enzymatic hydrolysis and ionic liquid recycle. Biotechnol. Bioeng. 2011, 108, 511–520. [DOI] [PubMed] [Google Scholar]
- 10. Hyeon, J. E. , You, S. K. , Kang, D. H. , Ryu, S. H. et al., Enzymatic degradation of lignocellulosic biomass by continuous process using laccase and cellulases with the aid of scaffoldin for ethanol production. Process Biochem. 2014, 49, 1266–1273. [Google Scholar]
- 11. Lu, Y. , Zhang, Y. H. , Lynd, L. R. , Enzyme‐microbe synergy during cellulose hydrolysis by Clostridium thermocellum . Proc. Natl. Acad. Sci. USA 2006, 103, 16165–16169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Inokuma, K. , Hasunuma, T. , Kondo, A. , Efficient yeast cell‐surface display of exo‐ and endo‐cellulase using the SED1 anchoring region and its original promoter. Biotechnol. Biofuels 2014, 7, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shin, S. K. , Hyeon, J. E. , Kim, Y. I. , Kang, D. H. et al., Enhanced hydrolysis of lignocellulosic biomass: Bi‐functional enzyme complexes expressed in Pichia pastoris improve bioethanol production from Miscanthus sinensis . Biotechnol. J. 2015, 10, 1912–1919. [DOI] [PubMed] [Google Scholar]
- 14. Garbett, D. , Bretscher, A. , The surprising dynamics of scaffolding proteins. Mol Biol Cell 2014, 25, 2315–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hyeon, J. E. , Yu, K. O. , Suh, D. J. , Suh, Y. W. et al., Production of minicellulosomes from Clostridium cellulovorans for the fermentation of cellulosic ethanol using engineered recombinant Saccharomyces cerevisiae . FEMS Microbiol. Lett. 2010, 310, 39–47. [DOI] [PubMed] [Google Scholar]
- 16. Takagi, M. , Hashida, S. , Goldstein, M. A. , Doi, R. H. , The hydrophobic repeated domain of the Clostridium cellulovorans cellulose‐binding protein (CbpA) has specific interactions with endoglucanases. J. Bacteriol. 1993, 175, 7119–7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wheeldon, I. , Minteer, S. D. , Banta, S. , Barton, S. C. et al., Substrate channelling as an approach to cascade reactions. Nat. Chem. 2016, 8, 299–309. [DOI] [PubMed] [Google Scholar]
- 18. Zhang, Y. H. P. , Substrate channeling and enzyme complexes for biotechnological applications. Biotechnol. Adv. 2011, 29, 715–725. [DOI] [PubMed] [Google Scholar]
- 19. You, C. , Myung, S. , Zhang, Y. H. , Facilitated substrate channeling in a self‐assembled trifunctional enzyme complex. Angew. Chem. Int. Ed. 2012, 51, 8787–8790. [DOI] [PubMed] [Google Scholar]
- 20. You, C. , Zhang, Y. H. P. , Self‐assembly of synthetic metabolons through synthetic protein scaffolds: One‐step purification, co‐immobilization, and substrate channeling. ACS Synth. Biol. 2013, 2, 102–110. [DOI] [PubMed] [Google Scholar]
- 21. You, C. , Zhang, Y. H. P. , Annexation of a high‐activity enzyme in a synthetic three‐enzyme complex greatly decreases the degree of substrate channeling. ACS Synth. Biol. 2014, 3, 380–386. [DOI] [PubMed] [Google Scholar]
- 22. Yang, Y. R. , Liu, Y. , Yan, H. , DNA Nanostructures as Programmable Biomolecular Scaffolds. Bioconjugate Chem. 2015, 26, 1381–1395. [DOI] [PubMed] [Google Scholar]
- 23. Fontes, C. M. , Gilbert, H. J. , Cellulosomes: Highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annu. Rev. Biochem. 2010, 79, 655–681. [DOI] [PubMed] [Google Scholar]
- 24. Doi, R. H. , Tamaru, Y. , The Clostridium cellulovorans cellulosome: An enzyme complex with plant cell wall degrading activity. Chem. Rec. 2001, 1, 24–32. [DOI] [PubMed] [Google Scholar]
- 25. Ha, S. J. , Galazka, J. M. , Kim, S. R. , Choi, J. H. et al., Engineered Saccharomyces cerevisiae capable of simultaneous cellobiose and xylose fermentation. Proc. Natl. Acad. Sci. USA 2011, 108, 504–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cho, W. , Jeon, S. D. , Shim, H. J. , Doi, R. H. , Han, S. O. , Cellulosomic profiling produced by Clostridium cellulovorans during growth on different carbon sources explored by the cohesin marker. J. Biotechnol. 2010, 145, 233–239. [DOI] [PubMed] [Google Scholar]
- 27. Stahl, S. W. , Nash, M. A. , Fried, D. B. , Slutzki, M. et al., Single‐molecule dissection of the high‐affinity cohesin‐dockerin complex. Proc. Natl. Acad. Sci. USA 2012, 109, 20431–20436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Doi, R. H. , Kosugi, A. , Murashima, K. , Tamaru, Y. , Han, S. O. , Cellulosomes from mesophilic bacteria. J. Bacteriol. 2003, 185, 5907–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fendri, I. , Tardif, C. , Fierobe, H. P. , Lignon, S. et al., The cellulosomes from Clostridium cellulolyticum: Identification of new components and synergies between complexes. FEBS J. 2009, 276, 3076–3086. [DOI] [PubMed] [Google Scholar]
- 30. Doi, R. H. , Kosugi, A. , Cellulosomes: Plant‐cell‐wall‐degrading enzyme complexes. Nat. Rev. Microbiol. 2004, 2, 541–551. [DOI] [PubMed] [Google Scholar]
- 31. Fox, J. M. , Jess, P. , Jambusaria, R. B. , Moo, G. M. et al., A single‐molecule analysis reveals morphological targets for cellulase synergy. Nat. Chem. Biol. 2013, 9, 356–361. [DOI] [PubMed] [Google Scholar]
- 32. Han, S. O. , Yukawa, H. , Inui, M. , Doi, R. H. , Regulation of expression of cellulosomal cellulase and hemicellulase genes in Clostridium cellulovorans . J. Bacteriol. 2003, 185, 6067–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sakka, K. , Sugihara, Y. , Jindou, S. , Sakka, M. et al., Analysis of cohesin‐dockerin interactions using mutant dockerin proteins. FEMS Microbiol. Lett. 2011, 314, 75–80. [DOI] [PubMed] [Google Scholar]
- 34. Yaron, S. , Shimon, L. J. , Frolow, F. , Lamed, R. et al., Expression, purification and crystallization of a cohesin domain from the cellulosome of Clostridium thermocellum . J. Biotechnol. 1996, 51, 243–249. [DOI] [PubMed] [Google Scholar]
- 35. Peer, A. , Smith, S. P. , Bayer, E. A. , Lamed, R. , Borovok, I. , Noncellulosomal cohesin‐ and dockerin‐like modules in the three domains of life. FEMS Microbiol. Lett. 2009, 291, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gilbert, H. J. , Cellulosomes: Microbial nanomachines that display plasticity in quaternary structure. Mol. Microbiol. 2007, 63, 1568–1576. [DOI] [PubMed] [Google Scholar]
- 37. Jeon, S. D. , Lee, J. E. , Kim, S. J. , Kim, S. W. , Han, S. O. , Analysis of selective, high protein‐protein binding interaction of cohesin‐dockerin complex using biosensing methods. Biosens. Bioelectron. 2012, 35, 382–389. [DOI] [PubMed] [Google Scholar]
- 38. Miras, I. , Schaeffer, F. , Beguin, P. , Alzari, P. M. , Mapping by site‐directed mutagenesis of the region responsible for cohesin‐dockerin interaction on the surface of the seventh cohesin domain of Clostridium thermocellum CipA. Biochemistry 2002, 41, 2115–2119. [DOI] [PubMed] [Google Scholar]
- 39. Tokatlidis, K. , Dhurjati, P. , Beguin, P. , Properties conferred on Clostridium‐thermocellum endoglucanase CelC by grafting the duplicated segment of endoglucanase CelD. Protein Eng. 1993, 6, 947–952. [DOI] [PubMed] [Google Scholar]
- 40. Murashima, K. , Kosugi, A. , Doi, R. H. , Synergistic effects of cellulosomal xylanase and cellulases from Clostridium cellulovorans on plant cell wall degradation. J. Bacteriol. 2003, 185, 1518–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bayer, E. A. , Morag, E. , Lamed, R. , The cellulosome‐‐a treasure‐trove for biotechnology. Trends Biotechnol. 1994, 12, 379–386. [DOI] [PubMed] [Google Scholar]
- 42. Cho, H. Y. , Yukawa, H. , Inui, M. , Doi, R. H. , Wong, S. L. , Production of minicellulosomes from Clostridium cellulovorans in Bacillus subtilis WB800. Appl. Environ. Microbiol. 2004, 70, 5704–5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mateo, C. , Fernandez‐Lorente, G. , Pessela, B. C. , Vian, A. et al., Affinity chromatography of polyhistidine tagged enzymes. New dextran‐coated immobilized metal ion affinity chromatography matrices for prevention of undesired multipoint adsorptions. J. Chromatogr. A 2001, 915, 97–106. [DOI] [PubMed] [Google Scholar]
- 44. Hyeon, J. E. , Jeon, W. J. , Whang, S. Y. , Han, S. O. , Production of minicellulosomes for the enhanced hydrolysis of cellulosic substrates by recombinant Corynebacterium glutamicum . Enzyme Microb. Technol. 2011, 48, 371–377. [DOI] [PubMed] [Google Scholar]
- 45. Oliveira, C. , Carvalho, V. , Domingues, L. , Gama, F. M. , Recombinant CBM‐fusion technology ‐ Applications overview. Biotechnol. Adv. 2015, 33, 358–369. [DOI] [PubMed] [Google Scholar]
- 46. Hyeon, J. E. , Kang, D. H. , Kim, Y. I. , Jeon, S. D. et al., Production of functional agarolytic nano‐complex for the synergistic hydrolysis of marine biomass and its potential application in carbohydrate‐binding module‐utilizing one‐step purification. Process Biochem. 2012, 47, 877–881. [Google Scholar]
- 47. Liang, Y. , Si, T. , Ang, E. L. , Zhao, H. , Engineered pentafunctional minicellulosome for simultaneous saccharification and ethanol fermentation in Saccharomyces cerevisiae . Appl. Environ. Microbiol. 2014, 80, 6677–6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jeon, S. D. , Yu, K. O. , Kim, S. W. , Han, S. O. , A celluloytic complex from Clostridium cellulovorans consisting of mannanase B and endoglucanase E has synergistic effects on galactomannan degradation. Appl. Microbiol. Biotechnol. 2011, 90, 565–572. [DOI] [PubMed] [Google Scholar]
- 49. Bayer, E. A. , Lamed, R. , Himmel, M. E. , The potential of cellulases and cellulosomes for cellulosic waste management. Curr. Opin. Biotechnol. 2007, 18, 237–245. [DOI] [PubMed] [Google Scholar]
- 50. Stern, J. , Morais, S. , Lamed, R. , Bayer, E. A. , Adaptor scaffoldins: An original strategy for extended designer cellulosomes, inspired from nature. MBio 2016, 7, e00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schuurmann, J. , Quehl, P. , Festel, G. , Jose, J. , Bacterial whole‐cell biocatalysts by surface display of enzymes: Toward industrial application. Appl. Microbiol. Biotechnol. 2014, 98, 8031–8046. [DOI] [PubMed] [Google Scholar]
- 52. Matano, Y. , Hasunuma, T. , Kondo, A. , Cell recycle batch fermentation of high‐solid lignocellulose using a recombinant cellulase‐displaying yeast strain for high yield ethanol production in consolidated bioprocessing. Bioresour. Technol. 2013, 135, 403–409. [DOI] [PubMed] [Google Scholar]
- 53. Kim, S. J. , Hyeon, J. E. , Jeon, S. D. , Choi, G. W. , Han, S. O. , Bi‐functional cellulases complexes displayed on the cell surface of Corynebacterium glutamicum increase hydrolysis of lignocelluloses at elevated temperature. Enzyme Microb. Technol. 2014, 66, 67–73. [DOI] [PubMed] [Google Scholar]
- 54. Hyeon, J. E. , Kim, S. W. , Park, C. , Han, S. O. , Efficient biological conversion of carbon monoxide (CO) to carbon dioxide (CO2) and for utilization in bioplastic production by Ralstonia eutropha through the display of an enzyme complex on the cell surface. Chem. Commun. 2015, 51, 10202–10205. [DOI] [PubMed] [Google Scholar]
- 55. Yang, J. , Dang, H. , Lu, J. R. , Improving genetic immobilization of a cellulase on yeast cell surface for bioethanol production using cellulose. J. Basic Microbiol. 2013, 53, 381–389. [DOI] [PubMed] [Google Scholar]
- 56. Fan, L. H. , Zhang, Z. J. , Yu, X. Y. , Xue, Y. X. , Tan, T. W. , Self‐surface assembly of cellulosomes with two miniscaffoldins on Saccharomyces cerevisiae for cellulosic ethanol production. Proc. Natl. Acad. Sci. USA 2012, 109, 13260–13265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kosugi, A. , Amano, Y. , Murashima, K. , Doi, R. H. , Hydrophilic domains of scaffolding protein CbpA promote glycosyl hydrolase activity and localization of cellulosomes to the cell surface of Clostridium cellulovorans . J. Bacteriol. 2004, 186, 6351–6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lee, S. Y. , Choi, J. H. , Xu, Z. , Microbial cell‐surface display. Trends Biotechnol. 2003, 21, 45–52. [DOI] [PubMed] [Google Scholar]
- 59. Nakayama, Y. , Yoshimura, K. , Iida, H. , Electrophysiological characterization of the mechanosensitive channel MscCG in Corynebacterium glutamicum . Biophys. J. 2013, 105, 1366–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Melville, S. , Craig, L. , Type IV pili in Gram‐positive bacteria. Microbiol. Mol. Biol. Rev. 2013, 77, 323–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liu, J. , Peng, J. , Shen, S. , Jin, Q. et al., Enzyme entrapped in polymer‐modified nanopores: The effects of macromolecular crowding and surface hydrophobicity. Chemistry 2013, 19, 2711–2719. [DOI] [PubMed] [Google Scholar]
- 62. Bhalla, A. , Bansal, N. , Kumar, S. , Bischoff, K. M. , Sani, R. K. , Improved lignocellulose conversion to biofuels with thermophilic bacteria and thermostable enzymes. Bioresour. Technol. 2013, 128, 751–759. [DOI] [PubMed] [Google Scholar]
- 63. Tsai, S. L. , DaSilva, N. A. , Chen, W. , Functional display of complex cellulosomes on the yeast surface via adaptive assembly. ACS Synth. Biol. 2013, 2, 14–21. [DOI] [PubMed] [Google Scholar]
- 64. You, C. , Zhang, X. Z. , Sathitsuksanoh, N. , Lynd, L. R. , Zhang, Y. H. P. , Enhanced microbial utilization of recalcitrant cellulose by an ex vivo cellulosome‐microbe complex. Appl. Environ. Microbiol. 2012, 78, 1437–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chen, Y. P. , Hwang, I. E. , Lin, C. J. , Wang, H. J. , Tseng, C. P. , Enhancing the stability of xylanase from Cellulomonas fimi by cell‐surface display on Escherichia coli . J. Appl. Microbiol. 2012, 112, 455–463. [DOI] [PubMed] [Google Scholar]
- 66. Guzik, U. , Hupert‐Kocurek, K. , Wojcieszynska, D. , Immobilization as a strategy for improving enzyme properties‐application to oxidoreductases. Molecules 2014, 19, 8995–9018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Solanki, K. , Gupta, M. N. , Simultaneous purification and immobilization of Candida rugosa lipase on superparamagnetic Fe3O4 nanoparticles for catalyzing transesterification reactions. New J. Chem. 2011, 35, 2551–2556. [Google Scholar]
- 68. Chang, R. H. Y. , Jang, J. , Wu, K. C. W. , Cellulase immobilized mesoporous silica nanocatalysts for efficient cellulose‐to‐glucose conversion. Green Chem. 2011, 13, 2844–2850. [Google Scholar]
- 69. Li, Y. , Wang, X. Y. , Zhang, R. Z. , Zhang, X. Y. et al., Molecular imprinting and immobilization of cellulase onto magnetic Fe3O4@SiO2 nanoparticles. J. Nanosci. Nanotechnol. 2014, 14, 2931–2936. [DOI] [PubMed] [Google Scholar]
- 70. Rodrigues, R. C. , Ortiz, C. , Berenguer‐Murcia, A. , Torres, R. , Fernandez‐Lafuente, R. , Modifying enzyme activity and selectivity by immobilization. Chem. Soc. Rev. 2013, 42, 6290–6307. [DOI] [PubMed] [Google Scholar]
- 71. Mohanpuria, P. , Rana, N. K. , Yadav, S. K. , Biosynthesis of nanoparticles: Technological concepts and future applications. J. Nanopart. Res. 2008, 10, 507–517. [Google Scholar]
- 72. Mishra, A. , Sardar, M. , Alpha‐amylase mediated synthesis of silver nanoparticles. Sci. Adv. Mater. 2012, 4, 143–146. [Google Scholar]
- 73. Mishra, A. , Kaushik, N. K. , Sardar, M. , Sahal, D. , Evaluation of antiplasmodial activity of green synthesized silver nanoparticles. Colloids Surf., B 2013, 111, 713–718. [DOI] [PubMed] [Google Scholar]
- 74. Mishra, A. , Sardar, M. , Cellulase assisted synthesis of nano‐silver and gold: Application as immobilization matrix for biocatalysis. Int. J. Biol. Macromol. 2015, 77, 105–113. [DOI] [PubMed] [Google Scholar]
- 75. Gupta, M. N. , Kaloti, M. , Kapoor, M. , Solanki, K. , Nanomaterials as matrices for enzyme immobilization. Artif. Cells Blood Substit. Immobil. Biotechnol. 2011, 39, 98–109. [DOI] [PubMed] [Google Scholar]
- 76. Ansari, S. A. , Husain, Q. , Potential applications of enzymes immobilized on/in nano materials: A review. Biotechnol. Adv. 2012, 30, 512–523. [DOI] [PubMed] [Google Scholar]
- 77. Vertegel, A. A. , Siegel, R. W. , Dordick, J. S. , Silica nanoparticle size influences the structure and enzymatic activity of adsorbed lysozyme. Langmuir 2004, 20, 6800–6807. [DOI] [PubMed] [Google Scholar]
- 78. Takimoto, A. , Shiomi, T. , Ino, K. , Tsunoda, T. et al., Encapsulation of cellulase with mesoporous silica (SBA‐15). Microporous Mesoporous Mater. 2008, 116, 601–606. [Google Scholar]
- 79. Myung, S. W. , You, C. , Zhang, Y. H. P. , Recyclable cellulose‐containing magnetic nanoparticles: Immobilization of cellulose‐binding module‐tagged proteins and a synthetic metabolon featuring substrate channeling. J. Mater. Chem. B 2013, 1, 4419–4427. [DOI] [PubMed] [Google Scholar]
- 80. Padmavathy, B. , Patel, A. , Kumar, R. V. , Ali, B. M. J. , Superparamagnetic nanoparticles based immunomagnetic separation‐multiplex polymerase chain reaction assay for detection of Salmonella . Sci. Adv. Mater. 2012, 4, 114–120. [Google Scholar]
- 81. Ahmad, R. , Sardar, M. , Immobilization of cellulase on TiO2 nanoparticles by physical and covalent methods: A comparative study. Indian J. Biochem. Biophysi. 2014, 51, 314–320. [PubMed] [Google Scholar]
- 82. Liese, A. , Hilterhaus, L. , Evaluation of immobilized enzymes for industrial applications. Chem. Soc. Rev. 2013, 42, 6236–6249. [DOI] [PubMed] [Google Scholar]
- 83. Prabhakar, D. , Kumari, V. , Islam, S. S. , Nanoporous silicon based electrochemical immunosensor. Sci. Adv. Mater. 2012, 4, 121–125. [Google Scholar]
- 84. Zhang, Y. W. , Jeya, M. , Lee, J. K. , L‐Ribulose production by an Escherichia coli harboring L‐arabinose isomerase from Bacillus licheniformis . Appl. Microbiol. Biotechnol. 2010, 87, 1993–1999. [DOI] [PubMed] [Google Scholar]
- 85. Gunnoo, M. , Cazade, P. A. , Galera‐Prat, A. , Nash, M. A. et al., Nanoscale engineering of designer cellulosomes. Adv. Mater. 2016, 28, 5619–5647. [DOI] [PubMed] [Google Scholar]
- 86. Liu, F. , Banta, S. , Chen, W. , Functional assembly of a multi‐enzyme methanol oxidation cascade on a surface‐displayed trifunctional scaffold for enhanced NADH production. Chem. Commun. 2013, 49, 3766–3768. [DOI] [PubMed] [Google Scholar]


