Abstract
Phosphorus MRSI (31P–MRSI) using a spiral‐trajectory readout at 7 T was developed for high temporal resolution mapping of the mitochondrial capacity of exercising human skeletal muscle.
The sensitivity and localization accuracy of the method was investigated in phantoms. In vivo performance was assessed in 12 volunteers, who performed a plantar flexion exercise inside a whole‐body 7 T MR scanner using an MR‐compatible ergometer and a surface coil. In five volunteers the knee was flexed (~60°) to shift the major workload from the gastrocnemii to the soleus muscle.
Spiral‐encoded MRSI provided 16–25 times faster mapping with a better point spread function than elliptical phase‐encoded MRSI with the same matrix size. The inevitable trade‐off for the increased temporal resolution was a reduced signal‐to‐noise ratio, but this was acceptable. The phosphocreatine (PCr) depletion caused by exercise at 0° knee angulation was significantly higher in both gastrocnemii than in the soleus (i.e. 64.8 ± 19.6% and 65.9 ± 23.6% in gastrocnemius lateralis and medialis versus 15.3 ± 8.4% in the soleus).
Spiral‐encoded 31P–MRSI is a powerful tool for dynamic mapping of exercising muscle oxidative metabolism, including localized assessment of PCr concentrations, pH and maximal oxidative flux with high temporal and spatial resolution.
Keywords: dynamic 31P–MRS, high energy phosphate, MRSI, skeletal muscle, spiral spectroscopic imaging, ultra‐high field
Abbreviations used
- 31P–MRI
phosphorus MRI
- 31P–MRS
phosphorus MRS
- 31P–MRSI
phosphorus MRSI
- ADP
adenosine diphosphate
- ATP
adenosine triphosphate
- FFT
fast Fourier transform
- FOV
field of view
- FWHM
full width at half maximum
- GL
gastrocnemius lateralis
- GM
gastrocnemius medialis
- GPC
glycerophosphocholine
- NA
number of averages
- PAD
peripheral arterial disease
- PCr
phosphocreatine
- PSF
point spread function
- SAR
specific absorption rate
- SNR
signal‐to‐noise ratio
- SOL
soleus
- SVS
single‐voxel spectroscopy
- TE*
acquisition delay
1. INTRODUCTION
Impaired energy metabolism in skeletal muscle mitochondria is indicative of muscular disorders (e.g. Duchenne muscular dystrophy1 or mitochondrial myopathy2), systemic metabolic diseases (e.g. diabetes mellitus3, 4) and cardiovascular diseases (e.g. peripheral arterial disease (PAD)5, 6, 7). These observations, together with the fact that evaluation of muscle oxidative metabolism provides insight into personal training status,8, 9 have led to a considerable methodological development in medical imaging for non‐invasive assessment of muscle energy metabolism.
In particular, phosphorus MRS (31P–MRS) has been established as a powerful tool for studies of energy metabolism.10, 11, 12, 13, 14, 15 More specifically, dynamic 31P–MRS, during exercise and recovery, allows direct estimation of the oxidative adenosine triphosphate (ATP) synthesis rate in challenged muscle, which reflects maximal mitochondrial capacity.12 Since high temporal resolution (of the order of seconds) is required to map the phosphocreatine (PCr) recovery curve, coarse signal localization restricted only by the sensitive volume of a surface coil is often used to retain sufficient signal‐to‐noise ratio (SNR) for dynamic 31P–MRS experiments.16, 17 Such localization cannot distinguish between muscle groups that are recruited differently in the performed exercise (e.g. soleus (SOL) and gastrocnemius during plantar flexion18, 19, 20, 21, 22, 23).
Single‐voxel MRS24, 25 or slab‐selective MRS26, 27 have been implemented to overcome this limitation and enable single‐muscle localization. However, spatially defined injuries, myopathies, or functional deficits in PAD can be overlooked if only a single predefined muscle location is examined. Thus, actual mapping approaches allowing for coverage of several muscle groups simultaneously are desirable. Frequency‐selective phosphorus MRI (31P–MRI) has been shown to provide information on PCr recovery kinetics from several muscles simultaneously,28, 29, 30 and was recently extended to provide information about pH dynamics during exercise.31 So far, these 31P–MRI techniques do not provide the same amount of information as 31P–MRS; e.g., there are no data on ATP signal intensity typically used for concentration quantification, or on other 31P metabolites. The temporal resolution of phosphorus MRSI (31P–MRSI) with Cartesian phase encoding is, however, insufficient for estimating mitochondrial capacity with high spatial resolution unless a dedicated exercise protocol (i.e. gated MRSI32, 33) is applied, which considerably complicates and prolongs the examination and may impose limitations, such as the requirement of only mild pH changes.
In this context, our aim was to develop and test a 31P–MRSI sequence using spiral readout trajectories with high temporal resolution for spatially resolved quantification of maximal oxidative ATP‐synthase flux in the muscles of the human calf during plantar flexion exercise. The performance of the proposed sequence was tested in a localization phantom and in healthy volunteers.
2. METHODS
2.1. Hardware
All MR measurements were performed on a 7 T MR system (Magnetom, Siemens Healthcare, Erlangen, Germany) using a 10 cm circular, dual‐tuned (31P/1H) surface‐coil (Rapid Biomedical, Rimpar, Germany). The MR system was equipped with an SC72 gradient system featuring 70 mT/m nominal gradient strength and 200 mT/m/s maximum gradient slew rate. For the dynamic in vivo experiments a dedicated plantar flexion ergometer (Trispect, Ergospect, Innsbruck, Austria) was used.
2.2. Sequence design
To accelerate the acquisition and hence increase the temporal resolution of our dynamic 31P–MRSI scans, we implemented a constant‐density spiral spectroscopic readout.34, 35 Following each slice‐selective excitation (achieved by a Hamming filtered SINC pulse with 600 μs duration and 6.4 kHz bandwidth), we played out consecutive and identical spiral gradient waveforms (n = 512 divided by the number of temporal interleaves) in the read (x) and phase (y) gradient directions that fully covered the k‐t space. The spiral trajectories for all excitations were identical. Since the duration of the spiral trajectories for the given voxel size was longer than the temporal sampling required for the desired spectral bandwidth, temporal interleaving (i.e. shifting the gradient trajectory for consecutive T R by a fraction of the dwell time) was employed (Figure 1) (e.g., for five temporal interleaves at 1.45 kHz spectral bandwidth 103 spirals of about 3.4 ms length per spiral were played out per interleave, leading to a total sampling duration of about 353 ms, with each interleave being delayed by 0.69 ms relative to its predecessor). To optimize the SNR, gradient rewinders (i.e. deadtime during readout that is necessary to return to the k‐space center) were minimized (<6%). Hence, only temporal interleaves were used and not spatial interleaves. The number of temporal interleaves was adjusted to the targeted matrix size, making sure that the spectral bandwidth remained between 1.4 and 1.45 kHz (i.e. covering a range of at least 12 ppm from Pi to γ‐ATP resonances). The achieved spectral resolution, i.e. the spectral bandwidth over the number of acquired spirals, was therefore always 2.7–2.8 Hz. The entire spiral trajectory calculation and data reconstruction were implemented on the MR scanner. The following four steps were performed within the online processing pipeline: (i) gradient delays between the ADC readout and the x/y gradients of 8 μs were corrected by shifting the sampled data inside the array; (ii) the acquired data were gridded two‐dimensionally to a twofold oversampled Cartesian matrix with a Kaiser–Bessel kernel, (iii) a fast Fourier transform (FFT) was applied in the x/y dimension, and then (iv) an FFT was used in the frequency dimension.
Figure 1.
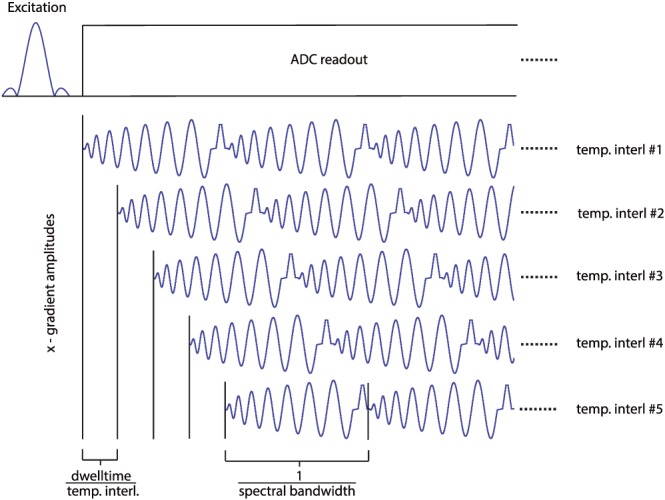
In‐plane constant density spiral readout for FOV 200 × 200 mm2 and 14 × 14 matrix with temporal interleaving. The slice‐selective excitation pulse is followed by the spiral gradient modulations (only x ‐direction shown). These spiral trajectories are played out repeatedly (512 times; only three shown for illustration) to cover the whole free induction decay in the time domain. As the duration of a spiral is too long to allow a sufficiently short spectral dwell time, temporal interleaves (five temporal interleaves are shown here for illustration) were acquired. Each temporal interleave is acquired after a separate excitation, and identical spiral gradients are played out with a predefined delay. This delay determines the actual dwell time and hence the spectral readout bandwidth. The number of temporal interleaves used in the sequence was derived as the minimum number required to cover the desired spectral bandwidth of 1.4 kHz
2.3. Phantom experiments
The point spread function (PSF) of the proposed MRSI sequence was measured in a two‐compartment phantom. The outer compartment (a cylinder with a diameter of 13 cm) was filled with tap water, and the inner compartment (a cube with 9 mm inner side length) was filled with a Pi solution (concentration = 1 mol/L). The cube was fixed in the center of the cylinder, 3 cm from its base. Three 2D–MRSI datasets (slice thickness = 30 mm) were collected in the transverse plane, perpendicular to the RF coil and covering the inner Pi‐filled compartment entirely, using (i) standard 2D–MRSI with fully sampled Cartesian k‐space (phase encoded), (ii) elliptically sampled Cartesian k space, and (iii) constant density spiral k‐space sampling. The acquisition parameters were matched for all three sequences: acquisition delay (T E*) = 1 ms; T R = 2 s; nominal flip angle =90°; field of view (FOV) = 200 × 200 mm2; acquisition matrix size =14 × 14. In the first step, the number of averages (NA) was set to 1. The corresponding acquisition times were 6 min 32 s for full phase encoding, 3 min 46 s for elliptical phase encoding, and 10 s for spiral encoding. To compare the spatial selectivities of all sampling approaches, the full width at half maximum (FWHM) of the PSF was measured. Additionally, to compare the SNRs of the MRSI sequences with elliptical phase‐encoding and spiral encoding, the NA of the spiral‐encoded sequence was increased to match the acquisition time of the elliptical Cartesian encoding (i.e., NA = 22, acquisition time = 3 min 40 s).
2.4. In vivo measurements
To test the potential temporal and spatial resolution of the proposed sequence in an in vivo situation, one healthy male volunteer underwent repeated measurements of the calf muscle. The volunteer lay supine on the MR table and the RF coil was placed under the right gastrocnemius muscle. The matrix sizes used and corresponding numbers of required temporal interleaves are listed in Table 1. Other parameters were set as follows: T E* = 1 ms; T R = 2 s; FOV = 200 × 200 mm2; slice thickness = 30 mm; vector size/total number of spirals for all temporal interleaves =512 points/spirals; NA = 1, 4, and 16. The SNR of PCr signal was calculated for each matrix size and NA.
Table 1.
The relation between the matrix size (i.e. spatial resolution) and the number of temporal interleaves required (i.e. temporal resolution). For comparison, the temporal resolution of an elliptically phase‐encoded MRSI (ePE) is also given. Note that the increase in temporal resolution comes at the cost of SNR, which is, however, comparable once the acquisition time is matched and the differences in PSF between spiral and elliptical encoding are corrected for
| Matrix size | Spatial resolution (mm3) | No of temporal interleaves | Temporal resolution (s) | |
|---|---|---|---|---|
| Spirals | ePE | |||
| 10 × 10 | 20 × 20 × 30 | 3 | 6 | 98 |
| 12 × 12 | 17 × 17 × 30 | 4 | 8 | 162 |
| 14 × 14 | 14 × 14 × 30 | 5 | 10 | 226 |
| 16 × 16 | 12 × 12 × 30 | 6 | 12 | 298 |
Twelve young, healthy individuals (nine males/three females, age (mean ± standard deviation) 28.7 ± 4.1 years, BMI (body mass index) 23.2 ± 2.6 kg/m2, no professional athletes) agreed to participate in the dynamic part of this study and signed a consent form approved by the institutional ethical board. Each volunteer lay supine on the ergometer, inside the MR system, with the RF coil strapped below their right calf. The knee of the volunteers was fully extended during the dynamic examination (2 min rest, 6 min exercise and 6 min recovery) to ensure a major involvement of the gastrocnemius muscles and only a minor contribution of the SOL muscle to the exercise performed.20, 21, 22 The volunteers performed plantar flexions at a workload set to about 25–35% of the maximal voluntary contraction force, once every T R (2 s). The exercise was synchronized with the data acquisition based on an audio signal, so that the MRSI data were acquired when the calf muscle was relaxed. In addition, five of the recruited male volunteers underwent a second dynamic examination with the knee flexed at about 60°. In this leg position the SOL muscle is expected to be involved more than the two gastrocnemii.20 This second examination was performed at least 20 min after the first one, to ensure sufficient metabolic recovery.36 Two additional spiral‐MRSI measurements were performed at rest in two of these subjects in order to acquire representative flip angle maps37 in both leg positions.
For both dynamic examinations, the proposed acquisition sequence was used, with the following parameters: T E* = 1 ms; T R = 2 s; nominal flip angle =52° (set about 2.5 cm deep from the coil, by recalculating the voltage that was required to obtain 90° excitation, i.e. maximum signal38); acquisition bandwidth =1450 Hz; vector size =512 points; four dummy scans. With matrix size =14 × 14, FOV = 200 × 200 mm2 and slice thickness = 30 mm, the nominal spatial resolution of the dynamic experiments was 14.3 × 14.3 × 30 mm3 (6.13 mL). The excitation frequency was centered at 180 Hz (~1.5 ppm) relative to PCr. The T R values used for the flip angle map acquisition were 2 s and 8 s. Two averages were acquired and the rest of the sequence parameters were kept the same as for the dynamic experiments.
2.5. Data analysis
All acquired MRSI data were interpolated to 16 × 16 matrixes, and voxels with SNR of the PCr signal above 8 in the resting state before exercise (no k‐space filtering was applied prior to data analysis) were pre‐selected for quantification of dynamic in vivo data. Custom software written in IDL (Exelis Visual Information Solutions, Boulder, CO, USA) was used for pre‐selection of these voxels. All selected spectra were analyzed using the jMRUI software (Version 5.0) with the AMARES (advanced method for accurate, robust, and efficient spectral fitting) time domain fitting routine.39 The γ‐ATP resonance was used as an internal concentration reference, assuming a stable cellular ATP concentration of 8.2 mM.40 To improve the SNR of the γ‐ATP peak for quantification purposes, the γ‐ATP signal intensity was averaged over the last six measurements (1 min) of the recovery period. For the saturation correction, the actual flip angle was calculated for each muscle group as the average of the measured datasets, and the previously measured T 1 values41 were used. The chemical shift of Pi relative to PCr was used to calculate the intracellular pH,42 according to the modified Henderson‐Hasselbalch equation.40
During the recovery period, a mono‐exponential function was fitted to the PCr signal time course using custom routine in MATLAB (MathWorks, Natick, MA, USA), yielding the time constant of the PCr recovery rate (τ PCr). The τ PCr was used to calculate the initial PCr recovery rate (V PCr), as reported before.15 The maximal rate of oxidative phosphorylation (Q max) was calculated according to the adenosine diphosphate (ADP)‐based model of Michaelis and Menten, for which the ADP concentration was calculated according to Kemp et al.43
The SNR was calculated based on the magnitude of the signal from the voxel covering the cubic phantom for the phantom experiments, comparing the SNR of elliptical and spiral encodings matched for acquisition time, and similarly by using the magnitude of PCr signal in voxels within a 5 × 3 area covering the skeletal muscle tissue for the in vivo experiment that compared the SNR for different spatial resolutions. The standard deviation of noise evaluated from 100 points of the spectra far from the metabolite signals was used for SNR calculation.
To compare the oxidative metabolism between the three muscle groups of interest—gastrocnemius medialis (GM), gastrocnemius lateralis (GL) and SOL—the muscles were manually segmented using 1H localizer images. Each segmented volume of interest contained 3–5 voxels, leaving at least one voxel free between the adjoining muscles. One way analysis of variance (ANOVA) with a Tukey post hoc test was consecutively applied for statistical analysis, considering p < 0.05 statistically significant. A paired t test was used to compare the measures of mitochondrial function determined for the exercise with straightened (0°) and bent (60°) knee, with the same level of significance (p < 0.05).
3. RESULTS
Spiral k‐space‐sampled MRSI provided 27% lower SNR than the elliptical‐encoded MRSI with the same acquisition time, i.e. 279.26 compared with 384.25 for spiral sampling (NA = 22) and elliptical phase encoding (NA = 1), respectively. However, the effective voxel size of the spiral‐encoded MRSI was 30% smaller, as the PSF area at FWHM was 3.5 cm2 versus 4.9 cm2 for the spiral and elliptical encoding, respectively. The PSF area at FWHM of the main lobe was 1.6 cm2 for the fully encoded k‐space. The results of the phantom PSF experiments are depicted in Figure 2.
Figure 2.
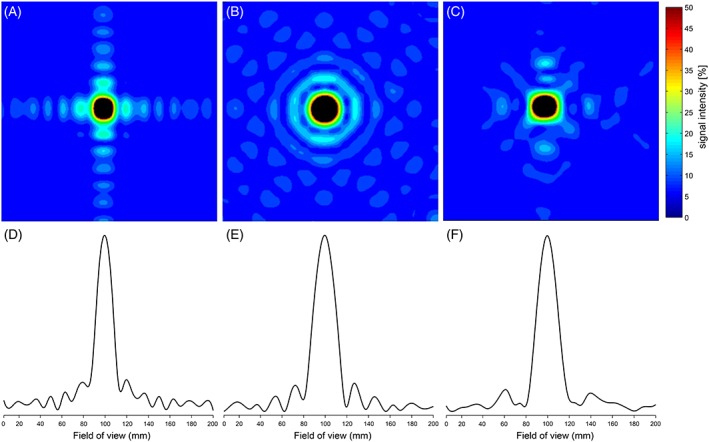
Experimentally determined PSFs for different k‐space sampling approaches for slice‐selective 2D–MRSI with a 14 × 14 matrix using a point source (i.e. inorganic phosphate solution) depicted as 2D maps (A‐C) and 1D plots (D‐F): A,D, full phase‐encoded k‐space sampling; B,E, elliptically phase‐encoded k‐space sampling; C,F, constant‐density spiral sampling with no spatial, but five temporal, interleaves. Constant‐density spirals featured a reduced number of side‐lobes in comparison with full phase encoding acquisition and provided smaller PSF than the elliptical encoding alternative (3.5 cm2 versus 4.9 cm2). All signal intensities are cut at 50% of the maximum signal intensity in the 2D maps to emphasize the FWHM. Note that all maps and plots are given in absolute values, i.e. all side‐lobes are depicted as positive
The SNR comparison between different matrix sizes and temporal resolutions measured in vivo at rest is given in Table 2. Depending on the chosen matrix size, spiral MRSI encoding improved the minimum temporal resolution compared with elliptical phase‐encoded MRSI by a factor of 16 (10 × 10 matrix) or up to 25 (16 × 16 matrix). For more details see Table 1. A 14 × 14 matrix size was selected for the dynamic experiments, as a good compromise between spatial (effective voxel size 7.56 mL, i.e. cylinder with a base equal to 64% of the PSF area44 and height equal to the slice thickness) and temporal (10 s) resolution with sufficient SNR.
Table 2.
The average SNR of PCr peak in a 5 × 3 area within the skeletal muscle for different matrix sizes for the spiral‐encoded MRSI
| NA | 10 × 10 | 12 × 12 | 14 × 14 | 16 × 16 |
|---|---|---|---|---|
| 1 | 26.9 ± 8.6 | 21.8 ± 6.7 | 14.7 ± 6.5 | 11.6 ± 4.8 |
| 4 | 50.8 ± 17.2 | 42.5 ± 13.3 | 26.8 ± 13.9 | 23.1 ± 10.3 |
| 16 | 101.4 ± 31.5 | 82.0 ± 27.7 | 52.9 ± 23.6 | 45.4 ± 20.9 |
Representative spectra acquired at rest and at the end of exercise from each investigated muscle group are depicted in Figure 3. To demonstrate the importance of 31P signal localization in dynamic experiments, a voxel containing a mixture of GM and SOL tissue is also visualized (green). Note that in such voxels of mixed tissue splitting of the Pi signal, which corresponds to two compartments with different pH values, can be observed.
Figure 3.
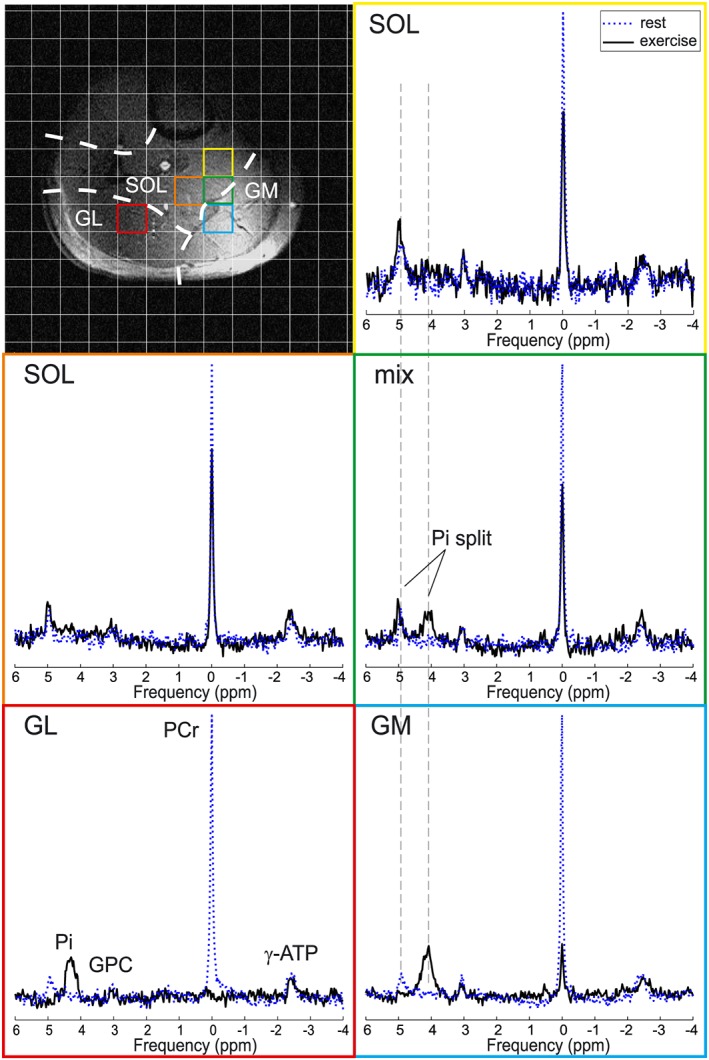
Representative 31P MR spectra acquired at rest (blue dotted line) and at the end of exercise (black solid line) in voxels representing single muscles, i.e. GL, GM and SOL, or a mixture of GM and SOL tissue. Note the Pi splitting in the mixed (green) voxel. A matched filter (6 Hz Lorentzian) was applied for visualization purposes only
The time evolution of PCr and Pi signals—in GM, GL and SOL—during plantar flexion exercise with a straightened knee is depicted in Figure 4 (top panel). The time evolution of pH in these muscles is also included in Figure 4 (bottom panel). The mean PCr depletion of 15.3 ± 8.4% measured in the SOL muscle was significantly lower (p < 0.01) than that in GM and GL (65.9 ± 23.6% and 64.8 ± 19.6%, respectively). Similarly, significantly lower V PCr and Q max values were calculated for the SOL muscle in comparison with both gastrocnemii in this type of exercise. All measured and evaluated measures of mitochondrial function are listed in Table 3.
Figure 4.
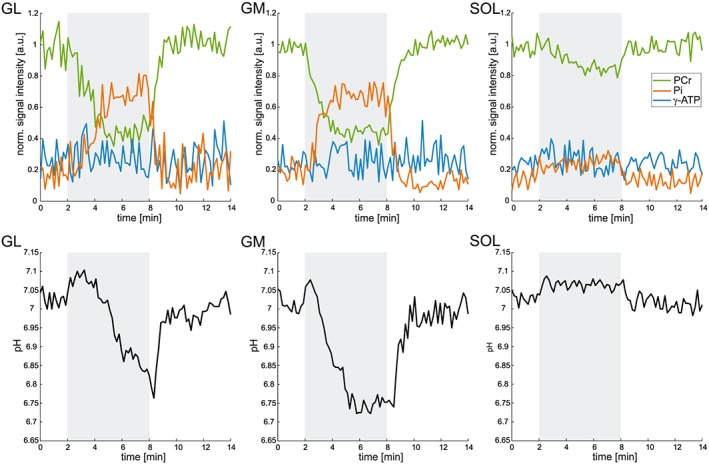
Time courses of the PCr (green), Pi (orange) and ATP (blue) signal intensities, normalized to the resting PCr signal intensity (top panel), and of pH (bottom panel), measured in all investigated muscles (2–4 voxels). The higher PCr depletion and pH drop in GL and GM indicate the prevailing involvement of these muscles in the plantar flexion exercise (0° flexion of the knee). The exercise period (6 min) is shaded grey
Table 3.
31P–MR measures of mitochondrial function measured and derived for GL, GM and SOL from the dynamic examination with straightened knee (n = 12)
| GL | GM | SOL | |
|---|---|---|---|
| PCr [mM] | 34.4 ± 4.2 | 37.0 ± 7.1 | 39.3 ± 5.4 |
| PCr drop [%] | 64.8 ± 19.6 | 65.9 ± 23.6 | 15.3 ± 8.4* , § |
| τ PCr [s] | 60.1 ± 21.4 | 63.1 ± 20.8 | 66.2 ± 16.0 |
| pH end_exercise | 6.79 ± 0.13 | 6.77 ± 0.18 | 6.93 ± 0.10# |
| V PCr [mM/s] | 0.41 ± 0.12 | 0.42 ± 0.16 | 0.10 ± 0.05* , § |
| Q max [mM/s] | 0.58 ± 0.17 | 0.57 ± 0.20 | 0.31 ± 0.10* , § |
Data are given as mean ± standard deviation. Significant differences between muscle groups are depicted as follows:
GL versus SOL (p < 0.01),
GM versus SOL (p < 0.01) and
GM versus SOL (p < 0.05).
Figure 5 depicts PCr depletion maps measured at the end of the exercise period for both knee positions (0° versus ~60° flexion). A clear difference in the involvement of the investigated muscle groups can be observed. While in the first case (0° flexion) the level of PCr in the SOL muscle is almost unaffected, during the second exercise (~60° flexion) the SOL muscle is the one with the highest PCr depletion. The difference in exercise‐induced drop in PCr signal between the two dynamic experiments was found to be statistically significant (p < 0.05) for all three muscle groups (i.e. lower for both gastrocnemii and higher for SOL in the second exercise). Significantly shorter τ PCr and higher Qmax were calculated for the SOL muscle in the second experiment; however, there were no statistical differences in these parameters found between the two experiments for either GM or GL. Table 4 summarizes the direct comparison of the measured and interpreted measures of mitochondrial function using these two exercise protocols.
Figure 5.
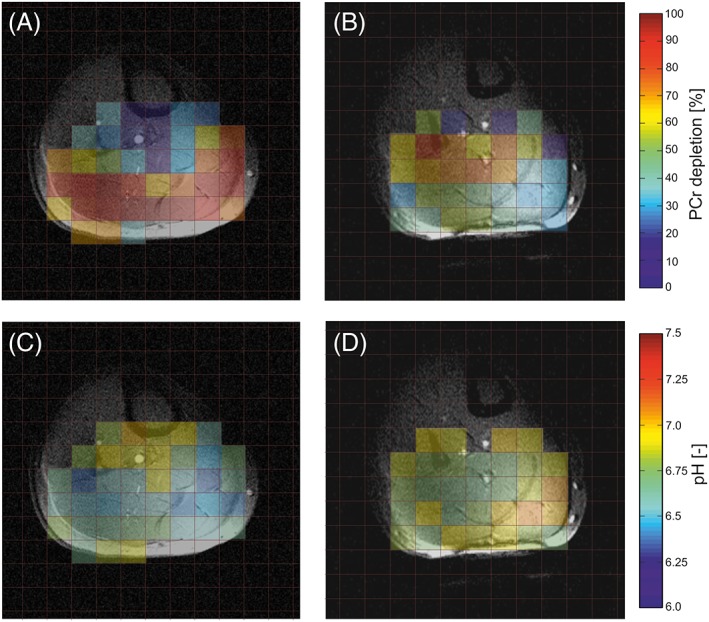
PCr depletion A,B and pH C,D maps of the same subject as a result of a plantar flexion exercise performed with a straight knee A,C and with the knee flexed by about 60° B,D. Note that in A,C most work is performed by the two superficially located gastrocnemii and the deeper situated SOL muscle is not much involved, as demonstrated by higher PCr depletions and lower pH values, whereas in B,D these roles are reversed
Table 4.
31P–MR measures of mitochondrial function measured and derived from the dynamic examinations in five subjects with different knee angulations (0° and ~60°) (n = 5)
| GL | GM | SOL | ||||
|---|---|---|---|---|---|---|
| Knee flexion [°] | 0 | 60 | 0 | 60 | 0 | 60 |
| PCr drop [%] | 75.4 ± 17.0 | 24.2 ± 12.7§ | 68.4 ± 24.9 | 20.4 ± 9.2§ | 18.0 ± 8.5 | 43.2 ± 19.1* |
| τ PCr [s] | 60.8 ± 10.1 | 57.7 ± 24.9 | 58.5 ± 21.7 | 58.7 ± 11.9 | 70.3 ± 12.0 | 42.5 ± 6.9§ |
| pH end_exercise | 6.76 ± 0.15 | 6.94 ± 0.07* | 6.76 ± 0.20 | 7.00 ± 0.05* | 6.93 ± 0.14 | 6.93 ± 0.14 |
| V PCr [mM/s] | 0.47 ± 0.08 | 0.24 ± 0.13* | 0.43 ± 0.18 | 0.21 ± 0.09* | 0.12 ± 0.07 | 0.47 ± 0.27* |
| Q max [mM/s] | 0.60 ± 0.13 | 0.44 ± 0.18 | 0.59 ± 0.22 | 0.45 ± 0.15 | 0.33 ± 0.14 | 0.80 ± 0.34* |
Data are given as mean ± standard deviation. Significant differences between exercise protocols are depicted as follows:
p < 0.05 and
p < 0.01.
4. DISCUSSION
In our study, we propose dynamic spiral‐accelerated 31P–MRSI as an efficient method for simultaneous mapping of the temporal changes in PCr and intracellular pH during plantar flexion exercise of the calf muscle at 7 T. This method allows the assessment of muscle‐specific oxidative metabolism during and after exercise. The performance of our rapid spiral‐encoded 31P–MRSI method and its feasibility for dynamic experiments was investigated in phantom experiments and in vivo in a group of 12 healthy volunteers. Using this rapid technique we were also able to show the shift of the workload from the gastrocnemius to the SOL muscle resulting from different knee angulation during plantar flexion exercise. This is demonstrated by differences in the PCr drop between respective muscle groups in our two exercise set‐ups.
The conventional approach to dynamic 31P–MRS of using the sensitive volume of the RF coil for signal localization16, 17 restricts investigations to protocols with equal muscle involvement or to studies on superficial muscle groups only. Previous reports indicated that acquisition of signals from several muscles with different levels of recruitment risks biasing the results, as it does not reflect the real metabolic status of the individual muscles27, 45 and can affect the comparability of results between sites.46 Thus, several 31P–MR localization methods attempting to improve spatial specificity have recently emerged. On one hand, this includes 31P–MRS techniques, focused on single muscles.24, 25, 45 On the other hand, 31P–MRI techniques aim to map oxidative metabolism over the whole FOV simultaneously.28, 29, 30, 31, 47
Single‐voxel spectroscopy (SVS) provides an accurate localization of a cuboid volume as long as selective RF‐pulses with broad bandwidth are applied.48 However, SVS typically targets only a single muscle at a time. An interleaved technique providing information from two voxels within one dynamic experiment was suggested recently,23 but this still lacks the high spatial resolution of our MRSI approach. Furthermore, accurate spatial selection of SVS at 7 T requires relatively long T E, thus reducing sensitivity for important 31P metabolites such as ATP.25 The proposed MRSI approach is FID based, and therefore does not require such long T E.
31P–MRI techniques, recently proposed for mapping of muscle oxidative metabolism,28, 29, 30, 31, 47 sacrifice spectral selectivity in favor of spatial specificity. The disadvantage of mapping of single‐metabolite (i.e. PCr) resonances is that information about pH is lost. A simultaneous frequency‐selective 31P–MRI of PCr and Pi can utilize the phase maps of Pi to calculate pH.31 However, such pH measurements are slightly biased if the phase information is not derived from Pi only. Our sampling approach benefits from the excellent spectral resolution that is maintained thanks to the use of temporal interleaving, and thus pH can be accurately determined from the frequency difference between PCr and Pi 40, 42 in the same way as in conventional 31P–MRSI. Even the ATP signal is mapped simultaneously, which can serve as an important concentration reference to directly calculate Q max, and thus obtain an interpretation of mitochondrial capacity.
Earlier dynamic 31P–MRSI experiments were limited by slow Cartesian phase encoding and, therefore, required lengthy exercise protocols (e.g. gated 31P–MRSI with 16 min of exercise) and enabled only relatively low spatial resolution using an 8 × 8 matrix (10.5 mL nominal voxel size).32 On the other hand, the effective temporal resolution achieved with such protocols is equal to the T R used, i.e. 2.1 s in the aforementioned study. Here we report an MRSI sequence with spiral‐trajectory encoding with a 14 × 14 matrix (6.1 mL nominal and 7.56 mL effective spatial resolution) and 10 s temporal resolution. Besides the increased encoding speed of our spiral‐MRSI, which supported a 23 times higher temporal resolution in comparison with the elliptical phase‐encoded MRSI, the spiral‐MRSI also led to an improved PSF (~30% smaller area at FWHM). Although the increased temporal resolution comes at the cost of SNR, after matching the acquisition times and correcting for the differences in PSF the SNRs achieved with the spiral‐encoded MRSI and with the elliptical‐encoded MRSI were comparable. The difference in PSF area measured in the phantom experiments is in good agreement with the results of Andronesi et al.,35 even though that the experimentally determined PSF for spiral encoding looks a little different from the simulations recently provided for 13C–MRSI.49 This was probably caused by the gridding process.
Comparing our spatial and temporal resolution with 31P–MRI studies, Schmid et al. proposed interleaved PCr/Pi mapping using a 3D spoiled gradient echo sequence with a 16 × 16 × 4 matrix (3 mL nominal spatial resolution) and 10 s temporal resolution.31 Parasoglou et al. suggested a PCr‐only frequency‐selective compressed sensing accelerated 3D turbo‐spin echo sequence with a 24 × 24 × 8 matrix (1.6 mL nominal spatial resolution) and 12 s temporal resolution.30 Greenman and Smithline reported 2D PCr imaging with a 20 × 20 matrix (5.6 mL nominal spatial resolution) and 6 s temporal resolution at 3 T.28 Our spectral‐spatial encoding approach via spiral spectroscopic imaging, therefore, merges the spectral specificity of 31P–MRS and spatial selectivity of 31P–MRI. In particular, our spiral‐MRSI sequence: (i) provides full signal intensity detection for all metabolites within the spectral bandwidth via direct acquisition of the FID; (ii) features the same high spectral resolution as conventional non‐localized 31P–MRS sequences; and (iii) provides accelerated spatial encoding similar to 31P–MRI for metabolic mapping.
Spectral bandwidth and temporal resolution can be balanced as required, while maintaining the full SNR per unit time (SNR/t). Theoretically, the use of rewinder gradients (i.e. gradients that are necessary to return to the center of k space) can reduce SNR/t efficiency. However, we have minimized this source of SNR loss by maximizing the number of spectral interleaves and eliminating the need for spatial interleaves. Thus, the necessary rewinder is very short, which restricts the SNR/t loss to well below 5%. Our phantom measurements also validate that the SNR/t values for elliptically phase‐encoded and spiral‐encoded MRSI were nearly identical. The entire image reconstruction is implemented online. Thus, spectra could be directly inspected/processed on the MR scanner.
The PCr depletions and τ PCr values obtained for the different muscles in the scope of this study are in good agreement with those reported previously in healthy volunteers.21, 22, 31 The significantly lower PCr drop in the SOL muscle, in comparison with both gastrocnemii, during typical plantar flexion exercise with straightened knee points towards its low involvement in this type of exercise. Minimal muscle recruitment in the performed exercise, alongside with the substantial distance of SOL from the RF coil used, yielded less reliable V PCr and Q max values for this muscle. A trend towards lower Q max in SOL for plantar flexion with a straight knee was also observed by Fiedler et al.21 This is also supported by our finding that once the recruitment of SOL was substantially increased through the flexion of the knee, as suggested by Price et al.20 and as confirmed by the significantly higher PCr depletion (43.2 ± 19.1% at ~60° flexion versus 18.0 ± 8.5% at 0°) in our study, the Q max measured in SOL rose significantly. Although both GL and GM were recruited to a lesser extent and experienced significantly lower PCr depletion in the second experiment, Q max was not found to be significantly different between the measurements for either of these muscle groups.
The use of a surface coil does introduce FA variability within the MRSI matrix. To overcome this, B 1 +‐insensitive adiabatic excitation could be used. However, slice‐selective adiabatic excitation pulses needed for 2D–MRSI would be extremely long and specific absorption rate (SAR) demanding, requiring prolonged T R. Another option would be to use non‐selective adiabatic excitation combined with slice‐selective adiabatic refocusing or inversion pulses. These approaches would lead to undesirably prolonged T E and increased SAR, or towards doubled acquisition time.50 Therefore, none of these possibilities constitutes a good option for dynamic examinations. Alternatively, it is possible to compensate for the B 1 + inhomogeneity of a surface coil by acquiring a map of the actual flip angle distribution, as was performed in this study using the method of Chmelík et al.37 The use of RF coils with a more homogeneous B 1 + distribution29, 51 would also be beneficial.
The spiral spectroscopic imaging readout is demanding for the scanner gradient hardware. This will induce slow frequency drifts over time, as reported previously.52 This is not a severe limitation for dynamic 31P–MRSI with high temporal resolution, because accurate investigation of the creatine kinase metabolism does not rely on good frequency stability, but frequency drift could be a problem if this readout is combined with frequency selective pulses. This should be kept in mind when spiral spectroscopic imaging or other fast spectral‐spatial encoding methods are combined with saturation transfer techniques.38, 53, 54 The use of interleaved navigators was shown to be an excellent way of correcting frequency drifts for spiral spectroscopic imaging when combined with frequency‐selective MEGA‐editing pulses.55 With a recently suggested approach it might be possible to use interleaved 1H navigators also for 31P–MRSI on common MRI systems.56 The spiral MRSI readout proposed in our study could potentially lead to small artifacts when the changes in metabolite concentrations occurring during the dynamic protocol happen in between the acquisitions of individual temporal interleaves. These would originate from the largest signal in the spectrum, which is in our case PCr. However, we did not observe any during our test measurements in vivo, thus these theoretically present artifacts seem to be at or below the level of noise. Nevertheless, the selected bandwidth and number of temporal interleaves should ensure that, even if present, they would not overlap with any visible 31P resonances. Another potential source of artifacts relates to the narrow acquisition bandwidth used. Metabolite signals that are outside the acquisition bandwidth of the spiral MRSI sequence, but are excited by the excitation pulse, such as α‐ATP, could be aliased into the acquired spectra. However, these aliased signals would have only a fraction of the original signal amplitude, due to PSF blurring of signals from outside the spectral width,57 and therefore were not observed in our in vivo data.
We conclude that spiral‐encoded 31P–MRSI is a powerful tool for dynamic mapping of the muscle oxidative metabolism, including localized assessment of PCr concentration, pH and ATP reference concentration with a high temporal and spatial resolution. This not only allows the assessment of muscle‐specific mitochondrial capacity for all muscles within a single slice in physiological studies, but has also great potential for identifying small, i.e. early staged, localized pathologic changes in injuries, myopathies or PAD.
ACKNOWLEDGEMENTS
This work was supported by the Christian Doppler Society—Clinical Molecular MR Imaging (MOLIMA to S.T.), by the OeNB Jubilaeumsfond (Grant 15363 to M.K., 15455 to L.V. and 16133 to W.B.), by the Austrian Science Fund FWF (Grant I1743‐B13 to M.M.), by the Slovak Grant Agency (VEGA Grant 2/0013/14 and APVV Grant 0431–12) and by a Sir Henry Dale Fellowship from the Wellcome Trust and the Royal Society (Grant 098436/Z/12/Z to C.T.R.).
Valkovič, L. , Chmelík, M. , Meyerspeer, M. , Gagoski, B. , Rodgers, C. T. , Krššák, M. , Andronesi, O. C. , Trattnig, S. , and Bogner, W. (2016) Dynamic 31P–MRSI using spiral spectroscopic imaging can map mitochondrial capacity in muscles of the human calf during plantar flexion exercise at 7 T. NMR Biomed., 29: 1825–1834. doi: 10.1002/nbm.3662.
REFERENCES
- 1. Newman RJ, Bore PJ, Chan L, et al. Nuclear magnetic resonance studies of forearm muscle in Duchenne dystrophy. Br Med J (Clin Res Ed). 1982;284:1072–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Taivassalo T, Shoubridge EA, Chen J, et al. Aerobic conditioning in patients with mitochondrial myopathies: physiological, biochemical, and genetic effects. Ann Neurol. 2001;50:133–141. [DOI] [PubMed] [Google Scholar]
- 3. Chibalin AV, Yu M, Ryder JW, et al. Exercise‐induced changes in expression and activity of proteins involved in insulin signal transduction in skeletal muscle: differential effects on insulin‐receptor substrates 1 and 2. Proc Natl Acad Sci U S A. 2000;97:38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scheuermann‐Freestone M, Madsen PL, Manners D, et al. Abnormal cardiac and skeletal muscle energy metabolism in patients with type 2 diabetes. Circulation. 2003;107:3040–3046. [DOI] [PubMed] [Google Scholar]
- 5. Pipinos II, Shepard AD, Anagnostopoulos PV, Katsamouris A, Boska MD. Phosphorus 31 nuclear magnetic resonance spectroscopy suggests a mitochondrial defect in claudicating skeletal muscle. J Vasc Surg. 2000;31:944–952. [DOI] [PubMed] [Google Scholar]
- 6. Greiner A, Esterhammer R, Messner H, et al. High‐energy phosphate metabolism during incremental calf exercise in patients with unilaterally symptomatic peripheral arterial disease measured by phosphor 31 magnetic resonance spectroscopy. J Vasc Surg. 2006;43:978–986. [DOI] [PubMed] [Google Scholar]
- 7. Pedersen BL, Baekgaard N, Quistorff B. Muscle mitochondrial function in patients with type 2 diabetes mellitus and peripheral arterial disease: implications in vascular surgery. Eur J Vasc Endovasc Surg. 2009;38:356–364. [DOI] [PubMed] [Google Scholar]
- 8. Layec G, Bringard A, Le Fur Y, et al. Comparative determination of energy production rates and mitochondrial function using different 31P MRS quantitative methods in sedentary and trained subjects. NMR Biomed. 2011;24:425–438. [DOI] [PubMed] [Google Scholar]
- 9. Valkovič L, Ukropcová B, Chmelík M, et al. Interrelation of 31P‐MRS metabolism measurements in resting and exercised quadriceps muscle of overweight‐to‐obese sedentary individuals. NMR Biomed. 2013;26:1714–1722. [DOI] [PubMed] [Google Scholar]
- 10. Chance B, Eleff S, Leigh JS Jr, Sokolow D, Sapega A. Mitochondrial regulation of phosphocreatine/inorganic phosphate ratios in exercising human muscle: a gated 31P NMR study. Proc Natl Acad Sci U S A. 1981;78:6714–6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bottomley PA, Charles HC, Roemer PB, et al. Human in vivo phosphate metabolite imaging with 31P NMR. Magn Reson Med. 1988;7:319–336. [DOI] [PubMed] [Google Scholar]
- 12. Kemp GJ, Radda GK. Quantitative interpretation of bioenergetic data from 31P and 1H magnetic resonance spectroscopic studies of skeletal muscle: an analytical review. Magn Reson Q. 1994;10:43–63. [PubMed] [Google Scholar]
- 13. Prompers JJ, Jeneson JA, Drost MR, Oomens CC, Strijkers GJ, Nicolay K. Dynamic MRS and MRI of skeletal muscle function and biomechanics. NMR Biomed. 2006;19:927–953. [DOI] [PubMed] [Google Scholar]
- 14. Rodgers CT, Clarke WT, Snyder C, Vaughan JT, Neubauer S, Robson MD. Human cardiac 31P magnetic resonance spectroscopy at 7 Tesla. Magn Reson Med. 2014;72:304–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Valkovič L, Chmelík M, Ukropcová B, et al. Skeletal muscle alkaline Pi pool is decreased in overweight‐to‐obese sedentary subjects and relates to mitochondrial capacity and phosphodiester content. Sci Rep. 2016;6:20087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Russ DW, Elliott MA, Vandenborne K, Walter GA, Binder‐Macleod SA. Metabolic costs of isometric force generation and maintenance of human skeletal muscle. Am J Physiol Endocrinol Metab. 2002;282:E448–E457. [DOI] [PubMed] [Google Scholar]
- 17. Edwards LM, Tyler DJ, Kemp GJ, et al. The reproducibility of 31‐phosphorus MRS measures of muscle energetics at 3 Tesla in trained men. PLoS One. 2012;7: e37237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vandenborne K, McCully K, Kakihira H, et al. Metabolic heterogeneity in human calf muscle during maximal exercise. Proc Natl Acad Sci U S A. 1991;88:5714–5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bottinelli R, Reggiani C. Human skeletal muscle fibres: molecular and functional diversity. Prog Biophys Mol Biol. 2000;73:195–262. [DOI] [PubMed] [Google Scholar]
- 20. Price TB, Kamen G, Damon BM, et al. Comparison of MRI with EMG to study muscle activity associated with dynamic plantar flexion. Magn Reson Imaging. 2003;21:853–861. [DOI] [PubMed] [Google Scholar]
- 21. Fiedler GB, Meyerspeer M, Schmid AI, et al. Localized semi‐LASER dynamic P magnetic resonance spectroscopy of the soleus during and following exercise at 7 T. Magn Reson Mater Phys Biol Med. 2015;28:493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fiedler GB, Schmid AI, Goluch S, et al. Skeletal muscle ATP synthesis and cellular H+ handling measured by localized 31P‐MRS during exercise and recovery. Sci Rep. 2016;6:32037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Niess F, Fiedler GB, Schmid AI, et al. Interleaved multivoxel 31P MR spectroscopy. Magn Reson Med. 2016. doi: 10.1002/mrm.26172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meyerspeer M, Krssak M, Kemp GJ, Roden M, Moser E. Dynamic interleaved 1H/31P STEAM MRS at 3 Tesla using a pneumatic force‐controlled plantar flexion exercise rig. Magn Reson Mater Phys Biol Med. 2005;18:257–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meyerspeer M, Scheenen T, Schmid AI, Mandl T, Unger E, Moser E. Semi‐LASER localized dynamic 31P magnetic resonance spectroscopy in exercising muscle at ultra‐high magnetic field. Magn Reson Med. 2011;65:1207–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Walter G, Vandenborne K, McCully KK, Leigh JS. Noninvasive measurement of phosphocreatine recovery kinetics in single human muscles. Am J Physiol. 1997;272:C525–C534. [DOI] [PubMed] [Google Scholar]
- 27. Valkovič L, Chmelík M, Just Kukurová I, et al. Depth‐resolved surface coil MRS (DRESS)‐localized dynamic 31P‐MRS of the exercising human gastrocnemius muscle at 7 T. NMR Biomed. 2014;27:1346–1352. [DOI] [PubMed] [Google Scholar]
- 28. Greenman RL, Smithline HA. The feasibility of measuring phosphocreatine recovery kinetics in muscle using a single‐shot 31P RARE MRI sequence. Acad Radiol. 2011;18:917–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Parasoglou P, Xia D, Chang G, Regatte RR. Dynamic three‐dimensional imaging of phosphocreatine recovery kinetics in the human lower leg muscles at 3 T and 7 T: a preliminary study. NMR Biomed. 2013;26:348–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Parasoglou P, Feng L, Xia D, Otazo R, Regatte RR. Rapid 3D‐imaging of phosphocreatine recovery kinetics in the human lower leg muscles with compressed sensing. Magn Reson Med. 2012;68:1738–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schmid AI, Meyerspeer M, Robinson SD, et al. Dynamic PCr and pH imaging of human calf muscles during exercise and recovery using P gradient‐echo MRI at 7 Tesla. Magn Reson Med. 2016;75:2324–2331. [DOI] [PubMed] [Google Scholar]
- 32. Slade JM, Towse TF, Delano MC, Wiseman RW, Meyer RA. A gated 31P NMR method for the estimation of phosphocreatine recovery time and contractile ATP cost in human muscle. NMR Biomed. 2006;19:573–580. [DOI] [PubMed] [Google Scholar]
- 33. Forbes SC, Slade JM, Francis RM, Meyer RA. Comparison of oxidative capacity among leg muscles in humans using gated 31P 2‐D chemical shift imaging. NMR Biomed. 2009;22:1063–1071. [DOI] [PubMed] [Google Scholar]
- 34. Adalsteinsson E, Irarrazabal P, Topp S, Meyer C, Macovski A, Spielman DM. Volumetric spectroscopic imaging with spiral‐based k‐space trajectories. Magn Reson Med. 1998;39:889–898. [DOI] [PubMed] [Google Scholar]
- 35. Andronesi OC, Gagoski BA, Sorensen AG. Neurologic 3D MR spectroscopic imaging with low‐power adiabatic pulses and fast spiral acquisition. Radiology. 2012;262:647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Forbes SC, Raymer GH, Kowalchuk JM, Thompson RT, Marsh GD. Effects of recovery time on phosphocreatine kinetics during repeated bouts of heavy‐intensity exercise. Eur J Appl Physiol. 2008;103:665–675. [DOI] [PubMed] [Google Scholar]
- 37. Chmelík M, Považan M, Jiru F, et al. Flip‐angle mapping of 31P coils by steady‐state MR spectroscopic imaging. J Magn Reson Imaging. 2014;40:391–397. [DOI] [PubMed] [Google Scholar]
- 38. Tušek Jelenc M, Chmelík M, Bogner W, Krššák M, Trattnig S, Valkovič L. Feasibility and repeatability of localized 31P‐MRS four‐angle saturation transfer (FAST) of the human gastrocnemius muscle using a surface coil at 7 T. NMR Biomed. 2016;29:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vanhamme L, Van Huffel S, Van Hecke P, van Ormondt D. Time‐domain quantification of series of biomedical magnetic resonance spectroscopy signals. J Magn Reson. 1999;140:120–130. [DOI] [PubMed] [Google Scholar]
- 40. Taylor DJ, Bore PJ, Styles P, Gadian DG, Radda GK. Bioenergetics of intact human muscle. A 31P nuclear magnetic resonance study. Mol Biol Med. 1983;1:77–94. [PubMed] [Google Scholar]
- 41. Bogner W, Chmelik M, Schmid AI, Moser E, Trattnig S, Gruber S. Assessment of 31P relaxation times in the human calf muscle: a comparison between 3 T and 7 T in vivo. Magn Reson Med. 2009;62:574–582. [DOI] [PubMed] [Google Scholar]
- 42. Moon RB, Richards JH. Determination of intracellular pH by 31P magnetic resonance. J Biol Chem. 1973;248:7276–7278. [PubMed] [Google Scholar]
- 43. Kemp GJ, Taylor DJ, Thompson CH, et al. Quantitative analysis by 31P magnetic resonance spectroscopy of abnormal mitochondrial oxidation in skeletal muscle during recovery from exercise. NMR Biomed. 1993;6:302–310. [DOI] [PubMed] [Google Scholar]
- 44. Pohmann R, von Kienlin M. Accurate phosphorus metabolite images of the human heart by 3D acquisition‐weighted CSI. Magn Reson Med. 2001;45:817–826. [DOI] [PubMed] [Google Scholar]
- 45. Meyerspeer M, Robinson S, Nabuurs CI, et al. Comparing localized and nonlocalized dynamic 31P magnetic resonance spectroscopy in exercising muscle at 7 T. Magn Reson Med. 2012;68:1713–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Šedivý P, Kipfelsberger MC, Dezortová M, et al. Dynamic 31P MR spectroscopy of plantar flexion: influence of ergometer design, magnetic field strength (3 and 7 T), and RF‐coil design. Med Phys. 2015;42:1678–1689. [DOI] [PubMed] [Google Scholar]
- 47. Steinseifer IK, Wijnen JP, Hamans BC, Heerschap A, Scheenen TW. Metabolic imaging of multiple X‐nucleus resonances. Magn Reson Med. 2013;70:169–175. [DOI] [PubMed] [Google Scholar]
- 48. Bogner W, Chmelík M, Andronesi OC, Sorensen AG, Trattnig S, Gruber S. In vivo 31P spectroscopy by fully adiabatic extended image selected in vivo spectroscopy: a comparison between 3 T and 7 T. Magn Reson Med. 2011;66:923–930. [DOI] [PubMed] [Google Scholar]
- 49. Durst M, Koellisch U, Frank A, et al. Comparison of acquisition schemes for hyperpolarised 13C imaging. NMR Biomed. 2015;28:715–725. [DOI] [PubMed] [Google Scholar]
- 50. Chmelík M, Just Kukurová I, Gruber S, et al. Fully adiabatic 31P 2D‐CSI with reduced chemical shift displacement error at 7 T—GOIA‐1D‐ISIS/2D‐CSI. Magn Reson Med. 2013;69:1233–1244. [DOI] [PubMed] [Google Scholar]
- 51. Goluch S, Kuehne A, Meyerspeer M, et al. A form‐fitted three channel 31P, two channel 1H transceiver coil array for calf muscle studies at 7 T. Magn Reson Med. 2015;73:2376–2389. [DOI] [PubMed] [Google Scholar]
- 52. Bogner W, Hess AT, Gagoski B, et al. Real‐time motion‐ and B0‐correction for LASER‐localized spiral‐accelerated 3D‐MRSI of the brain at 3 T. Neuroimage. 2013;88C:22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Valkovič L, Chmelík M, Just Kukurová I, et al. Time‐resolved phosphorous magnetization transfer of the human calf muscle at 3 T and 7 T: a feasibility study. Eur J Radiol. 2013;82:745–751. [DOI] [PubMed] [Google Scholar]
- 54. Parasoglou P, Xia D, Chang G, Regatte RR. Three‐dimensional saturation transfer 31P‐MRI in muscles of the lower leg at 3.0 T. Sci Rep. 2014;4:5219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bogner W, Gagoski B, Hess AT, et al. 3D GABA imaging with real‐time motion correction, shim update and reacquisition of adiabatic spiral MRSI. Neuroimage. 2014;103:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Meyerspeer M, Magill AW, Kuehne A, Gruetter R, Moser E, Schmid AI. Simultaneous and interleaved acquisition of NMR signals from different nuclei with a clinical MRI scanner. Magn Reson Med. 2016;76:1636–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mayer D, Levin YS, Hurd RE, Glover GH, Spielman DM. Fast metabolic imaging of systems with sparse spectra: application for hyperpolarized 13C imaging. Magn Reson Med. 2006;56:932–937. [DOI] [PubMed] [Google Scholar]


