Abstract
Diffuse large B cell lymphoma (DLBCL) is the most common form of non-Hodgkin lymphoma worldwide. We describe the establishment and molecular characteristics of the DLBCL cell line U-2946. This cell line was derived from a 52-year-old male with DLBCL. U-2946 cells carried the chromosomal translocation t(8;14) and strongly expressed MYC, but not the mature B-cell lymphoma associated oncogenes BCL2 and BCL6. Instead, U-2946 cells expressed the antiapoptotic BCL2 family member MCL1 which was highly amplified genomically (14n). MCL1 amplification is recurrent in DLBCL, especially in the activated B cell (ABC) variant. Results of microarray expression cluster analysis placed U-2946 together with ABC-, but apart from germinal center (GC)-type DLBCL cell lines. The 1q21.3 region including MCL1 was focally coamplified with a short region of 17p11.2 (also present at 14n). The MCL1 inhibitor A-1210477 triggered apoptosis in U-2946 (MCL1pos/BCL2neg) cells. In contrast to BCL2pos DLBCL cell lines, U-2946 did not respond to the BCL2 inhibitor ABT-263. In conclusion, the novel characteristics of cell line U-2946 renders it a unique model system to test the function of small molecule inhibitors, especially when constructing a panel of DLBCL cell lines expressing broad combinations of antiapoptotic BCL2-family members.
Introduction
Continuous cell lines have become indispensable tools in leukemia/lymphoma research. Self -renewing and transportable in the frozen state, cell lines provide unlimited supplies of cellular tumor material worldwide. Descendent from single cells of a patient´s tumor, they retain oncogenomic aberrations and gene expression patterns characteristic of the primary tumor. Tumor congruence is a key reason why cell lines are used as models to study the function of aberrantly expressed genes or to test the efficiency of novel anti-cancer drugs [1,2].
Diffuse large B cell lymphoma (DLBCL) is the most common form of lymphoma in adults. Various genes are recurrently affected by mutations and chromosomal aberrations therein, including the germinal center oncogenes BCL2, BCL6 and MYC [3]. Expression array analysis has identified two molecularly distinct forms of the tumor, termed germinal center (GC) and activated B-cell (ABC) [4]. DLBCL-derived cell lines also show correspondingly distinct expression profiles allowing classification according to the GC- and ABC-scheme [5–9]. In contrast to GC-type DLBCL, ABC-type cells rely on the constitutive activation of the NF-kB pathway to block apoptosis [10]. Cell lines have been widely used to determine the effect of recurrent mutations or overexpressed genes on signaling pathways in ABC DLBCL and other lymphoma entities and to develop drugs for targeted therapies [5,7,10]. One important step in tumorigenesis is the loss of functional apoptosis, explaining why overexpression of antiapoptotic genes can contribute to tumorigenesis [11]. In DLBCL, the antiapoptotic genes BCL2 and MCL1 are recurrently overexpressed, as result of chromosomal translocations, amplification or other mechanisms [12–14]. We describe the establishment and molecular characteristics of the DLBCL-derived cell line U-2946. Due to an amplification on chr. 1q21.3, this cell line overexpresses MCL1. Several DLBCL cell lines have been used to validate novel drugs targeting antiapoptotic BCL2 family members [13–18]. We propose U-2946 as auspicious model cell line which shows the rare combination of MCL1 positivity and BCL2 negativity.
Materials and Methods
Human cell lines
Authenticated stocks of cell line U-2946 were grown in RPMI 1640 (Invitrogen, Darmstadt, Germany) containing 10% fetal bovine serum (FBS) (Sigma-Aldrich, Taufkirchen, Germany). Cell lines applied in this study are all held by the DSMZ—German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany (www.dsmz.de) or were supplied by the originators for research purpose. Detailed references and cultivation protocols have been described previously [19]. The BCL2 family inhibitors ABT-263 and A-1210477 were obtained from Selleckchem (München, Germany).
Cytogenetic analysis
Cells were harvested and fixed as described previously [20]. Spectral karyotyping (SKY) and fluorescence in situ hybridization (FISH) were performed as described previously [21]. Tilepath bacterial artificial chromosome (BAC) clones were sourced from BAC-PAC Resources (Children´s Hospital, Oakland, CA, USA), and SKY probes from Applied Spectral Imaging (Edingen, Germany). Probe selection was performed using the UCSC Genome Browser (https://genome-euro.ucsc.edu/) guided by the results of combined conventional cytogenetic, SKY and Cytoscan copy number array data (see below). For colocalization experiments commercial chromosome painting probes were mixed 1:1 with labelled BACs. Probe preparation and labelling were as described previously [21]. Briefly, BAC clone DNA was labelled by nick translation with dUTPs contrastingly labelled with fluors DY-495/547/590 purchased from Dyomics (Jena, Germany) and slide preparations counterstained with DAPI (4′,6-Diamidine-2′-phenylindole dihydrochloride) in Vector antifade mountant (Biozol, Eching, Germany). Imaging and analysis were performed using an Axioimager D1 microscope system equipped with an alpha-Plan Apochromat 100x objective (Zeiss, Goettingen, Germany) configured to a Spectral Imaging analysis system (Applied Spectral Imaging). For FISH, monochromatic fluor and DAPI signals were captured, merged and the pseudocolored images aligned automatically to generate reverse G-banding as described previously [21].
Numerical aberrations
CytoScan HD Array (Affymetrix, Santa Clara, CA, USA) hybridization analysis was performed to identify numerical aberrations. DNA was prepared using the Qiagen Gentra Puregene Kit (Qiagen, Hilden, Germany). Data were analyzed using the Chromosome Analysis Suite software version 2.0.1.2 (Affymetrix).
Cell surface marker analysis
Immunophenotyping was conducted on a FACSCalibur (Becton Dickinson, Heidelberg, Germany). Antibodies (Abs) against CD3 (Leu4), CD5 (UCHT2), CD10 (HI10a), CD19 (Leu12), CD20 (Leu16), CD34 (clone 581), CD37 (M-B371), CD38 (HIT2), CD138 (MI15) and HLA-DR (Tu39) were purchased from Becton Dickinson. Ab CD13 (My7) was from Beckman Coulter (Krefeld, Germany). Optimal dilutions of the Abs were determined with positive and negative control cell lines. FITC-labeled Abs against IgG, IgM, Ig kappa and Ig lambda were obtained from Southern Biotech/Biozol (Eching, Germany).
Screening for immunoglobulin rearrangements and hypermutations
Rearrangement of the immunoglobulin (Ig) heavy chain was determined by genomic PCR using primers described by van Dongen et al. [22]. The PCR was performed for 35 cycles at 60°C annealing temperature. The level of Ig hypermutation was determined by sequencing the cloned PCR products. Ten clones were sequenced to screen for existence of U-2946 subclones.
DNA microarray hybridization
Five hundred ng total RNA were used for biotin labelling according to the 3´ IVT Express Kit (Affymetrix); 7.5 μg of biotinylated cDNA were fragmented and placed in a hybridization cocktail containing four biotinylated hybridization controls (BioB, BioC, BioD, and Cre). Samples were hybridized to an identical lot of Affymetrix GeneChip HG-U133 Plus 2.0 for 16 h at 45°C. Steps for washing and SA-PE staining were processed on the fluidics station 450 using the recommended FS450 protocol (Affymetrix). Image analysis was performed on GCS3000 Scanner and GCOS1.2 Software Suite (Affymetrix). Data processing and analysis (probe summarisation, RMA-background correction and quantile normalization of spot intensities) were performed via R/Bioconductor using limma and affy packages [23–25].
Quantitative real-time PCR analysis
Quantitative PCR was performed on a 7500 Applied Biosystems real-time PCR system (Darmstadt, Germany). RNA was prepared using the RNeasy Mini kit (Qiagen). Reverse transcription was performed using the SuperScript II reverse transcriptase kit (Invitrogen, Karlsruhe, Germany). TaqMan probes (Applied Biosystems) were used to quantify human BCL2 (Hs00153350_m1), BCL6 (Hs00153368_m1), MAP2K3 (Hs00177127_m1), MCL1 Exon 1/2 (Hs03043899_m1), MYC exon 1/2 (Hs00905030_m1) and MYC exon 2/3 (Hs00153408_m1) expression levels with TBP as endogenous control. Relative expression levels were calculated using the ΔΔCt method.
Ploidy levels of BCL2, MAP2K3 and MCL1 were determined by genomic PCR applying the SYBR green assay system (Applied Biosystems) with ABL1 as internal control and NC-NC as diploid reference cell line. DNA was prepared using the High pure PCR template preparation kit (Roche Life Science, Mannheim, Germany). The applied primers are listed in S1 Table.
Western blot analysis
Samples were prepared as described previously [26]. Anti BCL2 (1:500 dilution) and anti MCL1 Abs (1:250) were purchased from Becton Dickinson. Abs against the phosphorylated and unphosphorylated forms of p38 and MAP2K3 (1:10,000 each) and against BCL6 (1:1,000), BCLXL (1:1,000) and MYC (1:2,000) were obtained from Cell Signaling Technology (Leiden, The Netherlands), the GAPDH mouse monoclonal Ab (mAb) (1:5,000) was from Abcam (Cambridge, UK). Specific bands on nitrocellulose membranes were visualized with the biotin/streptavidin-horseradish peroxidase system (GE Healthcare, Little Chalfont, UK) in combination with the “Renaissance Western Blot Chemoluminescence Reagent” protocol (Perkin Elmer, Waltham, MA, USA).
3H-thymidine uptake
Assays of 3H-thymidine incorporation were executed as follows: 1.25 x 104 cells (in 100 μl) were seeded in triplicate in 96-well flat-bottom microtiter cell culture plates. Inhibitors were added as 2x concentrated solution in a 100 μl volume. For the last 3 h of the incubation period, 1 μCi 3H-thymidine (Hartmann Analytic, Braunschweig, Germany) was added to each well.
Detection of apoptotic cells
Apoptotic cells were detected and quantified with the annexin-V/propidium iodide method using the TACS Annexin-V-FITC kit (R&D Systems, Wiesbaden, Germany) according to the manufacturer’s instructions. Binding of fluorescein isothiocyanate-labeled annexin-V and propidium iodide staining of the cells were determined by flow cytometry on the FACSCalibur (Becton Dickinson).
Results
Clinical history
The patient was a previously healthy man, age 52 years who sought medical care due to abdominal pain and tiredness. Computer tomography examination revealed an abdominal tumor. In July 1996 he underwent a right-sided hemicolectomy due to a tumor of the ascending colon. The tumor, 15 cm in diameter, removed macroscopically radically. Pathological examination showed DLBCL. Diagnostic work-up revealed lymphomatous engagement in the right testicle and in paratesticular tissue with a diameter of >10 cm, in lymph nodes and in the bone marrow (stage IVB). International Prognostic Index was retrospectively assigned to 3 (Stage IV, >1 extranodal site and elevated S-LDH (7 x upper normal limit)). At diagnosis he also had a palsy of the left N. abducens but examination of cerebrospinal fluid showed no signs of lymphoma and a CT of the brain was normal.
He was treated with VACOP-B [27]: a weekly schedule of alternating chemotherapy routinely used for aggressive lymphomas at our department at that time. He also received intrathecal injections of methotrexate 12.5 mg. At week 7 he had progressive disease and the treatment was changed to MIME [28], which was given for two courses before the patient progressed and received palliative radiotherapy. His disease progressed rapidly and he developed pleural effusion and leukemic lymphoma with a WBC count of > 100 x 109/l. The patient gave informed consent to use the pleural effusion for research including the possible establishment of a cell line. He died of progressive lymphoma three months after diagnosis. The overall picture of the lymphoma was an extremely aggressive disease with fast progression and very limited sensitivity to chemotherapy.
Large blast cells with the appearance of both immunoblasts and centroblasts grew in a diffuse and infiltrative pattern in the intestine. Several mitotic figures and apoptotic cells were present. Immunohistochemical stainings were performed and the lymphoma cells were positive for CD20, CD79a, CD10, BCL-6, MYC, p53 and negative for BCL-2, CD5, CD30, cyclin D1, SOX11 with a high proliferation index of Ki67. Partial expression of MUM1 and FOXP1 was detected. According to the WHO classification and update [29,30] the lymphoma would be classified as a DLBCL with a germinal-center derived phenotype according to the Hans et al algorithm (S1 Fig) [31,32]. In addition, MYC translocation was detected by in situ hybridization.
Cell culture
Tumor cells from pleural effusion were obtained by Ficoll-Isopaque (Lymphoprep, Nycomed) gradient separation and cultured in RPMI 1640 containing 10% of FBS, glutamine and antibiotics (100 IU/ml penicillin and 50 μg/ml streptomycin) at 37°C in 5% CO2 humidified environment. Cells were split every 72 h with an optimal density of 0.3 x 105 cells/ml and a doubling time of 48 h. After one month, the cell line U-2946 was considered established and grew in continuous culture in RPMI 1640 with 10% FBS for another 2 months. U-2946 cells tested EBV-negative excluding a lymphoblastoid origin. May-Grünwald-Giemsa stain of the cell line is shown in S1 Fig.
U-2946: immunoglobulin rearrangement and immunophenotype
We tested the Ig rearrangement status of cell line U-2946 using previously described primers [22]. The cell line was found to carry an IGHV4-59 rearrangement with 6% hypermutation. Consistent with the patient´s tumor, U-2946 cells showed the typical immunophenotype of a B-lymphoma cell line (Table 1, S2 Fig).
Table 1. Immunoprofile of patient and–derived cell line U-2946.
| patient | U-2946 | patient | U-2946 | ||
|---|---|---|---|---|---|
| CD3 | neg | neg | CD37 | n.d. | pos |
| CD5 | neg | neg | CD38 | n.d. | pos |
| CD10 | pos | pos | CD138 | n.d. | neg |
| CD13 | n.d. | neg | HLA-DR | n.d. | pos |
| CD19 | pos | pos | IgG | n.d. | neg |
| CD20 | pos | pos | IgM | n.d. | pos |
| CD22 | pos | n.d. | kappa | n.d. | neg |
| CD34 | n.d. | neg | lambda | n.d. | pos |
Expression of surface markers was assessed by flow cytometry with the appropriate isotype controls. pos, ≥ 15% express antigen
neg, < 15% express antigen; n.d., not done.
Karyotyping
The consensus karyotype, performed using classical (Fig 1A) and spectral karyotyping (Fig 1B), together with FISH covering the IGH and MYC loci (Fig 1C–1G) was resolved to: 47<2n>XY, +7, der(9)t(2;9)(p25;p21), der(8)inv(8)(p22;q24), der(9)t(2;9)(p25;p22), add(17)(p11). FISH revealed canonical juxtaposition of IGH and MYC, typical of B-cell neoplasms plus an additional non IGH rearrangement, der(8)t(8;8)(p23;q24), effecting copy number increase of the terminal 8q region just including PVT1, but excluding MYC.
Fig 1. Cytogenetic analysis of U-2946 cells.
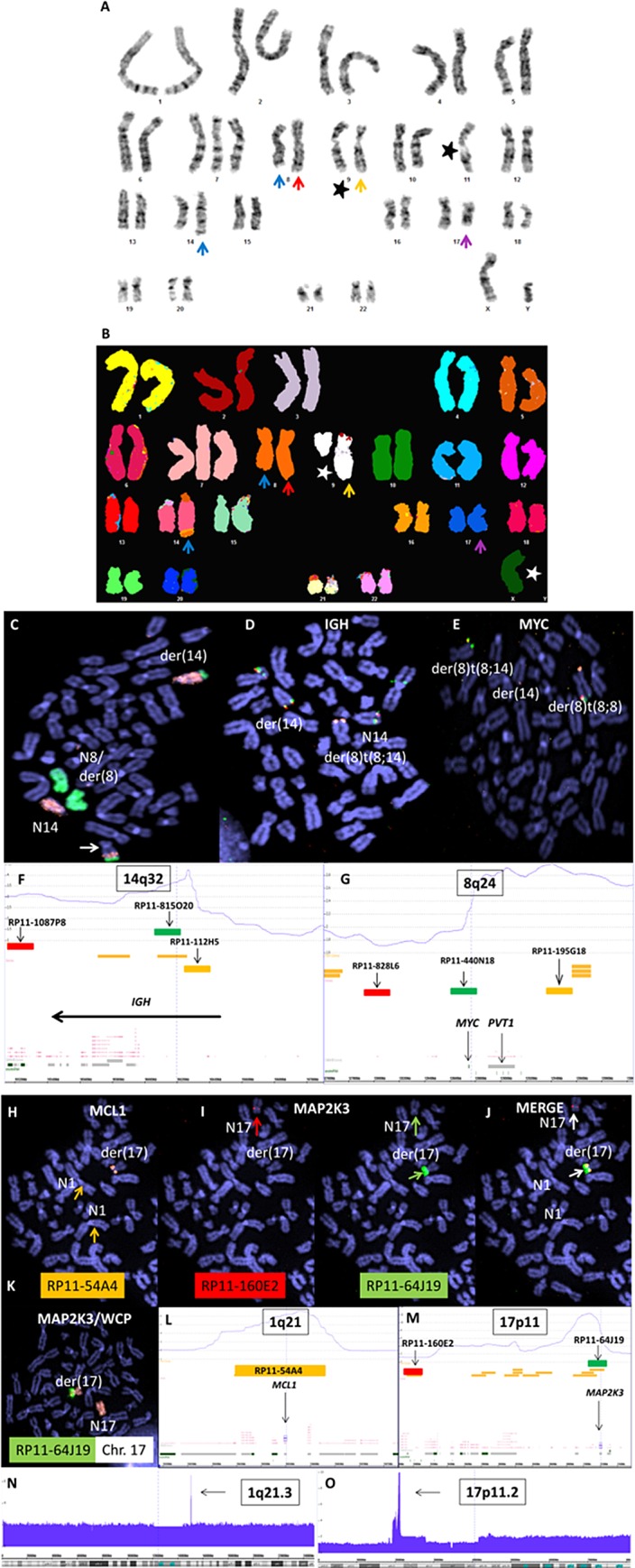
A) Classical and B) Spectral Karyotyping shows key oncogenomic rearrangements present in U-2946 cells. BAC clones used for FISH are shown along with corresponding genomic copy number plots. Blue arrows show t(8;14)(q24;q32), red arrows der(8)t(8;8)(p22;q24), yellow arrows der(9)t(2;9) and purple arrows add(17)(p11). Note differences between cells in A and B reflecting clonal heterogeneity indicated by stars. C-G) Image depicts analysis of t(8;14) by chromosome painting (C), and single locus FISH using probes for IGH (D) and MYC (E). Positions of BAC clones are shown for IGH (F) and MYC loci (G). Note partial duplication of the terminal der(14) long arm region including IGH/MYC (arrow) present in a subclone (C). H-M) Analysis of MCL1/MAP2K3 coamplification: gold signals from MCL1 clone (RP11-54A4), barely visible at native loci (arrows), are intense on der(17) due to amplification (H). Similarly, green signals from 17p11 clone (RP11-64J19) covering MAP2K3 are amplified, while red signals (RP11-160E2) from the neigboring hemizygously deleted region are present on the unrearranged chromosome only. Chromosome painting shows localization of amplicon to 17p (K). Genomic copy number plots show consistency with FISH amplification data for both MCL1 (L) and MAP2K3 (M). N-O) Cytoscan HD array analysis revealed prominent amplification of 1q21.3 (N) and 17p11.2 (O).
Cytogenetic analysis also revealed a novel rearrangement, focal co-amplification of 1q21 and 17p11.2 replacing a deleted segment of chromosome 17p (Fig 1H–1M). The ca. 67 kbp–amplicon at 1q21 covered two loci (MCL1, ENSA), while that at 1p11 extending some 461 kbp covered four loci (USP22, DHRS7B, TMEM11, MAP2K3). Coamplification of gene pairs normally arises when a genomically remote gene pair, ectopically juxtaposed or fused by chromosome rearrangement is locally subjected to linear copy number amplification, e.g. following selection. Such copy number aberrations are hallmarks of oncogenes and are thought to arise by conferral of proliferation or antiapoptopic advantage.
U-2946 with ABC marker expression
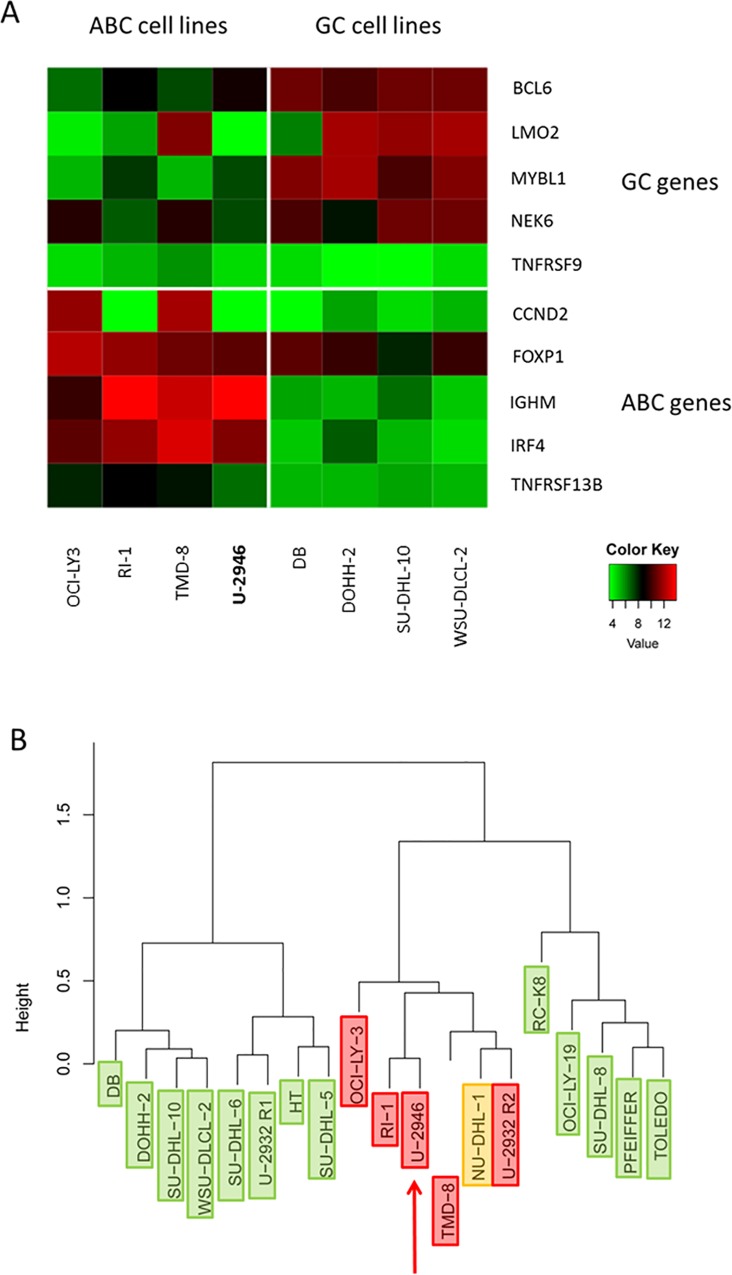
Expression array analysis allows distinction between two forms of DLBCL, the GC and the ABC forms [4]. Based on five ABC marker genes and five GC markers [33], cell line U-2946 was assigned to the ABC group (Fig 2A). Unsupervised clustering of 19 DLBCL cell lines confirmed the ABC character of this cell line (Fig 2B).
Fig 2. U-2946: an ABC DLBCL cell line.
A) heatmap of ABC/GC gene expression, data from expression array analysis. The majority of ten ABC and GC markers described by Ruminy et al. [33] class U-2946 as ABC DLBCL cell line. B) Applying the same set of marker genes, cell line U-2946 clustered together with described ABC DLBCL cell lines (red), not with GC cell lines (green) [5–9]. Cell line NU-DHL-1 (orange) was discordantly categorized by different authors [6,8]. We showed that subclone R1 of the ABC cell line U-2932 highly expressed various GC markers, absent from subclone R2 [34].
BCL2, BCL6 and MYC
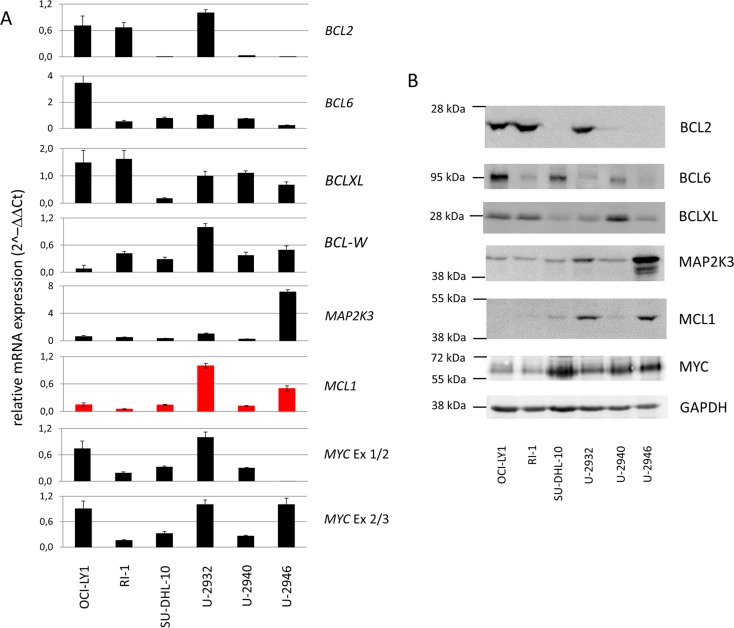
Mature B cell lymphomas often carry rearrangements and numerical aberrations leading to aberrant expression of BCL2, BCL6 and MYC. These aberrations can occur in the same tumors, which are then called “double-hit” or “triple-hit” lymphoma according to their aberration scores. Cell lines representing double-hit and triple-hit lymphomas have recently been reviewed in detail [35]. Amplifications or canonical translocations affecting chromosomes 18q21 (BCL2), 3q27 (BCL6) were not detected in U-2946 cells. As shown in Fig 1C–1G, MYC on 8q24 was fused to IGH on 14q32. In accordance with the genomic findings, cell line U-2946 was BCL2neg/BCL6low/MYCpos (Fig 3A and 3B). 5´RACE revealed that the MYC transcript began in intron 1/2, not in exon 1 (data not shown). Accordingly qRT-PCR showed MYC transcript in exon 2 but not in exon 1 suggesting (i) that the genomic break was located in intron 1/2, and (ii) that translation started with the alternative ATG start codon in exon 2 and not with the “internal ribosome entry site” (IRES)-mediated CTG codon in exon 1 (Fig 3A).
Fig 3. Expression of BCL2, BCL6, BCLXL, MAP2K3, MCL1, and MYC in DLBCL cell lines.
A) qRT-PCR and B) Western blot analysis was performed to assess the expression of oncogenes in U-2946 and other DLBCL cell lines. The bars in A) indicate means with standard deviation (n = 3). The ct values for BCL-W in qRT-PCR were high (>30) in all cell lines tested explaining absence of specific bands in Western blot analysis (not shown). Note the high-level expression of MCL1 (red) in cell lines U-2946 and U-2932.
Numerical aberrations are responsible for aberrant gene expression in U-2946
To find genes conspicuously expressed in U-2946, we compared expression array data of this cell line with those of 55 other certified B-lymphoma cell lines spanning the whole spectrum of B-NHL. Thirty-nine of the 46 marker genes (85%) were upregulated in U-2946; another 7 genes (15%) lay below the average of B-lymphoma cell lines (S3 Fig). High density genomic array analyses revealed that 6/7 underexpressed genes mapped to haploid regions (S2 Table). Five genes were amplified, all of them overexpressed (S2 Table). Thus, we found an almost perfect correlation between deletion and lack of expression on the one side, and amplification and overexpression on the other. The most prominent amplifications were at regions 1q21.3 (6n) (Fig 1N) and 17p11.2 (4n-10n) (Fig 1O). The amplified genes are listed in S3 Table.
MAP2K3: amplified but inactive
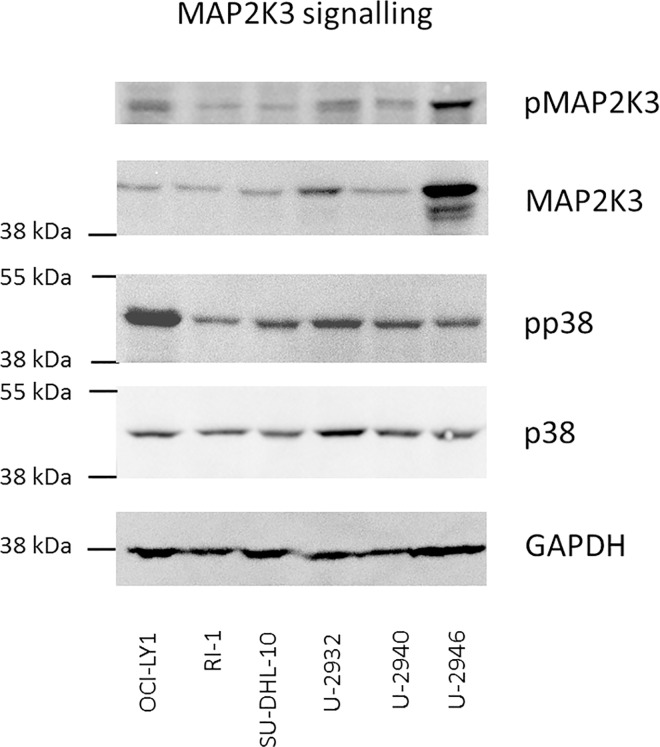
Located on 17p11.2, MAP2K3 (MEK3) was amplified in cell line U-2946 (Fig 1O, S3 Table). Amplification of the gene was accompanied by overexpression of MAP2K3, at the mRNA and protein levels (Fig 3A and 3B). This kinase is known to activate the p38 MAPK pathway [36]. Sequencing of MAP2K3 cDNA showed that the gene was not mutated. Nevertheless, cell lines with low expression of MAP2K3 showed the same p38 MAPK phosphorylation level as cell line U-2946 (Fig 4). Thus, MAP2K3 was highly expressed in cell line U-2946, but did not aberrantly trigger the p38 MAPK pathway.
Fig 4. Phosphorylation of the MAP2K3/p38 pathway.
Expression and phosphorylation of MAP2K3 and p38 in U-2946 and five other DLBCL cell lines was determined by Western blot analysis. Note that overexpression of MAP2K3 was not accompanied by high phosphorylation of the MAP2K3 target in cell line U-2946.
MCL1 amplified on der chr17
Like BCL2, MCL1 is an antiapoptotic gene and recurrently overexpressed in DLBCL [13,14]. Accordingly, the 1q21 region including the MCL1 locus is recurrently amplified in DLBCL [37]. A positive correlation of MCL1 amplification and gene expression was specifically observed in ABC DLBCL [14]. Thus 1q21.3 amplification (including the MCL1 gene locus) is consistent with the ABC character proposed here for cell line U-2946 (Fig 1N, S3 Table). FISH analysis revealed that MCL1 was translocated to chr17, specifically to the amplified region on 17p11.2 including MAP2K3 (Fig 1H–1M).
To elucidate whether MCL1 amplification might correlate with the gene´s overexpression, we performed qRT-PCR and Western blot analysis for U-2946 in comparison with other DLBCL cell lines (Fig 3A and 3B). Cell line U-2946 was MCL1pos/BCL2neg. All other cell lines showed different combinations of MCL1 and BCL2 expression (y3A and 3B). Overexpression of both genes was always accompanied by genomic amplification, except in OCI-LY1, where we observed BCL2 mRNA overexpression without genomic amplification (S4 Fig). This cell line is known to carry the t(14;18) effecting IGH/BCL2 juxtaposition, another well-described cause of aberrant BCL2 expression [12,38].
MCL1 with antiapoptotic function in cell line U-2946
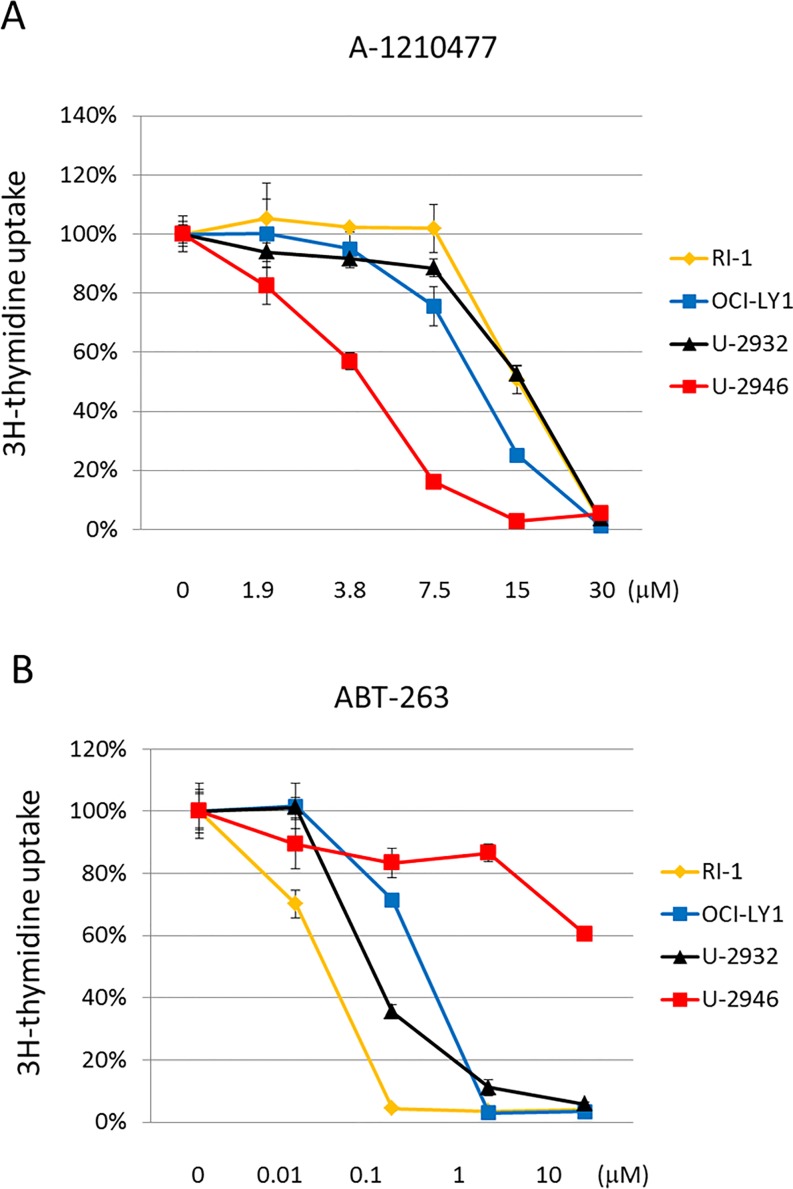
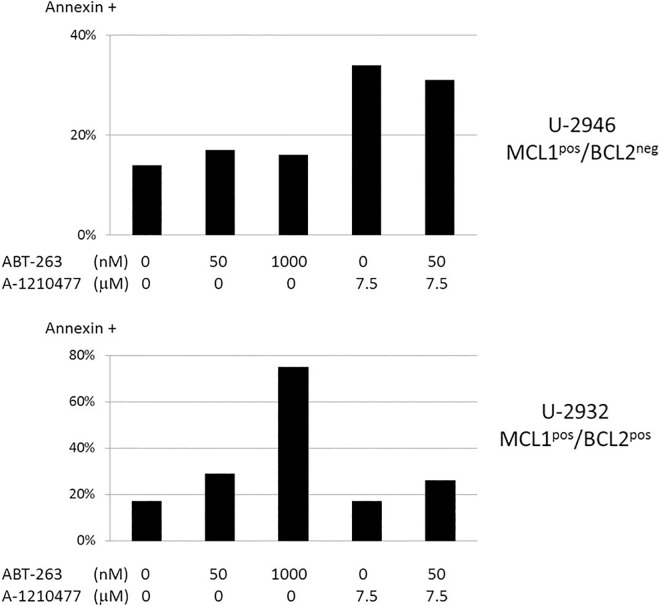
To find out whether MCL1 and BCL2 were critical for cell survival in our DLBCL cell lines, we incubated the cell lines with inhibitors of MCL1 (A-1210477) or BCL2, BCL-XL and BCL-W (ABT-263). The MCL1 inhibitor (A-1210477, 7.5 μM) substantially reduced 3H-thymidine uptake and triggered apoptosis in the MCL1pos cell line U-2946, while MCL1neg cell lines were not affected (Figs 5A and 6). All BCL2pos cell lines (OCI-LY1, RI-1, U-2932) were responsive to low doses of the BCL2 inhibitor (Figs 5B and 6). Noteworthingly, the pharmacological MCL1 inhibitor left the MCL1pos/BCL2pos cell line U-2932 unperturbed. Even in the presence of low concentrations of the BCL2 inhibitor (50 nM ABT-263), the MCL1 inhibitor (7.5 μM A-1219477) did not have any effect on U-2932 cells, suggesting that BCL2 was dominant over MCL1 in the U-2932 cell system (Fig 6 and S5 Fig).
Fig 5. Inhibition of BCL2 and MCL1: effect on proliferation.
The effect of A) A-1210477 (MCL1 inhibitor) and B) ABT-263 (BCL2 inhibitor) on proliferation of DLBCL cell lines was assessed by 3H-thymidine uptake (d2). Cell lines OCI-LY1 and RI-1 are MCL1neg/BCL2pos, cell line U-2932 is MCL1pos/BCL2pos and cell line U-2946 is MCL1pos/BCL2neg. Especially note the significant reduction of 3H-thymidine incorporation in cell line U-2946 after addition of the MCL1 inhibitor.
Fig 6. Inhibition of BCL2 and MCL1: effect on apoptosis.
Apoptosis was assessed by Annexin-V staining, 24 h after addition of a BCL2 inhibitor (ABT-263) or an MCL1 inhibitor (A-1210477). The MCL1 inhibitor triggered apoptosis in the MCL1pos/BCL2neg cell line U-2946, BCL2 inhibition induced apoptosis in the MCL1pos/BCL2pos cell line U-2932. Although expressing MCL1, cell line U-2932 was not responsive to MCL1 inhibition, suggesting dominant function of BCL2. The data were reproduced in two independent experiments.
Discussion
Cell line U-2946 was established in 1996 from a 52-year-old male patient with terminal DLBCL following standard chemotherapy. Immunohistochemical staining classified the tumor into a GC-derived phenotype according to the Hans classification [39]. However, the only data published so far on cell line U-2946 describe the strong expression of ZAP70 and PKC-ϐ II, a feature occurring in the more aggressive clinical form of non-GC DLBCL [39]. Expression profiling confirmed that ZAP70 levels were much higher in U-2946 than in the average of 56 B-lymphoma cell lines tested. Also consistent with the non-GC character of this cell line, U-2946 clustered together with other ABC DLBCL cell lines, as evidenced by gene expression analysis, applying a short-list of five ABC marker genes and five GC markers.
Translocations and/or numerical aberrations affecting the germinal center oncogenes BCL2 and BCL6 were not detected in U-2946 cells. FISH revealed the canonical juxtaposition of IGH and MYC, leading to overexpression of the oncogene in this cell line. The most prominent numerical abnormalities in U-2946 were amplifications on chromosomes 1q21.3 and 17p11.2. Both sites were coamplified on a derivative chromosome 17. MAP2K3, on 17p21.3, and MCL1, located on 1q21.3, were highly expressed. The MAP2K3 overexpression did not lead to unusual phosphorylation of the downstream target p38 MAPK, suggesting that amplification of the MAP2K3 was a passenger or helper mutation and that amplification of MCL1 might be the true oncogenic event. MCL1 is frequently expressed in DLBCL, in the ABC subtype more often than in GC and also at higher expression levels, which correlates with higher frequency of 1q21 amplification in ABC cases [13,14]. MCL1 rescues cells from apoptosis by binding and sequestering proapoptotic BCL2-family members. Small-molecule inhibitors, developed to trigger apoptosis, have been tested in cell lines expressing antiapoptotic BCL2-family proteins [13,15–18]. Recently, it has been shown that DLBCL can be divided into BCL2-dependent and MCL1-dependent groups [40]. Cell line U-2946 expresses MCL1, but not BCL2 and only low levels of BCL-XL. Accordingly, apoptosis was induced by the MCL1 inhibitor A-1210477, while the BCL2/BCL-XL inhibitor ABT-263 (Navitoclax) had no effect. Other DLBCL cell lines expressed BCL2 and responded to ABT-263. U-2932 was one of these BCL2pos cell lines. This cell line also expressed MCL1, but did not respond to the MCL1 inhibitor. Even with low doses of the BCL2 inhibitor, A-1210477 was inefficient which suggests that the effect of BCL2 is dominant over MCL1, at least in this cell system.
We conclude that the MCL1pos/BCL2neg cell line U-2946 promises to be a valuable model for inhibitor studies, useful especially in combination with cell lines expressing other antiapoptotic BCL2 family members. Cell line U-2946 will be available from the public repository DSMZ (www.dsmz.de).
Conclusions
We describe the establishment and characteristics of the DLBCL cell line U-2946. According to gene expression data, the cell line clusters together with ABC DLBCL cell lines. The cell line carries an amplification of the antiapoptotic BCL2 family member MCL1. The MCL1 chromosomal region (1q21.3) was involved in a translocation to chromosome 17. The amplified MCL1 gene is highly expressed in the cell line (MCL1pos/BCL2neg). Inhibitor experiments revealed the importance of MCL1 for growth and survival of U-2946 cells. Cell line U-2946 will prove to be an important model system for the functional analysis of small BCL2-family inhibitors, especially in a panel of certified DLBCL cell lines showing other MCL1 and BCL2 expression profiles.
Supporting Information
A-E) Immunohistochemical stainings of lymphoma cells. Positive: BCL6 (A), CD10 (B), CD20 (C), CD79a, FOXP1, MUM1 (partial), MYC (D), p53 and negative: BCL2 (E), CD3, CD5, CD30, cyclin D1. (F) May-Grünwald-Giemsa stain of cell line U-2946.
(TIF)
Cells were stained with Abs against the antigens indicated and analyzed by flow cytometry.
(TIF)
The 47 genes with strongest expression differences between U-2946 and median of 55 B-lymphoma cell lines in relation to variance (IQR, interquartile range)–highest differences on the left. Data base on expression array analyses. Red dots, cell line U-2946.
(TIF)
Quantitative genomic PCR (upper) and qRT-PCR (lower) detecting correlation between amplification and RNA expression in BCL2, MAP2K3 and MCL1. The bars indicate means with standard deviation (n = 3). NC-NC as diploid reference cell line for genomic PCR.
(TIF)
3H-thymidine uptake after 48 h. The MCL1 inhibitor A-1210477 (7.5 μM) inhibits growth of the MCL1pos/BCL2neg cell line U-2946, but has no effect on the MCL1pos/BCL2pos cell line U-2932 –neither alone nor together with suboptimal doses of the BCL2 inhibitor ABT-263 (50 nM). The bars indicate means with standard deviation (n = 3).
(TIF)
(XLSX)
Outliers are listed according to chromosomal position. Note the perfect correlation between genomic imbalances and expression in cell line U-2946, 6/6 hemiploid genes (black and bold) being repressed, 5/5 amplified genes (red and bold) being overexpressed. Up, higher than average; down, lower than average.
(XLSX)
Amplified genes on 1q21.3 are listed from centromere to telomere, genes on 17p11.2 from telomere to centromere. Bold: strongly expressed genes as assessed by expression array analysis.
(XLSX)
Data Availability
All relevant data are within the paper and its supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Drexler HG. Establishment and culture of leukemia-lymphoma cell lines In: Cree I editor. Cancer Cell Culture: Methods and Protocols, 2nd edition, Springer, Berlin; 2011. pp. 181–200. [DOI] [PubMed] [Google Scholar]
- 2.Drexler HG, Quentmeier H, MacLeod RAF. Cell line models of leukemia. Drug Discovery Today—Disease Models 2005;2:51–56. [Google Scholar]
- 3.Pasqualucci L, Dalla-Favera R. SnapShot: diffuse large B cell lymphoma. Cancer Cell 2014;25:132 10.1016/j.ccr.2013.12.012 [DOI] [PubMed] [Google Scholar]
- 4.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000;403:503–511. 10.1038/35000501 [DOI] [PubMed] [Google Scholar]
- 5.Davis RE, Ngo VN, Lenz G, Tolar P, Young RM, Romesser PB, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature 2010;463:88–92. 10.1038/nature08638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lenz G, Nagel I, Siebert R, Roschke AV, Sanger W, Wright GW, et al. Aberrant immunoglobulin class switch recombination and switch translocations in activated B cell-like diffuse large B cell lymphoma. J Exp Med. 2007;204:633–643. 10.1084/jem.20062041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim KH, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature 2011;470:115–118. 10.1038/nature09671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laursen MB, Falgreen S, Boedker JS, Schmitz A, Kjeldsen MK, Soerensen S, et al. Human B-cell cancer cell lines as a preclinical model for studies of drug effect in diffuse large B-cell lymphoma and multiple myeloma. Exp Hematol. 2014;42:927–938. 10.1016/j.exphem.2014.07.263 [DOI] [PubMed] [Google Scholar]
- 9.Pfeifer M, Zheng B, Erdmann T, Koeppen H, McCord R, Grau M, et al. Anti-CD22 and anti-CD79B antibody drug conjugates are active in different molecular diffuse large B-cell lymphoma subtypes. Leukemia 2015;29:1578–1586. 10.1038/leu.2015.48 [DOI] [PubMed] [Google Scholar]
- 10.Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J Exp Med;194:1861–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–674.. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 12.Monni O, Joensuu H, Franssila K, Klefstrom J, Alitalo K, Knuutila S. BCL2 overexpression associated with chromosomal amplification in diffuse large B-cell lymphoma. Blood 1997;90:1168–1174. [PubMed] [Google Scholar]
- 13.Li L, Pontompipat P, Tiutan T, Kendrick SL, Park S, Persky DO et al. Synergistic induction of apoptosis in high-risk DLBCL by BCL2 inhibition with ABT-199 combined with pharmacologic loss of MCL1. Leukemia 2015;29:1702–1712. 10.1038/leu.2015.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wenzel SS, Grau M, Mavis C, Hailfinger S, Wolf A, Madle H, et al. MCL1 is deregulated in subgroups of diffuse large B-cell lymphoma. Leukemia 2013;27:1381–1390. 10.1038/leu.2012.367 [DOI] [PubMed] [Google Scholar]
- 15.Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity whil sparing platelets. Nature Med. 2013;19:202–208. 10.1038/nm.3048 [DOI] [PubMed] [Google Scholar]
- 16.Phillips DC, Xiao Y, Lam LT, Litvinovich E, Roberts-Rapp L, Souers AJ, et al. Loss in MCL-1 function sensitiztes non-Hodgkin´s lymphoma cell lines to the BCL-2-selective inhibitor venetoclax (ABT-199). Blood Cancer Journal 2015;5:e368 10.1038/bcj.2015.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leverson JD, Zhang H, Chen J, Tahir SK, Phillips DC, Xue J, et al. Potent and selective small-molecule MCL-1 inhibitors demonstrate on-target cancer killing activity as single agents and in combination with ABT-263 (navitoclax). Cell Death and Disease 2015;6:e1590 10.1038/cddis.2014.561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klanova M, Andera L, Branzina J, Svadlenka J, Benesova S, Soukup J, et al. Targeting of BCL2 family proteins with ABT-199 and homoharringtonine reveals BCL1- and MCL1-dependent subgroups of difuse large B-cell lymphoma. Clin Cancer Res. 2016;22:1138–1149. 10.1158/1078-0432.CCR-15-1191 [DOI] [PubMed] [Google Scholar]
- 19.Drexler HG. Guide to leukemia-lymphoma cell lines 2nd Edn., Braunschweig, 2010. [Google Scholar]
- 20.MacLeod RA, Kaufmann M, Drexler HG. Cytogenetic harvesting of commonly unsed tumour cell lines. Nat Protoc. 2007;2:372–382. 10.1038/nprot.2007.29 [DOI] [PubMed] [Google Scholar]
- 21.MacLeod RA, Kaufmann M, Drexler HG. Cytogenetic analysis of cancer cell lines. Methods Mol Biol. 2011;731:57–78. 10.1007/978-1-61779-080-5_6 [DOI] [PubMed] [Google Scholar]
- 22.Van Dongen JJM, Langerak AW, Brüggemann M, Evans PAS, Hummel M, Lavender FL, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 concerted action BMH4-CT98-3936. Leukemia 2003;17:2257–2317. 10.1038/sj.leu.2403202 [DOI] [PubMed] [Google Scholar]
- 23.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. 10.1093/bioinformatics/btg405 [DOI] [PubMed] [Google Scholar]
- 25.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, et al. Evolving gene/transcript definitions significantly alert the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175 10.1093/nar/gni179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quentmeier H, Schneider B, Röhrs S, Romani J, Zaborski M, MacLeod RAF, et al. SET-NUP214 fusion in acute myeloid leukemia- and T-cell acute lymphoblastic leukemia-derived cell lines. J Hematol Oncol. 2009;2:3 10.1186/1756-8722-2-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pichert G, Peters J, Stahel RA, Dommann C, Joss R, Gebbers JO, et al. Chemotherapy with MACOP-B and VACOP-B for intermediate- and high-grade non-Hodgkin´s lymphoma: clinical results and analysis of prognostic factors. Ann Oncol. 1992;3:645–649. [DOI] [PubMed] [Google Scholar]
- 28.Cabanillas F, Hagemeister FB, McLaughlin P, Velasques WS, Riggs S, Fuller L, et al. Results of MIME salvage regimen for recurrent or refractory lymphoma. J Clin Oncol. 1987;5:407–412. 10.1200/jco.1987.5.3.407 [DOI] [PubMed] [Google Scholar]
- 29.Swerdlow SH, Campo E, Harris N, Jaffe E, Pileri S, Stein H, et al. WHO classification of tumours of hematopoietic and lymphoid tissues In: Bosman FT, Jaffe ES, Lakhani SR, Ohgaki H, eds. World Health Organization Classification of Tumours. Lyon, France: IARC; 2008. [Google Scholar]
- 30.Swerdlow SH, Campo E, Pileri SA, Harris NL, Stain H, Siebert R, et al. The 2016 revision of the World health organization classification of lymphoid neoplasms. Blood 2016;127:2375–2390. 10.1182/blood-2016-01-643569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hans CP, Weisenburger DD, Greiner TC, Greiner TC, Gascoyne RD, Delabie J, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 2004;103:275–282. 10.1182/blood-2003-05-1545 [DOI] [PubMed] [Google Scholar]
- 32.Visco C, Li Y, Xu-Monette ZY, Miranda RN, Green TM, Li Y, et al. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lympnoma: a report from the international DLBCL rituximab-CHOP consortium program study. Leukemia 2012;26:2103–2113. 10.1038/leu.2012.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruminy P, Mareschal S, Bagacean C, Rainville V, Leroy K, Jais JP, et al. Accurate classification of GCB/ABC and MYC/BCL2 diffuse large B-cell lymphoma with a 14 genes expression signature and a simple and robust RT-MLPA assay. Blood 2013;122(21):84. [Google Scholar]
- 34.Ding J, Dirks WG, Ehrentraut S, Geffers R, MacLeod R, MacLeod RAF, et al. BCL6 –regulated by AhR/ARNT and wild-type MEF2B –drives expression of germinal center markers MYBL1 and LMO2. Haematologica 2015;100:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drexler HG, Eberth S, Nagel S, MacLeod RAF. Malignant hematopoietic cell lines: in vitro models for double-hit B-cell lymphomas. Leukemia Lymphoma 2016;57:1015–1020. 10.3109/10428194.2015.1108414 [DOI] [PubMed] [Google Scholar]
- 36.Herlaar E, Brown Z. p38 MAPK signalling cascades in inflammatory disease. Mol Med Today 1999;5:439–447. [DOI] [PubMed] [Google Scholar]
- 37.Palanisamy N, Abou-Elella AA, Chaganti SR, Houldsworth J, Offit K, Louie DC, et al. Similar patterns of genomic alterations characterize primary mediastinal large B-cell lymphoma and diffuse large B cell lymphoma. Genes, Chromosomes & Cancer 2002;33:114–122. [DOI] [PubMed] [Google Scholar]
- 38.Farrugia MM, Duan LJ, Reis MD, Ngan BY, Berinstein NL. Alterations of the p53 tumor suppressor gene in diffuse large cell lymphomas with translocations of the c-MYC and BCL-2 proto-oncogenes. Blood 1994;83:191–198. [PubMed] [Google Scholar]
- 39.Fridberg M, Servin A, Anagnostaki L, Linderoth J, Berglund M, Soederberg O, et al. Protein expression and cellular localization in two prognostic subgroups of diffuse large B-cell lymphoma: higher expression of ZAP70 and PKC-b II in the non-germinal center group and poor survival in patients deficient in nuclear PTEN. Leukemia Lymphoma 2007;48:2221–2232. 10.1080/10428190701636443 [DOI] [PubMed] [Google Scholar]
- 40.Klanova M, Andera L, Brazina J, Svadlenka J, Benesova S, Soukup J, et al. Targeting of BCL2 family proteins with ABT-199 and homoharringtonine reveals BCL2- and MCL1-dependent subgroups of diffuse large B-cell lymphoma. Clin Cancer Res. 2016;22:1138–1149. 10.1158/1078-0432.CCR-15-1191 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A-E) Immunohistochemical stainings of lymphoma cells. Positive: BCL6 (A), CD10 (B), CD20 (C), CD79a, FOXP1, MUM1 (partial), MYC (D), p53 and negative: BCL2 (E), CD3, CD5, CD30, cyclin D1. (F) May-Grünwald-Giemsa stain of cell line U-2946.
(TIF)
Cells were stained with Abs against the antigens indicated and analyzed by flow cytometry.
(TIF)
The 47 genes with strongest expression differences between U-2946 and median of 55 B-lymphoma cell lines in relation to variance (IQR, interquartile range)–highest differences on the left. Data base on expression array analyses. Red dots, cell line U-2946.
(TIF)
Quantitative genomic PCR (upper) and qRT-PCR (lower) detecting correlation between amplification and RNA expression in BCL2, MAP2K3 and MCL1. The bars indicate means with standard deviation (n = 3). NC-NC as diploid reference cell line for genomic PCR.
(TIF)
3H-thymidine uptake after 48 h. The MCL1 inhibitor A-1210477 (7.5 μM) inhibits growth of the MCL1pos/BCL2neg cell line U-2946, but has no effect on the MCL1pos/BCL2pos cell line U-2932 –neither alone nor together with suboptimal doses of the BCL2 inhibitor ABT-263 (50 nM). The bars indicate means with standard deviation (n = 3).
(TIF)
(XLSX)
Outliers are listed according to chromosomal position. Note the perfect correlation between genomic imbalances and expression in cell line U-2946, 6/6 hemiploid genes (black and bold) being repressed, 5/5 amplified genes (red and bold) being overexpressed. Up, higher than average; down, lower than average.
(XLSX)
Amplified genes on 1q21.3 are listed from centromere to telomere, genes on 17p11.2 from telomere to centromere. Bold: strongly expressed genes as assessed by expression array analysis.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its supporting Information files.