Abstract
Fibroblast growth factor receptor (FGFR) gene alterations are relatively frequent in lung squamous cell carcinoma (LSCC) and are a potential targets for therapy with FGFR inhibitors. However, little is known regarding the clinicopathologic features associated with FGFR alterations. The angiokinase inhibitor nintedanib has shown promising activity in clinical trials for non‐small cell lung cancer. We have now applied next‐generation sequencing (NGS) to characterize FGFR alterations in LSCC patients as well as examined the antitumor activity of nintedanib in LSCC cell lines positive for FGFR1 copy number gain (CNG). The effects of nintedanib on the proliferation of and FGFR signaling in LSCC cell lines were examined in vitro, and its effects on tumor formation were examined in vivo. A total of 75 clinical LSCC specimens were screened for FGFR alterations by NGS. Nintedanib inhibited the proliferation of FGFR1 CNG‐positive LSCC cell lines in association with attenuation of the FGFR1–ERK signaling pathway in vitro and in vivo. FGFR1 CNG (10.7%), FGFR1 mutation (2.7%), FGFR2 mutation (2.7%), FGFR4 mutation (5.3%), and FGFR3 fusion (1.3%) were detected in LSCC specimens by NGS. Clinicopathologic features did not differ between LSCC patients positive or negative for FGFR alterations. However, among the 36 patients with disease recurrence after surgery, prognosis was significantly worse for those harboring FGFR alterations. Screening for FGFR alterations by NGS warrants further study as a means to identify patients with LSCC recurrence after surgery who might benefit from nintedanib therapy.
Keywords: Copy number gain, FGFR1, lung squamous cell cancer, next‐generation sequencing, nintedanib
Lung cancer is the leading cause of cancer‐related to cancer worldwide, with NSCLC being the most common form of this disease1 and LSCC accounting for 30–35% of NSCLC cases.2 Although recent progress in molecular targeted therapy, including the development of small‐molecule drugs that target oncogene products, has improved the outlook for patients with lung adenocarcinoma,3 no “druggable” targets have been established to date for LSCC.4 Multikinase inhibitors such as sunitinib,5 sorafenib,6 pazopanib,7 and vandetanib8 that target proangiogenic signaling pathways in addition to that triggered by VEGF have been approved as anti‐angiogenic agents for the treatment of various solid tumors. The molecular targets of these inhibitors include VEGFR1–3, PDGFRα and β, and FGFR1–3. Nintedanib (formerly known as BIBF 1120) is also an angiokinase inhibitor that is highly active against VEGFR1–3, FGFR1–3, and PDGFRα/β with IC50 values of 13–34, 37–108, and 59–65 nmol/L, respectively.9 This agent has also shown promise in clinical trials for patients with various solid tumors including NSCLC,10, 11 renal cell cancer,12 ovarian cancer,13 and prostate cancer.14
Fibroblast growth factor receptor is a potential therapeutic target for several types of cancer in which FGFR signaling plays an important role in tumor growth.15 The four members of the FGFR family (FGFR1–4) are encoded by different genes. FGFR gene alterations such as mutations and amplification were found to be most common in bladder carcinoma, uterine cancer, and LSCC.16 Gene amplification and overexpression of FGFR1 or FGFR2 have also been identified in breast17 and gastric18 cancer, respectively, and mutation of FGFR3 or FGFR4 has been detected in bladder cancer19 and rhabdomyosarcoma,20 respectively. However, the consequences of FGFR genetic alterations for nintedanib treatment in LSCC patients after surgery remain unclear.
We have now characterized FGFR alterations in LSCC patients as well as evaluated the clinicopathologic features of patients positive for such gene alterations and the impact of the genetic changes on patient survival after disease recurrence. In addition, we examined the effects of nintedanib on human LSCC cell lines harboring FGFR1 CNG.
Materials and Methods
Cell culture
The human NSCLC cell line PC‐9 was provided by Tokyo Medical University (Tokyo, Japan),21, 22 and the LK‐2, A549, H520, H1299, and H1581 lines were obtained from ATCC (Manassas, VA, USA) and authenticated by short tandem repeat‐based DNA profiling (Takara Bio, Shiga, Japan). All cells were cultured under a humidified atmosphere of 5% CO2 at 37°C in RPMI‐1640 (Sigma, St. Louis, MO, USA) supplemented with 10% heat‐inactivated FBS (Equitech‐Bio, Kerrville, TX, USA).
Cell proliferation assay
Nintedanib was obtained from Selleck Chemicals (Houston, TX, USA). To assay the effect of nintedanib on cell proliferation, cells (1000–3000/well) were transferred to 96‐well flat‐bottomed plates and cultured for 24 h before the addition of various concentrations of nintedanib and incubated for an additional 72 h. TetraColor One (5 mmol/L tetrazolium monosodium salt and 0.2 mmol/L 1‐methoxy‐5‐methylphenazinium methylsulfate; Seikagaku, Tokyo, Japan) was then added to each well, and the cells were incubated for 3 h at 37°C before measurement of absorbance at 490 nm with a Multiskan Spectrum instrument (Thermo Labsystems, Boston, MA, USA). Absorbance values were expressed as a percentage of that for untreated cells, and IC50 values were calculated.
Immunoblot analysis
Protein extraction was carried out using cell lysis buffer (Cell Signaling Technology, Danvers, MA, USA) for cells and Lysing Matrix D (MP Biomedicals, Santa Ana, CA, USA) for tissues. Lysates were fractionated by SDS‐PAGE, transferred onto a nitrocellulose membrane, blocked with 5% skim milk, and incubated overnight at 4°C with primary antibodies including: p‐FGFR, ERK, AKT, and p‐AKT (Cell Signaling Technology); FGFR and p‐ERK (Santa Cruz Biotechnology, Santa Cruz, CA, USA); and β‐actin (Sigma). Immune complexes were detected by incubating the membrane for 1 h at room temperature with corresponding HRP‐conjugated goat antibodies (Amersham Biosciences, Little Chalfont, UK) and exposed to enhanced chemiluminescence reagents (Perkin‐Elmer, Boston, MA, USA).
Fluorescence in situ hybridization
FGFR copy number per cell was determined by FISH with the use of an FGFR1 Split FISH Probe (FS0025; GSP Lab, Kanagawa, Japan). Gene CNG was strictly defined on the basis of a mean FGFR1 copy number of >4. Fluorescence signals were evaluated by at least two independent observers.
Xenograft model
Mice were maintained in accordance with the Recommendations for the Handling of Laboratory Animals for Biomedical Research compiled by the Committee on Safety and Ethical Handling Regulations for Laboratory Animal Experiments (Kindai University, Osaka‐Sayama, Japan). Ethical procedures met the guidelines established by the UK Coordinating Committee on Cancer Research. Six‐week‐old female BALB/c nu/nu (nude) mice (Clea Japan, Tokyo, Japan) were injected s.c. with a suspension of H520 or LK‐2 cells (5 × 106 cells) in 100 μL PBS. After 1 week, the mice were assigned to three groups in such a manner as to ensure a similar mean tumor size in each group. Saline vehicle or nintedanib were given orally at 30 or 50 mg/kg per day for 15 days. Tumor volume (length × width2 × 0.5) was measured twice a week.
Immunohistochemistry and analysis
For immunohistochemistry, FFPE tissue sections were steamed in Dako antigen retrieval solution (Dako North America, Carpinteria, CA, USA) and incubated overnight with the following antibodies: p‐FGFR (Cell Signaling Technology), CD31 (BD Biosciences San Jose, CA, USA) and Ki‐67 (Thermo Fisher Scientific, Waltham, MA, USA). Slides were then labelled using the avidin‐biotin complex (ABC) method (Vector Laboratories, Burlingame, CA, USA) following the manufacturer's protocols, developed in 3,3’‐diaminobenzidine‐tetrachloride and counterstained with hematoxylin. Quantification was undertaken on a minimum of 10 random fields of viable tumor tissue/sample at 10× magnification. Ki‐67 expression was assessed using Aperio's ImageScope version 12.3 (Leica Biosystems, Buffalo Grove, IL, USA) and microvessel analysis was carried out using ImageJ (http://imagej.nih.gov/ij/).
Clinical specimens
A total of 75 LSCC tumor specimens were collected at Kindai University Faculty of Medicine between 2005 and 2011. All patients provided written informed consent for participation in the study, including the collection of tumor tissue for analysis. The protocol for the clinical aspect of the study was approved by the institutional ethics committee of Kindai University Faculty of Medicine (approval no. 26‐250). After pulmonary resection, the patients were examined at 3‐ or 6‐month intervals. Evaluations included physical examinations, chest radiography, or computed tomography scan, and detection of tumor markers. When recurrence was suspected, brain magnetic resonance imaging, bone scintigraphy, or positron emission tomography was performed.
Tissue processing
Formalin‐fixed, paraffin‐embedded tumor specimens underwent histological review, and only those containing sufficient tumor cells (at least 75%) as revealed by H&E staining were subjected to nucleic acid extraction. DNA and RNA were isolated from the tissue with the use of an AllPrep DNA/RNA FFPE Kit (Qiagen, Valencia, CA, USA). The quality and quantity of the nucleic acid were verified with the use of a NanoDrop 2000 device, PicoGreen dsDNA Reagent, and RiboGreen RNA Reagent (Thermo Fisher Scientific).
Next‐generation DNA sequencing
Tumor DNA was subjected to analysis with NGS panels for detection of mutations and CNG. A panel for the entire coding sequences of FGFR1, FGFR2, FGFR3, and FGFR4 was designed with the use of Ion AmpliSeq Designer (Life Technologies, Carlsbad, CA, USA) (Table S1). For evaluation of copy number gain, regions of TERT and RPPH1 were added to the panel as reference genes. The panel thus consisted of 40 amplicons of FGFR1, 43 amplicons of FGFR2, 30 amplicons of FGFR3, 34 amplicons of FGFR4, 11 amplicons of TERT, and 3 amplicons of RPPH1 in two pools. For library preparation, tumor DNA (10 ng) was subjected to multiplex PCR amplification with the use of an Ion AmpliSeq Library Kit 2.0 (Life Technologies). Polymerase chain reaction products were ligated to Ion Xpress Barcode Adapters (Life Technologies) and purified with the use of Agencourt AMPure XP beads (Beckman Coulter, Brea, CA, USA). Purified libraries were pooled and then sequenced with an Ion Torrent Proton instrument, Ion PI IC 200 Kit, and Ion PI version 2 Chip (all from Life Technologies). DNA sequencing data were accessed through the Torrent Suite version 4.4 program (Life Technologies). Reads were aligned with the hg19 human reference genome, and potential mutations were called with the use of Variant Call Format version 4.4. (Waltham, MA, USA) For detection of mutations, raw variant calls were filtered with the following annotations: quality score of <100 and depth of coverage of <19. Nucleotide and amino acid numbers refer to the following GenBank accession numbers: FGFR1, NM_023110 or NM_001174067; FGFR2, NM_000141; FGFR3, NM_000142; and FGFR4, NM_022963. Germline mutations were excluded with the use of the Human Genetic Variation Database (http://www.genome.med.kyoto-u.ac.jp/SnpDB).23 For detection of CNG, the read depth of target genes (FGFR1–4) was divided by the average depth for the reference genes (TERT and RPPH1). Adjusted read depth was log2‐transformed, and the median log2 value per gene was used for copy number analysis. The log2 ratio cut‐off value for CNG was set at 1.25 with reference to a previous study.24 The mutation and copy number data were visualized with the use of OncoPrinter.25, 26
Next‐generation RNA sequencing
A panel of FGFR fusion genes (Table S2) was designed by the Ion Torrent White Glove Team (Life Technologies). For RNA sequencing, RNA (10 ng) was subjected to reverse transcription with the use of a SuperScript VILO cDNA Synthesis Kit (Life Technologies) followed by library generation with the use of an Ion AmpliSeq Library Kit 2.0 (Life Technologies), the latter of which allows detection of transcripts derived from two FGFR1, eight FGFR2, and four FGFR3 fusion genes as well as from five housekeeping genes (HMBS, TBP, ITGB7, MYC, and LMNA). After multiplex PCR amplification, Ion Xpress Barcode Adapters were ligated to the PCR products, which were then purified with the use of Agencourt AMPure XP beads. The purified libraries were pooled and then sequenced with the use of an Ion Torrent Proton instrument, Ion PI IC 200 Kit, and Ion PI version 2 Chip. RNA sequencing data were accessed through Torrent Suite version 4.4. Read data were aligned to GRCh37/hg19 in BAM format and were analyzed for the number of counts per amplicon in each sample with a coverage analysis plugin (version 4.4; Life Technologies). Fusion genes were judged positive if the fusion read count was >100.
Real‐time PCR‐based copy number assay
DNA copy numbers for FGFR1, FGFR2, FGFR3, and FGFR4 were determined with the use of TaqMan Copy Number Assays (Applied Biosystems, Foster City, CA, USA) and primers Hs02164585_cn, Hs05208783_cn, Hs00113109_cn, and Hs01949336_cn, respectively. The TERT locus was used as an internal reference, and human genomic DNA (Promega, Madison, WI, USA) was used as a normal control. Polymerase chain reaction analysis was undertaken with an StepOnePlus Real‐Time PCR System (Applied Biosystems), and the gene copy number was calculated using the method. The cut‐off value for CNG was set at 5.0 with reference to a previous study.18
Statistical analysis
Data were reported as mean values ± SD and were analyzed using the Student's t‐test and one‐way anova. Categorical data for patient characteristics were evaluated with Fisher's exact test. Patient survival was analyzed by the Kaplan–Meier method and log–rank test. All statistical analyses were carried out with the use of Prism software (version 5.01; GraphPad Software, San Diego, CA, USA). A P‐value of <0.05 was considered statistically significant.
Results
Nintedanib inhibits the proliferation of LSCC cells harboring FGFR1 copy number gain
To investigate the antitumor activity of nintedanib for NSCLC cells harboring FGFR alterations, we examined two human LSCC cell lines (LK‐2 and H520) and one large‐cell carcinoma cell line (H1581). All three cell lines were positive for FGFR1 CNG; conversely, NSCLC cell lines (H1299, A549, and PC‐9) were negative for FGFR1 CNG, as confirmed by FISH analysis (Figs 1a,S1). Immunoblot analysis revealed increased levels of both FGFR1 and p‐FGFR1 in H520, H1581, and LK‐2 cells compared with H1299, A549, and PC‐9 (Fig. 1b). These results are consistent with previous findings.27 Nintedanib potently inhibited proliferation of all three cell lines positive for FGFR1 CNG with IC50 values of 0.76 ± 0.04, 0.55 ± 0.24, and 0.09 ± 0.01 μM for H520, H1581, and LK‐2 cells, respectively (Fig. 1c). In contrast, the IC50 values of nintedanib for lung cancer cells without FGFR1 CNG were approximately an order of magnitude larger (6.14 ± 1.28, 3.98 ± 0.16, and 5.37 ± 2.38 μM for H1299, A549, and PC‐9 cells, respectively), indicating that nintedanib selectively inhibits the proliferation of cells positive for FGFR1 CNG. Nintedanib inhibited the p‐FGFR1 and its downstream signaling molecule ERK in a concentration‐dependent manner in H520 and LK‐2 cells (Fig. 1d). It also completely inhibited the phosphorylation of the downstream kinase AKT at a concentration of 1 μM in H520 cells but not in LK‐2 cells (Fig. 1d). Nintedanib had no effect on the phosphorylation of these various signaling molecules in cells negative for FGFR1 CNG (data not shown). These results suggest that nintedanib inhibits the proliferation of FGFR1 CNG‐positive lung cancer cells, at least in part, by blocking the FGFR1–ERK signaling pathway.
Figure 1.
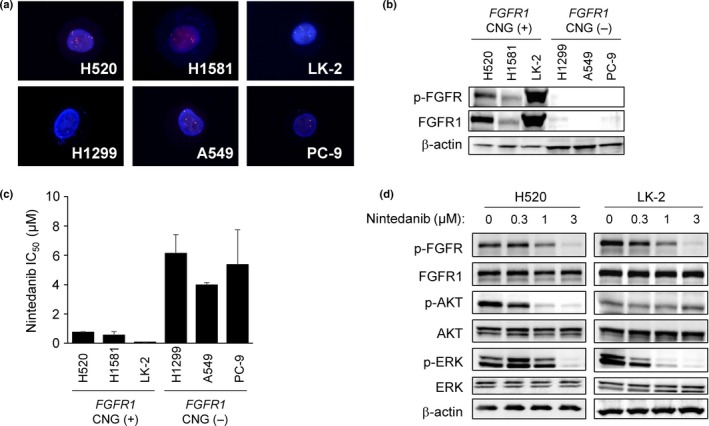
Sensitivity of lung cancer cell lines positive for FGFR1 copy number gain (CNG) to nintedanib. (a) FISH analysis of FGFR1 copy number in lung cancer cell lines. The 5′ and 3′ probe signals for FGFR1 appear green and red, respectively. Nuclei are stained blue with DAPI. (b) Immunoblot analysis of phosphorylated fibroblast growth factor receptor (p‐FGFR), FGFR1, and β‐actin (loading control) in lung cancer cell lines positive (H520, H1581, LK‐2) or negative (H1299, A549, PC‐9) for FGFR1 CNG. (c) Effects of nintedanib on the proliferation of lung cancer cell lines according to FGFR1 copy number status. The IC 50 values are means ± SD from three independent experiments. (d) Effects of nintedanib on FGFR1, ERK, and AKT phosphorylation in FGFR1 CNG‐positive lung squamous cell carcinoma cell lines. H520 and LK‐2 cells were incubated for 6 h in the presence of the indicated concentrations of nintedanib, after which cell lysates (25 μg soluble protein) were subjected to immunoblot analysis with antibodies to the indicated proteins.
Nintedanib inhibits the growth of FGFR1 CNG‐positive LSCC xenografts
We examined the preclinical efficacy of nintedanib treatment on H520 and LK‐2 LSCC cells harboring FGFR1 alterations. Nintedanib was given orally at a 30 or 50 mg/kg per day for 15 days. Nintedanib was well tolerated and no body weight loss was observed (Fig. 2a). Treatment with nintedanib inhibited the growth of H520 tumors in a dose‐dependent manner by 26.7% and 41.4% (P = 0.0082, 50 mg/kg vs vehicle) at 30 and 50 mg/kg, respectively (Fig. 2b). Treatment of LK‐2 tumors with nintedanib yielded a ~34% reduction of tumor growth regardless of dose. Although nintedanib showed some suppression of LK‐2 xenograft growth, the differences were not statistically significant (Fig. 2b).
Figure 2.
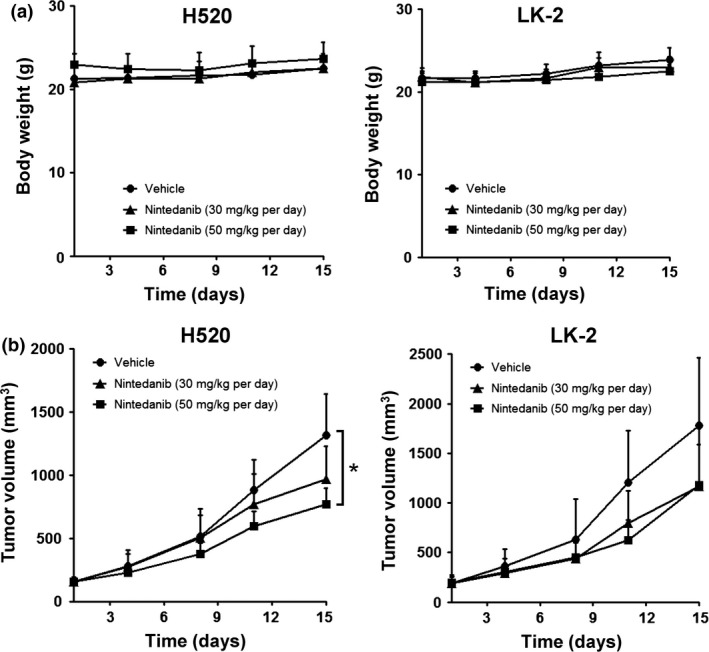
Inhibition of FGFR1 copy number gain‐positive lung squamous cell carcinoma tumor growth in vivo by nintedanib. Nude mice with s.c. tumors formed by injected H520 or LK‐2 cells were treated orally with nintedanib at a dose of 30 or 50 mg/kg per day or with vehicle alone for 15 days. (a) Effects of nintedanib on body weight change of mice harboring xenograft tumors. (b) Antitumor effect of nintedanib treatment. Tumor volume was determined at the indicated times after the onset of treatment. Data are means ± SD. *P < 0.05 versus corresponding value for vehicle‐treated mice (Student's t‐test).
To determine whether the antitumor activity of nintedanib observed on FGFR1 CNG‐positive LSCC tumors was a direct result of FGFR1 signal inhibition, we characterized FGFR1 signaling in H520 and LK‐2 xenografts. Indeed, immunoblot analyses revealed that nintedanib inhibited the FGFR1 phosphorylation and its downstream molecule ERK in H520 tumors (Fig. 3a). However, low levels of p‐FGFR1 were detected in LK‐2 cells at basal levels (vehicle), and remained unchanged after treatment. Notably, p‐ERK decreased in LK‐2 tumors treated with high dose nintedanib. Both VEGFR and PDGFR are also possible targets of nintedanib and may contribute to decreased ERK activity. However, due to technical limitations, we were unable to measure VEGFR and PDGFR phosphorylation in our samples. We confirmed FGFR1 activity in LSCC xenografts by immunohistochemistry. In the H520 tumor xenografts, p‐FGFR1 was localized in both human cancer and mouse stromal/vascular cells. Treatment with nintedanib clearly reduced p‐FGFR1 in both cell types (Fig. 3b). However, in LK‐2 tumors, p‐FGFR1 was detected in the mouse stromal/vascular cells but not the LK‐2 cells. Still, nintedanib suppressed p‐FGFR1 in mouse stroma (Fig. 3b).
Figure 3.
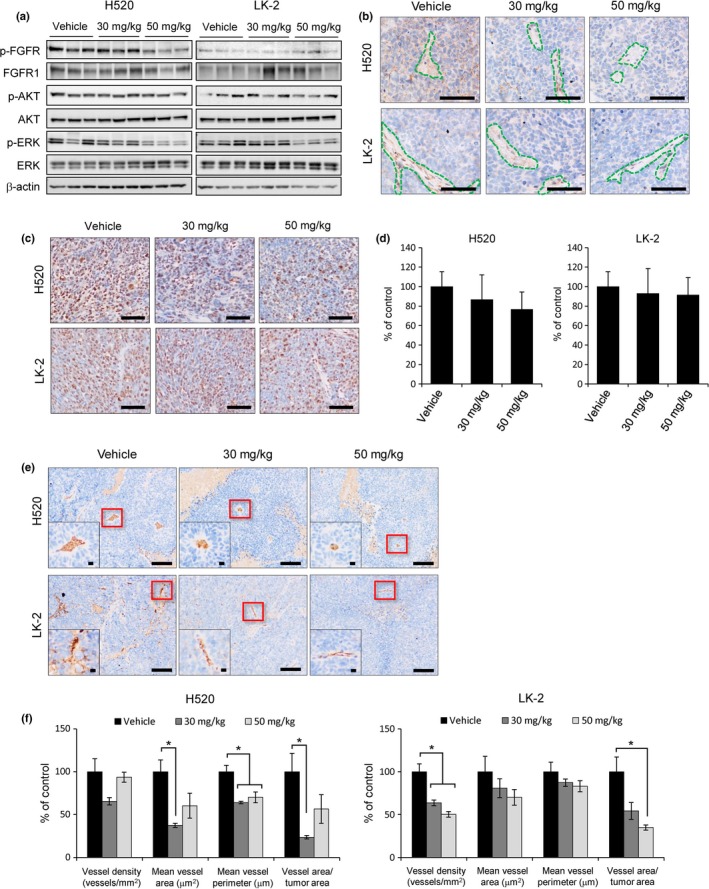
Effects of nintedanib therapy on molecular markers in vivo. Tumor samples were collected from mice 2 h after the final gavage following 15 days of treatment as indicated and subjected to molecular marker analysis. (a) Immunoblot analysis of the effects of nintedanib therapy on FGFR1, ERK, and AKT phosphorylation in the H520 and LK‐2 xenograft tumor model. Cell lysates (25 μg soluble protein) were subjected to immunoblot analysis with antibodies to the indicated proteins. (b) Representative images of p‐FGFR1 immunostaining in each group. Stromal/vascular regions are denoted by the green dotted line. Scale bar = 100 μm. (c) Representative images of Ki‐67 immunostaining in each group. Scale bar = 100 μm. (d) Quantitative analyses of the number of Ki‐67‐positive cells in each group. (e) Representative images of CD31 immunostaining in each group. Insets show higher‐magnification images of the blood vessel shown by the red frame. Scale bar = 100 μm (whole image) and 10 μm (inset image). (f) Quantitative analyses of vessel density, vessel area, vessel perimeter, and the ratio of vessel area/tumor area determined with CD31‐positive cells. *P < 0.05 versus corresponding value for vehicle‐treated mice (Student's t‐test).
Fibroblast growth factor receptor 1 signaling has been implicated in a wide range of cellular processes, including proliferation.15 Therefore, we examined the influence of FGFR1 signal inhibition with nintedanib on cancer cell proliferation by Ki‐67 expression (Fig. 3c,d). A clear but statistically insignificant dose‐dependent trend of reduced of Ki‐67‐positive cells was observed in H520 xenografts (Fig. 3c,d). In contrast, no changes in Ki‐67‐positive cells were observed in nintedanib‐treated LK‐2 xenografts. Changes in tumor cell proliferation were similar to changes in p‐FGFR1, suggesting that inhibition of FGFR1 phosphorylation by nintedanib may have contributed to suppress tumor growth.
To further determine the antitumor activity of nintedanib on LSCC tumors, we examined its effects on neovascularization. Figure 3e shows representative images of CD31‐positive microvessels in tumor sections. Treatment with nintedanib reduced overall vessel size (vessel area, vessel perimeter) in H520 xenografts, resulting in a diminished blood supply to the tumors (ration of vessel area/tumor area) (Fig. 3f). In contrast, nintedanib decreased blood supply to LK‐2 xenografts by reducing vessel density (Fig. 3f). Differences in the patterns of tumor microvasculature between the two models may be due to variances in tumor cell proliferation, metabolic demands, and complex interactions with host stromal cells.28 Still, it is important to note that in both models, treatment with nintedanib increased the intercapillary distance, indicating anti‐angiogenic activity. Interestingly, a dose‐dependent effect was not observed in either case, suggesting that low‐dose nintedanib may be sufficient to induce anti‐angiogenic effects, but higher doses might be required to inhibit cancer cell proliferation.
Overall, we showed that nintedanib produces a potent antitumor effect on H520 xenografts and is characterized by decreased p‐FGFR1 signaling, cancer cell proliferation, neovascularization, and tumor volume. Hence, our findings suggest that FGFR1 CNG‐positive LSCC showing increased FGFR1 signaling may be susceptible to therapy with multikinase inhibitors such as nintedanib.
FGFR alterations in clinical LSCC specimens
We next investigated FGFR alteration status in 75 clinical LSCC specimens by NGS. DNA and RNA sequencing was carried out to detect copy number variation, mutations, and fusion genes. Copy number variation for FGFR1, FGFR2, FGFR3, and FGFR4 was analyzed with a log2 ratio cut‐off value for copy number gain of 1.25. An increased FGFR1 copy number was detected in 8/75 (10.7%) clinical LSCC specimens (Fig. 4a), whereas CNG for FGFR2, FGFR3, or FGFR4 was not apparent in any of the tumor specimens. To evaluate the accuracy of CNG determined by NGS, we also used a real‐time PCR‐based copy number assay with a cut‐off value of 5.0. An increased FGFR1 copy number was detected in 10/75 (13.3%) LSCC specimens (Fig. 4b), with no CNG again being apparent for FGFR2, FGFR3, or FGFR4 in any of the tumor samples. Comparison of FGFR1 copy number status, determined by the detection of FGFR1 CNG with NGS and real‐time PCR assay, revealed a sensitivity and specificity of 80% (8/10) and 100% (65/65), respectively, with an accuracy of 97.3% (73/75) (Table S3). Matched specimens for all eight tumors found to be positive for FGFR1 copy number gain by NGS were also judged to be positive for FGFR1 CNG by FISH (Fig. 4c), suggesting that the power of NGS for detection of copy number gain is similar to that of standard FISH analysis. Somatic missense mutations in the coding regions of FGFR1, FGFR2, FGFR3, and FGFR4 were detected in 2/75 (2.7%), 2/75 (2.7%), 0/75 (0%), and 4/75 (5.3%) specimens, respectively, with one tumor showing missense mutations of both FGFR1 and FGFR2. The NGS analysis of FGFR1, FGFR2, and FGFR3 fusion genes in the 75 LSCC specimens revealed only the presence of FGFR3‐TACC3 in one case, which was also positive for a somatic mutation of FGFR4. In total, FGFR alterations were detected by NGS in 15/75 (20.0%) clinical LSCC specimens (Fig. 5), consistent with data from The Cancer Genome Atlas.29 Moreover, FGFR gene copy number and mutations were mutually exclusive. The results of NGS and real‐time PCR analysis for all 75 LSCC specimens are shown in Table S4.
Figure 4.
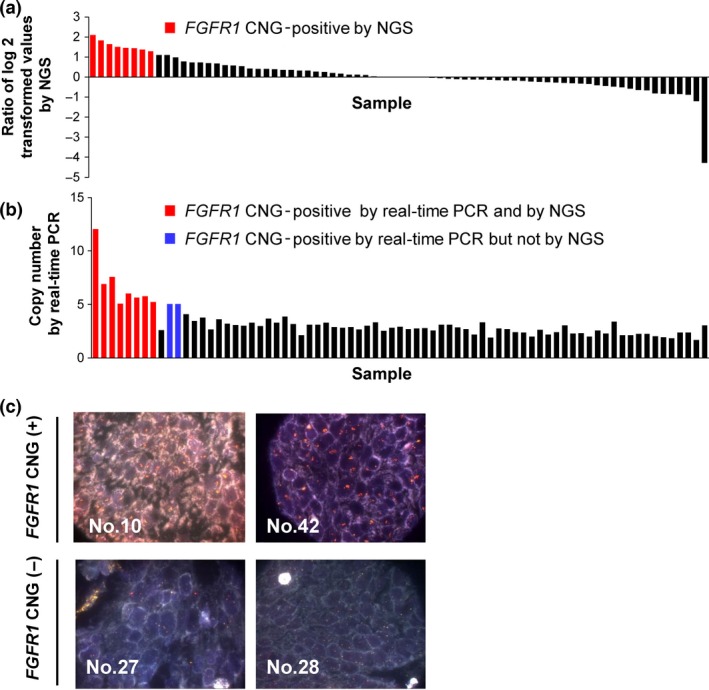
FGFR1 copy number gain in clinical lung squamous cell carcinoma specimens. (a,b) Distribution of FGFR1 copy number determined by NGS (a) or real‐time PCR analysis (b) for 75 lung squamous cell carcinoma specimens. (c) Representative FISH images for specimens positive or negative for FGFR1 copy number gain. The 5′ and 3′ probe signals for FGFR1 appear green and red, respectively.
Figure 5.
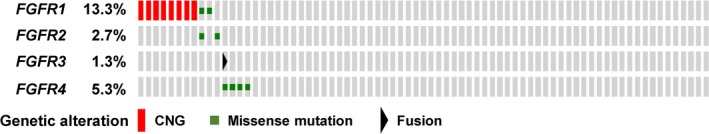
Distribution of FGFR alterations in 75 clinical lung squamous cell carcinoma specimens as determined by NGS and visualized by OncoPrinter. CNG, copy number gain.
Prognostic value of FGFR alterations in LSCC patients
The clinicopathologic features of the LSCC cohort are summarized in Table 1. Tumor tissues were collected from 75 LSCC patients at surgical resection. Among all patients, there was no significant difference in age, sex, smoking status, clinical stage, or recurrence between patients classified as FGFR alteration‐positive or ‐negative by NGS. The OS rates of the FGFR alteration‐positive and ‐negative patients were investigated. There was no difference in median survival for both groups (Fig. 6a). We further analyzed the association between FGFR alteration status and survival in the subgroup of patients with recurrence. Thirty‐six of the 75 patients relapsed at a median of 6.9 months after surgery, with the most frequent sites for metastasis being bone, liver, and brain. Among the recurrent cases, 34 of 36 patients received radiotherapy (12/34), chemotherapy (12/34), or chemoradiation (10/34). Of the other two patients, one patient died before starting treatment. The remaining patient underwent surgical resection for the recurrent lung tumor. There was no significant difference in OS among the treatment choice. Subset analysis of these 36 patients revealed that the FGFR alteration‐positive group had a shorter median OS than did the FGFR alteration‐negative group (17.5 vs 37.7 months, P = 0.0025) (Fig. 6b). FGFR alteration‐positive patients tended to have shorter recurrence‐free survival compared with the alteration‐negative patients (6.7 vs 7.1 months, P = 0.1393) (Fig. 6c), although this difference was not statistically significant. These results suggest that FGFR alteration might be a prognostic marker for LSCC patients with recurrence after surgery.
Table 1.
Clinical characteristics of patients with lung squamous cell carcinoma classified according to FGFR alteration status as determined by NGS
| Characteristic | Total cohort (n = 75) | FGFR alteration positive (n = 15) | FGFR alteration negative (n = 60) | P‐value |
|---|---|---|---|---|
| No. (%) | No. (%) | No. (%) | ||
| Age, years (mean ± SD, 70.6 ± 8.3) | ||||
| <65 | 16 (21.3) | 1 (6.7) | 15 (25.0) | 0.121 |
| ≥65 | 59 (78.7) | 14 (93.3) | 45 (75.0) | |
| Sex | ||||
| Male | 69 (92.0) | 14 (93.3) | 55 (91.7) | 0.832 |
| Female | 6 (8.0) | 1 (6.7) | 5 (8.3) | |
| Smoking | ||||
| Yes | 73 (97.3) | 15 (100.0) | 58 (96.7) | 0.474 |
| No | 2 (2.7) | 0 (0.0) | 2 (3.3) | |
| Stage | ||||
| I | 26 (34.7) | 6 (40.0) | 20 (33.3) | 0.672 |
| II | 27 (36.0) | 6 (40.0) | 21 (35.0) | |
| III | 22 (29.3) | 3 (20.0) | 19 (31.7) | |
| Recurrence | ||||
| Yes | 36 (48.0) | 6 (40.0) | 30 (50.0) | 0.488 |
| No | 39 (52.0) | 9 (60.0) | 30 (50.0) | |
P‐values were determined by Fisher's exact test.
Figure 6.
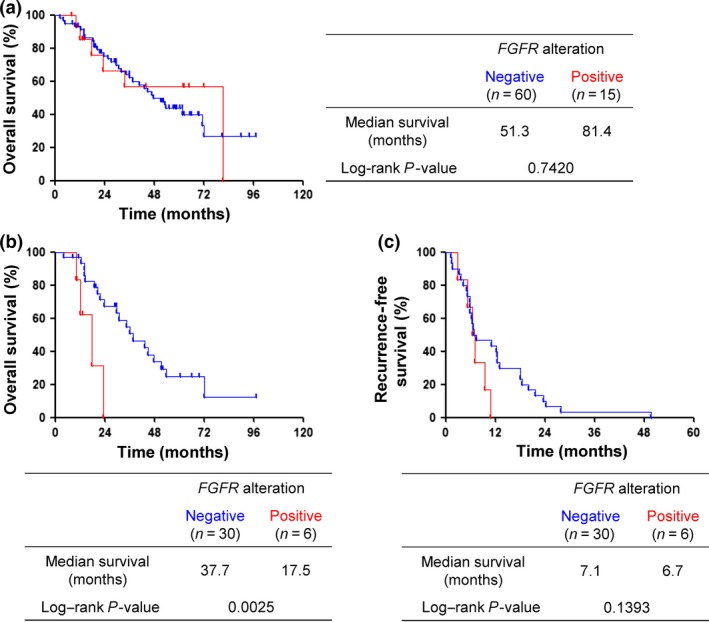
Survival analysis for overall survival and recurrence‐free survival according to FGFR alteration status as determined by NGS. (a) Overall survival of patients with or without FGFR alteration for the whole population (n = 75). (b) Subset analysis of overall survival according to FGFR alteration status for lung squamous cell carcinoma patients with recurrence (n = 36). (c) Recurrence‐free survival of relapsed patients with or without FGFR alteration (n = 36).
Discussion
We have here shown the utility of FGFR alteration screening using NGS. The overall frequency of FGFR alterations in LSCC detected by NGS was 20.0% in the present study. Among the relapsed patients in this study, the OS time in FGFR alteration‐positive patients was significantly shorter than that in negative patients. Thus, FGFR alteration is considered a potential target for therapy with FGFR inhibitors. Nintedanib is a triple angiokinase inhibitor that simultaneously acts on VEGFR, PDGFR, and FGFR.9 There is no previous study reporting the antitumor activity of nintedanib to LSCC with FGFR1 CNG. The screening for FGFR alteration in order to select patients whose tumors may be sensitive to nintedanib thus warrants further investigation as a potential new therapeutic approach for LSCC.
We have also shown that nintedanib inhibited proliferation of the FGFR1 CNG‐positive LSCC cell lines H520 and LK‐2 in vitro as well as the growth of tumors formed by these cells in vivo. Additionally, nintedanib exerted stronger antitumor activity in H520 cells in vivo through the suppression of tumor cell proliferation and neovascularization. Accordingly, these effects were consistent with inhibition of FGFR signaling, which has previously been identified as a potential therapeutic target in LSCC.30 It is important to note that there were discrepancies between in vivo and in vitro FGFR1 activity of LK‐2 cells. Notably, LK‐2 cells showed high basal levels of FGFR1 phosphorylation in vitro and were effectively inhibited by nintedanib. However, LK‐2 xenografts showed low FGFR1 activity (Fig. 3a,b). Although the precise reason for this inconsistency is unknown, it is not uncommon for tumor cells to show different molecular phenotypes between 2‐D in vitro culture and in vivo models. Nevertheless, this phenomenon afforded us an opportunity to compare FGFR1‐specific activity of nintedanib. The difference in FGFR1 phosphorylation between the two tumor models could account for the difference in sensitivity to nintedanib. In essence, our findings showed that inhibition of the FGFR1 signal pathway targets both cancer and vascular cells in tumors and provided preclinical evidence that supports the targeting of FGFR1 with nintedanib in LSCC tumors with FGFR1 alterations.
Nintedanib monotherapy provides modest preclinical activity and will warrant further studies to determine whether rational combinations with other therapeutic methods may improve treatment responses. In the clinical setting, a phase III clinical trial carried out in patients with advanced NSCLC reported that nintedanib combined with docetaxel significantly improved progression‐free survival compared to docetaxel alone in squamous populations, but did not prolong OS.11 The combination of nintedanib with platinum doublet (cisplatin plus gemcitabine) was also evaluated in a phase I/II study (NCT01346540). Combination with platinum doublet is considered to be a potential strategy for clinical development of nintedanib.
Aberrant activation of FGFR signaling can result from FGFR alterations including gene amplification, somatic missense mutation, and chromosomal translocation. Next‐generation sequencing technology encompassing the sequencing of both DNA and RNA allows the simultaneous detection of copy number variation, mutations, and gene fusions. We have also now applied this approach to evaluate FGFR copy number, mutation, and fusion in clinical specimens of LSCC. Copy number gain for FGFR1 was the most frequently detected FGFR alteration in our LSCC cohort. The results for FGFR1 CNG obtained by NGS showed a 97.3% concordance with those obtained with a real‐time PCR‐based copy number assay. Previous studies also identified FGFR1 amplification as a frequent FGFR alteration in LSCC.27, 31 The frequency of FGFR1 amplification among LSCC patients who were current smokers was also previously found to be 15.8–28.9%.32, 33 In the present study, 97.3% of LSCC patients had a smoking history and the frequency of FGFR1 CNG was 13.3% (10/75) as determined by the real‐time PCR assay, consistent with these previous results.
We detected FGFR missense mutations at a frequency of only 9.3% (7/75) in our LSCC cohort, consistent with the results of a previous study.29 Fibroblast growth factor receptor proteins consist of an extracellular Ig‐like domain, a transmembrane domain, and a cytoplasmic kinase domain. We detected FGFR1, FGFR2, and FGFR4 mutations in 2.7%, 2.7%, and 5.3% of LSCC specimens. These mutations included: (i) G70R located in the Ig‐like domain of FGFR1 (NM_023110), which has previously been detected in lung adenocarcinoma;34 (ii) P582S located in the tyrosine kinase domain of FGFR2, with mutations at this site also having been detected in cancer cell lines;35 and (iii) T27I of FGFR1 (NM_001174067), T14I of FGFR2, as well as E381K, S382L, and G408S of FGFR4, all of which are located in alternative exons and have not been previously reported. FGFR fusion genes including BAG4–FGFR1, SLC45A3–FGFR2, and FGFR3–TACC3 have been detected in various cancer types including LSCC, prostate, and bladder tumors.36, 37, 38 We detected only one FGFR fusion gene, FGFR3–TACC3, in our LSCC cohort by NGS.
The association between the nintedanib and other FGFR gene alterations, other than FGFR1 CNG, remains unclear and should be clarified in future studies. However, there are some reports regarding FGFR kinase or multikinase inhibitors against cancers with other FGFR gene alterations. AZD4547, a selective inhibitor of FGFR1, FGFR2, and FGFR3, has shown potent antitumor activity against an FGFR2 amplified xenograft model.39 Tumors, including LSCC harboring FGFR2, FGFR3, or FGFR4 mutations, were found to be sensitive to the FGFR selective inhibitor BGJ398 and a multikinase inhibitor (ponatinib) in vitro and in vivo.40, 41 Tumors harboring FGFR fusions also showed increased sensitivity to the FGFR selective inhibitor JNJ‐42756493 and the multikinase inhibitor pazopanib.42, 43 Thus, FGFR gene alterations other than FGFR1 CNG are considered to be a potential targets for cancer therapy.
Of the 75 patients in the present cohort, 36 individuals showed relapse at a median interval of 6.9 months after surgery. No difference in relapse frequency was apparent between patients whose tumors were positive or negative for FGFR alteration, suggesting that FGFR alteration is not a risk factor for recurrence. In contrast, FGFR1 amplification was associated with a greater risk of recurrence in individuals with esophageal cancer.44 This difference between LSCC and esophageal cancer may be due to biological differences between the two tumor types. Among the relapsed patients in the present study, the OS time of those positive for FGFR alterations was significantly shorter than in those without such genetic changes. The difference in recurrence‐free survival between the two groups did not reach statistical significance; this is likely due to small sample size.
In summary, we have screened LSCC specimens for FGFR alterations including mutations, copy number variation, and fusions by NGS. The detection rate of NGS for CNG was similar to that of real‐time PCR, and the NGS results were validated by FISH analysis. The presence of FGFR aberrations as detected by NGS was associated with a significantly worse prognosis among patients with disease recurrence after surgery. Together with our finding that nintedanib showed antitumor activity both in vitro and in vivo against LSCC cells positive for FGFR1 CNG, these results suggest that clinical investigation of nintedanib as a potential targeted therapy for recurrent LSCC positive for FGFR alteration is warranted.
Disclosure Statement
M. Hibi is an employee of SRL Inc. (Tokyo, Japan). H. Kaneda has received lecture fees from Chugai Pharmaceutical and Pfizer. Y. Togashi has received lecture fees from Boehringer‐Ingelheim. M.A. De Velasco has received research funding from AstraZeneca. T. Mitsudomi has received lecture fees and research funding from AstraZeneca, Boehringer‐Ingelheim, Chugai Pharmaceutical, and Pfizer. K. Nakagawa has received lecture fees and research funding from Chugai Pharmaceutical, Eli Lilly Japan, Daiichi Sankyo, AstraZeneca, Pfizer, Astellas Pharma, Boehringer‐Ingelheim, Bristol Myers Squibb, Taiho Pharmaceutical, Ono Pharmaceutical, Oncotherapy Science, MSD, EPS Associates, Eisai, Takeda Pharmaceutical, Japan Clinical Research Operations, and Quintiles. I. Okamoto has received lecture fees and research funding from Chugai, Boehringer‐Ingelheim, Eli Lilly, Pfizer, and AstraZeneca. K. Nishio has received lecture fees from Chugai Pharmaceutical, Daiichi Sankyo, and Sumitomo Bakelite. The other authors have no conflict of interest.
Abbreviations
- AKT
protein kinase B
- CNG
copy number gain
- FGFR
fibroblast growth factor receptor
- IC50
half maximal (50%) inhibitory concentration
- LSCC
lung squamous cell carcinoma
- NGS
next‐generation sequencing
- NSCLC
non‐small cell lung cancer
- OS
overall survival
- p‐
phosphorylated
- PDGFR
platelet‐derived growth factor receptor
- VEGF
vascular endothelial growth factor
- VEGFR
VEGF receptor
Supporting information
Fig. S1. Fluorescence in situ hybridization analysis of FGFR1 copy number in lung cancer cell lines.
Table S1. Primer sequence for next‐generation DNA sequencing.
Table S2. Primer sequence for next‐generation RNA sequencing.
Table S3. Methods correlation between next‐generation sequencing (NGS) and copy number assay.
Table S4. FGFR alterations in lung squamous cell carcinoma specimens detected by next‐generation sequencing and real‐time PCR.
Acknowledgment
We thank Y. Hosono, H. Sakamoto, T. Miyazaki, A. Kurumatani, M. Tsukihara, and M. Kitano for their technical assistance.
Cancer Sci 107 (2016) 1667–1676
Funding InformationApplied Research for Innovative Treatment of Cancer (14525177) Ministry of Health, Labor, and Welfare of Japan.
References
- 1. Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin 2014; 64: 9–29. [DOI] [PubMed] [Google Scholar]
- 2. Travis WD, Brambilla E, Nicholson AG et al The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol 2015; 10: 1243–60. [DOI] [PubMed] [Google Scholar]
- 3. Shea M, Costa DB, Rangachari D. Management of advanced non‐small cell lung cancers with known mutations or rearrangements: latest evidence and treatment approaches. Ther Adv Respir Dis 2016; 10: 113–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scagliotti GV, Parikh P, von Pawel J et al Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy‐naive patients with advanced‐stage non‐small‐cell lung cancer. J Clin Oncol 2008; 26: 3543–51. [DOI] [PubMed] [Google Scholar]
- 5. Motzer RJ, Hutson TE, Tomczak P et al Sunitinib versus interferon alfa in metastatic renal‐cell carcinoma. N Engl J Med 2007; 356: 115–24. [DOI] [PubMed] [Google Scholar]
- 6. Escudier B, Eisen T, Stadler WM et al Sorafenib in advanced clear‐cell renal‐cell carcinoma. N Engl J Med 2007; 356: 125–34. [DOI] [PubMed] [Google Scholar]
- 7. Sternberg CN, Davis ID, Mardiak J et al Pazopanib in locally advanced or metastatic renal cell carcinoma: results of a randomized phase III trial. J Clin Oncol 2010; 28: 1061–8. [DOI] [PubMed] [Google Scholar]
- 8. Wells SA Jr, Robinson BG, Gagel RF et al Vandetanib in patients with locally advanced or metastatic medullary thyroid cancer: a randomized, double‐blind phase III trial. J Clin Oncol 2012; 30: 134–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Awasthi N, Hinz S, Brekken RA, Schwarz MA, Schwarz RE. Nintedanib, a triple angiokinase inhibitor, enhances cytotoxic therapy response in pancreatic cancer. Cancer Lett 2015; 358: 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reck M, Kaiser R, Eschbach C et al A phase II double‐blind study to investigate efficacy and safety of two doses of the triple angiokinase inhibitor BIBF 1120 in patients with relapsed advanced non‐small‐cell lung cancer. Ann Oncol 2011; 22: 1374–81. [DOI] [PubMed] [Google Scholar]
- 11. Reck M, Kaiser R, Mellemgaard A et al Docetaxel plus nintedanib versus docetaxel plus placebo in patients with previously treated non‐small‐cell lung cancer (LUME‐Lung 1): a phase 3, double‐blind, randomised controlled trial. Lancet Oncol 2014; 15: 143–55. [DOI] [PubMed] [Google Scholar]
- 12. Eisen T, Loembe AB, Shparyk Y et al A randomised, phase II study of nintedanib or sunitinib in previously untreated patients with advanced renal cell cancer: 3‐year results. Br J Cancer 2015; 113: 1140–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ledermann JA, Hackshaw A, Kaye S et al Randomized phase II placebo‐controlled trial of maintenance therapy using the oral triple angiokinase inhibitor BIBF 1120 after chemotherapy for relapsed ovarian cancer. J Clin Oncol 2011; 29: 3798–804. [DOI] [PubMed] [Google Scholar]
- 14. Droz JP, Medioni J, Chevreau C et al Randomized phase II study of nintedanib in metastatic castration‐resistant prostate cancer postdocetaxel. Anticancer Drugs 2014; 25: 1081–8. [DOI] [PubMed] [Google Scholar]
- 15. Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell 2000; 103: 211–25. [DOI] [PubMed] [Google Scholar]
- 16. Helsten T, Elkin S, Arthur E, Tomson BN, Carter J, Kurzrock R. The FGFR landscape in cancer: analysis of 4,853 tumors by next‐generation sequencing. Clin Cancer Res 2016; 22: 259–67. [DOI] [PubMed] [Google Scholar]
- 17. Turner N, Pearson A, Sharpe R et al FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res 2010; 70: 2085–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matsumoto K, Arao T, Hamaguchi T et al FGFR2 gene amplification and clinicopathological features in gastric cancer. Br J Cancer 2012; 106: 727–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gust KM, McConkey DJ, Awrey S et al Fibroblast growth factor receptor 3 is a rational therapeutic target in bladder cancer. Mol Cancer Ther 2013; 12: 1245–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Taylor JG, Cheuk AT, Tsang PS et al Identification of FGFR4‐activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J Clin Invest 2009; 119: 3395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kawamura‐Akiyama Y, Kusaba H, Kanzawa F, Tamura T, Saijo N, Nishio K. Non‐cross resistance of ZD0473 in acquired cisplatin‐resistant lung cancer cell lines. Lung Cancer 2002; 38: 43–50. [DOI] [PubMed] [Google Scholar]
- 22. Nishio K, Arioka H, Ishida T et al Enhanced interaction between tubulin and microtubule‐associated protein 2 via inhibition of MAP kinase and CDC2 kinase by paclitaxel. Int J Cancer 1995; 63: 688–93. [DOI] [PubMed] [Google Scholar]
- 23. Narahara M, Higasa K, Nakamura S et al Large‐scale East‐Asian eQTL mapping reveals novel candidate genes for LD mapping and the genomic landscape of transcriptional effects of sequence variants. PLoS One 2014; 9: e100924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lonigro RJ, Grasso CS, Robinson DR et al Detection of somatic copy number alterations in cancer using targeted exome capture sequencing. Neoplasia 2011; 13: 1019–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cerami E, Gao J, Dogrusoz U et al The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2: 401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gao J, Aksoy BA, Dogrusoz U et al Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Weiss J, Sos ML, Seidel D et al Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med 2010; 2: 62ra93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hlatky L, Hahnfeldt P, Folkman J. Clinical application of antiangiogenic therapy: microvessel density, what it does and doesn't tell us. J Natl Cancer Inst 2002; 94: 883–93. [DOI] [PubMed] [Google Scholar]
- 29. Cancer Genome Atlas Research Network . Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012; 489: 519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tiseo M, Gelsomino F, Alfieri R et al FGFR as potential target in the treatment of squamous non small cell lung cancer. Cancer Treat Rev 2015; 41: 527–39. [DOI] [PubMed] [Google Scholar]
- 31. Heist RS, Mino‐Kenudson M, Sequist LV et al FGFR1 amplification in squamous cell carcinoma of the lung. J Thorac Oncol 2012; 7: 1775–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Goke F, Franzen A, Menon R et al Rationale for treatment of metastatic squamous cell carcinoma of the lung using fibroblast growth factor receptor inhibitors. Chest 2012; 142: 1020–6. [DOI] [PubMed] [Google Scholar]
- 33. Kim HS, Mitsudomi T, Soo RA, Cho BC. Personalized therapy on the horizon for squamous cell carcinoma of the lung. Lung Cancer 2013; 80: 249–55. [DOI] [PubMed] [Google Scholar]
- 34. Ding L, Getz G, Wheeler DA et al Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008; 455: 1069–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Abaan OD, Polley EC, Davis SR et al The exomes of the NCI‐60 panel: a genomic resource for cancer biology and systems pharmacology. Cancer Res 2013; 73: 4372–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang R, Wang L, Li Y et al FGFR1/3 tyrosine kinase fusions define a unique molecular subtype of non‐small cell lung cancer. Clin Cancer Res 2014; 20: 4107–14. [DOI] [PubMed] [Google Scholar]
- 37. Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet 2013; 22: 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu YM, Su F, Kalyana‐Sundaram S et al Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov 2013; 3: 636–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xie L, Su X, Zhang L et al FGFR2 gene amplification in gastric cancer predicts sensitivity to the selective FGFR inhibitor AZD4547. Clin Cancer Res 2013; 19: 2572–83. [DOI] [PubMed] [Google Scholar]
- 40. Li SQ, Cheuk AT, Shern JF et al Targeting wild‐type and mutationally activated FGFR4 in rhabdomyosarcoma with the inhibitor ponatinib (AP24534). PLoS One 2013; 8: e76551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liao RG, Jung J, Tchaicha J et al Inhibitor‐sensitive FGFR2 and FGFR3 mutations in lung squamous cell carcinoma. Cancer Res 2013; 73: 5195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Capelletti M, Dodge ME, Ercan D et al Identification of recurrent FGFR3‐TACC3 fusion oncogenes from lung adenocarcinoma. Clin Cancer Res 2014; 20: 6551–8. [DOI] [PubMed] [Google Scholar]
- 43. Di Stefano AL, Fucci A, Frattini V et al Detection, characterization, and inhibition of FGFR‐TACC fusions in IDH wild‐type glioma. Clin Cancer Res 2015; 21: 3307–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim HS, Lee SE, Bae YS et al Fibroblast growth factor receptor 1 gene amplification is associated with poor survival in patients with resected esophageal squamous cell carcinoma. Oncotarget 2015; 6: 2562–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Fluorescence in situ hybridization analysis of FGFR1 copy number in lung cancer cell lines.
Table S1. Primer sequence for next‐generation DNA sequencing.
Table S2. Primer sequence for next‐generation RNA sequencing.
Table S3. Methods correlation between next‐generation sequencing (NGS) and copy number assay.
Table S4. FGFR alterations in lung squamous cell carcinoma specimens detected by next‐generation sequencing and real‐time PCR.


