Abstract
We recently found that the product of the AES gene functions as a metastasis suppressor of colorectal cancer (CRC) in both humans and mice. Expression of amino‐terminal enhancer of split (AES) protein is significantly decreased in liver metastatic lesions compared with primary colon tumors. To investigate its downregulation mechanism in metastases, we searched for transcriptional regulators of AES in human CRC and found that its expression is reduced mainly by transcriptional dysregulation and, in some cases, by additional haploidization of its coding gene. The AES promoter‐enhancer is in a typical CpG island, and contains a Yin‐Yang transcription factor recognition sequence (YY element). In human epithelial cells of normal colon and primary tumors, transcription factor YY2, a member of the YY family, binds directly to the YY element, and stimulates expression of AES. In a transplantation mouse model of liver metastases, however, expression of Yy2 (and therefore of Aes) is downregulated. In human CRC metastases to the liver, the levels of AES protein are correlated with those of YY2. In addition, we noticed copy‐number reduction for the AES coding gene in chromosome 19p13.3 in 12% (5/42) of human CRC cell lines. We excluded other mechanisms such as point or indel mutations in the coding or regulatory regions of the AES gene, CpG methylation in the AES promoter enhancer, expression of microRNAs, and chromatin histone modifications. These results indicate that Aes may belong to a novel family of metastasis suppressors with a CpG‐island promoter enhancer, and it is regulated transcriptionally.
Keywords: AES, colon cancer, CpG island, transcription, YY2
Metastasis is responsible for most cancer deaths.1, 2 The metastasis cascade consists of local invasion, intravasation, transport, extravasation, formation of micrometastases, and colonization.3 Studies over the past two decades showed that approximately 20 proteins function as metastasis suppressors in various types of cancer.4, 5 We recently identified AES as a novel metastasis suppressor in human CRC, and it is an endogenous inhibitor of Notch signaling.6, 7 Namely, expression of Aes inhibits metastasis of CRC cells in an orthotopic transplantation model in mice, whereas Aes gene knockout causes local invasion and intravasation of tumors in intestinal adenomatosis mice.7, 8, 9 The expression of Aes/AES protein is decreased in liver metastases as compared with primary colon tumors in both mice and humans; however, its downregulation, the key mechanism for metastasis, has not been investigated.
Several lines of evidence indicate that expression of metastasis suppressors is controlled at the transcription level.10 For example, expression of KAI1 is transcriptionally regulated by β‐catenin and p53, negatively and positively, respectively.11, 12 While KiSS1 is stimulated by the DRIP‐130–SP1 transcription complex,13 reversion‐inducing cysteine‐rich protein with RECK is transcriptionally suppressed downstream of the RAS signaling.14
In this study, we aimed to identify regulators of AES expression. Possible mechanism(s) include transcriptional suppression, point or indel mutations in the AES coding or regulatory regions of the gene, CpG methylation in the AES promoter enhancer, transcriptional pausing, selective protein degradation, expression of miRNAs, and/or chromatin histone modifications. We present here the results showing that AES is stimulated by transcription factor YY2 in the colonic epithelium, and that expression of YY2 is decreased in liver metastatic lesions of CRC. In addition, we found that the AES gene is heterozygously deleted in 12% of CRC cell lines. We have also ruled out most other conceivable mechanisms mentioned above.
Materials and Methods
Materials and methods are provided in Data S1.
Results
Transcription of AES mRNA is initiated from a CpG island promoter at multiple sites
We focused on transcriptional regulation of the AES gene because other mechanisms did not appear to affect expression of AES protein, as described later (Fig. 1, final section). First, we noticed that the 1st exon of the major AES mRNA is in a CpG island (86.0% GC content, 17.5 CpG and 19.4 GpC doublets/100 bp) (Fig. 2a). To determine the transcription start sites of the AES gene, we used 5′ RACE. The results revealed clustered AES transcription start sites producing the major (short form of the) mRNA, with multiple 5′‐ends in human CRC cells (Fig. 2a,b). In addition, we found another minor (long form of the) mRNA initiated from an upstream promoter. We designate here the short mRNA as AES and the long one as AES‐L (Figs 2a,S1a), because the level of the short form was much higher (>15 times) than that of the long form in the colonic epithelium and in CRC cell lines (Fig. 2c). The high level expression of AES mRNA may be attributable to the strong CpG island promoter, in contrast to the AES‐L mRNA that is driven by a weaker TATA‐box‐like promoter (Fig. 2b,d). Consistently, we failed to detect AES‐L protein by Western blotting of CRC cells, although forced expression of AES‐L mRNA produced a corresponding protein band in 293T cells (Fig. S1b). In addition, exogenous expression of AES‐L mRNA only weakly inhibited Notch activities in RKO and HCT116 that expressed endogenous Notch receptors, whereas AES did inhibit all Notch1–4‐mediated transcription significantly (Fig. 2e). These results suggest that AES promoter and mRNA are responsible for expression of AES protein, whereas AES‐L mRNA has little significance in its expression.
Figure 1.
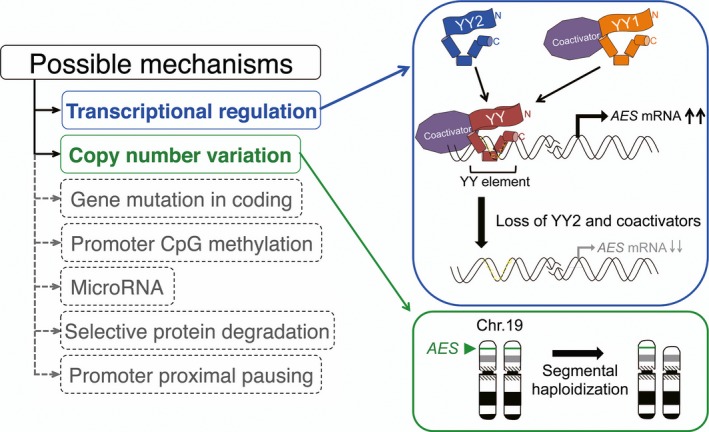
Schematic representation of possible regulatory mechanisms for amino‐terminal enhancer of split (AES). We propose that expression of AES is suppressed by transcriptional regulation including loss of Yin Yang 2 transcription factor (YY2) and/or co‐activators (blue rounded rectangle); and in some cases, by segmental haploidization of the AES‐coding chromosome (green rounded rectangles). We have excluded other conceivable mechanisms such as mutations in the coding or regulatory regions of the AES gene, promoter CpG methylation, microRNA expression, selective protein degradation, and promoter‐proximal transcriptional pausing (dashed rounded rectangles). Green arrowhead and bands indicate the position of the AES gene in chromosome (Chr.) 19p13.3.
Figure 2.
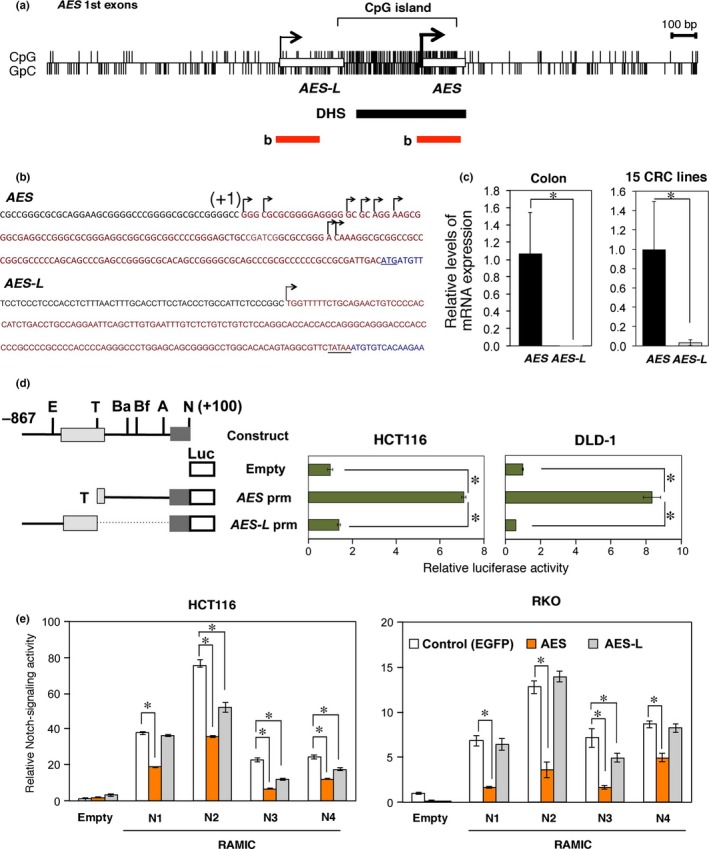
Promoter analysis of the human AES gene. (a) Schematic representation of the CpG island in the AES promoter region. Open rectangles indicate the 1st exons of AES and AES‐L. Upper and lower vertical lines show CpG and GpC dinucleotides, respectively. Black and red bars represent the DNase I hypersensitive site (DHS) and the regions shown in (b), respectively. (b) Transcription start sites of the AES and AES‐L genes in human colorectal cancer (CRC) cell line T84 determined by 5′ RACE. Transcribed genomic sequences are shown in red, with the coding sequence in blue. The hooked arrows show clustered multiple start sites for AES mRNA with (+1) indicating the most upstream one, and single start site for AES‐L. (c) Quantitative RT‐PCR analysis of the endogenous AES and AES‐L mRNA levels in human colonic mucosa and CRC cell lines. (d) Promoter activities of AES and AES‐L as determined by luciferase reporter transfections. The numbers (+100 and −867) indicate the nucleotide positions counted from the most upstream transcription start site (+1). Recognition sites by restriction enzymes are shown on top: A, AscI; Ba, BamHI; Bf, BfuAI; E, EcoRI; N, NotI; T, TaqI. Filled bars on the right indicate relative luciferase activities calibrated to those of the promoter‐less construct (Empty). Error bars show the standard deviations of triplicated samples (P < 0.01). (e) Suppression of Notch reporter activity by AES in HCT116 and RKO human CRC cells. Expression vectors encoding EGFP (control) or AES, and Notch1–4 (N1–4) intracellular domain (RAMIC)42 were cotransfected with the Notch‐reporter plasmid pGa981‐6. Luciferase activities are shown relative to those of the control vector‐transfected cells. *P < 0.01 compared with controls.
Yin Yang element plays essential roles in transcription of AES gene
Because multiple transcription factors cooperatively activate CpG island promoter genes,15 we hypothesized that AES is one of these genes. To test this hypothesis, we first searched through the AES promoter enhancer sequence for regulatory elements using ENCODE,16 Harr plot,17 and promoter reporter assays.
By ENCODE analysis, we found DNase I hypersensitive sites in the segment of −285 to +195, suggesting its actively transcribed state (Fig. 2a). To determine the subregions important for expression of AES, we constructed two series of luciferase reporters that carried the partial segments of the AES promoter enhancer with or without additional CMV‐minimal promoter (Fig. 3). The reporters carrying the Ba to A (−230 to −22) and Bf to A fragment (−172 to −22) showed the strongest luciferase activities (Figs 3a,b,S2a), suggesting that the enhancer activity resides between Bf and A. This enhancer sequence (downloaded from the University California, Santa Cruz Genome Browser http://genome.ucsc.edu) contains some distinct elements that can bind transcription factors such as Enhancer box binding protein, YY, and SOX (Fig. S2b). In addition, a Harr plot analysis revealed that these recognition sequences are conserved between humans and mice (Fig. 3c,e), and that the YY element is found even in frogs, such as Xenopus tropicalis (Fig. 3d,e). Interestingly, the Xenopus aes promoter was not associated with a CpG island, likely because of the low density of CpG islands in the species (Fig. S2b).18
Figure 3.
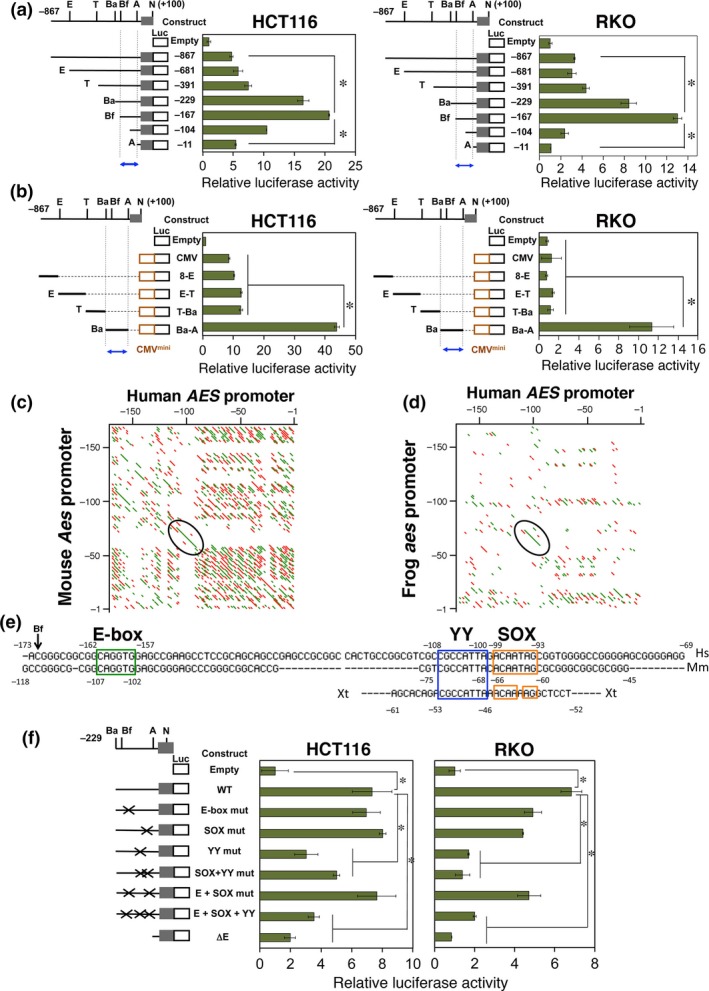
Evolutionarily conserved Yin Yang (YY) element in AES enhancer is essential for transcription. (a, b) Localization of regulatory elements in the AES enhancer promoter region. (a) Luciferase reporter constructs driven by the endogenous AES promoter. (b) Additional constructs containing exogenous CMV‐minimal promoter. The nucleotide numbers are based on the most upstream transcription start site (+1). Green bars indicate relative luciferase activities calibrated to those of the promoter‐less construct on top (Empty). Error bars show standard deviations of triplicated samples (P < 0.01). Blue arrows indicate the restriction fragments that contained the enhancer sequence shown in (e) below. (c, d) Harr‐plot analysis of the human, mouse, and frog AES/Aes/aes promoters. Evolutionarily conserved sequences between human and mouse (c), and human and frog (d) are shown as line segments. Encircled regions contain YY and SRY‐related HMG box (SOX) elements (c), and YY element (d). Both green and red lines show the conserved elements between the species. Two colors were used for clarity. (e) Nucleotide sequences of the AES enhancer promoter segments in humans (Hs), mice (Mm), and frogs (Xt) aligned for the best matches between the species. Possible binding motifs for relevant transcription factors are framed in colors. The nucleotide numbers are counted from the upstream‐most transcription start site (+1) (Fig. 2b). (f) Mutagenesis analysis of the putative transcription factor binding motifs in the AES enhancer promoter. Inactivating point mutations (mut) were introduced into the respective binding sites. Green bars show the relative luciferase activities calibrated to those of the basal vector (Empty). Error bars show standard deviations of triplicated samples. *P < 0.01. ΔE, reporter lacking upstream of the AscI (A) site (for enhancer deletion); Ba, Bam HI; Bf, Bfu AI; E, Eco RI; N, NotI; T, TaqI.
To determine whether these transcription factor binding elements actually participate in expression of AES in vivo, we introduced inactivating mutations into three motif sequences, and carried out reporter assays (Fig. 3f). The constructs carrying a point mutation in the YY element, either with or without additional ones in the SOX binding and/or E box elements, significantly decreased the AES promoter activity in the CRC cell lines. Taken together, these results indicate that the YY element is essential for the AES promoter activities.
Transcription factor YY2 stimulates expression of AES from AES promoter YY element
To test whether the transcription factors that can recognize the AES promoter enhancer affect transcription of AES, we next expressed them exogenously in HCT116 and RKO CRC cell lines. Consistent with the mutagenesis analysis (Fig. 3f), YY2 that binds to the YY element increased expression of the AES reporter significantly (Fig. 4a),19 although no other transcription factors (TFs) for the three elements did. Furthermore, YY2 did not affect transcription from other tumor‐suppressor genes, such as TP53 or SLC5A8, although it increased the transcription from AES promoter up to approximately six times that of the vector control in a dose‐dependent manner (Fig. 4b).20, 21, 22 Deletion of the enhancer region or a point mutation in the YY element sequence reduced the YY2‐dependent transcriptional activation to 18–25% of the wild‐type control (Fig. 4c). These results indicate that YY2 stimulates transcription from the AES promoter by binding to the YY element.
Figure 4.
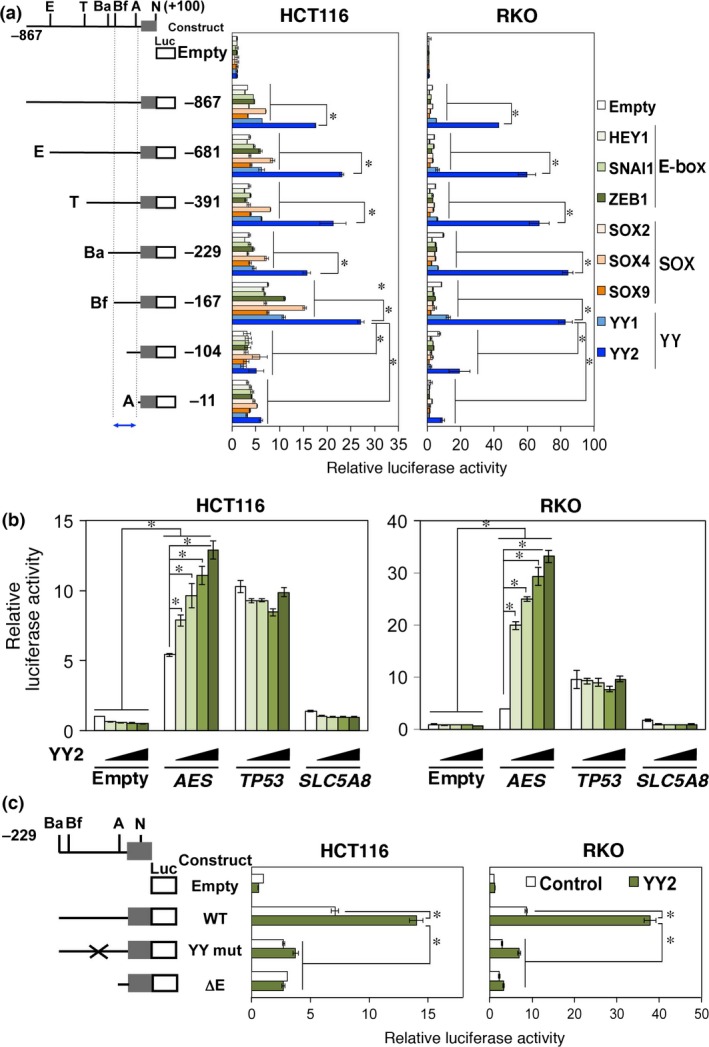
Yin Yang 2 transcription factor (YY2) stimulates transcription from AES enhancer promoter. (a) Effects of exogenously introduced transcription factors that recognize E‐box, SRY‐related HMG box (SOX), and YY elements of the AES enhancer promoter. Colored bars on the right show the relative luciferase activities calibrated to those of the basal luciferase vector and those without expression of exogenous transcription factors (Empty). Blue arrow indicates the enhancer‐containing fragment. (b) Effects of exogenously introduced YY2 on the activity of AES,TP53 and SLC5A8 reporters. Expression vectors were transfected together with the luciferase reporter plasmid. (c) Activation of AES transcription mediated by the YY element and transcription factor YY2. YY mut contains the point mutation that abolishes YY2 binding to the YY element. Luciferase activities are shown relative to those of the empty vector‐transfected cells. *P < 0.01 compared with controls. ΔE, reporter lacking upstream of the AscI (A) site (for enhancer deletion); Ba, Bam HI; Bf, Bfu AI; E, Eco RI; Luc, luciferase; N, NotI; T, TaqI.
It is reported that transcription factors YY1, YY2, and reduced expression protein 1/Zinc finger protein 42 can bind to the YY element, and activate or suppress transcription from their target promoters.23 We therefore evaluated YY1 and YY2 for the regulation of AES expression (Figs 5, 6,S3). Here, we excluded reduced expression protein 1, an embryonic stem cell marker from the evaluation, because of its lack of expression in CRC cell lines.24 To determine whether YY1 and YY2 bound differentially to the YY element of AES promoter enhancer, we next performed EMSA using recombinant YY proteins. The binding affinity of YY2 was far stronger than that of YY1 (Figs 5a,6a,S3a). As described above (Fig. 4a), YY1 showed much fewer effects on AES promoter activity than YY2.
Figure 5.
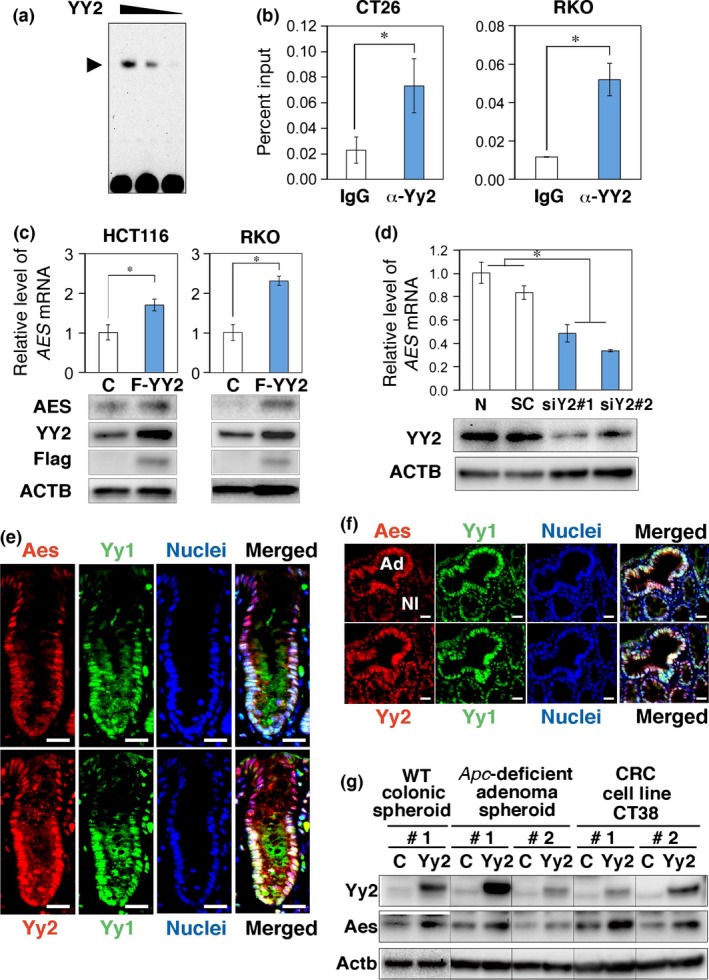
Yin Yang 2 transcription factor (YY2) directly binds to and activates the endogenous AES enhancer promoter. (a) EMSA using the YY element of AES enhancer fragment and the recombinant YY2 protein. Arrowhead indicates the position of the YY2 protein–DNA complexes. (b) ChIP–quantitative (q)PCR analysis of Yy2/YY2 binding to the Aes/AES promoter in CT26 and RKO colorectal cancer cells, respectively. α‐Yy2 and α‐YY2 show anti‐Yy2 and anti‐YY2 antibodies, respectively. (c) Summary of qRT‐PCR determinations of the AES mRNA levels on exogenous expression of Flag‐YY2 in HCT116 and RKO cells. Bottom photographs show Western blotting for protein expression. ACTB, β‐actin; C, control empty vector. (d) Summary of qRT‐PCR determinations of the AES mRNA levels on expression of siRNA against YY2 in HCT116 cells. siY2#1 and siY2#2 indicate independent siRNA constructs with scramble siRNA (SC) and mock transfection. Bottom photographs show Western blotting for protein expression. N, none. (e, f) Immunofluorescence localization of Aes, Yy1, and Yy2 proteins in mouse colonic crypts (e) and Apc‐deficient adenomas (Ad) (f). Left photographs indicate Aes (top) and Yy2 (bottom) staining, respectively (red). Center photographs show Yy1 (green) and nuclear staining with DAPI (blue). Right photographs show merged images of the left and center. Nl, normal epithelium. (g) Induction of AES by exogenously introduced Flag‐Yy2 in mouse cells analyzed by Western blotting. Spheroid cultures of normal colonic epithelium (left) and Apc‐deficient adenomas (center) as well as a colorectal cancer (CRC) cell line CT38 (right) were analyzed. Columns #1 and #2 indicate that the Yy2 coding region was driven by the promoter enhancer segments for PGK and MSCV, respectively. *P < 0.01 compared with controls (b–d). Scale bar = 20 μm (e, f). C, empty control vector.
Figure 6.
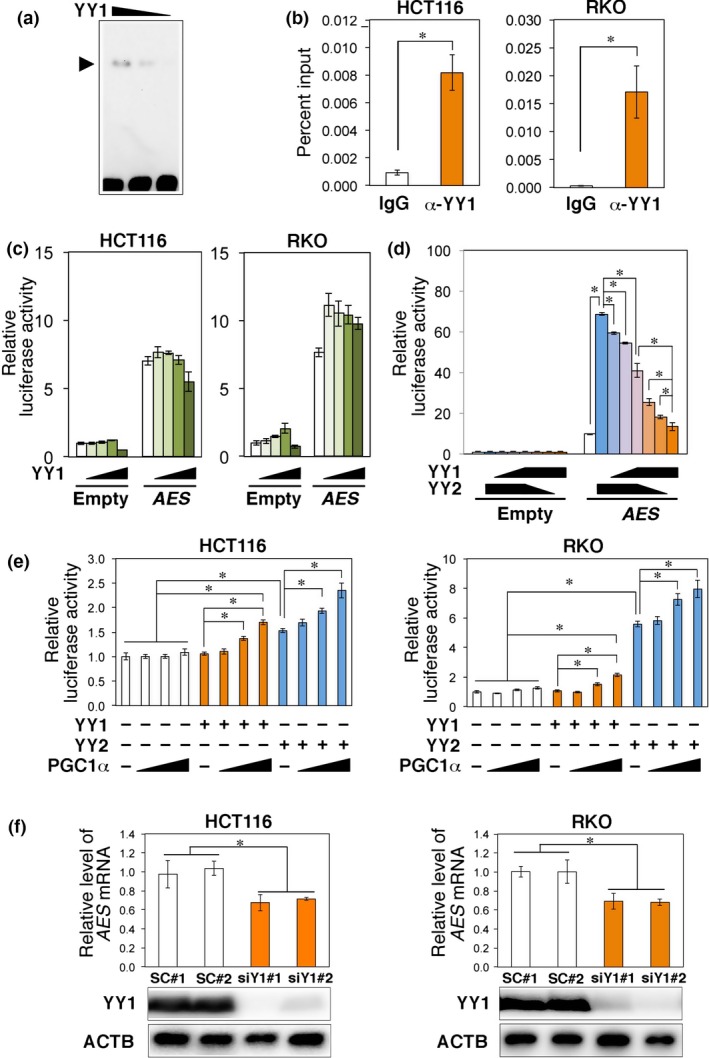
Yin Yang 1 transcription factor (YY1) stimulates expression of AES in the presence of co‐activators. (a) EMSA using the YY element of AES enhancer fragment and recombinant YY1 protein. Arrowhead indicates the position of the YY1 protein–DNA complex. (b) ChIP–quantitative PCR analysis of YY1 binding to the AES promoter using HCT116 and RKO colorectal cancer cell extracts. The column α‐YY1 shows immunoprecipitates with the anti‐YY1 antibody. (c) Effects of exogenously introduced YY1 on the activity of AES reporters. Expression vectors were transfected together with the luciferase reporter plasmid. (d) Competitive effects of exogenously introduced YY1 and/or YY2 on the activity of AES reporters. Expression vectors were transfected together with the luciferase reporter plasmid. (e) Activation of AES promoter by exogenously introduced PGC1α in HCT116 and RKO cells in the presence of YY proteins. (f) Summary of quantitative RT‐PCR determinations of AES mRNA levels on expression of siRNA against YY1 in HCT116 and RKO cells. siY1#1 and siY1#2 indicate independent siRNA constructs compared with scramble siRNA (SC#1 and SC#2). Bottom photographs show Western blotting for protein expression. *P < 0.01 compared with controls (b–f).
To test whether YY2 bound to the genomic AES promoter, we carried out ChIP‐PCR using sonicated extracts of mouse CT26 and human RKO CRC cells that endogenously expressed YY2 (Fig. S4a–c). We confirmed that each antibody for YY proteins specifically reacted to the target antigen (Fig. S3b) and that the two independent anti‐YY2 antibodies successfully immunoprecipitated YY2 protein in this analysis (Fig. S3c,d,f). As shown in Figure 5(b), YY2 was enriched in the AES promoter region. To further study whether YY2 affected expression of the endogenous AES gene, we then determined the levels of AES mRNA in RKO and HCT116 cells in which expression of YY2 was altered (Fig. 5c,d). Overexpression of YY2 significantly increased the level of AES mRNA by 1.7–2.3 times (Fig. 5c). Consistently, knockdown of YY2 in HCT116 decreased the level of AES mRNA to 35–50% of the controls (Fig. 5d). These results indicate that YY2 can bind directly to and activate the genomic AES promoter in CRC cells.
We next analyzed expression of Aes and Yy2 proteins in mouse intestine using anti‐Aes and anti‐Yy2 antibodies. We first confirmed that the Yy2 signal detected by immunoblotting was weakened by siRNA against Yy2 in CT26 cells (Fig. S4a,b) and that the protein bound by anti‐Yy2 antibody included Yy2‐specific fragments by mass‐spectrometric analysis (Fig. S4d). A Western blot analysis revealed that Yy2 as well as Aes was expressed in various mouse organs including the brain, liver, kidney, testis, stomach, and intestines at wide levels (Fig. S4e,f).
Using mouse colonic epithelial cell spheroids,25 we further found that both Aes and Yy2 proteins are expressed in the mouse colonic epithelium at significant levels (Fig. S4g). We also confirmed their expression in the normal and adenomatous colonic epithelia by immunohistochemistry, as shown in Figure 5(e,f). Notably, the Yy2 protein was localized both in the cytoplasm and nucleus of the colonic crypt epithelia that also expressed Aes. Likewise, we confirmed upregulation of Aes by overexpression of Yy2 protein in the normal and adenomatous colonic epithelial spheroids, and another mouse CRC cell line CT38 (Fig. 5g). Taken together, these results suggest that expression of Aes is stimulated by Yy2 in the normal colonic, adenomatous, and cancer epithelial cells of the mouse.
Yin Yang 1 stimulates expression of AES in the presence of co‐activators
Electrophoretic mobility‐shift assay using recombinant YY proteins showed that the binding affinity of YY2 was far stronger than that of YY1 (Figs 5a,6a,S3a). However, YY1 protein is abundantly expressed in mammalian normal and tumor cells (Fig. S4b,e,f)26 and shares the common target DNA sequence motif with YY2.23 We therefore tested whether YY1 can bind to and stimulate the AES promoter in vivo. A ChIP‐PCR analysis using sonicated extracts of HCT116 and RKO CRC cells showed that YY1 was enriched in the AES promoter region (Figs 6b,S3e,f). To further test whether the binding enhances transcription from the AES promoter, we undertook luciferase reporter assays. Although YY1 hardly affected the AES promoter by itself (Fig. 6c), it could disturb the YY2‐mediated transcription from the AES promoter (Fig. 6d). In contrast, YY1 stimulated expression of AES in the presence of co‐activators (Fig. 6e).27 To further analyze the role of YY1 in stimulating expression of AES in vivo, we determined the effects of knocking down YY1 expression in CRC cell lines. Knockdown of YY1 in HCT116 or RKO cells decreased the expression levels of AES mRNA to approximately 70% of that of the controls (Fig. 6f). We also confirmed that a simultaneous knockdown of YY1 and YY2 further decreased expression of AES (Fig. S3g). These results indicate that YY1 can enhance AES expression in the presence of co‐activators.
Levels of Aes are correlated with those of YY2 in liver metastases
By microarray analysis, we previously found that expression of Aes is downregulated in liver metastases of mouse CRC cells transplanted into mouse rectal mucosa.7 Accordingly, we re‐examined the array data and found that Yy2 mRNA level was also decreased in metastatic lesions both in the liver and lung (Fig. S3h), which was consistent with the immunohistopathology (Fig. S3i). A qRT‐PCR analysis confirmed reduced expression of Yy2 and Aes mRNA in mouse liver metastases, compared with the primary tumors (Fig. 7a). Furthermore, we analyzed expression of AES and YY2 proteins by immunohistopathology in human CRC lesions metastasized to the liver, and found that the levels of AES were positively correlated with those of YY2 (Figs 7b,S5a, Table 1; P = 1.78 × 10–6). We also confirmed the positive correlation between them even in human primary CRC (Fig. S5b, Table S1; P = 0.0002). In addition, we found a tendency for positive correlation between YY1 and AES expression in CRC primary tumors (P = 0.077) and in their liver metastases (P = 0.002) (Tables S2,S3). Notably, the statistical significance for YY2 (primary tumors, P = 0.0002; liver metastases, P = 1.78 × 10–6) is far stronger than that for YY1 (P = 0.077 for primary tumors and P = 0.002 for liver metastases) (Tables 1,S1–S3). Taken together, these results strongly suggest that expression of AES is driven chiefly by YY2 in both human and mouse CRC lesions.
Figure 7.
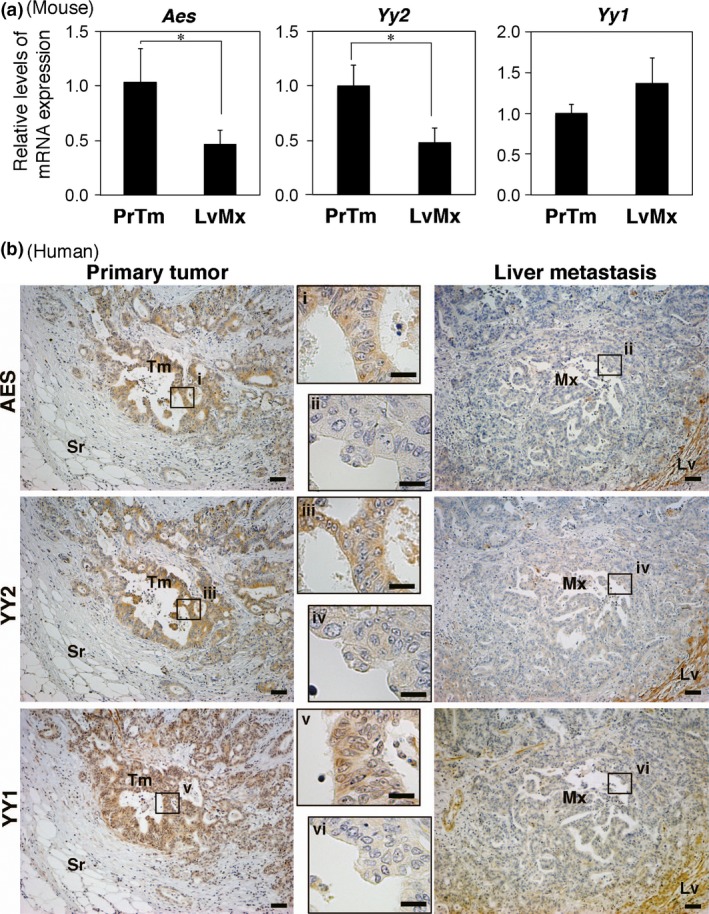
Reduced expression of amino‐terminal enhancer of split (AES) and Yin Yang 2 transcription factor (YY2) in liver metastases of colorectal cancer (CRC). (a) Summary of quantitative RT‐PCR quantifications of Aes, Yy2, and Yy1 in liver metastases (LvMx) of mouse CRC cells transplanted to the mouse rectum. *P < 0.05. PrTm, primary tumor. (b) Immunohistochemical staining for AES, YY1, and YY2 in primary and liver metastasis lesions obtained from the same CRC patient. Framed areas i–iv are magnified in the middle panels. Scale bar = 20 μm and 100 μm for high and low magnifications, respectively. Lv, liver parenchyma; Mx, metastasis; Sr, serosa; Tm, tumor.
Table 1.
Relationship of expression between amino‐terminal enhancer of split (AES) and Yin Yang 2 (YY2) in metastatic liver lesions of human colorectal cancer patients
| Factors | AES expression | P‐value | |
|---|---|---|---|
| + (n = 24) | − (n = 31) | ||
| YY2 expression | 1.78 × 10−6 | ||
| + (n = 28) | 21 | 7 | |
| − (n = 27) | 3 | 24 | |
Segmental haploidization of AES‐coding chromosome in CRC cells
Recently, array‐based comparative genomic hybridization analyses were carried out to study the genomic landscape of CRC, and the results were compiled in public databases. We looked into them for copy number alterations in the genomic region of chromosome 19p near the telomere where AES is located (Fig. S6a,b). Notably, we found segmental haploidization of 19p13.3 in approximately 12% (5/42) of CRC cell lines, and in approximately 3% of tumor tissues in the CRC patients (Cancer Genome Project database at Sanger Institute http://www.sanger.ac.uk/cgi-bin/genetics/CGP/cghviewer/CghHome.cgi and SKY/M‐FISH and CGH Database at NCBI http://www.ncbi.nlm.nih.gov/sky/) (Fig. S6a–c).28 Three of these five CRC cell lines (DLD‐1, SW837, and WiDr) that were haploid for the AES gene hardly expressed AES protein (Fig. S6d, right), suggesting that losing one of the alleles was partly responsible for the extremely reduced AES levels. Expression of AES mRNA was decreased also in other CRC cell lines without copy number alterations (e.g., HCT116, HT29, and RKO) when compared with normal colonic tissues and adenoma cell line AA/C1 (Fig. S6d,e). These results suggest that LOH contributed to the reduced expression of AES in a subset of CRC cases in addition to transcriptional repression (Fig. 1).
Downregulation of AES is not associated with gene mutations, promoter CpG methylation, or miRNA expression
We found that the levels of AES protein and AES mRNA were reduced significantly in human CRC cell lines compared with an adenoma cell line or colonic epithelium (Fig. S6d,e). Consistently, one can find in cancer tissue databases that AES mRNA levels are lower than in the normal colonic mucosa (Fig. S6f). To find additional causes of AES downregulation, we searched for mutations in the AES coding and promoter regions in CRC. However, we failed to find any mutations in AES cDNA by sequencing 14 human CRC cell lines (Fig. S7a). Likewise, we found no mutations in the AES coding sequence in The Cancer Genome Atlas database.29 We did not find any mutations in the AES promoter region either, including in the YY element (Fig. S7a).
Having excluded the mutational changes in the coding and promoter sequences of AES (Fig. S7a), we tested the possibilities of AES protein destabilization, CpG methylation in the AES promoter enhancer, promoter‐proximal transcriptional pausing,30 and expression of miRNAs, all with negative data (Figs 1,S7–S10). We evaluated the protein stability of AES using translation inhibitor cycloheximide. AES protein was as stable as the bulk of other proteins in human CRC cells (Fig. S7d). As shown in Figure 2(a), AES promoter enhancer is in a typical CpG island. Accordingly, we suspected possible CpG methylation in the region. However, expression of AES was unaffected by DNA demethylating reagent 5‐aza‐2′‐deoxycytidine in eight CRC cell lines (Fig. S8). Although we also assessed the possibility of epigenetic suppression by histone modification, histone deacetylase inhibitors did not increase the expression of AES in any of six human CRC cell lines (Fig. S9). The 3′‐UTR of the AES gene contains an miRNA recognition motif for mir‐124, which is evolutionarily conserved in mammals (Fig. S7b). Yet, no members of the mir‐124 family were expressed in the CRC cell lines at any detectable levels by qPCR (Fig. S7b,c), consistent with a previous report.31 We also examined the possibility of promoter‐proximal transcriptional pausing,30 but it did not appear to be taking place at levels that affected AES expression significantly (Fig. S10a–c). These results ruled out the above mechanisms commonly found in tumor‐suppressor genes, and suggested that expression of AES is regulated by other mechanisms such as specific transcription factor YY2, and by copy number reduction as presented above (Fig. 1).
Discussion
In human CpG island promoters, binding motifs are enriched for transcription factors Sp1, E2F, and ETS,32 as well as for BoxA, CRE, and E‐box binding factors.33 In addition, the YY element is also found in CpG island promoters far more frequently than in non‐CpG island regions of the genome, driving expression of, for example, mitochondrial genes.34 Consistently, our search through the ENCODE ChIP‐seq data revealed that YY1 transcription factors are preferentially bound to CpG island promoters.16 However, information on YY2 appears to be relatively rare.
Expression of YY2 was downregulated in 49% (27/55) of liver metastasis lesions of CRC (Fig. 7b, Table 1). We noticed two possible mechanisms: (i) in 2% of CRC cases, point or indel mutations were present in the YY2 coding region, which could destabilize YY2 protein (COSMIC database, http://cancer.sanger.ac.uk/cosmic); and (ii) epigenetic changes including histone modifications appeared important for transcription of the YY2 promoter (Fig. S10d).
We recently found that AES suppresses invasion and metastasis of CRC through Notch signaling inhibition, and forms NM foci together with Rbpj transcription complex, TLE1, and HSC70.7, 35 It is known that transcription factor YY1 is associated with NM and nucleoli.36 It remains to be determined whether YY2 modifies the AES–TLE1–HSC70 NM foci and thereby affects Notch signaling through transcriptional induction of AES. One of the Notch target genes inhibited by AES in CRC is DAB1.8 DAB1 protein is phosphorylated by ABL tyrosine kinase, which activates ABL reciprocally by stimulating its autophosphorylation, causing strong activation of ABL in a manner quite localized to the sites of invasion including intravasation. One of the targets of phosphorylation by ABL is the RAC/RHO‐GEF protein TRIO at its Tyr residue 2681 (pY2681), resulting in RHO activation in CRC cells.8 Importantly, the extent of phosphorylation at TRIO Y2681 correlates with significantly poorer prognosis of patients with CRC after surgery.8 Accordingly, these results indicate that the transcriptional activation of YY2 followed by expression of AES leads to suppression of CRC invasion and metastasis mediated by the NOTCH–DAB1–ABL–TRIO–RHO cascade.
It has been reported that mutation profiles are hardly changed between metastatic lesions and the primary tumor, and that epigenetic changes appear to be important for metastatic progression.37 Consistently, metastasis suppressor AES had no mutations in CRC, and its positive regulator YY2 is likely to be induced by epigenetic changes (Fig. S10d).38 It is also possible that the copy‐number reduction at the AES locus plays a role in metastatic progression in a subset of cancer cases. Namely, the chromosomal region 19p13.3 including AES is deleted often in other types of cancer such as lung cancer, lymphoma, and breast carcinoma,39, 40, 41 although its frequency in CRC is only approximately 3% (7/280; http://www.ncbi.nlm.nih.gov/sky/). Accordingly, it is possible that partial haploidization at 19p13.3 contributes to the progression of multiple types of cancer. Further investigations will be needed to elucidate the precise mechanisms that affect AES expression upstream of YY2.
Disclosure Statement
The authors have no conflict of interest.
Abbreviations
- AES
amino‐terminal enhancer of split
- CRC
colorectal cancer
- ENCODE
Encyclopedia of DNA Elements
- indel
insertion–deletion
- miRNA
microRNA
- NM
nuclear‐matrix
- qRT‐PCR
quantitative RT‐PCR
- SOX
SRY‐related HMG box
- YY2
Yin Yang 2 transcription factor
Supporting information
Fig. S1. Information on AES‐L mRNA.
Fig. S2. AES promoter analysis.
Fig. S3. Evaluation of anti‐Yin Yang 2 transcription factor (YY2) antibodies and expression of Yy2 in liver metastases of mouse colorectal cancer.
Fig. S4. Expression analysis of Yin Yang 2 transcription factor (Yy2)/YY2 in tissues and colorectal cancer cell lines.
Fig. S5. Expression of amino‐terminal enhancer of split (AES) and Yin Yang 2 transcription factor (YY2) in human primary colorectal cancer and its loss in liver metastasis.
Fig. S6. AES expression is reduced by copy number alteration and transcriptional regulation.
Fig. S7. Sequence analysis for coding and 3′‐UTR regions of AES, and protein stability analysis.
Fig. S8. Demethylation analysis of AES promoter.
Fig. S9. Histone acetylation analysis of AES promoter.
Fig. S10. Testing for promoter‐proximal transcriptional pausing in the AES promoter, and induction of YY2 transcription by histone deacetylase inhibitors.
Table S1. Relationship of expression between amino‐terminal enhancer of split (AES) and Yin Yang 2 transcription factor (YY2) in primary tumors of colorectal cancer patients.
Table S2. Relationship of expression between amino‐terminal enhancer of split (AES) and Yin Yang 1 transcription factor (YY1) in primary tumors of colorectal cancer patients.
Table S3. Relationship of expression between amino‐terminal enhancer of split (AES) and Yin Yang 1 transcription factor (YY1) in metastatic liver lesions of colorectal cancer patients.
Data S1. Supplementary materials and methods.
Acknowledgments
We thank B. Aronow, T. Honjo, S. Nagata, M. Yoshida, H. Handa, K. Biyajima, and S. Yamashita for helpful suggestions and discussions; T. Honjo for the NICD constructs; M. Okabe for the pCX vector; S. Nagata for the pEFBosneo vector; C. Paraskeva for the AA/C1 colon adenoma cell line; and T. Yamori for the CT38 CRC cell line.
Cancer Sci 107 (2016) 1622–1631
Funding Information
Ministry of Education, Culture, Sports, Science and Technology of Japan; Japan Agency for Medical Research and Development.
References
- 1. American Cancer Society . Cancer Facts & Figures. Atlanta: American Cancer Society, 2015. [Google Scholar]
- 2. Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med 2006; 12: 895–904. [DOI] [PubMed] [Google Scholar]
- 3. Fidler IJ. The pathogenesis of cancer metastasis: “the seed and soil” hypothesis revisited. Nat Rev Cancer 2003; 3: 453–8. [DOI] [PubMed] [Google Scholar]
- 4. Steeg PS. Metastasis suppressors alter the signal transduction of cancer cells. Nat Rev Cancer 2003; 3: 55–63. [DOI] [PubMed] [Google Scholar]
- 5. Bodenstine MT, Welch RD. Metastasis suppressors and the tumor microenvironment. Cancer Microenviron 2008; 1: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beagle B, Johnson GB. AES/GRG5: more than just a dominant‐negative TLE/GRG family member. Dev Dyn 2010; 239: 2795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sonoshita M, Aoki M, Fuwa H et al Suppression of colon cancer metastasis by Aes through inhibition of Notch signaling. Cancer Cell 2011; 19: 125–37. [DOI] [PubMed] [Google Scholar]
- 8. Sonoshita M, Itatani Y, Kakizaki F et al Promotion of colorectal cancer invasion and metastasis through activation of NOTCH‐DAB1‐ABL‐RHOGEF protein TRIO. Cancer Discov 2015; 5: 198–211. [DOI] [PubMed] [Google Scholar]
- 9. Taketo MM, Edelmann W. Mouse models of colon cancer. Gastroenterology 2009; 136: 780–98. [DOI] [PubMed] [Google Scholar]
- 10. Smith CS, Theodorescu D. Learning therapeutic lessons from metastasis suppressor proteins. Nat Rev Cancer 2009; 9: 253–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim JH, Kim B, Cai L et al Transcriptional regulation of a metastasis suppressor gene by Tip60 and beta‐catenin complexes. Nature 2005; 434: 921–6. [DOI] [PubMed] [Google Scholar]
- 12. Mashimo T, Watabe M, Hirota S et al The expression of the KAI1 gene, a tumor metastasis suppressor, is directly activated by p53. Proc Natl Acad Sci USA 1998; 95: 11307–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mitchell DC, Stafford LJ, Li D, Bar‐Eli M, Liu M. Transcriptional regulation of KiSS‐1 gene expression in metastatic melanoma by specificity protein‐1 and its coactivator DRIP‐130. Oncogene 2007; 26: 1739–47. [DOI] [PubMed] [Google Scholar]
- 14. Chang HC, Cho CY, Hung WC. Silencing of metastasis suppressor RECK by RAS oncogene is mediated by DNA methyltransferase 3b‐induced promoter methylation. Cancer Res 2006; 66: 8413–20. [DOI] [PubMed] [Google Scholar]
- 15. Deaton AM, Bird A. CpG islands and the regulation of transcription. Gene Dev 2011; 25: 1010–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. The ENCODE Project Consortium . An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harr R, Hagblom P, Gustafsson P. Two‐dimensional graphic analysis of DNA sequence homologies. Nucleic Acids Res 1982; 10: 365–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sharif J, Endo TA, Toyoda T, Koseki H. Divergence of CpG island promoters: a consequence or cause of evolution? Dev Growth Differ 2010; 52: 545–54. [DOI] [PubMed] [Google Scholar]
- 19. Nugyen N, Zhang X, Olashaw N, Seto E. Molecular cloning and functional characterization of the transcription factor YY2. J Biol Chem 2004; 279: 25927–34. [DOI] [PubMed] [Google Scholar]
- 20. Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53‐mediated tumour suppression. Nat Rev Cancer 2014; 14: 359–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li H, Myeroff L, Smiraglia D et al SLC5A8, a sodium transporter, is a tumor suppressor gene silenced by methylation in human colon aberrant crypt foci and cancers. Proc Natl Acad Sci USA 2003; 100: 8412–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kakizaki F, Aoki K, Miyoshi H, Carrasco N, Aoki M, Taketo MM. CDX transcription factors positively regulate expression of solute carrier family 5, member 8 in the colonic epithelium. Gastroenterology 2010; 138: 627–35. [DOI] [PubMed] [Google Scholar]
- 23. Kim JD, Faulk C, Kim J. Retroposition and evolution of the DNA‐binding motifs of YY1, YY2 and REX1. Nucleic Acids Res 2007; 35: 3442–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hosler BA, LaRosa GJ, Grippo JF, Gudas LJ. Expression of REX‐1, a gene containing zinc finger motifs, is rapidly reduced by retinoic acid in F9 teratocarcinoma cells. Mol Cell Biol 1989; 9: 5623–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miyoshi H, Stappenbeck TS. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc 2013; 8: 2471–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pilarsky C, Wenzig M, Spech T, Saeger HD, Grützmann R. Identification and validation of commonly overexpressed genes in solid tumors by comparison of microarray data. Neoplasia 2004; 6: 744–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1‐PGC‐1alpha transcriptional complex. Nature 2007; 450: 736–40. [DOI] [PubMed] [Google Scholar]
- 28. Knutsen T, Gobu V, Knaus R et al The interactive online SKY/M‐FISH & CGH database and the Entrez cancer chromosomes search database: linkage of chromosomal aberrations with the genome sequence. Genes Chromosom Cancer 2005; 44: 52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Network Cancer Genome Atlas . Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012; 487: 330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chiba K, Yamamoto J, Yamaguchi Y, Handa H. Promoter‐proximal pausing and its release: molecular mechanisms and physiological functions. Exp Cell Res 2010; 316: 2723–30. [DOI] [PubMed] [Google Scholar]
- 31. Lagos‐Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschi T. Identification of tissue‐specific microRNA from mouse. Curr Biol 2002; 12: 735–9. [DOI] [PubMed] [Google Scholar]
- 32. Landolin JM, Johnson DS, Trinklein ND et al Sequence features that drive human promoter function and tissue specificity. Genome Res 2010; 20: 890–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rozenberg JM, Shlyakhtenko A, Glass K et al All and only CpG containing sequences are enriched in promoters abundantly bound by RNA polymerase II in multiple tissues. BMC Genom 2008; 9: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schug J, Schuller WP, Kappen C, Salbaum JM, Bucan M, Stoechert CJ Jr. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol 2005; 6: R33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Itatani Y, Sonoshita M, Kakizaki F et al Characterization of Aes nuclear foci in colorectal cancer cells. J Biochem 2016; 159: 133–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guo B, Odgren PR, van Wijnen AJ et al The nuclear matrix protein NMP‐1 is the transcription factor YY1. Proc Natl Acad Sci USA 1995; 92: 10526–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jones S, Chen WD, Parmigiani G et al Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci USA 2008; 105: 4283–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Klar M, Drews D, Dame C. Transcriptional activity of the novel identified human yy2 promoter is modified by DNA methylation. Gene 2009; 430: 58–63. [DOI] [PubMed] [Google Scholar]
- 39. Wang X, Zhang Y, Nilsson CL et al Association of chromosome 19 to lung cancer genotypes and phenotypes. Cancer Metastasis Rev 2015; 34: 217–26. [DOI] [PubMed] [Google Scholar]
- 40. Tagawa H, Karnan S, Suzuki R et al Genome‐wide array‐based CGH for mantle cell lymphoma: identification of homozygous deletions of the proapoptotic gene BIM. Oncogene 2005; 24: 1348–58. [DOI] [PubMed] [Google Scholar]
- 41. Yang TL, Su YR, Huang CS et al High‐resolution 19p13.2‐13.3 allelotyping of breast carcinomas demonstrates frequent loss of heterozygosity. Genes Chromosom Cancer 2004; 41: 250–6. [DOI] [PubMed] [Google Scholar]
- 42. Kato H, Sakai T, Tamura K et al Functional conservation of mouse Notch receptor family members. FEBS Lett 1996; 395: 221–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Information on AES‐L mRNA.
Fig. S2. AES promoter analysis.
Fig. S3. Evaluation of anti‐Yin Yang 2 transcription factor (YY2) antibodies and expression of Yy2 in liver metastases of mouse colorectal cancer.
Fig. S4. Expression analysis of Yin Yang 2 transcription factor (Yy2)/YY2 in tissues and colorectal cancer cell lines.
Fig. S5. Expression of amino‐terminal enhancer of split (AES) and Yin Yang 2 transcription factor (YY2) in human primary colorectal cancer and its loss in liver metastasis.
Fig. S6. AES expression is reduced by copy number alteration and transcriptional regulation.
Fig. S7. Sequence analysis for coding and 3′‐UTR regions of AES, and protein stability analysis.
Fig. S8. Demethylation analysis of AES promoter.
Fig. S9. Histone acetylation analysis of AES promoter.
Fig. S10. Testing for promoter‐proximal transcriptional pausing in the AES promoter, and induction of YY2 transcription by histone deacetylase inhibitors.
Table S1. Relationship of expression between amino‐terminal enhancer of split (AES) and Yin Yang 2 transcription factor (YY2) in primary tumors of colorectal cancer patients.
Table S2. Relationship of expression between amino‐terminal enhancer of split (AES) and Yin Yang 1 transcription factor (YY1) in primary tumors of colorectal cancer patients.
Table S3. Relationship of expression between amino‐terminal enhancer of split (AES) and Yin Yang 1 transcription factor (YY1) in metastatic liver lesions of colorectal cancer patients.
Data S1. Supplementary materials and methods.


