Abstract
Caenorhabditis elegans is a small nematode that can be maintained at low cost and handled using standard in vitro techniques. Unlike toxicity testing using cell cultures, C. elegans toxicity assays provide data from a whole animal with intact and metabolically active digestive, reproductive, endocrine, sensory and neuromuscular systems. Toxicity ranking screens in C. elegans have repeatedly been shown to be as predictive of rat LD50 ranking as mouse LD50 ranking. Additionally, many instances of conservation of mode of toxic action have been noted between C. elegans and mammals. These consistent correlations make the case for inclusion of C. elegans assays in early safety testing and as one component in tiered or integrated toxicity testing strategies, but do not indicate that nematodes alone can replace data from mammals for hazard evaluation. As with cell cultures, good C. elegans culture practice (GCeCP) is essential for reliable results. This article reviews C. elegans use in various toxicity assays, the C. elegans model's strengths and limitations for use in predictive toxicology, and GCeCP. Published 2016. This article is a U.S. Government work and is in the public domain in the USA. Journal of Applied Toxicology published by John Wiley & Sons Ltd.
Keywords: alternative model, predictive toxicology, toxin ranking, screening, good C. elegans culture practice
Short abstract
Toxicity ranking screens in Caenorhabditis elegans have repeatedly been shown to be as predictive of rat LD50 ranking as mouse LD50 ranking. Additionally, many instances of conservation of mode of toxic action have been noted between C. elegans and mammals. These consistent correlations make the case for inclusion of C. elegans assays in early safety testing and as one component in tiered or integrated toxicity testing strategies. Good C. elegans culture practice (GCeCP) is essential for reliable results.
Introduction
Much of our knowledge within the field of biology is based on scientific experimentation using in vivo and in vitro models. Toxicity testing is done with the expectation that information acquired in a particular model will apply to other biological systems, with each model presenting strengths and limitations depending on the information required. Mammalian laboratory animals share similar developmental pathways and most organs with humans, making toxicity testing in mammals the current ‘gold standard‘ in toxicology. However, no model is perfect, and even human trials do not always predict outcomes in the population at large.
Toxicity studies using mammalian models are expensive and time‐consuming (Nass and Hamza, 2007; Tralau et al., 2012), and meta‐analyses indicate that rodent models predict specific toxic effects in humans only about 50% of the time (Hartung, 2009; Knight et al., 2009; Olson et al., 2000). Using more than one mammalian species can increase predictivity (Olson et al., 2000) but will also increase cost and decrease throughput. Predictive toxicology seeks to use alternative methods to improve prediction of human outcomes while reducing the cost, time and use of mammals in toxicity assessments. Chemicals of concern can be tested far more rapidly and at a much larger range of concentrations if in vitro assays are used to assess perturbations in toxicity pathways. While the use of primary human cells does have the potential to more accurately reflect human‐specific metabolism and modes of action than testing in lab animals (Li et al., 1999; Miranda et al., 2009; Scott et al., 2013), results cannot be used to predict a response at the organismal level using current techniques. Additionally, testing using immortalized cell lines can have high rates of false positives (Kirkland et al., 2005; Bhattacharya et al., 2011; Pfuhler et al., 2010) or false‐negatives (Knight et al., 2009) depending on the assay type. Thus, the use of in vitro tests alone for hazard assessment gives rise to the possibility that compounds that are harmless in vivo will be unnecessarily restricted and that harmful compounds will be incorrectly presumed to be safe.
Another option is to utilize a small model organism such as the nematode Caenorhabditis elegans, which can be handled using in vitro techniques. Unlike in vitro testing, C. elegans toxicity assays provide data from a whole animal with intact and metabolically active digestive, reproductive, endocrine, sensory and neuromuscular systems (Fig. 1). As government‐sponsored efforts to improve toxicity screening and predictive toxicology progress (Tice et al., 2013; Casey et al., 2015), we may eventually find that testing strategies which use multiple types of in silico, in vitro and non‐mammalian small animal model‐based assays together has the potential to inform risk evaluation as well or better than in vivo toxicity studies using mammals, but much more work is still required (Krewski et al., 2009; Hartung et al., 2013). For now, mammalian models will continue to be used to predict safe human exposure levels because no combination of current alternative assays can replicate the complexity of interacting metabolism, homeostasis and signaling mechanisms that are present in mammals (Tice et al., 2013). However, given the high percentage of commercially available chemicals for which there is little or no toxicity data available (Dix et al., 2007; Judson et al., 2009), toxicity screens that can at least flag for further study those chemicals with the most potential for harm are urgently needed. As an intermediate between in vitro and mammalian testing, toxin ranking using various C. elegans assays has consistently predicted toxicity ranking in mammals.
Figure 1.
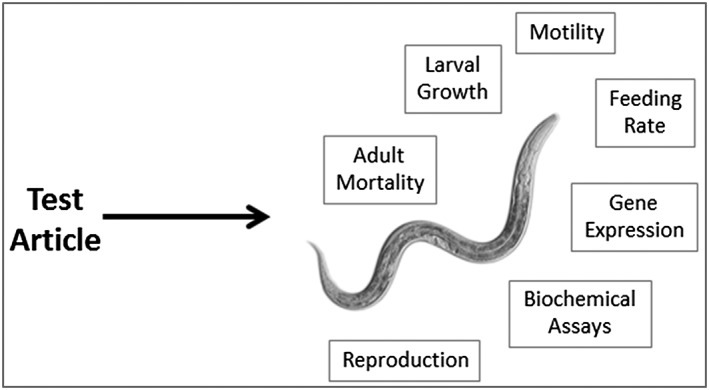
Toxicity testing in C. elegans can provide a bridge between in vitro and mammalian in vivo testing.
Caenorhabditis elegans
C. elegans is a nematode that feeds on fungi and bacteria in soil and rotting fruit. At a bit over 1 mm long, adults are just visible by eye. Since Sydney Brenner's initial characterization of the model in the 1960's, C. elegans research has been essential in the elucidation of several basic aspects of biology, including apopotosis, RNA interference, and miRNA function. The majority of these studies were carried out using E. coli as a feeder organism. For toxicology purposes however, the use of axenic media is preferred to avoid confounding issues of xenobiotic metabolism. In the lab, C. elegans small size means that thousands of animals can be maintained in nutrient media in multi‐well plates, so studies assessing multiple compounds or mixtures at a wide range of concentrations can be carried out in a small space. With a reproductive capacity of about 300 progeny per hermaphrodite adult by self‐fertilization, and a life cycle of approximately 3 days, millions of animals can be rapidly generated, and most experiments can be completed by one person in a week or less. C. elegans has a tough but transparent cuticle, which allows for visualization of internal structures without dissection and facilitates tracking of organellar dyes and structure‐specific gene expression in transgenic strains. Importantly, C. elegans is non‐hazardous to lab workers, and does not reproduce at temperatures above 25°C.
C. elegans somatic cell locations and lineages as well as neural networks have been mapped (White et al., 1986), allowing morphological assessments of toxin induced abnormalities and in depth neurological/behavior correlations. Given that (a) C. elegans was the first multicellular organism to have its genome completely sequenced (C. elegans Sequencing Consortium, 1998), (b) genes and signaling pathways are well conserved between C. elegans and humans (Kaletta and Hengartner, 2006, Leung et al., 2008), (c) studies to understand C. elegans genetics have been underway for over 40 years (Brenner, 1974; Corsi et al., 2015; Nass and Hamza, 2007), and (d) mutant and transgenic C. elegans strains are readily available for many many genes, this model has great potential for the assessment of human‐relevant pathways of toxicity.
Toxicity screening in C. elegans
Studies designed to rank toxicity in C. elegans have consistently shown good correlation with rodent oral LD50 ranking. In an early ranking study, using C. elegans maintained on plates with test articles dissolved in agar, it was found that the toxicity order for eight metal salts based on C. elegans adult mortality correlated with rat and mouse oral LD50 ranking at one‐tenth the cost of rodent testing (Williams and Dusenbery, 1988). The authors also demonstrated that that LC50 ranking in C. elegans was as predictive of acute toxicity in mammals, other than rat and mouse, as LD50 ranking in rat or mouse (Williams and Dusenbery, 1988). In this first assessment comparing LC50 ranking in C. elegans to LD50 ranking in mammals, it was noted that LC50s in C. elegans were high when compared to corresponding mammalian LD50s (Williams and Dusenbery, 1988). It was the relative order of toxicity that correlated rather than the concentration, as subsequent studies have also found. This is probably due to robust genetic mechanisms that allow C. elegans to exist and reproduce in a wide variety of harsh environments.
The Complex Object Parametric Analyzer and Sorter™ (COPAS; Union Biometrica, Holliston, MA, USA) uses microfluidics and laser‐based technologies to automate the analysis of multiple endpoints on hundreds of C. elegans per minute. In my lab, using a novel COPAS‐based assay to assess the fraction of live C. elegans in dosed populations, we also found that LC50 ranking in C. elegans matched LD50 ranking in rat for five metal salts (Hunt et al., 2012). These two studies used very different techniques (agar plates vs. axenic liquid media, manual manipulation with microscopy inspection vs. automated assessment), but in both C. elegans ranking for lethality predicted the relative rodent toxicity for metal salts.
The COPAS has also been used to assess larval growth and reproductive output, and studies assessing six or seven water‐soluble compounds have shown that ranking for these endpoints in C. elegans also correlates with rodent LD50 ranking for the same compounds (Boyd et al., 2010; Ferguson et al., 2010). In a much larger, COPAS‐based study of hundreds of compounds from the ToxCast™ libraries, C. elegans larval growth identified rabbit or rat developmental toxins with a balanced accuracy of 52–53%, whereas the concordance for developmental toxicity between rat and rabbit was 58% (Boyd et al., 2016). These levels of predictivity are consistent with an earlier meta‐study which found that a single‐species rodent study alone predicted human toxicity less than 50% of the time and that using one rodent and one non‐rodent mammalian species together, predictivity increased to 71% (Olson et al., 2000). Unlike a study in mammals, however, a small‐scale C. elegans larval growth assay can be conducted by a single technician in less than a week with an incubator, a basic microscope and a few simple tools. Interestingly, the sensitivity of the larval growth assay for mammalian developmental toxins was high, but the balanced accuracy was brought down by low specificity (Boyd et al., 2016). In contrast, in a recent study evaluating C. elegans egg viability using 72 compounds of known developmental activity in mammals, the specificity of the egg viability test was high whereas the sensitivity was low (Harlow et al., 2016). This begs the question of how the C. elegans larval growth and egg viability assays might be used together, or perhaps in conjunction with mammalian cell based in vitro assays, to better detect mammalian developmental toxins.
In a screen of 21 chemicals in which C. elegans were individually assessed by eye for viability, LC50 ranking in C. elegans and LD50 ranking in mouse predicted rat LD50 ranking equally well, but only when the four compounds that reduced pH to below 3.2 were excluded (Li et al., 2013). In another study using motility as an endpoint to assess 15 organophosphate pesticides and herbicides, the ranking of EC50 values in C. elegans was statistically consistent with the order of rat LD50s (Cole et al., 2004). However, for the two organophosphates that reduced pH to below 3.5, altered motility tracked more consistently with pH than the test article concentration. In this same study, five out of six organophosphate insecticides tested, which were known to inhibit cholinesterase in mammals, also inhibited cholinesterase activity in C. elegans, indicating conservation of the toxicity mechanism (Cole et al., 2004). While factors such as pH and solubility limit C. elegans toxicity studies, they are also limitations for in vitro and zebrafish embryo testing.
In a study comparing the C. elegans endpoints of lethality vs. reproduction for five metabolic poisons, reproduction was suppressed at far lower concentrations than that required for lethality (Middendorf and Dusenbery, 1993). The authors of this study did not compare their data to toxicity in mammals, yet an analysis of the published results relative to rat oral LD50s shows that for the five metabolic poisons tested, C. elegans reproductive EC50 ranking differed from rat ranking only in that the toxins ranked two and three are inverted between the two species. In this study (Middendorf and Dusenbery, 1993), C. elegans LC50 ranking did not correlate with the mammalian data as well as reproductive EC50 ranking, indicating that some endpoints may detect certain categories of toxins better than others, and that in testing unknowns, the assessment of multiple C. elegans endpoints will probably increase the sensitivity of a screen. Blinded validation studies comparing the success of specific C. elegans assays at detecting various types of toxins have not yet been conducted, however. It is likely that rather than relying strictly on ranking, combining the results from a few different C. elegans assay endpoints into broader categories of nontoxic, mildly toxic, toxic, and very toxic will be more useful for tiered testing and predictive toxicology.
In addition to ranking studies, targeted C. elegans screens have also been successful in detecting specific types of toxins. One example is a recently developed elegant assay for the detection of aneugens, toxins that induce alterations in the number of chromosomes per cell. Self‐fertilizing C. elegans XX hermaphrodites normally make up over 99.8% of the wild‐type population, whereas rare XO males arise spontaneously by chromosomal non‐disjunction. Using a male‐specific promoter to drive green fluorescent protein expression, transgenic C. elegans males will fluoresce. When this transgenic strain was exposed to chemicals and assessed for the number of male offspring, seven of eight known mammalian aneugens significantly increased the number of XO male progeny, whereas five known non‐aneugens tested negative (Allard et al., 2013). Cadmium is known to induce intestinal pathology in mammals and fish at a low concentration. In a test of four metal chlorides on adult C. elegans morphology in my lab, we found that at low concentrations which did not alter viability or other evaluated physical parameters, only cadmium reduced intestinal diameter and opacity, a change that was measurable both by light microscopy and by COPAS (Hunt et al., 2012). This indicates a conserved mode of action for cadmium and the possibility that other intestinal toxins can be identified in C. elegans using higher throughput microfluidics techniques.
Predictive toxicology using C. elegans: strengths, limitations and GCeCP
Caenorhabditis elegans can be used to predict nanosilver (AgNP) toxicity in other models. In orally exposed C. elegans and rats, ionic silver is more toxic and is taken up into tissues more extensively than 10‐nm citrate‐coated silver nanospheres (Cit‐AgNP) (Hunt et al., 2013). Polyvinylpyrrolidone (PVP)‐coated AgNP are associated with higher silver uptake and higher toxicity relative to Cit‐AgNP in C. elegans (Hunt et al., 2014) and zebrafish embryos (Kim et al., 2013), whereas in a subsequent study of orally exposed mice, no nanosilver toxicity was observed at study dosing levels, but there was a trend of greater intestinal uptake for PVP‐AgNP over Cit‐AgNP (Bergin et al., 2016). In my lab, we have quickly identified toxicity in specific batches of nanomaterials that had been carefully evaluated for quality and consistency by the synthesizing lab. In one instance, the AgNP producer later identified endotoxin contamination in one of their supplied reagents (Hunt et al., 2013), and in another, a switch from centrifugation‐based nanomaterial wash steps to the use of filters for washing led to the retention of unbound, positively charged coat material that should have been removed from the final product (Hunt et al., 2014). Had these batches been tested directly in mammals, toxicity might have been attributed to the nanomaterial itself rather than a contaminant. This underscores both the importance of assessing all component materials plus the soluble fraction as controls in nanomaterial toxicity testing, and the utility of C. elegans assays which are rapid and inexpensive enough to be used on each batch of nanomaterial produced.
Given the success of the studies highlighted here, it is interesting to speculate why C. elegans is not more widely used in toxicity screening. It may be that the majority of researchers specializing in studying C. elegans are more focused on genetics, development and cell signaling than on toxicity screening, whereas most toxicologists and risk assessors are simply unfamiliar with the model. Certainly, many people, including those on scientific funding committees, mistakenly believe that worms are slimy things with no connection to humans. However, the high conservation of genes and signaling pathways between C. elegans and mammals (Kaletta and Hengartner, 2006, Leung et al., 2008) indicate that the model is likely to continue to do well when used to predict broad measures of toxicity, if good C. elegans culture practice (GCeCP) is followed.
Just as good cell culture practice is essential for reliability and reproducibility in in vitro studies (Hartung et al., 2002), C. elegans used in toxicity testing must be maintained in a highly consistent manner, with temperature, salt concentration and sufficient nutrient supply held constant to ensure reliable, repeatable results (Table 1). Alterations of even a short duration in any of these variables will lead to altered gene expression and toxin response, potentially for multiple generations. Even the brief contact of a human finger applied to a row of wells for a few seconds by picking up a multi‐well plate from the bottom can alter the growth rate of the larvae in those wells.
Table 1.
C. elegans culture standardization factors for toxicology and GCeCP
| Factor | Details |
|---|---|
| Temperature | Small temperature differences have a large effect on C. elegans growth rate, motility, lifecycle, lifespan, and gene expression. The method and duration of vessel contact with human hands and metal surfaces can significantly affect many endpoints. Handling culture vessels by edges that are not in contact with medium, and a layer of Styrofoam on work surfaces will reduce heat exchange. |
| Humidity | Low humidity will alter test article and nutrient concentration, the smaller the volume the larger the effect. Unless equipment is enclosed and carefully climate controlled, it is unlikely that the small volumes used in HTS will work well with C. elegans assays. For larger test volumes, incubators can be humidified with an open vessel of water that is cleaned and refilled regularly. |
| pH | Extreme pH is required to alter adult C. elegans viability, but other endpoints are more sensitive to pH. The appropriate pH range should be determined for each assay, and the pH of test articles in assay medium must be assessed and reported. |
| Worm Density | C. elegans gene expression and life cycle respond to nutrient availability and secreted hormones. Given a 3‐day generation time and ~300 progeny per worm, cultures can easily outstrip nutrients if they are not consistently monitored. Conversely, C. elegans do not grow well if maintained too sparsely. Note that the worms will not necessarily die in these conditions, instead they will adapt epigenetically (Hall et al., 2010), potentially resulting in altered toxicity test results for several generations. |
| Cohort Synchronization | A cohort of 1st larval stage (L1) C. elegans can be isolated by hypochlorite treatment of gravid hermaphrodites (an egg prep) followed by hatching of the released eggs in non‐nutrient buffer. In the absence of nutrients, these L1 s halt development just after hatching. At 20 °C, about 12 hours are required for all the eggs to hatch. After more than 18 hours in buffer, gene expression is altered resulting in delayed and unsynchronized development, and increased stress resistance (Jobson et al., 2015, Nass and Hamza, 2007). Some genetics protocols state that hatched L1 s can be maintained in non‐nutrient buffer and used for a week or more, but this will result in variable toxicity outcomes. |
| Dauers | The C. elegans dauer larva is a stress resistant, long‐lived alternate to the 3rd larval stage (L3). Dauers must revert to the normal lifecycle in order to grow and reproduce. C. elegans dauers secrete dauer pheromone, which promotes conversion to the dauer state in other larvae and induces increased stress resistance in exposed adults. This will both reduce apparent growth rates as measured by worm length (dauers can remain at the L3 length for months or even years), and increase viability in the presence of many toxins. Dauers are thinner and darker than L3s of similar length, and lack the clearly defined gonadal region and visible intestinal lumen identifiable in developing C. elegans. Liquid cultures are unlikely to produce dauers if they are consistently maintained with adequate nutrient supply and are started from agarose cultures that were well fed for at least 3 generations. Daily media exchange along with a few sequential egg preps as soon as each generation becomes gravid can sometimes free a culture of dauers. |
| Genetic Drift | Genetics labs often maintain commonly used C. elegans strains at room temperature as dauers. If C. elegans cultures are consistently well fed for optimal toxicity studies, use of frozen stocks must be scheduled in order to prevent genetic drift. |
| Males | Non‐disjunction of the X chromosome results in XO males. This happens rarely in nature, and is induced by toxins and stress. C. elegans males can be identified by their flared tail and single gonad arm. Males are smaller than the XX hermaphrodites, which will result in apparent reduced growth if automated methods are used and technicians are not trained to recognize males. Mating with males more than doubles the progeny per hermaphrodite relative to selfing, so males in a culture will increase reproductive output. Removing males from a culture requires isolating developing hermaphrodites away from the males. |
| Solid vs. Liquid Medium | When the test article is mixed into molten agar, or spread in solution onto solidified agar, and then the E. coli feeder organism is grown in a lawn on top of the dosed agar, the true exposure will depend on many factors such as humidity, compound solubility, compound‐agar interaction, and feeder organism uptake and metabolism. Dosing in liquid medium provides a measureable exposure, but limits the test to water‐soluble compounds. |
| E. coli vs. Axenic Medium | Axenic medium avoids the complicating factor of the metabolic response of the feeder organism, which is especially important for test articles with antibiotic activity. Killed E. coli are sometimes used (usually heat or UV), but the method and exposure time must be carefully controlled to avoid the bacteria producing toxins which will alter assay results. |
Males normally make up 0.2% of the C. elegans population, although males can be induced by exposure to elevated temperatures or toxins (Corsi et al., 2015). Self‐fertilization maintains the population as nearly all hermaphrodites, while mating produces half male and half hermaphrodite progeny. Caenorhabditis elegans males are smaller than the hermaphrodites, and mating produces more offspring than self‐fertilization. Thus the introduction of males into a culture will skew larval growth and reproductive assays in opposite directions. Additionally, C. elegans have an alternative developmental stage called dauer larva that can be induced by stressful conditions such as low nutrient availability and some toxins. Caenorhabditis elegans exposed to dauer pheromone have higher levels of stress resistance gene expression and are more likely to become dauers themselves. In the dauer state, the cuticle grows over the buccal and anal openings, making dauers resistant to harsh environments. This makes dauer formation a likely explanation for many published C. elegans studies that beautifully demonstrate a mechanism of action or pathway of toxicity, but fail to show a dose response beyond a threshold. Thus, in contrast to some well‐established genetics and cell signaling protocols which utilize dauers and males, C. elegans cultures containing dauers or males should not be used for standard toxicity assays.
The differences in morphology among hermaphrodites, dauers and males are subtle and likely to go unrecognized without training. This training is quickly and easily acquired, however. It is not necessary to check every worm in a start culture for phenotype, because if dauers or males are present, they will likely make up a significant portion of the population within a week. A scan of a few microscopy fields of view per flask is sufficient, with the caveat that if males or dauers are identified, assays completed with that culture within the past week are suspect. Once consistent handling and workflow for GCeCP is established, a single C. elegans technician, using just a few pieces of equipment, can assess a dozen compounds or mixtures at multiple concentrations in a week. While this type of medium‐throughput screening cannot replace a descriptive toxicology study in lab mammals, given the massive backlog of compounds in consumer products and in the environment that have received no toxicological assessment at all (Tice et al., 2013), it makes sense to utilize a small model organism such as C. elegans as one component within a battery of tests to help prioritize compounds of unknown toxicity for further assessment.
Some companies and government agencies are beginning to utilize C. elegans for rapid toxicity evaluations. For example, in response to the 2014 West Virginia Elk River chemical spill, the National Toxicology Program examined the effects of the major constituents of the spill using several toxicity assessment models. No effects were detected for the 4 most prevalent spill compounds in a panel of 27 different in vitro assays (NTP, 2015). Similarly, 12 different compounds and mixtures identified in the spill had no effects on growth, pharyngeal pumping or reproduction in C. elegans (NTP, 2015). While mild dermal irritation and sensitization was detected in mice, and prenatal developmental toxicity was detected in rats, doses were high, and rat studies are planned to assess the effects of lower doses that are more relevant to likely human exposures (NTP, 2015). This set of experiments demonstrates some of the strengths and weaknesses of the C. elegans model. In C. elegans, 12 compounds and mixtures were assessed in a quick study that evaluated three endpoints, whereas a maximum of 3 compounds and mixtures were assessed in the more lengthy and costly mouse and rat assays. The C. elegans data were in accord with the in vitro assessments indicating no deleterious biological activity at tested concentrations, but did not predict mouse dermal irritation response at high concentration, nor a rat reproductive response to high oral gavage doses that corresponded to over 20 g for a 70‐kg person (NTP, 2015). Thus, the C. elegans model predicted the absence of general acute toxicity for the spill components at lower doses but failed to detect specific high‐dose mammalian toxic outcomes.
In theory, C. elegans ease of culture, small size and ability to grow in axenic liquid media indicate that the model would lend itself to the type of high‐throughput screening (HTS) now in use by multi‐agency efforts to modernize toxicity testing such as Tox21 (Tice et al., 2013). Current HTS equipment may not have the level of temperature, humidity and sterility control that reliable C. elegans testing requires, however. In the very small volumes that are used in HTS, small changes in ambient humidity can dramatically alter test article, nutrient and salt concentrations, which will in turn alter assessed endpoints. Additionally, because C. elegans are highly responsive to their environment, small temperature differences among metal robotic arms, plastic tubing and mechanical shelving will have a large effect on C. elegans metabolism and growth rates. These factors will lead to variable test results that depend on weather (humid or dry), well selection (close or far from mechanical arm grips) and plate location (local air flow, proximity to motors) that are avoided when larger volumes and low‐ to medium‐throughput techniques are used. Consistency in HTS could dramatically improve if factors such as nutrient availability, temperature and humidity are carefully controlled and start cultures are monitored for consistent reproductive capacity and life cycle time, as well as the absence of dauers and males.
Homologous genes and concordant pathways
The first completely sequenced genome of a multicellular organism was that of C. elegans, and many genes and signaling pathways are conserved between nematodes and humans (C. elegans Sequencing Consortium, 1998). For example, apoptosis, or programmed cell death, an essential process in normal animal development and adult homeostasis, is now known to be a well‐conserved process (Vaux et al., 1992), but it was in C. elegans that the first genes in the apoptosis pathway were identified (Ellis and Horvitz, 1986). Additionally, many elements of the insulin/IGF‐1 signaling (IIS) pathway, which uses both the endocrine system and cellular mechanisms to regulate glucose utilization and metabolism, are shared in C. elegans and mammals. DAF‐16, the sole C. elegans homolog of the mammalian FOXO forkhead transcription factors, was found to be a negative regulator of insulin signaling 5 years before a mouse FOXO was identified as a regulator of the same pathway (Ogg et al., 1997; Nakae et al., 2002). It was first discovered in C. elegans that IIS is involved in aging (Kenyon et al., 1993), and later that corresponding genetic variants within this pathway in humans and C. elegans have analogous effects on lifespan (Kenyon, 2011). Additionally, dietary restriction‐induced increases in healthspan and lifespan are mediated by IIS and other conserved pathways in both nematodes and mammals (Fontana et al., 2010). Drugs such as metformin that are used to alter IIS activity to improve human health can increase the healthspan and lifespan in C. elegans (Onken and Driscoll, 2010) and mice (Martin‐Montalvo et al., 2013), whereas diets high in glucose are associated with poor health in humans and a decreased lifespan in C. elegans (Lee et al., 2009). Thus, a 4‐week screening assay in C. elegans using lifespan as an endpoint has the potential to identify compounds that can improve or worsen human health.
The majority of familial early‐onset Alzheimer's disease (AD) patients have mutations in presenilin‐1 (PSEN1), a gene required for processing of amyloid precursor protein. Four out of the initial five disease‐causing missense mutations found in PSEN1 alter amino acids that are conserved between PSEN1 and the C. elegans presenilin gene sel‐12 (Levitan and Greenwald, 1995). As with PSEN1 in humans, mutations in sel‐12 induce altered neuronal morphology and memory defects in C. elegans (Levitan and Greenwald, 1995), and the memory defects in C. elegans sel‐12 mutants can be rescued by the normal human PSEN1 gene (Wittenburg et al., 2000). Caenorhabditis elegans also has close homologs in several other genes associated with AD etiology, potentially making the worm a useful model in which to understand and treat AD (Alexander et al., 2014; Lu et al., 2014).
An early example of the mechanism of toxicity identification that was supported by research in C. elegans is that of phorbol esters (PE), potent mammalian tumor promoters that induce developmental arrest and altered motility in C. elegans (Miwa et al., 1982). In a single assay screening for mutants generated by exposure to ethyl methane sulfonate for reversion to control phenotype in the presence of PE, a C. elegans protein kinase C (PKC) homolog tpa‐1 was identified as required for PE toxicity (Tabuse and Miwa, 1983; Tabuse et al., 1989). In contrast, PKC was identified as the major receptor for PE in mammals in a far more complex and incremental manner involving multiple labs over several years (Blumberg, 1988). Thus, phorbol ester toxicity acts via PKC in mammals and worms, although the endpoints (growth and motility vs. tumorigenesis) differ in the two models. Additionally, PEs induce lysosomal degradation in mammalian cell cultures (Daniels and Amara, 1999), and the major pathway change detected in the analysis of gene expression in PE‐exposed C. elegans is upregulation of lysosome degradation (Ana DePina and Xiugong Gao, personal communication) indicating at least one conserved mode of toxic action. The somatic cells of C. elegans adults are post‐mitotic (Gartner et al., 2008), so the species is not used as a direct model for tumorigenesis. However, the example of PE toxicity demonstrates (a) how research using a small organism that lacks many systems and phenotypes present in more complex animals can still provide useful toxicological information, (b) how screening for endpoints without direct correlations to human outcomes such as cancer can still be used to detect conserved perturbations in toxicity pathways, and (c) how a relatively simple mutagenesis experiment in C. elegans can be used to identify a conserved mechanism of toxicity. Moreover, given that the majority of cancer genes have a C. elegans ortholog (Rubin et al., 2000) and that conserved processes such as apoptosis are required for evading tumorigenesis, it is possible that many human carcinogens could be identified in C. elegans by screening for alterations in the activity of genes in related pathways.
Two‐thirds of human proteins have C. elegans homologs, and nearly 80% of genes for human inborn errors of metabolism have C. elegans homologs (Sonnhammer and Durbin, 1997; Kuwabara and O'Neil, 2001;). The C. elegans ortholog of TRPML1, the gene for mucolipidosis type IV, is cup‐5 (coelomocyte‐uptake defective) (Campbell and Fares, 2010). In humans, mutations in TRPML1 result in psychomotor retardation and ophthalmological abnormalities, whereas C. elegans homozygous for mutations in cup‐5 are sterile, yet in both species absence of TRPML1/cup‐5 activity at the cellular level results in inappropriate accumulation of lysosomes. Additionally, the normal human TRPML1 gene rescues C. elegans cup‐5 reproductive and lysosomal abnormalities (Hersh et al., 2002). In a similar way, toxicity endpoints in C. elegans such as reduced reproductive capacity, altered motility patterns, or developmental defects may detect toxins that elicit very different organismal endpoints in humans, but subsequent analyses of more subtle effects at the molecular level have the potential indicate shared pathways of toxicity. For example, homologs of genes associated with neural tube defects in humans were recently found to be required for C. elegans gastrulation (Sullivan‐Brown et al., 2016). Vertebrate neural tube closure and C. elegans gastrulation involve many of the same processes such as internalization of surface cells, actomyosin‐driven apical constriction and adhesions between specific cells, suggesting that screening in C. elegans for early embryonic abnormalities may detect toxins that induce neural tube and other developmental defects.
Caenorhabditis elegans utilize most of the same neurotransmitters and neuronal signaling pathways as vertebrates (Kaletta and Hengartner, 2006; Peterson et al., 2008). From worms to mice, dopamine and serotonin play similar roles in the transition from one form of locomotion to another (Vidal‐Gadea et al., 2011). Caenorhabditis elegans share several nicotinic acetylcholine receptor‐dependent behavioral responses to nicotine with mammals (Feng et al., 2006), and C. elegans mutagenesis screens to detect worms with altered motility has led to the identification of human genes for specific myosins and neurotransmitter transporters (Beron et al., 2015). However, the C. elegans genome lacks voltage‐gated sodium channels (Bargmann, 1998), so the model is not likely to detect excitotoxins and suppressors that exert their effects via this class of ion channel. Conversely, nematodes and insects have inhibitory glutamate‐gated chloride channels that mammals do not, allowing for the selective action of some pesticides (Peterson et al., 2008). These differences underlie the importance of fully understanding any model prior to using it for hazard evaluations.
Promising avenues for toxicity testing in C. elegans
Methods to assess lethality in C. elegans range from simple and inexpensive for a non‐worm lab to set up, to those requiring costly automated motility tracking or microfluidics equipment. Adult C. elegans are amazingly hearty, so while more screens for adult mortality have been published showing mammalian‐correlated toxicity ranking, screens for larval growth, reproduction and motility have also shown ranking correlations and can be more sensitive by an order of magnitude or more (Anderson et al., 2004; Dhawan et al., 1999; Boyd et al., 2010), allowing for reduced test article concentrations and, therefore, fewer solubility issues. Additionally, assays for motility, enzyme activity, reactive oxygen species, RNAi response and gene expression can provide information on mechanisms of action as well as relative toxicity. Other endpoints to assess chemical toxicity include intestinal morphology (Hunt et al., 2012; Stutz et al., 2015), accumulation of autofluorescence (Gerstbrein et al., 2005), gonadal morphology (Hunt et al., 2012) and internal hatching (Hunt et al., 2013).
The C. elegans alimentary system has many facets that are comparable to that of mammals, including an acidified lumen, microvilli that form a brush border, secretion of digestive enzymes, uptake of digested components and peristalsis (Hall and Altun, 2008; Chauhan et al., 2013; Stutz et al., 2015). Mammalian and C. elegans intestinal dysmorphology in response to cadmium and toxic lectins is comparable (Hunt et al., 2012; Stutz et al., 2015). In contrast to mammals, however, the damage induced by intestinal toxins can also be visualized in live worms by light microscopy or COPAS detection (Hunt et al., 2012). Caenorhabditis elegans feed by pulling liquid phase components from their environment into their pharynx by pumping and peristalsis. Thus, they can be used as an oral toxicity model by adding test articles to their nutrient supply. Additionally, pharyngeal pumping rates, which decrease in the presence of several known mammalian neurotoxins, can be observed by light microscopy in individual worms. A higher throughput method of evaluating the effect of toxins on feeding rates involves exposure to fluorescent microbeads followed by an assessment of internal fluorescence (Boyd et al., 2007).
Over 70% of human lipid genes are conserved in C. elegans, and 20% of C. elegans lipid genes are orthologs of human metabolic disease genes (Zhang et al., 2013). Distinct long‐ and short‐term fat storage vesicles are easily visualized in C. elegans (O'Rourke et al., 2009) allowing for screening of compounds that alter lipid metabolism and storage. Similarly, 90% of human lysosome‐associated non‐disease genes, as well as 70% of human lysosomal storage disorder genes, have C. elegans homologs (de Voer et al., 2008). There are several methods to stain specific subsets of endosome and lysosome‐related vesicles in live and fixed worms (Clokey and Jacobson, 1986; Hersh et al., 2002; Chen et al., 2006; Roh et al., 2012), allowing for rapid evaluation of potential lysosomal toxins.
Adult C. elegans somatic cells are post‐mitotic, making the model seem at first glance to be a poor one in which to assess carcinogenicity. The cellular machinery for DNA replication and repair is highly conserved between C. elegans and mammals (Leung et al., 2008) however, and related pathways that prevent the propagation of carcinogenic mutations, such as apoptosis and cell cycle checkpoints, also have conserved elements (Stergiou and Hengartner, 2004). In addition to the previously discussed assay for the detection of aneugens (Allard et al., 2013), C. elegans‐based assays for the detection of DNA damage include DNA sequencing, reporter assays, lethality assays and gene expression assessments (Leung et al., 2008). Specific DNA lesions can also be detected in C. elegans by gas chromatography/tandem mass spectrometry (Arczewska et al., 2013; Hunt et al., 2013). In another promising new assay, a green fluorescent protein labeled ZTF‐8 altered localization in C. elegans specifically in response to agents that induced double‐stranded DNA breaks but not other types of DNA damage (Kim and Colaiacovo, 2014).
On a much larger scale, screening of thousands of potential drugs with the COPAS using C. elegans pre‐infected with human pathogens has been used to identify compounds with antimicrobial activity. This method is a departure from traditional screens for antibiotics, which test compounds for their ability to directly kill microorganisms or inhibit their growth. By using infected C. elegans, drug efficacy and host toxicity are simultaneously assessed in vivo, while making it possible to identify compounds that require metabolic activation for antimicrobial activity, stimulate innate immune activity, and/or inhibit host‐pathogen interactions for the establishment of infection, without harming the host (Moy et al., 2009). This approach was recently used to evaluate 300 000 compounds to identify drug candidates that might improve outcomes for patients infected with multi‐drug‐resistant B. pseudomallei (Lakshmanan et al., 2014). On a much smaller scale, C. elegans have also been used as part of a two‐tiered approach to identify new non‐toxic compounds for the treatment of B. cenocepacia K56–2 infections in cystic fibrosis patients (Selin et al., 2015). It is too early to tell if the findings from these studies will result in safe treatments for patients, but the strategy appears promising.
Summary: The C. elegans Toxicology Model
Model Strengths
The first completely sequenced genome of a multicellular organism, plus over 40 years of genetics, neuroscience, and cell signaling research make C. elegans a well understood model
Extensive homology to mammals at the genetic level
Many key cellular metabolic and signaling pathways are conserved
Many elements of neuronal function are conserved
Conserved alimentary features make C. elegans a good oral toxicity model
Unlike cell and tissue cultures, C. elegans has neuronal, motor, digestive, and reproductive systems, endocrine signaling, and sensory/behavioral responses to stimuli
C. elegans can be maintained at 15 to 25°C, using relatively simple techniques and without the need for CO2 incubators
Inexpensive and compact platform allows multiple concentrations and exposure times to be assessed simultaneously
Short life cycle allows for low‐dose lifespan and multi‐generation testing in a couple of weeks rather than years
Transparent tissues and fully mapped body and neuronal plans allow for rapid morphology and transgene expression assessment at the tissue, cellular and sub‐cellular levels
Multiple types of toxicity ranking screens have demonstrated good correlation of endpoints in C. elegans to rat LD50s, with C. elegans data and mouse data predicting toxicity ranking in rat equally well
Model Limitations
C. elegans lack many mammalian organs such as eyes, lungs, heart, kidney, and liver
As with the zebrafish embryo model, C. elegans have a functioning innate immune system, but lack adaptive immunity
In contrast to zebrafish embryos, C. elegans is not a good absorption model due to its tough cuticle
As with cell‐based models, pH range is wide but still limited, and liquid culture testing requires soluble test compounds
Small changes in temperature, nutrient, or salt concentration elicit adaptive responses that can significantly alter assay results, sometimes for multiple generations – this currently limits the number of compounds that can be screened by a single lab, but could change with enclosed, climate‐controlled microfluidic HTS equipment
Incorrect handling of stock cultures can result in altered gene expression patterns and the accumulation of dauers or males – phenotype recognition training and good C. elegans culture practice (GCeCP, Table 1) is essential for reliable, repeatable data
Some C. elegans systems act/respond in similar ways as analogous mammalian systems, but lack homology at the genetic level (e.g. the innate immune system) – development of microarray analysis software that is specific for C. elegans using data that is already available would fix this problem
Conclusions
Currently accepted standards for hazard assessment using conventional laboratory animals are hampered by duration and cost, as well as species specificity. The need for more affordable, rapid and predictive toxicity testing paradigms has led to several coordinated national and international efforts to test and validate novel assays and toxicity evaluation strategies (Casey et al., 2015). These efforts have focused mainly on high‐throughput in vitro testing, which can reveal molecular and cellular mechanisms of toxicity, but cannot currently provide information required for hazard assessment, such as the effects of exposure route, metabolism by and transport through multiple tissues, and communication among interacting tissues (Tice et al., 2013). Caenorhabditis elegans can provide a bridge between in vitro assays and mammalian toxicity testing by combining established in vitro handling techniques and cost ratios with oral toxicity test data from an intact organism, although standardized culture practices are required to achieve consistent results. Given that nematodes lack most mammalian organs, it is unrealistic to expect that any combination of C. elegans assays alone will replace in‐depth descriptive toxicology analyses in mammals. However, although organismal toxicity endpoints often differ, many pathways of toxicity and modes of toxic action are conserved between worms and humans. Caenorhabditis elegans screening assays have consistently predicted LD50 ranking in mammals, indicating the utility of the model for screening at an early step in tiered testing strategies. The use of C. elegans assays within integrated testing strategies that include other models such as zebrafish or human cell cultures have yet to be evaluated.
Conflict of interest
The authors do not report any conflict of interest.
Acknowledgements
The author wishes to thank Dr. Robert Sprando for many helpful discussions and suggestions for improving this manuscript, and Dr. Ann Heinzer for careful reading of the final draft.
Hunt, P. R. (2017) The C. elegans model in toxicity testing. J. Appl. Toxicol., 37: 50–59. doi: 10.1002/jat.3357.
References
- Alexander AG, Marfil V, Li C. 2014. Use of Caenorhabditis elegans as a model to study Alzheimer's disease and other neurodegenerative diseases. Front. Genet. 5: 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard P, Kleinstreuer NC, Knudsen TB, Colaiacovo MP. 2013. A C. elegans screening platform for the rapid assessment of chemical disruption of germline function. Environ . Health Perspect. 121: 717–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GL, Cole RD, Williams PL. 2004. Assessing behavioral toxicity with Caenorhabditis elegans . Environ. Toxicol. Chem. 23: 1235–1240. [DOI] [PubMed] [Google Scholar]
- Arczewska KD, Tomazella GG, Lindvall JM, Kassahun H, Maglioni S, Torgovnick A, Henriksson J, Matilainen O, Marquis BJ, Nelson BC, Jaruga P, Babaie E, Holmberg CI, Burglin TR, Ventura N, Thiede B, Nilsen H. 2013. Active transcriptomic and proteomic reprogramming in the C. elegans nucleotide excision repair mutant xpa‐1. Nucleic Acids Res. 41: 5368–5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI. 1998. Neurobiology of the Caenorhabditis elegans genome. Science 282: 2028–2033. [DOI] [PubMed] [Google Scholar]
- Bergin IL, Wilding LA, Morishita M, Walacavage K, Ault AP, Axson JL, Stark DI, Hashway SA, Capracotta SS, Leroueil PR, Maynard AD, Philbert MA. 2016. Effects of particle size and coating on toxicologic parameters, fecal elimination kinetics and tissue distribution of acutely ingested silver nanoparticles in a mouse model. Nanotoxicology 10: 352–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beron C, Vidal‐Gadea AG, Cohn J, Parikh A, Hwang G, Pierce‐Shimomura JT. 2015. The burrowing behavior of the nematode Caenorhabditis elegans: a new assay for the study of neuromuscular disorders. Genes Brain Behav. 14: 357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Zhang Q, Carmichael PL, Boekelheide K, Andersen ME. 2011. Toxicity testing in the 21 century: defining new risk assessment approaches based on perturbation of intracellular toxicity pathways. PLoS One 6: e20887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg PM. 1988. Protein kinase C as the receptor for the phorbol ester tumor promoters: sixth Rhoads memorial award lecture. Cancer Res. 48: 1–8. [PubMed] [Google Scholar]
- Boyd WA, McBride SJ, Freedman JH. 2007. Effects of genetic mutations and chemical exposures on Caenorhabditis elegans feeding: evaluation of a novel, high‐throughput screening assay. PLoS One 2: e1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd WA, McBride SJ, Rice JR, Snyder DW, Freedman JH. 2010. A high‐throughput method for assessing chemical toxicity using a Caenorhabditis elegans reproduction assay. Toxicol. Appl. Pharmacol. 245: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd WA, Smith MV, Co CA, Pirone JR, Rice JR, Shockley KR, Freedman JH. 2016. Developmental Effects of the ToxCast™ Phase I and II chemicals in Caenorhabditis elegans and corresponding responses in zebrafish, rats, and rabbits. Environ. Health Perspect. 124: 586–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans . Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium . 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018. [DOI] [PubMed] [Google Scholar]
- Campbell EM, Fares H. 2010. Roles of CUP‐5, the Caenorhabditis elegans orthologue of human TRPML1, in lysosome and gut granule biogenesis. BMC Cell Biol. 11: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey W, Jacobs A, Maull E, Matheson J, Clarke C, Lowit A. 2015. A new path forward: the Interagency Coordinating Committee on the Validation of Alternative Methods (ICCVAM) and National Toxicology Program's Interagency Center for the Evaluation of Alternative Toxicological Methods (NICEATM). J. Am. Assoc. Lab. Anim. Sci. 54: 170–173. [PMC free article] [PubMed] [Google Scholar]
- Chauhan VM, Orsi G, Brown A, Pritchard DI, Aylott JW. 2013. Mapping the pharyngeal and intestinal pH of Caenorhabditis elegans and real‐time luminal pH oscillations using extended dynamic range pH‐sensitive nanosensors. ACS Nano 7: 5577–5587. [DOI] [PubMed] [Google Scholar]
- Chen CC, Schweinsberg PJ, Vashist S, Mareiniss DP, Lambie EJ, Grant BD. 2006. RAB‐10 is required for endocytic recycling in the Caenorhabditis elegans intestine. Mol. Biol. Cell 17: 1286–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clokey GV, Jacobson LA. 1986. The autofluorescent "lipofuscin granules" in the intestinal cells of Caenorhabditis elegans are secondary lysosomes. Mech. Ageing Dev. 35: 79–94. [DOI] [PubMed] [Google Scholar]
- Cole RD, Anderson GL, Williams PL. 2004. The nematode Caenorhabditis elegans as a model of organophosphate‐induced mammalian neurotoxicity. Toxicol. Appl. Pharmacol. 194: 248–256. [DOI] [PubMed] [Google Scholar]
- Corsi AK, Wightman B, Chalfie M. 2015. A Transparent Window into Biology: A Primer on Caenorhabditis elegans . WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.177.1, http://www.wormbook.org (accessed March 2, 2016).
- Daniels GM, Amara SG. 1999. Regulated trafficking of the human dopamine transporter. Clathrin‐mediated internalization and lysosomal degradation in response to phorbol esters. J. Biol. Chem. 274: 35794–35801. [DOI] [PubMed] [Google Scholar]
- de Voer G, Peters D, Taschner PE. 2008. Caenorhabditis elegans as a model for lysosomal storage disorders. Biochim. Biophys. Acta 1782: 433–446. [DOI] [PubMed] [Google Scholar]
- Dhawan R, Dusenbery DB, Williams PL. 1999. Comparison of lethality, reproduction, and behavior as toxicological endpoints in the nematode Caenorhabditis elegans . J. Toxicol. Environ. Health A 58: 451–462. [DOI] [PubMed] [Google Scholar]
- Dix DJ, Houck KA, Martin MT, Richard AM, Setzer RW, Kavlock RJ. 2007. The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol. Sci. 95: 5–12. [DOI] [PubMed] [Google Scholar]
- Ellis HM, Horvitz HR. 1986. Genetic control of programmed cell death in the nematode C. elegans . Cell 44: 817–829. [DOI] [PubMed] [Google Scholar]
- Feng Z, Li W, Ward A, Piggott BJ, Larkspur ER, Sternberg PW, Xu XZ. 2006. A C. elegans model of nicotine‐dependent behavior: regulation by TRP‐family channels. Cell 127: 621–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson M, Boyer M, Sprando R. 2010. A method for ranking compounds based on their relative toxicity using neural networking, C. elegans, axenic liquid culture, and the COPAS parameters TOF and EXT. Open Access Bioinformatics 2010: 139–144. [Google Scholar]
- Fontana L, Partridge L, Longo VD. 2010. Extending healthy life span‐‐from yeast to humans. Science 328: 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A, Boag PR, Blackwell TK. 2008. Germline Survival and Apoptosis. WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.145.1, http://www.wormbook.org (accessed May 16, 2016). [DOI] [PMC free article] [PubMed]
- Gerstbrein B, Stamatas G, Kollias N, Driscoll M. 2005. In vivo spectrofluorimetry reveals endogenous biomarkers that report healthspan and dietary restriction in Caenorhabditis elegans . Aging Cell 4: 127–137. [DOI] [PubMed] [Google Scholar]
- Hall DH, Altun ZF. 2008. C. elegans Atlas. Cold Spring Harbor Laboratory Press: Cold Spring Harbor. [Google Scholar]
- Hall SE, Beverly M, Russ C, Nusbaum C, Sengupta P. 2010. A cellular memory of developmental history generates phenotypic diversity in C. elegans . Curr. Biol. 20: 149–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow PH, Perry SJ, Widdison S, Daniels S, Bondo E, Lamberth C, Currie RA, Flemming AJ. 2016. The nematode Caenorhabditis elegans as a tool to predict chemical activity on mammalian development and identify mechanisms influencing toxicological outcome. Sci. Rep. 6: 22965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung T. 2009. Toxicology for the twenty‐first century. Nature 460: 208–212. [DOI] [PubMed] [Google Scholar]
- Hartung T, Balls M, Bardouille C, Blanck O, Coecke S, Gstraunthaler G, Lewis D. 2002. Good Cell Culture Practice. ECVAM Good Cell Culture Practice Task Force Report 1. Altern. Lab. Anim. 30: 407–414. [DOI] [PubMed] [Google Scholar]
- Hartung T, Luechtefeld T, Maertens A, Kleensang A. 2013. Integrated testing strategies for safety assessments. ALTEX 30: 3–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersh BM, Hartwieg E, Horvitz HR. 2002. The Caenorhabditis elegans mucolipin‐like gene cup‐5 is essential for viability and regulates lysosomes in multiple cell types. Proc. Natl. Acad. Sci. U. S. A. 99: 4355–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt PR, Keltner Z, Gao X, Oldenburg SJ, Bushana P, Olejnik N, Sprando RL. 2014. Bioactivity of nanosilver in Caenorhabditis elegans: Effects of size, coat, and shape. Toxic. Rep. 1: 923–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt PR, Marquis BJ, Tyner KM, Conklin S, Olejnik N, Nelson BC, Sprando RL. 2013. Nanosilver suppresses growth and induces oxidative damage to DNA in Caenorhabditis elegans . J. Appl. Toxicol. 33: 1131–1142. [DOI] [PubMed] [Google Scholar]
- Hunt PR, Olejnik N, Sprando RL. 2012. Toxicity ranking of heavy metals with screening method using adult Caenorhabditis elegans and propidium iodide replicates toxicity ranking in rat. Food Chem. Toxicol. 50: 3280–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobson MA, Jordan JM, Sandrof MA, Hibshman JD, Lennox AL, Baugh LR. 2015. Transgenerational Effects of Early Life Starvation on Growth, Reproduction, and Stress Resistance in Caenorhabditis elegans . Genetics 201: 201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson R, Richard A, Dix DJ, Houck K, Martin M, Kavlock R, Dellarco V, Henry T, Holderman T, Sayre P, Tan S, Carpenter T, Smith E. 2009. The toxicity data landscape for environmental chemicals. Environ. Health Perspect. 117: 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletta T, Hengartner MO. 2006. Finding function in novel targets: C. elegans as a model organism. Nat. Rev . Drug Discov. 5: 387–398. [DOI] [PubMed] [Google Scholar]
- Kenyon C. 2011. QnAs with Cynthia Kenyon. Proc. Natl. Acad. Sci. U. S. A. 108: 16875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. 1993. A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464. [DOI] [PubMed] [Google Scholar]
- Kim HM, Colaiacovo MP. 2014. ZTF‐8 interacts with the 9‐1‐1 complex and is required for DNA damage response and double‐strand break repair in the C. elegans germline. PLoS Genet. 10: e1004723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K‐T, Truong L, Wehmas L, Tanguay RL. 2013. Silver nanoparticle toxicity in the embryonic zebrafish is governed by particle dispersion and ionic environment. Nanotechnology 24: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland D, Aardema M, Henderson L, Muller L. 2005. Evaluation of the ability of a battery of three in vitro genotoxicity tests to discriminate rodent carcinogens and non‐carcinogens I. Sensitivity, specificity and relative predictivity. Mutat. Res. 584: 1–256. [DOI] [PubMed] [Google Scholar]
- Knight AW, Little S, Houck K, Dix D, Judson R, Richard A, McCarroll N, Akerman G, Yang C, Birrell L, Walmsley RM. 2009. Evaluation of high‐throughput genotoxicity assays used in profiling the US EPA ToxCast chemicals. Regul. Toxicol. Pharmacol. 55: 188–199. [DOI] [PubMed] [Google Scholar]
- Krewski D, Andersen ME, Mantus E, Zeise L. 2009. Toxicity testing in the 21st century: implications for human health risk assessment. Risk Anal. 29: 474–479. [DOI] [PubMed] [Google Scholar]
- Kuwabara PE, O'Neil N. 2001. The use of functional genomics in C. elegans for studying human development and disease. J. Inherit. Metab. Dis. 24: 127–138. [DOI] [PubMed] [Google Scholar]
- Lakshmanan U, Yap A, Fulwood J, Yichun L, Hoon SS, Lim J, Ting A, Sem XH, Kreisberg JF, Tan P, Tan G, Flotow H. 2014. Establishment of a novel whole animal HTS technology platform for melioidosis drug discovery. Comb. Chem . High Throughput Screen 17: 790–803. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Murphy CT, Kenyon C. 2009. Glucose shortens the life span of C. elegans by downregulating DAF‐16/FOXO activity and aquaporin gene expression. Cell Metab. 10: 379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung MCK, Williams PL, Benedetto A, Au C, Helmcke KJ, Aschner M, Meyer JN. 2008. Caenorhabditis elegans: an emerging model in biomedical and environmental toxicology. Toxicol. Sci. 106: 5–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitan D, Greenwald I. 1995. Facilitation of lin‐12‐mediated signalling by sel‐12, a Caenorhabditis elegans S182 Alzheimer's disease gene. Nature 377: 351–354. [DOI] [PubMed] [Google Scholar]
- Li AP, Lu C, Brent JA, Pham C, Fackett A, Ruegg CE, Silber PM. 1999. Cryopreserved human hepatocytes: characterization of drug‐metabolizing enzyme activities and applications in higher throughput screening assays for hepatotoxicity, metabolic stability, and drug–drug interaction potential. Chem. Biol. Interact. 121: 17–35. [DOI] [PubMed] [Google Scholar]
- Li Y, Gao S, Jing H, Qi L, Ning J, Tan Z, Yang K, Zhao C, Ma L, Li G. 2013. Correlation of chemical acute toxicity between the nematode and the rodent. Toxicol. Res. 2: 403–412. [Google Scholar]
- Lu T, Aron L, Zullo J, Pan Y, Kim H, Chen Y, Yang TH, Kim HM, Drake D, Liu XS, Bennett DA, Colaiacovo MP, Yankner BA. 2014. REST and stress resistance in ageing and Alzheimer's disease. Nature 507: 448–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin‐Montalvo A, Mercken EM, Mitchell SJ, Palacios HH, Mote PL, Scheibye‐Knudsen M, Gomes AP, Ward TM, Minor RK, Blouin MJ, Schwab M, Pollak M, Zhang Y, Yu Y, Becker KG, Bohr VA, Ingram DK, Sinclair DA, Wolf NS, Spindler SR, Bernier M, de Cabo R. 2013. Metformin improves healthspan and lifespan in mice. Nat. Commun. 4: 2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middendorf PJ, Dusenbery DB. 1993. Fluoroacetic Acid Is a Potent and Specific Inhibitor of Reproduction in the Nematode Caenorhabditis elegans . J. Nematol. 25: 573–577. [PMC free article] [PubMed] [Google Scholar]
- Miranda JP, Leite SB, Muller‐Vieira U, Rodrigues A, Carrondo MJ, Alves PM. 2009. Towards an extended functional hepatocyte in vitro culture. Tissue Eng. Part C Methods 15: 157–167. [DOI] [PubMed] [Google Scholar]
- Miwa J, Tabuse Y, Furusawa M, Yamasaki H. 1982. Tumor promoters specifically and reversibly disturb development and behavior of Caenorhabditis elegans . J. Cancer Res. Clin. Oncol. 104: 81–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy TI, Conery AL, Larkins‐Ford J, Wu G, Mazitschek R, Casadei G, Lewis K, Carpenter AE, Ausubel FM. 2009. High‐throughput screen for novel antimicrobials using a whole animal infection model. ACS Chem. Biol. 4: 527–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakae J, Biggs WH, 3rd , Kitamura T, Cavenee WK, Wright CV, Arden KC, Accili D. 2002. Regulation of insulin action and pancreatic beta‐cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nat. Genet. 32: 245–253. [DOI] [PubMed] [Google Scholar]
- Nass R, Hamza I. 2007. The nematode C. elegans as an animal model to explore toxicology in vivo: solid and axenic growth culture conditions and compound exposure parameters. Curr. Protoc. Toxicol. Chapter 1: Unit1 9 doi: 10.1002/0471140856.tx0109s31. [DOI] [PubMed] [Google Scholar]
- NTP . 2015. West Virginia Chemical Spill: NTP Studies and Results . National Toxicology Program http://ntp.niehs.nih.gov/results/areas/wvspill/studies/index.html [accessed 4 June 2015].
- O'Rourke EJ, Soukas AA, Carr CE, Ruvkun G. 2009. C. elegans major fats are stored in vesicles distinct from lysosome‐related organelles. Cell Metab. 10: 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. 1997. The Fork head transcription factor DAF‐16 transduces insulin‐like metabolic and longevity signals in C. elegans . Nature 389: 994–999. [DOI] [PubMed] [Google Scholar]
- Olson H, Betton G, Robinson D, Thomas K, Monro A, Kolaja G, Lilly P, Sanders J, Sipes G, Bracken W, Dorato M, Van Deun K, Smith P, Berger B, Heller A. 2000. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul. Toxicol. Pharmacol. 32: 56–67. [DOI] [PubMed] [Google Scholar]
- Onken B, Driscoll M. 2010. Metformin induces a dietary restriction‐like state and the oxidative stress response to extend C. elegans Healthspan via AMPK, LKB1, and SKN‐1. PLoS One 5: e8758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RT, Nass R, Boyd WA, Freedman JH, Dong K, Narahashi T. 2008. Use of non‐mammalian alternative models for neurotoxicological study. Neurotoxicology 29: 546–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfuhler S, Kirst A, Aardema M, Banduhn N, Goebel C, Araki D, Costabel‐Farkas M, Dufour E, Fautz R, Harvey J, Hewitt NJ, Hibatallah J, Carmichael P, Macfarlane M, Reisinger K, Rowland J, Schellauf F, Schepky A, Scheel J. 2010. A tiered approach to the use of alternatives to animal testing for the safety assessment of cosmetics: genotoxicity. A COLIPA analysis. Regul. Toxicol. Pharmacol. 57: 315–324. [DOI] [PubMed] [Google Scholar]
- Roh HC, Collier S, Guthrie J, Robertson JD, Kornfeld K. 2012. Lysosome‐related organelles in intestinal cells are a zinc storage site in C. elegans . Cell Metab. 15: 88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, Cherry JM, Henikoff S, Skupski MP, Misra S, Ashburner M, Birney E, Boguski MS, Brody T, Brokstein P, Celniker SE, Chervitz SA, Coates D, Cravchik A, Gabrielian A, Galle RF, Gelbart WM, George RA, Goldstein LS, Gong F, Guan P, Harris NL, Hay BA, Hoskins RA, Li J, Li Z, Hynes RO, Jones SJ, Kuehl PM, Lemaitre B, Littleton JT, Morrison DK, Mungall C, O'Farrell PH, Pickeral OK, Shue C, Vosshall LB, Zhang J, Zhao Q, Zheng XH, Lewis S. 2000. Comparative genomics of the eukaryotes. Science 287: 2204–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott CW, Peters MF, Dragan YP. 2013. Human induced pluripotent stem cells and their use in drug discovery for toxicity testing. Toxicol. Lett. 219: 49–58. [DOI] [PubMed] [Google Scholar]
- Selin C, Stietz MS, Blanchard JE, Gehrke SS, Bernard S, Hall DG, Brown ED, Cardona ST. 2015. A Pipeline for Screening Small Molecules with Growth Inhibitory Activity against Burkholderia cenocepacia. PLoS One 10: e0128587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, Durbin R. 1997. Analysis of protein domain families in Caenorhabditis elegans . Genomics 46: 200–216. [DOI] [PubMed] [Google Scholar]
- Stergiou L, Hengartner MO. 2004. Death and more: DNA damage response pathways in the nematode C. elegans . Cell Death Differ. 11: 21–28. [DOI] [PubMed] [Google Scholar]
- Stutz K, Kaech A, Aebi M, Kunzler M, Hengartner MO. 2015. Disruption of the C. elegans Intestinal Brush Border by the Fungal Lectin CCL2 Phenocopies Dietary Lectin Toxicity in Mammals. PLoS One 10: e0129381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan‐Brown JL, Tandon P, Bird KE, Dickinson DJ, Tintori SC, Heppert JK, Meserve JH, Trogden KP, Orlowski SK, Conlon FL, Goldstein B. 2016. Identifying Regulators of Morphogenesis Common to Vertebrate Neural Tube Closure and Caenorhabditis elegans Gastrulation. Genetics 202: 123–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuse Y, Miwa J. 1983. A gene involved in action of tumor promoters is identified and mapped in Caenorhabditis elegans . Carcinogenesis 4: 783–786. [DOI] [PubMed] [Google Scholar]
- Tabuse Y, Nishiwaki K, Miwa J. 1989. Mutations in a protein kinase C homolog confer phorbol ester resistance on Caenorhabditis elegans . Science 243: 1713–1716. [DOI] [PubMed] [Google Scholar]
- Tice RR, Austin CP, Kavlock RJ, Bucher JR. 2013. Improving the human hazard characterization of chemicals: a Tox21 update. Environ. Health Perspect. 121: 756–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tralau T, Riebeling C, Pirow R, Oelgeschlager M, Seiler A, Liebsch M, Luch A. 2012. Wind of change challenges toxicological regulators. Environ. Health Perspect. 120: 1489–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux DL, Weissman IL, Kim SK. 1992. Prevention of programmed cell death in Caenorhabditis elegans by human bcl‐2. Science 258: 1955–1957. [DOI] [PubMed] [Google Scholar]
- Vidal‐Gadea A, Topper S, Young L, Crisp A, Kressin L, Elbel E, Maples T, Brauner M, Erbguth K, Axelrod A, Gottschalk A, Siegel D, Pierce‐Shimomura JT. 2011. Caenorhabditis elegans selects distinct crawling and swimming gaits via dopamine and serotonin. Proc. Natl. Acad. Sci. U. S. A. 108: 17504–17509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. 1986. The Structure of the Nervous System of the Nematode Caenorhabditis elegans (the Mind of a Worm). Philos. Trans. R. Soc. Lond. B Biol. Sci. 314: 1–340. [DOI] [PubMed] [Google Scholar]
- Williams PL, Dusenbery DB. 1988. Using the nematode Caenorhabditis elegans to predict mammalian acute lethality to metallic salts. Toxicol. Ind. Health 4: 469–478. [DOI] [PubMed] [Google Scholar]
- Wittenburg N, Eimer S, Lakowski B, Rohrig S, Rudolph C, Baumeister R. 2000. Presenilin is required for proper morphology and function of neurons in C. elegans . Nature 406: 306–309. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zou X, Ding Y, Wang H, Wu X, Liang B. 2013. Comparative genomics and functional study of lipid metabolic genes in Caenorhabditis elegans . BMC Genomics 14: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]


