SUMMARY
Chinese hamster ovary (CHO) cells dominate biotherapeutic protein production and are widely used in mammalian cell line engineering research. To elucidate metabolic bottlenecks in protein production and to guide cell engineering and bioprocess optimization, we reconstructed the metabolic pathways in CHO and associated them with >1,700 genes in the Cricetulus griseus genome. The genome-scale metabolic model based on this reconstruction, iCHO1766, and cell line-specific models for CHO-K1, CHO-S, and CHO-DG44 cells, provide the biochemical basis of growth and recombinant protein production. The models accurately predict growth phenotypes and known auxotrophies in CHO cells. With the models, we quantify the protein synthesis capacity of CHO cells and demonstrate that common bioprocess treatments, such as histone deacetylase inhibitors, inefficiently increase product yield. However, our simulations show the metabolic resources in CHO are >3 times more efficiently utilized for growth or recombinant protein synthesis following targeted efforts to engineer the CHO secretory pathway. This model will further accelerate CHO cell engineering and help optimize bioprocesses.
Graphical abstract
INTRODUCTION
Since their first commercial use in the late 1980s to produce tissue plasminogen activator, Chinese hamster ovary (CHO) cell lines have remained the platform of choice for producing proteins requiring complex post-translational modifications for therapeutic activity and regulatory approval (Kildegaard et al., 2013). Over the years, dramatic increases in product titer have been achieved in CHO cells as the result of bioprocess optimizations that increased cell culture density and longevity (Jayapal et al., 2007), resulting in CHO being the dominant host cell line for biotherapeutic production (Walsh, 2014). Despite these achievements, the molecular basis of protein production in CHO cells remains poorly characterized. Recent access to genome sequences (Brinkrolf et al., 2013; Lewis et al., 2013; Xu et al., 2011) and advances in systems biology (Gutierrez and Lewis, 2015) now enable the construction of a mechanistic basis for growth and protein production in CHO cells.
Three key cellular processes drive recombinant protein production: transgene expression, metabolism, and protein secretion. Metabolism is particularly important and inexorably linked to the others. For example, metabolic enzymes, including dihydrofolate reductase (Kaufman and Sharp, 1982) and glutamine synthetase (Bebbington et al., 1992), have served as selection systems for transfecting and amplifying transgenes in CHO cells. Additionally, metabolism provides the building blocks for the protein product and the secretory machinery needed to secrete it. Cell metabolism has been modulated extensively in the enhancement of CHO-based bioprocessing. Specifically, the balance of cellular metabolic demands has been targeted through media optimization to improve cell density, growth, and product yields (Castro et al., 1992). Efforts have also reduced the secretion of undesirable byproducts (e.g., lactate and NH3) to ameliorate the impact on cell growth (Lao and Toth, 1997), product quality (Chen and Harcum, 2006), and the cellular metabolic state (Yang and Butler, 2000). Additionally, metabolism influences product quality attributes (e.g., drug efficacy and compatibility with the human immune system) including glycosylation (Fan et al., 2015), oxidation, acetylation, and disulfide bridge formation (Lorendeau et al., 2015). Intuitive modifications of metabolic enzyme levels have improved protein production and quality (Altamirano et al., 2013); however, since each enzyme contributes to pathways, imbalances of components and interactions between pathways can yield unexpected results. Thus, a more complete understanding of CHO metabolism is vital to identify metabolic bottlenecks in CHO cell culture and to rationally guide complex cell engineering efforts.
To cope with the complexity of CHO metabolism, computational models have been applied to study CHO under various conditions (Carinhas et al., 2013; Nolan and Lee, 2011; Selvarasu et al., 2012; Sengupta et al., 2011; Templeton et al., 2013; Zamorano et al., 2010). Studies have focused primarily on central metabolism (Templeton et al., 2013) or used models extrapolated from mice (Martínez et al., 2015; Selvarasu et al., 2012; Smallbone, 2013). However, CHO-specific genome-scale metabolic models (GeMs) are now within reach, given the recent sequencing of the CHO-K1 and Chinese hamster genomes (Brinkrolf et al., 2013; Lewis et al., 2013; Xu et al., 2011). GeMs (Lewis et al., 2012) contain detailed information about all known biochemical reactions in a specific organism based on its genome and physiological information. Since metabolic pathways synthesize the components necessary for growth and survival, these models link the genetic basis of a cell to phenotypic capabilities, allowing more precise and complex metabolic engineering efforts (Curran et al., 2013; Gutierrez and Lewis, 2015).
Here we present a genome-scale metabolic network reconstruction for CHO cells that specifically links the genes encoded by the CHO-K1 and hamster genome to growth and recombinant protein production. This network was constructed and carefully curated by dozens of researchers in the community, and delineates the genetic basis of the metabolic pathways fueling all cell functions in CHO. We further built specific models for the CHO-K1, -S, and -DG44 cell lines and demonstrate that the models accurately predict important phenotypes such as growth rate, and unique metabolic features of CHO cells (e.g., auxotrophies). Using these models we analyzed the metabolic impact of common bioprocess treatments that aim to increase cell productivity. We found that while the treatments increased product titers by liberating cell resources, the cells failed to efficiently redirect metabolic precursors towards recombinant protein production after treatment. However, targeted engineering efforts showed more efficient redirection. Differences between treatments in redirection efficacy highlight potential avenues for further engineering efforts. Thus, the genome-scale network of CHO metabolism is an invaluable tool for data analysis, bioprocess optimization, and CHO cell engineering efforts.
RESULTS
A community genome-scale metabolic network reconstruction of Cricetulus griseus
Draft C. griseus metabolic models were reconstructed independently by multiple research groups. Subsequently, these models were merged using a systematic reconciliation process to capture all the careful curation that had gone into each model by groups representing diverse areas of expertise (Figure 1). As a result, we constructed and present a community consensus model for CHO metabolism and protein secretion.
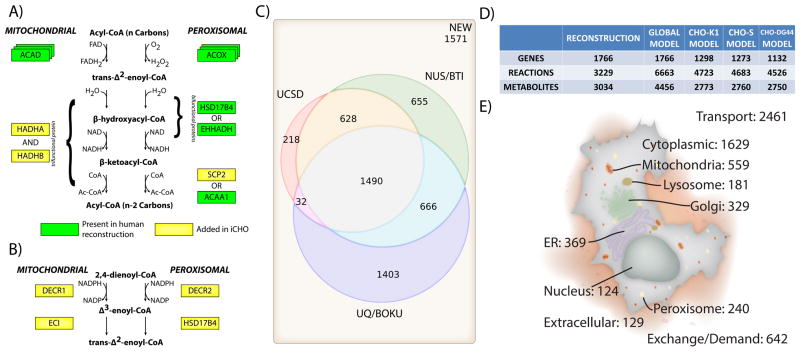
Figure 1. A multi-step process was used to reconcile a few existing unpublished models and to generate the final community reconstruction and models.
(A) The manually-curated human metabolic network reconstructions (Recon 1 (Duarte et al., 2007) and Recon 2 (Quek et al., 2014)) were used to define an initial set of reactions catalyzed in C. griseus and the genes and proteins involved in each reaction (GPRs). Specifically, Recon 1 and Recon 2 were combined and all enzyme-catalyzed reactions that differed between the two were manually curated and reconciled to obtain consistent GPRs. C. griseus homologs were obtained for each human gene to obtain a set of draft GPRs linked specifically to genes in the CHO-K1 genome annotation. (B) The draft CHO GPRs were then compared with the GPRs from three independently-reconstructed and unpublished CHO genome-scale models, thus leveraging the manual curation invested in each input model. By manually verifying all GPRs and adding additional CHO-specific reactions present in the input CHO genome-scale models, we obtained a more comprehensive community reconstruction for C. griseus. To enable computation with this network, orphan reactions from Recon 2 were added, and omics data were used to build a global and cell-line specific models.
The reconciliation and reconstruction effort first identified and curated the biochemical relationships linking genes, proteins, and reactions (GPRs) for human metabolism from established genome-scale reconstructions for Homo sapiens (Duarte et al., 2007; Quek et al., 2014; Swainston et al., 2016). CHO homologs to human genes were found using three different methods: reciprocal BLAST, the standalone InParanoid program v4.1 (Ostlund et al., 2010), and gene name matching. Based on this effort, putative CHO GPRs were constructed. These GPRs were then compared against the GPRs from each of the earlier independently-developed genome-scale metabolic network reconstructions. Manual curation of the reactions ensured accuracy of the GPRs, subcellular compartmentalization, and reaction stoichiometry. Additional CHO-specific reactions were included based on literature support and updates were made to mitochondrial and peroxisomal beta oxidation to reflect biochemical GPRs (Figure 2A–B).
Figure 2. Outcomes from the reconciliation process.
GPRs were updated and novel reactions were added in the reconstruction process, as shown for example in (A) mitochondrial and peroxisomal beta oxidation. Specifically, the trifunctional enzyme necessary for the 2nd, 3rd, and 4th steps of mitochondrial beta oxidation has been added to oxidation reactions occurring in the mitochondria while the GPRs of peroxisomal beta oxidation reactions have been updated to reflect additional catalytic activity of the SCP2 protein. (B) Additional enzymes necessary for catabolism of various unsaturated fatty acids have been added for both peroxisomal and mitochondrial beta oxidation. (C) The starting models had considerable differences in content, and following reconciliation, 1,571 new reactions were added to the model that had not been included in any of the starting CHO models. UCSD, NUS, and UQ/BOKU indicate the various groups contributing models to the initial reconciliation effort (see STAR Methods for additional details for each model). (D) The reconstruction refers solely to gene-associated content. To convert the reconstruction into a computable model, a global C. griseus model was built by including orphan reactions from Recon 2. These additions enable the activity of known enzymes in C. griseus and serve as hypotheses for enzyme discovery in CHO. After removing unexpressed genes, cell-line specific models were constructed for CHO-K1, -S, and -DG44 cells. (E) The reactions in the global C. griseus model account for pathways in multiple subcellular compartments.
The resulting community genome-scale metabolic network reconstruction, based on literature support from over 1,300 publications, includes 1,766 genes and 3,229 reactions associated with those genes. As is common when building genome-scale metabolic models, an additional 3,434 reactions without gene associations were added to convert the reconstruction to a computable model. These include boundary reactions defining metabolite uptake and secretion rates, as well as reactions where the specific gene responsible is unknown (e.g., transporters). In total, the global C. griseus metabolic model, iCHO1766, contains 6,663 metabolic reactions involving 2,341 unique metabolites (4,456 metabolites when accounting for subcellular compartmentalization). Only 22% (1,490) of the reactions were present in all three input models. Furthermore, almost 25% (1,571) of the reactions were newly added, in that these reactions were not present in any of the initial genome-scale CHO models. A more detailed breakdown of reaction sources is shown in Figure 2C.
To formulate a GeM that can simulate growth and recombinant protein production, we used known compositional data to define the relative amounts of each metabolite needed for synthesizing all cell components (i.e., biomass (Feist and Palsson, 2010)) and the recombinant proteins erythropoietin (EPO) and IgG. Due to differences between calculated gross cell composition in nonproducing CHO cell lines and measured values for IgG-producing hybridoma lines (i.e. measured protein fraction was more than 70% of cell dry weight in a producing cell, while calculated to be 55% in a non-producing cell), two separate biomass reactions were formulated (see STAR Methods). Cell-line specific models for CHO-K1, -S, and -DG44 were constructed using the GIMME algorithm (Becker and Palsson, 2008) and contained 4,718, 4,672, and 4,502 reactions, respectively (Figure 2D). The CHO-S and CHO-K1 models were built using RNA-Seq and proteomic data for both cell lines. In the proteomic data, 1,326 and 3,200 proteins were detected in CHO-S and CHO-K1, respectively. The CHO-DG44 model was built based on microarray data for 13,504 genes (see STAR Methods for data generation protocols, and Supplemental Data S4 for data used for model construction). Additional information on reconstruction and model content is available in Supplemental Data S1, including supporting literature associated with reactions, metabolites, and/or genes. The model accounts for 9 compartments (cytosol, mitochondria, nucleus, endoplasmic reticulum, Golgi complex, lysosome, peroxisome, mitochondrial intermembrane space, and extracellular space) with the subcellular localization of reactions summarized in Figure 2E. The global model and reconstruction are available to browse and download at the BiGG Models Database (King et al., 2015) (http://bigg.ucsd.edu), while the three cell line models and global model are additionally hosted at http://www.CHOgenome.org (Kremkow et al., 2015).
CHO cell-line specific models recapitulate experimental growth rates
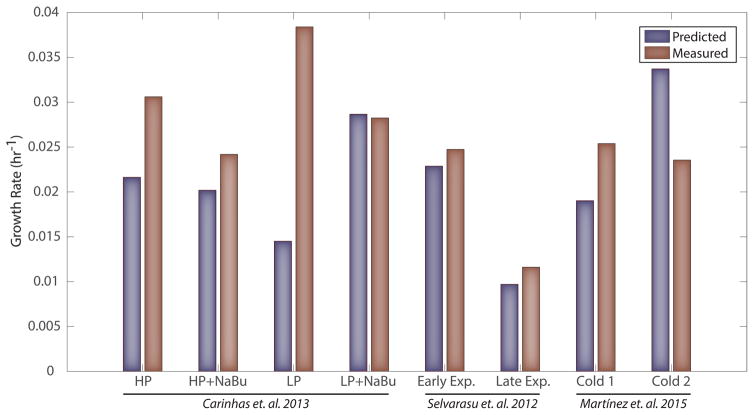
Genome-scale models can be used to predict phenotypes, including growth rates, when uptake and secretion rates are provided for major metabolites. Thus, to evaluate the models, we simulated growth rates for several different cell lines producing recombinant proteins. Data were acquired from literature (Ahn and Antoniewicz, 2013, 2011; Carinhas et al., 2013; Martínez et al., 2015; Selvarasu et al., 2012) and from new experiments presented here (see STAR Methods). Measured uptake and secretion rates for major nutrients and recombinant proteins were applied as constraints to the appropriate cell-line specific model, and flux balance analysis was used to predict growth rates by optimizing for flux through the appropriate biomass reaction (see Supplemental Data S3 for constraints). These computationally predicted growth rates were—on average—within 25% of the measured growth rates for cells grown in serum free conditions (Figure 3). Additional predictions were made for datasets exhibiting high uncertainty in calculated uptake and secretion rates, as well as cells grown in serum containing media (Supplemental Figures S3–5).
Figure 3. The CHO cell-line models can compute growth rates for various IgG-producing cell lines.
All cell lines are grown in serum-free media and are producing IgG. HP and LP refer to high and low producers respectively while NaBu indicates the presence of sodium butyrate addition to the media (Carinhas et al., 2013). Early exp./Late exp. refer to the early and late exponential phase, respectively, (Selvarasu et al., 2012). Two cell lines are from cultures exposed to a temperature shift (Martínez et al., 2015); however the data points used come from the time period prior to the temperature shift. Simulations for the Selvarasu study utilize the CHO-DG44 cell line model. Other simulations use the CHO-K1 cell line model.
Cell-line specific models recapitulate known amino acid auxotrophies
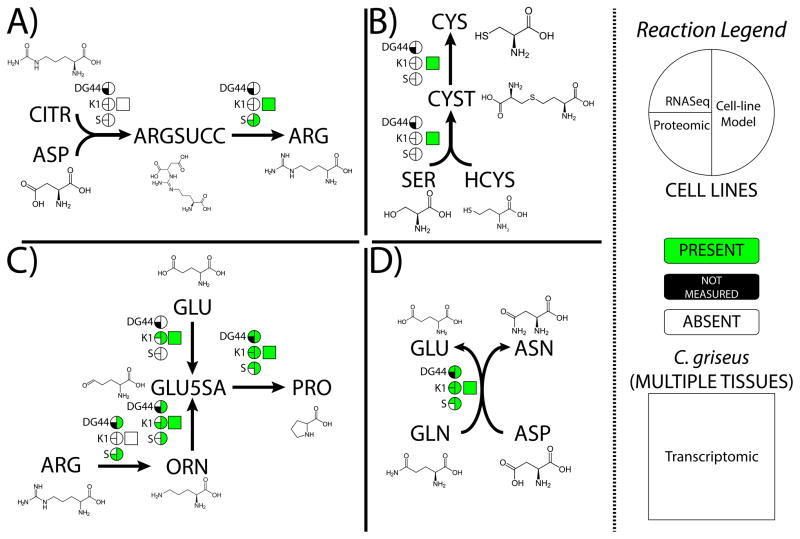
CHO cells exhibit several amino acid auxotrophies (Kao and Puck, 1967; Naylor et al., 1979; Valle et al., 1973) beyond the nine essential amino acids in human (His, Ile, Leu, Lys, Met, Phe, Thr, Trp, and Val) and arginine, which is additionally essential in rats for normal growth (Borman et al., 1946). The CHO-specific auxotrophies include cysteine, proline, and at least one report of an asparagine auxotrophy (Duarte et al., 2014). While cysteine, proline, asparagine, and arginine are essential based on experimental evidence, homologs for all genes required for their biosynthesis are in the C. griseus genome. Thus, we tested the cell line specific models for agreement with the reported amino acid auxotrophies and investigated the cell line specific transcriptomic and proteomic data (see Supplemental Data S4) to understand the underlying mechanisms.
All the cell line models reproduced the arginine and cysteine auxotrophies. Arginine biosynthesis was inhibited due to low or absent gene expression of one or both arginine biosynthetic genes (Figure 4A), and cysteine synthesis was disabled since cystathionine lyase and synthase were not expressed (Figure 4B). The asparagine synthase reaction, on the other hand, has strong transcriptomic and proteomic evidence for its presence in CHO cell lines (Figure 4D). Therefore, we experimentally tested if our CHO-S and CHO-K1 lines were auxotrophic for asparagine. Consistent with our models and the expression data, the cells could grow without asparagine (Supplemental Table S1). The reported asparagine auxotrophy was likely unique to the CHOK1SV cell line used in the earlier study (Duarte et al., 2014), and not a general characteristic of CHO cells.
Figure 4. The models provide insights into the molecular basis of CHO-specific amino acid auxotrophies.
(A) Arginine, (B) Cysteine, (C) Proline, (D) Asparagine. For each reaction, information in the circles indicates whether the enzyme catalyzing the reaction is seen in transcriptomic (upper left quarter-circle) and/or proteomic (lower left quarter-circle) data, as well as its presence or absence in the cell line specific models generated by GIMME (right semi-circle). The square shows whether the enzyme catalyzing the reaction is seen in transcriptomic data from a mix of C. griseus tissues. Data used is available in Supplemental Data S4. Metabolite abbreviations are as follows: CITR-citrulline, ASP-aspartate, ARGSUCC-argininosuccinate, ARG-arginine, SER-serine, HCYS-homocysteine, CYST-cystathionine, ORN-ornithine, GLU-glutamate, GLU5SA-glutamate 5 semialdehyde, PRO-proline, GLN-glutamine, ASN-asparagine.
We next analyzed the source of the proline auxotrophy in CHO cells (Figure 4C). Mammals can synthesize proline from arginine or glutamate; however, previous reports show that CHO-K1 cells do not incorporate glutamate into proline, and exhibit a drastically decreased (yet present) rate of ornithine transaminase activity (<10% of the activity in C. griseus lung cells) (Valle et al., 1973). Both biosynthetic pathways normally converge upon glutamate-5-semialdehyde, which is then converted to proline after spontaneous decomposition to 1-pyrroline-5-carboxylate. In the CHO-S and CHO-DG44 models, the pathway from glutamate is missing due to a lack of the necessary protein and/or mRNA expression. The GIMME algorithm incorrectly included the pathway from arginine to proline (via ornithine) in the CHO-S and CHO-DG44 models (the CHO-K1 model was missing the arginase reaction); however, careful inspection of the transcriptomic and proteomic data demonstrate that this pathway is missing expression of at least one enzyme in all of the cell lines. The gene responsible for the final step in proline biosynthesis, reduction of pyrroline-5-carboxylate, is expressed in all three cell lines, consistent with previous reports showing activity of the enzyme in CHO-K1 (Kao and Puck, 1967). Thus, the model-predicted proline synthesis capabilities show some inconsistencies with experimental observations due to limitations of the GIMME algorithm (i.e., it erroneously added the pathway linking arginine to proline); however, by overlaying transcriptomic and proteomic data, the reconstruction provided an explanation for the proline auxotrophy characteristic of CHO cells (Figure 4C), guiding further model curation and enabling more accurate model predictions.
An examination of C. griseus transcriptomic data from multiple tissues (Lewis et al., 2013) (see Supplemental Data S4) further elucidates which auxotrophies are characteristic of the organism and which are cell line-specific. The enzymes needed for cysteine and asparagine synthesis were expressed in the hamster tissues, consistent with their nonessentiality in rat and human (Figure 4B, D). In the hamster tissues, the arginine biosynthetic pathway has high expression in one of two necessary reactions. The second reaction, argininosuccinate synthase, exhibits low expression levels (Figure 4A). This is perhaps unsurprising since rats can synthesize arginine, but not in levels required to support normal growth (Borman et al., 1946). Finally, synthesis of proline via ornithine shows expression in the hamster tissues, consistent with reports of higher ornithine transaminase expression in C. griseus lung cells (Valle et al., 1973); the pathway via glutamate also shows expression in the hamster tissues (Figure 4C). These data suggest that the cysteine and proline auxotrophies are specific to the CHO cell lines and the arginine auxotrophy is—at least partially—shared between the hamster and the CHO cell lines. Thus, by analyzing transcriptomic and proteomic data in the context of the cellular pathways, we are able to elucidate the functional basis for known amino acid auxotrophies in CHO cells.
Cell engineering enhances the efficiency of resource utilization compared to common bioprocess treatments
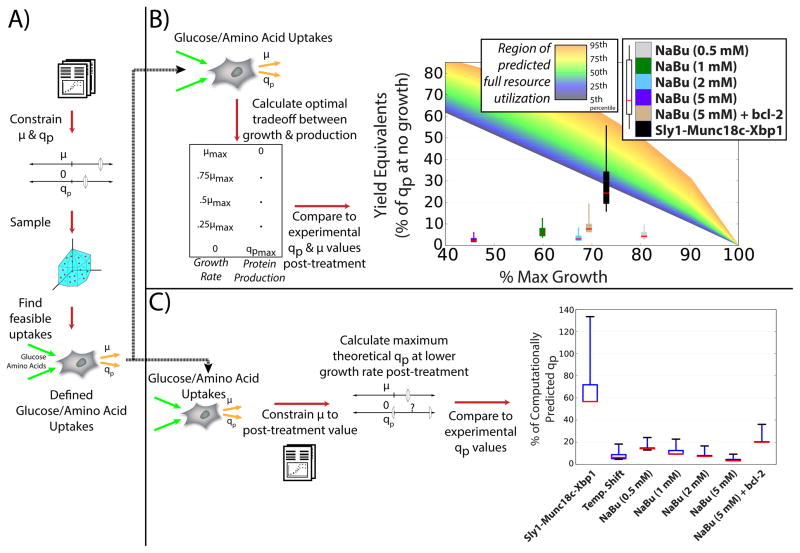
Substantial increases have been achieved in recombinant protein yields in CHO-based bioprocessing in part by balancing nutrient concentrations in media. Since nutrients can be used for growth and other purposes, chemical treatments and temperature shifts have been employed to increase product synthesis at the expense of growth. However, it is unclear how well these treatments redirect resources from growth to protein product. To test this, we used the cell-line models to simulate cell metabolism and recombinant protein production before and after culture perturbations. This approach allows us to analyze how efficiently cellular resources were redirected to protein synthesis following bioprocess treatments or targeted cell engineering efforts.
Specifically, we studied temperature shifts (Kim and Lee, 2009), sodium butyrate (NaBu) addition with (Kim and Lee, 2000) and without (De Leon Gatti et al., 2007; Mimura et al., 2001) BCL2 overexpression, and overexpression of secretory pathway genes (Peng and Fussenegger, 2009). In each case, estimates for uptake and secretion rates of major nutrients were predicted for the control culture from each study (Figure 5A) by sampling feasible uptake and secretion rates that supported the measured growth and protein secretion rates (see STAR Methods and Supplemental Data S5). These limits on nutrient availability were then applied as constraints to the models exhibiting the lower growth rates and higher protein secretion rates observed in the studies after bioprocess treatments or cell engineering. Using this approach, we calculated the maximum protein production rate as a function of growth rate for each treatment by optimizing for protein production after constraining growth to a fixed value. When compared to experimentally measured changes in growth and protein synthesis following each treatment, we quantified how well the cells made use of resources for both growth and production (Figure 5B). Furthermore, we assessed how efficiently the cells use newly available resources at lower growth rates for protein production (Figure 5C), as detailed in the following sections.
Figure 5. Resource utilization efficiency in CHO cell lines is greater after cell line engineering.
(A) Feasible uptake fluxes were generated for nutrient utilization efficiency analyses. Growth rates and specific productivities were obtained and used to constrain the model. By sampling the constrained model, a set of feasible metabolite uptake rates and secretion rates was calculated that support growth and production at the specified values. (B) The efficacy of resource utilization following common growth-inhibiting treatments in protein-producing CHO cell lines was quantified. Uptake and secretion rates from (A) were used to predict maximum growth (i.e., no protein production) and maximum protein yield (i.e., no growth), as well as yields at various fractions of maximum growth. These were used to predict a range of optimal protein production rates (i.e. making full use of resources) as growth rate decreases (indicated as a region of full resource utilization), thus showing the theoretical maximal protein secretion rates at decreased growth rates. The 5th, 25th, 50th, 75th, and 95th percentile of theoretical maximal protein production rates at each growth rate are indicated by the gradient in color from black to orange. After several cell treatments, the experimentally measured increased protein yield and decreased cell growth rate were compared to the predicted optimal protein secretion rates to assess how successfully each treatment utilized available resources (e.g., amino acids and sugars) for growth and protein production. Boxes span the 25th and 75th percentiles, whiskers represent the 5th and 95th percentiles, and a red line denotes the median value of overall resource utilization efficiency for each treatment, calculated as the ratio of experimentally measured protein production to the theoretical maximum protein yield (i.e., no growth). (C) The efficiency of diversion of resources toward protein production following common treatments was assessed. Uptake and secretion rates from (A) were used to compute the theoretical maximum specific productivity after cell line or process modifications yielding a range of theoretical optimal qp values were computed. Experimentally measured production rates were compared to the computational predictions to assess how effectively the cells are able to make use of resources gained from growing slower. Boxes span the 25th and 75th percentiles, whiskers represent the 5th and 95th percentiles, and a red line denoting the median value.
Bioprocess treatments, such as histone deacetylase inhibitors and temperature shifts can improve specific productivities and product titers. However, our analysis shows that overall resource utilization was inefficient post-treatment, independent of the decrease in growth rate (Figure 5B). To quantify the inefficiency, we compared the amount of protein that was produced experimentally to the amount of protein that could be produced if growth was halted (as calculated via model simulation). We simulated maximum protein production across the range of feasible growth rates, and defined a region of optimal resource utilization for protein production—an area where neither additional growth nor protein production is possible with the resources available (rainbow region in Figure 5B). For several treatments, we examined proximity to optimal resource utilization by comparing the measured growth and protein production rates following bioprocess or cell engineering treatments to the simulated maximum growth rate and protein production rate. Cells treated with NaBu produced protein at 2–6% of the predicted maximum rate, irrespective of the decrease in growth rate, and were far from the region of full resource utilization. BCL2 overexpression (relieving the apoptotic effect of NaBu) still only achieved 8% of the maximum protein production rate. Meanwhile, overexpressing the secretory pathway proteins XBP1, STXBP3 (MUNC18C), and SCFD1 (SLY1) allowed protein production at 24% of the maximum possible rate—3 fold higher than the best performance seen with cells overexpressing BCL2 and treated with NaBu. Thus, yields are low, even following bioprocess treatments. However, the increases seen with targeted engineering, as cells move closer to the region of optimal resource utilization, suggest that there is considerable potential for increasing cell-specific productivity to near optimal levels.
The inherent trade-off between growth and protein production suggests that resources for protein production may be liberated following growth-inhibiting bioprocess treatments. Thus, we simulated how efficiently cells, after suffering a decrease in growth rate from the different treatments, redirected newly available resources to protein production (Figure 5C). We found that while culture at lower temperatures (33°C vs. 37°C) often increases final titers, the specific productivity (qp) only increased to 6% of the theoretically possible level at the measured growth rate. Similarly, NaBu treated cells produced only 3–14% of their computationally predicted qp. Furthermore, simulating increases in NaBu concentration resulted in even less efficient redirection of resources to protein production. While these bioprocess treatments failed to efficiently redirect resources specifically to protein production, targeted cell engineering efforts improved the shift of resources. For example, overexpressing BCL2 during NaBu treatment resulted in a modest improvement in resource utilization (roughly 20% of the predicted qp). The greatest improvement in resource utilization was observed when the secretory pathway proteins were overexpressed, producing at close to 57% of computationally predicted maximum specific productivity.
DISCUSSION
Here we present a genome-scale metabolic network reconstruction for the Chinese hamster, C. griseus. This resource enables the enumeration of true genotype-phenotype links by connecting the functions of more than 1,700 genes to CHO cell phenotypes such as growth, metabolic pathway activity, and protein production. It presents many opportunities for analyzing bioprocess performance and guiding cell engineering. First, the reconstruction provides a platform for managing and interpreting CHO-relevant big data. Second, the models demonstrated potential for improving recombinant protein production by engineering cells to efficiently shift resources to protein production. The reconstruction will guide further strategies for cell line development as advances are made in modeling and relevant techniques. Third, the reconstruction will serve as a resource, in which up-to-date knowledge on the biochemistry of CHO cells can be maintained and made available to the entire CHO community.
To gain greater control of product yields and quality attributes of biopharmaceuticals, efforts to engineer CHO cells must consider the activities of cell pathways and associated genes. Thus, CHO-based bioprocessing is adopting data-rich technologies to quantify the cellular parts, using next-generation sequencing, high-throughput omics techniques, high-content imaging, and online bioprocess measurement techniques. To effectively analyze the data and deploy interventions, powerful statistical and modeling methods are needed (Clarke et al., 2014). This reconstruction serves as a platform for analyzing many types of molecular and phenotypic data using a variety of algorithms (Lewis et al., 2012). Furthermore, since this reconstruction provides a mechanistic link between the genotype and phenotype of CHO cells (via enumeration of enzymes underlying metabolic pathways), it allows for the effective integration of orthogonal data types such as metabolomics, transcriptomics, genetic variants, and growth rates (Cardoso et al., 2015; Hyduke et al., 2013). We demonstrated this with our cell line specific models for the CHO-K1, -S, and -DG44 lines. These models were built by integrating transcriptomic, proteomic, and metabolomic data with the global genome-scale metabolic model, validated by predicting growth rates and auxotrophies, and ultimately used to predict protein production capabilities. As the scope and quality of data expands, the CHO genome-scale network reconstruction will continue to enable the diagnosis of the molecular basis of different phenotypes, by serving as a platform to analyze the interplay of diverse data types.
The models provided quantitative evidence that targeted engineering of the secretory pathway allows for more efficient use of liberated resources as growth decreases. The models will enable further analyses of cell lines to help design mutants that provide desired protein quality attributes. The accuracy of such cell designs will further improve as five challenges are addressed. First, different CHO cell lines have accumulated hundreds of mutations in metabolic pathways (Lewis et al., 2013), which may contribute to differences seen in phenotypic traits across cell lines (Golabgir et al., 2016). However, more research is required to identify which mutations impact the activity of pathways. Second, growth and protein yield predictions require accurate measurements of metabolite concentrations, and advances in analytical chemistry will provide improved constraints on simulations with CHO metabolic models (see Supplemental Figure S4). Third, gaps remain in our understanding of mammalian metabolism, but algorithmic developments (Thiele et al., 2014) and biochemical assays will continue to refine the CHO metabolic network. Fourth, our biomass objective functions were defined based on experimental measurements in CHO and hybridoma cells and provided accurate predictions of cell growth; however, biomass composition is not static, which may impact quantitative predictions of growth and protein production. Thus, there is a need for comprehensive measurements of the composition of CHO cells (e.g. protein, DNA, RNA, lipids, etc.) under different conditions to formulate more accurate biomass objective functions. Finally, metabolism is only part of the system that controls the quantity and quality of recombinant proteins. Efforts to model other processes such as glycosylation (Sha et al., 2016; Spahn et al., 2016) and the secretory pathway (Feizi et al., 2013) can be integrated with metabolic networks for a more comprehensive view of protein production; further addition of signaling and regulatory networks could reveal mechanisms behind inefficiencies in protein production. These five advances will further improve the predictive power of the genome-scale CHO models.
Lastly, the development of this resource involved the concerted effort of many groups in the community with interest in genome-scale metabolic modeling for mammalian bioprocesses and CHO cells. Together, these combined efforts enabled the careful curation of this community resource. The models are available at http://www.CHOgenome.org (Kremkow et al., 2015). Additionally, the global model can be browsed and downloaded at the BiGG Models database (King et al., 2015) (accessible at http://bigg.ucsd.edu), where literature information and experimental evidence is provided for each gene, reaction, and metabolite in the model. Updates will be made with continued research into CHO cells, including improvements in the C. griseus genome sequence and annotation, and experimental tests of model predictions (Chowdhury et al., 2015). As such studies accumulate, the network reconstruction, as a community resource, will be maintained and improved over time, thus enabling researchers throughout the CHO cell community to conduct many studies to enhance the value of the primary host for biotherapeutic production.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for datasets and/or protocols may be directed to, and will be fulfilled by the corresponding author Nathan Lewis (nlewisres@ucsd.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cultures for Transcriptomic, Proteomic, and/or Metabolomic Data
CHO-S (Life Technologies # A11557-01) cells were cultured in CD CHO medium (Life Technologies # 10743-029) supplemented with 8 mM L-glutamine (Life Technologies # 25030-024), Anti-clumping agent 1:500 (Life Technologies # 0010057 AE), and Pen-Strep 1:100 (Life Technologies # 15140-122) in 2 L Corning shake flasks (Sigma # 431255) with 400 mL medium. All cultures were maintained in an incubator kept at 37°C, 5% CO2, 70% humidity and 25 mm throw, shaking at 120 rpm.
Adherent CHO-K1 (ATCC CCL-61) cells were grown in F-12K medium, supplemented with 10% FBS, 1% non-essential amino acids, and 2 mM L-glutamine (Gibco) incubated at 37°C and 5% CO2. Spent medium was sampled from the culture at regular intervals and metabolite concentrations were measured. Cell-free medium was also sampled at regular intervals to control for metabolite degradation. Samples were taken from the CHO-K1 and cell-free control every 2 hours between 24–36 hours, and every 4 hours subsequently (see Supplemental Data S4).
Cultures for Determining Asparagine Essentiality
CHO-S (Life Technologies) and CHO-K1 (ATCC CCL-61 adapted to grow in suspension, serum-free conditions) cells were grown in CD-CHO media with and without asparagine at a seeding concentration of 0.4–0.5 × 106 cells/mL. Asparagine was either added via MEM NEAA solution (Control 1) or as an individual amino acid supplementing MEM NEAA solution lacking asparagine (Control 2). Both cell lines were able to grow successfully in media lacking asparagine, as measured by viability and VCD 72 hours after seeding (Supplemental Table S1). Cell growth and viability were monitored using the NucleoCounter NC-200 Cell Counter (ChemoMetec, Allerod, Denmark) based on two fluorescent dyes, acridine orange and DAPI for the total and dead cell populations, respectively.
METHOD DETAILS
Input genome-scale model construction
In the preliminary stages of this effort, three research groups contributing to this study independently developed unpublished genome-scale models of CHO cell metabolism. These were supplemented by an additional independent reconstruction of amino acid metabolism.
University of California San Diego metabolic model
The initial draft of the UCSD genome-scale CHO metabolic network was reconstructed using GIMMEp (Bordbar et al., 2012). The algorithm requires an initial metabolic network, a core set of reactions that must be operational, a set of required metabolic functions, and an optional set of additional reactions that may or may not be included in the final model.
First, we used the global human metabolic network (Recon 1 (Duarte et al., 2007)) as the initial network for the algorithm to build the CHO metabolic network. Recon 1 served as a template for which metabolic reactions were initially chosen for the CHO metabolic network.
Second, a core set of reactions was defined based on homology between the CHO and human genomes. A bidirectional BLASTP was done by comparing human RefSeq proteins (downloaded on 12/13/2011) against the CHO genome sequence (Xu et al., 2011). Metabolic proteins from Recon 1 with a high identity (>70%) and high overlap (>70%) with a CHO protein in the bidirectional BLAST were marked as the core set of homologous proteins. A small set of proteins met the inclusion criteria in only one direction and was subsequently manually curated for inclusion. The gene-protein-reaction rules for Recon 1 reactions were updated with CHO annotations for homologous proteins, and removed if no homologous protein was present. The set of optional reactions provided as an input to GIMMEp was not included in the final model.
Third, a set of metabolic functions was defined including: 1) the ability for the metabolic model to produce biomass, 2) the ability to produce a glycosylated protein (EPO), and 3) the ability to uptake and secrete known metabolites characteristic of CHO-K1 cells. The necessary reactions to accomplish these three tasks were added to the initial model before the model building algorithm was run and were included in the core set of reactions. A biomass objective function (Feist and Palsson, 2010) was constructed accounting for RNA, DNA, protein content, free amino acids, glycogen, free fatty acids, lipids, phospholipids, triglycerides, and associated ATP maintenance costs using primary literature. The ratio of nucleotide content in DNA was set based on the GC content of the genome (Xu et al., 2011). Ratios of nucleotide content in RNA and amino acid content in proteins were estimated based on RNA-Seq data (Xu et al., 2011). In addition, metabolic reactions were reconstructed that translated the EPO protein and added the necessary N- and O-glycans, as well as degraded the protein. Also, the exchange reactions of metabolites known to be taken up or secreted by CHO cells were included in the core set of reactions.
The resultant draft model contained all CHO metabolic proteins that had high homology with the human metabolic proteins, while minimizing the number of metabolic reactions with low homology that were added to enable the model to accomplish a set of key metabolic functions including growth, protein production, and uptake/secretion of documented metabolites.
This was followed by manual curation of the model to ensure that gene associations, reaction localization, and other reaction contents were accurate.
National University of Singapore/Bioprocessing Technology Institute metabolic model
The H. sapiens reconstruction, Recon 1 (Duarte et al., 2007), served as the starting point of the NUS CHO-K1 genome scale metabolic model reconstruction. This was motivated by the high-quality manual curation performed on Recon 1, and the large number of genes (81%) that were shared between the human and CHO-K1 genomes (Xu et al., 2011). To create a draft list of reactions for the new model, a protein BLAST search of human genes was conducted against CHO-K1 genes. This allowed the mapping of metabolic genes from human to CHO-K1. Of the 1496 metabolic genes in the Recon 1 model, 1441 (96%) corresponding matches were found in the CHO-K1 genome. Based on the list of 55 missing genes, 39 relevant reactions were removed from the Recon 1 model. The number of reactions removed is less than the number of genes due to reactions associated with isozymes. The 1514 non-gene associated reactions in the original model were retained, since they were necessary to produce a functioning model.
The removal of missing genes introduced gaps within the model that prevented biomass production and removed metabolic functions. By examining the list of removed reactions, SQLEr (squalene epoxidase) and C3STKR2r (C-3 sterol keto reductase zymosterol) were found to be necessary to ensure biomass formation. We also found that the removal of the three reactions catalyzed by 2-oxoisovalerate dehydrogenase prevented the catabolism of branched amino acids (valine, leucine, and isoleucine). While the removal of TPI1 (triosephosphate isomerase 1) strictly did not impair biomass formation, there are strong physiological arguments for its existence, and there is evidence that the enzyme exists in the ancestral CHO cell line (Daar et al., 1986). Based on experimental evidence (Selvarasu et al., 2012), the GGLUCT (gamma-glutamylcyclotransferase) reaction was not also removed.
As the Recon 1 model was constructed to represent the global human metabolic model, the model includes tissue specific reactions that are not relevant to CHO-K1. Consequently, we removed these reactions from the reaction list. The reactions removed include 26 demand reactions for micronutrients such as vitamins, and 36 duplicate reactions for liver and uterus tissue (e.g., in dolichol biosynthesis). The R Group synthesis reactions were also removed as the lipid requirement for CHO-K1 biomass formation was subsequently reconstructed based on literature sources. Finally, 80 reactions and relevant metabolites with the “hs” suffix were relabeled to the “cho” suffix.
To further refine the model, we utilized the KEGG PATHWAY (Kanehisa and Goto, 2000) and BRENDA Enzyme Database (Chang et al., 2009) databases to identify CHO metabolic reactions based on physiological evidence. Based on curated databases of BRENDA and KEGG, 85 additional reactions were added to subsystems such as arginine and proline metabolism, IMP biosynthesis, and inositol phosphate metabolism. These include two reactions previously removed based on the gene mapping using BLAST.
The model reconstruction was originally conceived for biotechnological applications. Therefore, relevant biomass formation reactions were constructed and added. As the R Group synthesis reactions were removed, 15 reactions were added for the production of fatty acid and cholesterol components of the cellular biomass. Reversible transport via proton symport reactions were also added to facilitate the independent exchange of amino acids. Finally, based on recent evidence obtained by LC-MS experiments (Selvarasu et al., 2012), 13 reactions were added to account for the production and secretion of the detected amino acid derivatives. The reconstructed CHO-K1 model consists of 3718 reactions and 2774 compartment specific metabolites (1523 non compartment specific).
The University of Queensland/University of Natural Resources and Life Sciences, Vienna metabolic model
An updated version (Quek et al., 2014) of the H. sapiens reconstruction Recon 2, was used as a starting point for generation of a manually curated CHO specific model by the UQ/BOKU groups. Curation focused on the identification of inconsistencies in the naming conventions, annotations, removal of duplicated reactions and metabolites, as well as correction of the mass and energy balance.
Homologous genes between CHO and H. sapiens were identified from RefSeq Release 66 using the standalone InParanoid program v4.1 (Ostlund et al., 2010). The identified homologies were then used to convert the human specific Recon 2 metabolic network to a CHO specific network. This provided an initial basis for a CHO-specific metabolic network based on Recon 2, which later was considered in the curation of the community-level network.
Technical University of Denmark amino acid subnetwork
A metabolic network reconstruction of amino acid metabolism in CHO cells was generated at DTU using mouse genomic and biochemical pathway information from the KEGG database as a starting point. To identify orthologous metabolic genes in CHO, a protein BLAST search of amino acid metabolic genes from mouse was manually conducted against the CHO-K1 genome (Xu et al., 2011), hosted at http://www.CHOgenome.org. The resulting list of CHO genes was manually curated for inclusion based on information from literature.
Reconciliation process
To merge these into one final community reconstruction, we developed a workflow to reconcile similarities and differences among the existing models (Figure 1).
The final metabolic network reconstruction was initially based on knowledge from human metabolism, as detailed in Recon 1 (Duarte et al., 2007) and an updated version of Recon 2 (Quek et al., 2014) (Figure 1A). Additional curation of Recon 2 was conducted to refine the reconstruction (Swainston et al., 2016). These two reconstructions were first compared to determine a baseline set of gene-protein-reaction (GPR) relationships for all reactions in the models. If GPRs showed discrepancies between the two reconstructions, manual curation was carried out to determine a consensus GPR. This combined human reconstruction was subsequently used to identify C. griseus homologs. Specifically, three different homology methods were used to determine CHO-human protein homologs. First, two-way BLAST was conducted between the CHO proteome and the human proteome. Reciprocal top matches for both C. griseus and human proteins were identified and retained. Second, the standalone InParanoid program v4.1 (Ostlund et al., 2010) was used to find orthologs between human and C. griseus. Third, a search was conducted to identify genes with identical gene names.
For each human gene, the union of these three methods was used to map from human GPRs to putative CHO GPRs, resulting in 3701 reactions with gene associations. In this set of reactions, 733 had GPRs that were identical between all 3 preliminary reconstructions and the putative CHO GPR. For the remaining reactions (2968), careful manual curation was carried out to determine the most accurate GPR for each reaction in the final model (Figure 1B). In each case, primary literature was searched to find CHO-specific information about the reaction (e.g. substrate specificity, subcellular localization, protein complex composition, gene association, etc.). When such information was unavailable, information from other mammals was used. Through the curation process, additional CHO-specific reactions were identified and added.
The curated gene-associated reactions were then combined with all the non-gene associated reactions from Recon 1 and the updated Recon 2 to give a base CHO metabolic network. Mass imbalances were corrected and the network was tested for biomass functionality. Further refinements included the removal of opposing irreversible reactions (in favor of a single reversible form), the replacement of lumped reactions, and the addition of pathways for synthesizing IgG and EPO.
As a final comparison, the model content was compared against another unpublished and independently reconstructed CHO metabolic reconstruction, based on Recon 1 (Duarte et al., 2007) (see below for details on this model’s construction) and any differences manually curated.
University of Chile metabolic model
For the generation of a preliminary CHO genome-scale metabolic model we used Pantograph, a tool to reconstruct genome-scale models for eukaryotic organisms (Loira et al., 2015). This tool uses a template metabolic model and annotated genomes for both template and objective organisms, automatically rewriting the template’s GPRs in terms of the genes of the target organism, inheriting knowledge from the template model and producing a draft metabolic model well suited for manual curation.
Three models were considered as templates for network reconstruction. A hybridoma metabolic reconstruction based on Mus musculus genomic and biochemical information (Selvarasu et al., 2010) was modified and upgraded to include new genomic information available in databases such as KEGG and NCBI-gene by manual curation and by using a script that connects to the KEGG database to download new GPR candidates to be added to the model.
A metabolic reconstruction for Mus musculus based on Recon 1 (Duarte et al., 2007), iMM1415, was also used as template for the generation of a model for CHO cell metabolism. This model includes 1,415 genes, 2,212 gene-associated reactions and 1,514 non-gene-associated reactions. Finally, the human reconstruction, Recon 1, was also used as a template for the generation of a CHO genome-scale model (Duarte et al., 2007).
Orthologous genes often exhibit similar biological activities. Thus, they can often reliably be used to build novel reconstructions from a template organism. The search for orthologs was performed using the standalone InParanoid program v4.1 (Ostlund et al., 2010), which finds clusters of orthologous genes based on similarity scores calculated by NCBI-Blast between proteomes of the analyzed species. The protein sequences for CHO were retrieved from http://www.CHOgenome.org and Ensembl, and these sequences were used to find orthologs between CHO and Mus musculus and CHO and Homo sapiens.
The template model and ortholog information for Mus musculus, Homo sapiens, and CHO were used to generate a preliminary genome-scale model for CHO cells using Pantograph. Critical components for biomass synthesis were identified by analyzing metabolic pathways that lead to their synthesis using the COBRA toolbox (Schellenberger et al., 2011).
GapFind (Satish Kumar et al., 2007) was used to find dead-end metabolites, which were subsequently studied using information from databases such as CHOgenome.org, KEGG, and Virtual Metabolic Human in order to fill the gaps present in the initial reconstruction. Model validation was performed using Pantograph (Loira et al., 2015) which tests the ability of the obtained genome-scale models to replicate experimental data, such as the effect of known gene deletions and use of alternate carbon sources for CHO cells in culture.
While the University of Chile model was not used as a base model in the reconciliation, it was compared to the global model and all discrepancies manually curated. All validated reactions suggested by this secondary curation process were added to the final global model.
Biomass reaction
Two biomass reactions were built here: one for a recombinant-protein producing cell line (biomass_cho_producing) and one for a nonproducing cell line (biomass_cho). The overall cell composition (e.g., protein, DNA, RNA, lipid fraction) of a protein producing cell line was averaged from previously reported values for mouse hybridoma cell lines. The nonproducing cell line overall cell composition was calculated based on literature values for different cellular components in CHO. Both biomass reactions used experimentally determined amino acid compositions of the protein fraction, obtained by averaging the composition of 5 cell lines (Selvarasu et al., 2012). The ratios of different nucleotides for RNA and DNA composition were determined from genome and transcript sequences. Phospholipid composition was taken from previously reported values for CHO-K1. A detailed formulation of the biomass equations (including all associated references) is available in Supplemental Data S2.
Choice of objective function
An objective function should encapsulate what the cell is “trying” to do. What that entails remains a matter of great scientific interest. In bacterial systems, the use of a biomass objective function (Feist and Palsson, 2010) has come to dominate, as experimental work shows cells evolving toward optimal states as predicted by maximization of biomass production (Edwards et al., 2001; Ibarra et al., 2002; Lewis et al., 2010).
Mammalian cells, as a whole, are more difficult to ascribe a one-size-fits-all objective function for, since many (e.g., terminally differentiated cells) do not rapidly proliferate; their ‘objective’ may be another biological activity such as antibody production (plasma cells), maintaining structural integrity (red blood cells), or generation of energy and signaling molecules (neurons). As CHO cells are proliferative, we selected a biomass objective function for simulation of cell growth. In a study comparing experimental fluxomic data to model flux predictions (Schuetz et al., 2007) in core metabolism in Escherichia coli, alternative objective functions were evaluated for their accuracy in recapitulating experimental measurements. This study found that maximization of ATP or biomass both led to the highest consistency between FBA predictions of metabolic flux and experimental flux measurements. Subsequent research provided further support for biomass optimization (Lewis et al., 2010), but found that maximization of ATP yield showed less consistency in genome-scale models. The authors noted that “in a genome-scale model, the maximization of ATP selects against the usage of biosynthetic pathways, since the end products are not specified in the objective function”. A similar conclusion can be drawn for minimizing redox load as an objective function, which also does not require the use of many biosynthetic pathways, and so such an objective function would not capture the activities of all pathways.
It is possible that in mammalian cell culture, that some combination of objective functions is the most biologically accurate; however, as our line of investigation largely focuses on predictions of growth rate, a gross phenotypic characteristic which definitionally has an upper limit set by cellular composition (e.g., biomass generation), a simplification to a biomass objective function for growth simulations is a reasonable approximation. Furthermore, the ability to recapitulate experimentally-measured growth rates provides further support for this assumption and that the measurements used the biomass objective functions are within a reasonable range.
Construction of cell-line specific models
We used GIMME (Becker and Palsson, 2008) to generate cell-line specific models for CHO-S, CHO-K1, and CHO-DG44. Transcriptomic and proteomic data were generated for CHO-S, while existing data were used for CHO-K1 and CHO-DG44 (see Method Details for information on data used or generated). Genes were called as present if their FPKM was greater than 1 or they were found in the proteomic analysis. For microarray data, genes were called as present if their normalized log2-transformed value was greater than 10. Blocked reactions were removed from the global model and the algorithm was used while optimizing for growth, IgG production, and erythropoietin production. The union of reactions present in these models served as the base cell-line model in each case.
Since algorithmic generation of the cell-line specific models ensures only functionality for user-defined objectives (here we used growth, IgG production, and erythropoietin production), additional manual curation was done to incorporate other biological functionalities important for simulation. We added eight reactions to all models: GLYCt and SUCCt4_3 were added to permit secretion of glycerol and succinate, respectively. HISD, URCN, IZPN, GluForTx, FTCD and DM_trp_L[c] were added to account for histidine and tryptophan uptake rates being significantly higher than needed for growth and protein production. Histidine catabolic reactions were added to permit conversion to glutamate, since evidence for histamine production (from histidine) in CHO could not be found. The tryptophan demand reaction was added as tryptophan is converted to many metabolites with diverse functions (e.g., hormones), at least some of which have been detected in CHO cell culture (Hiller and Mulukutla, 2015); however, we were uncertain whether the production of specific tryptophan-derived metabolites is cell line specific or a characteristic of the CHO cell in general, thus the inclusion of a generic demand reaction rather than specific biosynthetic pathways. Three additional reactions were added to the DG44 model, the mitochondrial and endoplasmic reticulum localized phosphatidylethanolamine N-methyltransferase and methionine adenosyltransferase (PETOHMm_cho, PETOHMr_cho, and METAT, respectively) to allow growth in the absence of measured choline uptake. The ORNTArm, GLU5Km, and G5SDym reactions were constrained to be off (lower bound=upper bound=0) to reflect the fact that—even if present—the enzymes do not appear to carry flux, leading to the experimentally observed proline auxotrophy in CHO cell lines (Valle et al., 1973).
Omic data generation
Transcriptomics and Proteomics
CHO-S
Starting at 72 hours into culture and every 12 hours after, cells were collected for proteomic (5 time points) and transcriptomic analysis (10 time points). 5 × 106 cells were harvested for transcriptomic analysis via RNA-Seq. Cells were centrifuged and the pellet resuspended in 2%/98% DTT/RLT buffer and stored at −80°C. RNA was extracted with Qiagen’s RNeasy mini kit (Qiagen #74104) according to manufacturer’s protocol with on-column DNase digestion. RNA libraries for sequencing were prepared using TruSeq Stranded mRNA Sample prep kit with 96 dual indexes (Illumina, CA, USA) according to the manufacturer’s instructions with the following changes. The protocols were automated using an Agilent NGS workstation (Agilent, CA, USA) using purification steps as previously described (Borgström et al., 2011; Lundin et al., 2010). Libraries were clustered using cBot and sequenced on HiSeq2500 (HiSeq Control Software 2.2.38/RTA 1.18.61) with a 2×101 setup in RapidHighOutput mode. The raw reads are available at GEO (Accession GSE77800). Bcl to Fastq conversion was performed using bcl2Fastq v1.8.3 from the CASAVA software suite. The quality scale is Sanger / phred33 / Illumina 1.8+.
FastQC (Andrews, 2010) was used to assess read quality. Trimmomatic v0.32 (Bolger et al., 2014) was used to trim reads with adapters or low quality scores. STAR2.4.0a (Dobin et al., 2013) was used to align trimmed reads to the CHO-K1 genome (Xu et al., 2011). Mapping results were stored using SAMtools 1.0(Li et al., 2009). Cufflinks 2.2.1 (Trapnell et al., 2010) was used to assemble mapped reads and quantify expression levels. FPKM levels for all time points are available in Supplemental Data S4.
For proteomic analysis, 1 × 107 cells were harvested and centrifuged. Pelleted CHO cells were lysed in 100 μl urea (8M, 75 mM NaCl, 50 mM Tris-HCl pH 8.2). To assist the rupture of the cells, two 3 mm Zirconium oxide beads were added and samples were placed in a mixer mill (Glen Mills Inc, NJ, USA). Two rounds of mixing were applied, with the first 2 min at 25 Hz in the mixer mill followed by 30 min at 4 °C and additional 2 min at 25 Hz in the mixer mill. Between the two rounds, an additional 100 μl of urea were added. Samples were then centrifuged for 15 minutes at 15000 g, after which the supernatant was collected. Then 400 μl 25 mM ammonium bicarbonate was added and the volume reduced to 100 μl using a 3 kDa cutoff filter. Five μl DTT were added to samples containing 100 ug of protein and then kept at 37 °C for 45 minutes, after which 100 μl of iodoacetamide was added and samples were kept in the dark for 45 minutes. Tryptic digestion of the proteins was done for 8 hours at 37 °C. Digestion was terminated with the addition of 10 μl 10% TFA and finally samples were staged tipped using C18 filters (Empore, 3M Company, USA) following an established protocol (Rappsilber et al., 2007).
Each sample was trapped on a precolumn (Symmetry C18 5μm, 180μm × 20mm, Waters, Manchester UK) and washed for 4 min after which it was loaded on the analytical column. The analytical setup consists of a nanoACQUITY™ System (Waters, Manchester UK) equipped with a nanoACQUITY™ BEH130 C18 1.7 μm, 75 μm × 250 mm analytical reversed-phase column (Waters, Manchester UK). The reverse phase elution profile included mobile phase A (0.1 formic acid in water) and mobile phase B (0.1% formic acid in acetonitrile), during which B was increased from 5–40% over 90 minutes with a flow rate of 250 nL min−1 and a column temperature of 35°C. To minimize carry over, the method included a 30 minutes wash phase to clear the column.
Data acquisition was accomplished on a Synapt G2 (Waters, Manchester UK) Q-TOF instrument using ESI with a NanoLock-spray source. The mass spectrometer was operated in positive and resolution mode with continuum spectra being acquired. Data were continuously calibrated using leucine encephalin as lock mass. Data were acquired using MSE, during which the mass spectrometer alternated between low- and high-energy mode using a scan time of 0.8 s for each mode over a 50–2000 Da interval. In the low-energy MS mode, data were collected at constant collision energy of 4 eV. In the elevated-energy MS mode, the collision energy was increased from 15 to 40 eV.
Raw data files for protein identification were obtained by using the ProteinLynxGlobalServer software v2.5.3 (Water corporation) using the in-build MSE search function against the Chinese hamster UniProt proteome database (UP000001075). The search parameters were trypsin as enzyme, carboxamidomethyl on cysteine as a fixed modification and oxidation of methionine as partial modification, while allowing one missed cleavage.
CHO-K1
Additional proteomic and RNA-Seq data were obtained from existing studies (Baycin-Hizal et al., 2012; Xu et al., 2011). Briefly, for RNA-Seq, CHO-K1 cells were grown in F-12K medium (Invitrogen) supplemented with 10% FBS at 37°C with RNA extraction during exponential phase. Sequencing was carried out using Illumina GA2 technology with paired-end reads. Quantification of expression levels was carried out in an identical manner as for CHO-S. For proteomics, CHO-K1 cells were grown in F-12K medium, supplemented with 10% FBS, 1% non-essential amino acids, and 2 mM L-glutamine (Gibco) and gathered at 70–80% confluence for analysis.
CHO-DG44
Microarray data for an IgG-producing CHO-DG44 derivative were obtained from literature (Courtes et al., 2013). Briefly, the CHO-DG44 cells expressing IgG, known as CHO M250-9, were grown in a proprietary protein free and chemically defined medium. The total RNAs were extracted using the Qiagen RNeasy Plant Mini Prep kit during the exponential phase of cell culture. Subsequently, the gene expression was profiled with a NimbleGen CHO microarray chip containing 135,883 probes corresponding to a total of 13,514 annotated CHO genes. Scanned microarray signals were then analyzed by the NimbleScan V2.6 (Nimblegen, U.S.A.) and quantile normalized using the R package AffyPLM (Bolstad et al., 2005).
Metabolomics
CHO-K1
For each media sample, polar extracellular metabolites were analyzed by ultra performance liquid chromatography (UPLC) (Acquity, Waters, Manchester, UK) coupled in line with a quadrupole-time-of flight hybrid mass spectrometer (Synapt G2, Waters, Manchester, UK) as previously reported (Paglia et al., 2012).
For the analysis of targeted metabolites, data were processed using TargetLynx (Waters) while for untargeted analysis MarkerLynx (Waters) was used to integrate and align MS data points and convert them into exact mass retention time pairs. Extracted ion chromatograms were obtained by using a 0.02 mDa window centered on the expected m/z for each targeted compound. Quantitation was performed by external calibration with reference standards (Paglia et al., 2012).
The identity of each metabolite was established by verifying retention time, accurate mass measurements and collision induced dissociation information against our in-house database and/or online databases, including HMDB and METLIN (Smith et al., 2005; Wishart et al., 2013).
All materials used in the UPLC-MS experiments were purchased from Sigma-Aldrich (Germany) and were of analytical grade or higher purity.
Model simulation
When optimizing for growth, solutions were obtained by maximizing flux through the biomass_cho or biomass_cho_producing reaction, depending on if the simulation was of a non-producing or producing cell line, respectively. For predicting maximum protein production, solutions were obtained by maximizing flux through the DM_igg[g] or DM_epo[g] reaction, for production of IgG or EPO, respectively.
Growth-rate prediction discrepancies
Serum containing media formulations
When applied as constraints, the uptakes from two CHO-K1 MFA studies were unable to recapitulate the experimentally observed growth rate for the CHO-K1 cells grown in serum. The cause of this discrepancy is due to the low uptake of proline in one study (Ahn and Antoniewicz, 2011) and the production of proline by the cells in another study (Ahn and Antoniewicz, 2013), despite reports of the proline auxotrophy being a hallmark of the cell line. It is possible that the presence of serum (and associated protease activity or peptide uptake and catabolism) may ‘mask’ a higher uptake rate of proline required to sustain the higher observed growth. However, we also investigated whether spontaneous proline prototroph revertants (Kao and Puck, 1967) could also explain the difference in calculated vs. observed growth rates. To do so, we first added an extracellular arginase reaction to the models (Hölttä and Pohjanpelto, 1982), and then simulated growth with individual or both proline biosynthetic pathways active, compared to simulated proline auxotrophy. Inclusion of active proline biosynthetic pathways in the model greatly improved the consistency of model predictions with measured growth (Supplemental Figure S3), indicating that increased proline availability, either via biosynthetic routes or via unmeasured uptake from serum components, can explain the discrepancies between model predicted and experimental growth rates.
Metabolomic measurement uncertainty impacts growth rate prediction accuracy
We also examined how variability in uptake and secretion rate calculations impacts the accuracy of model predictions. To do this test, we focused on three data sets in which computed uptake and secretion rates for some metabolites demonstrated large standard deviations. These included three lower quality cultures generated by the authors of this study. In these experiments, the calculated range (between 5th and 95th percentile confidence intervals) for more than half of the metabolite uptake or secretion rates was larger than the predicted best fit value (Supplemental Figure S4). For completeness, we show this metric compared to two cultures from a recent temperature shift study (Martínez et al., 2015) (labeled “Cold 1” and “Cold 2” in Supplemental Figure S4) that exhibited lower variability and are the same as those included in the main text (Figure 3).
We took the calculated values for amino acids, glucose, lactate, and IgG (if produced) and generated a family of uptake and secretion values (within the 5th–95th percentile confidence intervals) for each metabolite based on its predicted directionality of flow. For example, essential amino acids were forced to be taken up; alanine, traditionally secreted, was forced to be secreted. More formally, we generated 3000 sets of uptake and secretion values satisfying the following criteria. If metabolite is forced to be taken up, we generated random uptake values between 5th percentile value and min(0, 95th percentile value). If metabolite is forced to be secreted, than we generated random secretion values between max(0, 5th percentile value) and 95th percentile value. Otherwise, we generated a random uptake or secretion value between the 5th and 95th percentile values. Each set of values was applied as constraints to the model and used to predict growth, and the results compared to the growth rate prediction obtained using just the calculated best-fit uptake/secretion values (Naive Uptakes bars in Supplemental Figure S5). This approach led to predictions more in agreement with experimental measurements (Supplemental Figure S5), highlighting the importance of accurate metabolomic measurements.
Uptake flux generation
Lower limits for amino acids and glucose were defined based on experimental measurements from the studies used for phenotype validation. The growth rate and production rate from the non-treated culture were set as constraints on the producing cell biomass reaction and IgG/EPO production (as appropriate) in the appropriate cell-line specific model. Iteratively, the following procedure was followed. First, an amino acid or glucose was randomly selected. If an uptake rate for the metabolite is not known, then we found the minimum and maximum allowable uptake/secretion rate for the nutrient that permits growth and production at the experimentally determined rate. Then the uptake or secretion of the nutrient was set to a randomly selected value within that range. This was then repeated until all amino acids and glucose had uptake or secretion values. For each study, 3000 sets of uptake fluxes were generated. Each set of uptake fluxes was checked by ensuring that removal of full constraints on protein production or growth resulted in a production/growth rate within 1% of the experimentally measured value. Uptake fluxes which satisfied this criterion were used for further analysis and are available in Supplemental Data S5.
Algorithmically generated uptake and secretion rates are consistent with experimental measurements
Results for nutrient uptake and secretion flux generation were validated using data from a previous study (Selvarasu et al., 2012). The algorithm was applied at the growth rate and specific productivity measured during early exponential phase and the range of resultant nutrient fluxes (available in Supplemental Data S5) is compared to the experimental values (Supplemental Figure S2). For 19 out of 20 measured metabolites (all except phenylalanine), the experimental uptake or secretion value was within the bounds of our algorithm-generated values. For phenylalanine, the deviation from the calculated range was very small: approximately 4.4×10−4 mmol gDW−1 hr−1 (2.8% of the median predicted value). Thus, it is clear that the predicted fluxes were consistent with experimentally measured fluxes.
Metabolite uptake and secretion rates are consistent before and after treatment
The resource redirection efficiency analysis assumes that metabolite uptake and secretion rates do not significantly change after treatment. This assumption was assessed by analyzing data from a study examining treatment of cells with sodium butyrate (NaBu) (Carinhas et al., 2013). In this study, essential amino acid fluxes (which are limiting for growth and protein production) remain fairly stable before and after NaBu treatment (Supplemental Figure S1). In fact, the majority of changes were actually in the direction of increased metabolite uptake, which—if extrapolated to all simulated uptakes for NaBu treated cell lines—would further decrease the calculated efficiencies and yields for those treatments.
Redirection analysis
For each ‘treatment’, the model growth rate was constrained to the experimentally measured value while protein production was unconstrained. After applying a set of uptake flux values, the model was simulated while optimizing for maximum protein production. This was repeated for all flux values passing the check for growth/production rate mentioned previously. The experimentally observed production rate was compared to the family of simulated production rates.
For the data stemming from a temperature shift treatment (Kim and Lee, 2009), fluxes were scaled by a random factor between 0.517 and 0.798 based on the difference in uptake rates observed for glucose and glutamine between cultures with and without a temperature shift.
Calculation of growth and protein production tradeoff
For each set of uptake fluxes passing the check for growth and production rates, the simulated maximum possible growth rate was calculated by not forcing any protein production and optimizing for biomass production (i.e. flux through biomass_cho_producing). Maximum simulated protein production was calculated by setting the growth rate to 0 and optimizing for protein production. The fraction of maximum protein production at various fractions of maximum growth rate were calculated by constraining growth rate to a fraction of the simulated maximum growth rate for a specific uptake flux and then optimizing for protein production (normalized by maximum protein production for that uptake flux). The growth rates and production rates from the treatment papers were normalized by the family of maximum growth rates and production rates, respectively, to evaluate production efficiency.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of model constraints
Metabolite uptake and secretion rates were quantified and used as constraints for the model simulations (see Supplemental Data S3). The workflow for quantification and integration of these exchange rates is presented below. Experimental growth rates were calculated by determining the slope of the linear polynomial fit to the natural log of the viable cell densities.
Serum-grown CHO-K1, non-producing (In house)
Uptake and secretion rates were computed by calculating the slope of a linear polynomial fit to metabolite concentrations in the spent and control media vs. the integral of cell concentration (with respect to time) for exponential phase. The sample at 36 h was excluded due to poor quality (i.e., there were spurious jumps in some metabolite concentrations, which was inconsistent with measurements of preceding and subsequent time points). The final exchange rates were calculated by subtracting the control media rate from the spent media rate. Cell dry weight was set at 216.1 pg/cell, based on component weights (see Supplemental Data S2). The metabolite exchange rates were consistent with the expected rates for growth, except for arginine and cysteine equivalents, which showed a net efflux, despite being essential in CHO (Naylor et al., 1979) (also see Supplemental Figure S5).
CHO M250-9 cells grown in protein-free, chemically defined (PFCD) media
Previously published data were acquired for the metabolite uptake and secretion rates (Selvarasu et al., 2012). Since no information was available on tryptophan uptake, a previously published (Carinhas et al., 2013) uptake rate of 0.0032 mmol gDW−1 hr−1 was used.
Butyrate-treated CHO cells
Previously published uptake and secretion rates were obtained for exponential phase of CHO cells producing high and low quantities of protein under control and butyrate treatment while growing on CD CHO medium (Carinhas et al., 2013). Since cysteine uptake was not measured, an uptake rate of 0.0052 mmol gDW−1 hr−1 for cysteine was obtained from a previously reported value (Selvarasu et al., 2012). This value is qualitatively consistent with a previous report (Gorfien et al., 2003) showing that cysteine is taken up.
CHO-K1 from MFA studies
Exchange rates were taken from previous studies (Ahn and Antoniewicz, 2013, 2011) and supplemented with the values used in the associated metabolic flux analysis (MFA) model simulations for metabolites for which experimental data were not available. As both studies reported uptakes on a per-cell basis, fluxes were calculated after scaling to a cell dry weight of 216.1 pg/cell, 315 pg/cell, and 350 pg/cell since dry weight composition was not measured. These flux values thus cover the range of observed cell weights for CHO (Altamirano et al., 2001; Martínez et al., 2015). As the cells were grown in 10% FBS, an extracellular arginase reaction was added to the models (Hölttä and Pohjanpelto, 1982).
CHO-K1 derivative producing IgG in serum-free medium
Data were taken from a CHO XL99-Ab2 cell line producing an IgG1 antibody (Martínez et al., 2015). Uptake and secretion rates were calculated by fitting a linear polynomial to metabolite concentration vs. the integral of cell concentration (with respect to time, prior to temperature shift) and scaled using a cell dry weight of 350 pg/cell. We discuss the control cultures (no temperature shift) in the supplemental information (see Supplemental Figure S4–5). Since cysteine uptake was not measured, an uptake rate of 0.0052 mmol gDW−1 hr−1 for cysteine was obtained from a previously reported value (Selvarasu et al., 2012).
DATA AND SOFTWARE AVAILABILITY
Raw data files for CHO-S RNA sequencing have been deposited in the NCBI Gene Expression Omnibus under accession number GSE77800. All models are available at http://www.CHOgenome.org. Additionally, the global model can be browsed and downloaded at the BiGG Models database (http://bigg.ucsd.edu).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Critical Commercial Assays | ||
| Qiagen’s RNeasy Mini Kit | Qiagen | #74104 |
| TruSeq Stranded mRNA Sample prep kit with 96 dual indexes | Illumina | RS-122-2103 |
| Deposited Data | ||
| Raw RNA-Seq data | This paper | GEO: GSE77800 |
| Global model of C. griseus metabolism | This paper | http://bigg.ucsd.edu; http://CHOgenome.org |
| Cell-line models of CHO-S, K1, and DG44 | This paper | http://CHOgenome.org |
| Experimental Models: Cell Lines | ||
| Hamster: CHO-S | Life Technologies | A11557-01 |
| Hamster: CHO-K1 | ATCC | CCL-61 |
| Experimental Models: Organisms/Strains | ||
| Recombinant DNA | ||
| Sequence-Based Reagents | ||
| Software and Algorithms | ||
| FastQC | Andrews 2010 | http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Trimmomatic | Bolger et al. 2014 | http://www.usadellab.org/cms/index.php?page=trimmomatic |
| STAR | Dobin et al. 2013 | https://github.com/alexdobin/STAR |
| SAMtools | Li et al. 2009 | http://www.htslib.org/ |
| Cufflinks | Trapnell et al. 2010 | https://github.com/cole-trapnelllab/cufflinks |
| COBRA Toolbox | Schellenberger et al., 2011 | https://github.com/opencobra/cobratoolbox/ |
| GIMME | Becker and Palsson 2008 | https://github.com/opencobra/cobratoolbox/ |
| Other | ||
Supplementary Material
Acknowledgments
We acknowledge funding from the National Institutes of Health (R35 GM119850) and generous funding through the Novo Nordisk Foundation Center for Biosustainability at the Technical University of Denmark (NNF16CC0021858). J.M.G. was provided funding from CONACYT and a graduate fellowship from the University of California Institute for Mexico and the United States. S.L. received funding from the Frontiers of Innovation Scholars Program at UCSD. D.-Y.L. was supported by the Synthetic Biology Initiative of the National University of Singapore (DPRT/943/09/14) and the Next-Generation BioGreen 21 Program of the Rural Development Administration, Republic of Korea (SSAC, PJ01109405). ACIB (M.H., D.R., J.Z., and N.B.) is supported by the COMET-K2 funding program managed by the Austrian Research Promotion Agency FFG. The authors would like to acknowledge support from the Science for Life Laboratory, the National Genomics Infrastructure (NGI), and Uppmax for providing assistance in massive parallel sequencing and computational infrastructure.
Footnotes
AUTHOR CONTRIBUTIONS
HH, KSA, and MH led construction of input CHO models. AB, HN, HH, JMG, SL, NEL, NEJ, NL, ZPG, MH, DR, JZ, NB, LEQ, LKN, VSM, BOP, KSA, ML, SK, and DYL helped to build preliminary CHO models or conceived of and oversaw the efforts. HH led the reconciliation effort and conducted the analyses. NEL, NB, DYL, and LKN conceived of and oversaw the reconciliation effort. HH, KSA, MH, DR, ML, CAO, YH, DL, VSM, SK, LEQ, SL, PNS, LEP, JMG, AT, AMAH, HFK, MRA, LKN, and NEL curated CHO reconstruction content. HH, DCZ, JMG, SL, NEL, KSA, ML, SK, CAO, VSM, LKN, LEQ, and MH curated human reconstruction content. DBH, GP, and MJB oversaw and generated metabolomic data for CHO-K1. DL, AML, and MRA constructed a reconstruction of amino acid metabolism in CHO. ZAK performed database checks and model transfer to the BiGG database. JSL validated asparagine auxotrophy predictions in CHO-K1 and CHO-S. TW, JA, and BGV oversaw and generated transcriptomic and proteomic data for the CHO-S cell line. SL processed RNA-Seq data. HH and NEL wrote the manuscript. HH and ML built the cell-line specific models. All authors read and approved of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn WS, Antoniewicz MR. Parallel labeling experiments with [1,2-(13)C]glucose and [U-(13)C]glutamine provide new insights into CHO cell metabolism. Metab Eng. 2013;15:34–47. doi: 10.1016/j.ymben.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Ahn WS, Antoniewicz MR. Metabolic flux analysis of CHO cells at growth and non-growth phases using isotopic tracers and mass spectrometry. Metab Eng. 2011;13:598–609. doi: 10.1016/j.ymben.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Altamirano C, Berrios J, Vergara M, Becerra S. Advances in improving mammalian cells metabolism for recombinant protein production. Electron J Biotechnol. 2013:16. doi: 10.2225/vol16-issue3-fulltext-2. [DOI] [Google Scholar]
- Altamirano C, Illanes A, Casablancas A, Gámez X, Cairó JJ, Gòdia C. Analysis of CHO cells metabolic redistribution in a glutamate-based defined medium in continuous culture. Biotechnol Prog. 2001;17:1032–41. doi: 10.1021/bp0100981. [DOI] [PubMed] [Google Scholar]
- Andrews S. FastQC: a quality control tool for high throughput sequence data 2010 [Google Scholar]
- Baycin-Hizal D, Tabb DL, Chaerkady R, Chen L, Lewis NE, Nagarajan H, Sarkaria V, Kumar A, Wolozny D, Colao J, Jacobson E, Tian Y, O’Meally RN, Krag SS, Cole RN, Palsson BO, Zhang H, Betenbaugh M. Proteomic analysis of Chinese hamster ovary cells. J Proteome Res. 2012;11:5265–76. doi: 10.1021/pr300476w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebbington CR, Renner G, Thomson S, King D, Abrams D, Yarranton GT. High-level expression of a recombinant antibody from myeloma cells using a glutamine synthetase gene as an amplifiable selectable marker. Biotechnology (N Y) 1992;10:169–75. doi: 10.1038/nbt0292-169. [DOI] [PubMed] [Google Scholar]
- Becker SA, Palsson BO. Context-specific metabolic networks are consistent with experiments. PLoS Comput Biol. 2008;4:e1000082. doi: 10.1371/journal.pcbi.1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Collin F, Brettschneider J, Simpson K, Cope L, Irizarry RA, Speed TP. Quality Assessment of Affymetrix GeneChip Data. In: Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor SE - 3, Statistics for Biology and Health. Springer; New York: 2005. pp. 33–47. [DOI] [Google Scholar]
- Bordbar A, Mo ML, Nakayasu ES, Schrimpe-Rutledge AC, Kim YMM, Metz TO, Jones MB, Frank BC, Smith RD, Peterson SN, Hyduke DR, Adkins JN, Palsson BO. Model-driven multi-omic data analysis elucidates metabolic immunomodulators of macrophage activation. Mol Syst Biol. 2012;8:558. doi: 10.1038/msb.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgström E, Lundin S, Lundeberg J. Large scale library generation for high throughput sequencing. PLoS One. 2011;6:e19119. doi: 10.1371/journal.pone.0019119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borman A, Wood TR, Black HC, Anderson EG, Oesterling MJ, Womack M, Rose WC. The role of arginine in growth with some observations on the effects of argininic acid. J Biol Chem. 1946;166:585–594. [PubMed] [Google Scholar]
- Brinkrolf K, Rupp O, Laux H, Kollin F, Ernst W, Linke B, Kofler R, Romand S, Hesse F, Budach WE, Galosy S, Müller D, Noll T, Wienberg J, Jostock T, Leonard M, Grillari J, Tauch A, Goesmann A, Helk B, Mott JE, Pühler A, Borth N. Chinese hamster genome sequenced from sorted chromosomes. Nat Biotechnol. 2013;31:694–5. doi: 10.1038/nbt.2645. [DOI] [PubMed] [Google Scholar]
- Cardoso JGR, Andersen MR, Herrgård MJ, Sonnenschein N. Analysis of genetic variation and potential applications in genome-scale metabolic modeling. Front Bioeng Biotechnol. 2015;3:13. doi: 10.3389/fbioe.2015.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carinhas N, Duarte TM, Barreiro LC, Carrondo MJT, Alves PM, Teixeira AP. Metabolic signatures of GS-CHO cell clones associated with butyrate treatment and culture phase transition. Biotechnol Bioeng. 2013;110:3244–57. doi: 10.1002/bit.24983. [DOI] [PubMed] [Google Scholar]
- Castro PL, Hayter P, Ison A, Bull A. Application of a statistical design to the optimization of culture medium for recombinant interferon-gamma production by Chinese hamster ovary cells. Appl Microbiol Biotechnol. 1992:38. doi: 10.1007/BF00169424. [DOI] [PubMed] [Google Scholar]
- Chang A, Scheer M, Grote A, Schomburg I, Schomburg D. BRENDA, AMENDA and FRENDA the enzyme information system: new content and tools in 2009. Nucleic Acids Res. 2009;37:D588–92. doi: 10.1093/nar/gkn820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Harcum SW. Effects of elevated ammonium on glycosylation gene expression in CHO cells. Metab Eng. 2006;8:123–32. doi: 10.1016/j.ymben.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Chowdhury R, Chowdhury A, Maranas CD. Using Gene Essentiality and Synthetic Lethality Information to Correct Yeast and CHO Cell Genome-Scale Models. Metabolites. 2015;5:536–70. doi: 10.3390/metabo5040536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke C, Barron N, Meleady P, Clynes M. Statistical methods for mining Chinese hamster ovary cell ‘omics data: from differential expression to integrated multilevel analysis of the biological system. Pharm Bioprocess. 2014;2:469–481. doi: 10.4155/pbp.14.50. [DOI] [Google Scholar]
- Courtes FC, Lin J, Lim HL, Ng SW, Wong NSC, Koh G, Vardy L, Yap MGS, Loo B, Lee DY. Translatome analysis of CHO cells to identify key growth genes. J Biotechnol. 2013;167:215–24. doi: 10.1016/j.jbiotec.2013.07.010. [DOI] [PubMed] [Google Scholar]
- Curran KA, Leavitt JM, Karim AS, Alper HS. Metabolic engineering of muconic acid production in Saccharomyces cerevisiae. Metab Eng. 2013;15:55–66. doi: 10.1016/j.ymben.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Daar IO, Artymiuk PJ, Phillips DC, Maquat LE. Human triose-phosphate isomerase deficiency: a single amino acid substitution results in a thermolabile enzyme. Proc Natl Acad Sci U S A. 1986;83:7903–7. doi: 10.1073/pnas.83.20.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leon Gatti M, Wlaschin KF, Nissom PM, Yap M, Hu WS. Comparative transcriptional analysis of mouse hybridoma and recombinant Chinese hamster ovary cells undergoing butyrate treatment. J Biosci Bioeng. 2007;103:82–91. doi: 10.1263/jbb.103.82. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson BØ. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci U S A. 2007;104:1777–82. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte TM, Carinhas N, Barreiro LC, Carrondo MJT, Alves PM, Teixeira AP. Metabolic responses of CHO cells to limitation of key amino acids. Biotechnol Bioeng. 2014;111:2095–106. doi: 10.1002/bit.25266. [DOI] [PubMed] [Google Scholar]
- Edwards JS, Ibarra RU, Palsson BO. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nat Biotechnol. 2001;19:125–130. doi: 10.1038/84379. [DOI] [PubMed] [Google Scholar]
- Fan Y, Jimenez Del Val I, Müller C, Wagtberg Sen J, Rasmussen SK, Kontoravdi C, Weilguny D, Andersen MR. Amino acid and glucose metabolism in fed-batch CHO cell culture affects antibody production and glycosylation. Biotechnol Bioeng. 2015;112:521–35. doi: 10.1002/bit.25450. [DOI] [PubMed] [Google Scholar]
- Feist AM, Palsson BO. The biomass objective function. Curr Opin Microbiol. 2010;13:344–349. doi: 10.1016/j.mib.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feizi A, Österlund T, Petranovic D, Bordel S, Nielsen J. Genome-scale modeling of the protein secretory machinery in yeast. PLoS One. 2013;8:e63284. doi: 10.1371/journal.pone.0063284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golabgir A, Gutierrez JM, Hefzi H, Li S, Palsson BO, Herwig C, Lewis NE. Quantitative feature extraction from the Chinese hamster ovary bioprocess bibliome using a novel meta-analysis workflow. Biotechnol Adv. 2016 doi: 10.1016/j.biotechadv.2016.02.011. [DOI] [PubMed] [Google Scholar]
- Gorfien SF, Paul W, Judd D, Tescione L, Jayme DW. Optimized Nutrient Additives for Fed-Batch Cultures. BioPharm Int. 2003;16:34. [Google Scholar]
- Gutierrez JM, Lewis NE. Optimizing eukaryotic cell hosts for protein production through systems biotechnology and genome-scale modeling. Biotechnol J. 2015;10:939–49. doi: 10.1002/biot.201400647. [DOI] [PubMed] [Google Scholar]
- Hiller GW, Mulukutla BC. Method of cell culture 2015 [Google Scholar]
- Hölttä E, Pohjanpelto P. Polyamine dependence of Chinese hamster ovary cells in serum-free culture is due to deficient arginase activity. Biochim Biophys Acta. 1982;721:321–7. doi: 10.1016/0167-4889(82)90085-4. [DOI] [PubMed] [Google Scholar]
- Hyduke DR, Lewis NE, Palsson BØ. Analysis of omics data with genome-scale models of metabolism. Mol Biosyst. 2013;9:167–74. doi: 10.1039/c2mb25453k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra RU, Edwards JS, Palsson BO. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature. 2002;420:186–189. doi: 10.1038/nature01149. [DOI] [PubMed] [Google Scholar]
- Jayapal K, Wlaschin K, Hu WS, Yap M. Recombinant Protein Therapeutics from Cho Cells - 20 Years and Counting. CHO Consort. SBE Spec Ed. 2007:40–47. [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao FT, Puck TT. Genetics of somatic mammalian cells. IV Properties of Chinese hamster cell mutants with respect to the requirement for proline. Genetics. 1967;55:513–24. doi: 10.1093/genetics/55.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman RJ, Sharp PA. Amplification and expression of sequences cotransfected with a modular dihydrofolate reductase complementary dna gene. J Mol Biol. 1982;159:601–21. doi: 10.1016/0022-2836(82)90103-6. [DOI] [PubMed] [Google Scholar]
- Kildegaard HF, Baycin-Hizal D, Lewis NE, Betenbaugh MJ. The emerging CHO systems biology era: harnessing the ’omics revolution for biotechnology. Curr Opin Biotechnol. 2013;24:1102–7. doi: 10.1016/j.copbio.2013.02.007. [DOI] [PubMed] [Google Scholar]
- Kim NS, Lee GM. Overexpression of bcl-2 inhibits sodium butyrate-induced apoptosis in Chinese hamster ovary cells resulting in enhanced humanized antibody production. Biotechnol Bioeng. 2000;71:184–93. doi: 10.1002/1097-0290(2000)71:3<184::aid-bit1008>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Kim YG, Lee GM. Bcl-xL overexpression does not enhance specific erythropoietin productivity of recombinant CHO cells grown at 33 degrees C and 37 degrees C. Biotechnol Prog. 2009;25:252–6. doi: 10.1002/btpr.91. [DOI] [PubMed] [Google Scholar]
- King ZA, Lu J, Dräger A, Miller P, Federowicz S, Lerman JA, Ebrahim A, Palsson BO, Lewis NE. BiGG Models: A platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 2015;44:D515–22. doi: 10.1093/nar/gkv1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremkow BG, Baik JY, MacDonald ML, Lee KH. CHOgenome.org 2.0: Genome resources and website updates. Biotechnol J. 2015;10:931–8. doi: 10.1002/biot.201400646. [DOI] [PubMed] [Google Scholar]
- Lao MS, Toth D. Effects of ammonium and lactate on growth and metabolism of a recombinant Chinese hamster ovary cell culture. Biotechnol Prog. 1997;13:688–91. doi: 10.1021/bp9602360. [DOI] [PubMed] [Google Scholar]
- Lewis NE, Hixson KK, Conrad TM, Lerman JA, Charusanti P, Polpitiya AD, Adkins JN, Schramm G, Purvine SO, Lopez-Ferrer D, Weitz KK, Eils R, Konig R, Smith RD, Palsson BO. Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models. Mol Syst Biol. 2010;6:390. doi: 10.1038/msb.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis NE, Liu X, Li Y, Nagarajan H, Yerganian G, O’Brien E, Bordbar A, Roth AM, Rosenbloom J, Bian C, Xie M, Chen W, Li N, Baycin-Hizal D, Latif H, Forster J, Betenbaugh MJ, Famili I, Xu X, Wang J, Palsson BO. Genomic landscapes of Chinese hamster ovary cell lines as revealed by the Cricetulus griseus draft genome. Nat Biotechnol. 2013;31:759–65. doi: 10.1038/nbt.2624. [DOI] [PubMed] [Google Scholar]
- Lewis NE, Nagarajan H, Palsson BO. Constraining the metabolic genotype-phenotype relationship using a phylogeny of in silico methods. Nat Rev Microbiol. 2012;10:291–305. doi: 10.1038/nrmicro2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loira N, Zhukova A, Sherman DJ. Pantograph: A template-based method for genome-scale metabolic model reconstruction. J Bioinform Comput Biol. 2015;13:1550006. doi: 10.1142/S0219720015500067. [DOI] [PubMed] [Google Scholar]
- Lorendeau D, Christen S, Rinaldi G, Fendt SM. Metabolic control of signalling pathways and metabolic auto-regulation. Biol Cell. 2015;107:251–72. doi: 10.1111/boc.201500015. [DOI] [PubMed] [Google Scholar]
- Lundin S, Stranneheim H, Pettersson E, Klevebring D, Lundeberg J. Increased throughput by parallelization of library preparation for massive sequencing. PLoS One. 2010;5:e10029. doi: 10.1371/journal.pone.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez VS, Buchsteiner M, Gray P, Nielsen LK, Quek LE. Dynamic metabolic flux analysis using B-splines to study the effects of temperature shift on CHO cell metabolism. Metab Eng Commun. 2015;2:46–57. doi: 10.1016/j.meteno.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura Y, Lund J, Church S, Dong S, Li J, Goodall M, Jefferis R. Butyrate increases production of human chimeric IgG in CHO-K1 cells whilst maintaining function and glycoform profile. J Immunol Methods. 2001;247:205–16. doi: 10.1016/s0022-1759(00)00308-2. [DOI] [PubMed] [Google Scholar]
- Naylor SL, Townsend JK, Klebe RJ. Characterization of naturally occurring auxotrophic mammalian cells. Somatic Cell Genet. 1979;5:271–7. doi: 10.1007/BF01539166. [DOI] [PubMed] [Google Scholar]
- Nolan RP, Lee K. Dynamic model of CHO cell metabolism. Metab Eng. 2011;13:108–24. doi: 10.1016/j.ymben.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Ostlund G, Schmitt T, Forslund K, Köstler T, Messina DN, Roopra S, Frings O, Sonnhammer ELL. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2010;38:D196–203. doi: 10.1093/nar/gkp931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paglia G, Hrafnsdóttir S, Magnúsdóttir M, Fleming RMT, Thorlacius S, Palsson BØ, Thiele I. Monitoring metabolites consumption and secretion in cultured cells using ultra-performance liquid chromatography quadrupole-time of flight mass spectrometry (UPLC-Q-ToF-MS) Anal Bioanal Chem. 2012;402:1183–98. doi: 10.1007/s00216-011-5556-4. [DOI] [PubMed] [Google Scholar]
- Peng RW, Fussenegger M. Molecular engineering of exocytic vesicle traffic enhances the productivity of Chinese hamster ovary cells. Biotechnol Bioeng. 2009;102:1170–81. doi: 10.1002/bit.22141. [DOI] [PubMed] [Google Scholar]
- Quek LE, Dietmair S, Hanscho M, Martínez VS, Borth N, Nielsen LK. Reducing Recon 2 for steady-state flux analysis of HEK cell culture. J Biotechnol. 2014;184:172–8. doi: 10.1016/j.jbiotec.2014.05.021. [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- Satish Kumar V, Dasika MS, Maranas CD. Optimization based automated curation of metabolic reconstructions. BMC Bioinformatics. 2007;8:212. doi: 10.1186/1471-2105-8-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberger J, Que R, Fleming RM, Thiele I, Orth JD, Feist AM, Zielinski DC, Bordbar A, Lewis NE, Rahmanian S, Kang J, Hyduke DR, Palsson BO. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat Protoc. 2011;6:1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz R, Kuepfer L, Sauer U. Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli. Mol Syst Biol. 2007;3:119. doi: 10.1038/msb4100162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvarasu S, Ho YS, Chong WPK, Wong NSC, Yusufi FNK, Lee YY, Yap MGS, Lee DY. Combined in silico modeling and metabolomics analysis to characterize fed-batch CHO cell culture. Biotechnol Bioeng. 2012;109:1415–29. doi: 10.1002/bit.24445. [DOI] [PubMed] [Google Scholar]
- Selvarasu S, Kim DY, Karimi IA, Lee DY. Combined data preprocessing and multivariate statistical analysis characterizes fed-batch culture of mouse hybridoma cells for rational medium design. J Biotechnol. 2010;150:94–100. doi: 10.1016/j.jbiotec.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Sengupta N, Rose ST, Morgan JA. Metabolic flux analysis of CHO cell metabolism in the late non-growth phase. Biotechnol Bioeng. 2011;108:82–92. doi: 10.1002/bit.22890. [DOI] [PubMed] [Google Scholar]
- Sha S, Agarabi C, Brorson K, Lee DY, Yoon S. N-Glycosylation Design and Control of Therapeutic Monoclonal Antibodies. Trends Biotechnol. 2016 doi: 10.1016/j.tibtech.2016.02.013. [DOI] [PubMed] [Google Scholar]
- Smallbone K. Standardized network reconstruction of CHO cell metabolism 2013 [Google Scholar]
- Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–51. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- Spahn PN, Hansen AH, Hansen HG, Arnsdorf J, Kildegaard HF, Lewis NE. A Markov chain model for N-linked protein glycosylation - towards a low-parameter tool for model-driven glycoengineering. Metab Eng. 2016;33:52–66. doi: 10.1016/j.ymben.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swainston N, Smallbone K, Hefzi H, Dobson PD, Brewer J, Hanscho M, Zielinski DC, Ang KS, Gardiner NJ, Gutierrez JM, Kyriakopoulos S, Lakshmanan M, Li S, Liu JK, Martinez VS, Orellana CA, Quek LE, Thomas A, Zanghellini J, Borth N, Lee DY, Nielsen LK, Kell DB, Lewis NE, Mendes P. Recon 2.2: from reconstruction to model of human metabolism. Metabolomics. 2016 doi: 10.1007/s11306-016-1051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton N, Dean J, Reddy P, Young JD. Peak antibody production is associated with increased oxidative metabolism in an industrially relevant fed-batch CHO cell culture. Biotechnol Bioeng. 2013;110:2013–24. doi: 10.1002/bit.24858. [DOI] [PubMed] [Google Scholar]
- Thiele I, Vlassis N, Fleming RMT. fastGapFill: efficient gap filling in metabolic networks. Bioinformatics. 2014;30:2529–31. doi: 10.1093/bioinformatics/btu321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle D, Downing SJ, Harris SC, Phang JM. Proline biosynthesis: Multiple defects in Chinese hamster ovary cells. Biochem Biophys Res Commun. 1973;53:1130–1136. doi: 10.1016/0006-291X(73)90582-2. [DOI] [PubMed] [Google Scholar]
- Walsh G. Biopharmaceutical benchmarks 2014. Nat Biotechnol. 2014;32:992–1000. doi: 10.1038/nbt.3040. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelnikov I, Arndt D, Xia J, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R, Scalbert A. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–7. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Nagarajan H, Lewis NE, Pan S, Cai Z, Liu X, Chen W, Xie M, Wang W, Hammond S, Andersen MR, Neff N, Passarelli B, Koh W, Fan HC, Wang J, Gui Y, Lee KH, Betenbaugh MJ, Quake SR, Famili I, Palsson BO, Wang J. The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat Biotechnol. 2011;29:735–41. doi: 10.1038/nbt.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Butler M. Effects of ammonia on CHO cell growth, erythropoietin production, and glycosylation. Biotechnol Bioeng. 2000;68:370–80. doi: 10.1002/(SICI)1097-0290(20000520)68:4<370::AID-BIT2>3.0.CO;2-K/full. [DOI] [PubMed] [Google Scholar]
- Zamorano F, Wouwer, Vande A, Bastin G. A detailed metabolic flux analysis of an underdetermined network of CHO cells. J Biotechnol. 2010;150:497–508. doi: 10.1016/j.jbiotec.2010.09.944. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.