Abstract
Hybridization and incomplete lineage sorting are common confounding factors in phylogeny and speciation resulting in mitonuclear disparity. Mitochondrial introgression, a particular case of hybridization, may, in extreme cases, lead to replacement of the mitochondrial genome of one species with that of another (mitochondrial capture). We investigated mitochondrial introgression involving two species of the cyprinid genus Squalius in the western Peloponnese region of Greece using molecular and morphological data. We found evidence of complete mitochondrial introgression of Squalius keadicus into two populations recognized as Squalius peloponensis from the Miras and Pamissos River basins and a divergence of mitochondrial genomes of S. keadicus from the Evrotas basin from that of the introgressed populations dating from the Pleistocene. Secondary contact among basins is a possible factor in connection of the species and the introgression event. Morphological analyses support the hypothesis of mitochondrial introgression, as S. keadicus was different from the other three populations recognized as S. peloponensis, although significant differences were found among the four populations. Isolation by geographical barriers arose during Pleistocene in the western Peloponnese were the source of the evolution of the two reciprocally monophyletic subclades found in the S. keadicus mitochondrial clade, and the morphological differences found among the four populations. Along with the lack of structure in the nuclear genome in the three populations ascribed to S. peloponensis, this suggests an incipient speciation process occurring in these Squalius species in the western Peloponnese.
Introduction
The use of mitochondrial DNA for inferring phylogenetic or phylogeographic relationships has been traditionally preferred over the use of the nuclear genome, as mtDNA accurately reflects recent divergence patterns than do nuclear markers [1]. Nevertheless, concordant patterns between mtDNA and nuclear DNA are not always observed in nature [2–4].
Evolutionary processes such as hybridization and incomplete lineage sorting are common confounding factors in phylogeny and speciation and primary causes of mitonuclear discordance [3–5]. Incomplete lineage sorting occurs when the coalescence time of genes and speciation differ, i.e. when lineages fail to sort out at the same time at speciation happens [6]. Therefore, gene trees do not always represent the true relationships among taxa because different genes may have not branched at the same time [7–9]. Distinguishing incomplete lineage sorting from hybridization can be difficult as these two processes may originate the same phylogenetic tree [10–12]. Nonetheless, among others, any biogeographic pattern is expected when incomplete lineage sorting is the confounding factor [4].
Hybridization between species involves mating between unrelated organisms regardless the taxonomic status and, in some cases, may lead to gene transfer, a common process in plants [13], or to very complex evolutionary processes occurring in freshwater fishes [14–15]. Introgression is a particular case of hybridization and occurs when gene flow exists between populations that hybridize and hybrids backcross to one or both parental populations [16]. Within this framework, mitochondrial capture is defined as complete mitochondrial introgression, a situation in which the mitochondrial genome of one species is replaced with that of another in an entire population, as a consequence of selective backcrossing of hybrids with one of the parent species [2–3, 17–19]. In hybridization, the complete replacement of the mitochondrial DNA of one species with that of another is more common than is replacement of a portion of the nuclear genome, due to maternal inheritance and the four-fold smaller effective population size of mitochondrial DNA compared to nuclear DNA, implying that mtDNA will complete the process of lineage sorting more rapidly than will nuclear DNA [1, 3, 20–21]. Understanding evolutionary processes such as hybridization, mitochondrial introgression, and incomplete lineage sorting is of vital importance to inference reliable phylogenies and reconstruction of evolutionary history and speciation processes, especially in currently allopatric species. Revealing mitochondrial introgression may be the only way to identify past hybridization of allopatric species [17].
In general, mitonuclear discordance due to mitochondrial introgression occurs more frequently in certain taxonomic groups, including mammals and fish; whereas, in groups such as birds and butterflies, in which the female is heterogametic, mitochondrial introgression is usually reduced, conforming to Haldane’s rule [4]. Examples of the complete replacement of mitochondrial DNA are found in various taxa [18–19, 22–25], including freshwater fishes [26–28].
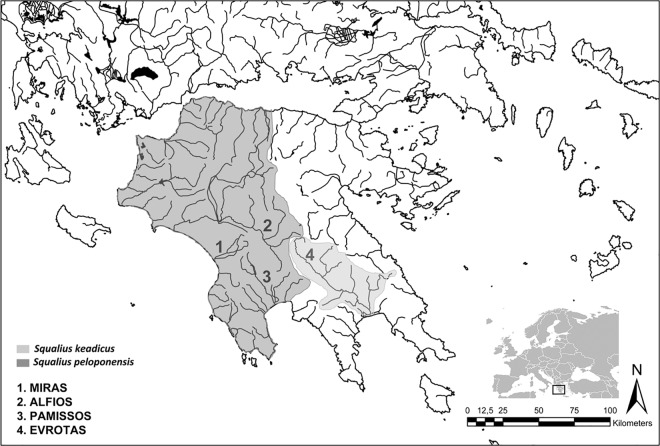
Within freshwater fishes the preponderance of hybridization is greater in cyprinids than is in other freshwater fish families such as salmonids or cichlids and several examples are observed [29–32]. A clear example of mitonuclear discordance has been described for the genus Squalius in the Greek Peloponnese. Previous studies focusing on a more general framework of phylogenetic relationships of Squalius hypothesised the presence of mitochondrial introgression to explain discrepancy found in the mitochondrial (DNA sequences) and nuclear (allozyme) phylogenetic position of S. keadicus (Stephanidis, 1971) and S. peloponensis (Valenncienes, 1844) [33–34]. These two species, currently showing allopatric distribution, belong to two different evolutionary lineages recognized within the genus Squalius; S. peloponesis belongs to the Euroasiatic group while S. keadicus is included together with other small-sized species within the Mediterranean group. These two evolutionary lineages have been isolated since the Middle Miocene [35–36]. Squalius keadicus is a small- to medium-sized fish endemic to the Evrotas Basin in the southwestern Peloponnese. Squalius peloponensis is a medium to large fish and has a broader distribution range in the western Peloponnese, encompassing several hydrological basins (Fig 1) [37]. The morphological distinction between both species is clear and several diagnostic morphometric and meristic characters are identified to distinguishing them from other Balkan Squalius species. Head length is 24–27% standard length in S. peloponensis. The posterior margin of the anal fin is different in both species, being straight in S. keadicus and slightly convex in S. peloponensis. Meristic diagnostic characters of S. keadicus show 44–49 scales on lateral line whereas in S. peloponensis this range is 40–44. The colouration of both species also differs; S. keadicus shows in live a conspicuous blackish stripe on flank from eye to caudal fin base and very dark scales in upper half of flank, which are not present in S. peloponensis [37].
Fig 1. Locality samples and distribution range of S. keadicus and S. peloponensis according to [37].
With regards to the paleogeographical context in which the two species have evolved, the southwestern Peloponnese lies near the Hellenic Trench in the Eastern Mediterranean region, one of the most active areas in the Euroasiatic-African convergence zone owing to the northward subduction of the African plate beneath the Aegean plate [38]. The region has a complex tectonic history and numerous currently active northwestern-southeastern tectonic faults [38–41]. The formation of the hydrological network of the Greek Peloponnese was complex, characterized by Late Miocene-Quaternary compressional structures as well as Pliocene lacustrine-marine and Pleistocene terrestrial-fluvial sedimentary successions leading to the formation of fluvial terraces [38, 42–43]. Such paleogeographic complexity may have promoted ancient contact among freshwater fauna inhabiting the basins of the western Peloponnese, resulting in hybridization events.
Within the framework of mitonuclear disparity, the aim of this study was to investigate the mitochondrial capture between two species of Squalius from the Greek Peloponnese using molecular and morphological analyses of three populations of S. peloponensis and one of S. keadicus. We also establish a temporal and biogeographical framework for the introgression episode.
Material and Methods
Sampling
Specimens of four populations of Squalius were collected by electrofishing from basins in the western Peloponnese region of Greece following the European regulations (EN ISO 14011:200. Water quality—Sampling of fish with electrophising) (Table 1; Fig 1). Sampling was performed with authorised permission from the Greek Ministry of Environment, Energy and Climatic Change and after consulting the Animal Care and Use Committee of the National Museum of Prague (č.j.NM/2014/258). Immediately after capture, all individuals were anesthetized using Tricaine methanesulfonate (MS-222) to alleviate suffering. A piece of ventral fin sample (~3 mm2 fin clips) was obtained from 61 fish belonging to the four Greek populations analysed (nine to S. keadicus and 52 to S. peloponensis). All tissues were preserved in 95% ethanol and stored at 4°C until is processing at laboratory for molecular analyses. These 61 specimens analysed for molecular markers were sacrificed for morphological analyses following humanely euthanization with overdose of MS-222 according to European Commission regulations (Directive 2010/63/EU). Death was confirmed after no gill movement was observed for at least 10 minutes. These new sampled specimens used for morphological purposes were preserved in a solution of 5% formalin and after a week they were transferred to a 70% ethanol solution. These new-collected specimens have been deposited in the Fish Collection of the National Museum of Prague (Czech Republic). The remaining specimens used for morphological analyses in this study came from historical deposits of the Fish Collections of the Museo Nacional de Ciencias Naturales, CSIC, in Madrid (Spain). Voucher tissues of new collected specimens are deposited in the DNA Collections of the Museo Nacional de Ciencias Naturales in Madrid, CSIC, (Spain) and in the National Museum of Prague (Czech Republic).
Table 1. Specimens used in molecular and morphological analyses and their voucher numbers.
GenBank accession numbers for all analysed genes and labels in phylogenetic trees. All these specimens except those marked with an * symbol, proceeding from GenBank database, have been used in morphological analyses.
| Species/Population (Voucher number) | Geographic coordinates | MT-CYB | RAG1 (phased alleles) | S7 (phased alleles) | Label in phylogenetic trees |
|---|---|---|---|---|---|
| Squalius keadicus/Evrotas (AJ252820)* | - | AJ252820 | - | - | Squalius keadicus 1 |
| Squalius keadicus/Evrotas (AF090760)* | 36° 51' 15.6" N, 22° 40' 29.8" E | AF090760 | - | - | Squalius keadicus 2 |
| Squalius keadicus/Evrotas (MNCN_ICTIO 123.875) | 36° 51' 15.6" N, 22° 40' 29.8" E | HM560185 | HM560583 | HM560572 | Squalius keadicus 3 |
| Squalius keadicus/Evrotas (NMP PV6 F1556) | 37° 5' 28.71" N, 22°25'38.74" E | KY070419 | KY070433 (a); KY070434 (b) | KY070547 (a); KY070548 (b) | Squalius keadicus 4 |
| Squalius keadicus/Evrotas (NMP PV6 F1557) | 37° 5' 28.71" N, 22°25'38.74" E | KY070420 | KY070435 (a); KY070436 (b) | KY070549 (a); KY070550 (b) | Squalius keadicus 5 |
| Squalius keadicus/Evrotas (NMP PV6 F1566) | 37° 5' 28.71" N, 22°25'38.74" E | KY070421 | KY070425 (a); KY070426 (b) | KY070503 (a); KY070504 (b) | Squalius keadicus 6 |
| Squalius keadicus/Evrotas (NMP PV6 F1567) | 37° 5' 28.71" N, 22°25'38.74" E | KY070422 | KY070427 (a); KY070428 (b) | KY070505 (a); KY070506 (b) | Squalius keadicus 7 |
| Squalius keadicus/Evrotas (NMP PV6 F1570) | 37° 5' 28.71" N, 22°25'38.74" E | KY070423 | KY070429 (a); KY070430 (b) | KY070551 (a); KY070552 (b) | Squalius keadicus 8 |
| Squalius keadicus/Evrotas (NMP PV6 F1571) | 37° 5' 28.71" N, 22°25'38.74" E | KY070424 | KY070431 (a); KY070432 (b) | KY070553 (a); KY070554 (b) | Squalius keadicus 9 |
| Squalius peloponensis/Alfios (MNCN_ICTIO 120.548) | 37° 28' 21.7" N, 22° 04' 33.9" E | KY070368 | - | KY070515 (a); KY070516 (b) | Alfios 1 |
| Squalius peloponensis/Alfios (MNCN_ICTIO 120.559) | 37° 28' 21.7" N, 22° 04' 33.9" E | KY070372 | KY070497 (a); KY070498 (b) | - | Alfios 2 |
| Squalius peloponensis/Alfios (MNCN_ICTIO 120.563) | 37° 28' 21.7" N, 22° 04' 33.9" E | KY070374 | KY070501 (a); KY070502 (b) | KY070523 (a); KY070524 (b) | Alfios 3 |
| Squalius peloponensis/Alfios (MNCN_ICTIO 120.553) | 37° 28' 21.7" N, 22° 04' 33.9" E | KY070369 | KY070491 (a); KY070492 (b) | KY070517 (a); KY070518 (b) | Alfios 4 |
| Squalius peloponensis/Alfios (MNCN_ICTIO 120.555) | 37° 28' 21.7" N, 22° 04' 33.9" E | KY070370 | - | - | Alfios 5 |
| Squalius peloponensis/Alfios (MNCN_ICTIO 120.556) | 37° 28' 21.7" N, 22° 04' 33.9" E | KY070371 | KY070493(a); KY070494 (b) | KY070519 (a); KY070520 (b) | Alfios 6 |
| Squalius peloponensis/Alfios (MNCN_ICTIO 120.560) | 37° 28' 21.7" N, 22° 04' 33.9" E | KY07037 | KY070499 (a); KY070500 (b) | KY070521 (a); KY070522 (b) | Alfios 7 |
| Squalius peloponensis/Alfios (MNCN_ICTIO 120.557) | 37° 28' 21.7" N, 22° 04' 33.9" E | - | KY070495 (a); KY070496 (b) | - | Alfios 8 |
| Squalius peloponensis/Miras (NMP PV6 F1470) | 37° 17' 34.2" N, 21° 42' 7.86" E | KY070407 | KY070467 (a); KY070468 (b) | KY070525 (a); KY070526 (b) | Miras 1 |
| Squalius peloponensis/Miras (NMP PV6 F1471) | 37° 17' 34.2" N, 21° 42' 7.86" E | KY070408 | KY070469 (a); KY070470 (b) | KY070527 (a); KY070528 (b) | Miras 2 |
| Squalius peloponensis/Miras (NMP PV6 F1472) | 37° 17' 34.2" N, 21° 42' 7.86" E | KY070409 | KY070471 (a); KY070472 (b) | - | Miras 3 |
| Squalius peloponensis/Miras (NMP PV6 G210) | 37° 17' 34.2" N, 21° 42' 7.86" E | KY070410 | KY070473 (a); KY070474 (b) | KY070555 (a); KY070556 (b) | Miras 4 |
| Squalius peloponensis/Miras (NMP PV6 G211) | 37° 17' 34.2" N, 21° 42' 7.86" E | KY070411 | KY070475 (a); KY070476 (b) | KY070557 (a); KY070558 (b) | Miras 5 |
| Squalius peloponensis/Miras (NMP PV6 G212) | 37° 17' 34.2" N, 21° 42' 7.86" E | KY070412 | KY070477 (a); KY070478 (b) | KY070559 (a); KY070560 (b) | Miras 6 |
| Squalius peloponensis/Miras (NMP PV6 G213) | 37° 17' 34.2" N, 21° 42' 7.86" E | KY070413 | KY070479 (a); KY070480 (b) | KY070561 (a); KY070562 (b) | Miras 7 |
| Squalius peloponensis/Miras (NMP PV6 G214) | 37° 17' 34.2" N, 21° 42' 7.86" E | KY070414 | KY070481 (a); KY070482 (b) | KY070563 (a); KY070564 (b) | Miras 8 |
| Squalius peloponensis/Miras (NMP PV6 G215) | 37° 17' 34.2" N, 21° 42' 7.86" E | KY070415 | KY070483 (a); KY070484 (b) | KY070565 (a); KY070566 (b) | Miras 9 |
| Squalius peloponensis/Miras (NMP PV6 G219) | 37° 17' 34.2" N, 21° 42' 7.86" E | KY070416 | KY070485 (a); KY070486 (b) | KY070567 (a); KY070568 (b) | Miras 10 |
| Squalius peloponensis/Miras (NMP PV6 G220) | 37° 17' 34.2" N, 21° 42' 7.86" E | KY070417 | KY070487 (a); KY070488 (b) | KY070569 (a); KY070570 (b) | Miras 11 |
| Squalius peloponensis/Miras (NMP PV6 G222) | 37° 17' 34.2" N, 21° 42' 7.86" E | KY070418 | KY070489 (a); KY070490 (b) | KY070571 (a); KY070572 (b) | Miras 12 |
| Squalius peloponensis/Miras (MNCN_ ICTIO 94.744) | 37° 00' 00.0" N, 21° 44' 23.5" E | KY070404 | KY070461 (a); KY070462 (b) | KY070507 (a); KY070508 (b) | Miras 13 |
| Squalius peloponensis/Miras (MNCN_ICTIO 94.745) | 37° 00' 00.0" N, 21° 44' 23.5" E | KY070405 | KY070463 (a); KY070464 (b) | KY070509 (a); KY070510 (b) | Miras 14 |
| Squalius peloponensis/Miras (MNCN_ICTIO 94.746) | 37° 00' 00.0" N, 21° 44' 23.5" E | KY070406 | KY070465 (a); KY070466 (b) | KY070511 (a); KY070512 (b) | Miras 15 |
| Squalius peloponensis/Pamissos (NMP PV6 F1514) | 37° 9'25.21" N, 21°58'43.16" E | KY070387 | - | - | Pamissos 1 |
| Squalius peloponensis/Pamissos (NMP PV6 F1515) | 37° 9'25.21" N, 21°58'43.16" E | KY070388 | - | - | Pamissos 2 |
| Squalius peloponensis/Pamissos (NMP PV6 F1516) | 37° 9'25.21" N, 21°58'43.16" E | KY070389 | - | - | Pamissos 3 |
| Squalius peloponensis/Pamissos (NMP PV6 F1517) | 37° 9'25.21" N, 21°58'43.16" E | KY070390 | - | - | Pamissos 4 |
| Squalius peloponensis/Pamissos (NMP PV6 F1518) | 37° 9'25.21" N, 21°58'43.16" E | KY070391 | - | - | Pamissos 5 |
| Squalius peloponensis/Pamissos (NMP PV6 F1519) | 37° 9'25.21" N, 21°58'43.16" E | KY070392 | KY070437 (a); KY070438 (b) | KY070529 (a); KY070530 (b) | Pamissos 6 |
| Squalius peloponensis/Pamissos (NMP PV6 F1520) | 37° 9'25.21" N, 21°58'43.16" E | KY070393 | KY070439 (a); KY070440 (b) | KY070531 (a); KY070532 (b) | Pamissos 7 |
| Squalius peloponensis/Pamissos (NMP PV6 F1521) | 37° 9'25.21" N, 21°58'43.16" E | KY070394 | KY070441 (a); KY070442 (b) | - | Pamissos 8 |
| Squalius peloponensis/Pamissos (NMP PV6 F1522) | 37° 9'25.21" N, 21°58'43.16" E | KY070395 | KY070443 (a); KY070444 (b) | KY070533 (a); KY070534 (b) | Pamissos 9 |
| Squalius peloponensis/Pamissos (NMP PV6 F1523) | 37° 9'25.21" N, 21°58'43.16" E | KY070396 | KY070445 (a); KY070446 (b) | KY070535 (a); KY070536 (b) | Pamissos 10 |
| Squalius peloponensis/Pamissos (NMP PV6 F1524) | 37° 9'25.21" N, 21°58'43.16" E | KY070397 | KY070447 (a); KY070448 (b) | KY070535 (a); KY070536 (b) | Pamissos 11 |
| Squalius peloponensis/Pamissos (NMP PV6 F1525) | 37° 9'25.21" N, 21°58'43.16" E | KY070398 | KY070449 (a); KY070450 (b) | KY070537 (a); KY070538 (b) | Pamissos 12 |
| Squalius peloponensis/Pamissos (NMP PV6 F1526) | 37° 9'25.21" N, 21°58'43.16" E | KY070399 | KY070451 (a); KY070452 (b) | KY070539 (a); KY070540 (b) | Pamissos 13 |
| Squalius peloponensis/Pamissos (NMP PV6 F1527) | 37° 9'25.21" N, 21°58'43.16" E | KY070400 | KY070433 (a); KY070454 (b) | KY070541 (a); KY070542 (b) | Pamissos 14 |
| Squalius peloponensis/Pamissos (NMP PV6 F1541) | 37° 9'25.21" N, 21°58'43.16" E | KY070401 | KY070455 (a); KY070456 (b) | KY070543 (a); KY070544 (b) | Pamissos 15 |
| Squalius peloponensis/Pamissos (G244) | 37°15'17.39"N, 21°53'45.15" E | KY070402 | KY070457 (a); KY070458 (b) | KY070545 (a); KY070546 (b) | Pamissos 16 |
| Squalius peloponensis/Pamissos (G245) | 37°15'17.39"N, 21°53'45.15" E | KY070403 | KY070459 (a); KY070460 (b) | KY070573 (a); KY070574 (b) | Pamissos 17 |
The study included the Evrotas River population, this being the type locality of S. keadicus and currently the only known locality for the species, and three populations recognized as S. peloponensis from the Alfios, Miras, and Pamissos basins (Fig 1). The type locality of S. peloponensis is uncertain, and only the Peloponnese peninsula (= Morea) is cited in its description [44]. Additional species of Squalius along its distribution range were included in the molecular analyses to assess the phylogenetic position of the four investigated populations (Table A in S1 File). The genus Petroleuciscus was used as the outgroup based on previous phylogenetic studies [35–36].
DNA extraction, amplification and sequencing
Eight to 17 specimens per population were analysed (Table 1). Total genomic DNA was isolated using the commercial kit Biosprint 15 for blood and tissue (Qiagen). For each specimen the complete mitochondrial cytochrome b (MT-CYB; 1140bp) gene and two nuclear genes, the first intron of the ribosomal S7 gene (S7; final alignment including gaps = 977bp) and the third exon of the recombination activating protein gene (RAG-1; 1473bp), were amplified. The PCR protocols and primers followed [36]. The PCR products were purified by Exo-SAP-IT (USB, Cleveland, OH, USA) and directly sequenced by Macrogen Europe (Amsterdam, The Netherlands; http://www.macrogen.com) using a 3730XL DNA sequencer. All new sequences of haplotypes and alleles obtained in this study were deposited in the GenBank database (Accession Numbers: MT-CYB: KY070368-KY070424; RAG1: KY070425-KY070574; S7: KY070503-KY070574).
Phylogenetic analyses
For the nuclear markers, phylogenetic inference of independent alleles was also conducted on gamete phases using the PHASE algorithm [45–46] implemented in DnaSP v. 5.0. [47] with a probability threshold of 0.9 to resolve alleles. Sequences were aligned using the default pairwise and multiple alignment parameters in Clustal W [48] implemented in MEGA v.7 [49]. Alignments were later revised. Recombination of nuclear genes was assessed by the PHI test [50] implemented in SplitsTree v. 4.13 [51]. No traces of recombination were found in either RAG-1 or S7 genes (p = 1.0 for both genes).
For phylogenetic analyses, the best-fit model of evolution for each independent gene was estimated using jModelTest v2 [52] (Table B in S1 File). Model parameters were used for subsequent phylogenetic analyses. Phylogenies based on Bayesian inference (BI) and maximum likelihood (ML) were constructed for independent genes in order to assess the phylogenetic position of each population based on mitochondrial and nuclear markers. BI was performed in MrBayes 3.2 [53]. Two simultaneous analyses were run for 10 million generations, each with four MCMC chains sampling every 1000 generations. Convergence was assessed using Tracer v.1.6 [54]. The 50% majority rule consensus tree was constructed after discarding the first 10% of generations as burn-in. For ML analyses, we used RaxML software implemented in the Trex-online server [55], employing the substitution model GTR+G+I for MT-CYB and GTR+G for nuclear genes and the rapid bootstrapping algorithm to estimate node confidence using different random seeds (1000 replicates) [56].
Uncorrected-p genetic distances between mitochondrial haplotypes and between nuclear phased alleles were estimated using the MEGA v.7 software [49].
Molecular clock
To establish a temporal framework for the introgression event, we estimated divergence time among the populations of Squalius keadicus (Evrotas R.) and the non-introgressed and introgressed populations of S. peloponensis using an uncorrelated relaxed molecular clock based on the MT-CYB gene implemented in BEAST v.2.0. [57]. To incorporate a speciation-model tree prior into the analyses, the birth-death model with incomplete taxon sampling [58], we used one specimen per species to estimate divergence time. For S. peloponensis, we used a specimen from the Alfios River (non-introgressed population) and one from the Miras River (introgressed population). As in the phylogenetic analysis, several species of the genus Petroleuciscus were used as outgroup. We calibrated the molecular clock using two fossil species of the genus Squalius (formerly within the genus Leuciscus) as lognormal prior: Leuciscus antunesi Gaudant 1977, found in a Portuguese deposit of the Middle Miocene period (13.5–14.5 Ma; [59–60]) and Leuciscus aff. cephalus from near the Aliakmon River in Greece (Lava 2 deposit) from Upper Miocene (6.56 Ma) [61]. The calibration point based on L. antunesi was allocated in the stem group of the Iberian Squalius species belonging to the Mediterranean lineage, whereas the age of the fossil Leuciscus aff. cephalus was placed in the crown of the current Squalius cephalus group. The MCMC analyses were run for 100 million generations, with parameters logged every 10000 generations. The remaining parameters were default parameters of the software. Convergence of parameters was evaluated using Tracer v.1.6 [54] and results were summarized in TreeAnnotator v.2.0 [57].
Morphological analyses
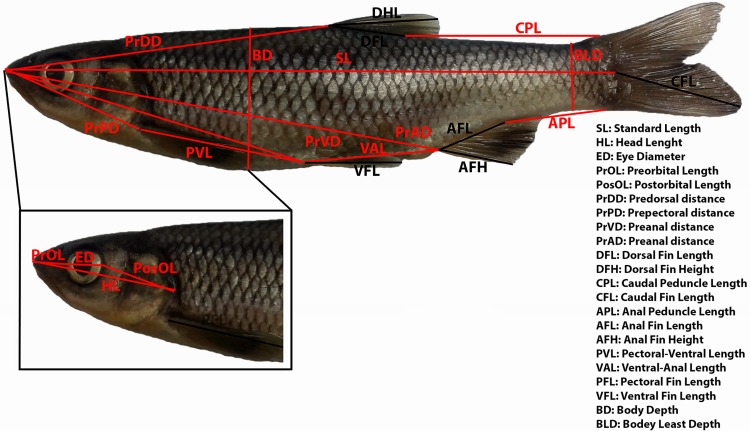
Morphological analyses included specimens from the Alfios, Miras, Pamissos and Evrotas basins. For each specimen, 23 morphometric and five meristic variables were recorded. All morphometric measurements were performed using digital callipers. Measurements and counts followed [62] and included standard length (SL), head length (HL), eye diameter (ED), interorbital width (IW), preorbital length (PrOL), postorbital length (PosOL), pre-dorsal distance (PrDD), pre-pectoral distance (PrPD), pre-ventral distance (PrPV), pre-anal distance (PrPA), caudal peduncle length (CPL), anal peduncle length (APL), pectoral-ventral length (PVL), ventral-anal length (VAL), body depth (BD), body least depth (BLD), dorsal fin height (DFH), dorsal fin length (DFL), anal fin height (AFH), anal fin length (AFL), pectoral fin length (PFL), ventral fin length (VFL), caudal fin length (CFL), lateral line scales (LLS), upper transverse scales (UTS), lower transverse scales (LTS), dorsal fin rays (D) and anal fin rays (A) (Fig 2).
Fig 2. Morphometric variables measured in the Squalius populations analysed.
After constructing the measurement matrix, Burnaby’s method was used to correct for size effect in morphometric variables [63–64]. A canonical variate analysis (CVA) was conducted to identify the variables most explanatory of the morphological variation among the analysed populations with the aim to maximize differences among populations. Non-parametric Kruskal–Wallis and Mann–Whitney post hoc comparisons were used to test for significant differences of morphological variables among populations. All analyses were performed using the Burnaby’s corrected matrix. Statistical analyses were carried out in PAST v.3.12 software [65].
Results
Molecular analyses
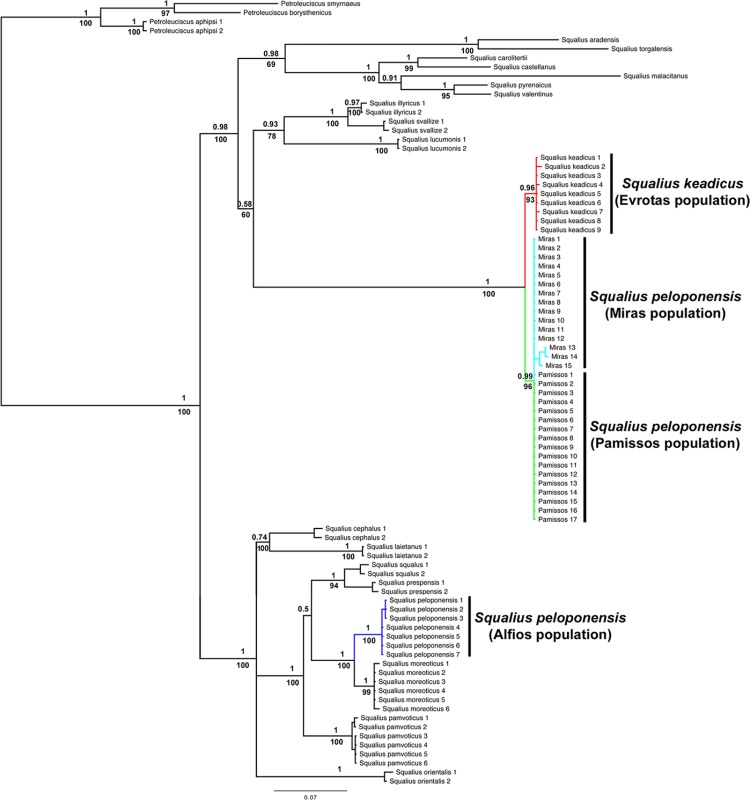
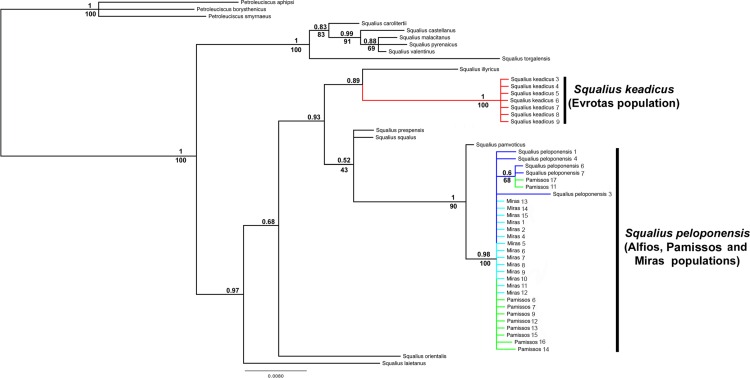
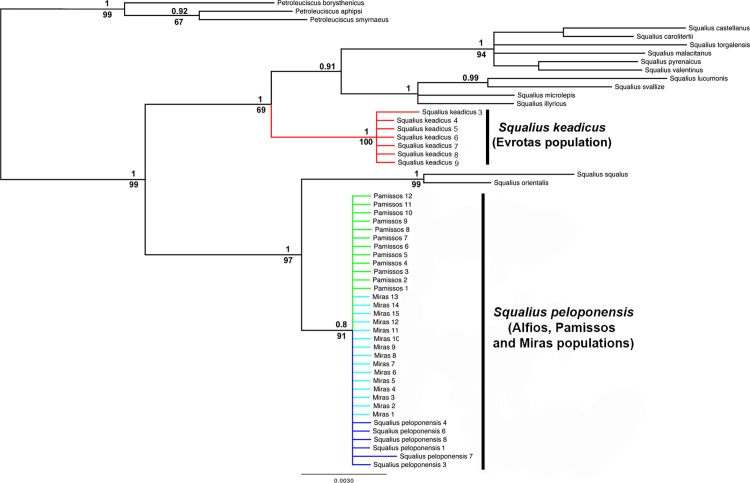
The result of mitochondrial phylogenetic analyses is shown in Fig 3. The Squalius populations from the Pamissos and Miras basins clustered together, forming a monophyletic clade, sister to S. keadicus. These two clades (S. keadicus and Pamissos/Miras S. peloponensis) were highly supported as monophyletic in the BI and ML analyses. Nuclear genes yielded tree topology inconsistent with that of the mitochondrial marker, with the Pamissos and Miras populations showing a highly supported cluster with S. peloponensis from the Alfios Basin, forming a monophyletic clade (Figs 4, 5 and 6). All phased alleles for the nuclear genes RAG1 and S7 from the Miras and Pamissos populations were nested within the clade of S. peloponensis and none of them was grouped with S. keadicus. No evidence of genetic structure was observed in the nuclear clade comprising the Alfios, Miras, and Pamissos populations.
Fig 3. Phylogenetic tree based on the MT-CYB gene.
Bootstrap values for ML analysis below branches. Posterior probability values for Bayesian inference above branches.
Fig 4. Phylogenetic tree based on the nuclear S7 gene.
Bootstrap values for ML analysis below branches. Posterior probability values for Bayesian inference above branches.
Fig 5. Phylogenetic tree based on the nuclear RAG1 gene.
Bootstrap values for ML analysis below branches. Posterior probability values for Bayesian inference above branches.
Fig 6.
Phylogenetic tree based on the phased nuclear RAG1 (A) and S7 (B) genes. Posterior probability values for Bayesian inference above branches. Blue = Alfios (S. peloponensis); Red = Evrotas (S. keadicus); Green = S. keadicus-like mitochondrial (Miras and Pamissos populations).
Uncorrected-p genetic distances between S. keadicus and the introgressed populations (Miras and Pamissos) for the three analysed genes are presented in Table 2. Table C in S1 File shows diagnostic autapomorphies characteristic of each population. For the MT-CYB, 115 autapomorphies (21 transversions) were recognized in the Alfios Basin population relative to the three other populations, five (1 transversion) were observed in S. keadicus and three in the introgressed populations, one of them present only in the Pamissos river system. For the RAG1 gene, 13 autapomorphies (4 transversions) were found in S. keadicus in relation to the three populations recognized as S. peloponensis. For the S7 gene, 23 autapomorphies (9 transversions) appeared in S. keadicus, as well as three insertions and three deletions not found in the other three populations.
Table 2. Uncorrected-p genetic distances (%) between (below diagonal) and within (in diagonal) populations for the three genes analysed.
| MT-CYB | ||||
| Evrotas | Pamissos | Miras | Alfios | |
| Evrotas | 0.01 | |||
| Pamissos | 0.6 | 0.00 | ||
| Miras | 0.6 | 0.1 | 0.01 | |
| Alfios | 11.0 | 10.7 | 10.8 | 0.01 |
| S7 | ||||
| Evrotas | Pamissos | Miras | Alfios | |
| Evrotas | 0.0 | |||
| Pamissos | 2.8 | 0.1 | ||
| Miras | 2.7 | 0.1 | 0.0 | |
| Alfios | 2.9 | 0.2 | 0.2 | 0.3 |
| RAG1 | ||||
| Evrotas | Pamissos | Miras | Alfios | |
| Evrotas | 0.0 | |||
| Pamissos | 1.0 | 0.1 | ||
| Miras | 1.0 | 0.2 | 0.1 | |
| Alfios | 1.0 | 0.1 | 0.2 | 0.1 |
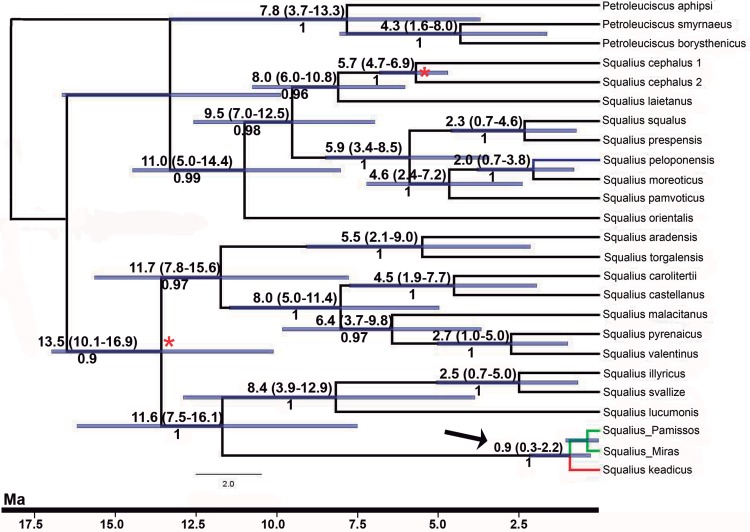
Estimated divergence times of the mitochondrial lineage of Squalius keadicus from that of the introgressed populations of S. peloponensis was approximately 0.9 Ma (CI 95% HPD: 0.3–2.2 Ma; Fig 7). The divergence of S. peloponensis from closely related Squalius spp occurred in Middle-Upper Pliocene. However, this phylogenetic relationship was not highly supported, probably as a consequence of an incomplete sampling of the Euroasiatic lineage of this genus sensu [35] (see also [37]).
Fig 7. Chronogram obtained for the MT-CYB gene for several Squalius species.
Divergence time estimates and their HPD 95% confidence intervals, in Ma, for the main cladogenetic events between Squalius species above branches. Posterior probability values of Bayesian inference below branches (** indicates posterior probability = 1). Black arrow highlights the divergence time estimated for the separation of mitochondrial lineages of S. keadicus and the introgressed populations. Red asterisk indicates the position of fossil ages used to calibrate the molecular clock.
Morphological analyses
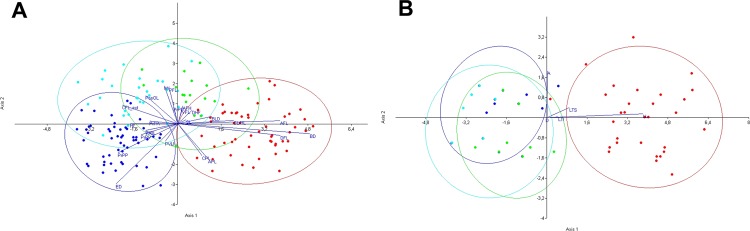
Kruskal–Wallis and Mann–Whitney post hoc analyses demonstrated significant differences among the four analysed Squalius populations in morphometric and meristic variables (Table 3; Fig 8). Canonical variate analysis (CVA) also supported morphological differences, showing four isolated groups with partial overlap between the populations from the Miras and Pamissos basins and between each of these populations with both the Evrotas (S. keadicus) and Alfios (S. peloponensis) populations. As expected, overlap between S. keadicus and the Alfios S. peloponensis population was not observed (Fig 9). The most contributory morphometric variables to the ordination in the CVA were AFL, BD, and DFL (Table 4).
Table 3. Kruskal-Wallis test and non-parametric Mann-Whitney pairwise post hoc comparisons for all populations.
Ev = Evrotas, Al = Alfios, Pa = Pamissos, Mi = Miras, Significant differences p < 0.01. Acronyms of variables are defined in the Material and Methods section.
| Variable | H | P-value | Significant Mann-Whitney pairwise comparisons |
|---|---|---|---|
| SL | 25.57 | <0.0001 | Al-Ev; Al-Pa |
| HL | 78.95 | <0.0001 | Al-Ev; Al-Pa; Ev-Mi; Ev-Pa |
| ED | 63.72 | <0.0001 | Al-Ev; Al-Pa; Al-Mi |
| IW | 56.48 | <0.0001 | Al-Ev; Al-Pa; Al-Mi; Ev-Pa; |
| PrOL | 19.51 | 0.0002 | Al-Ev |
| PosOL | 30.77 | <0.0001 | Al-Ev; Pa-Ev; Mi-Ev |
| PrPD | 11.38 | 0.0099 | - |
| PrPP | 92.4 | <0.0001 | Al-Ev; Al-Pa; Al-Mi; Ev-Mi; Pa-Mi |
| PrPV | 86.22 | <0.0001 | Al-Ev; Al-Pa; Al-Mi; Ev-Mi; Ev-Pa |
| PrPA | 59.86 | <0.0001 | Ev-Al; Ev-Pa; Ev-Mi |
| DHF | 13.39 | 0.0039 | Al-Ev |
| DFL | 84.98 | <0.0001 | Ev-Al; Ev-Pa; Ev-Mi; Al-Pa |
| CPL | 16.06 | 0.0011 | Ev-Mi |
| CFL | 46.87 | <0.0001 | Ev-Al; Ev-Mi |
| BLD | 16.6 | 0.0009 | Ev-Al; Ev-Mi |
| BD | 100.8 | <0.0001 | Ev-Al; Ev-Pa; Ev-Mi; Al-Pa |
| APL | 21.72 | <0.0001 | Ev-Mi |
| AFL | 56.89 | <0.0001 | Al-Ev; Al-Pa; Ev-Mi |
| AFH | 7.91 | 0.0479 | - |
| VAL | 17.37 | 0.0006 | Al-Ev |
| PVL | 4.794 | 0.1875 | - |
| VFL | 10.42 | 0.0154 | Al-Mi |
| PFL | 11.64 | 0.0087 | Mi-Al; Mi-Ev |
| LL | 106.6 | <0.0001 | Ev-Al; Ev-Pa; Ev-Mi |
| UTS | 74.29 | <0.0001 | Ev-Al; Ev-Pa; Ev-Mi; Al-Mi |
| LTS | 40.74 | <0.0001 | Ev-Al; Ev-Pa; Ev-Mi |
| D | 0.5093 | 0.9169 | - |
| A | 19.1 | 0.0002 | Al-Pa; Al-Mi |
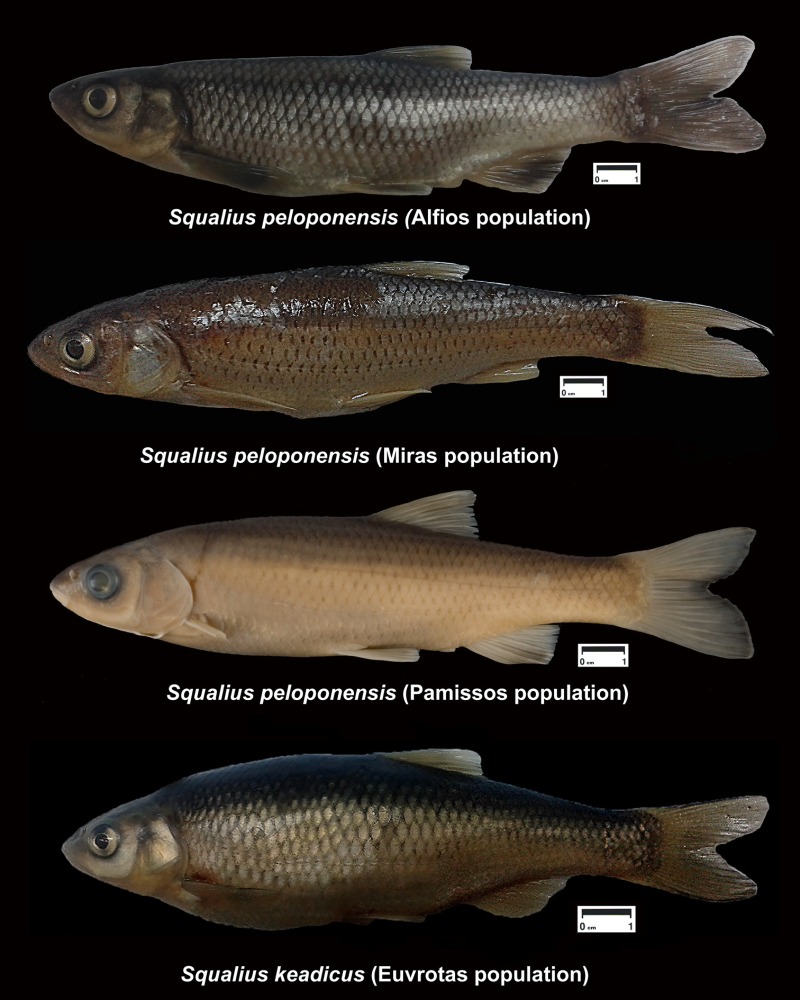
Fig 8. Specimens of the four analysed populations.
Voucher specimen numbers: Squalius peloponensis from Alfios River (MNCN_ICTIO 94.982); Squalius peloponensis from Pamissos River (NMP PV6 F1227); Squalius peloponensis from Miras River (MNCN_ICTIO 94.747); Squalius keadicus from Evrotas River (MNCN_ICTIO 123.875).
Fig 9. Canonical Variate Analysis of the four Squalius populations.
A) Morphometric measurements. B) Meristic measurements. Red: Evrotas population (S. keadicus), blue: Alfios population (S. peloponensis); light blue: Miras population (S. peloponensis); green: Pamissos population (S. peloponensis).
Table 4. Canonical Variate Analysis values for the first three canonical axes of analyses of morphometric and meristic variables.
Acronyms of variables are defined in the Material and Method section.
| VARIABLE | AXIS 1 | AXIS 2 | AXIS 3 |
|---|---|---|---|
| Eigenvalue 1 = 4.861 (77.76%) | |||
| Eigenvalue 2 = 0.9997 (15.99%) | |||
| SL | 0.0009 | 0.0008 | 0.0028 |
| HL | -0.0055 | 0.0003 | 0.0002 |
| ED | -0.0069 | -0.0094 | 0.0031 |
| IW | -0.0016 | 0.0060 | -0.0002 |
| PrOL | -0.0038 | -0.0014 | -0.0049 |
| PosOL | -0.0037 | 0.0048 | 0.0015 |
| PrPD | -0.0005 | 0.0029 | 0.0003 |
| PrPP | -0.0069 | -0.0043 | -0.0046 |
| PrPV | -0.0042 | -0.0015 | 0.0013 |
| PrPA | -0.0032 | 0.0006 | 0.0018 |
| DFH | 0.0017 | 0.0029 | -0.0030 |
| DFL | 0.0119 | -0.0015 | -0.0075 |
| CPL | 0.0027 | -0.0051 | 0.0019 |
| CFL | -0.0068 | 0.0021 | -0.0028 |
| BLD | 0.0040 | 0.0008 | 0.0029 |
| BD | 0.0153 | -0.0009 | -0.0042 |
| APL | 0.0032 | -0.0046 | 0.0085 |
| AFL | 0.0123 | -0.0002 | 0.0070 |
| AFH | 0.0006 | 0.0032 | -0.0044 |
| VAL | -0.0037 | -0.0012 | 0.0056 |
| VFL | 0.0004 | 0.0026 | -0.0064 |
| PVL | -0.0014 | -0.0033 | -0.0014 |
| PFL | -0.0009 | 0.0051 | -0.0028 |
| Eigenvalue 1 = 8.176 (97.3%) | |||
| Eigenvalue 2 = 0.21 (2.5%) | |||
| LL | 0.9484 | 0.0370 | -0.0734 |
| UTS | 0.2201 | 0.0981 | 0.4004 |
| LTS | 0.1131 | 0.0053 | -0.1368 |
| D | -0.0126 | -0.0023 | 0.0229 |
| A | 0.0051 | 0.4609 | -0.0231 |
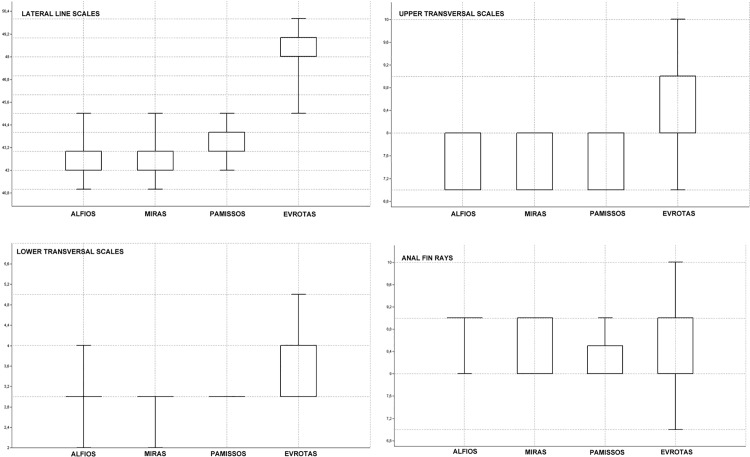
Meristic variables in the Alfios, Pamissos, and Miras populations overlapped with the Evrotas population (S. keadicus) forming a differentiated group in CVA (Fig 9). Kruskal–Wallis and Mann–Whitney post hoc comparisons were significant for LL, LTS, LTI, and A, reaching higher values in S. keadicus relative to the other three populations (Fig 10).
Fig 10. Box-plot of meristic variables.
The 25–75 percent quartiles are drawn inside the box. Short horizontal lines represent minimal and maximal values.
The Evrotas population showed greater BD and longer APL, DFL and AFL fins than the other three populations. It also exhibited significantly shorter HL relative to SL than in other populations (Table 5). The Pamissos population exhibited the significantly smaller ED relative to HL and SL than seen in the other three populations. The largest ED relative to the standard length was found in the S. peloponensis from Alfios (Table 5). Although the raw data matrix was corrected by Burnaby’s method, differences in ED may be attributed to allometric growth rather than to real population differences. The Alfios population had the narrowest head in relation to HL (Table 5), a ratio significantly different from the Evrotas and Miras populations, but not from that of the Pamissos.
Table 5. Significant proportions (%) relative to standard length (SL) or head length (HL) of several morphometric variables differentiating the four analysed populations.
| VARIABLE | POPULATION | % RELATIVE TO SL OR TO HL |
|---|---|---|
| BD relative to SL | Evrotas | 25.9 (24.0–28.7) |
| Alfios | 21.6 (21.2–21.5 | |
| Miras | 22.4 (21.1–22.7) | |
| Pamissos | 22.5 (23.0–24.7) | |
| DFL relative to SL | Evrotas | 12.6 (12.0–13.4) |
| Alfios | 11.1 (10.4–11.4) | |
| Miras | 11.3 (10.5–11.4) | |
| Pamissos | 11.5 (10.8–12.0) | |
| AFL relative to SL | Evrotas | 11.4 (8.9–12.6) |
| Alfios | 10.4 (8.7–12.3) | |
| Miras | 10.2 (9.3–10.0) | |
| Pamissos | 11.5 (10.5–12.7) | |
| APL relative to SL | Evrotas | 21.4 (21.0–21.7) |
| Alfios | 20.8 (20.9–21.1) | |
| Miras | 20.2 (18.0–18.9) | |
| Pamissos | 20.5 (19.9–19.9) | |
| HL relative to SL | Evrotas | 23.5 (24.4–24.6) |
| Alfios | 25.1 (24.9–26.3) | |
| Miras | 24.9 (23.5–26.8) | |
| Pamissos | 23.8 (23.6–26.0) | |
| ED relative to HL | Evrotas | 24.6 (24.0–25.5) |
| Alfios | 27.0 (24.8–28.9) | |
| Miras | 27.0 (24.4–29.5) | |
| Pamissos | 24.6 (24.0–25.5) | |
| ED relative to SL | Evrotas | 6.3 (6.0–7.2) |
| Alfios | 6.8 (6.2–7.6) | |
| Miras | 6.5 (5.5–6.6) | |
| Pamissos | 5.8 (5.9–6.6) | |
| IW relative to HL | Evrotas | 42.5 (39.2–40.2) |
| Alfios | 36.0 (35.4–39.6) | |
| Miras | 42.0 (39.7–41.6) | |
| Pamissos | 41.6 (41.2–41.9) |
Discussion
In this study we tested through morphological and molecular analyses the hypothesis of mitochondrial introgression between two species of the genus Squalius from Greece, S. keadicus and S. peloponensis, formulated on the basis of the discordance between mitochondrial (MT-CYB; [34]) and nuclear (allozymes; [33]) genomes found in one population from the Greek Peloponnese (from Miras Basin).
The populations from the Miras and Pamissos basins were morphologically similar to S. peloponensis from Alfios Basin, as expected based on sharing of the nuclear genome and the lack of diagnostic nuclear autapomorphies among the three populations (Table C in S1 File), as well as on meristic characters, which fall within the range of S. peloponensis (Fig 10). However, conserved meristic measurements are observed in Squalius species of the Euroasiatic lineage sensu [35] [37]. Although the populations from Miras and Pamissos were recognized as S. peloponensis significant differences between these two populations were found in some morphometric measurements, as well as between S. peloponensis and S. keadicus (Tables 3 and 5; Fig 9).
Whereas the Miras and Pamissos populations showed the nuclear genome and morphology typical of Squalius peloponensis, their mitochondrial genome clustered with S. keadicus (Evrotas population), but not with the S. peloponensis (Alfios population). This mitonuclear discordance supports the idea of mitochondrial introgression of S. keadicus in the Miras and Pamissos populations. Sharing of similar morphology and the nuclear genome allowed us to reject the hypothesis of Miras and Pamissos populations being hybrids of S. keadicus and S. peloponensis, since they should show similarity to both hypothetical parent species in the nuclear genome as well as in the mitochondrial genome. Therefore, the hypothesis of mitochondrial DNA introgression cannot be rejected based on our results. Hybridization is common in fish, and the boundaries between species may be obscure. In cyprinids, hybridization is a common evolutionary process [26, 66–69]. In our analyses, the two nuclear markers were S. peloponensis-like in introgressed populations, and we did not find evidence of recombination between species (PHI = 1 for both nuclear markers), as demonstrated by the phylogenetic relationships of phased alleles (Fig 6), and low (0.1–0.2%) genetic distance between populations in both nuclear markers. Hence, we accept the hypothesis of mitochondrial introgression and reject the hypothesis of hybrid speciation.
All analysed specimens from Miras and Pamissos were mitochondrially S. keadicus-like and nuclearly S. peloponensis-like, suggesting unidirectional complete mitochondrial introgression (mitochondrial capture) of S. keadicus to S. peloponensis [34]. Nonetheless, they formed an independent highly supported phylogenetic clade, sister to S. keadicus from the Evrotas population (Fig 3). These two highly supported reciprocal monophyletic groups found in the mitochondrial tree within the phylogenetic lineage of S. keadicus indicate that the introgression event was ancient, but it is not clear whether divergence of these populations, allowing mitochondrial DNA to sort, occurred before or after the introgression event. Ancient mitochondrial captures have also been described for reptiles [70–71], insects [18], and fishes [27–28]. Since these four Squalius populations currently occur in allopatry, a geographical barrier may be considered the primary source of their morphological differences possibly through a process of incipient speciation.
Isolation of populations and incipient speciation in sympatry can be attributed to sexual selection [72–73], divergent selection, and phenotypic plasticity [74], or segregation of ecological niches [75–76]. The morphological and molecular differentiation found among the analysed populations of Squalius from western Peloponnese probably occurred after their allopatric separation linked to the formation of the hydrological network in the region during the Upper Pliocene-Pleistocene periods [38, 43]. Incipient speciation or occurrence of differentiated lineages during the Pleistocene as a consequence of geographic barriers has been suggested for several taxa [77–79]. Examples of isolation-driven divergence to new lineages after ancient hybridization have been reported [19, 80]. This is demonstrated here by the lack of shared mitochondrial haplotypes among populations in the rivers of the western Peloponnese. The sharing of the nuclear genome among the Alfios, Miras, and Pamissos populations ascribed to S. peloponensis, clustering in the same clade, also supports the scenario of incipient speciation.
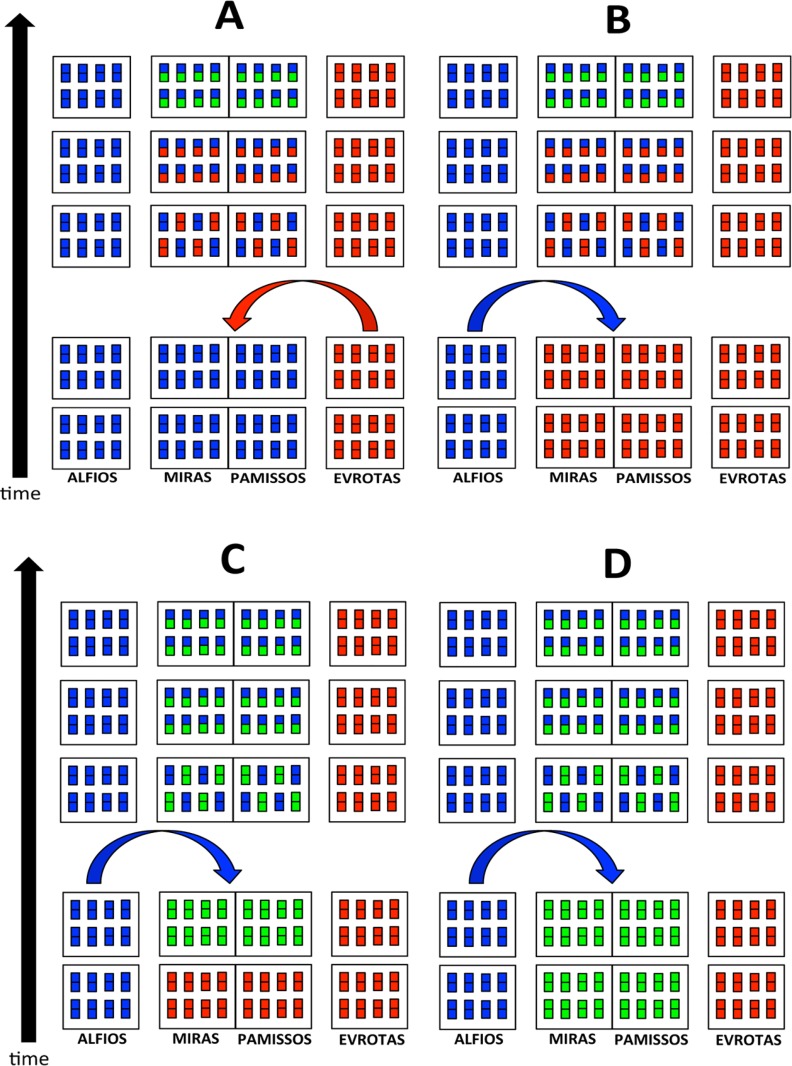
Several biogeographic scenarios may be proposed to explain the mitochondrial introgression event and the current molecular configuration of the four analysed populations (Fig 11): A) The lineage of Squalius peloponensis inhabited the Alfios, Miras, and Pamissos basins, and at some time the lineage of S. keadicus invaded the Miras and Pamissos basins. The introgression took place, and subsequently mitogenomes of the mitochondrial lineages of S. keadicus from the Evrotas basin and the introgressed populations of the Miras and Pamissos diverged. B) The lineage of Squalius keadicus inhabited the Evrotas, Miras, and Pamissos basins, and at some time the lineage of S. peloponensis invaded the Miras and Pamissos basins. The introgression event took place, mitogenomes of the lineages of S. keadicus from the Evrotas basins and the introgressed Miras and Pamissos populations subsequently diverged. C) Miras and Pamissos basins were inhabited by the lineage of Squalius keadicus and the mitogenome of these populations diverged from that of the Evrotas population. The lineage of S. peloponensis then invaded the Miras and Pamissos basins and introgression occurred. D) A case similar to scenario C is that in which it was not the mitochondrial lineage of Squalius keadicus that inhabited the Miras and Pamissos basins prior to the invasion by the lineage of S. peloponensis, but a phylogenetically closely related species.
Fig 11. Hypothetical biogeographical scenarios explaining the mitochondrial capture event from Squalius keadicus to Squalius peloponensis.
Each rectangle symbolizes a hypothetical individual within the analysed population. The two squares within each rectangle symbolize the mitochondrial and the nuclear genome of this hypothetical individual in the population. Colours indicate populations: Blue = S. peloponensis lineage; Red = S. keadicus lineage; Green = S. keadicus-like mitochondrial lineage (Miras and Pamissos populations).
The main difference in these biogeographical scenarios is the timing of the introgression event as before (scenarios A and B) or after (scenarios C and D) the divergence of the S. keadicus from the S. keadicus-like mitogenome. The common factor among the hypothetical scenarios is the biogeographical discordance between mitochondrial and nuclear DNA [4]. Most patterns of biogeographic mitonuclear discordance are characterized by previously isolated lineages that came into contact and generated hybrid zones in which gene flow patterns in mitochondrial and nuclear DNA differed [4]. This hypothesis of secondary contact is consistent with the hypothetical biogeographical scenarios that we propose, which differ only whether the invader species is S. keadicus (A) or S. peloponensis (B, C, and D). Differences in effective population size of the resident and the invader species could be responsible for the unidirectionality of the introgression event found in the present study [2, 4, 81]. In the initial stages of an invasion event, a small number of invader specimens are expected in comparison to the resident population, and simulated studies have revealed unidirectional introgression of neutral loci from the resident to the invader population [21]. This would imply that S. keadicus (B, C) or S. keadicus-like (D) was the resident species and S. peloponensis the invader. Nevertheless, if the invader becomes more numerous as invasion continues, the direction of the introgression will depend of the demographic balance of the resident and invader populations [21, 82]. Mitochondrial DNA introgression as a consequence of extreme differences in population size has been described in other freshwater fish species [75]. In our hypothetical scenarios, S. keadicus or an extinct species phylogenetically similar to S. keadicus, regardless of whether it was the resident or the invader species, must have had a population large enough to erode the mitochondrial genome of S. peloponensis to the point of elimination. Another possible source of mitonuclear discordance may be sex-biased reproduction or sex-biased offspring production during the hybridization event [4]. Empirical experiments are necessary to assess this hypothesis.
Differences in distribution range of two species involved in an introgression event when a hybrid zone is formed explain asymmetric mitochondrial introgression, associated with differences in population size and competition between their mitochondrial genomes [4]. It is not possible to estimate the range of S. keadicus or the size of the hybrid zone in the Miras and Pamissos basins, because S. keadicus, or the S. keadicus-like species, is currently extinct in this region and no fossil evidence exists, but it is clear that the distribution range of the studied species overlapped in the past.
Although the mitochondrial genome is generally considered to be neutral [11], positive mitochondrial selection has been described for this marker [20], and is another possible explanation of the observed unidirectional mitochondrial introgression. Thus, the mitochondrial genome with the highest fitness may be introgressed into the other species, regardless of whether it is resident or invader [81]. These selective sweeps have been attributed to climatic adaptation, especially to temperature [83]. Our study area, however, was restricted to a small geographical region with no significant climatic differences in river systems. Furthermore, the HKA test of selection [84] performed on the MT-CYB gene (p>0.05) suggested the potential for neutral evolution in this gene, and a selective sweep is not indicated in the studied species.
The most recent common ancestor of the mitochondrion of the Evrotas S. keadicus and the introgressed populations of the Miras and Pamissos basins dated from 0.9 Ma (CI 95% HPD 0.3–2.2 Ma) in the Pleistocene, and introgression may have occurred before or after divergence. Mitochondrial captures during Pleistocene have been described for other freshwater fish species [28]. The divergence of the mitochondrial clades in the present study may be attributed to Pleistocene tectonic movements related to uplift and river system formation. The paleogeographical history of western Greece and the Peloponnese is complex; the area comprises a network of active faults that constitutes a multifractured neotectonic macrostructure [38]. Tectonic movements occurred throughout the Upper Pliocene-Pleistocene period and continue to the present causing earthquakes and tsunamis [39, 85]. The faults had great influence on the drainage network formation [38]. The current hydrological basins of the western Peloponnese were formed during the Pleistocene as a consequence of marine or continental sediment infilling when fault systems were activated. Marine sediments found at several hundred metres elevation in some mountainous areas are evidence of vertical tectonic movements throughout this period [38, 86]. These sedimentary events have contributed to configuration of the main fluvial basins of western Peloponnese, which show a radial pattern commonly found in areas of rapid uplifting [43].
The recent paleogeographic events may be responsible for the divergence of mitochondrial lineages of S. keadicus and the introgressed populations (mitochondrial S. keadicus–like) of S. peloponensis as well as morphological differences among the four populations that evolved following the isolation of the hydrological basins (Fig 11). Complex tectonic movements configuring the hydrological network of western Peloponnese may also have been the source of contact between species and the introgression event, through river piracy.
Conclusions
Molecular and morphological evidence supports the hypothesis of mitochondrial introgression of S. keadicus to S. peloponensis. We found unidirectional mitochondrial introgression from S. keadicus into populations from two basins of the western Peloponnese, the Miras and Pamissos River systems, but not into that from the Alfios Basin, and a sorting of the mitochondrial genome of introgressed populations from that of S. keadicus, likely as a consequence of isolation by biogeographical barriers, leading to a process of incipient speciation. Recent secondary contact among basins is the most plausible hypothesis to explain the mitochondrial introgression.
The identification of mitochondrial capture is crucial to understanding the evolutionary history of living organisms, and adequate molecular and morphological analyses are essential to distinguish among evolutionary processes leading to mitonuclear discordance, such as the case of mitochondrial introgression.
Supporting Information
Table A. Additional species used in phylogenetic performance. GenBank accession numbers and labels in phylogenetic trees. Table B. Evolutionary models estimated by jModelTest for the three analysed genes. Table C. Autapomorphies in the three analysed genes. * transversions.
(DOCX)
Acknowledgments
We thank I. Karakousis, P. Economidis, P. Garzón, A. Perdices and J.A. Carmona for field assistance. The Lucidus Consultancy helped us with English editing of the manuscript.
Data Availability
All new sequences of haplotypes and alleles obtained in this study were deposited in the GenBank database (Accession numbers: MT-CYB: KY070368-KY070424; RAG1: KY070425-KY070574; S7: KY070503-KY070574). All other relevant data are included within the manuscript and its Supporting Information files.
Funding Statement
This study was financially sponsored by the Spanish Ministry of Economy and Competitiveness (project CGL2013-41375-P) to ID and SP, the Czech Science Foundation (project GA15-19382S) to JV and RS, and by the SYNTHESYS Programme project ES-TAF-5255 at the Museo Nacional de Ciencias Naturales (CSIC), financed by European Community Research Infrastructure Action under the FP7 "Capacities" Programme to RS.
References
- 1.Zink RM, Barraclough G. Mitochondrial DNA under siege in avian phylogeography. Mol. Ecol. 2008; 17: 2107–2121. 10.1111/j.1365-294X.2008.03737.x [DOI] [PubMed] [Google Scholar]
- 2.Funk DJ, Omland KE. Species-level paraphyly and polyphyly: Frequency, causes, and consequences, with insights from Animal Mitochondrial DNA. Annu. Rev. Ecol. Evol. S. 2003; 34: 397–423. [Google Scholar]
- 3.Chan KMA, Levin SA. Leaky prezygotic isolation and porous genomes: rapid introgression of maternally inheritance DNA. Evolution, 2005; 59: 720–729. [PubMed] [Google Scholar]
- 4.Toews DPL, Brelsford A. The biogeography of mitochondrial and nuclear discordance in animals. Mol. Ecol. 2012; 21: 3907–3930. 10.1111/j.1365-294X.2012.05664.x [DOI] [PubMed] [Google Scholar]
- 5.Joly S, McLenachan PA, Lockhart PJ. A statistical approach for distinguishing hybridization and incomplete lineage sorting. Am. Nat. 2009; 174: 54–70. [DOI] [PubMed] [Google Scholar]
- 6.Maddison WP. Gene trees in species trees. Syst. Biol. 1997; 46(3): 523–536. [Google Scholar]
- 7.Degnan JH, Rosenberg NA. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol. Evol. 2009; 24(6): 332–340. 10.1016/j.tree.2009.01.009 [DOI] [PubMed] [Google Scholar]
- 8.Edwards S, Bensch S. Looking forwards or looking backwards in avian phylogeography? A comment on. Mol. Ecol. 2009; 18(14): 2930–2933. 10.1111/j.1365-294X.2009.04270.x [DOI] [PubMed] [Google Scholar]
- 9.Rosenberg NA. 2013. Discordance of species trees with their most likely gene trees: a unifying principle. Mol. Biol. Evol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng C, Kubatko LS. Detecting hybrid speciation in the presence of incomplete lineage sorting using gene tree incongruence: A model. Theor. Popul. Biol. 2009; 75(1): 35–45. 10.1016/j.tpb.2008.10.004 [DOI] [PubMed] [Google Scholar]
- 11.McKay BD, Zink RM. The causes of mitochondrial DNA gene tree paraphyly in birds. Mol. Phylogenet. Evol. 2010; 54: 647–650. 10.1016/j.ympev.2009.08.024 [DOI] [PubMed] [Google Scholar]
- 12.Yu Y, Than C, Degnan JH, Nakleh L. Coalescent histories on phylogenetic networks and detection of hybridization despite incomplete lineage sorting. Syst. Biol. 2011; 60(2): 138–149. 10.1093/sysbio/syq084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jan CC, Fernández-Martínez JM. Interespecific hybridization, gene transfer and the development of resistance to the broomrape race F in Spain. Helia. 2002; 25(36): 123–136. [Google Scholar]
- 14.Cunha C, Coelho MM, Carmona JA, Doadrio I. Phylogeographical insights into the origins of the Squalius alburnoides complex via multiple hybridization events. Mol. Ecol. 2004; 13(9): 2807–2817. 10.1111/j.1365-294X.2004.02283.x [DOI] [PubMed] [Google Scholar]
- 15.Sousa-Santos C, Collares-Pereira MJ, Almada VC. Evidence of extensive mitocondrial introgression with nearly complete substitution of the typical Squalius pyrenaicus-like mtDNA of the Squalius alburnoides complex (Cyprinidae) in an independent Iberian lineage. J. Fish. Biol. 2006; 68(SB): 292–301. [Google Scholar]
- 16.Rhymer JM, Simberloff D. Extinction by hybridization and introgression. Annu. Rev. Ecol. Syst. 1996; 27: 83–109. [Google Scholar]
- 17.Weisrock DWK, Hozak H, Larson A. Phylogeographic analysis of mitochondrial gene flow and introgression in the salamander Plethodon shermani. Mol. Ecol. 2005; 14: 1457–1472. 10.1111/j.1365-294X.2005.02524.x [DOI] [PubMed] [Google Scholar]
- 18.Linnen CR, Farrell BD. Mitonuclear discordance is caused by rampant mitochondrial introgression in Neodiprion (Hymenoptera: Diprionidae) sawflies. Evolution, 2007; 61(6): 1417–1438. 10.1111/j.1558-5646.2007.00114.x [DOI] [PubMed] [Google Scholar]
- 19.Bryson RW Jr, Smith BT, Nieto-Montes de Oca A, García-Vázquez UO, Riddle BR. The role of mitochondrial introgression in illuminating the evolutionary history of Neartic treefrogs. Zool. J. Linn. Soc. 2014; 172: 103–116. [Google Scholar]
- 20.Ballard JWO, Whitlock MC. The incomplete natural history of mitochondria. Mol. Ecol. 2004; 13: 729–744. [DOI] [PubMed] [Google Scholar]
- 21.Currat M, Ruedi M, Petit RJ, Excoffier L. The hidden side of invasions: massive introgression by local genes. Evolution, 2008; 62(8): 1908–1920. 10.1111/j.1558-5646.2008.00413.x [DOI] [PubMed] [Google Scholar]
- 22.Good JM, Hird S, Reid N, Demboski JR, Steppan SJ, Martin-Nims TR, et al. Ancient hybridization and mitochondrial capture between two species of chipmunks. Mol. Ecol. 2008; 17: 1313–1327. 10.1111/j.1365-294X.2007.03640.x [DOI] [PubMed] [Google Scholar]
- 23.Rabosky DL, Talaba AL, Donnellan SC. Lovette IJ. Molecular evidence for hybridization between two Australian desert skinks, Ctenotus leonhardii and Ctenotus quattuordecimlineatus (Scindidae: Squamata). Mol. Phylogenet. Evol. 2009; 53: 368–377. 10.1016/j.ympev.2009.06.020 [DOI] [PubMed] [Google Scholar]
- 24.Marková S, Dufresne F, Manca M, Kotlík P. Mitochondrial capture misleads about ecological speciation in Daphnia pulex complex. PLoS One, 2013; 8(7): e69497 10.1371/journal.pone.0069497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willis SC, Farias IP, Ortí G. Testing mitochondrial capture and deep coalescence in Amazonian cichlid fishes (Cichlidae: Cichla). Evolution 2014; 68: 256–268. 10.1111/evo.12230 [DOI] [PubMed] [Google Scholar]
- 26.Freyhof J, Dietmar L, Pitra C, Ludwig A. Molecules and morphology: Evidence for introgression of mitochondrial DNA in Dalmatian cyprinids. Mol. Phylogenet. Evol. 2005; 37: 347–354. 10.1016/j.ympev.2005.07.018 [DOI] [PubMed] [Google Scholar]
- 27.Nevado B, Koblmüller S, Sturmbauer C, Snoeks J, Usano-Alemany J, Verheyen E. Complete mitochondrial DNA replacement in a Lake Tanganyia cichlid fish. Mol. Ecol. 2009; 18: 4240–4255. 10.1111/j.1365-294X.2009.04348.x [DOI] [PubMed] [Google Scholar]
- 28.Tang Q-Y, Liu S-Q, Yu D, Liu H-Z, Danley PD. Mitochondrial capture and incomplete lineage sorting in the diversification of balitorine loaches (Cypriniformes, Balitoridae) revealed by mitochondrial and nuclear genes. Zool. Scr. 2012; 41(4): 233–247. [Google Scholar]
- 29.Costedoat C, Pech N, Chappaz R, Gilles A. Novelties in hybrid zones: Crossroads between population genomic and ecological approaches. 2007. PLoS One. 2007; 2(4): e357 10.1371/journal.pone.0000357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aboim MA, Mavárez J, Bernatchez L, Coelho MM. Introgressive hybridization between two Iberian endemic cyprinid fish: a comparison between two independent hybrid zones. J. Evol. Biol. 2010; 23(4): 817–828. 10.1111/j.1420-9101.2010.01953.x [DOI] [PubMed] [Google Scholar]
- 31.Sinama M, Gilles A, Costedoat C, Corse E, Olivier J- M, Chappaz R, et al. Non-homogeneus combination of two porous genomes induces complex body shape trajectories in cyprinid hybrids. Front. Zool. 2013; 10:22 10.1186/1742-9994-10-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geiger M, Schreiner C, Delmastro GB, Herder F. Combining geometric morphometrics with molecular genetics to investigate a putative hybrid complex: a case study with barbels Barbus spp. (Teleostei: Cyprinidae). J. Fish Biol. 2016; 88(3): 1038–1055. 10.1111/jfb.12871 [DOI] [PubMed] [Google Scholar]
- 33.Doadrio I, Carmona JA. Genetic diversity in Greek of the genus Leuciscus and its evolutionary and biogeographical implications. J. Fish Biol. 1998; 53(3): 591–613. [Google Scholar]
- 34.Durand JD, Ünlu E, Doadrio I, Pipoyan S, Templeton AR. Origin, radiation, dispersion and allopatric hybridization in the chub Leuciscus cephalus. Proc. R. Soc. Biol. Sci. 2000; 267: 1687–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanjur O, Carmona JA, Doadrio I. Molecular phylogeny of Iberian chub (genus Squalius, Cyprinidae) inferred from molecular data. Mol. Phylogenet. Evol. 2003; 29: 20–30. [DOI] [PubMed] [Google Scholar]
- 36.Perea S, Böhme M, Zupančič P, Freyhof J, Šanda R, Özulug M, et al. Phylogenetic relationships and biogeographical patterns in Circum-Mediterranean subfamily Leuciscinae (Teleostei, Cyprinidae) inferred from both mitochondrial and nuclear data. BMC Evol. Biol. 2010; 10: 265 10.1186/1471-2148-10-265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kottelat M, Freyhof J. Handbook of the European Freshwater Fishes. Cornol. Berlin. 2007.
- 38.Fountoulis I, Mariolakos I, Ladas I. Quaternary basin sedimentation and geodynamics in SW Peloponnese (Greece) and late stage uplift of Taygetos Mt. B. Geofis. Teor. Appl. 2014; 55(2): 303–324. [Google Scholar]
- 39.Kokkalas S, Xypolias P, Koukovelas I, Doutsos T. Postcollisional contractional and extensional deformation in the Aegean region. In: Dilek Y, Pavlides S, eds. Post Collisional Tectonics & Magmatism in the Mediterranean Region and Asia. Geol. S. Am. S. 2006; 409, 97–123. [Google Scholar]
- 40.Papanikolau D, Fountoulis I, Metaxas C. 2007. Active faults, deformation rates and Quaternary paleogeography at Kyparissiakos Gulf (SW Greece) deduced from onshore and offshore data. Quat. Int. 172: 14–30. [Google Scholar]
- 41.Fountoulis I, Mariolakos I. Neotectonic folds in the central-western Peloponnese, Greece. Z. Dtsch. Ges. Geowiss 2008; 159(3): 485–494. [Google Scholar]
- 42.Koukouvelas I, Kokkalas S, Xypolias P. Surface deformation during the Mw 6.4 (8 June 2008) Movri Mountain earthquarke in the Peloponnese, and its implications for the seismotectonics of western Greece. Inter. Geol. Rev. 2009; 52(2–3): 249–268. [Google Scholar]
- 43.Athanassas C, Fountoulis I. Quaternary neotectonic configuration of the southwestern Peloponnese, Greece, based on luminescence ages of marine terraces. J. Earth Sci. 2013; 24(3): 410–427. [Google Scholar]
- 44.Cuvier G, Valenciennes A. Histoire naturelle des poisons. Tome dix-septième. Suite du livre dix-huitième. Cyprinoïdes. v.17: i-xxiii + 1–497 + 2 pp.; 1844. p. 487–519. [Google Scholar]
- 45.Stephens M, Smith N, Donelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001; 68: 978–989. 10.1086/319501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stephens M, Donelly P. 2003. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. American Journal of Human Genetics, 73: 1162–1169. 10.1086/379378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Librado P, Rozas J. DnaSP v.5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 2009; 25: 1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 48.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994; 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar S, Stecher G, Tamura K. MEGA v.7: Molecular Evolutionary Genetics Analysis Version 7. Mol. Biol. Evol. 2015. (submitted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bruen TC, Phillipe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics 2006; 172(4): 2665–2681. 10.1534/genetics.105.048975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huson DH, Bryant D. Application of phylogenetic networks in Evolutionary Studies. Mol. Biol. Evol. 2006; 23(2): 254–267. 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- 52.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 2012; 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012; 61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rambaut A, Drummond AJ. Tracer v.1.6. Available from http://beast.bio.ed.ac.uk./Tracer. 2007.
- 55.Stamakis A. RaxML-VI-HPC: Maximum Likelihood-based phylogenetic Analyses with thousands of taxa and Mixed Models. Bioinformatics, 2006; 22(21): 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- 56.Stamakis A, Blagojevic F, Nikolopoulos D, Antonopoulos C. Exploring new search algorithms and hadware for phylogenetics: RaxML Meets the IBM Cell. The Journal of VLSI Signal Processing, 2007; 48: 271–286. [Google Scholar]
- 57.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, et al. BEAST 2: A Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2014; 10(4): e1003537 10.1371/journal.pcbi.1003537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stadler T. On incomplete taxon sampling under birth-death models and connection to the sampling-based coalescent. J. Theor. Biol. 2009; 261: 58–66. 10.1016/j.jtbi.2009.07.018 [DOI] [PubMed] [Google Scholar]
- 59.Gaudant J. Contributions à la Paléontologie du Miocène moyen continental du bassin du Tage. II-Observations sur les dents pharyngiennes de Poissons Cyprinidés-Póvoa de Santarém. Ciencias da Terra, 1977; 3: 129–141. [Google Scholar]
- 60.Cabrera L, Gaudant J. 1985. Los Ciprínidos (Pisces) del sistema lacustre Oligocénico-Miocénico de los Monegros (secto SE de la cuenca del Ebro, provincias de Lleida, Tarragona, Huesca y Zaragoza). Acta Geológica Hispánica, 1985; 20(3–4): 219–226. [Google Scholar]
- 61.Böhme, M, Ilg, A. 2003. fosFARbase, www.wahre-staerke.com/ (Accessed April 2016).
- 62.Doadrio I, Perea S, Alonso F. A new species of the genus Squalius Bonaparte, 1837 in the Tagus Basin (Central Spain). Graellsia, 2007; 63(1): 89–100. [Google Scholar]
- 63.Burnaby TP. Growth-invariant discriminant functions and generalized distances. Biometrics, 1966; 22: 96–110. [Google Scholar]
- 64.Rohlf FJ, Bookstein FL. A comment on shearing as a method for “size correction”. Syst. Zool. 1987; 36: 357–367. [Google Scholar]
- 65.Hammer Ø, Harper DAT, Ryan D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Paleontological Electronica, 2001. 41(1): 9pp. [Google Scholar]
- 66.Machordom A, Berrebi P, Doadrio I. Spanish barbell hybridization detected using enzymatic markers: Barbus meridionalis Risso x Barbus haasi Mertens (Osteichthyes, Cyprinidae). Aquat. Living Resour. 1990: 3(4): 295–303. [Google Scholar]
- 67.Cunha C, Bastir M, Coelho MM, Doadrio I. Body shape evolution among ploidy levels of the Squalius alburnoides hybrid complex (Teleostei, Cyprinidae). J. Evol. Biol. 2004; 22: 718–728. [DOI] [PubMed] [Google Scholar]
- 68.Aguilar A, Jones WJ. Nuclear and mitochondrial diversification in two native California minnows: insights into taxonomic identity and regional Phylogeography. Mol. Phylogenet. Evol. 2009; 51: 373–381. [DOI] [PubMed] [Google Scholar]
- 69.Broughton RE, Vedala KC, Crowl TM, Ritterhouse LL. Current and historical hybridization with differential introgression among three species of cyprinid fishes (genus Cyprinella). Genetica 2011; 139: 699–707. 10.1007/s10709-011-9578-9 [DOI] [PubMed] [Google Scholar]
- 70.Leaché AD, McGuire JA. Phylogenetic relationships of horned lizards (Phrynosoma) based on nuclear and mitochondrial data: Evidence for a misleading mitochondrial gene tree. Mol. Phylogenet. Evol. 2006; 39(3): 628–644. 10.1016/j.ympev.2005.12.016 [DOI] [PubMed] [Google Scholar]
- 71.Barbanera F, Zuffi MAL, Guerrini M, Gentilli A, Tofanelli S, Fasola M, et al. Molecular Phylogeography of the asp viper Vipera aspis (Linnaeus, 1758) in Italy: evidence for introgressive hybridization and mitochondrial DNA capture. Mol. Phylogenet. Evol. 2009; 52: 103–114. 10.1016/j.ympev.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 72.Alexander HJ, Breden F. Sexual isolation and extreme morphological divergence in the Cumaná guppy: a possible case of incipient speciation. J. Evol. Biol. 2004; 17: 1238–1254. 10.1111/j.1420-9101.2004.00788.x [DOI] [PubMed] [Google Scholar]
- 73.Meyer A, Salzburger W, Schartl M. Hybrid origin of a swordtail species (Teleostei: Xiphophorus clemenciae) driven by sexual selection. Mol. Ecol. 2006; 15: 721–730. 10.1111/j.1365-294X.2006.02810.x [DOI] [PubMed] [Google Scholar]
- 74.Magalhaes IS, Mwaiko S, Schneider V, Seehaunsen O. Divergent selection and phenotypic plasticity during incipient speciation in Lake Victoria cichlid fish. J. Evol. Biol. 2009; 22: 260–274. 10.1111/j.1420-9101.2008.01637.x [DOI] [PubMed] [Google Scholar]
- 75.Knudsen R, Klemetsen A, Amundsen P-A, Hermansen B. Incipient speciation through niche expansion: an example from the artic charr in a subartic lake. Proc. R. Soc. Biol. Sci. 2006; 273: 2291–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nosil P, Harmon LJ, Seehausen O. Ecological explanations for (incomplete) speciation. Trends Ecol. Evol. 2009; 24(3): 145–156. 10.1016/j.tree.2008.10.011 [DOI] [PubMed] [Google Scholar]
- 77.Perdices A, Doadrio I, Economidis PS, Bohlen J, Doadrio I. Pleistocene effects on the European freshwater fish fauna: double origin of the cobitid genus Sabanejewia in the Danube basin (Osteichthyes, Cobitidae). Mol. Phylogenet. Evol. 2003. 26: 289–299. [DOI] [PubMed] [Google Scholar]
- 78.Vila M, Vidal-Romaní JR, Björklund M. The importance of time scale and multiple refugia: incipient speciation and admixture of lineages in the butterfly Erebia triaria (Nymphalidae). Mol. Phylogenet. Evol. 2005; 36: 249–260. 10.1016/j.ympev.2005.02.019 [DOI] [PubMed] [Google Scholar]
- 79.Bohlen J, Perdices A, Doadrio I, Economidisi PS. Vicariance, colonisation, and fast local speciation in Asia Minor and the Balkans as revealed from the phylogeny of spined loaches (Osteichthyes; Cobitidae). Mol. Phylogenet. Evol. 2006; 39: 552–561. 10.1016/j.ympev.2005.12.007 [DOI] [PubMed] [Google Scholar]
- 80.Bryson RW Jr, Nieto-Montes de Oca A, Jaeger JR, Riddle BR. Elucidation of cryptic diversity in a widespread Neartic treefrog reveals episodes of mitochondrial gene capture as frogs diversified across a dynamic landscape. Evolution, 2010; 64: 2315–2330. 10.1111/j.1558-5646.2010.01014.x [DOI] [PubMed] [Google Scholar]
- 81.Rheindt FE, Edwards SV. Genetic introgression: an integral but neglected component of speciation in birds. The Auk 2011; 128: 620–632. [Google Scholar]
- 82.Ludwig A, Congiu L, Pitra C. Nonconcordant evolutionary history of maternal and paternal lineages in Adriatic sturgeon. Mol. Ecol. 2003. 12: 3253–3264. [DOI] [PubMed] [Google Scholar]
- 83.Cheviron ZA, Brumfield RT. Migration-selection balance and local adaptation of mitochondrial haplotypes in Rufous-collared sparrows (Zonotrichia capensis) along an elevational gradient. Evolution 2009; 63: 1593–1605. 10.1111/j.1558-5646.2009.00644.x [DOI] [PubMed] [Google Scholar]
- 84.Hudson RR, Kreitman M, Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics 1987; 116: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ntageretzis K, Vött A, Fischer P, Hadler H, Emde K, Röloke BR, et al. Paleotsunami history of the Eos Plain (Evrotas River delta, Peloponnese, Greece). Z. Geomorphol. 2015; 59(4): 253–273. [Google Scholar]
- 86.Athanassas C. Optically stimulated luminescence dating of Late Quaternary marine terraces, sea-level changes and coastal neotectonics in W. Messinia (SW Peloponnese, Greece). Unpublished D. Phil. Thesis. Faculty of Geology and Geoenvironment. National and Kapodistrian University of Athens. Greece. (In Greek). 2010.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table A. Additional species used in phylogenetic performance. GenBank accession numbers and labels in phylogenetic trees. Table B. Evolutionary models estimated by jModelTest for the three analysed genes. Table C. Autapomorphies in the three analysed genes. * transversions.
(DOCX)
Data Availability Statement
All new sequences of haplotypes and alleles obtained in this study were deposited in the GenBank database (Accession numbers: MT-CYB: KY070368-KY070424; RAG1: KY070425-KY070574; S7: KY070503-KY070574). All other relevant data are included within the manuscript and its Supporting Information files.