Abstract
PURPOSE
Whole genome sequencing (WGS) can be used as a powerful diagnostic tool which could also be used for screening but may generate anxiety, unnecessary testing and overtreatment. Current guidelines suggest reporting clinically actionable secondary findings when diagnostic testing is performed. We estimated preferences for receiving WGS results.
METHODS
A US nationally representative survey (n=410 adults) was used to rank preferences for who decides (expert panel, your doctor, you) which WGS results are reported. We estimated the value of information about variants with varying levels of clinical usefulness using willingness-to-pay contingent valuation questions.
RESULTS
43% preferred to decide themselves what information is included in the WGS report. 38% (95% CI:33–43%) would not pay for actionable variants, and 3% (95% CI:1–5%) would pay more than $1000. 55% (95% CI:50–60%) would not pay for variants in which medical treatment is currently unclear, and 7% (95% CI:5–9%) would pay more than $400.
CONCLUSION
Most people prefer to decide what WGS results are reported. Despite valuing actionable information more, some respondents perceive that genetic information could negatively impact them. Preference heterogeneity for WGS information should be considered in the development of policies, particularly to integrate patient preferences with personalized medicine and shared decision making.
Keywords: Whole genome sequencing, willingness-to-pay, human genome, secondary findings, attitude to health, health care costs
INTRODUCTION
Whole genome sequencing (WGS) determines the complete DNA sequence of an individual and can be used as a powerful diagnostic tool.1,2 Applied in the general population it could be used for screening.2–4 Unlike whole exome sequencing, which targets the protein coding regions of the genome (<2% of the genome), WGS involves sequencing the whole genome (coding, non-coding and mitochondrial DNA).5 Results from WGS include primary findings (variants in a gene(s) relevant to the diagnostic indication for which sequencing was ordered) and secondary findings (also termed incidental findings, variants in genes not apparently relevant to a diagnostic indication for which sequencing was ordered).6 In 2013, the American College of Medical Genetics and Genomics (ACMG) recommended that only secondary finding variants that are currently determined to be clinically actionable be reported when ordering sequencing for a primary indication.6
In 2015 the recommendations were updated to address the critical issue of who decides what results should be included in the WGS report. The new guidelines recommend that “patients should be able to opt out of the analysis of genes unrelated to the indication for testing.”7 Reporting secondary findings that are currently determined to have unclear medical treatment could generate anxiety and unnecessary medical tests, but patients could miss valuable information if not reported.8,9 Further, over-treatment may occur by acting on findings prematurely, potentially causing harm and unnecessary health care resource use.8,9 However, individuals may want to learn about these findings to reassess personal priorities and/or get affairs in order if their chance of death is increased, or they may hope information on long-term risk becomes actionable in the future. Clinical genome or exome sequencing are currently indicated for the detection of rare variants in patients seeking a diagnosis for a potential medelian genetic disorder and several thousand tests have already been ordered for this population.4 As costs decrease, it is possible that WGS will become more routine and possibly used for screening in the general population.1,2,4,10 The ACMG recommends that when WGS is used for diagnostic purposes, the individual tested (or their guardian) should undergo informed consent regarding possible secondary findings in 56 genes for conditions that “have a high likelihood of severe disease that is preventable if identified before symptoms occur.”.7 Costs for genome sequencing data can range from $4000-$15,000 USD and vary depending on interpretation and volume of data.4,11
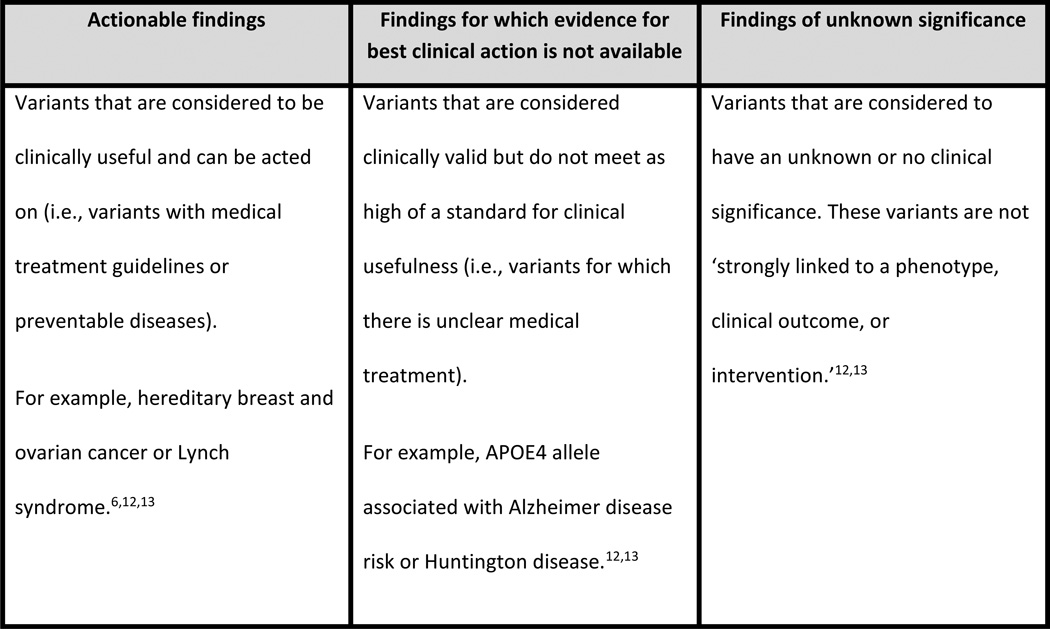
Whether WGS can achieve its potential to improve patient outcomes will depend on what information is given to patients, and how patients and providers respond to and value the information provided. From a healthcare system perspective, the benefits and costs of WGS will depend on short- and long-term consequences associated with receiving WGS information, what is recommended about ordering WGS in guidelines, and whether the cost of WGS will be covered by payers. Assessing the value of WGS is complex because it provides not just one test result, but multiple results that have varying levels of clinical usefulness (Figure 1). Actionable findings are variants that are considered to be clinically useful and can be acted on (i.e., variants with medical treatment guidelines or preventable diseases).12,13 Findings for which evidence for best clinical action is not available are variants that are considered clinically valid but do not meet as high a standard for clinical usefulness (i.e., variants for which there is unclear medical treatment). Findings of unknown significance are variants that are considered to have an unknown or no clinical significance. These variants are not ‘strongly linked to a phenotype, clinical outcome, or intervention.’12,13
Figure 1.
Summary and examples of actionable findings, findings for which evidence for best clinical action is not available, and findings of unknown significance.
There are multiple methods to estimate value and personal utility. In genomics, personal utility is defined as the meaning and worth an individual gives to a genomic or genetic test from their personal perspective.10,14 Cost-effectiveness, using an incremental cost-effectiveness ratio, is one approach to capturing value typically from a health care system and payer perspective to inform resource allocation decisions.15 Contingent valuation is a survey based method used for valuation of non-market services (e.g. health treatments) to estimate willingness to pay in monetary units (with a range and maximum amount).16 Contingent valuation can be used to estimate personal utility and the value of WGS information.
Previous research suggests that individuals, regardless of health status, value having choices about the WGS information they receive and many clinicians believe that people should have a choice about the results they receive.17–20 However, there is limited evidence on willingness-to-pay for WGS information depending on whether or not that information is medically actionable, or who should decide what results are included and returned in a WGS report. In this study, we elicit preferences for who defines which WGS results are included in a WGS report to help inform health policy, and estimate the value of WGS information using contingent valuation methods to elicit willingness-to-pay in a nationally representative sample of adults from the United States general population.
MATERIALS AND METHODS
Study Design
We designed an online survey to elicit stated preferences for WGS information in the context of screening for pathogenic variants. The survey included the following sections:
A ranking exercise to elicit preferences for who defines which WGS results are included in a genomic report;
Contingent valuation questions to elicit willingness-to-pay for a basic genomic report that includes only actionable genomic status information (based on ACMG recommendations);6
Contingent valuation questions to elicit willingness-to-pay for genomic information for which the medical treatment is currently unclear and would not be included in a basic genomic report (based on ACMG recommendations).
Our survey development was informed by: a literature review of attitudes, values and preferences in genomic/genetic testing; pre-test interviews and cognitive testing (using one-on-one ‘think aloud’ methods) with 13 consecutive adults who were enrolled in the MedSeq study (U01-HG006500) at the time of baseline data collection;2,21 and consultation with our team genetics experts. Risks were explained in words as: chance of having a gene variant that leads to health problems; chance of having additional gene variants that are not included in a genome report; and chance of death in the next 10 years if you have one of the additional gene variants. Each risk contained a probability expressed as a number and a diagram (e.g. chance of having any of the additional gene variants is 1 out of 100 (1%)). The diagram of the probability had 100 dots with a colored dot(s) to express the probability out of 100 and the remaining dots were grey. The costs presented to respondents in the contingent valuation questions were based on a review of the cost of WGS in the United States in 2014 and the upper levels were informed by maximum willingness-to-pay stated in pre-test interviews.
For the ranking exercise, respondents were asked to rank three mutually exclusive options from 1 to 3 (where 1 was the most desirable option and 3 was the least desirable) for defining the results included in the genomic report:
Option 1) A panel of experts decides which variants to include: a report that only includes results for gene variants that could lead to health problems that a panel of experts are sure can be prevented or treated.
Option 2) Your doctor decides which variants to include: a report your doctor chooses based on your family history and the information your doctor thinks you would find useful. Your doctor could explain the results that you get with this report, but you would not necessarily know how this report is different from the experts’ genome report.
Option 3) You decide which variants to include: a report that includes results for gene variants you choose based on your own understanding and concerns about your chance of health problems and possible treatments.
The survey included the following description about the information that the respondents would receive from WGS in a basic genomic report in the context of using WGS as a screening tool for pathogenic variants:
Based on the recommendations of experts, people who sequence their genome receive a report on whether they have any one of hundreds of gene variants that lead to health problems. About 1% (1 out of 100) of people learn that they have at least one of the gene variants included in the genome report. If you have any of the gene variants included in the genome report, you can get information about your chance of getting health problems that can be prevented or treated, and information about the chance that your children or family members have health problems.
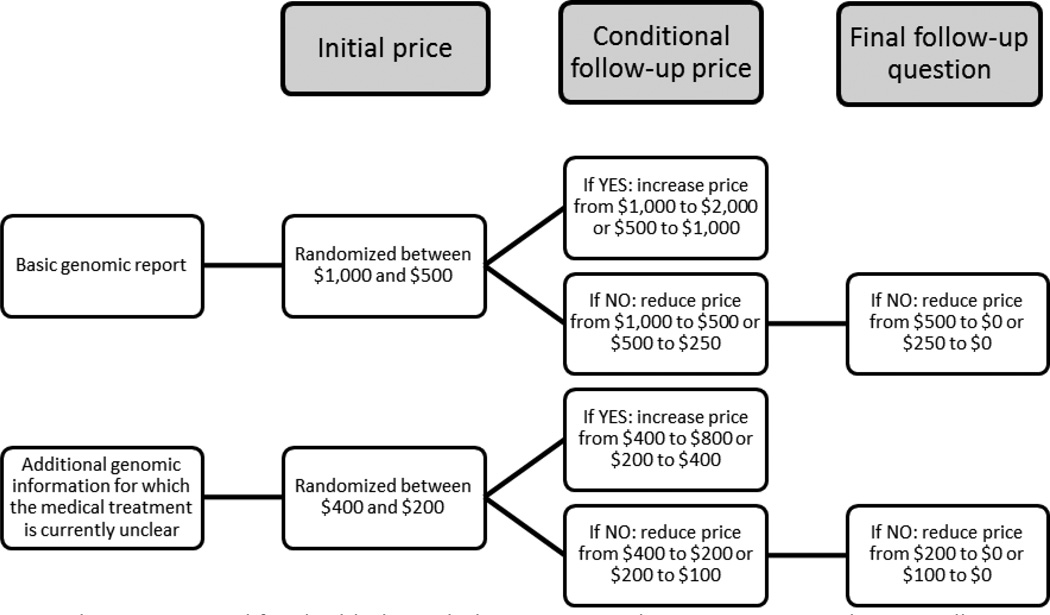
Following the WGS report description, respondents were asked contingent valuation questions to elicit their willingness-to-pay for genomic information. We used double-bounded dichotomous choice elicitation format varying the initial WGS report cost across respondents (two-question bidding game; Figure 2 and Appendix A), which has been widely used to value public goods, including health.16,22
Figure 2.
Bid structure used for double-bounded contingent valuation questions eliciting willingness-to-pay for: a basic genomic report that includes only actionable genomic status information; and additional genomic information for which the medical treatment is currently unclear.
In the first set of DBCV questions, respondents were randomly assigned to one of two initial basic report costs ($1,000 or $500). The cost doubled if the respondent answered ‘yes’ to the initial report cost, and was halved if the respondent answered ‘no’ to the initial cost. When a respondent rejected the report cost a second time, the cost was set to $0. In the second set of DBCV questions, respondents were randomly assigned to one of two initial costs ($400 or $200) for the additional genomic information for which the medical treatment is currently unclear. The cost doubled if the respondent answered ‘yes’ to the initial cost, and was halved if the respondent answered ‘no’ to the initial cost. If a respondent rejected the additional genomic information cost a second time, the cost was set to $0.
Statistical Methods
We calculated the proportion of respondents (and 95% confidence interval (CI)) who chose which WGS results are reported as their first preference (expert panel, your doctor, you). We used a rank-ordered logit (ROL) regression model to analyse the ranking of options.23
Only answers from respondents who stated they would be willing to accept genomic information were considered in the analysis, implicitly conditioning willingness-to-pay estimation to represent the value of information among those who were in the market for WGS reports. To analyze the DBCV questions, we used an interval regression (IR) model.24 The IR model conditions respondents’ willingness-to-pay on the attributes of the information respondents would be acquiring with their genomic status and respondents’ personal characteristics. The IR model uses answers to the DBCV questions to identify intervals within which respondents’ willingness-to-pay is expected. Results from the IR model can be interpreted as the contribution of the characteristics of genomic information and respondents’ personal characteristics to their willingness-to-pay for WGS information.
To understand which respondents were willing to acquire genomic information, we estimated a probit regression model relating a participation variable to personal characteristics.
Study Sample
The survey was administered online to a general population sample of adults (21 years and older) in the United States. Participant recruitment occurred in two stages. First, GfK recruited respondents by invitation through their internet-based panel, KnowledgePanel.25 KnowledgePanel is a probability-based web panel designed to be representative of the United States population. This panel has been used extensively in over 400 papers, articles and books, including several studies on genetic testing and is validated by the American Association for Public Opinion Research.25–27 GfK uses an ongoing modest incentive program for panel members to encourage participation, including entry into raffles or special sweepstakes with both cash rewards and other prizes. Households are provided with access to the Internet and a laptop if needed. Second, panel members were randomly invited to participate in our survey using residential address-based sampling methods. Email reminders were sent to non-responders four times.
A total of 873 individuals were invited to participate in the survey, 410 gave consent to participate and completed the survey in full (47% response rate, Appendix B). This allowed for robust statistical analyses with a minimum acceptable level of statistical precision (standard error less than 0.05).
Ethics and Consent
Informed consent was obtained from respondents prior to beginning the survey. Ethics approval was obtained through University of California – San Francisco Human Research Protection Program Committee on Human Research (12-09652) and the University of Calgary Conjoint Health Research Ethics Board (13-1231).
RESULTS
Overall, respondents were similar to the general population of the United States, except median age was higher (50 vs 37 years), the proportion of males was higher (53% vs 49%), more were married or living with a partner (64% vs 51%), levels of post-secondary education were higher (64% vs 58%), and incomes were higher (Table 1).28 The median duration to complete the survey was 22 minutes.
Table 1.
Demographics and other respondent characteristics (n=410)
| Characteristic | Unweighted summarya |
Weighted summary |
|
|---|---|---|---|
| Age (years) | |||
| Median (SD) | 50 (17) | 48 (17) | |
| Range | 21–91 | 21–91 | |
| Gender, n (%) | |||
| Male | 218 (53%) | 197 (48%) | |
| Female | 192 (47%) | 213 (52%) | |
| Marital status, n (%) | |||
| Married/living with partner | 261 (64%) | 252 (61%) | |
| Widowed/divorced/separated/never married | 149 (36%) | 158 (39%) | |
| Education, n (%) | |||
| Less than high school/high school | 148 (36%) | 169 (41%) | |
| Some college | 121 (30%) | 115 (28%) | |
| Bachelor's degree or higher | 141 (34%) | 126 (31%) | |
| Employment, n (%) | |||
| Employed | 242 (59%) | 236 (58%) | |
| Not employed | 168 (41%) | 174 (42%) | |
| Race/ethnicity, n (%) | |||
| White, non-Hispanic | 287 (70%) | 271 (66%) | |
| Black, non-Hispanic | 44 (11%) | 47 (11%) | |
| Other, non-Hispanic | 31 (8%) | 32 (8%) | |
| Hispanic | 48 (12%) | 61 (15%) | |
| Has children, n (%) | |||
| Yes | 133 (32%) | 138 (34%) | |
| No | 277 (68%) | 272 (66%) | |
| Medical conditions, n (%)b | |||
| Yes | 263 (64%) | 261 (64%) | |
| None of the conditions listed | 147 (36%) | 149 (36%) | |
| Health insurance, n (%)c | |||
| Yes | 377 (92%) | 372 (91%) | |
| No/don’t know/unsure | 31 (8%) | 36 (9%) | |
| Missing | 2 | 2 | |
| Household internet access, n (%) | |||
| Yes | 340 (83%) | 321 (78%) | |
| No | 70 (17%) | 89 (22%) | |
| Income, n (%) | |||
| <$20,000 | 41 (10%) | 51 (12%) | |
| $20,000 – $49,999 | 112 (27%) | 114 (28%) | |
| $50,000 – $84,999 | 103 (25%) | 106 (26%) | |
| ≥$85,000 | 154 (38%) | 138 (34%) | |
Data were weighted to represent the general population of the United States.
Could select ≥1 of the following medical conditions: arthritis, asthma or allergies, cancer, diabetes, gastrointestinal conditions, heart disease, high blood pressure, high cholesterol, migraines, osteoporosis, stroke.
Could select ≥1 of the following health insurance options: private insurance paid for by self, private insurance paid for by employer (spouse or self), Medicaid, Medicare, Veteran’s health insurance, other.
Regarding experiences with genomic testing (Appendix C), 7% reported ever having a genetic test and only 2% reported ever having their genome sequenced. When respondents were asked if they would want to learn if they had a gene variant that could lead to a fatal disease for which the medical treatment is currently unclear, 34% would want to learn about this information. These individuals were mostly males, an average of 47 years old (SD: 16 years), white (non-Hispanic), had at least some college education and a household income of $50,000 or more. When respondents were asked if they would want to learn if they had a gene variant that could lead to severe, progressive memory loss that is not treatable, 48% would want to learn about this information. These individuals were an even split of males and females, an average of 47 years old (SD: 17 years), white (non-Hispanic), had at least some college education and a household income of $50,000 or more.
Deciding themselves about what information is included in the basic genomic report was the most preferred option for 43% of respondents, and 34% would prefer that their primary care physicians decided (Table 2). Only 23% of respondents would prefer that a panel of experts defined the information contained in the basic genomic report. We found no statistically significant effect of any individual characteristics that helped explain differences in the ranking of options.
Table 2.
Summary of rankings: most and least preferred option of who defines results included in the basic genomic report (n=410)
| Chose as most preferred | Total (least preferred) | ||||
|---|---|---|---|---|---|
| Panel | Doctor | You | |||
|
Chose as least preferred |
Panel | 0 | 68 | 133 | 201 |
| Doctor | 27 | 0 | 44 | 71 | |
| You | 66 | 72 | 0 | 138 | |
| Total (most preferred) | 93 | 140 | 177 | 410 | |
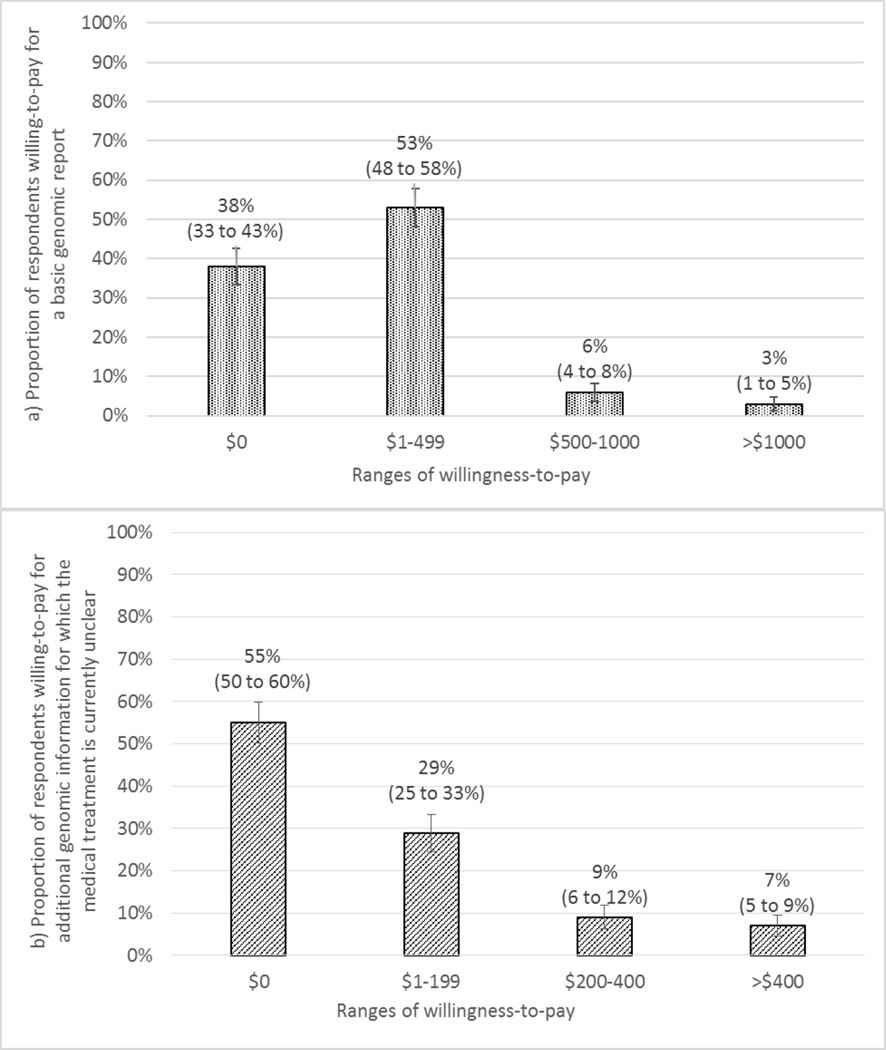
A substantial proportion of respondents did not value obtaining genomic information included in the basic report (38%, 95% CI: 33–43%) or genomic information for which the medical treatment is currently unclear (55%, 95%CI: 50–60%), even if this information were free (Figure 3). For the basic WGS report, most respondents (53%, 95% CI: 48–58%) were willing-to-pay up to $500 with only 9% willing-to-pay more than $500 (Figure 3a). For respondents who expressed interest in obtaining genomic information included in the basic report (n=253), the average willingness-to-pay estimate was $299 USD (SD: $86, p<0.01). None of the respondent characteristic variables included in the regression model (preference for who decides what results to include in a report, interest in knowing about gene variants related to fatal health problems for which the medical treatment is currently unclear or memory loss and function, having children <18 years old, household income, gender, race, previous genetic testing and time it took to complete the survey) were significantly associated with willingness-to-pay for the basic report.
Figure 3.
Distribution of respondents’ (n=410) willingness-to-pay for: a) a basic genomic report that includes only actionable genomic status information; and b) additional genomic information for which the medical treatment is currently unclear.
For the genomic information for which the medical treatment is currently unclear, 29% (95% CI: 25–33%) were willing-to-pay up to $200 and 16% were willing-to-pay more than $200 (Figure 3b) for this information. For those respondents who expressed interest in obtaining this genomic information (n=184), the average willingness-to-pay was $180 USD (SD: $83, p<0.05). Respondent characteristics, such as interest in knowing about fatal health problems for which the medical treatment is currently unclear (p<0.05) and history of previous genetic testing (p<0.01) were associated with higher willingness-to-pay. No other respondent characteristic variables included in the regression model (chance of having a gene variant for which the medical treatment is currently unclear, chance of death in the next 10 years from a health problem, preference for who decides what results to include in a report, interest in knowing about gene variants related to memory loss and function for which the medical treatment is currently unclear, having children <18 years old, household income, gender, race, time it took to complete the survey and interaction terms) were significantly associated with willingness-to-pay for this additional genomic information.
Respondents’ willingness to obtain genomic information was positively associated with interest in knowing about gene variants related to fatal health problems for which the medical treatment is currently unclear (p<0.01), and problems that would affect quality of life significantly (e.g. permanent loss of memory and function; p<0.01).Respondents who reported preferring control over the type of genomic information included in the basic genomic report were more likely to acquire information on gene variants for which the medical treatment is currently unclear, if offered (p<0.01). Being male was significantly associated with interest in obtaining additional genomic information for which the medical treatment is currently unclear (p<0.05) but not significantly associated with willingness to obtain the basic report. All other variables included in the regression model (preference for doctor deciding what results to include in a report, having children <18 years old, household income, previous genetic testing and race) were not significantly associated with respondents’ interest in obtaining WGS information. The model controlled for the influence of respondents (n=29) who completed the survey more quickly than expected if they were reading and considering all the responses.
DISCUSSION
Assessing the value of WGS is complex because it is a technology that provides multiple results with varying levels of clinical usefulness. Willingness-to-pay, as measured by contingent valuation, can be used to reflect the value of WGS information. There is limited evidence on willingness-to-pay for WGS information depending on whether or not that information is medically actionable, or who should decide what results are included and returned in a WGS report. We address this gap by eliciting preferences for who defines which WGS results are included in a WGS report and estimating the value of WGS information using contingent valuation methods.
In exploring who decides which results are included in a basic WGS report, we found that most individuals (43%) would prefer to decide themselves. Although our study uniquely addressed the issue of who should decide, our findings are supported by other findings that people, regardless of their health status, would generally like a choice about the WGS information they receive and many clinicians believe that people should have a choice about the results they receive (actionable or not).17–19,29 Concerns have been raised regarding the feasibility of sharing WGS results, specifically with regard to the large amount of time and resources required to review and discuss all possible results in a genomic report,30,31 as well as the difficulty counselling patients about all results in a report, or choosing a subset of actionable genes for analysis, when this information is constantly evolving.7 These are important considerations given that the general population has limited knowledge and understanding of personalized medicine32 and genomic policy experts have raised concerns regarding genomic literacy.33 Furthermore, although people are currently able to opt out of the analysis of genes unrelated to the indication for testing, they are not allowed to opt in.7 Our findings suggest some people would be better off, in terms of personality utility (as reflected by their willingness-to-pay), if they were also allowed to make an informed decision to opt in to receive genomic information for which the medical treatment is currently unclear. This may be associated with getting their affairs in order if a gene variant were to increase the chance of death, or hope that information on long-term risks may be actionable in the future.
In our nationally representative sample, we found that some respondents would not want genomic information, even if it were free. If they were interested in obtaining genomic information, respondents were willing-to-pay more for the basic WGS report ($299; actionable findings) than for the additional genomic findings for which the medical treatment is currently unclear ($180), suggesting that respondents valued the prophylactic or therapeutic benefits of the information in the WGS report. The characteristics of the genomic information (prevalence, severity of the health problems the variant might cause, and risk of death in the next 10 years associated with having the gene variant) did not seem to influence the amount respondents were willing-to-pay for additional genomic information, suggesting that the information itself, not the outcomes associated with the genomic information, was considered to be of value.
Previous research has demonstrated varying levels of interest in undergoing WGS34 and a wide range of estimates for willingness-to-pay to undergo genetic or genomic testing. In Canada and the United Kingdom, 30–80% of individuals are willing to undergo genetic or genomic testing if it is free of charge and 5–37% are willing-to-pay up to approximately $500 to undergo testing with few willing-to-pay more than $500.35–37 Age and interest in undergoing genetic or genomic testing influence willingness-to-pay.35,36
Using contingent valuation methods to reduce the potential bias of eliciting willingness-to-pay directly, our findings are generally consistent in the context of previous studies on preferences and value of complex genetic information. For example, research by Ries et al explored willingness-to-pay for genetic testing in a Canadian general population sample using a cost scale ($0, $1–499, $500–1,999, $2000+), and found that most people are not willing-to-pay more than $500 to learn about manageable diseases.36 Additionally, 48% would not pay anything to learn about their risk of developing a serious condition and 32% would pay up to $500 for this information.36
Research by Regier et al, focussed on information derived from secondary genomic findings, estimated willingness-to-pay using discrete choice experiment methods in a sample of adults from the general population.20 In a scenario that explores willingness-to-pay for information that may or may not be actionable, mean willingness-to-pay was $280.20 These methods were different from our study, and willingness-to-pay was greater than our findings, but similar in magnitude.
Implementation of WGS in a clinical setting has been limited partly due to lack of coverage of test costs by insurance companies and inability of some patients to pay the out-of-pocket costs.11 There is variation in cost coverage depending on insurance company and type of testing. However, coverage policies are being updated as use of this technology increases.11
Although our study has several strengths, there are some limitations. First, there are limitations associated with online research panels, such as possible incentive bias. However, this method and use of incentives are common. Research exploring online surveys has demonstrated validity and reliability that is comparable to traditional methods.38,39 Second, although contingent valuation is well-established, this approach does not inform questions about what specific aspects of the service are valued. To understand how different aspects of services are valued, decompositional approaches such as conjoint analysis are needed. Conjoint analysis characterizes a service based on multiple attributes and elicits trade-offs amongst these attributes.40
Our results provide evidence that individuals in the general population value actionable genetic information more than genomic information for which the medical treatment is currently unclear as reflected by their willingness-to-pay, but some respondents did not value any genetic information based on their lack of interest even if it were free. Furthermore, although the original ACMG guidelines for genomic testing were developed based on input from experts, we found that people prefer to decide themselves what WGS information is reported. These findings support the more recent ACMG policy statement that patients should be able to opt out of genetic analysis unrelated to the indication for testing. Our findings indicate that some people would be better off, in terms of personal utility, if they were also allowed to make an informed decision to opt in to receive genomic information for which the medical treatment is currently unclear, but for some, there may be negative value associated with the information generated from these results. However, if a person chooses to opt in to receive this information, decision makers need to consider who should pay and which individuals would be eligible to opt in given the potential financial impact on the healthcare system. This suggests that patient preferences should be used to inform policies and consent processes about WGS testing and how results are reported in the future since there may be negative value associated with information generated from these results.
Supplementary Material
Acknowledgments
This study was partially funded by a NHGRI grant to Dr. Phillips (R01HG007063), a NCI grant to the UCSF Helen Diller Family Comprehensive Cancer Center (5P30CA082013-15), and the UCSF Mount Zion Health Fund. We thank Denise Lautenbach, Jill Oliver Robinson and the MedSeq study team (U01-HG006500) for their advice and guidance with developing our survey and recruiting participants for pre-test interviews. We thank GfK for their assistance programming and hosting our online survey.
Deborah A Marshall is supported by a Canada Research Chair, Health Services and Systems Research and the Arthur J.E. Child Chair in Rheumatology Outcomes Research. She undertakes ad hoc consulting to support health economics and outcomes research for Optum Insight.
Footnotes
CONFLICT OF INTEREST NOTIFICATION
All other authors declare no conflicts of interest.
REFERENCES
- 1.Green RC, Berg JS, Berry GT, et al. Exploring concordance and discordance for return of incidental findings from clinical sequencing. Genet. Med. 2012;14(4):405–410. doi: 10.1038/gim.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vassy J, Lautenbach D, McLaughlin H, et al. The MedSeq Project: a randomized trial of integrating whole genome sequencing into clinical medicine. Trials. 2014;15(1):85. doi: 10.1186/1745-6215-15-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green RC, Rehm HL, Kohane IS. Chapter 9 - Clinical Genome Sequencing. In: Willard GSGF, editor. Genomic and Personalized Medicine. Second. Academic Press; 2013. pp. 102–122. [Google Scholar]
- 4.Biesecker LG, Green RC. Diagnostic Clinical Genome and Exome Sequencing. N. Engl. J. Med. 2014;370(25):2418–2425. doi: 10.1056/NEJMra1312543. [DOI] [PubMed] [Google Scholar]
- 5.Bamshad MJ, Ng SB, Bigham AW, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12(11):745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 6.Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet. Med. 2013;15(7):565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.ACMG Board of Directors. ACMG policy statement: updated recommendations regarding analysis and reporting of secondary findings in clinical genome-scale sequencing. Genet. Med. 2014;17:68–69. doi: 10.1038/gim.2014.151. [DOI] [PubMed] [Google Scholar]
- 8.McGuire AL, Burke W. An unwelcome side effect of direct-to-consumer personal genome testing: Raiding the medical commons. JAMA. 2008;300(22):2669–2671. doi: 10.1001/jama.2008.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohane IS, Masys DR, Altman RB. The incidentalome: a threat to genomic medicine. JAMA. 2006;296(2):212–215. doi: 10.1001/jama.296.2.212. [DOI] [PubMed] [Google Scholar]
- 10.Gray SW, Martins Y, Feuerman LZ, et al. Social and behavioral research in genomic sequencing: approaches from the Clinical Sequencing Exploratory Research Consortium Outcomes and Measures Working Group. Genet. Med. 2014;16(10):727–735. doi: 10.1038/gim.2014.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakradhar S. Insurance companies are slow to cover next-generation sequencing. Nat. Med. 2015;21(3):204–205. doi: 10.1038/nm0315-204. [DOI] [PubMed] [Google Scholar]
- 12.Berg JS, Khoury MJ, Evans JP. Deploying whole genome sequencing in clinical practice and public health: Meeting the challenge one bin at a time. Genet. Med. 2011;13(6):499–504. doi: 10.1097/GIM.0b013e318220aaba. [DOI] [PubMed] [Google Scholar]
- 13.Ramos EM, Din-Lovinescu C, Berg JS, et al. Characterizing genetic variants for clinical action. American Journal of Medical Genetics Part C: Seminars in Medical Genetics. 2014;166(1):93–104. doi: 10.1002/ajmg.c.31386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feero W, Wicklund C, Veenstra DL. The economics of genomic medicine: Insights from the iom roundtable on translating genomic-based research for health. JAMA. 2013;309(12):1235–1236. doi: 10.1001/jama.2013.113. [DOI] [PubMed] [Google Scholar]
- 15.Douglas MP, Ladabaum U, Pletcher MJ, Marshall DA, Phillips KA. Economic evidence on identifying clinically actionable findings with whole-genome sequencing: a scoping review. Genet. Med. 2015 doi: 10.1038/gim.2015.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell RC, Carson RT. Using Surveys to Value Public Goods - The Contingent Valuation Method. New York, New York: Resources for the Future; 1993. [Google Scholar]
- 17.Schneider J, Goddard KB, Davis J, et al. “Is It Worth Knowing?” Focus Group Participants’ Perceived Utility of Genomic Preconception Carrier Screening. Journal of Genetic Counseling. 2015:1–11. doi: 10.1007/s10897-015-9851-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holm IA, Savage SK, Green RC, et al. Guidelines for return of research results from pediatric genomic studies: deliberations of the Boston Children's Hospital Gene Partnership Informed Cohort Oversight Board. Genet. Med. 2014;16(7):547–552. doi: 10.1038/gim.2013.190. [DOI] [PubMed] [Google Scholar]
- 19.Klitzman R, Appelbaum PS, Fyer A, et al. Researchers' views on return of incidental genomic research results: qualitative and quantitative findings. Genet. Med. 2013;15(11):888–895. doi: 10.1038/gim.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Regier DA, Peacock SJ, Pataky R, et al. Societal preferences for the return of incidental findings from clinical genomic sequencing: a discrete-choice experiment. Can. Med. Assoc. J. 2015 doi: 10.1503/cmaj.140697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Presser S, Couper MP, Lessler JT, et al. Methods for Testing and Evaluating Survey Questions. Public Opin. Q. 2004;68(1):109–130. [Google Scholar]
- 22.Klose T. The contingent valuation method in health care. Health Policy. 1999;47(2):97–123. doi: 10.1016/s0168-8510(99)00010-x. [DOI] [PubMed] [Google Scholar]
- 23.Long JS, Freese J. Regression models for categorical dependent variables using Stata. 2nd. College Station, TX: Stata Press; 2006. [Google Scholar]
- 24.Amemiya T. Regression Analysis when the Dependent Variable Is Truncated Normal. Econometrica. 1973;41(6):997–1016. [Google Scholar]
- 25.Baker L, Bundorf M, Singer S, Wagner T. Validity of the Survey of Health and the Internet, and Knowledge Networks’ Panel and Sampling. Stanford, CA: 2003. [Google Scholar]
- 26.Baker L, Wagner TH, Singer S, Bundorf M. Use of the internet and e-mail for health care information: Results from a national survey. JAMA. 2003;289(18):2400–2406. doi: 10.1001/jama.289.18.2400. [DOI] [PubMed] [Google Scholar]
- 27.Baker R, Blumberg SJ, Brick JM, et al. Research Synthesis: AAPOR Report on Online Panels. Public Opin. Q. 2010;74(4):711–781. [Google Scholar]
- 28.U.S. Census Bureau. Current Population Survey - Annual Social and Economic Supplement. 2012 [Google Scholar]
- 29.Graves KD, Sinicrope PS, McCormick JB, Zhou Y, Vadaparampil ST, Lindor NM. Public Perceptions of Disease Severity but Not Actionability Correlate with Interest in Receiving Genomic Results: Nonalignment with Current Trends in Practice. Public Health Genomics. 2015;18(3):173–183. doi: 10.1159/000375479. [DOI] [PubMed] [Google Scholar]
- 30.Dewey FE, Grove ME, Pan C, et al. Clinical interpretation and implications of whole-genome sequencing. JAMA. 2014;311(10):1035–1045. doi: 10.1001/jama.2014.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dorschner Michael O, Amendola Laura M, Turner Emily H, et al. Actionable, Pathogenic Incidental Findings in 1,000 Participants’ Exomes. The American Journal of Human Genetics. 2013;93(4):631–640. doi: 10.1016/j.ajhg.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garfield S, Douglas MP, MacDonald KV, Marshall DA, Phillips KA. Consumer familiarity, perspectives and expected value of personalized medicine with a focus on applications in oncology. Per. Med. 2015;12(1):13–22. doi: 10.2217/pme.14.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson KJ, Gehlert S. Return of results from genomic sequencing: A policy discussion of secondary findings for cancer predisposition. Journal of Cancer Policy. 2014;2(3):75–80. doi: 10.1016/j.jcpo.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dodson DS, Goldenberg AJ, Davis MM, Singer DC, Tarini BA. Parent and Public Interest in Whole-Genome Sequencing. Public Health Genomics. 2015;18(3):151–159. doi: 10.1159/000375115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bosompra K, Ashikaga T, Flynn BS, Worden JK, Solomon LJ. Psychosocial factors associated with the public's willingness to pay for genetic testing for cancer risk: a structural equations model. Health Educ. Res. 2001;16(2):157–172. doi: 10.1093/her/16.2.157. [DOI] [PubMed] [Google Scholar]
- 36.Ries NM, Hyde-Lay R, Caulfield T. Willingness to pay for genetic testing: a study of attitudes in a Canadian population. Public Health Genomics. 2010;13(5):292–300. doi: 10.1159/000253120. [DOI] [PubMed] [Google Scholar]
- 37.Cherkas LF, Harris JM, Levinson E, Spector TD, Prainsack B. A Survey of UK Public Interest in Internet-Based Personal Genome Testing. PLoS One. 2010;5(10):e13473. doi: 10.1371/journal.pone.0013473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evans JR, Mathur A. The value of online surveys. Internet Research. 2005;15(2):195–219. [Google Scholar]
- 39.Eysenbach G, Wyatt J. Using the Internet for Surveys and Health Research. J. Med. Internet Res. 2002;4(2):e13. doi: 10.2196/jmir.4.2.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bridges JF, Hauber AB, Marshall D, et al. Conjoint analysis applications in health--a checklist: a report of the ISPOR Good Research Practices for Conjoint Analysis Task Force. Value Health. 2011;14(4):403–413. doi: 10.1016/j.jval.2010.11.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.