Abstract
PURPOSE
Using SNP chip and exome sequence data from individuals participating in the NIH Undiagnosed Diseases Program (UDP), we evaluated the number and therapeutic informativeness of incidental pharmacogenetic variants.
METHODS
Pharmacogenomics Knowledgebase (PharmGKB) annotated sequence variants were identified in 1,101 individuals. Medication records of participants were used to identify individuals prescribed medications for which a genetic variant might alter efficacy.
RESULTS
395 sequence variants, including 19 PharmGKB 1A and 1B variants, were identified in SNP chip sequence data and 388 variants, including 21 PharmGKB 1A and 1B variants, were identified in the exome sequence data. Nine participants had incidental pharmacogenetic variants associated with altered efficacy of a prescribed medication.
CONCLUSIONS
Despite the small size of the NIH UDP patient cohort, we identified pharmacogenetic incidental findings potentially useful for guiding therapy. Consequently, groups conducting clinical genomic studies might consider reporting of pharmacogenetic incidental findings.
Keywords: Incidental findings, Secondary findings, SNP genotypes, exome sequencing, NIH Undiagnosed Diseases Program, personalized medicine, precision medicine
INTRODUCTION
Incidental or secondary genetic findings are variants with medical or social implications discovered during genetic testing for an unrelated indication1. Recent discussions and the report by the ACMG Working Group on Incidental Findings in Clinical Exome and Genome Sequencing have focused on disease-associated genes and not genetic determinants of drug metabolism2. Given that genomic variation influences human responsiveness to many drugs and contributes to phenotypes ranging from life-threatening adverse drug reactions to lack of therapeutic efficacy3, the return of pharmacogenetic incidental findings has potentially significant medical benefit4.
The Clinical Pharmacogenetics Implementation Consortium (CPIC), which develops guidelines for incorporating pharmacogenomics findings into clinical practice, has identified variant-drug associations of high concern for clinicians, and provides drug-dosing guidelines based on patient genotype. We hypothesized, therefore, that designing and implementing a process to identify pharmacological incidental findings in the genomic data generated by the NIH Undiagnosed Diseases Program5 (UDP) provides information to the medical community on the quantity and quality of pharmacogenetic incidental findings.
The NIH UDP routinely conducts SNP chip and exome sequencing analyses on probands and their family members. These data can be analyzed for pharmacogenetic incidental findings based on variant-drug associations listed in the Pharmacogenomics Knowledgebase (PharmGKB). The typical number of such pharmacogenetic incidental findings has not been widely studied, particularly when family members other than the proband are included in diagnostic studies. Consequently, more data are needed to assess the possible impact and need for resources.
To delineate the impact of identifying pharmacogenetic incidental findings, we analyzed SNP chip data from 1,101 individuals derived from 308 families and research exome sequence data from 645 individuals derived from 158 families. For the 868 pharmacogenetic loci listed in the Pharmacogenomics Knowledgebase (PharmGKB), we identified 949 independent pharmacogenetic findings using the SNP chip and exome sequence data and found that each individual had at least one. For nine individuals, these constituted incidental findings relevant to a medication that they were taking. These data refine strategies for reporting of pharmacogenetic incidental findings.
MATERIALS AND METHODS
Methods and Materials are available online
RESULTS
The PharmGKB database includes 868 annotated SNPs with published pharmacological implications. PharmGKB has six levels of evidence based on published evidence and the drug-dosing guidelines of the Clinical Pharmacogenetics Implementation Consortium (CPIC)6. The top two levels (1A and 1B) have substantive evidence for clinical relevance and CPIC guidelines or known clinical implementation.
The SNP chips used in our study provided 46% coverage of the SNPs annotated in the PharmGKB database and 53% coverage of high priority SNPs (PharmGKB 1A and 1B categories). Variants identified through whole exome sequencing conducted on the patients in the UDP covered 45% of the SNPs in the PharmGKB database and 58% of PharmGKB 1A and 1B SNPs. Combining the SNPs from both the SNP chip genotyping and exome sequencing covered 65% of the SNPs in the PharmGKB database and 81% of the PharmGKB 1A and 1B SNPs.
All UDP patients with SNP genotypes or exome sequencing data had potential pharmacogenetic incidental findings. Within the SNP chip data, there were 696 potential incidental findings per patient that were associated with 276 different drugs. These included 19 variants ranked within PharmGKB categories 1A and 1B, and these 19 variants were associated with the efficacy of 17 drugs. Within the exome sequencing data, there were 728 detectable variants per patient that were associated with 283 different drugs related to 388 PharmGKB documented SNPs. These included 21 variants within PharmGKB categories 1A and 1B that were associated with 14 different drugs. Combined SNP chip and exome data detected 949 variants per patient, and 29 were variants ranked in the PharmGKB 1A and 1B categories.
To determine if these variants constituted incidental findings, i.e., were of therapeutic relevance, we focused on the 359 individuals for whom we had medication records. None of these individuals had been prescribed a medication for which they carried a PharmGKB 1A or 1B category variant. We therefore tested whether any of the pharmacogenetic variants they carried might provide insight into their response to a prescribed medication. This identified five pharmacogenetically relevant variants or incidental findings among nine individuals (Table 1). Three of the variants were detectable by both the SNP chip and exome sequencing, and two were detectable only by exome sequencing.
Table 1.
Medically relevant pharmacogenomics variants in the NIH Undiagnosed Diseases Program cohort
| UDP Patient |
SNP | Genotype | Age, Sex, Race |
Phenotype | Drug | Pharm GKB Rank |
Pharmacological Implication |
|---|---|---|---|---|---|---|---|
| 1 | rs6318 | CG | 46, Female, Caucasian | Fibromyalgia, Depression, Hypertension | escitalopram | 3 | CG associated with greater efficacy for treatment of nerve pain in males |
| 2 | rs1051740 | CT | 14, Male, Caucasian | Episodic Ataxia Type 1, Seizures | carbamazepine | 2B | CT associated with increased metabolism of carbamazepine in people with Epilepsy |
| 3 | rs1799971 | AG | 27, Female, Caucasian | Facial and abdominal pain, lung nodules, facial masses, recurrent pericarditis and pleuritis | morphine | 3 | AG associated with increased metabolism and decreased efficacy for pain reduction |
| 4 | rs5985 | AC | 62, Female, Caucasian | Autonomic dysfunction, ataxia | acetylsalicylic acid | 3 | Associated with inhibition of FXIII activation |
| 5 | rs1799983 | GT | 57, Male, Caucasian | Neurological, no cardiovascular disease risks | acetylsalicylic acid | 3 | Associated with increased in-stent restenosis in men with Coronary Artery Disease |
| 6 | rs1799983, rs5985 | GT, AC | 52, Male, Caucasian | Fevers, CSF Pleocytosis | acetylsalicylic acid | 3 | Associated with increased in-stent restenosis in men with Coronary Artery Disease and inhibition of FXIII activation |
| 7 | rs5985 | AC | 68, Male, Caucasian | Fever, pulmonary | acetylsalicylic acid | 3 | Associated with inhibition of FXIII activation |
| 8 | rs1799983 | GT | 53, Male, Caucasian | Cardiac, heart murmur, aortic valve replacement, myopathy | acetylsalicylic acid | 3 | Associated with increased in-stent restenosis in men with Coronary Artery Disease |
| 9 | rs5985 | AC | 60, Male, Caucasian | Myopathy, fatigue, Myocardial infarction, bicuspid aortic value, pulmonary embolism | acetylsalicylic acid | 3 | Associated with inhibition of FXIII activation |
The five variants were relevant to efficacy of four different medications (Table 1). These medications were escitalopram, carbamazepine, morphine, and acetylsalicylic acid. First, a female adult (Patient 1) prescribed escitalopram had a PharmGKB category 3 association: rs6318 genotype CG, which has been associated with altered function of the X-linked serotonin 5-HTR2C receptor and various phenotypes ranging from altered cortisol response to stress7 to increased risk for cardiovascular disease mortality and morbidity8. Second, a child (Patient 2) who had seizures refractory to carbamazepine had a PharmGKB category 2B association: rs1051740 genotype CT, which has been associated with a requirement for increased dosage of carbamazepine9. Third, a female adult (Patient 3), who had been prescribed morphine and reported that ibuprofen gave better pain control, had a PharmGKB category 3 association: rs1799971 genotype AG, which has been associated with decreased efficacy of morphine in Caucasians10–12. Fourth, a male adult (Patient 8) prescribed acetylsalicylic acid and with a cardiomyopathy had a PharmGKB category 3 association: rs1799983 genotype GT, which has been associated with acetylsalicylic acid promoting in-stent restenosis13. Fifth, a male adult (Patient 9) was prescribed acetylsalicylic acid, had a history of myocardial infraction, and had a PharmGKB category 3 association: rs5985 genotype AC, which has been associated with an increased effectiveness of acetylsalicylic acid inhibition of Factor XIII activation, ultimately lowering an individual’s risk of myocardial infarction and death14.
DISCUSSION
By using the SNP chip data to analyze genotypes for 696 polymorphic variants and exome sequence data to define genotypes for 728 polymorphic variants, we were able to interrogate the genotype of 949 pharmacogenetically relevant variants for each individual enrolled in the NIH UDP. Focusing on the 359 individuals for whom we had medication information, we discovered five reportable variants among nine individuals from nine different families (eight probands and one unaffected family member). For five (56%) of these individuals, the pharmacogenetic incidental findings were potentially relevant to their medical management.
Although the sequencing conducted by the NIH UDP was not designed to screen pharmacologically relevant variants, 65% of the annotated variants in the PharmGKB databases were represented in the UDP sequencing data. All individuals in the study received SNP chip genotyping; exome sequencing was conducted on 59% of these individuals. Both SNP chip genotyping and exome sequencing were important in identifying the pharmacogenetically relevant variants. Although 50% of the variants that were discovered in the UDP dataset could be identified with either the SNP chip genotyping data or the exome sequencing data, 23% were found only within the SNP chip genotyping data and 27% were found only within the exome sequencing data.
Four additional issues arising during our analysis were 1) defining the level of relative risk warranting reporting of a potential variant, 2) determining how to weight variants identified in an ethnic group different from that of the subject, 3) the need for clinical correlation, and 4) obligations to family members. Relevant to the first issue, clinicians expressed a need for prioritization of the findings since there were such a large number of potential incidental findings per patient. To address this request, we used the clinical annotation levels of evidence of PharmGKB. Additionally, we reported the ethnicity within which the variant-drug associations were identified and highlighted any differences in ethnicity between the patient and that population and prioritized variant-drug associations accordingly.
Although filtering the incidental findings for PharmGKB 1A and 1B categories produces a manageable list of 21 medications, none of the individuals for whom we had medication data had a PharmGKB category 1A or 1B association. We therefore tested for associations ranked lower in PharmGKB and identified five PharmGKB category 2 or 3 associations. Five of these were potentially relevant to the care of the individuals, three patients and one family member (Table 1).
A one-sentence summary of the PharmGKB variant with the associated drug information was incorporated into the report entered into the NIH UDP database. Entry of the findings into the database also enabled sorting of the results and searching by individual, drug name and priority. Primary clinicians’ discussions regarding whether to inform individuals enrolled under the NIH UDP protocol about the identified variants focused on the delineated and perceived obligations defined by the language of the consent document and the process by which the consent was explained. The choice of whether to report a given variant to a given study subject was deferred to the relevant clinical team. No clinical team elected to return a PharmGKB category 2 or 3 variant detected by this study for at least two reasons. Firstly, the study consent reflected routine practice from the early days of the application of genome-scale sequencing to medical diagnostics. Only DNA variants that might contribute to the test indication were to be returned. Secondly, current ACMG guidelines do not include such variants among those recommended to be returned2. The UDP recognizes that these are areas of intense debate in the literature and elsewhere; the program is prepared to adjust its practice as the standard of care evolves.
Our analysis had some limitations. Some of the variant annotations in PharmGKB associate a variant to a class of drugs (e.g., bisphosphonates or antidepressants) instead of a specific drug. In contrast, UDP medication records list the specific drug name; therefore, our simple matching algorithm did not identify all associations. This problem can be readily addressed with drug ontology that would allow drug class matches. We also did not have access to the complete list of medications administered to our patients prior to their participation in our study. Despite these limitations, a small percentage of individuals with relevant medication prescriptions was identified in the cohort, indicating that there were likely an even larger percentage of medically relevant findings if the computational filtering mechanisms were refined.
The results of this study indicate that, by looking for pharmacogenetically relevant findings in SNP genotypes or exome sequencing data, incidental findings can be identified and potentially provide a valuable resource for patient care. The interpretation, use in medical practice, and returning of pharmacogenomics secondary findings are, however, different than the non-pharmacogenomic incidental findings listed in the original ACMG publication2. These pharmacogenomic variants different from non-pharmacogenomic incidental findings, in that, they are only relevant in the presence of an environmental factor, i.e., the drug that they have an effect on. This means that a pharmacogenomic variant may never be relevant in one person’s lifetime but absolutely important for another person who is prescribed the drug in question. For example, in a recent study of 48 pharmacogenomically relevant variants among a 94 patient cohort15, the electronic medical record decision support system gave no alerts during these patients’ hospital stay.
Our described methodology and findings, nonetheless, suggest that identifying and reporting pharmacogenetic incidental findings can improve patient care and personalized medicine. Additionally, if medically relevant findings can be found in the small UDP cohort with sequencing strategies designed for diagnostic purposes and not pharmacogenetic purposes, we hypothesize that such incidental findings occur in the sequencing data for other medical programs.
In summary, using SNP chip genotyping and exome sequencing, the NIH UDP identified pharmacogenetic incidental findings in its cohort. We established a framework for evaluating and selecting pharmacogenomic variants hypothetically useful for therapeutic management.
Supplementary Material
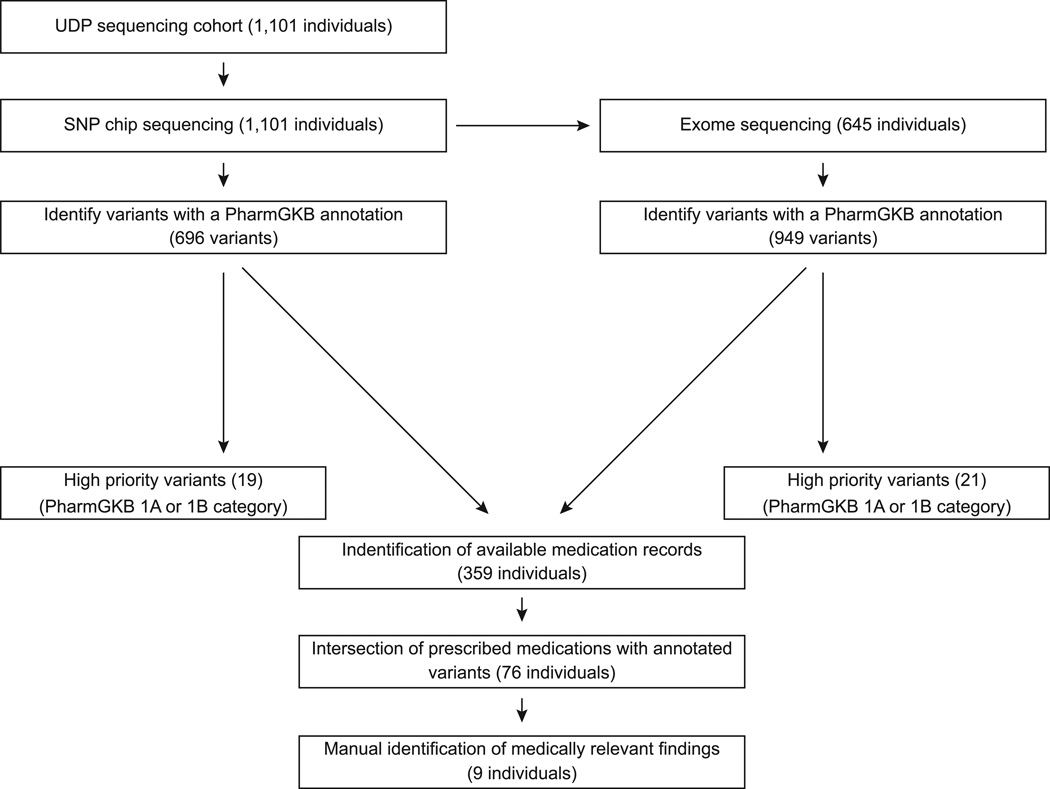
Figure 1.
Flow chart summarizing the NIH Undiagnosed Diseases Program analysis of and observations for the pharmacogenetic variants listed in PharmGKB. The observations were derived from analysis of SNP chip and exome sequence data. The SNP chip data were derived from a cohort of 308 families consisting of 355 affected individuals, 278 unaffected siblings, 459 unaffected parents, and 9 other unaffected family members; this cohort included 564 females and 537 males. The exome sequence data were from a subset of this cohort, 158 families consisting of 182 affected individuals, 150 unaffected siblings, 313 unaffected parents, 326 females and 319 males.
Acknowledgments
We thank the NHGRI Intramural Sequencing Center and Appistry, Inc. for their sequencing, alignment, genotyping and annotation services. This work was supported in part by the Common Fund, Office of the Director, and the Intramural Research Program of the National Human Genome Research Institute (NIH, Bethesda, Maryland)
REFERENCES
- 1.Wolf SM, Crock BN, Van Ness B, et al. Managing incidental findings and research results in genomic research involving biobanks and archived data sets. Genetics in medicine : official journal of the American College of Medical Genetics. 2012 Apr;14(4):361–384. doi: 10.1038/gim.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genetics in medicine : official journal of the American College of Medical Genetics. 2013 Jul;15(7):565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leeder JS. Developmental and pediatric pharmacogenomics. Pharmacogenomics. 2003 May;4(3):331–341. doi: 10.1517/phgs.4.3.331.22693. [DOI] [PubMed] [Google Scholar]
- 4.Hawcutt DB, Thompson B, Smyth RL, Pirmohamed M. Paediatric pharmacogenomics: an overview. Archives of disease in childhood. 2013 Mar;98(3):232–237. doi: 10.1136/archdischild-2012-302852. [DOI] [PubMed] [Google Scholar]
- 5.Gahl WA, Tifft CJ. The NIH Undiagnosed Diseases Program: lessons learned. Jama. 2011 May 11;305(18):1904–1905. doi: 10.1001/jama.2011.613. [DOI] [PubMed] [Google Scholar]
- 6.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clinical pharmacology and therapeutics. 2011 Mar;89(3):464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brummett BH, Babyak MA, Williams RB, et al. A putatively functional polymorphism in the HTR2C gene is associated with depressive symptoms in white females reporting significant life stress. PloS one. 2014;9(12):e114451. doi: 10.1371/journal.pone.0114451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brummett BH, Babyak MA, Jiang R, et al. A functional polymorphism in the 5HTR2C gene associated with stress responses also predicts incident cardiovascular events. PloS one. 2013;8(12):e82781. doi: 10.1371/journal.pone.0082781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makmor-Bakry M, Sills GJ, Hitiris N, Butler E, Wilson EA, Brodie MJ. Genetic variants in microsomal epoxide hydrolase influence carbamazepine dosing. Clinical neuropharmacology. 2009 Jul-Aug;32(4):205–212. doi: 10.1097/WNF.0b013e318187972a. [DOI] [PubMed] [Google Scholar]
- 10.Campa D, Gioia A, Tomei A, Poli P, Barale R. Association of ABCB1/MDR1 and OPRM1 gene polymorphisms with morphine pain relief. Clinical pharmacology and therapeutics. 2008 Apr;83(4):559–566. doi: 10.1038/sj.clpt.6100385. [DOI] [PubMed] [Google Scholar]
- 11.Matic M, Simons SH, van Lingen RA, et al. Rescue morphine in mechanically ventilated newborns associated with combined OPRM1 and COMT genotype. Pharmacogenomics. 2014 Jul;15(10):1287–1295. doi: 10.2217/pgs.14.100. [DOI] [PubMed] [Google Scholar]
- 12.Reyes-Gibby CC, Shete S, Rakvag T, et al. Exploring joint effects of genes and the clinical efficacy of morphine for cancer pain: OPRM1 and COMT gene. Pain. 2007 Jul;130(1–2):25–30. doi: 10.1016/j.pain.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shuvalova YA, Kaminnyi AI, Meshkov AN, Shirokov RO, Samko AN. Association between polymorphisms of eNOS and GPx-1 genes, activity of free-radical processes and in-stent restenosis. Molecular and cellular biochemistry. 2012 Nov;370(1–2):241–249. doi: 10.1007/s11010-012-1419-3. [DOI] [PubMed] [Google Scholar]
- 14.Undas A, Sydor WJ, Brummel K, Musial J, Mann KG, Szczeklik A. Aspirin alters the cardioprotective effects of the factor XIII Val34Leu polymorphism. Circulation. 2003 Jan 7;107(1):17–20. doi: 10.1161/01.cir.0000047062.03282.a3. [DOI] [PubMed] [Google Scholar]
- 15.Nishimura AA, Shirts BH, Dorschner MO, et al. Development of clinical decision support alerts for pharmacogenomic incidental findings from exome sequencing. Genetics in medicine : official journal of the American College of Medical Genetics. 2015 Mar 5; doi: 10.1038/gim.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.