Abstract
Though toxic in excess, iron is vital for life. Thus, its use in all cells is tightly regulated. Analysis of Saccharomyces cerevisiae, which has been used extensively as a model system, has revealed layers of regulation of cellular iron trafficking and utilization. This regulation is based on the availability of both elemental iron and functionality of the Fe–S cluster biogenesis system. Here, we discuss a possible “first responder” regulatory mechanism centered on the stability of the scaffold protein on which Fe–S clusters are built.
Keywords: Fe–S cluster biogenesis, Mitochondrial dysfunction, Posttranslational regulation, Lon proteases, Mitochondria
Introduction
Either in its elemental form or as a part of biologically synthesized cofactors, such as heme and iron-sulfur clusters, iron participates in many types of reactions in cells (Wofford and Lindahl 2015). These include enzymatic catalysis, electron transfer and conformational changes of regulatory proteins. Through these avenues, iron is central to vital cellular processes including DNA synthesis and repair, amino acid metabolism, ribosome assembly and oxidative phosphorylation (Lill et al. 2012). However, free iron is toxic to cells, as its reactivity induces production of reactive oxygen species, which damage biologically relevant macromolecules (Mena et al. 2015). Thus, iron uptake, trafficking, storage and utilization is tightly controlled. As part of this regulation, a specific response is triggered by iron deprivation, leading to an increase in iron uptake, release of stored iron and a profound remodeling of cellular metabolic pathways, thereby minimizing the cell’s need for iron (Outten and Albetel 2013). In S. cerevisiae this response is in good part under the control of the Aft1 and Aft2 transcription factors (Rutherford et al. 2003), which are inactive when coordinating an Fe–S cluster (Poor et al. 2014). Thus cellular iron sensing is tied to the status of Fe–S cluster production (Outten and Albetel 2013; Wofford and Lindahl 2015). The mitochondrial matrix can be considered the hub of cellular Fe–S cluster biogenesis. The system present in the mitochondria for Fe–S cluster biogenesis not only synthesizes clusters for mitochondrial proteins, it is also critical for generation of clusters on cytosolic and nuclear proteins (Lill et al. 2014). Thus, maintaining Aft1/2 transcription factors inactive requires a robust mitochondrial Fe–S cluster biogenesis system.
The mitochondrial process of Fe–S cluster biogenesis can be divided into two sequential steps: first, the assembly of a Fe–S cluster on a highly conserved scaffold protein (called Isu in S. cerevisiae), and second, the transfer of the cluster from the scaffold to a recipient protein (Fig. 1). Several protein factors in addition to Isu are required both for the assembly process and for the transfer process. Since Fe–S cluster biogenesis is essential, abolishing either step results in cell death. Partial impairment not only results in activation of Aft1/2 (Hausmann et al. 2008), but also an increase in the level of Isu (Rutherford et al. 2003; Knieszner et al. 2005; Andrew et al. 2008; Yoon et al. 2014). Most of this, approximately 15-fold, increase is due to an increase in the stability of the Isu protein—that is the regulation occurs at the posttranslational level. Here, we discuss both the mechanism behind this regulation and its possible biological significance. We put forth the idea that this response to the partial disruption of Fe–S cluster biogenesis may act as a rapid, specific, stop-gap compensatory mechanism. Such an increase may allow the cell to ride out temporary disruptions in Fe–S cluster biogenesis without resorting to the massive remodeling of iron-utilizing metabolic pathways that occurs upon Aft1/2 activation.
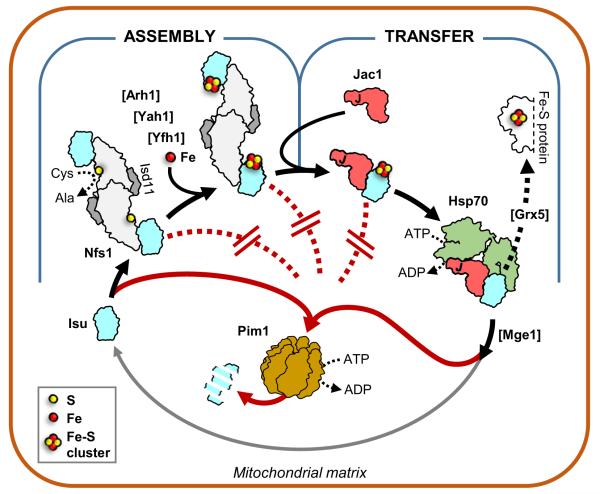
Fig. 1.
Mitochondrial Fe–S cluster biogenesis and Isu degradation. Model of regulation of Isu degradation in the mitochondrial matrix. Isu protein, a key component for Fe–S cluster biogenesis (black arrows), is susceptible to degradation by the Lon-type protease Pim1 (red arrows). However, Isu becomes protected from Pim1 driven degradation (red dotted lines) during both the assembly and transfer steps of the process through a combination of partner binding and Fe–S cluster coordination. Isu becomes susceptible to degradation upon completion of the transfer step. Assembly step. Fe–S cluster assembly is initiated by interaction of the scaffold Isu with Nfs1 (Lill et al. 2012). In the yeast S. cerevisiae the scaffold is encoded by two paralogous genes, ISU1 and ISU2; Isu1 and Isu2 are functionally redundant, with Isu1 being predominant and Isu2 less abundant (Schilke et al. 1999; Garland et al. 1999). As a cysteine desulfurase, Nfs1 provides sulfur for synthesis of the cluster. It functions as a Nfs1(Isd11) complex composed of two monomers of Nfs1 and two monomers of Isd11. Isd11 is an accessory protein required for Nfs1 stability and/or function (Wiedemann et al. 2006). Other factors needed for cluster assembly include: Yeast frataxin Yfh1, proposed to serve as an iron donor and/or regulator (Pastore and Puccio 2013), and yeast ferredoxin Yah1, which is functionally coupled with ferredoxin reductase Arh1, to provide electrons required for sulfur reduction (Webert et al. 2014). Transfer step. Following its assembly, the Fe–S cluster is transferred from Isu to a recipient protein (Lill et al. 2012). Transfer requires Hsp70 chaperone machinery composed of Hsp70, and its partnering co-chaperones Jac1 and Mge1 (Dutkiewicz et al. 2003). Initially Jac1 binds and targets the Isu coordinated Fe–S cluster to Hsp70. Interaction with Isu, coupled with Hsp70 ATPase activity stimulation by the J-domain of Jac1, results in Hsp70:Isu complex formation. As a result, the Fe–S cluster can be transferred via potential carrier proteins such as Grx5 (Uzarska et al. 2013) to the mitochondrial subset of recipient proteins or, through a yet elusive pathway, can be utilized for maturation of cytosolic and nuclear proteins (Lill et al. 2014). Isu is released from Hsp70, due to the nucleotide exchange activity of Mge1 protein (Dutkiewicz et al. 2003). Upon release, Isu becomes once more susceptible to degradation by Pim1, but can also be recycled and serve as a scaffold for cluster assembly again (gray arrow). Disruption at either the assembly or transfer step leads to increase in Isu levels as shown by analysis of yeast strains having reduced or perturbed function of Yah1, Yfh1, Jac1, Hsp70 or Grx5 protein (Andrew et al. 2008; Song et al. 2012)
Increase in Isu levels in response to Fe–S cluster biogenesis disorder due to decreased proteolysis
The increase in Isu levels occurs when the functionality of components that act in either the assembly or transfer steps of the biogenesis process are reduced (see Fig. 1). This upregulation is specific to Isu, as the levels of the other factors that function during cluster biogenesis are not affected (Andrew et al. 2008). An approximately two-fold increase in ISU mRNA, driven by activation of the Aft1/2 transcription factors, does occur under such circumstances (Rutherford et al. 2003). But much of the increase is independent of the Aft1/2 regulated pathways (Andrew et al. 2008). The half-life of Isu measured by in vivo pulse-chase experiments upon reduction in activity of one of the transfer factors revealed that most of the difference can be attributed to an increase in stability of Isu (Song et al. 2012). The change takes place quickly after perturbation. For example, when levels of a factor important for transfer dropped to 25 % of normal levels, significant upregulation was observed (Andrew et al. 2008). Most importantly, lowering of Isu levels in a strain with compromised Fe–S cluster biogenesis to levels present under normal conditions, severely impairs cell growth (Andrew et al. 2008). Thus, elevation is not only a specific effect, directly correlated with Fe–S cluster biogenesis disruption, it is beneficial to the cell.
Interplay between degradation and partner binding regulates Isu abundance in vivo
In vivo and in vitro results demonstrate that the Lon-type protease of the mitochondrial matrix, Pim1, is responsible for degradation of Isu. Its absence, but not the absence of other mitochondrial proteases, in vivo results in a dramatic increase in Isu levels (Ciesielski et al. 2016). Remarkably, in a pim1Δ strain Isu is so stable that its half-life is practically unmeasurable (Song et al. 2012). In an in vitro system, Lon-protease degrades purified Isu (Ciesielski et al. 2016).
What is the basis of this remarkable range of Isu stability? Evidence to date suggests that two factors play a role in protection. First, physical interaction of biogenesis factors is protective. Two factors, Nfs1 and Jac1, whose sites of interaction on Isu overlap, are capable of protecting Isu from degradation both in vivo and in vitro (Ciesielski et al. 2016; Song et al. 2012). Nfs1 and Jac1 act in different steps of the biogenesis process; Nfs1 is the sulfur donor for cluster formation, initiating the assembly step, while Jac1 is a J-protein co-chaperone that initiates the Hsp70 driven transfer of the cluster from Isu (see Fig. 1). What these two factors have in common is their interaction with a hydrophobic patch on the surface of Isu (Fig. 2). The second factor is the presence of a cluster on Isu. Isu coordinating a Zn ion, which is known to mimic Fe–S cluster binding, is degraded less effectively by Lon in vitro than apo-Isu (Ciesielski et al. 2016).
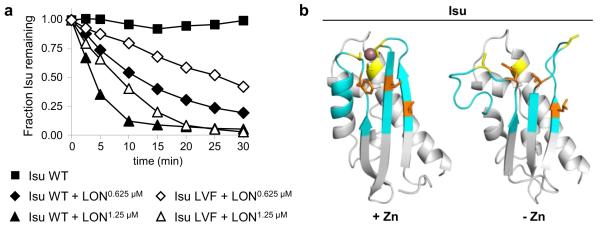
Fig. 2.
Recognition site for Pim1: evidence for a role of the hydrophobic region of Isu that is important for interaction of Nfs1 and Jac1. a Residues Leu63, Val72 and Phe94 form a hydrophobic patch on the surface of Isu, which is a critical part of the interface for both the Nfs1:Isu and Jac1:Isu complexes (Majewska et al. 2013). Using an established in vitro degradation assay (Ciesielski et al. 2016), we examined an Isu variant with serine substitutions of these residues (Isu LVF) to test the importance of this region for recognition of Isu as a Pim1 substrate. Briefly 7.5 μM of purified Isu1 wild type (WT) or Isu1 variant was incubated alone or with one of two different concentrations (0.625 and 1.25 μM) of mitochondrial LON protease, a human homologue of Pim1, in reaction buffer (50 mM HEPES–NaOH pH 8.0, 150 mM NaCl, 10 mM MgCl2, 1 mM dithiotreitol, 5 mM ATP, 0.15 mg/ml bovine serum albumin) for 30 min, at 30 °C. At indicated times, aliquots were removed, separated by SDS-PAGE and stained. Amounts of full-length Isu were quantitated by densitometry using ImageJ (Schindelin et al. 2012) and plotted as relative units with the time zero value set at 1. b Similar to Fe–S cluster coordination, binding of a Zn ion engages three evolutionarily conserved cysteine residues of Isu (Iannuzzi et al. 2014). Comparison of models of Zn-bound Isu (left) and apo-Isu (right) showing local structural rearrangement (cyan) in regions containing the LVF residues (orange) involved in Nfs1/Jac1 interaction, and cysteine residues (yellow), involved in zinc ion (purple) coordination. Models of Isu were obtained using SWISS-MODEL server (Bordoli et al. 2009) and visualized using PyMOL (Schrodinger, LLC). For Zn-bound Isu and apo-Isu, NMR solved structures for H. influenza IscU (PDB ID: 1R9P; Ramelot et al. 2004) and E. coli IscU (PDB ID: 2L4X; Kim et al. 2012) were used as templates, respectively. For modeling, Isu lacking the mitochondrial targeting sequence (residues 35–165) was used. In the final model the 16 N-terminal and 7 C-terminal residues are not depicted due to high flexibility of these segments
Insights into the molecular mechanism of Isu degradation and protection
Though the specific mechanism(s) of substrate recognition by mitochondrial Lon-type proteases is not known (Venkatesh et al. 2012), surface exposed hydrophobic regions have been shown to contribute significantly to substrate recognition in some cases (Ondrovicova et al. 2005). Intriguingly, analysis of interaction of Isu with Nfs1 and Jac1 revealed that the overlapping region of their binding sites contains the three hydrophobic residues Leu63, Val72 and Phe94 (Majewska et al. 2013). To assess whether this hydrophobic region plays a role in Isu degradation, we tested a variant of Isu having each of these residues changed to the polar residue, serine (Isu LVF_SSS) in the in vitro degradation assay. These alterations significantly decreased Isu’s susceptibility to degradation (Fig. 2a), supporting the idea that effective recognition by Pim1 is decreased by these changes and suggesting that these hydrophobic sequences may be part of the recognition site.
It is important to emphasize that degradation of Isu LVF_SSS was not inhibited completely (Fig. 2a), thus, these three residues are certainly not the sole determinant of susceptibility to Lon. That ligand (i.e. the Fe–S cluster) binding affects Isu degradation (Ciesielski et al. 2016) also suggests a more nuanced mechanism of Isu recognition and degradation by Pim1. Isu contains a flexible site dedicated to Fe–S cluster assembly and binding. The presence of a Fe–S cluster or zinc ion not only makes this site more rigid, but also affects the conformation of the nearby hydrophobic region (Fig. 2b). This change in Isu structural compactness upon zinc binding (Ramelot et al. 2004; Iannuzzi et al. 2014; Kim et al. 2012), which results in slower Isu degradation, fits well with observations that the presence of less structured regions contributes significantly to recognition and degradation by Lon-like proteases (von Janowsky et al. 2005; Koodathingal et al. 2009).
Increased Isu abundance as a compensatory mechanism for compromised Fe–S cluster biogenesis
Taken together, available data points to a model in which Isu abundance is regulated by the interplay between Pim1 dependent degradation and protection provided by binding to its partners that drive the biogenesis process (see Fig. 1). Employing Pim1 dependent degradation of Isu fits well with the emerging appreciation of a more profound role of Lon-like proteases in regulation of mitochondrial physiology, in addition to their well established quality control function (Quiros et al. 2014; Pinti et al. 2015, 2016). In the case of Isu regulation, binding of Nfs1 to Isu diminishes its susceptibility to Pim1 degradation, while concurrently initiating the first step of Fe–S cluster biogenesis. In the transition between the assembly and the transfer steps, Isu is continuously protected from Pim1 degradation upon complex formation with Jac1, which is enhanced by the presence of the Fe–S cluster. After interaction with Hsp70, which displaces Jac1 and initiates Fe–S cluster transfer, Isu becomes susceptible to degradation by Pim1. A slowdown at any point in the pathway following the initial Nfs1:Isu binding will produce a bottleneck in the general flux of the process and, in turn, result in Isu accumulation. Accumulation of Isu prior to the rate limiting step could be beneficial by shifting the chemical equilibrium in the productive direction, helping to overcome the bottleneck.
Such a mechanism could provide a self-regulated, localized and immediate response to a perturbation in the mitochondrial Fe–S cluster biogenesis process. Such short term perturbations might occur in the cell due to environmental changes, such as iron shortage, or upon disruption of mitochondrial metabolism (Ho and Gasch 2015; Mishra and Chan 2016; Wai and Langer 2016). This type of compensatory regulation may not only restore sufficient Fe–S cluster production, but also avoid activation of the Aft1/2 pathway and the consequences of cellular metabolism remodeling. Such a response would be reserved for extreme conditions such as prolonged iron deprivation. Consistent with this model, the stability of Isu in yeast cells expressing a transfer factor variant of only mildly reduced functionality is increased, but the Aft1/2 pathway was not activated in these cells, which grow as well as wild type cells (Andrew et al. 2008).
Under some conditions “high” levels of Isu are clearly beneficial. For example, yeast cells with levels similar to those found in pim1Δ cells, maintain viability for a significantly longer time after reaching stationary phase than wild type cells (Song et al. 2012). Then, why is not Isu normally expressed at these high levels? The answer likely lies in the fact that although Fe–S clusters are critically important, so are other cellular uses of iron. Thus, unbridled Isu levels may unfavorably tilt the balance of iron utilization. Indeed, constitutively high Isu levels results in lower activity of the heme-requiring enzyme ferric reductase. Cell growth upon iron depletion is negatively affected by maintaining Isu levels artificially constitutively high, rather than allowing normal regulation of Isu levels (Song et al. 2012). Thus this regulation of proteolysis may serve to exquisitely regulate Isu levels, balancing key needs of the cell for iron metabolism.
Acknowledgments
We thank Brenda Schilke for helpful comments on the manuscript. This work was supported by National Institutes of Health Grant GM27870 (E. A. C.).
References
- Andrew AJ, Song JY, Schilke B, Craig EA. Posttranslational regulation of the scaffold for Fe–S cluster biogenesis, Isu. Mol Biol Cell. 2008;19:5259–5266. doi: 10.1091/mbc.E08-06-0622. doi: 10.1091/mbc.E08-06-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2009;4:1–13. doi: 10.1038/nprot.2008.197. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- Ciesielski SJ, Schilke B, Marszalek J, Craig EA. Protection of scaffold protein Isu from degradation by the Lon protease Pim1 as a component of Fe–S cluster biogenesis regulation. Mol Biol Cell. 2016;27:1060–1068. doi: 10.1091/mbc.E15-12-0815. doi: 10.1091/mbc.E15-12-0815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutkiewicz R, Schilke B, Knieszner H, Walter W, Craig EA, Marszalek J. Ssq1, a mitochondrial Hsp70 involved in iron-sulfur (Fe/S) center biogenesis Similarities to and differences from its bacterial counterpart. J Biol Chem. 2003;278:29719–29727. doi: 10.1074/jbc.M303527200. doi: 10.1074/jbc.M303527200. [DOI] [PubMed] [Google Scholar]
- Garland SA, Hoff K, Vickery LE, Culotta VC. Saccharomyces cerevisiae ISU1 and ISU2: members of a well-conserved gene family for iron-sulfur cluster assembly. J Mol Biol. 1999;294:897–907. doi: 10.1006/jmbi.1999.3294. doi: 10.1006/jmbi.1999.3294. [DOI] [PubMed] [Google Scholar]
- Hausmann A, Samans B, Lill R, Muhlenhoff U. Cellular and mitochondrial remodeling upon defects in iron-sulfur protein biogenesis. J Biol Chem. 2008;283:8318–8330. doi: 10.1074/jbc.M705570200. doi: 10.1074/jbc.M705570200. [DOI] [PubMed] [Google Scholar]
- Ho YH, Gasch AP. Exploiting the yeast stress-activated signaling network to inform on stress biology and disease signaling. Curr Genet. 2015;61:503–511. doi: 10.1007/s00294-015-0491-0. doi: 10.1007/s00294-015-0491-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannuzzi C, Adrover M, Puglisi R, Yan R, Temussi PA, Pastore A. The role of zinc in the stability of the marginally stable IscU scaffold protein. Prot Sci Publ Prot Soc. 2014;23:1208–1219. doi: 10.1002/pro.2501. doi: 10.1002/pro.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Tonelli M, Kim T, Markley JL. Three-dimensional structure and determinants of stability of the iron–sulfur cluster scaffold protein IscU from Escherichia coli. Biochemistry. 2012;51:5557–5563. doi: 10.1021/bi300579p. doi: 10.1021/bi300579p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knieszner H, Schilke B, Dutkiewicz R, D’Silva P, Cheng S, Ohlson M, Craig EA, Marszalek J. Compensation for a defective interaction of the hsp70 ssq1 with the mitochondrial Fe–S cluster scaffold isu. J Biol Chem. 2005;280:28966–28972. doi: 10.1074/jbc.M503031200. doi: 10.1074/jbc.M503031200. [DOI] [PubMed] [Google Scholar]
- Koodathingal P, Jaffe NE, Kraut DA, Prakash S, Fishbain S, Herman C, Matouschek A. ATP-dependent proteases differ substantially in their ability to unfold globular proteins. J Biol Chem. 2009;284:18674–18684. doi: 10.1074/jbc.M900783200. doi: 10.1074/jbc.M900783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R, Hoffmann B, Molik S, Pierik AJ, Rietzschel N, Stehling O, Uzarska MA, Webert H, Wilbrecht C, Muhlenhoff U. The role of mitochondria in cellular iron–sulfur protein biogenesis and iron metabolism. Biochim Biophys Acta. 2012;1823:1491–1508. doi: 10.1016/j.bbamcr.2012.05.009. doi: 10.1016/j.bbamcr.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Lill R, Srinivasan V, Muhlenhoff U. The role of mitochondria in cytosolic-nuclear iron-sulfur protein biogenesis and in cellular iron regulation. Curr Opin Microbiol. 2014;22:111–119. doi: 10.1016/j.mib.2014.09.015. doi: 10.1016/j.mib.2014.09.015. [DOI] [PubMed] [Google Scholar]
- Majewska J, Ciesielski SJ, Schilke B, Kominek J, Blenska A, Delewski W, Song JY, Marszalek J, Craig EA, Dutkiewicz R. Binding of the chaperone Jac1 protein and cysteine desulfurase Nfs1 to the iron–sulfur cluster scaffold Isu protein is mutually exclusive. J Biol Chem. 2013;288:29134–29142. doi: 10.1074/jbc.M113.503524. doi: 10.1074/jbc.M113.503524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena NP, Urrutia PJ, Lourido F, Carrasco CM, Nunez MT. Mitochondrial iron homeostasis and its dysfunctions in neurode-generative disorders. Mitochondrion. 2015;21:92–105. doi: 10.1016/j.mito.2015.02.001. doi: 10.1016/j.mito.2015.02.001. [DOI] [PubMed] [Google Scholar]
- Mishra P, Chan DC. Metabolic regulation of mitochondrial dynamics. J Cell Biol. 2016;212:379–387. doi: 10.1083/jcb.201511036. doi: 10.1083/jcb.201511036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondrovicova G, Liu T, Singh K, Tian B, Li H, Gakh O, Perecko D, Janata J, Granot Z, Orly J, Kutejova E, Suzuki CK. Cleavage site selection within a folded substrate by the ATP-dependent lon protease. J Biol Chem. 2005;280:25103–25110. doi: 10.1074/jbc.M502796200. doi: 10.1074/jbc.M502796200. [DOI] [PubMed] [Google Scholar]
- Outten CE, Albetel AN. Iron sensing and regulation in Saccharomyces cerevisiae: ironing out the mechanistic details. Curr Opin Microbiol. 2013;16:662–668. doi: 10.1016/j.mib.2013.07.020. doi: 10.1016/j.mib.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore A, Puccio H. Frataxin: a protein in search for a function. J Neurochem. 2013;126(Suppl 1):43–52. doi: 10.1111/jnc.12220. doi: 10.1111/jnc.12220. [DOI] [PubMed] [Google Scholar]
- Pinti M, Gibellini L, Liu Y, Xu S, Lu B, Cossarizza A. Mitochondrial lon protease at the crossroads of oxidative stress, ageing and cancer. Cell Mol Life Sci CMLS. 2015;72:4807–4824. doi: 10.1007/s00018-015-2039-3. doi: 10.1007/s00018-015-2039-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinti M, Gibellini L, Nasi M, De Biasi S, Bortolotti CA, Iannone A, Cossarizza A. Emerging role of lon protease as a master regulator of mitochondrial functions. Biochim Biophys Acta. 2016 doi: 10.1016/j.bbabio.2016.03.025. doi: 10.1016/j.bbabio.2016.03.025. [DOI] [PubMed] [Google Scholar]
- Poor CB, Wegner SV, Li H, Dlouhy AC, Schuermann JP, Sanishvili R, Hinshaw JR, Riggs-Gelasco PJ, Outten CE, He C. Molecular mechanism and structure of the Saccharomyces cerevisiae iron regulator Aft2. Proc Natl Acad Sci USA. 2014;111:4043–4048. doi: 10.1073/pnas.1318869111. doi: 10.1073/pnas.1318869111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiros PM, Espanol Y, Acin-Perez R, Rodriguez F, Barcena C, Watanabe K, Calvo E, Loureiro M, Fernandez-Garcia MS, Fueyo A, Vazquez J, Enriquez JA, Lopez-Otin C. ATP-dependent lon protease controls tumor bioenergetics by reprogramming mitochondrial activity. Cell Rep. 2014;8:542–556. doi: 10.1016/j.celrep.2014.06.018. doi: 10.1016/j.celrep.2014.06.018. [DOI] [PubMed] [Google Scholar]
- Ramelot TA, Cort JR, Goldsmith-Fischman S, Kornhaber GJ, Xiao R, Shastry R, Acton TB, Honig B, Montelione GT, Kennedy MA. Solution NMR structure of the iron–sulfur cluster assembly protein U (IscU) with zinc bound at the active site. J Mol Biol. 2004;344:567–583. doi: 10.1016/j.jmb.2004.08.038. doi: 10.1016/j.jmb.2004.08.038. [DOI] [PubMed] [Google Scholar]
- Rutherford JC, Jaron S, Winge DR. Aft1p and Aft2p mediate iron-responsive gene expression in yeast through related promoter elements. J Biol Chem. 2003;278:27636–27643. doi: 10.1074/jbc.M300076200. doi: 10.1074/jbc.M300076200. [DOI] [PubMed] [Google Scholar]
- Schilke B, Voisine C, Beinert H, Craig E. Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1999;96:10206–10211. doi: 10.1073/pnas.96.18.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Method. 2012;9:676–682. doi: 10.1038/nmeth.2019. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JY, Marszalek J, Craig EA. Cysteine desulfurase Nfs1 and Pim1 protease control levels of Isu, the Fe–S cluster bio-genesis scaffold. Proc Natl Acad Sci USA. 2012;109:10370–10375. doi: 10.1073/pnas.1206945109. doi: 10.1073/pnas.1206945109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzarska MA, Dutkiewicz R, Freibert SA, Lill R, Muhlenhoff U. The mitochondrial Hsp70 chaperone Ssq1 facilitates Fe/S cluster transfer from Isu1 to Grx5 by complex formation. Mol Biol Cell. 2013;24:1830–1841. doi: 10.1091/mbc.E12-09-0644. doi: 10.1091/mbc.E12-09-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh S, Lee J, Singh K, Lee I, Suzuki CK. Multitasking in the mitochondrion by the ATP-dependent lon protease. Biochim Biophys Acta. 2012;1823:56–66. doi: 10.1016/j.bbamcr.2011.11.003. doi: 10.1016/j.bbamcr.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Janowsky B, Knapp K, Major T, Krayl M, Guiard B, Voos W. Structural properties of substrate proteins determine their proteolysis by the mitochondrial AAA + protease Pim1. Biol Chem. 2005;386:1307–1317. doi: 10.1515/BC.2005.149. doi: 10.1515/BC.2005.149. [DOI] [PubMed] [Google Scholar]
- Wai T, Langer T. Mitochondrial dynamics and metabolic regulation. Trend Endocrinol Metab TEM. 2016;27:105–117. doi: 10.1016/j.tem.2015.12.001. doi: 10.1016/j.tem.2015.12.001. [DOI] [PubMed] [Google Scholar]
- Webert H, Freibert SA, Gallo A, Heidenreich T, Linne U, Amlacher S, Hurt E, Muhlenhoff U, Banci L, Lill R. Functional reconstitution of mitochondrial Fe/S cluster synthesis on Isu1 reveals the involvement of ferredoxin. Nat Commun. 2014;5:5013. doi: 10.1038/ncomms6013. doi: 10.1038/ncomms6013. [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Urzica E, Guiard B, Muller H, Lohaus C, Meyer HE, Ryan MT, Meisinger C, Muhlenhoff U, Lill R, Pfanner N. Essential role of Isd11 in mitochondrial iron-sulfur cluster synthesis on Isu scaffold proteins. EMBO J. 2006;25:184–195. doi: 10.1038/sj.emboj.7600906. doi: 10.1038/sj.emboj.7600906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wofford JD, Lindahl PA. Mitochondrial iron–sulfur cluster activity and cytosolic iron regulate iron traffic in Saccharomyces cerevisiae. J Biol Chem. 2015;290:26968–26977. doi: 10.1074/jbc.M115.676668. doi: 10.1074/jbc.M115.676668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H, Knight SA, Pandey A, Pain J, Zhang Y, Pain D, Dancis A. Frataxin-bypassing Isu1: characterization of the bypass activity in cells and mitochondria. Biochem J. 2014;459:71–81. doi: 10.1042/BJ20131273. doi: 10.1042/BJ20131273. [DOI] [PMC free article] [PubMed] [Google Scholar]