Abstract
Genome editing technologies such as Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) systems have ushered in a new era of targeted DNA manipulation. The easy programmability of CRISPR using short oligonucleotides enables rapid synthesis of large-scale libraries for functional genetic screens. Here we present fundamental concepts and methods for pooled CRISPR screens and review biological results from recent genome-scale loss-of-function and gain-of-function screens. We also discuss new frontiers in pooled screens, including novel effector domains for functional screens and applications in the noncoding genome.
Introduction
Pooled genetic screens, where each cell receives a different genetic perturbation prior to a phenotype-based selection, are a powerful technique for rapidly identifying specific genome elements that influence a selected phenotype. A single successful screen may provide a treasure trove of new genotype-phenotype interactions that can be used as a launching point for follow-up studies on the most promising gene targets. The key idea behind pooled screens is that phenotypic selection results in an enrichment (or depletion) of genetic perturbations relevant to the phenotype. An example of a selected phenotype can be resistance to a drug or expression of a particular cell surface receptor. In this way, pooled screens can search over a space of thousands of perturbations (e.g. genome-scale), providing a substantially simpler technique than arrayed-format screens where each perturbation is delivered in a separate well.
Traditionally, to generate the pool of perturbations, these screens have relied on DNA mutagenesis (induced through chemical mutagens, mobile genetic element insertion or radiation) or manipulations at the transcript-level, such as RNA interference (RNAi). However, pooled screens using DNA mutagenesis can require painstaking work to map the mutation sites. Pooled screening at the transcript level with RNAi also presents challenges, such as incomplete knock-down and large off-target effects [1,2].
Recently, the easy programmability of microbial CRISPR systems has created a new opportunity for performing pooled screens at the DNA-level in a targeted fashion [3,4]. In their native context, CRISPR nucleases function as a prokaryotic immune system, cleaving foreign DNA from phage or plasmids before they can damage the host cell [5–7]. The nuclease is guided to a specific DNA sequence using a short, single-stranded RNA that is complementary to the target DNA. Genome editing applications using S. pyogenes Cas9 [8,9], the most commonly used CRISPR system, replace the endogenous prokaryotic RNA components with a synthetic single guide RNA (sgRNA) that has a 20-bp sequence complementary to the target site [10]. After the Cas9-sgRNA ribonucleoprotein complex recognizes the target sequence in the genome, it creates a double-strand break (DSB). DSB repair mechanisms, such as non-homologous end-joining (NHEJ), can delete or add a few bases during the repair process. When the NHEJ-mediated repair occurs in a coding region, this can introduce a frameshift mutation where the net result is gene loss-of-function. As reviewed elsewhere, the Cas9 system results in high efficiency genome modification in diverse genetic model (and non-model) organisms [11].
Using this RNA-guided system to produce targeted loss-of-function mutations, it is possible to build large, genome-scale libraries of CRISPR reagents for high-throughput pooled screens. The combination of DNA-level manipulation with easy targeting/programmability presents a new avenue for understanding the function of genome (and epigenome) elements in a wide variety of functional assays.
Technological underpinnings of CRISPR screens
Within months of the initial use of Cas9 for mammalian genome editing [8,9], several groups (including our own) took advantage of existing pooled synthesis and cloning techniques to construct large Cas9-sgRNA lentiviral libraries using similar methods to those previously used for large RNAi lentiviral libraries [3,4,12]. Typically, libraries are designed with multiple sgRNAs targeting each gene. Consistent changes in multiple sgRNAs can be used to increase confidence in a particular candidate gene. Briefly, libraries are synthesized as DNA and cloned into plasmids to produce lentivirus. After designing sgRNA guide sequences to target different genes, the oligonucleotides are synthesized in a pool of typically 104–105 different guides. Several different commercial platforms exist for producing pooled oligonucleotides (e.g. CustomArray, Twist Bioscience, Agilent Technologies) and synthesis fidelity and length is constantly improving. The pooled oligos are cloned into the sgRNA cassette of the backbone and the resulting lentivirus expresses the Cas9 protein and target-specific sgRNA (Figure 1a); alternatively, for higher viral titer, the small sgRNA can be delivered on a separate virus from the large Cas9 protein [13].
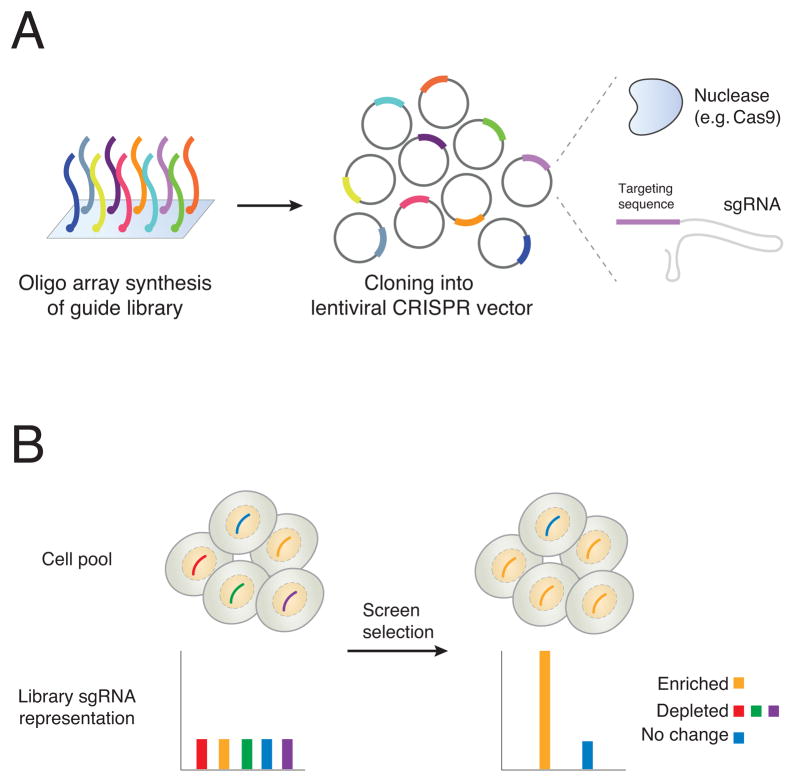
Figure 1. Synthesis of CRISPR libraries and changes in single-guide RNA (sgRNA) representation after enrichment or depletion.
(a) Single-stranded DNA oligonucleotides are synthesized on an array. Each one contains a specific targeting sequence (e.g. ~20 bp spacer sequence) flanked by universal sequences for PCR amplification. After amplification, they are cloned into a lentiviral vector that contains a cassette for the ~20 bp spacer (expressed as part of the sgRNA) and that also encodes the nuclease (e.g. Cas9). After viral production and transduction, the nuclease and the sgRNA are expressed in the target cells. (b) Library representation in a pool of cells before and after phenotypic selection. Initial library representation after viral transduction is uniform across different CRISPR reagents. After screen selection, the representation of the library has changed; 1 sgRNA (yellow) is enriched, 3 sgRNAs (red, green, purple) are depleted and 1 sgRNA (blue) did not change in abundance. At each timepoint, the representation of the library is assayed by taking a sample of the cell population, extracting genomic DNA, and PCR amplifying from the genomically-integrated lentiviral cassettes. After next-generation sequencing of the pool, the frequency of each sgRNA is counted.
Lentivirus is produced from the cloned plasmid pool and then used to transduce mammalian cells for screening. Because lentiviruses integrate into the genome, the viral integrant serves as a tag for readout of which sgRNA construct is delivered to a particular cell. Viruses are introduced at a low multiplicity-of-infection (MOI) in order to ensure that cells receive only a single sgRNA from the library pool. The frequency of observing any particular tag (sgRNA) before and after phenotypic selection is the key parameter measured during a pooled screen. This frequency can be computed by taking a representative sample of the population, PCR-amplifying the lentiviral cassettes and their sgRNAs, and then counting the frequency of each sgRNA after next-generation sequencing (Figure 1b). Ideally, the initial distribution of the library is as uniform as possible, so that, after selection, any depletion or enrichment of specific sgRNAs is readily identifiable. In reality, biases can be introduced at many different stages, such as library synthesis, cloning, viral transduction, cell sampling and PCR amplification, which makes it important to carefully control each stage of library handling before sequencing.
After selection, changes in sgRNA representation (enrichment or depletion) indicate potential candidates for further study (Figure 1b). A key concept in screen analysis is that the strongest gene candidates are those which have multiple sgRNAs simultaneously enriched/depleted. In enrichment screens, cells carrying specific sgRNAs are selected for. Representative examples of enrichment are drug resistance or toxin screens, where a gene knock-out promotes cell survival after exposure to a drug/toxin, or FACS-based selection of cells with a fluorescent gene reporter to find mutations that modulate gene expression. By contrast, depletion screens can identify essential genes, where loss-of-function is incompatible with continued cell survival, and synthetic lethal interactions, where typically nonessential genes become essential when cells carry a particular (usually oncogenic) mutation in a different gene. Depletion analysis in a drug or toxin screen can also be used to find gene targets that enhance sensitivity to the drug or toxin.
Applications of pooled CRISPR screens
Over the past two years, pooled CRISPR screens have been deployed in diverse phenotypic assays: cancer drug resistance [3,4,14], bacterial toxin resistance [12], West Nile virus-induced cell death [15], mitochondrial metabolism [16], identifying essential and synthetic lethal genes in cancer cell lines [17–19], identifying genetic drivers of metastasis in an in vivo screen [20], and understanding gene networks in immune cells [21,22]. In addition, two catalytically inactive versions of Cas9 with arrays of transcriptional activation domains (in conjunction with sgRNA libraries targeting promoter regions) have facilitated genome-scale gain-of-function screens (Figure 2) [23,24]. To illustrate the utility of pooled CRISPR screens, I focus here on two examples of loss-of-function screens: an enrichment screen and a set of related depletion screens.
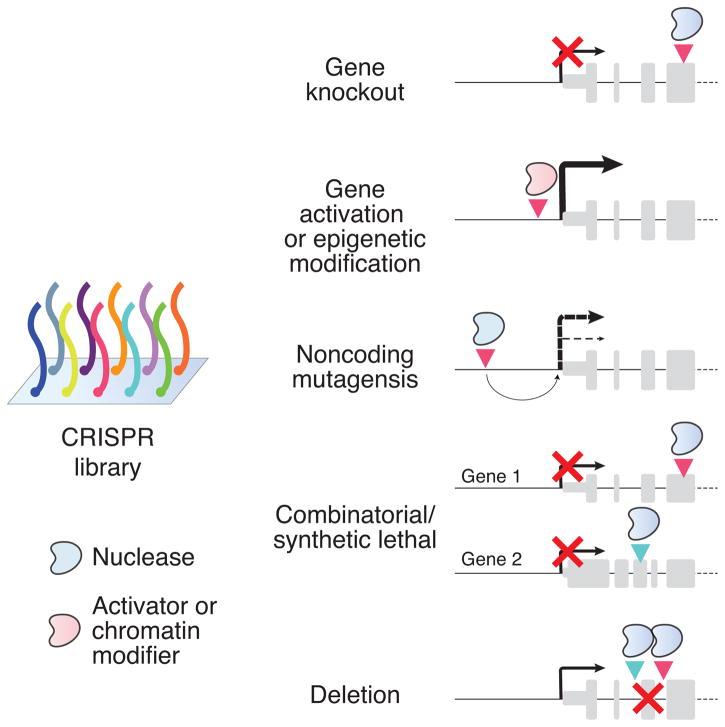
Figure 2. Applications of CRISPR libraries for gene manipulation.
Targeting of CRISPR nuclease/activator combined with sgRNAs from the libraries to specific gene features can be used for different kinds of screens. The specific screening application dictates the library sgRNA design and the type of CRISPR enzyme (e.g. nuclease or activator) used. Gene knockout: The sgRNA targets the CRISPR nuclease (e.g. Cas9) to an exon, where resulting frameshift mutations can trigger nonsense-mediated decay of messenger RNA and block production of functional protein products. Gene activation or chromatin modification: The sgRNA targets the CRISPR activator/epigenetic modifier complex (e.g. a catalytically inactive form of Cas9 with other functional domains added directly or as part of a larger complex) near the promoter of the gene to drive overexpression from the endogenous gene locus. Noncoding mutagenesis: The sgRNA targets the CRISPR nuclease to a putative regulatory element in the noncoding genome. Combinatorial or synthetic lethal: Sets of sgRNAs delivered into the same cell target CRISPR nucleases to the exons of different genes to examine possible interactions between loss-of-function for multiple genes. Deletion: Pairs of sgRNAs delivered into the same cell target CRISPR nucleases to nearby regions that, at some frequency, delete the intervening segment. This can be used to remove either coding elements like exons (as depicted) or to delete larger noncoding elements (e.g. intergenic noncoding RNAs) that are more difficult to manipulate using single sgRNA mutagenesis.
One of the first pooled screens was a positive selection screen for drug resistance in a human BRAF mutant (V600E) melanoma cell line using a genome-scale library targeting ~18,000 genes with ~65,000 sgRNAs [3]. This BRAF gain-of-function mutation is found in more than 50% of malignant melanomas and vemurafenib, a FDA-approved BRAF inhibitor, was shown to induce apoptosis preferentially in cells with the mutant form of BRAF [25]. In the clinic, vemurafenib results in reduction of solid tumors but, within weeks, resistance to the drug develops, which is followed by tumor re-growth and a poor prognosis. Identifying mechanisms of resistance can help inform the design of new treatment strategies to combine targeted BRAF inhibitors with other drugs. In the BRAF mutant melanoma cells, the genome-scale CRISPR screen identified both of the previously established loss-of-function mechanisms of vemurafenib resistance and several novel gene targets [26,27]. Importantly, the consistency of different reagents targeting the same gene was higher in the CRISPR screen than in the same screen using a genome-scale lentiviral RNAi library [27]. For example, the 10 most highly enriched genes in the RNAi screen had, on average, only 20% of the reagents targeting each gene enriched, indicating little agreement between different RNAi reagents designed to target the same gene. The CRISPR screen had, on average, 80% of reagents targeting each gene enriched for the 10 most highly enriched genes. This large difference in consistency suggests that the false-positive rate is lower using CRISPR reagents (further confirmed in a recent detailed comparison between screening technologies [28]) and thus gives us greater confidence in the identified gene candidates. As expected, individual loss-of-function mutations in several candidates from the CRISPR screen were shown to confer vemurafenib resistance in follow-up experiments [3], whereas only one gene from the RNAi screen could be validated [27]. A practical advantage of the lower false-positive rate is that genome-wide CRISPR screens can be conducted with smaller libraries (and thus using fewer cells) than a comparable genome-wide RNAi screen.
Two recent papers used depletion screens in multiple cell lines to estimate the percentage of genes in the human genome that are essential [17,18]. By measuring sgRNAs that are consistently depleted across multiple cell lines, they estimate that about 1,700 genes (~10% of all protein coding genes) are essential, which is about 5-fold higher than previous estimates from RNAi-based screens [29,30]. One reason for this increase is that RNAi knock-down is incomplete, whereas CRISPR is able to modify all copies of a gene when delivered using lentivirus and expressed constitutively [3,17]. This advantage is also relevant when comparing CRISPR screens to other DNA-based screening technologies: Wang et al. used both CRISPR knock-out and a gene-trap technique using retroviral insertions in the near-haploid KBM7 cell line [17]. The gene-trap screen was unable to detect any essential genes on chromosome 8, which is the only diploid chromosome in KBM7 cells. Since gene-trap screens rely on random insertion in the genome, complete knock-out is only possible in haploid cells, which only have one copy of each gene. This issue is especially relevant in cancer cell lines where multi-copy gene amplifications are more common. Finally, by comparing essential genes across different cell lines, both papers identified core essential genes and a much smaller set of context-dependent essential genes that were essential only in particular lines [17,18].
New frontiers in CRISPR screens
Since their initial development, pooled CRISPR screens have delivered new insights in basic biology and in applied, clinically relevant domains, yet there are still many opportunities for further development of this technology. Although in vivo screens have established a first step toward understanding cell-cell interactions [20], most CRISPR screens have been performed with a single cell type in isolation. New screens could replace drug selection with a cell-based selective pressure (e.g. T cell-mediated targeting of cancer cells) or combine library-transduced cells with other disease-relevant cell types (e.g. cancer cells cultured with endothelial or immune cells). It also is possible to deliver different pooled libraries to different populations of cells, capitalizing on recent cell type-specific gene expression data from resources like the Genotype-Tissue Expression (GTEx) project and the Allen Brain Atlas [31,32]. For example, in a co-culture system of neurons and glia, each cell type could separately be transduced with a specific library (targeting either neuron- or glia-relevant genes) before being cultured together. Along similar lines, combinatorial delivery of two or more sgRNAs in a single construct would allow targeting of multiple genes within one cell (Figure 2). However, it will be necessary to use smaller, targeted libraries to avoid a combinatorial explosion: A genome-scale CRISPR library with 105 distinct sgRNAs would require (at 103 cells/construct) an impractical 10 trillion cells to screen all possible pairs of sgRNAs but a library with 102–103 distinct sgRNAs is feasible given current screen handling and cell culture capacities.
The success in repurposing CRISPR using transcriptional activation domains for gain-of-function screens suggests several other kinds of screens might be possible by attaching different epigenetic effector domains to a catalytically inactive CRISPR nuclease. These could act to modulate chromatin states or alter other epigenetic elements at specific genomic locations [33,34]. Another consideration is where in the genome to target, either with nuclease or other effector domains. Presently, there has been little work on using pooled CRISPR screening approaches to manipulate non-coding genome elements, such as non-coding RNAs (e.g. miRNAs, lincRNAs), enhancers, repressors, promoters, or structural elements (e.g. CTCF binding sites) [13,20,35]. Delivery of multiple guides with a nuclease may enable large deletions, which would be useful for assessing the importance of larger non-coding elements. Both dual sgRNA-based deletions and single sgRNA scanning mutagenesis will be helpful for locating and understanding functional elements in the non-coding genome. From a technological standpoint, most CRISPR screens have focused on loss-of-function because of the high efficiency of NHEJ-mediated DSB repair after nuclease cutting, but it would be more desirable to precisely knock-in specific human genetic variants (e.g. SNPs, CNVs) at predefined locations. Although doable at a single genomic locus (c.f. CRISPR-based pooled saturating mutagenesis of 3 residues in the gene BRCA1 [36]), this type of precision editing is not yet possible at multiple genomic loci — however, given the rapid pace of genome engineering technology development, this may change soon.
Compared with RNAi and other pooled screening technologies, CRISPR pooled screens have demonstrated higher consistency between different reagents targeting the same genetic element and, because of this, they have already found broad applicability and produced new biological insights. Given the many promising future directions, CRISPR pooled screens have enormous potential to improve our understanding of the genome and to identify the causal role of genome elements in both normal and disease states.
Acknowledgments
R. Macrae provided several helpful comments on an early draft. NES is supported by NIH NHGRI award R00-HG008171.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Echeverri CJ, Beachy PA, Baum B, Boutros M, Buchholz F, Chanda SK, et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- 2.Singh S, Wu X, Ljosa V, Bray MA, Piccioni F, Root DE, et al. Morphological Profiles of RNAi-Induced Gene Knockdown Are Highly Reproducible but Dominated by Seed Effects. PLoS ONE. 2015;10:e0131370. doi: 10.1371/journal.pone.0131370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mojica FJM, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 6.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology (Reading Engl) 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 7.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 8.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jinek MM, Chylinski KK, Fonfara I, Hauer MM, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koike-Yusa H, Li Y, Tan E-P, Velasco-Herrera MDC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2013 doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 13.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol. 2015;33:661–667. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma H, Dang Y, Wu Y, Jia G, Anaya E, Zhang J, et al. A CRISPR-Based Screen Identifies Genes Essential for West-Nile-Virus-Induced Cell Death. Cell Rep. 2015;12:673–683. doi: 10.1016/j.celrep.2015.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162:540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, et al. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 19.Toledo CM, Ding Y, Hoellerbauer P, Davis RJ, Basom R, Girard EJ, et al. Genome-wide CRISPR-Cas9 Screens Reveal Loss of Redundancy between PKMYT1 and WEE1 in Glioblastoma Stem-like Cells. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parnas O, Jovanovic M, Eisenhaure TM, Herbst RH, Dixit A, Ye CJ, et al. A Genome-wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. Cell. 2015;162:675–686. doi: 10.1016/j.cell.2015.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmid-Burgk JL, Chauhan D, Schmidt T, Ebert TS, Reinhardt J, Endl E, et al. A genome-wide CRISPR screen identifies NEK7 as an essential component of NLRP3 inflammasome activation. J Biol Chem. 2015 doi: 10.1074/jbc.C115.700492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das Thakur M, Stuart DD. The evolution of melanoma resistance reveals therapeutic opportunities. Cancer Res. 2013;73:6106–6110. doi: 10.1158/0008-5472.CAN-13-1633. [DOI] [PubMed] [Google Scholar]
- 26.Huang S, Hölzel M, Knijnenburg T, Schlicker A, Roepman P, McDermott U, et al. MED12 Controls the Response to Multiple Cancer Drugs through Regulation of TGF-β Receptor Signaling. Cell. 2012;151:937–950. doi: 10.1016/j.cell.2012.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whittaker SR, Theurillat JP, Van Allen E, Wagle N, Hsiao J, Cowley GS, et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov. 2013;3:350–362. doi: 10.1158/2159-8290.CD-12-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evers B, Jastrzebski K, Heijmans JPM, Grernrum W, Beijersbergen RL, Bernards R. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat Biotechnol. 2016 doi: 10.1038/nbt.3536. [DOI] [PubMed] [Google Scholar]
- 29.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcotte R, Brown KR, Suarez F, Sayad A, Karamboulas K, Krzyzanowski PM, et al. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov. 2012;2:172–189. doi: 10.1158/2159-8290.CD-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 33.Thakore PI, D’Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods. 2015;12:1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, et al. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods. 2015;12:401–403. doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014;513:120–123. doi: 10.1038/nature13695. [DOI] [PMC free article] [PubMed] [Google Scholar]